Abstract
Bulky cisplatin lesions are repaired primarily by nucleotide excision repair (NER), in which the structure specific endonuclease XPF–ERCC1 is a critical component. It is now known that the XPF–ERCC1 complex has repair functions beyond NER and plays a role in homologous recombination (HR). It has been suggested that expression of ERCC1 correlates with cisplatin drug resistance in non-small cell lung cancer (NSCLC). In our study, using NSCLC, ovarian, and breast cancer cells, we show that the XPF–ERCC1 complex is a valid target to increase cisplatin cytotoxicity and efficacy. We targeted XPF–ERCC1 complex by RNA interference and assessed the repair capacity of cisplatin intrastrand and interstrand crosslinks by ELISA and alkaline comet assay, respectively. We also assessed the repair of cisplatin-ICL-induced double-strand breaks (DSBs) by monitoring γ-H2AX focus formation. Interestingly, XPF protein levels were significantly reduced following ERCC1 downregulation, but the converse was not observed. The transcript levels were unaffected suggesting that XPF protein stability is likely affected. The repair of both types of cisplatin-DNA lesions was decreased with downregulation of XPF, ERCC1 or both XPF–ERCC1. The ICL-induced DSBs persist in the absence of XPF–ERCC1. The suppression of the XPF–ERCC1 complex significantly decreases the cellular viability which correlates well with the decrease in DNA repair capacity. A double knockdown of XPF–ERCC1 displays the greatest level of cellular cytotoxicity when compared with XPF or ERCC1 alone. The difference in cytotoxicity observed is likely due to the level of total protein complex remaining. These data demonstrate that XPF–ERCC1 is a valid target to enhance cisplatin efficacy in cancer cells by affecting cisplatin-DNA repair pathways.
Keywords: XPF–ERCC1, Cisplatin, DNA repair and cancer
1. Introduction
Cisplatin [cis-diammine-dichloroplatinum (II)] is one of the most widely used platinum containing chemotherapeutic agents. It is an alkylating agent that acts by forming cisplatin-DNA adducts which include monoadducts, intrastrand DNA adducts and DNA interstrand crosslinks (ICLs). These adducts inhibit and block DNA replication, which leads to cell death [1,2]. Cisplatin is used clinically to treat a wide variety of tumors such as ovarian, testicular, head and neck, and NSCLC. Randomized clinical trials have confirmed that adjuvant chemotherapy with platinum-based (carboplatin or cisplatin) drug combinations for NSCLC significantly increases survival [3].
Despite its success with testicular cancer, its effectiveness in the treatment of other cancers is limited. This is due to the development of resistance over time. There are multiple mechanisms of cisplatin resistance but increased DNA repair is proposed to be one of the major reasons of resistance development [4]. Studies with a series of cisplatin-resistant cells in ovarian cancer cell lines show a clear relation between adduct removal and cisplatin cytotoxicity in a cisplatin-resistant model [5]. A major pathway involved in the repair of cisplatin-DNA adducts is the nucleotide excision repair (NER) pathway [6]. While the intrastrand DNA lesions and monoadducts are repaired by NER, the exact mechanism and events occurring during ICL repair are poorly understood [7].
The NER pathway has several steps: DNA damage recognition, dual incision/excision, repair synthesis and ligation. An important member of NER, the excision repair cross-complementing group 1 protein (ERCC1), forms a heterodimer with XPF and together they perform a critical incision step in the NER reaction. The XPF–ERCC1 complex is responsible for the incision 5′ to the lesion to cleave the damaged strand during NER [8].
It has been suggested that ERCC1 levels influence and correlate with DNA repair capacity [9–11]. ERCC1 expression has been associated with cellular and clinical resistance to platinum-based chemotherapy. There is much speculation about the importance of ERCC1 as prognostic indicator of cisplatin response in different cancer types [12]. Testicular cancer is generally responsive to cisplatin and has low levels of XPF–ERCC1, raising the possibility that XPF–ERCC1 levels influence the response to platinum therapy [13]. XPF–ERCC1 also has specific roles in ICL repair, recombination and regulates telomere integrity [14–19]. It has been shown that XPF–ERCC1 facilitates the repair of DSBs induced by cisplatin-ICL processing, and it has been proposed that the key role of XPF–ERCC1 is unhooking the crosslink in ICL repair [14]. It has also been shown that the complex plays a role in completion of homologous recombination during ICL repair [20].
Thus, we rationalize that XPF–ERCC1 complex is an important molecular target in cancer chemotherapy to potentiate cisplatin cytotoxicity as decreased levels of XPF–ERCC1 can increase responses to cisplatin treatment. In our study, we used RNA interference to downregulate the XPF–ERCC1 complex in addition to individual suppression of XPF and ERCC1 proteins in cancer cell lines to compare their DNA repair capacity and test the effect on cisplatin sensitivity.
2. Methods
2.1. Chemicals
Cisplatin [cis-diammine-dichloroplatinum (II)] was purchased from Sigma–Aldrich. For StaRT-PCR, primers that amplify ERCC1, XPF and β-actin (control) were obtained from Gene Express (Toledo, OH). ERCC1 forward primer 5′-CTGGAGCCCCGAGGAAGC-3′; reverse primer 5′-CACTGGGGGTTTCCTTGG-3′. XPF forward primer 5′-AGTGCATCTCCATGTCCCGCTACTA-3′; reverse primer 5′-CGATGTTCTTAACGTGGTGCATCAA-3′. β-actin forward primer 5′-CCCAGATCATGTTTGAGACC-3′; reverse primer 5′-CCATCTCTTGC TCGAAGTCC-3′. TRIzol reagent was from Invitrogen. We used the Qiagen DNeasy blood and tissue kit for DNA isolation and Profoldin DNA binding plates for the ELISA. All other reagents and chemicals were from standard suppliers.
2.2. Antibodies
The antibodies were polyclonal ERCC1-fl-297 (sc-10798, Santa Cruz), monoclonal XPF (MS-1381-PIABX, Neomarker), monoclonal α-tubulin (T5168, Sigma), monoclonal anti-phosopho γ-H2AX (clone JBW301, Millipore), and Alexa 488-conjugated goat anti-mouse (Molecular Probes). ICR4 antibody was kindly provided by Michael J. Tilby, University of New Castle, UK.
2.3. Cell culture
NSCLC cell lines, H1299 (provided by Dr. Randall Ruch, University of Toledo) and H1355 (provided by Dr. James C. Willey, University of Toledo) and 2008, ovarian cancer cell line (provided by Stephen Howell, University of California, San Diego), were maintained in RPM1 1640 supplemented with 10% FBS in the presence of penicillin (100 IU/ml) and streptomycin (100µg/ml). MDA-MB-231, which is a triple negative breast cancer cell line (provided by Dr. Manohar Ratnam, University of Toledo) was maintained in DMEM high glucose with 10% FBS in the presence of glutamine and sodium pyruvate and antibiotics. Cells were grown at 37 °C in a 5% CO2 incubator.
2.4. siRNA sequence
siRNA smart pools designed to target human ERCC1 and XPF were purchased from Dharmacon RNA technologies, catalogue numbers L-006311-00 and L-019946-00-10, respectively. A non-targeting siRNA pool was used in control experiments (catalogue number D-001810-10-20).
2.5. Transfection with siRNAs
H1299, H1355, 2008, MDA-MB-231 cells were seeded in six-well plates (density 2.5 × 105 cells/well) in antibiotic free media. Two transfections were done at 24 h interval in each cell line to knockdown ERCC1, XPF or XPF–ERCC1 together according to the manufacturer’s protocol. Addition of lipid reagent (Dharmafect, Dharmacon) without the siRNA is described as mock transfection.
2.6. Western blot
At each indicated time points of 72, 96 and 120 h post-transfection one, the cells were centrifuged, washed with PBS, and lysed on ice for 30 min in lysis buffer (10mM Tris, pH 8.0, 120mM NaCl, 0.5% NP-40, 1mM EDTA) with protease inhibitors (0.5M phenyl methyl sulfonyl fluoride (PMSF), 1mg/ml leupeptin, 1mg/ml pepstatin A). Equal amounts of protein were loaded and electrophoresed on 10% SDS–polyacrylamide gels. The proteins were transferred onto PVDF membrane (Immobilon transfer membrane, Millipore Corporation). After electroblotting, the membranes were blocked with Tris-buffered saline with Tween-20 (1M Tris–HCl, pH 7.5, 150mM NaCl, and 0.5% Tween-20) containing 2% non-fat dry milk. Primary antibodies, recognizing ERCC1, XPF or α-tubulin were diluted in blocking buffer and incubated for 30 min. The membranes were then washed, incubated with the appropriate secondary antibodies in a blocking buffer for 30 min, and washed again. The blotted proteins were detected using chemiluminescence detection system. The level of protein knockdown was determined using an Alpha Innotech Fluoro Chem HD2.
2.7. RNA isolation, reverse transcription and transcript abundance
Cells were lysed with 1ml of TRIzol reagent and the total RNA was extracted following the manufacturer’s protocol. The total RNA extracted from each cell line was reverse transcribed with oligo dT primer and M-MLV-RT as described previously [21,22]. Transcript levels were quantified using the previously described StaRT-PCR protocol [21,22]. Briefly, amixture of internal standard competitive template (SYSTEM 1, Gene Express, Inc.) was included in a master mix with cDNA and PCR reagents (dNTPs, etc.). The use of internal standards allows comparing data from different experiments giving a highly reproducible, standardized, quantitative measurement of transcript levels. In these studies, β-actin (ACTB) was used as a loading control gene. The master mix was aliquoted into tubes containing each gene-specific primer (ACTB, ERCC1, and XPF). PCR was carried out in a Rapidcycler (Idaho Technology Inc.) with each reaction mixture subjected to 35 cycles each of 5 s denaturation at 94 °C, 10 s of annealing at 58 °C and 15 s of elongation at 72 °C. PCR products were separated and quantified electrophoretically by the Agilent 2100 Bioanalyzer (Agilent Technologies Inc.) using the DNA 1000 Assay kit. Following electrophoresis, a ratio of the endogenous PCR products (or native template, NT) to the internal standard (competitive template) was taken to calculate molecules of NT in the reaction. Each transcript abundance value was normalized to ACTB and values are reported as target gene mRNA/106 ACTB mRNA. All experiments were performed in triplicate.
2.8. Cisplatin intrastrand adduct measurement by ELISA
Repair of intrastrand adducts was assessed by ELISA as described with some modifications [23,24]. Cells were treated with cisplatin for 2 h in serum free medium. Cells were then washed with PBS and fresh medium was added. At various time points between 0 and 48 h after drug treatment, genomic DNA was isolated and sonicated for 30–60 s in a Cole Palmer ultrasonic processor. Equal amounts of DNA were coated on 96-well DNA binding ELISA plates in binding buffer (1M sodium chloride, 50mM sodium phosphate buffer, pH 7.4, 0.02% sodium azide) and incubated at 4 °C overnight. The wells were blocked with 1% BSA in PBS for 1 h at room temperature. ICR4 antibody diluted 1:2000 in dilution buffer (0.2% BSA, 90mM sodium chloride, 0.2% Tween-20 in PBS) was added to the wells and incubated at 37 °C for 1 h. Following three washes with washing buffer (0.1% Tween-20 in PBS), HRP conjugated goat anti-rat antibody diluted 1:2500 (1% BSA, 0.2% Tween-20 in PBS) was added to the wells and incubated at 37 °C for 30–60 min. After five washes with washing buffer, TMB (1 step ultra TMB-ELISA, Thermo Scientific) was added and kinetics of absorbance was measured at 650nm in a Spectramax M5 plate reader (Molecular Devices) for 15 min. The reaction was stopped by adding 1M sulfuric acid and absorbance was measured at 450 nm. All samples were assayed in triplicates. The mean background (antibody blank) was subtracted from all the readings and the % intrastrand adducts were calculated using OD 450nm where the 0 h time point was used as 100% intrastrand adducts in each cell line.
2.9. Cisplatin interstrand crosslink measurement by comet assay
Repair of interstrand crosslinks was assessed by alkaline comet assay with some modifications [25,26]. Cells were treated with cisplatin for 2 h in serum free medium. At the end of treatment, cells were washed with PBS and incubated in fresh medium for the required post-incubation time or assayed immediately (time 0 h). Cells were further treated with 100µM hydrogen peroxide for 15 min to induce DNA strand breaks. Cells were then trypsinized, pelleted and resuspended in 1% low melting point agarose. Cell samples (~10,000 cells) were embedded on a microscopic slide precoated with 1% normal melting point agarose. Another layer of 0.5% low melting point agarose was added and allowed to solidify. The slides were then incubated in lysis solution (2.5M NaCl, 10mM Tris, 100mM EDTA, pH 10, containing 1%, v/v Triton X-100) for 1 h at 4 °C in the dark. The slides were then transferred to an electrophoresis tank containing ice-cold alkaline solution (300mM NaOH, 1mM EDTA, pH > 13), incubated for 20 min to allow DNA unwinding to occur and electrophoresis was carried out for 30 min at 0.7 v/cm, 300 mA. Slides were removed and kept in neutralizing solution (0.4M Tris–HCl, pH 7.5) for 10 min. Slides were then stained with SYBR green (Trevigen) and comets were analyzed using a Nikon epifluorescence microscope at 200× magnification. Fifty cells were analyzed per slide using Komet Assay Software 5.5F (Kinetic Imaging, Liver-pool, UK). The degree of DNA interstrand crosslinking present in the cisplatin-treated sample was determined as described by comparing the tail moment of cisplatin +H2O2 treated samples with H2O2 treated samples and untreated control samples [24]. The level of interstrand crosslinking was calculated by the following formula: [1 − (TMpt − TMctl)/(TMH2O2 − TMctl)] × 100, where TMpt is the mean tail moment of the cisplatin +H2O2 treated sample, TMctl is the mean tail moment of the untreated control sample and TMH2O2 is the mean tail moment of H2O2 treated sample. The data was expressed as the percent of ICLs that remained at a particular time point where 0 h was normalized to 100%.
2.10. Immunofluorescence
H1299, H1355, 2008 and MDA-MB-231 untransfected and siRNA transfected (double knockdown cells at 48 h post-initial transfection) cells were trypsinized and seeded onto glass coverslips at 25% confluency. The next day the cells were treated with cisplatin in serum free medium and 2 h later fresh complete medium was added and the cells were incubated for the indicated time points. For all experiments, control and knockdown cells were treated with equitoxic dose of cisplatin, with doses yielding 50% survival for all cell lines. The cells were washed with Hank’s balanced salt solution, fixed and permeabilized with ice-cold methanol for 15 min and blocked with 10% goat serum in PBS. For detecting the phosphorylated form of γ-H2AX, cells were incubated for 1 h with the monoclonal anti-γ-H2AX (1:500, Millipore) antibody followed by incubation with Alexa-488 goat anti-mouse antibody (1:1000, Molecular Probes) diluted in 10% goat serum in PBS. Cells were washed and counterstained with DAPI for 5 min. Coverslips were mounted with DAKO mounting medium onto slides and the edges were sealed with nailpolish. Images were visualized using a Nikon Eclipse T2000-U microscope at 100× oil immersion objective. Foci were counted in 250 cells at each time point per condition in each cell line and results are expressed as % γ-H2AX foci per nuclei. The data was collected from two individual experiments.
2.11. Colony survival assay
H1299, H1355, 2008 and MDA-MB-231 cells following two transfections, were split and seeded at a density of 300–600 cells in 60mmplates and incubated overnight. The next day, the cells were treated with different concentrations of cisplatin for 2 h, and after treatment, fresh complete medium with antibiotics was added and the cells were then allowed to grow for 7–14 days. Fresh medium was added when needed. Colonies were fixed with 95% methanol and stained with 0.2% crystal violet. Colonies with ≥50 cells were counted using a light microscope. Cell survival was expressed as the ratio of the average number of colonies in drug treated cells versus control cells × 100. The experiment was done in triplicate for each drug concentration.
3. Results
3.1. siRNA mediated knockdown of ERCC1, XPF and XPF–ERCC1 in cancer cells
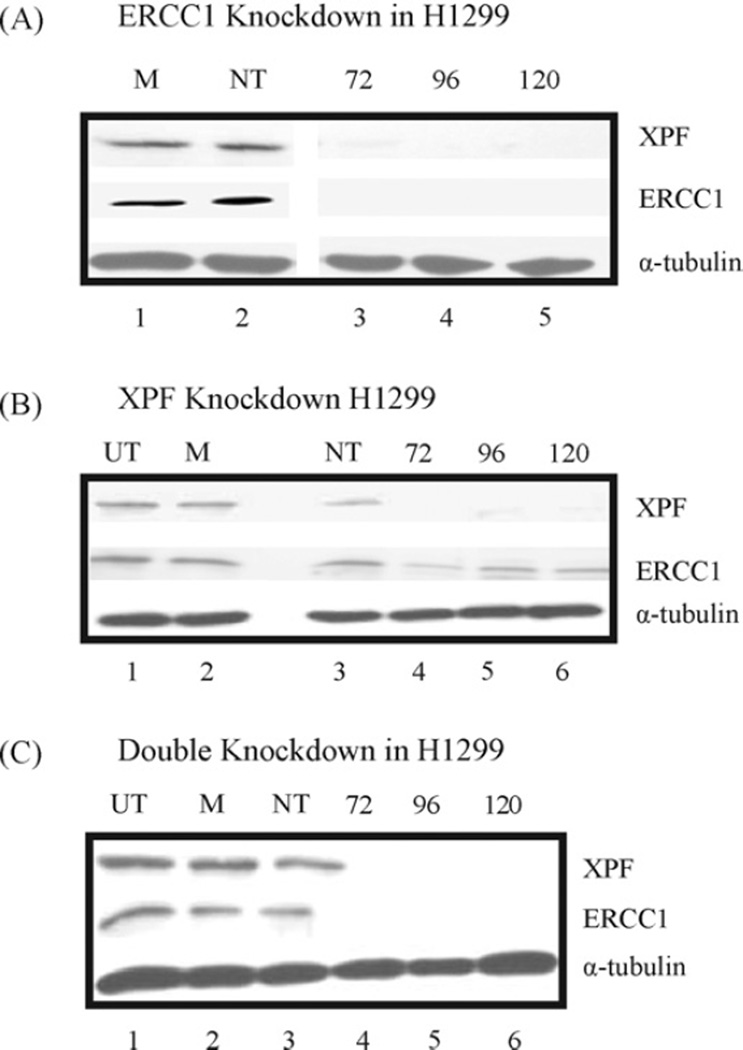
We have chosen different NSCLC, ovarian and breast cancer cells to downregulate XPF–ERCC1. NSCLC cells, H1299 and H1355 cells were transiently transfected with smartpool siRNA’s either directed individually against XPF and ERCC1 or at XPF–ERCC1 simultaneously (Fig. 1, Supplemental Fig. 1 and Table 1). Protein extracts from 72 to 120 h post-initial transfection were analyzed for ERCC1 and XPF expression with α-tubulin as a loading control in each cell line (Fig. 1, Supplemental Fig. 1 and Table 1). The results demonstrate that we achieve protein knockdown between 72 and 96 h. The knockdown for each protein is sustained past 120 h. Quantification of the protein levels indicate that we achieve >90% knockdown for ERCC1 and >80% knockdown for XPF. When both XPF and ERCC1 are knocked down simultaneously, we achieve the greatest level of protein knockdown for each. We also observed that when ERCC1 protein is knocked down, the XPF protein was also significantly reduced in both cell lines (Fig. 1A). However, ERCC1 protein levels showed a modest decrease as compared to controls when XPF is knockdown (Fig. 1B).
Figure 1.
Timecourse showing siRNA mediated downregulation of ERCC1 and XPF protein in H1299 cells. Cells were transiently transfected twice at 24 h intervals, each time with smartpool siRNAs (100 nM) directed against ERCC1 (A), XPF (B) and both proteins (C, double knockdown). Control cells were left untransfected (UT), or mock (M) – and non-targeting (NT) siRNA (100 nM) – transfected. Proteins were extracted at the indicated time points of 72, 96 and 120 h post-transfection, and probed with XPF, ERCC1 and α-tubulin as a loading control. The percent of knockdown was quantified using the Alpha Innotech HD2 and is represented for all cell lines tested in Table 1. Blots from (A) and (B) were also probed for XPF or ERCC1 on individual knockdown of ERCC1 or XPF, respectively.
Table 1.
Summary of cisplatin IC50s (represented as mean values from three independent experiments ± SEM) for knockdown and control cells with quantification of protein knockdown at the time of cisplatin treatment using the Alpha Innotech HD2 and transcript levels using StaRT-PCR represented as percent knockdown.
| Cell line | siRNA knockdown | Protein knockdown quantification (~%) |
Transcript knockdown (~%) |
Knockdown IC50 (µM) ± SEM |
siControl IC50 (µM) ± SEM |
Fold change (~) |
|---|---|---|---|---|---|---|
| H1299 | 1. XPF (X) | X - 81% | X - 80% | 1.54 ± 0.068 | 2.5 ± 0.059 | ~1.62 |
| 2. ERCC1 (E) | E - 95% | E - 93% | 0.74 ± 0.042 | ~3.5 | ||
| 3. XPF (X) + ERCC1 (E) | X - 90% and E - 90% | X - 88% and E - 99% | 0.42 ± 0.013 | ~6.0 | ||
| H1355 | 1. XPF (X) | X - 87% | X - 84% | 2.96 ± 0.036 | 4.32 ± 0.028 | ~1.5 |
| 2. ERCC1 (E) | E - 94% | E - 95% | 1.44 ± 0.476 | ~3.0 | ||
| 3. XPF (X) + ERCC1 (E) | X - 90% and E - 91% | X - 92% and E - 99% | 1.04 ± 0.040 | 4.0 | ||
| 2008 | 1. XPF (X) + ERCC1 (E) | X - 83% and E - 80% | X - 88% and E - 92% | 0.87 ± 0.03 | 1.61 ± 0.06 | ~2.0 |
| MDA-MB-231 | 1. XPF (X) + ERCC1 (E) | X - 83% and E - 90% | X - 90% and E - 89% | 3.03 ± 0.50 | 10.5 ± 0.94 | ~3.5 |
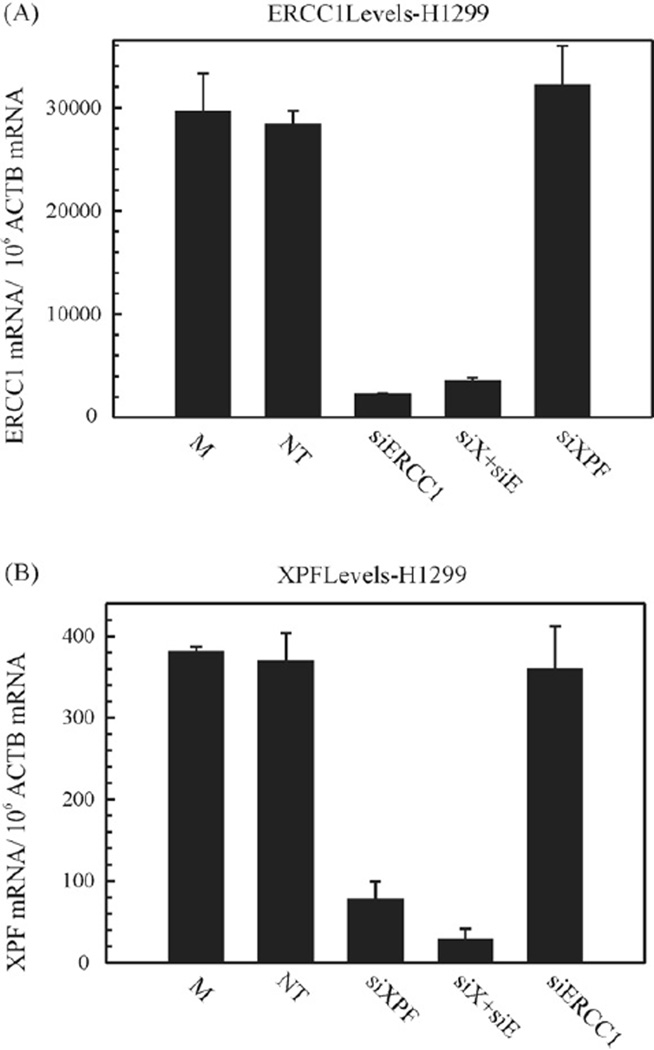
Next, the changes in protein expression were correlated with the change in transcript level in all cell lines tested (Fig. 2, Supplemental Fig. 2 and Table 1). StaRT-PCR [21,22] was employed to assess the basal target gene expression as well as the XPF and ERCC1 knockdowns in H1299 and H1355 cells (Fig. 2 and Supplemental Fig. 2). The 2008 and MDA-MB-231 cell lines are summarized in Table 1 after double knockdown. The mRNA levels were measured 48 h post-first transfection. The cells were either mock transfected, transfected with non-targeting (NT)-, XPF-, ERCC1- or XPF–ERCC1 siRNA. Experimental results show that ERCC1 siRNA (Fig. 2A, Supplemental Fig. 2A and Table 1) decreased the mRNA levels by ≥95% on individual and double knockdown, respectively, compared to the non-targeting control siRNA. XPF siRNA (Fig. 2B, Supplemental Fig. 2B and Table 1) reduced mRNA levels by ~80–85% on individual knockdown and ≥90% on double knockdown, as compared to the controls. The gene expression is expressed as a ratio of target gene mRNA/106 ACTB mRNA. To further assess the result that we observed with protein levels, we monitored XPF and ERCC1 transcript levels when ERCC1 or XPF was knocked down, respectively in both cell lines. We show that ERCC1 siRNA does not change XPF transcript levels and XPF siRNA has no effect on ERCC1 transcript levels (Fig. 2 and Supplemental Fig. 2). This shows that the decreased XPF protein levels are not due to non-specific effects caused by the siRNA transfection.
Figure 2.
Transcript levels on siRNA mediated downregulation. (A) represents ERCC1 transcript levels. H1299 cells were mock (M) treated or non-targeting (NT)-, XPF-, ERCC1-, and XPF–ERCC1 (denoted as siX + siE) siRNA transfected, twice at 24 h intervals each and harvested at 48 h post-transfection. Total RNA was extracted from cells and analyzed using StaRT-PCR, as described in Section 2. Each PCR was run in triplicate. The transcript levels are represented as ERCC1 mRNA/106 ACTB mRNA. The values are represented as mean±SEM from triplicate PCRs. (B) represents XPF transcript levels. H1299 cells were mock (M) treated or non-targeting (NT)-, XPF-, ERCC1-, and XPF–ERCC1 (denoted as siX + siE) siRNA transfected, twice at 24 h intervals each and harvested at 48 h post-transfection. Total RNA was extracted from cells and analyzed using StaRT-PCR, as described. Each PCR was run in triplicate. The transcript levels are represented as XPF mRNA/106 ACTB mRNA. The values are represented as mean±SEM from triplicate PCRs.
3.2. Repair kinetics of cisplatin intrastrand crosslinks
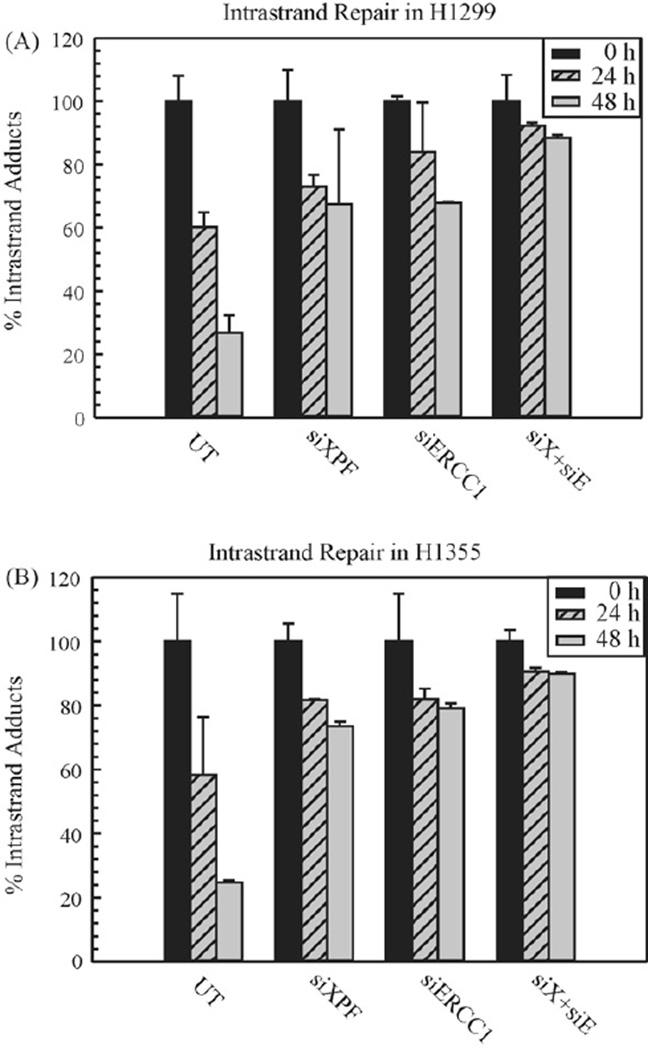
Considering that XPF–ERCC1 is required for NER and the cisplatin intrastrand adducts are repaired via NER [27], we assessed the repair efficiency of intrastrand adducts in cell lines following XPF, ERCC1 and XPF–ERCC1 downregulation. Fig. 3 shows the repair kinetics of cisplatin intrastrand adducts in H1299 and H1355 cell lines. We utilized an ELISA method to assess the repair of cisplatin 1,2 dGpG intrastrand adducts over time. This assay utilizes a monoclonal antibody specific for the major cisplatin dGpG intrastrand adduct [23,24]. The repair kinetics of cisplatin intrastrand adducts was calculated as the percent of adducts remaining over time, relative to the percent of adducts present at the 0 h treatment (100%). In untransfected cells, the intrastrand adducts were repaired gradually from 24 to 48 h, with ~75% of adducts were removed at 48 h in both cell lines. In XPF or ERCC1 individual siRNA transfected cells, the removal rate of these adduct was decreased. The double knockdown of XPF–ERCC1 results in the highest level of complex downregulation (summarized in Table 1) and concomitantly, results in the lowest level of cisplatin intrastrand adduct repair. This data illustrates the importance of the XPF–ERCC1 complex in the repair and removal of the major cisplatin-DNA adduct and is consistent with previous results, where NER mutants are defective in the repair of intrastrand adducts compared to wild type cells [24].
Figure 3.
Repair of cisplatin intrastrand adducts in H1299 (A) and H1355 (B) NSCLC cell lines. Untransfected (UT), siXPF, siERCC1 and both siXPF–siERCC1 (denoted as siX + siE) transfected cells were treated with cisplatin for 2 h and genomic DNA was isolated at different time intervals (0, 24, and 48 h). ELISAs were performed as described using cisplatin intrastrand adduct antibody and the percentage of intrastrand adducts remaining was calculated for H1299 (A) and H1355 (B) at the denoted times. The results are represented as mean±SEM of three independent experiments.
3.3. Repair of cisplatin interstrand crosslinks (ICLs)
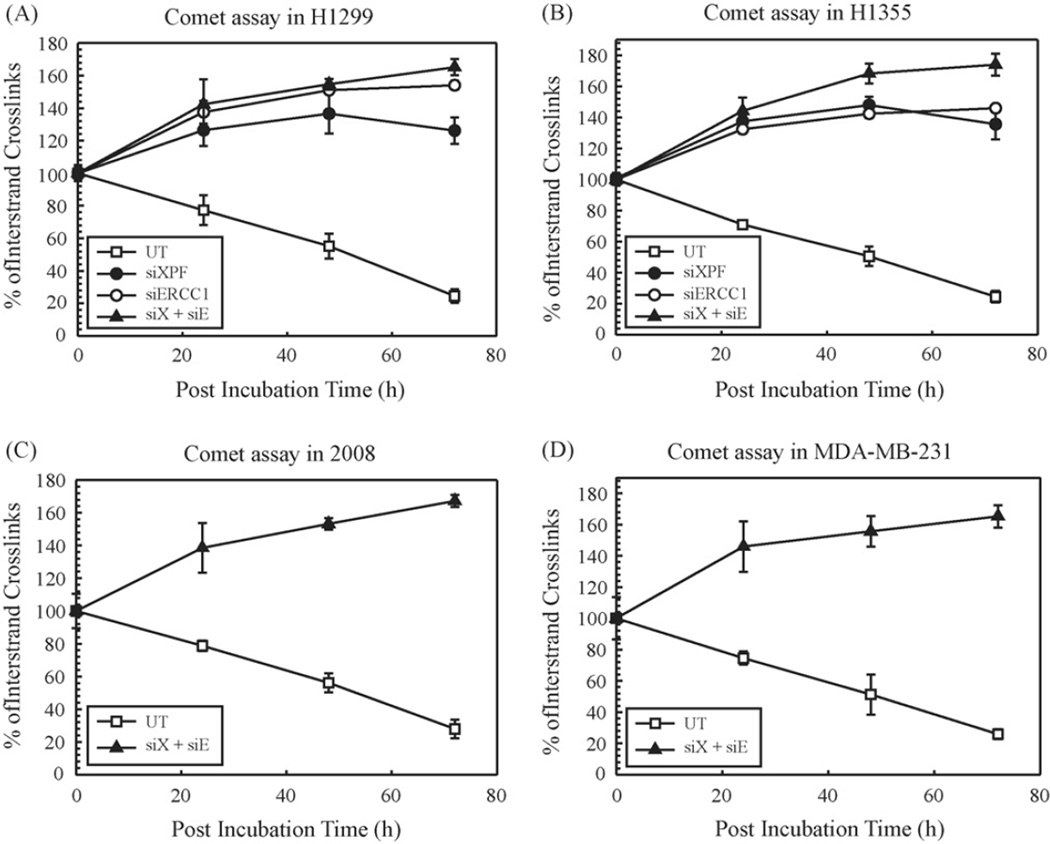
After demonstrating the role of XPF and ERCC1 on the repair of cisplatin intrastrand adducts, we then investigated the effect of silencing these two proteins on cisplatin-ICL DNA repair by alkaline comet assay. Comet assay has been used to evaluate DNA interstrand crosslink induction and repair in vivo at the single cell level [28]. The repair kinetics of cisplatin-ICLs in each cell line was evaluated after 0, 24, 48 and 72 h post-treatment with cisplatin and was expressed as the percentage of crosslinks remaining at the time points assessed. Fig. 4 shows the percentage of cisplatin-ICLs with increasing time in untransfected and siRNA transfected H1299 (Fig. 4A) and H1355 (Fig. 4B) NSCLC cells. Fig. 4C and D shows the percent of ICLs with increasing time in 2008 and MDA-MB-231, respectively in untransfected and double knockdown cells. Cisplatin treatment induced a similar extent of ICL formation at 0 h in untransfected and transfected cells for both cell lines. Cisplatin- ICLs were removed efficiently in untransfected cells with ~25% of the ICLs remaining at 72 h, whereas in transfected cells, significantly greater levels of ICLs still remained. Increased formation of cisplatin-ICLs in transfected cells at 24, 48 and 72 h indicates a possible conversion of monoadducts or intrastrand adducts to interstrand crosslinks. Of significance, no cisplatin-ICL repair is observed out to 72 h in the siXPF, siERCC1 or siXPF–ERCC1 (siX + siE) transfected cells. These data support previous reports of ICL repair in mammalian cells and show a requirement for XPF–ERCC1 in cisplatin-ICL repair [24,29].We speculate that there is a direct relation between the time required to repair a cisplatin-DNA lesion and the cytotoxic effect of the drug.
Figure 4.
Repair of cisplatin interstrand crosslinks in H1299 (A) and H1355 (B) 2008 (C) and MDA-MB-231 (D) cell lines. Untransfected (UT, open squares), siXPF (filled circles), siERCC1 (open circles) and siXPF–siERCC1 siRNA (filled triangles, denoted as siX + siE) transfected cells were treated with cisplatin for 2 h and the comet assay was performed as described at different time intervals (0, 24, 48 and 72 h). The percentage of interstrand crosslinks remaining was calculated using olive tail moment. Results are represented as mean ± SEM of three independent experiments.
3.4. Kinetics of γ-H2AX focus formation and repair of DSBs post-cisplatin-DNA damage
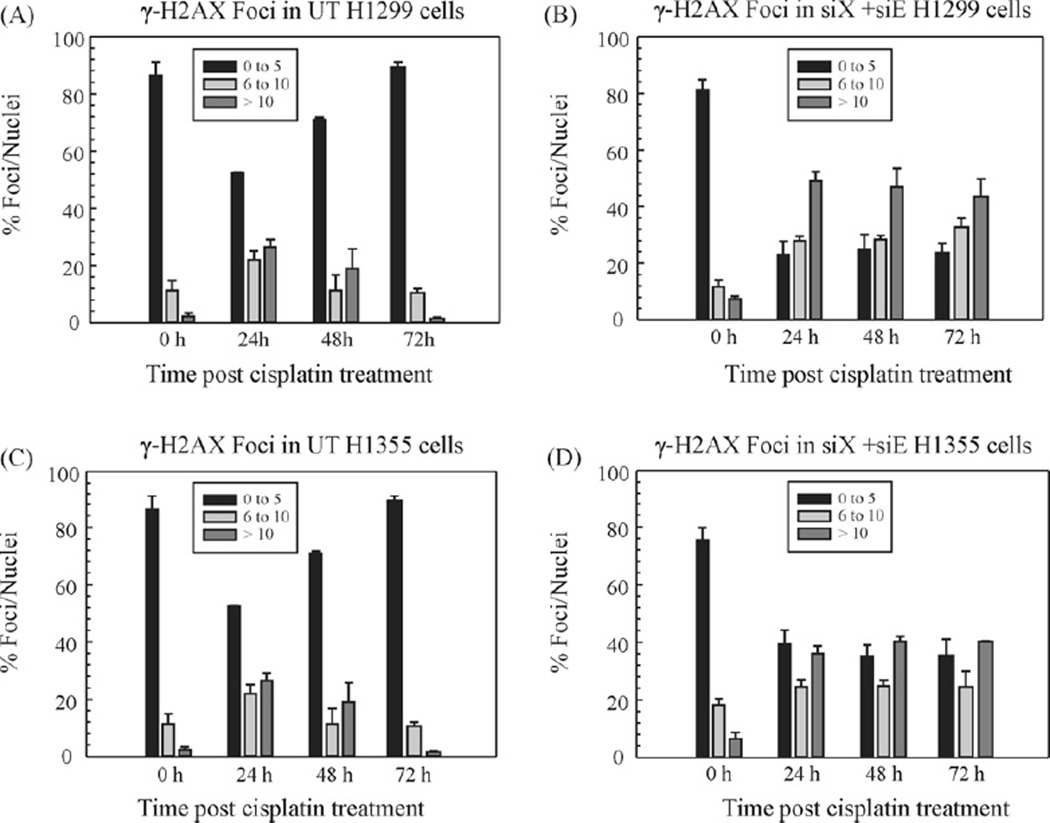
To further investigate and corroborate the results of the comet assay, we investigated the repair of ICL-induced DSBs in untransfected and XPF–ERCC1 double knockdown cells. The histone variant H2AX is phosphorylated at serine 139 upon exposure to ionizing radiation and forms distinct nuclear foci at sites of DSBs [30]. γ-H2AX foci also form upon exposure to cisplatin, although detection of DSBs in certain instances has been shown to be limited [31]. The nuclease processing at the sites flanking the ICLs leads to the generation of the DSBs [14]. Cisplatin-treated cells were categorized as having 0–5, 6–10 and >10 foci/nuclei (Fig. 5). Elevated levels of spontaneous endogenous γ-H2AX foci has been previously observed in cancer cells [32], and it is believed that these cryptic foci are a consequence of chromatin instability [33].
Figure 5.
Repair kinetics of γ-H2AX foci post-cisplatin treatment. H1299 (A and B) and H1355 (C and D) cell lines and quantitation of γ-H2AX focus formation at various time points post-cisplatin treatment in untransfected (A and C) and XPF + ERCC1 (B and D) double knockdown cells, respectively. The cells were seeded onto glass coverslips at 25% confluency. The next day they were treated with cisplatin for 2 h and then fresh complete medium was added. The cells were fixed and immunostained for γ-H2AX at the indicated time points post-cisplatin treatment. For each data point, foci were counted in 250 cells per condition in each cell line. The foci have been categorized as having 0–5, 6–10, or >10 foci per nucleus. The results are expressed as % γ-H2AX foci per nuclei. The data was collected from two individual experiments.
In comparison to untransfected cells, XPF–ERCC1 knockdown cells showed a higher frequency of γ-H2AX foci formation as well as more nuclei with >10 foci. This suggests that in the absence of the XPF–ERCC1 complex the cells retain a state of DNA damage which remains unrepaired. Fig. 5 shows kinetics of γ-H2AX focus formation in H1299 (Fig. 5A and B) and H1355 (Fig. 5C and D) untransfected and double knockdown cells. Under both conditions, cells with more than 10 foci peak at 12 h post-cisplatin treatment (data not shown). Untransfected cells consistently repaired DSBs in all the cell lines and the cells with a maximum number of foci started declining between 12 and 24 h (Fig. 5A and C and Supplemental Fig. 3). In contrast, DSBs were sustained even at 72 h in XPF–ERCC1 knockdown cells with persistence of >10 foci (Fig. 5B and D and Supplemental Fig. 3). This data demonstrates that the damage appears to be more pronounced in cancer cells that lack XPF–ERCC1. Foci in MDA-MB-231 cells were divided into 0, 1–10 and >10 foci/nuclei and the results for both the 2008 and MDA-MB-231 cells are in Supplementary Fig. 3. Overall, we show that in the absence of XPF–ERCC1, there is an accumulation of ICL-induced DSBs which remain persistent as the crosslinks are not “unhooked”/uncoupled. These results parallel results observed on Mitomycin C damage in Ercc1−/− null cells [14].
3.5. Downregulation of the XPF–ERCC1 complex sensitizes NSCLC cells to cisplatin
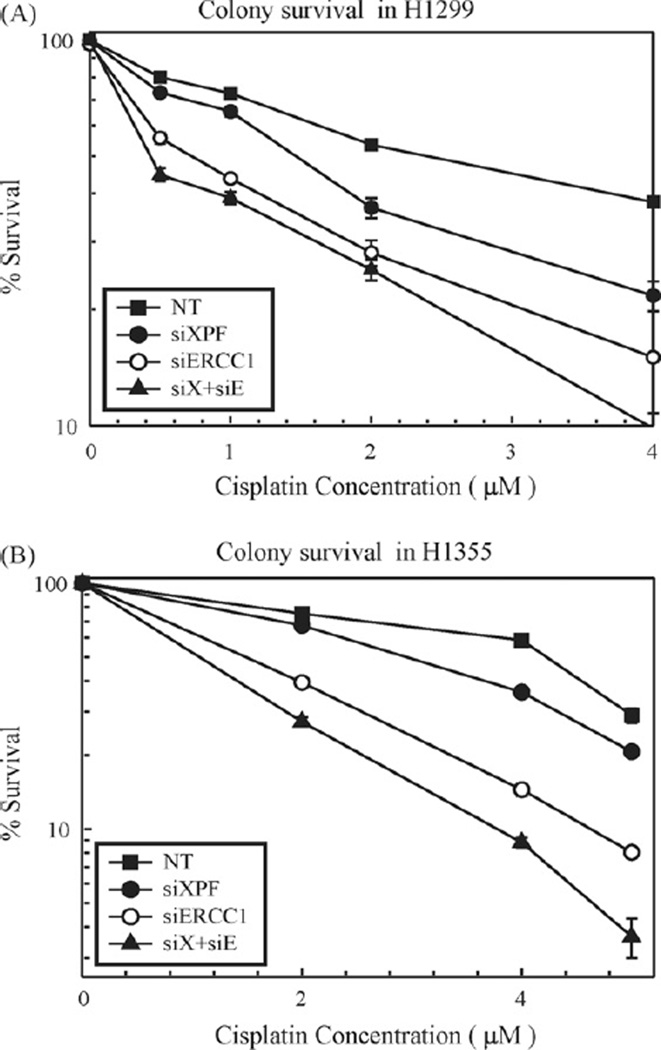
To assess the effect of XPF–ERCC1 downregulation on cell viability in response to cisplatin, clonogenic assays were performed in transfected and control cells. The results of the colony survival assays are shown in Fig. 6 and summarized in Table 1. XPF and ERCC1 individual knockdowns increased cisplatin cytotoxicity by 1.5–2-fold and 3–4-fold, respectively in H1299 (Fig. 6A) and H1355 (Fig. 6B) NSCLC cell lines. XPF–ERCC1 simultaneous downregulation results in 6-fold and 4-fold changes in IC50 values for H1299 and H1355 (Fig. 6A and B), respectively when compared to mock (data not shown) and non-targeting siRNA treated cells. 2008 cells showed ~2-fold change while MDA-MB-231 cells showed ~3.5-fold change in IC50 values following double knockdown (Table 1). The fold changes observed in IC50 values are likely a result of the level of protein downregulation (summarized in Table 1), where XPF–ERCC1 downregulation results in the best protein/transcript knockdown. This data supports our hypothesis that XPF–ERCC1 is a valid target for enhancing cisplatin efficacy for cancer chemotherapy.
Figure 6.
Colony survival in H1299 (A) and H1355 (B) cell lines. Non-targeting (NT, filled squares), siXPF (filled circles), siERCC1 (open circles) and siXPF–siERCC1 (filled triangles, denoted as siX + siE) siRNA transfected cells were treated with increasing doses of cisplatin for 2 h, and cell viability was determined by a clonogenic assay. The mock treated cells were left off of the graph for clarity reasons, but resulted in cytotoxicity nearly identical to non-targeting siRNA (data not shown). Calculated cisplatin IC50s for transfected cells are summarized in Table 1. Table 1 also summarizes the IC50s for control and knockdown cells for all other cell lines tested. Values are represented as mean±SEM from three independent experiments.
4. Discussion
Cisplatin cytotoxicity is believed to be attributed to the formation of platinum-DNA adducts which are primarily repaired by NER [5,7]. As NER is a major player in the mechanism of cisplatininduced DNA repair, targeting key NER components represents a molecular approach to enhance cisplatin efficacy for cancer treatment. Clinical investigations suggest that ERCC1 expression is a useful marker or predictor of response to cisplatin chemotherapy [34]. This has led to our evaluation of the XPF–ERCC1 complex, which could eventually mediate platinum resistance. Low levels of ERCC1 correlates with better clinical outcome and provides a strong rationale for our knockdown studies [35,36]. In our study, we targeted the XPF–ERCC1 complex through RNA interference to assess effects on DNA repair and cisplatin cytotoxicity in different cancer types. The set of timecourse experiments (Fig. 1) delineate time points to treat cells with cisplatin and also define optimal conditions to perform later experiments to assess DNA repair and cisplatin cytotoxicity in vivo.We also looked at the transcript levels which complement the protein expression studies. The XPF mRNA levels are relatively similar in all cell lines tested. However, the transcript levels for ERCC1 differ, wherein the cell line most sensitive to cisplatin had decreased abundance as compared to others in the panel (data not shown). Studies with more cell lines would be required to fully address this phenomenon.
Interestingly, when we knock down ERCC1, a significant reduction in XPF protein expression is observed. However, we observed no changes in XPF mRNA levels. Previous literature shows that in eukaryotic cells, formation of the heterodimeric complex is required for the stability of both components [37–41]. A recent study showed that XPF forms HhH homodimers in vitro which are more stable than the heterodimer with ERCC1. However, the same results are not observed in vivo with a low amount or no XPF present in the absence of ERCC1 [42]. In our study, we observed minimal loss of ERCC1 protein following XPF knockdown. The degradation or elimination of the binding partner must require an extremely low level or complete absence of the other. Studies with XPF cases and the first case of human ERCC1 deficiency show reduced but not complete absence of XPF–ERCC1, which showed only a mild impairment of NER in opposition to what we might expect [41]. It is possible that low levels of XPF–ERCC1 are still sufficient to perform critical NER functions. A previous study showed that ERCC1 mRNA is transcribed normally in XP2YO (XPF) cells [43], so the effect involves XPF protein stability due to the lack of complex formation in the absence of ERCC1. XPF-deficient and ERCC1 knockout mice have identical phenotypes which supports their function as a complex rather than separate functions in repair [44–46].
To address the effects on DNA repair, we studied the repair of the cisplatin 1,2-dGpG intrastrand adduct and ICLs. The intrastrand adducts represent the most abundant lesions and are repaired by the NER pathway [7]. In all cell lines, double knockdown of XPF–ERCC1 showed maximum reduction in repair of cisplatin-DNA intrastrand adducts (Fig. 3 and data not shown). ICL repair encompasses multiple mechanisms with important roles from the XPF–ERCC1 complex. ICLs are believed to play a significant role in the cytotoxicity and antitumor activity of anticancer drugs [47,48]. A recent study showed that there is an increase in the repair of cisplatin-DNA ICLs in ovarian cancer patients post-cisplatin treatment and this is an important factor that leads to acquired clinical resistance [49]. This strengthens the importance of increased DNA repair which is detrimental to drug cytotoxicity in the context of cancer chemotherapy. Our data reveals that ICL repair is significantly inhibited in a double knockdown of XPF–ERCC1. The data shows that there is no repair of cisplatin-ICLs. This has a major impact on platinum therapy as it increases the potential for the cisplatin lesion to persist longer on DNA and hence, can have a better cytotoxic effect. We also studied the kinetics of γ-H2AX foci formation in response to ICL-induced DSB formation. In the absence of XPF–ERCC1, the DSBs persist and remain unrepaired. DSB formation is detrimental to cell survival, and if they remain unrepaired, it can lead to cell death. Collectively, our data shows that XPF–ERCC1 mediates the repair of cisplatin-DNA adducts, and its absence significantly inhibits cisplatin-DNA repair.
The cellular capacity of the cells to repair cisplatin-DNA adducts is an important way for tumor cells to survive and resist the effects of chemotherapy. Hence, cells that are defective in uncoupling ICLs and cisplatin intrastrand adducts should show extreme sensitivity upon exposure to cisplatin. Also, ERCC1 and XPF–defective cells show UV sensitivity and high sensitivity to DNA crosslinking agents and are the most sensitive to crosslinking agents in comparison to other NER mutants [50,51]. Results from the clonogenic assay demonstrate a correlation between the decrease in DNA repair capacity and the enhancement of cellular cytotoxicity to cisplatin (Fig. 6 and Table 1). An XPF–ERCC1 double knockdown shows a particularly pronounced increase in cellular cytotoxicity in both H1299 and H1355. When we target XPF–ERCC1 simultaneously in H1299, we see a 6-fold change in cisplatin IC50 values while we observe a 4-fold change in H1355. It is important to note here that these two cell lines also differ in their p53 status. H1299 is p53 deficient while H1355 is a heterozygous mutant of p53. Following DNA damage, p53 is normally activated to induce cell cycle arrest and facilitate DNA repair [52]. The differences in p53 expression could affect cell survival and DNA repair efficiency following XPF–ERCC1 knockdown. Studies in mice have shown that a ~2-fold change in IC50 value is significant in regard to affecting tumor growth in response to cisplatin treatment [53]. The results suggest that targeting XPF–ERCC1 could bring about changes in chemotherapeutic regimens where decreased drug concentrations could have a greater cytotoxic effect. Overall our data contributes to the evidence that XPF–ERCC1 is vital to mediate the response against cisplatin-induced DNA damage. These results are consistent with previous work on ERCC1 disruption in cancer cell lines including ovarian, colon, and prostate [5,54,55].
In conclusion, we are the first to show that disrupting the XPF–ERCC1 complex instead of ERCC1 alone through a targeted mechanism results in significant enhancement in cisplatin cytotoxicity in cancer cell lines. XPF–ERCC1 is a broader therapeutic target than its counterparts in the NER pathway due to its repair roles in both intrastrand and ICL lesions. These studies support the idea that XPF–ERCC1 is a valid therapeutic target for enhancing cisplatin effectiveness not only in lung cancer, but other cancer types. Identification of small molecule inhibitors of the XPF–ERCC1 endonuclease complex could potentiate the effectiveness of cisplatin and yield better patient outcome and increase survival rates in lung cancer and other cancers which are quickly becoming resistant to conventional treatment regimens.
Supplementary Material
Acknowledgements
We gratefully acknowledge Erin L. Crawford and Dr. James C. Willey (University of Toledo) for helping with technical procedures for StaRT-PCR/the transcript abundance data. We also thank Akshada Sawant for helping with the γ-H2AX foci images.
Abbreviations
- NER
nucleotide excision repair
- NSCLC
non-small cell lung cancer
- ICL
interstrand crosslink
- DSBs
double-strand breaks
- ERCC1
excision repair cross-complementation group 1
- XPF
xeroderma pigmentation group F
- siRNA
small interfering RNA
- StaRT-PCR
standardized reverse transcription-polymerase chain reaction
- ACTB
β-actin
- ELISA
enzyme linked immuno-absorbent assay
- SEM
standard error of the mean
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.dnarep.2010.03.010.
Footnotes
Financial support: American Cancer Society - (ACS, RSG-06-163-01-GMC) to S.M.P.
Conflict of interest
None declared.
References
- 1.Reed E. Platinum-DNA adduct, nucleotide excision repair and platinum based anti-cancer chemotherapy. Cancer Treat. Rev. 1998;24:331–344. doi: 10.1016/s0305-7372(98)90056-1. [DOI] [PubMed] [Google Scholar]
- 2.Zamble DB, Lippard SJ. Cisplatin and DNA-repair in cancer-chemotherapy. Trends Biochem. Sci. 1995;20:435–439. doi: 10.1016/s0968-0004(00)89095-7. [DOI] [PubMed] [Google Scholar]
- 3.Le Chevalier T, Arriagada R, Le Pechoux C, Grunenwald D, Dunant A, Pignon JP, Tarayre M, Abratt R, Arriagada R, Bergman B, Gralla R, Grunenwald D, Le Chevalier T, Orlowski T, Papadakis E, Pinel MIS, Araujo C, la Torre H, de Solchaga MM, Abdi E, Blum R, Ball D, Basser R, De Boer R, Bishop J, Brigham B, Davis S, Fox D, Richardson G, Wyld D, Pirker R, Humblet Y, Delaunois L, Van Meerbeeck JP, Germonpre P, Vansteenkiste J, Nackaerts K, Pinel MIS, Vauthier G, Younes RN, Arriagada R, Baeza R, Carvajal P, Kleinman S, Orlandi L, Castro C, Godoy J, Kosatova K, Gaafar R, Azarian R, Benichou M, Boudoux L, Boyer G, Chirat E, Chomy F, Coetmeur D, Couderc LJ, Coudert B, Crequit J, David P, Delaval P, Dohollou N, Foucher P, Gonzalez G, Jeanfaivre T, Joveniaux A, Kanoui AA, Le Chevalier T, Le Pechoux C, Baldeyrou P, Debrosse D, Girard P, Grunenwald D, Ruffie P, Leclerc P, Milleron B, Monnet I, Monnot H, Morere JF, Naman H, Dides S, Noel JP, Otto J, Ozanne F, Ozenne G, Parent F, Peureux M, Picon B, Pujol JL, Quoix E, Rabut H, Riviere A, Gervais R, Icard P, Rixe O, Deplanque G, Sevin D, Spaeth D, Tourani JM, Toussaint C, Tuchais C, Vannetzel JM, VArflet F, Virally J, Zaegel M, Zalcman G, Livatorwski A, Eberhardt W, Alexopoulos C, Apostolopoulou F, Bachlitzanakis N, Bellenis I, Christaki P, Dosios T, Georgoulias V, Iliadis K, Kardamakis D, Konstantinou M, Kotsovoulou V, Pouliou M, Michalopoulou P, Papadakis E, Agelidou A, Zarogoulidis K, Gottfried M, Kovner F, Agostara B, Ascoli M, Brigandi A, Cigolari S, Colucci G, De Braud F, Dirosa T, Giglio A, Maiorino A, Masotti A, Rusca M, Ogawara M, Osada H, Park K, Jackevicius A, Smickoska S, El Gueddari BE, Aaesebo U, Desales R, Furman M, Kozlowski M, Laudansk J, Grodzki T, Orlowski T, Chabowski M, Pluzanska A, Gaspar L, Ciuleanu T, Baumohl J, Cerna M, Bitenc M, Abratt R, Chasen M, Alberola V, Azagra P, Garcia PB, Cardenal F, Curto CP, Salinas J, Serrate RS, Bergman B, Andersson L, Hero U, Mansson T, Martinsson JE, Hillerdal G, Agrenius V, Sederholm C, Arvidsson I, Ydreborg SO, Thaning L, Sjogren A, Jonsson P, Grip H, Singer J, Lundgren R, Brodin O, Andejani D, Branden E, Lamberg K, Riska H, Egli E, Joss R, Martinelli G, Roth A, Stahel R, Stupp R, El Kamel A, Gralla R, Cole JT, Jahanzeb M, Wiznitzer I, Masters GA, Radosavljevic G, Babiker A, Green M, Osterlind K, Taube A, Wedel H. Cisplatin-based adjuvant chemotherapy in patients with completely resected non-small-cell lung cancer. N. Engl. J. Med. 2004;350:351–360. doi: 10.1056/NEJMoa031644. [DOI] [PubMed] [Google Scholar]
- 4.McGurk CJ, Cummings M, Koberle B, Hartley JA, Oliver RT, Masters JR. Regulation of DNA repair gene expression in human cancer cell lines. J. Cell. Biochem. 2006;97:1121–1136. doi: 10.1002/jcb.20711. [DOI] [PubMed] [Google Scholar]
- 5.Selvakumaran M, Pisarcik DA, Bao R, Yeung AT, Hamilton TC. Enhanced cisplatin cytotoxicity by disturbing the nucleotide excision repair pathway in ovarian cancer cell lines. Cancer Res. 2003;63:1311–1316. [PubMed] [Google Scholar]
- 6.Chu G. Cellular responses to cisplatin. The roles of DNA-binding proteins and DNA Repair [Review] [74 Refs.] J. Biol. Chem. 1994;269:787–790. [PubMed] [Google Scholar]
- 7.Gillet LCJ, Scharer OD. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem. Rev. 2006;106:253–276. doi: 10.1021/cr040483f. [DOI] [PubMed] [Google Scholar]
- 8.Bessho T, Sancar A, Thompson LH, Thelen MP. Reconstitution of human excision nuclease with recombinant XPF–ERCC1 complex. J. Biol. Chem. 1997;272:3833–3837. doi: 10.1074/jbc.272.6.3833. [DOI] [PubMed] [Google Scholar]
- 9.Rosell R, Lord RVN, Taron M, Reguart N. DNA repair and cisplatin resistance in non-small-cell lung cancer. Lung Cancer. 2002;38:217–227. doi: 10.1016/s0169-5002(02)00224-6. [DOI] [PubMed] [Google Scholar]
- 10.Li QD, Yu JJ, Mu CJ, Yunmbam MK, Slavsky D, Cross CL, Bostickbruton F, Reed E. Association between the level of ERCC-1 expression and the repair of cisplatin-induced DNA damage in human ovarian cancer cells. Anticancer Res. 2000:645–652. [PubMed] [Google Scholar]
- 11.Park SY, Hong YC, Kim JH, Kwak SM, Cho JH, Lee HL, Ryu JS. Effect of ERCC1 polymorphisms and the modification by smoking on the survival of nonsmall cell lung cancer patients. Med. Oncol. 2006;23:489–498. doi: 10.1385/MO:23:4:489. [DOI] [PubMed] [Google Scholar]
- 12.Olaussen KA, Dunant A, Fouret P, Brambilla E, Andre F, Haddad V, Taranchon E, Filipits M, Pirker R, Popper HH, Stahel R, Sabatier L, Pignon J, Tursz T, Le Chevalier T, Soria J. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N. Engl. J. Med. 2006;355:983–991. doi: 10.1056/NEJMoa060570. [DOI] [PubMed] [Google Scholar]
- 13.Welsh C, Day R, McGurk C, Masters JRW, Wood RD, Koberle B. Reduced levels of XPA, ERCC1 and XPF DNA repair proteins in testis tumor cell lines. Int. J. Cancer. 2004;110:352–361. doi: 10.1002/ijc.20134. [DOI] [PubMed] [Google Scholar]
- 14.Niedernhofer LJ, Odijk H, Budzowska M, van Drunen E, Maas A, Theil AF, de Wit J, Jaspers NGJ, Beverloo HB, Hoeijmakers JHJ, Kanaar R. The structure-specific endonuclease ERCC1-XPF is required to resolve DNA interstrand crosslink-induced double-strand breaks. Mol. Cell. Biol. 2004;24:5776–5787. doi: 10.1128/MCB.24.13.5776-5787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuraoka I, Kobertz WR, Ariza RR, Biggerstaff M, Essigmann JM, Wood RD. Repair of an interstrand DNA cross-link initiated by ERCC1-XPF repair/recombination nuclease. J. Biol. Chem. 2000;275:26632–26636. doi: 10.1074/jbc.C000337200. [DOI] [PubMed] [Google Scholar]
- 16.Schiestl RH, Prakash S. RAD10, an excision repair gene of Saccharomyces cere-visiae, is involved in the RAD1 pathway of mitotic recombination. Mol. Cell. Biol. 1990;10:2485–2491. doi: 10.1128/mcb.10.6.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niedernhofer LJ, Essers J, Weeda G, Beverloo B, de Wit J, Muijtjens M, Odijk H, Hoeijmakers JHJ, Kanaar R. The structure-specific endonuclease ERCC1-XPF is required for targeted gene replacement in embryonic stem cells. EMBO J. 2001;20:6540–6549. doi: 10.1093/emboj/20.22.6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu XD, Niedernhofer L, Kuster B, Mann M, Hoeijmakers JHJ, de Lange T. ERCC1/XPF removes the 3′ overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol. Cell. 2003;12:1489–1498. doi: 10.1016/s1097-2765(03)00478-7. [DOI] [PubMed] [Google Scholar]
- 19.Munoz P, Blanco R, Flores JM, Blasco MA. XPF nuclease-dependent telomere loss and increased DNA damage in mice overexpressing TRF2 result in premature aging and cancer. Nat. Genet. 2005;37:1063–1071. doi: 10.1038/ng1633. [DOI] [PubMed] [Google Scholar]
- 20.Al-Minawi AZ, Lee YF, Hakansson D, Johansson F, Lundin C, Saleh-Gohari N, Schultz N, Jenssen D, Bryant HE, Meuth M, Hinz JM, Helleday T. The ERCC1/XPF endonuclease is required for completion of homologous recombination at DNA replication forks stalled by inter-strand cross-links. Nucleic Acids Res. 2009;37:6400–6413. doi: 10.1093/nar/gkp705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Willey JC, Crawford EL, Jackson CM, Weaver DA, Hoban JC, Khuder SA, Demuth JP. Expression measurement of many genes simultaneously by quantitative RT-PCR using standardized mixtures of competitive templates. Am. J. Respir. Cell Mol. Biol. 1998;19:6–17. doi: 10.1165/ajrcmb.19.1.3076. [DOI] [PubMed] [Google Scholar]
- 22.Weaver DA, Crawford EL, Warner KA, Elkhairi F, Khuder SA, Willey JC. ABCC5, ERCC2, XPA and XRCC1 transcript abundance levels correlate with cisplatin chemoresistance in non-small cell lung cancer cell lines. Mol. Cancer. 2005;4 doi: 10.1186/1476-4598-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tilby MJ, Johnson C, Knox RJ, Cordell J, Roberts JJ, Dean CJ. Sensitive detection of DNA modifications induced by cisplatin and carboplatin in vitro and in vivo using a monoclonal-antibody. Cancer Res. 1991;51:123–129. [PubMed] [Google Scholar]
- 24.De Silva IU, McHugh PJ, Clingen PH, Hartley JA. Defects in interstrand crosslink uncoupling do not account for the extreme sensitivity of ERCC1 and XPF cells to cisplatin. Nucleic Acids Res. 2002;30:3848–3856. doi: 10.1093/nar/gkf479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olive PL, Banath JP, Durand RE. Heterogeneity in radiation-induced DNA damage and repair in tumor and normal-cells measured using the comet assay. Radiat. Res. 1990;122:86–94. [PubMed] [Google Scholar]
- 26.Singh NP, Mccoy MT, Tice RR, Schneider EL. A simple technique for quantitation of low-levels of DNA damage in individual cells. Exp. Cell Res. 1988;175:184–191. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- 27.Zamble DB, Mu D, Reardon JT, Sancar A, Lippard SJ. Repair of cisplatin-DNA adducts by the mammalian excision nuclease. Biochemistry. 1996;35:10004–10013. doi: 10.1021/bi960453+. [DOI] [PubMed] [Google Scholar]
- 28.Hartley JM, Spanswick VJ, Gander M, Giacomini G, Whelan J, Souhami RL, Hartley JA. Measurement of DNA cross-linking in patients on ifosfamide therapy using the single cell gel electrophoresis (comet) assay. Clin. Cancer Res. 1999;5:507–512. [PubMed] [Google Scholar]
- 29.Friedmann B, Caplin M, Hartley JA, Hochhauser D. Modulation of DNA repair in vitro after treatment with chemotherapeutic agents by the epidermal growth factor receptor inhibitor gefitinib (ZD1839) Clin. Cancer Res. 2004;10:6476–6486. doi: 10.1158/1078-0432.CCR-04-0586. [DOI] [PubMed] [Google Scholar]
- 30.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 31.Olive PL, Banath JP. Kinetics of H2AX Phosphorylation after exposure to cisplatin. Cytometry Part B: Clin. Cytom. 2009;76B:79–90. doi: 10.1002/cyto.b.20450. [DOI] [PubMed] [Google Scholar]
- 32.Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 2000;10:886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 33.Yu T, MacPhail SH, Banath JP, Klokov D, Olive PL. Endogenous expression of phosphorylated histone H2AX in tumors in relation to DNA double-strand breaks and genomic instability. DNA Repair. 2006;5:935–946. doi: 10.1016/j.dnarep.2006.05.040. [DOI] [PubMed] [Google Scholar]
- 34.Reed E. ERCC1 measurements in clinical oncology. N. Engl. J. Med. 2006;355:1054–1055. doi: 10.1056/NEJMe068162. [DOI] [PubMed] [Google Scholar]
- 35.Lord RVN, Brabender J, Gandara D, Alberola V, Camps C, Domine M, Cardenal F, Sanchez JM, Gumerlock PH, Taron M, Sanchez JJ, Danenberg KD, Danenberg PV, Rosell R. Low ERCC1 expression correlates with prolonged survival after cisplatin plus gemcitabine chemotherapy in non-small cell lung cancer. Clin. Cancer Res. 2002;8:2286–2291. [PubMed] [Google Scholar]
- 36.Azuma K, Komohara Y, Sasada T, Terazaki Y, Ikeda J, Hoshino T, Itoh K, Yamada A, Aizawa H. Excision repair cross-complementation group 1 predicts progression-free and overall survival in non-small cell lung cancer patients treated with platinum-based chemotherapy. Cancer Sci. 2007;98:1336–1343. doi: 10.1111/j.1349-7006.2007.00557.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tripsianes K, Folkers G, Ab E, Das D, Odijk H, Jaspers NGJ, Hoeijmakers JHJ, Kaptein R, Boelens R. The structure of the human ERCC1/XPF interaction domains reveals a complementary role for the two proteins in nucleotide excision repair. Structure. 2005;13:1849–1858. doi: 10.1016/j.str.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 38.Choi YJ, Ryu KS, Ko YM, Chae YK, Pelton JG, Wemmer DE, Choi BS. Biophysical characterization of the interaction domains and mapping of the contact residues in the XPF–ERCC1 complex. J. Biol. Chem. 2005;280:28644–28652. doi: 10.1074/jbc.M501083200. [DOI] [PubMed] [Google Scholar]
- 39.Biggerstaff M, Szymkowski DE, Wood RD. Co-correction of the ERCC1, ERCC4 and Xeroderma-Pigmentosum group-F DNA-repair defects in-vitro. EMBO J. 1993;12:3685–3692. doi: 10.1002/j.1460-2075.1993.tb06043.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niedernhofer LJ, Bhagwat N, Wood RD. ERCC1 and non-small-cell lung cancer. N. Engl. J. Med. 2007;356:2538–2540. doi: 10.1056/NEJMc070742. [DOI] [PubMed] [Google Scholar]
- 41.Jaspers NGJ, Raams A, Silengo MC, Wijgers N, Niedernhofer LJ, Robinson AR, Giglia-Mari G, Hoogstraten D, Kleijer WJ, Hoeijmakers JHJ, Vermeulen W. First reported patient with human ERCC1 deficiency has cerebrooculo-facio-skeletal syndrome with amild defect in nucleotide excision repair and severe developmental failure. Am. J. Hum. Genet. 2007;80:457–466. doi: 10.1086/512486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Das D, Tripsianes K, Jaspers NGJ, Hoeijmakers JHJ, Kaptein R, Boelens R, Folkers GE. The HhH domain of the human DNA repair protein XPF forms stable homodimers. Proteins Struct. Funct. Bioinform. 2008;70:1551–1563. doi: 10.1002/prot.21635. [DOI] [PubMed] [Google Scholar]
- 43.Vanduin M, Vredeveldt G, Mayne LV, Odijk H, Vermeulen W, Klein B, Weeda G, Hoeijmakers JHJ, Bootsma D, Westerveld A. The cloned human DNA excision repair gene Errc-1 fails to correct Xeroderma Pigmentosum complementation group-A through group-I. Mutat. Res. 1989;217:83–92. doi: 10.1016/0921-8777(89)90059-1. [DOI] [PubMed] [Google Scholar]
- 44.Tian M, Shinkura R, Shinkura N, Alt FW. Growth retardation, early death, and DNA repair defects in mice deficient for the nucleotide excision repair enzyme XPF. Mol. Cell. Biol. 2004;24:1200–1205. doi: 10.1128/MCB.24.3.1200-1205.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weeda G, Donker I, deWit J, Morreau H, Janssens R, Vissers CJ, Nigg A, vanSteeg H, Bootsma D, Hoeijmakers JHJ. Disruption of mouse ERCC1 results in a novel repair syndrome with growth failure, nuclear abnormalities and senescence. Curr. Biol. 1997;7:427–439. doi: 10.1016/s0960-9822(06)00190-4. [DOI] [PubMed] [Google Scholar]
- 46.Mcwhir J, Selfridge J, Harrison DJ, Squires S, Melton DW. Mice with DNA repair gene (Ercc-1) deficiency have elevated levels of p53, liver nuclear abnormalities and die before weaning. Nat. Genet. 1993;5:217–224. doi: 10.1038/ng1193-217. [DOI] [PubMed] [Google Scholar]
- 47.Zheng HY, Wang X, Warren AJ, Legerski RJ, Nairn RS, Hamilton JW, Li L. Nucleotide excision repair- and polymerase eta-mediated error-prone removal of mitomycin C interstrand cross-links. Mol. Cell. Biol. 2003;23:754–761. doi: 10.1128/MCB.23.2.754-761.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bergstralh DT, Sekelsky J. Interstrand crosslink repair: can XPF–ERCC1 be let off the hook? Trends Genet. 2008;24:70–76. doi: 10.1016/j.tig.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 49.Wynne P, Newton C, Ledermann J, Olaitan A, Mould TA, Hartley JA. Enhanced repair of DNA interstrand crosslinking in ovarian cancer cells from patients following treatment with platinum-based chemotherapy. Br. J. Cancer. 2007;97:927–933. doi: 10.1038/sj.bjc.6603973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McHugh P, Spanswick V, Hartley J. Repair of DNA Interstrand crosslinks: molecular mechanisms and clinical relevance. Lancet Oncol. 2001;2(8):483–490. doi: 10.1016/S1470-2045(01)00454-5. [DOI] [PubMed] [Google Scholar]
- 51.Andersson BS, Sadeghi T, Siciliano MJ, Legerski R, Murray D. Nucleotide excision repair genes as determinants of cellular sensitivity to cyclophosphamide analogs. Cancer Chemother. Pharmacol. 1996;38:406–416. doi: 10.1007/s002800050504. [DOI] [PubMed] [Google Scholar]
- 52.Carvalho H, Ortolan TG, Depaula T, Leite RA, Weinlich R, Marante-Mendes GP, Menck CFM. Sustained activation of p53 in confluent nucleotide excision repair-deficient cells resistant to ultraviolet-induced apoptosis. DNA Repair. 2008;7:922–931. doi: 10.1016/j.dnarep.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 53.Fink D, Zheng H, Nebel S, Norris PS, Aebi S, Lin TP, Nehme A, Christen RD, Haas M, MacLeod CL, Howell SB. In vitro and in vivo resistance to cisplatin in cells that have lost DNA mismatch repair. Cancer Res. 1997;57:1841–1845. [PubMed] [Google Scholar]
- 54.Chang IY, Kim MH, Kim HB, Lee DY, Kim SH, Kim HY, You HJ. Small interfering RNA-induced suppression of ERCC1 enhances sensitivity of human cancer cells to cisplatin. Biochem. Biophys. Res. Commun. 2005;327:225–233. doi: 10.1016/j.bbrc.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 55.Cummings M, Higginbottom K, McGurk CJ, Wong OGW, Koberle B, Oliver RTD, Masters JR. XPA versus ERCC1 as chemosensitising agents to cisplatin and mitomycin C in prostate cancer cells: role of ERCC1 in homologous recombination repair. Biochem. Pharmacol. 2006;72:166–175. doi: 10.1016/j.bcp.2006.04.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.