Abstract
Epigenetic regulation of gene expression by DNA methylation is a central mechanism governing the silencing of tumor suppressor genes in many forms of cancer. Current methods have not proven optimal for the quantitative analysis of DNA methylation and corresponding in situ protein expression within cells in small specimens like skin biopsies. We have overcome this limitation by combining and modifying several techniques: target cell enrichment, DNA micro-isolation, one-step denaturation/bisulphite conversion/in-column desulphonation, specially designed PCR amplification, pyrosequencing and multispectral image analysis. Using this approach optimized for small samples, we can quantify minor alterations in gene methylation and protein expression using minimal amounts of tissue. Comparative studies of fresh and processed cells showed that our method is valid for DNA in both fresh and formalin-fixed, paraffin-embedded specimens. We can measure the effects of DNA methylation inhibitors, administered in vitro or in vivo, on the promoter methylation and protein expression of selected genes in specific cells. This novel approach should prove useful for a wide variety of investigative and clinical applications in dermatology and other specialties where the collection of small, routinely processed biopsy specimens is common. We refer to this method as Q-GAME (quantitative gene analysis of methylation and expression).
Keywords: FAS/CD95, laser capture microdissection, methylation, multispectral imaging, pyrosequencing
Introduction
Epigenetic regulation of gene expression is an area of intensive investigation relevant to the pathogenesis of many forms of neoplasia including skin cancers (1–3). One major focus of epigenetic regulation is the silencing of tumor suppressor genes by DNA methylation. This mechanism is relevant to the pathogenesis of a wide variety of cutaneous tumors including non-melanoma skin cancers, melanomas and cutaneous T-cell lymphomas (CTCL) (4–10). Analysis of promoter methylation has been important for clinical applications such as prognostication and therapy selection (11–19). Methods exist for genome-wide screening of the methylome. However, once a gene of interest is identified, there is no available method that allows detailed quantitative analysis of methylation at all relevant CpG sites coupled with quantitative analysis of corresponding protein expression in situ. This is especially relevant for clinical samples such as skin biopsy specimens whose small size, heterogeneous cellular composition and standard processing by formalin fixation and paraffin embedding (FFPE) pose special challenges for analysis of gene methylation and its effect on protein expression. No techniques described previously have adequately addressed these challenges (19–29).
In this report, we describe a method that we have named Quantitative Gene Analysis of Methylation and Expression or Q-GAME. It allows quantitative assessment of gene methylation and corresponding in situ protein expression. It involves target cell enrichment, DNA micro-isolation, one-step denaturation/bisulphite conversion/in-column desulphonation, a specially designed 50-cycle PCR amplification, pyrosequencing and multispectral image analysis. Q-GAME has been optimized for analysis of limited numbers of fresh or processed cells in suspensions or in small, archival biopsy specimens. This approach is proving valuable not only for documenting baseline epigenetic regulation of gene expression but also for monitoring alterations of gene expression induced by therapy.
Materials and methods
Cell lines and tissues
All clinical samples were obtained with institutional review board approval. All patients gave informed consent, and all protocols adhered to the Declaration of Helsinki principles. Archival skin biopsy specimens from 11 CTCL patients were the source of cells for laser capture microdissection. For validation studies, we also used human CTCL lines HH, SZ4, MyLa, Hut 78, human T-cell acute lymphoblastic leukaemia cell line Jurkat and human melanoma cell line A375, all of which have been described previously (30, 31). Cells were cultured and harvested by centrifugation. The cell pellets were processed in a clinical laboratory using standard procedures, that is, formalin fixation, paraffin embedding, microtome sectioning and eosin staining.
Targeted cell enrichment
Specific cells were enriched from 5-µm tissue sections of formalin-fixed, paraffin-embedded human skin biopsy specimens using a laser capture microdissection instrument (MDS Analytical Technologies, Ontario, Canada) as described previously (22). Based on the average diameter of the cells (11 µm) and the range in area of the microdissected patches (1000–24 000 µm2), there were approximately 10–300 cells per patch. The number of patches that can be harvested is limited only by density of the target cell population and the size of the specimen. A typical 6-mm-diameter biopsy specimen can provide at least several hundred 5-µm tissue sections. For this study, 500–5000 total cells were collected onto the inside surface of the cap of the microdissection collecting tube. All dissections were performed by an experienced dermatopathologist. Benign keratinocytes and neoplastic T cells were recognized by morphologic criteria and harvested separately for analysis.
DNA isolation
Cell lines or microdissected samples were suspended in 25 µl of lysis buffer containing proteinase K and were digested overnight in 56°C. Carrier RNA was added, and DNA was precipitated and purified through the QIAamp MinElute Column (DNA purification kit QIAamp DNA Micro, Qiagen, Valencia, CA, USA).
One-step denaturation/bisulphite conversion/in-column desulphonation
Bisulphite conversion was performed using the MethylCode Bisulfite Conversion kit (Invitrogen, Carlsbad, CA, USA), which integrates the denaturation and conversion into one step, with in-column desulphonation and without sodium hydroxide denaturing. Total DNA was added to the cytosine–uracil conversion reaction solution and run in a thermal cycler at 98°C for 10 min, 64°C for 2.5 h, and then harvested by column matrix. For microdissected cells, 2–100 ng of DNA was used for bisulphite conversion. For cell lines, 400 ng of DNA was used for each conversion reaction. DNA was then recovered and used for PCR amplification.
Polymerase chain reaction amplification
The promoter region of FAS gene was PCR amplified using 8 sets of forward primers and biotinylated reverse primers designed by PSQ Assay Design (Qiagen) [8]. The product sizes were between 100 and 200 bp. PCR was run on a DNA Engine thermal cycler (Bio-Rad, Hercules, CA, USA) as previously described (8). The promoter region of FAS ligand was amplified using 9 sets of primers biotinylated for forward or reverse according to software design (see Table S1), and the PCR products were generated using the same protocol as for FAS. Each PCR mix contained 1×PCR buffer, the forward and reverse primers at 0.4 µm, 200 µm of each of dNTP, 2.5 units of HotStar Taq polymerase (Qiagen) and ≤2 ng of bisulphite-treated template DNA in a total volume of 50 µl. Optimal PCR conditions were determined empirically: initial denaturing at 95°C for 15 min, 50 cycles at 94°C for 1 min, 55°C for 1 min and 72°C for 1 min; and final extension at 72°C for 10 min. For assessment of PCR products by electrophoresis, 20 µl of PCR reaction solution was added directly to a 1% agarose gel mixed with ethidium bromide. DNA bands were visualized by UV light.
Pyrosequencing
Pyrosequencing was performed on a PyroMark MD pyrosequencer (Qiagen) as previously described (8). Briefly, 5 µl of PCR product was immobilized by 2 µl of streptavidin–Sepharose beads (GE Healthcare, Piscataway, NJ, USA), washed and denatured using a vacuum prep workstation (Qiagen). The single-stranded PCR products were annealed with sequencing primers in a pyrosequencing plate, and the plate was placed in a Pyromark Q96 MD machine (Qiagen) for pyrosequencing reaction. Pyro Gold reagents were added to the reaction automatically by six dispensers. Eight sequencing primers specific to bisulphite-treated FAS promoter DNA, and nine for FAS-ligand promoter DNA, were designed using PyroMark Assay Design 2.0 software (Qiagen) (see Table S1). Each pyrosequencing primer set included one biotinylated primer, one unlabelled primer and one sequencing primer. Pyro Q-CpG™ software was used to analyse each reaction and to calculate the corresponding percentage of methylation at each CpG island.
In situ multispectral imaging
For quantitative analysis of protein expression in situ, we used the Nuance multispectral imaging system (Perkin-Elmer, Waltham, MA, USA) (32). This consists of a light microscope equipped with a liquid crystal tunable filter, high-resolution digital camera and a computer loaded with proprietary image analysis software. Specific detection of FAS/CD95 protein and CD30 protein was performed using anti-FAS/CD95 mAb (clone DX2) from Enzo Life Sciences, Farmingdale, NY, anti-CD30 mAb (clone Ber-H2) from Dako, Carpenteria, CA, and the MACH4 Universal HRP-Polymer detection system (Biocare Medical, Concord, CA) visualized with 3,3-diaminobenzidine (DAB). Sections 5 µm thick were counterstained with either methylene blue or haematoxylin. Controls included normal serum and isotype antibody as well as counterstain only and immunostain without counterstain. These single colour controls allowed acquisition of a multispectral signature for each chromogen in the staining system (here purple for haematoxylin and brown for DAB). After tissue section staining and digital image capture, cells of interest were circled individually within the image and the system measured the amount of DAB signal intensity, automatically converting these data into optical density (OD) units. Positive staining was adjusted by subtracting background control signals. The results were recorded on a cell-by-cell basis as avg. OD value/cell. They were expressed as histograms of mean/median staining intensity or spike plots of individual cell intensities arranged to depict the full range of antigen expression within the cell population being studied. Expression was also shown in photomicrographs of immunostains.
Flow cytometry immunostaining
For comparison of FAS expression by CTCL lines assessed by flow cytometry versus multispectral imaging, fresh cells were stained with FITC-conjugated anti-FAS/CD95 mAb (clone DX2) from Becton Dickinson, San Jose, CA. Isotype-matched mAbs of irrelevant specificity were used as negative controls. Briefly, 2 × 105 lymphoid cells from cultured cell lines were washed twice with phosphate-buffered saline (PBS) and blocked with 1:10 normal goat serum in phosphate-buffered saline for 20 min. Cells were immunostained for 30 min at room temperature, then washed twice with PBS, resuspended in FACS buffer (2% BSA inPBS), and analysed with a LSR II bench top flow cytometer (BD Biosciences, San Jose, CA, USA). Data analysis was performed by FlowJo software.
Statistic analysis
Statistical analysis was performed by Student’s t-test. A P-value < 0.05 was considered statistically significant. In the figures, (*) and (**) represent P < 0.05 and P < 0.01, respectively. The relative standard deviations of different samples were calculated as coefficient of variation (CV) values.
Results
Q-GAME overview
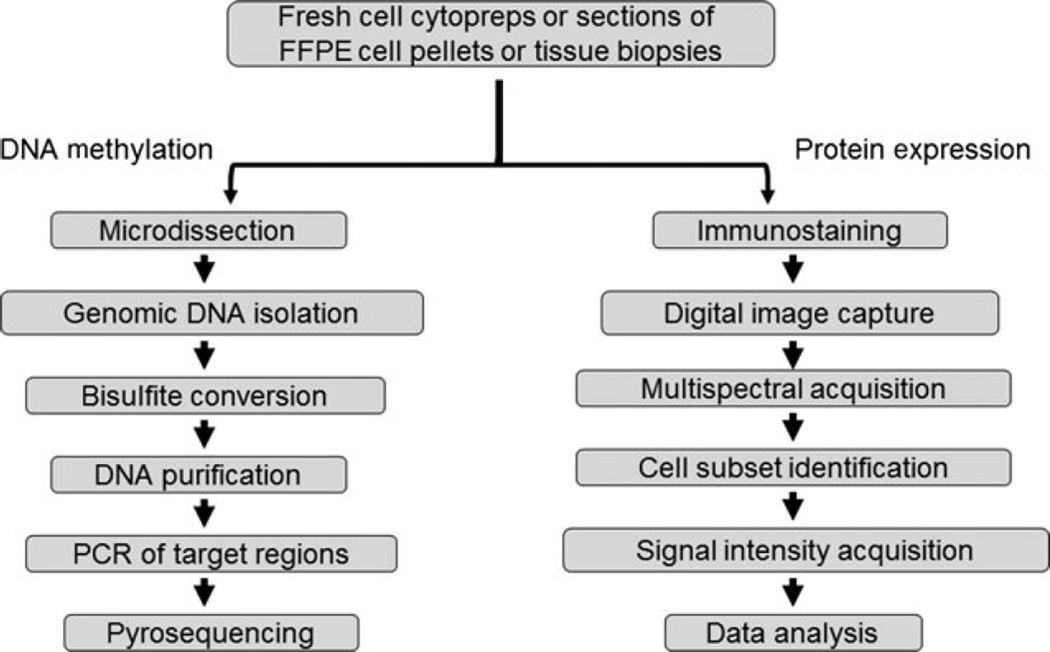
Our Q-GAME method consists of two arms summarized in Fig. 1. Details can be found in the Materials and Methods section. The DNA methylation arm includes the following steps: (i) microdissection, (ii) genomic DNA micro-isolation, (iii) one-step denaturation/ bisulphite conversion/in-column desulphonation, (iv) DNA purification, (v) PCR of target regions, (vi) pyrosequencing. The source of DNA can be fresh cell suspensions, FFPE cell pellets or cells microdissected from FFPE tissue specimens. The in situ protein expression arm includes the following steps: (i) immunostaining, (ii) digital image capture, (iii) acquisition of the multispectral signature for each chromogen, (iv) identification of cell subset to be analysed, (v) acquisition of signal intensity on a cell-by-cell basis and (vi) data analysis. Material suitable for image analysis includes cyto-preparations of fresh cell suspensions and sections of FFPE cell pellets and tissues. For comparison between pyrosequencing and multispectral imaging results in tissues, we used semiserial tissue sections.
Figure 1.
Q-GAME flow chart.
DNA requirements for fresh and FFPE samples
In tissue biopsy specimens, infiltrating lymphoid cells and constituent normal cells such as epidermal keratinocytes can be recognized by pathologists and harvested separately. For each cell type in each slide, we microdissected multiple patches of cells according to the availability of the pure cell regions on the slide. The microdissection software calculated the surface area of the patches which ranged from approximately 1000 to 24 000 µm2. Given a typical cell diameter of 10–12 µm, there were approximately 10–300 cells in each cell patch. Typically, 500–5000 cells of each type were collected from each specimen. Using these amounts of harvested cells, the yield of DNA before bisulphite conversion ranged between 10 and 100 ng. The quantity of DNA lost in bisulphite processing was generally <50%, and the range of total converted DNA yields for downstream PCR amplification was approximately 3–80 ng. To find the minimum quantity of DNA needed for pyrosequencing, we tested DNA samples ranging from 100 pg to 100 ng. The minimal amount of converted DNA needed for each PCR in our experiments was around 500 pg; however, the final result had less run-to-run variation when using amounts of DNA in the ng range. The minimal DNA amount extracted from microdissected archival FFPE cells that was suitable for pyrosequencing was similar to the minimal amount of DNA required from fresh unprocessed cells.
Quantitative DNA methylation assay
To optimize a method for the quantitative analysis of all CpG sites within a specific gene region of interest, the 2 kb FAS/CD95 promoter region immediately upstream of the start codon was selected as a model (8). Our choice was based on the well-established recognition that human CTCL exhibits interpersonal variation in the extent of DNA methylation of this gene segment. Furthermore, the extent of methylation among the 37 CpG sites within this region shows considerable variation, even within an individual case, as shown in our previous study of CTCL and other T-cell lines (8). Specifically, the extent of promoter methylation at six CpG sites (termed CpG 1, 2, 10, 14, 15 and 16) correlates inversely with FAS protein expression.
Optimal preparation and yield of DNA for pyrosequencing were achieved with one-step denaturation/bisulphite conversion/in-column desulphonation. PCR conditions were empirically adjusted to eliminate free biotinylated primers that could cause noise signals. To test this method across a region containing multiple CpG sites, we studied all 37 CpG sites in the 2 kb FAS/CD95 promoter region using DNA extracted from fresh CTCL lines as well as microdissected tumor cells and keratinocytes from FFPE CTCL skin biopsy specimens. The pyrosequencing detection was successful with clean pyrograms in all 8 amplicons from 4 CTCL lines and 19 CTCL biopsy samples obtained from 11 patients. A representative pyrogram is shown in Figure S1. The biopsy samples ranged in age from 1 to 11 years (median 6 years). This indicates that our protocols for DNA isolation and bisulphite conversion are suitable for both fresh and FFPE material, including archival pathology specimens.
To determine the validity of analysing FFPE DNA, we compared the status of the FAS promoter in DNA isolated from FFPE versus fresh samples of 4 different CTCL lines that vary in their level of FAS promoter methylation. FFPE specimens were prepared by processing cell pellets with the same formalin fixation and paraffin embedding used for processing tissue biopsy specimens. The results confirmed the validity of FFPE DNA results with methylation values differing <5% between fresh and fixed versions of the same CTCL line at 18 distinct CpG sites (Figure S2). These results also demonstrate the ability of our method to distinguish the higher FAS promoter methylation levels present in the HH and SZ4 CTCL lines compared with the MyLa and Hut-78 CTCL lines.
To determine the extent of run-to-run variations, we performed triplicate assays starting from DNA isolation for 19 microdissected CTCL and keratinocyte samples obtained from 8 FFPE lesional skin biopsies from 3 CTCL patients. Within a single sample, the per cent methylation showed only minimal variation in repeated runs. Figure S3 shows representative results for CpG site 2. The SD values ranged from 0.2% to 3.1% in three repeated pyrosequencing assays for each bar. The coefficient of variation (CV) values of the 19 samples ranged from 3.2% to 4.8%. Across all these 19 samples, the mean methylation level at CpG 2 ranged from 42% to 55%. Several of the higher values differed significantly from the lowest. Hence, the pyrosequencing results were highly reproducible within individual microdissection samples, and the method was able to detect significantly different methylation levels among different samples and cases.
Measuring changes in DNA methylation
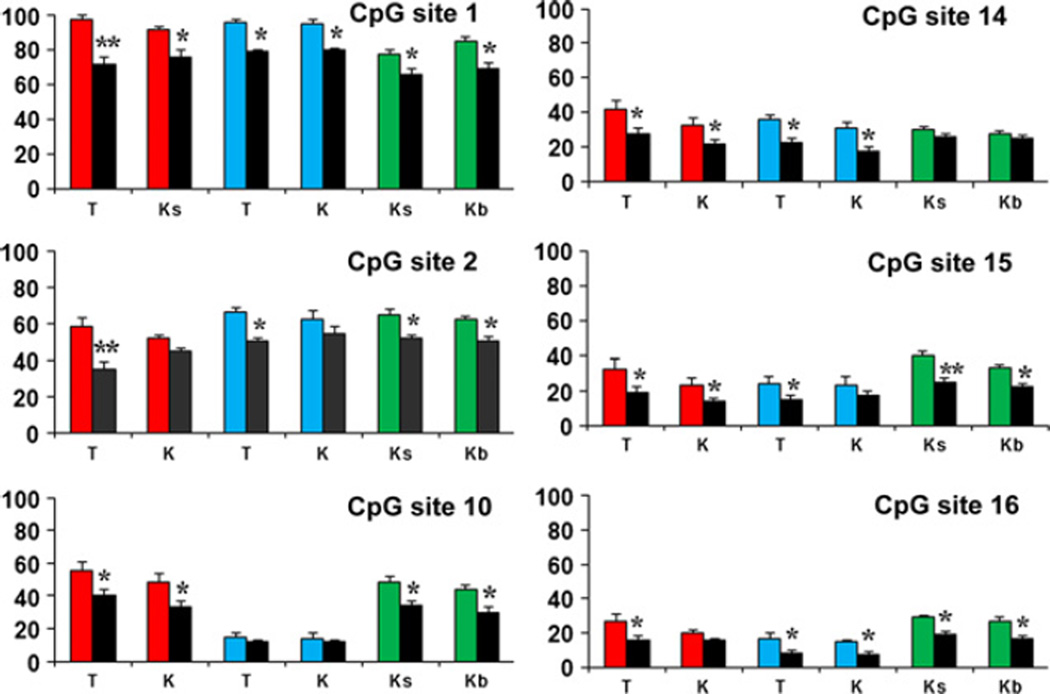
We reported previously that methotrexate (MTX) functions as a DNA methylation inhibitor by blocking synthesis of S-adenosylmethionine, the principal methyl donor for DNA methyltransferases [8]. In fresh CTCL lines, the level of FAS promoter methylation was reduced at 6 distinct CpG sites in response to MTX treatment in the two lines with high baseline methylation (HH, SZ4) but not the two with low baseline methylation (MyLa, Hut-78) (Figure S4). Consistent with these in vitro findings, the level of FAS promoter methylation was significantly reduced at these same six CpG sites within both tumor cells and normal keratinocytes microdissected from FFPE lesional skin biopsy specimens of CTCL patients treated with MTX (Figure 2).
Figure 2.
MTX-induced promoter demethylation detected in microdissected FFPE cells. Many CpG sites (each panel number represents a different site) show significantly reduced methylation in total keratinocytes (K), spinous keratinocytes (Ks), basal keratinocytes (Kb) and T cells (T) microdissected from 3 CTCL patients before (3 different coloured bars) and after (black bars) MTX therapy. Y-axis: per cent methylation. Statistical analysis was performed using Student’s t-test, and a P-value <0.05 was considered statistically significant. (*) and (**) represent P < 0.05 and P < 0.01 respectively.
Testing other cell lines and genes
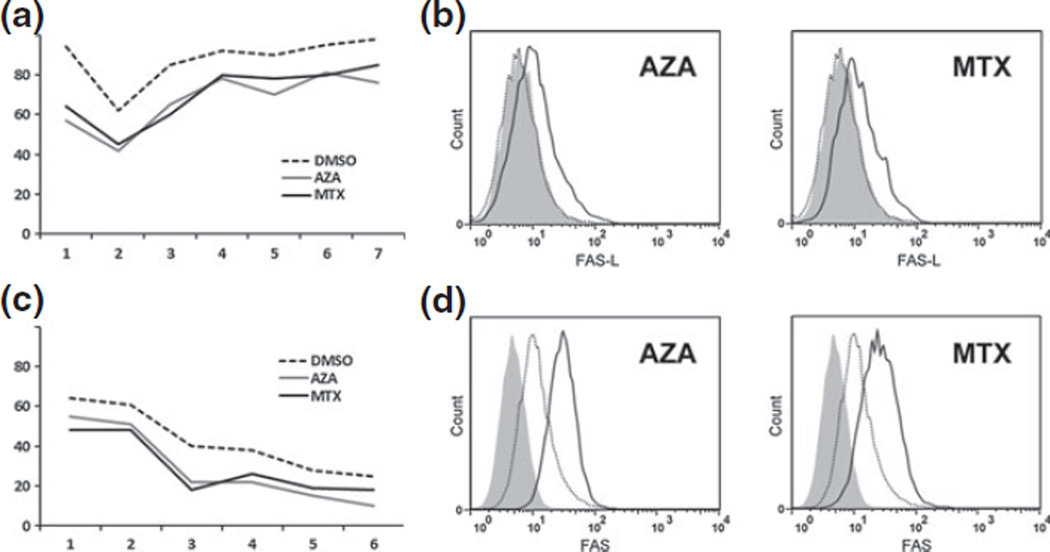
To test our method in other epigenetic models, we analysed the effects of demethylation on FAS in the A375 melanoma cell line and on FAS ligand (FASL) in the HH CTCL cell line. Figure 3 demonstrates that we can quantify alterations in promoter methylation and corresponding protein expression in these additional models just as we have carried out for FAS in multiple CTCL cell lines.
Figure 3.
Monitoring promoter methylation in other models. Pyrosequencing results demonstrate that our methods can monitor FAS-ligand promoter demethylation in CTCL line HH (panel a) as well as FAS promoter demethylation in melanoma cell line A375 (panel c). Two demethylators were used: 5-azacytidine and MTX. Flow cytometry demonstrates that promoter methylation is associated with increased expression of FASL (panel b) and FAS (panel d) in these systems. a and c: Y-axis shows per cent methylation. X-axis shows multiple distinct CpG sites. b and d: Y-axis shows relative number of cells. X-axis shows intensity of antigen expression. Curves show isotype control (solid grey), specific antibody before demethylator treatment (dotted line) and specific antibody after demethylator treatment (solid line).
Quantitative in situ protein expression assay
To validate the ability of multispectral imaging to measure protein expression in situ, we used 4 CTCL lines to compare the expression of FAS assessed by flow cytometry of cell suspensions to the expression assessed by multispectral imaging of cyto-preparations. The results were very similar and showed the same hierarchy of FAS expression levels (Figure S5).
Measuring changes in protein expression in situ
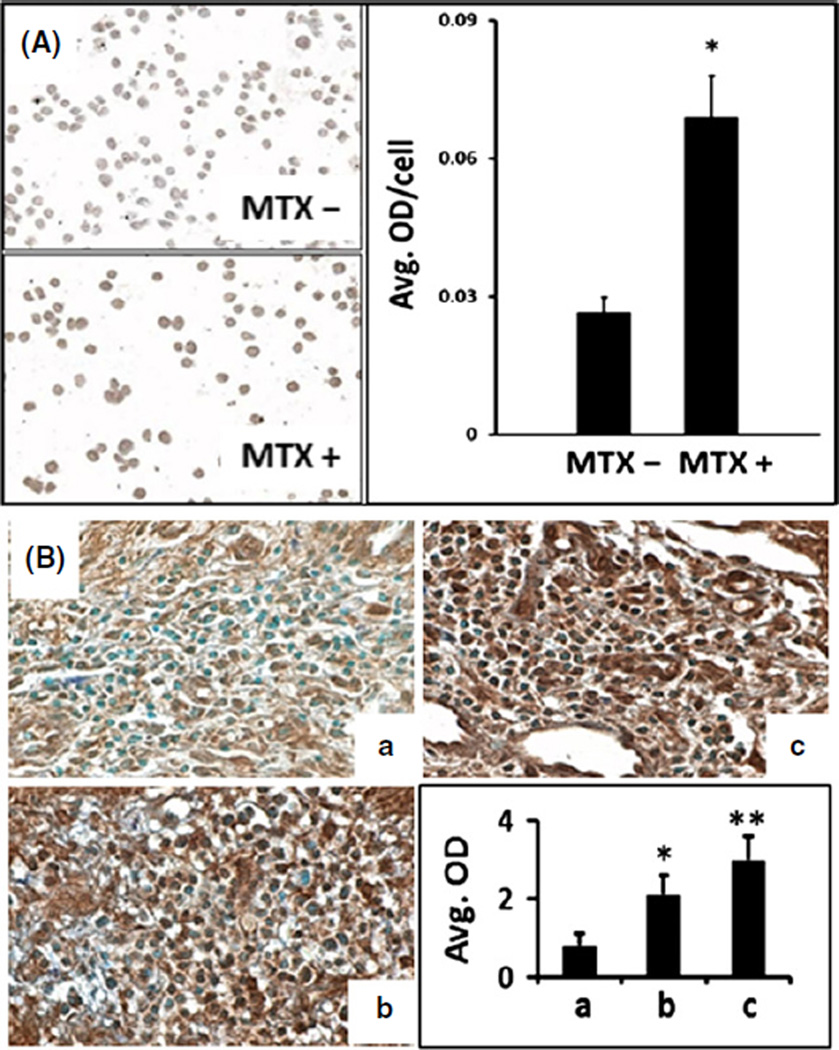
In CTCL lines, the demethylation of the FAS promoter by MTX led to a corresponding increase in FAS protein expression that could be appreciated by visual inspection of immunostained cytopreparations and quantified using multispectral imaging (Figure 4a). As shown in Figure 4b, tumor cells in lesional skin from CTCL patients treated with MTX also showed increased FAS protein expression that could be quantified by multispectral imaging and that correlated with clinical improvement.
Figure 4.
Multispectral imaging quantifies alterations in FAS protein expression induced by MTX. (A) Photomicrographs and corresponding multispectral imaging histograms show increased FAS expression (*P < 0.01) induced by MTX in CTCL line HH (A) and in serial lesional skin biopsies from a CTCL patient (B). Samples Ba, Bb and Bc represent pre-MTX, MTX for 2 years and MTX for 5 years, respectively. The progressive increase in lesional FAS expression was associated with a gradual improvement in the extent of skin lesions assessed by the percentage of body surface area (BSA) involved. Image magnification ×400. OD: optical density quantified by multispectral imaging. Statistical analysis was performed using Student’s t-test, and a P-value <0.05 was considered statistically significant. (*) and (**) represent P < 0.05 and P < 0.01, respectively.
Sensitivity of multispectral imaging for detecting low-level antigen expression
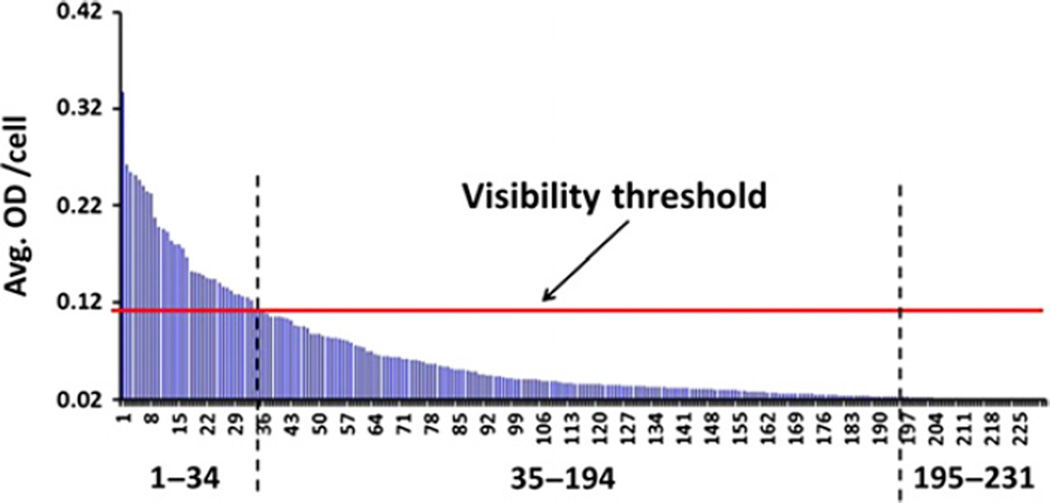
Multispectral imaging is superior to visual inspection via conventional light microscopy (LM) for detecting antigen expression because it is more sensitive than the human eye and allows a quantitative assessment of the protein amount that cannot be achieved by simple microscopic observation. As illustrated in Figure 5, multispectral imaging can detect low-level protein expression (in this case CD30) that is significantly above-background controls yet below the threshold of detectability by LM. This threshold, which we refer to as the visibility threshold (Figure 5), was established by viewing LM images of cells with previously determined OD values in 11 biopsies and ranged between 0.1 and 0.14 OD (median 0.11). Cells with OD values equal to or above the visibility threshold were identified as CD30 positive by both LM and multispectral imaging, whereas cells with values that were lower than the threshold but exceeded 0.02 (OD of DAB background stain, including 2 SD) were recognized as positive by multispectral imaging while appearing negative by LM. Although the threshold values varied somewhat from biopsy to biopsy (possibly due to differences in the intensity of the counterstain which might have visually obscured very weak yet above-background specific staining), these data indicate that multispectral imaging is approximately 5× more sensitive than the human eye (0.02 vs. 0.11 O.D.).
Figure 5.
Multispectral imaging is more sensitive than the human eye. A total of 231 CTCL cells in this biopsy were analysed for CD30 protein expression using multispectral imaging. Each blue spike represents the optical density (OD) value of an individual cell. Background OD (0.02, including 2 SD) was subtracted from all values, and any spike therefore signifies specific CD30 staining. The threshold of visibility indicates the OD below which CD30 stain was undetectable by visual inspection with a standard light microscope (LM), but positive per multispectral imaging analysis. In this example, CD30 stain was identifiable by standard LM in 34 of 231 cells (15%), whereas the majority of cells (cells 35–194, or 69%) showed low-level expression that was undetectable by LM but was captured by multispectral imaging. Because the OD values of cells 195–231 (16%) did not differ from non-specific background, they were assessed as CD30 negative by both LM as well as multispectral imaging.
Discussion
Using the FAS/CD95 promoter region as an example, we have shown how Quantitative Gene Analysis of Methylation and Expression or Q-GAME (which involves a combination of multiple techniques including pyrosequencing and multispectral imaging) can be used to quantitatively measure gene methylation and corresponding protein expression in fresh cells or archival skin biopsy specimens, either at baseline or in response to therapy. There are several aspects of Q-GAME methodology that make it novel and particularly well suited to analysis of both cells and biopsy specimens. It requires only small amounts of cells or tissues. DNA preparation and pyrosequencing have been optimized to yield reproducibly high-quality methylation results using only nanogram amounts of DNA. By combining this approach with multispectral image analysis, it is possible to determine the effects of gene methylation on corresponding protein expression. Multispectral imaging requires only a few standard tissue sections, preserves tissue architecture and allows in situ quantification of protein expression with a level of sensitivity superior to the human eye. Because multispectral imaging can be performed using multiple immunomarkers, specific cells can also be identified by co-expression of key antigens (32). Importantly, both fresh and FFPE samples are suitable for Q-GAME. This means that no special tissue processing is required and that retrospective studies can utilize the extensive archival tissue banks available at many academic medical centres. Our studies produced high-quality results using specimens ranging from 1 to 10 years in age.
These attributes make our method superior to several alternatives. Some CpG sites are not assessable using restriction endonuclease digestion and methylation-specific PCR (MSP). Bisulphite sequencing is laborious and requires about 10 clone reads. These approaches lack direct quantitative information, and there are potential challenges with reproducibility, time, expense and statistical analysis. The application of recently developed next-generation sequencing technology to methylation detection can provide screening information regarding the entire methylome (33). However, it is costly and inadequate for detailed quantitative analysis of all CpG sites in specific genes in small samples. The number of CpG sites analysed in a single assay using conventional methods is often limited. Therefore, the methylation levels at these CpG sites can be misinterpreted as representative of the whole promoter. These assays may not account for the fact that each CpG site in a specific gene promoter may be methylated to a different extent. Furthermore, most methylation assays require fresh as opposed to archival DNA or amounts of DNA that are too large to harvest from small tissue samples.
Some methods of cell harvesting for DNA methylation analysis, such as macrodissection, are imprecise and prone to cross-contamination with unwanted cell types. Prior attempts at combining laser capture microdissection with bisulphite conversion and pyrosequencing have proven suboptimal, yielding inconsistent methylation data (22, 28). This might be due to the use of conventional NaOH-based denaturing for bisulphite conversion which can cause considerable DNA degradation and incomplete conversion (29, 34, 35). In our method, the DNA lost in bisulphite processing was around 50%, compared with at least 84% in traditional methods (84–96%) (36). Others have used pyrosequencing to analyse DNA methylation in cells manually microdissected or laser microdissected from cancer tissues (19, 27); however, these studies sometimes combined captured cells from multiple tissue blocks, thus solving the problem of cell heterogeneity but not the problem of insufficient cell quantity, which exists for many small archival specimens.
Q-GAME is quantitative rather than qualitative. Therefore, alterations of gene methylation or protein expression can be precisely measured at baseline, over time and in response to therapeutic intervention without the subjectivity inherent in conventional immunohistopathological estimates of the percentage of positive cells or their staining intensities expressed on a semiquantitative scale. The sensitivity of multispectral image analysis is also highly advantageous. Low-level protein expression that might be misinterpreted as negative by visual inspection can still be detected and quantified. For example, this has proven relevant to explaining the efficacy of antibody therapy targeted at low-level CD30 antigen expression by cutaneous T-cell lymphomas that were judged to be CD30 negative by conventional immunohistology (37). Those interested in our method can compare and contrast it to several other studies published in Experimental Dermatology that involve analysis of gene methylation or protein expression in clinical samples (38–43).
In summary, Q-GAME allows highly sensitive and quantitative measurement of gene methylation and associated protein expression in specific cell subsets present in small, routinely processed biopsy specimens. As such, it is proving particularly valuable for the analysis of epigenetic regulation in both laboratory and clinical settings.
Supplementary Material
Acknowledgements
Dr. Wood designed the research study; Drs. Wu, Salva and Stutz performed the research; Drs. Longley and Spiegelman contributed essential reagents and tools; all authors analysed the data and wrote the manuscript. Supported by Merit Review funding from the Department of Veterans Affairs (Dr. Wood) and NIH T32 postdoctoral training Grant (AR055893, Dr. Salva).
Abbreviations
- CTCL
cutaneous T-cell lymphoma
- CpG
cytosine phosphoguanosine
Footnotes
Conflict of interest
None of the authors has any conflict of interest to disclose.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Table S1. Pyrosequencing primers FAS (FAS-1 to FAS-8) and FAS ligand (FASL-1 to FASL-9) primers.
Figure S1. Representative pyrogram. Bisulphite converted DNA from FFPE laser captured cells was amplified using primer sets designed for the region spanning the target CpGs. PCR products were used for pyrosequencing. The figure shows the high quality of the pyrogram. The shadowed columns highlight analysed T/C peaks indicating converted cytosine and unconverted cytosine respectively at each CpG site.
Figure S2. Pyrosequencing results are not affected by FFPE. Pyrosequencing of four CTCL lines demonstrated that FFPE samples showed levels of FAS promoter methylation that accurately reflected those seen in corresponding fresh samples regardless of whether the lines were inherently methylation high or low.
Figure S3. Per cent methylation at FAS promoter CpG site 2 in 19 different microdissected FFPE cell samples. The SD values ranged from 0.2% to 3.1% in three repeated pyrosequencing assays for each bar. The coefficient of variation (CV) values of the 19 samples ranged from 3.2% to 4.8%. Samples included CTCL tumor cells (black columns), basal keratinocytes (grey columns) and spinous keratinocytes (white columns) harvested from CTCL lesional skin. Several values were significantly different than the lowest value marked with an arrow. Statistical analysis was performed using Student’s t-test and a P-value <0.05 was considered statistically significant. (*) and (**) represent P < 0.05 and P < 0.01 respectively.
Figure S4. Methotrexate (MTX) demethylates the FAS promoter. Pyrosequencing results for fresh DNA isolated from four different CTCL lines before and after MTX treatment. Y-axis: per cent methylation. X-axis: CpG sites.
Figure S5. FAS protein expression by CTCL lines. Multispectral image analysis and flow cytometry yield similar results. MFI: mean fluorescence intensity quantified by flow cytometry. OD: optical density quantified by multispectral imaging.
References
- 1.Esteller M, Corn PG, Baylin SB, et al. Cancer Res. 2001;61:3225–3229. [PubMed] [Google Scholar]
- 2.Jones PA, Baylin SB. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 3.Herman JG, Baylin SB. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 4.Worm J, Bartkova J, Kirkin AF, et al. Oncogene. 2000;19:5111–5115. doi: 10.1038/sj.onc.1203891. [DOI] [PubMed] [Google Scholar]
- 5.Furuta J, Umebayashi Y, Miyamoto K, et al. Cancer Sci. 2004;95:962–968. doi: 10.1111/j.1349-7006.2004.tb03184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonazzi VF, Irwin D, Hayward NK. Genes Chromosom Cancer. 2009;48:10–21. doi: 10.1002/gcc.20615. [DOI] [PubMed] [Google Scholar]
- 7.Jayaraj P, Sen S, Sharma A, et al. Br J Dermatol. 2012;167:583–590. doi: 10.1111/j.1365-2133.2012.10968.x. [DOI] [PubMed] [Google Scholar]
- 8.Wu J, Wood GS. Arch Dermatol. 2011;147:443–449. doi: 10.1001/archdermatol.2010.376. [DOI] [PubMed] [Google Scholar]
- 9.Gallardo F, Esteller M, Pujol RM, et al. Haematologica. 2004;89:1401–1403. [PubMed] [Google Scholar]
- 10.van Doorn R, Zoutman WH, Dijkman R, et al. J Clin Oncol. 2005;23:3886–3896. doi: 10.1200/JCO.2005.11.353. [DOI] [PubMed] [Google Scholar]
- 11.Kawamoto K, Enokida H, Gotanda T, et al. Biochem Biophys Res Commun. 2006;339:790–796. doi: 10.1016/j.bbrc.2005.11.072. [DOI] [PubMed] [Google Scholar]
- 12.Laird PW. Nat Rev Cancer. 2003;3:253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- 13.Shi H, Wang MX, Caldwell CW. Expert Rev Mol Diagn. 2007;7:519–531. doi: 10.1586/14737159.7.5.519. [DOI] [PubMed] [Google Scholar]
- 14.Aggerholm A, Holm MS, Guldberg P, et al. Eur J Haematol. 2006;76:23–32. doi: 10.1111/j.1600-0609.2005.00559.x. [DOI] [PubMed] [Google Scholar]
- 15.Fischer JR, Ohnmacht U, Rieger N, et al. Lung Cancer. 2006;54:109–116. doi: 10.1016/j.lungcan.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 16.Müller HM, Widschwendter A, Fiegl H, et al. Cancer Res. 2003;63:7641–7645. [PubMed] [Google Scholar]
- 17.Wallner M, Herbst A, Behrens A, et al. Clin Cancer Res. 2006;12:7347–7352. doi: 10.1158/1078-0432.CCR-06-1264. [DOI] [PubMed] [Google Scholar]
- 18.Yu J, Cheng YY, Tao Q, et al. Gastroenterology. 2009;136:640–651. doi: 10.1053/j.gastro.2008.10.050. [DOI] [PubMed] [Google Scholar]
- 19.Buckingham L, Penfield FL, Kim A, et al. Int J Cancer. 2010;126:1630–1639. doi: 10.1002/ijc.24896. [DOI] [PubMed] [Google Scholar]
- 20.Moribe T, Iizuka N, Miura T, et al. Int J Cancer. 2009;125:388–397. doi: 10.1002/ijc.24394. [DOI] [PubMed] [Google Scholar]
- 21.Kerjean A, Vieillefond A, Thiounn N, et al. Nucleic Acids Res. 2001;29:e106. doi: 10.1093/nar/29.21.e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Irahara N, Nosho K, Baba Y, et al. J Mol Diagn. 2010;12:177–183. doi: 10.2353/jmoldx.2010.090106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tost J, Gut IG. Nat Protoc. 2007;2:2265–2275. doi: 10.1038/nprot.2007.314. [DOI] [PubMed] [Google Scholar]
- 24.Kim M, Kim JH, Jang HR, et al. Cancer Res. 2008;68:7147–7155. doi: 10.1158/0008-5472.CAN-08-0667. [DOI] [PubMed] [Google Scholar]
- 25.Bian YS, Yan P, Osterheld MC, et al. Biotechniques. 2001;30:66–72. doi: 10.2144/01301st02. [DOI] [PubMed] [Google Scholar]
- 26.Warnecke PM, Stirzaker C, Melki JR, et al. Nucleic Acids Res. 1997;25:4422–4426. doi: 10.1093/nar/25.21.4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodriguez-Canales J, Hanson JC, Tangrea MA, et al. J Pathol. 2007;211:410–419. doi: 10.1002/path.2133. [DOI] [PubMed] [Google Scholar]
- 28.Hiura H, Obata Y, Komiyama J, et al. Genes Cells. 2006;11:353–361. doi: 10.1111/j.1365-2443.2006.00943.x. [DOI] [PubMed] [Google Scholar]
- 29.Zon G, Barker MA, Kaur P, et al. Anal Biochem. 2009;392:117–125. doi: 10.1016/j.ab.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 30.Wu J, Nihal M, Siddiqui J, et al. J Invest Dermatol. 2009;129:1165–1173. doi: 10.1038/jid.2008.309. [DOI] [PubMed] [Google Scholar]
- 31.Nihal M, Roelke CT, Wood GS. Pharm Res. 2010;27:1103–1114. doi: 10.1007/s11095-010-0054-5. [DOI] [PubMed] [Google Scholar]
- 32.Mansfield JR, Hoyt C, Levenson RM. Curr Protoc Mol Biol. 2008;84:1–14. doi: 10.1002/0471142727.mb1419s84. [DOI] [PubMed] [Google Scholar]
- 33.Meaburn E, Schulz R. Semin Cell Dev Biol. 2012;23:192–199. doi: 10.1016/j.semcdb.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 34.Xu C, Houck JR, Fan W, et al. J Mol Diagn. 2008;2:129–134. doi: 10.2353/jmoldx.2008.070131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ogino S, Kawasaki T, Brahmandam M, et al. J Mol Diagn. 2006;8:209–217. doi: 10.2353/jmoldx.2006.050135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grunau C, Clark SJ, Rosenthal A. Nucleic Acids Res. 2001;29:E65–E65. doi: 10.1093/nar/29.13.e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krathen MS, Sundram U, Bashey S, et al. Blood. 2012;120:797. [Google Scholar]
- 38.Elder JT, Zhao X. Exp Dermatol. 2002;11:406–412. doi: 10.1034/j.1600-0625.2002.110503.x. [DOI] [PubMed] [Google Scholar]
- 39.Li Y, Liu ZL, Zhang KL, et al. Exp Dermatol. 2009;18:842–848. doi: 10.1111/j.1600-0625.2009.00922.x. [DOI] [PubMed] [Google Scholar]
- 40.Heitzer E, Bambach I, Dandachi N, et al. Exp Dermatol. 2010;19:926–928. doi: 10.1111/j.1600-0625.2010.01120.x. [DOI] [PubMed] [Google Scholar]
- 41.Bazhin AV, Smet CD, Golovastova MO, et al. Exp Dermatol. 2010;19:1023–1025. doi: 10.1111/j.1600-0625.2010.01126.x. [DOI] [PubMed] [Google Scholar]
- 42.Chen NN, Li Y, Wu ML, et al. Exp Dermatol. 2012;21:13–18. doi: 10.1111/j.1600-0625.2011.01392.x. [DOI] [PubMed] [Google Scholar]
- 43.Schäfer A, Gratchev A, Seebode C, et al. Exp Dermatol. 2013;22:486–489. doi: 10.1111/exd.12166. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.