Abstract
Objective
We aimed to examine the association of apolipoprotein E (APOE) ε4 genotype with neuroimaging markers of Alzheimer’s disease: hippocampal volume, brain amyloid deposition and cerebral metabolism.
Methods
We performed a systematic review and meta-analysis of 14 cross-sectional studies identified in Pubmed from 1996 to 2014 (n=1628). The pooled standard mean difference (SMD) was used to estimate the association between APOE and hippocampal volume and amyloid deposition. Meta-analysis was performed using effect size signed differential mapping using coordinates extracted from clusters with statistically significant difference in cerebral metabolic rate for glucose between APOE ε4+ and ε4− groups.
Results
APOE ε4 carrier status was associated with atrophic hippocampal volume (pooled SMD: −0.47; 95% CI −0.82 to −0.13; p=0.007) and increased cerebral amyloid positron emission tomography tracer (pooled SMD: 0.62, 95% CI 0.27 to 0.98, p=0.0006). APOE ε4 was also associated with decreased cerebral metabolism, especially in right middle frontal gyrus.
Conclusions
APOE ε4 was associated with atrophic hippocampal volume in MRI markers, increased cerebral amyloid deposition and cerebral hypometabolism. Theses associations may indicate the potential role of the APOE gene in the pathophysiology of Alzheimer’s disease.
INTRODUCTION
Alzheimer’s disease (AD) is the most common form of age-related dementia, accounting for nearly 80% of all cases. The ε4 allele of the apolipoprotein E (APOE) gene is by far the major risk factor for dementia, especially AD. The ε4 allele has been confirmed as playing a pivotal role in AD because it is less effective in breaking down the peptide amyloid-β, which consequently leads to an increased risk of formation of the characteristic AD plaques. However, whether the ε polymorphism is also associated with the neuroimaging markers is unclear. Indeed, the advances in neuroimaging technologies have allowed us to investigate the relationship in detail between the APOE ε4 allele and certain neuroimaging markers of AD, such as structural MRI, fluorodeoxyglucose positron emission tomography (FDG-PET) and PET-amyloid tracers capable of delineating the extent and distribution of amyloid-β (Aβ) deposits in the brain. Thus, with the discovery of this common susceptibility gene for late onset AD, numerous explorers became engrossed in using the imaging techniques to detect and track brain changes associated with the predisposition to AD in carriers of the ε4 allele of the APOE gene. Neuroimaging markers of AD, including hippocampal atrophy, Aβ burden and cerebral glucose hypometabolism, are important predictors of AD. Dissecting the relationship between the APOE ε4 allele and the neuroimaging markers of AD could give us new clues to the mechanisms underlying the association between APOE and risk of AD.
MRI morphological evaluation has been widely used to explore the effect of APOE on the brain in AD subjects. The close clinical/anatomical correlation between hippocampal atrophy and memory deficits makes hippocampal atrophy a candidate marker to monitor disease progression in clinical trials.1 Besides, according to a meta-analysis of MRI studies, a statistically significant volume reduction of about 12% can be detected even in the preclinical stage.2 A number of previous studies suggest that the APOE genotype has effects on the hippocampal size, atrophy and hemispherical lateralisation.3,4 FDG-PET measurements of the cerebral metabolic rate for glucose (CMRgl) provide a promising quantitative neuroimaging endo-phenotype of AD risk. To date, Aβ deposition is one of the main hallmarks of AD because it was thought to eventually cause neuronal death. The application of [11C]-Pittsburgh compound B (PiB) was regarded as an important tool in imaging Aβ fibrillar pathology in vivo,5 even if it is reported to be a non-specific marker of Aβ-peptide related cerebral amyloidosis.
The biological basis for the underlying effect of APOE ε4 as a risk factor for developing AD is unknown yet. It has been reported that the APOE ε4 allele was associated with a faster pathological progression of brain lesions, greater cerebral atrophy and lower regional CMRgl. To date, no meta-analysis of such studies has been conducted on the association between the APOE ε4 allele and the neuroimaging markers. Thus, our aim is to provide a systematic review and meta-analysis of studies evaluating the relationship of the APOE ε4 allele with the three neuroimaging markers of AD.
METHODS
Search strategy and selection criteria
The literature published from 1 January 1996 to 1 March 2014 was systematically screened in the PubMed, MEDLINE according to preferred reporting items for systematic reviews and meta-analyses (PRISMA) guidelines using the following terms in the title, abstract or descriptors: APOE, Apolipoprotein E, MRI, hippocampal, volume, PET, PiB, amyloid, glucose, Alzheimer disease, AD. We restricted the search to studies in humans.
We included studies testing the association of the APOE genotype with at least one of the neuroimaging markers of AD. The following were including criteria: (1) peer-reviewed; (2) original studies; (3) reported in English; (4) including at least 10 subjects; (5) mild cognitive impairment (MCI), AD (clinical diagnosis of AD was based on the National Institute of Neurologic and Communicative Disorders and Stroke-Alzheimer disease and Related Disorders Association criteria) or healthy normal were involved; (6) for MRI, only structural MRI of hippocampal volume was selected; (7) for FDG-PET studies, it used statistically significant thresholds for data that were either corrected for multiple comparisons or uncorrected with spatial extent thresholds.
Data extraction and quality assessment
Two authors independently extracted the following data from the studies: sample size, study population, mean age, MRI characteristics and sequences, PET characteristics and associated technical details. For measures of the association between APOE genotype and MRI markers, we recorded the mean, SD or SE for the continuous hippocampal volumes. If a study provided raw volume (ie, mm3) and normalised volume of the hippocampus,6 the latter would be chosen. Two studies provided left and right hippocampal volumes separately, and were all included in meta-analysis. For measures of the association between APOE genotype and florbetapir PET markers, the mean and SD on florbetapir standardised uptake values ratios (SUVRs) or the global [11C] Pittsburgh compound B non-displaceable binding potential were extracted for the included florbetapir [11C] and [18F] PET studies. For measures of the association between APOE genotype and FDG-PET marker, we recorded the coordinates and t values in each study independently according to the effect size signed differential mapping (ES-SDM) method.7 When none could be extracted, we contacted the authors to provide those via email.8 If measures of association remained unavailable thereafter, qualitative results were reported. Two authors extracted the above information from each study, resolving any disagreement by discussion.
Statistical analyses
Meta-analysis was performed when at least three studies were available for the same outcome. Associations of hippocampal atrophy with APOE ε4 genotype were summarised between APOE ε4 carriers (ε4+ group including ε4ε4, ε3ε4) and APOE ε4 non-carriers (ε4− group including ε3ε3). For studies that provide three groups including APOE ε4 homozygote genotype, heterozygote genotype (eg, ε3ε4, ε2ε4) and APOE ε4 non-carrier, we chose the APOE ε4 homozygote genotype to be the APOE ε4+ group first.
For hippocampal volume and PET-amyloid calculation, a meta-analysis of all statistics was performed using standard mean difference (SMD) methodology in Review Manager separately (V.5.2.3 for Windows Copenhagen: The Nordic Cochrane Centre, The Cochrane Collaboration, 2012). For hippocampal volume, we performed left hippocampal volume and right hippocampal volume analyses to explore the association between characteristics of studies and their results. A fixed-effects meta-analysis was used in the absence of heterogeneity and a random-effects model if the heterogeneity between studies was statistically significant. Overall heterogeneity was assessed using the Cochran Q (p value >0.10 on the Q test, which reflects a lack of heterogeneity among studies) and I2 (values of more than 50% was considered as ‘considerable heterogeneity’).9,10 We used the 95% CI to gauge the precision of the summary estimates. Publication bias was investigated using funnel plots, with a roughly symmetrical distributed on either side of the summary estimate suggesting a lack of bias.
Separate voxel-based morphometry (VBM) meta-analyses of CMRgl via FDG-PET were conducted with ES-SDM,7,9 which has already been used successfully to meta-analyse studies on neuropsychiatric disorders and AD.11–13 This method is based on using the peak coordinates to recreate, for each study, a map of the effect size of the differences between the two compared groups, and then conducting a standard random-effects variance-weighted meta-analysis in each voxel.7 Using the ES-SDM software, VBM was performed on the included studies to compare the CMRgl changes between the APOE ε4+ group and the APOE ε4− group. A systematic whole-brain voxel-based jackknife sensitivity analysis was carried out to test the replicability of the results. All these processes were referred to the ES-SDM tutorial and publications. The statistical threshold for this analysis was set to a p value of <0.0001. Sensitivity analysis is conducted to ensure that no single study will bias the combined results by removing one study each time and recalculating the stability of the remaining studies.
RESULTS
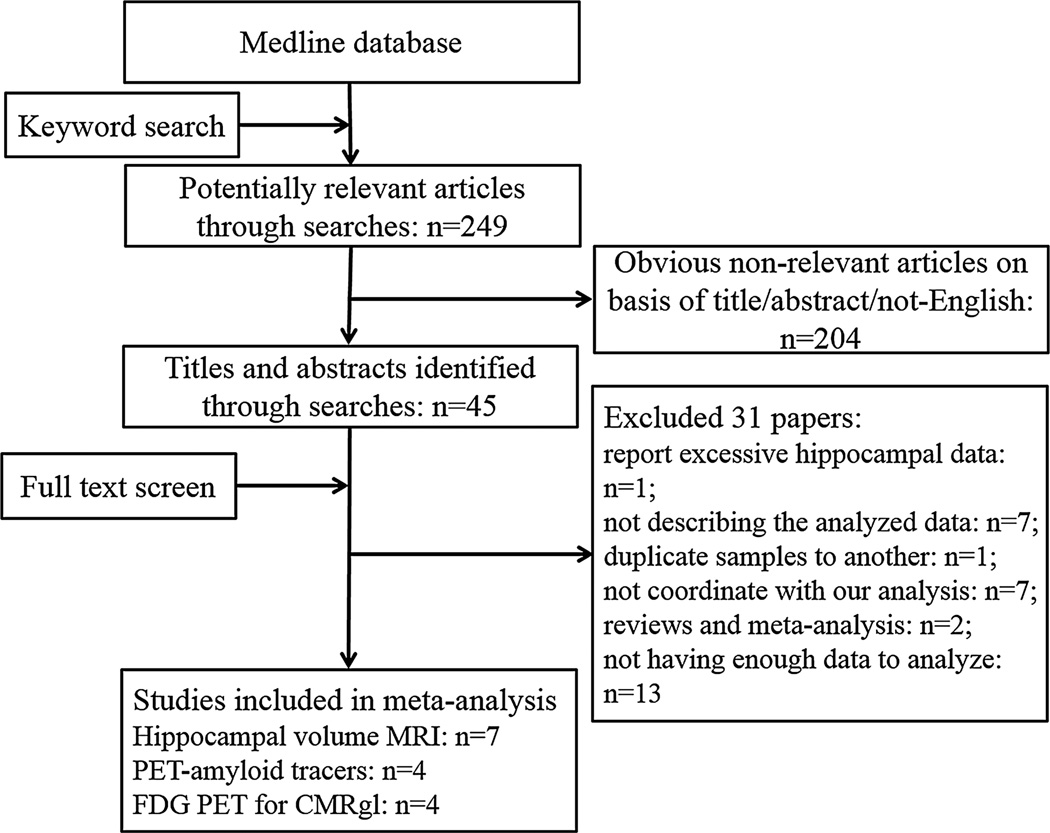
The initial literature search identified 249 potentially relevant articles, of which 45 met the inclusion criteria. After screening the full text, 31 articles were excluded because of different reasons (figure 1). Finally, 14 articles that were published between 1996 and 2014 met the selection criteria and had accessible information to study the association between APOE and the three neuroimaging biomarkers (figure 1). A total of 620 APOE ε4 carriers (APOE ε4+ group) and 1008 APOE ε4 non-carriers (APOE ε4− group) were included. In each study, no statistically significant difference was found in age, gender and mini-mental-state examination score between the APOE ε4 + group and the APOE ε4− groups. Sensitivity analysis was first used and no outliers were found in this study. Upon removal of any study (each study from AD, MCI and healthy controls), there was no statistically significant change in the pooled SMD for hippocampal volume analysis (SMD was between −0.31 and −0.56) or for the amyloid PET analysis (SMD was between 0.58 and 0.73). Thus, the overall results of this meta-analysis were statistically robust.
Figure 1.
Flow chart of studies through screening, inclusion and exclusion. CMRgl, cerebral metabolic rates for glucose; FDG PET, flurodeoxyglucose positron emission tomography
APOE ε4 and MRI marker of AD (hippocampal volume)
Description of studies
Fourteen studies investigated the association between APOE ε4 and hippocampal volume. Of the excluded studies, three studies reported structural MRI data which conducted voxel-based analysis; one study focused on the segmented and constructed hippocampal surface morphometry statistics14; four studies’ data could not be used for analysis.3 Hence, a total of seven studies which met the inclusion criteria were entered into our meta-analysis,4,6,8,15–18 including 171 APOE ε4 carriers (APOE ε4+ group) and 201 APOE ε4 non-carriers (APOE ε4− group). Of the six included studies, four studies reported the hippocampal volume was significantly smaller in APOE ε4+ subjects than in APOE ε4− subjects, whereas two studies didn’t find any statistically significant difference. The pooled subjects included healthy normal subjects, MCI subjects and subjects diagnosed with AD in Europe (one study), Asia (two studies), North America (two studies) and Oceania (one study). All the studies showed no statistically significant difference in age and mini-mental-state examination between APOE ε4 carriers and APOE ε4 non-carriers. Data details of the included studies are presented in table 1.
Table 1.
Characteristics of the included studies for the meta-analysis of MRI
| MRI operating parameters | Mean age (y) | Sex, Male (%) | MMSE | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Study name | Origin | Field (Tesla) |
TR/TE (ms) |
Field of view (mm) |
Matrix | Flip angle (°) |
Hippocampal volumetry |
Study used |
APOE+ | APOE− | APOE+ | APOE− | APOE+ | APOE− |
| Pievani et al16 | Italy | 1 | 5000/100 | 220 | 256×256 | 90 | Normalised HFV (R/L) | AD | 71.7±8.2 | 71.2±9.5 | 12 (86) | 8 (57) | 20.1±4.4 | 19.8±4.7 |
| Mori et al15 | Japan | 1.5 | 14/3 | 220 | 256×256 | 20 | HFV | AD | 72.4±5.5 | 72.4±5.5 | 6 (15.8) | 4 (6.4) | 19.2±5.1 | 19.1±6.3 |
| Rowe et al18 | Australia | NA | 2300/2.98 | 160 | 240×256 | 9 | The proportion of the total ICV | MCI | NA | NA | NA | NA | NA | NA |
| Hashimoto et al6 | Japan | 1.5 | 14/3 | 220 | 256×256 | 20 | Normalised HFV | AD | 69.5±6.0 | 69.5±6.6 | 16 (34.8) | 16 (34.8) | 19.9±4.5 | 19.9±4.2 |
| Reiman et al4 | USA | 1.5 | 33/5 | 240 | 256×192 | 30 | Normalised HFV (R/L) | Normal* | NA | NA | NA | NA | NA | NA |
| Protas et al17 | USA | 1.5 | 33/5 | 240 | 256×192 | 30 | Normalised HFV | Normal | 55.5±5.1 | 56.5±4.7 | 10 (32.2) | 28 (36.8) | 29.8±0.6 | 29.8±0.5 |
Cognitively normal and with a family history of Alzheimer’s dementia in at least one first-degree relative.
AD, Alzheimer’s disease; HFV, hippocampal formation volume (right-left averaged); ICV, intracranial volume; MCI, mild cognitive impairment; MMSE, mini-mental-state examination; NA, not available; R/L, right/left separated.
Hippocampal volume
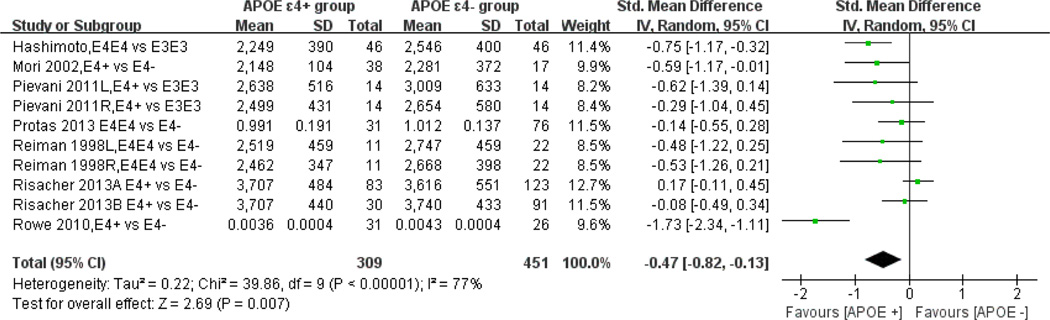
As shown in the forest plots (figure 2), combining the six studies using continuous hippocampal volume and providing weighted mean differences (n=309 APOE ε4+ population and n=451 APOE ε4− population), yielded a statistically significant association between the APOE ε4 allele and atrophic hippocampal volume: pooled standardised mean difference=−0.47 (95% CI −0.82 to −0.13) (I2=77%, Cochran’s Q=39.86, p<0.0001); the test for overall effect: Z=2.69, p=0.007. Four studies found a statistically significant association between the APOE ε4 allele and the hippocampal atrophy,6,8,16,18 and one study16 found a statistically significant association between the APOE ε4 allele and right hippocampal atrophy, while other studies did not.
Figure 2.
Meta-analysis of studies testing the association between APOE ε4+ and hippocampal volume. Data type: continuous, Effect measure: standard mean difference, Analysis model: random effects, Statistical method: inverse variance. Risacher 2013A, the early mild cognitive impairment (E-MCI) group; 2013B, the healthy control group.
APOE ε4 and PET markers of AD
Description of included studies
The search strategy identified four papers8,19–21 met the inclusion criteria for the PET-amyloid tracer study and four papers for the FDG-PET study.22–25 No additional articles were found in the reference list of the selected studies. For the PET-amyloid tracer study, data from two studies26,27 could not be used for analysis in this meta-analysis. The clinical, demographic and technical data of participants from all recruited studies and two excluded studies are presented in table 2. A total of 208 APOE ε4 carriers and 345 APOE ε4 non-carriers were included. For the FDG-PET study, four studies met the selection criteria, including a total of 212 APOE ε4 non-carriers and 103 APOE ε4 carriers. The detailed clinical and demographic data of participants from the four recruited studies are presented in table 3. The technical details of the included studies are shown in the table 4.
Table 2.
Demographic, clinical and technical characteristics of PET studies for amyloid deposition in meta-analysis
| Study | Included in the meta analysis |
Population | PET | VBM (p value) |
Analysis software for PET |
PET marker | Significant brain regions by VBM |
Subjects analysis |
APOE genotype |
Sample size |
Age, years* | Male (%) | MMSE* |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ossenkoppele et al20 | Yes† | NA | Dynamic [11C] PIB and static [18F] FDG-PET | No | PVE-lab | Global [11C] PIB BPND | – | AD | ε4 (−) | 22 | 61 (44–77) | 16 (72.7%) | 24±3 |
| ε4 ε4 | 22 | 65 (50–80) | 13 (59.9) | 24±4 | |||||||||
| Risacher et al8 | Yes | USA | [11C] Florbetapir PET | Yes, p<0.01 | SPM 8 Freesurfer version | Florbetapir SUVR | Left orbitofrontal cortex; medial frontal lobe/anterior cingulate cortex; right orbitofrontal cortex and the posterior cingulate/precuneus | HC | ε4 (−) | 93 | 74.1±0.72 | 49 (52.7%) | 29.05±0.14 |
| ε4 (+) | 30 | 73.67±1.27 | 13 (43.3%) | 28.91±0.25 | |||||||||
| E-MCI | ε4 (−) | 124 | 71.47±0.62 | 64 (51.6%) | 28.57±0.12 | ||||||||
| ε4 (+) | 85 | 70.26±0.75 | 52 (61.2%) | 28.06±0.15 | |||||||||
| Grimmer et al26 | No | Germany | [11C] PIB PET | No | – | [11C] PIB uptake ratio | All parts of the neocortex | AD | ε4 (−) | 38 | – | – | – |
| ε4ε4 | 19 | – | – | – | |||||||||
| Drzezga et al25 | No | Germany | [11C] PIB PET | Yes | SPM5 | [11C] PIB retention | Bilateral temporoparietal and frontal cortex | AD | ε4 (+) | 18 | 67.1±7.8 | 9 (50%) | 20.1±4.5 |
| ε4 (−) | 14 | 68.1±10 | 9 (64.28%) | 21.8±3.6 | |||||||||
| Fleisher et al19 | Yes | USA | Florbetapir PET | – | SPM5 | SUVR | Bilateral temporoparietal and frontal cortex | DAT | ε4 (−) | 22 | 76.41±10.19 | 11 (47.8%) | 21.05±3.97 |
| ε4 (+) | 23 | 73.09±7.65 | 11 (50%) | 20.26±4.65 | |||||||||
| MCI | ε4 (−) | 32 | 70.00±10.64 | 16 (50%) | 27.56±1.64 | ||||||||
| ε4 (+) | 21 | 72.76±10.11 | 6 (29%) | 27.38±1.96 | |||||||||
| oHC | ε4 (−) | 46 | 68.11±11.32 | 21 (45.7%) | 29.63±0.49 | ||||||||
| ε4 (+) | 15 | 67.27±11.71 | 6 (40%) | 29.40±0.51 | |||||||||
| Yi et al21 | Yes | Korea | [11C] PIB PET | Yes | SPM8 | Global cortical [11C] PIB retention | Left superior frontal gyrus; Left superior temporal gyrus; Right middle temporal gyrus | HC | ε4 (−) | 15 | 73.1±4.9 | 7 (46.7%) | 52.1±7.7 |
| ε4 (+) | 17 | 70.2±4.9 | 4 (23.5%) | 46.75±10.5 |
Data are presented as mean±SD or mean (range).
The data of Global [11C] PIB BPND was included in this meta-analysis.
AD, Alzheimer’s disease; BPND, non-displaceable binding potential; DAT, patients with AD dementia; E-MCI, early mild cognitive impairment; FDG, fluorodeoxyglucose; HC, healthy control; MMSE, mini mental-state examination; NA, not available; oHC, older healthy control; PET, positron emission tomography; PIB, Pittsburgh compound B; PVE-lab, a software program that uses a probability map based on 35 previously validated regions of interest (ROIs); SPM, statistical parametric mapping; SUVR, standardised uptake value ratio; VBM, voxel-based morphometry.
Table 3.
Demographic and clinical characteristics of VBM studies for hypometabolism in meta-analysis
| Number (male) | age | MMSE | |||||
|---|---|---|---|---|---|---|---|
| Study | Subjects | APOE ε4− | APOE ε4+ | APOE ε4− | APOE ε4+ | APOE ε4− | APOE ε4+ |
| Mosconi et al (2004)25 | AD | 46 (16) | 40 (12) | 73±13 | 76±9 | 21.8±3.5 | 23.4±3.4 |
| Jagust et al22 | HC | 135 (67) | 40 (20) | 77.3±6.1 | 76.8±5.6 | 29.1±1.2 | 28.6±1.3 |
| Reiman et al24 | HC | 15 (3) | 12 (3) | 31.2±5.0 | 30.7±5.4 | 29.9±0.3 | 29.9±0.3 |
| Langbaum et al23 | HC | 16 (4) | 11 (1) | 53.9±4.7 | 55.5±8.1 | 29.4±0.5 | 29.4±0.7 |
AD, Alzheimer’s disease; APOE ε4−, APOE ε4 non-carriers; APOE ε4, the ε 4 allele of the apolipoprotein E gene; APOE ε4+, APOE ε4 carriers; HC, cognitively normal and healthy control; MMSE, mini mental-state examination; VBM, voxel-based morphometry.
Table 4.
Technical details of VBM studies for CMRgl (regional CMRgl) in meta-analysis
| Study | PET scanner | Software | p Value | Brain areas (most significant correlation) | Coordinates* |
|---|---|---|---|---|---|
| Mosconi et al (2004)25 | 18F-FDG PET | SPM 99 | <0.001, uncorrected | Frontal and anterior cingulate | Talairach |
| Jagust et al22 | 18F-FDG PET | SPM5 | <0.001, uncorrected | Temporal, parietal and frontal cortex | Talairach |
| Reiman et al24 | 18F-FDG PET | SPM99 | <0.001, uncorrected | Posterior cingulate, parietal, temporal and prefrontal cortex | Talairach and Tournoux |
| Langbaum et al23 | 18F-FDG PET | SPM5 | <0.005, uncorrected | Middle and anterior cingulate cortex, hippocampus and thalamus | Talairach and Tournoux |
Each subject’s PET image was deformed according to the coordinates of a standard atlas of the brain. Measurements in each voxel were normalised to that in the pons. Atlas coordinates included x, y, z (mm). x is the distance in mm to the right (+) or left (−) of the midline, y is the distance anterior (+) or posterior (−) to the anterior commissure, and z is the distance superior (+) or inferior (−) to a horizontal plane through the anterior and posterior commissure.
CMRgl, cerebral glucose metabolism (cerebral metabolic rate for glucose); 18F-FDG, 18F-flurodeoxyglucose; PET, positron emission tomography; SPM, statistical parametric mapping; VBM, voxel-based morphometry.
APOE ε4 and PET-amyloid tracers
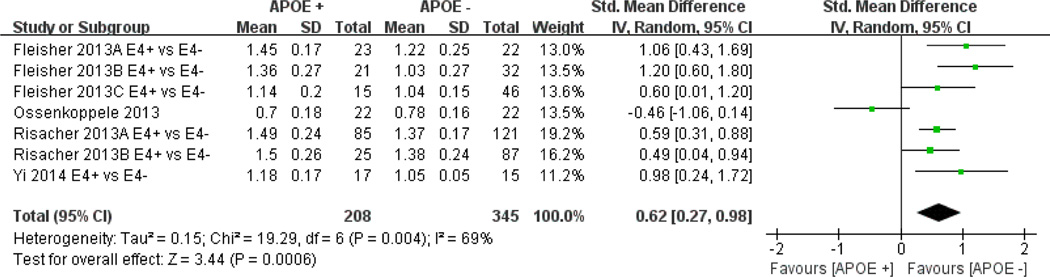
When meta-analysing four studies, including the subgroups of the two included studies, in a sample size-weighted meta-analysis, the APOE ε4 allele was significantly associated with increased amyloid deposition (test for overall effect: Z=3.44, p=0.0006). Combined analysis of the relationship between the cortical SUVR and APOE ε4 is shown in the forest plots (figure 3). The pooled SMD from a random-effects model was 0.14 (pooled SMD=0.62, 95% CI (0.27 to 0.98), I2=69%; Cochran’s Q=19.29, p=0.004). Of note, this meta-analysis included a total of AD (n+=45; n− = 44), MCI (n+=106; n− =153) and healthy controls (n+=67; n−=148).
Figure 3.
Meta-analysis of studies testing the association between APOE ε4+ and positron emission tomography-amyloid tracers. Data type: continuous, Effect measure: standard mean difference, Analysis model: random effects, Statistical method: inverse variance. (1) Fleisher 2013A, AD with Dementia (DAT) group; 2013B, mild cognitive impairment (MCI) group; 2013C, old healthy control group (2) Risacher 2013A, the early MCI (E-MCI) group; 2013B, the healthy control group.
APOE ε4 and FDG-PET for cerebral hypometabolism
The four included studies reported CMRgl reductions at coordinates in APOE ε4 carriers. A group comparison of APOE ε4 carriers and APOE ε4 non-carriers was carried out. In patients with the APOE ε4 allele, CMRgl reductions were found in the right cluster, that is, right middle frontal gyrus. As shown in table 5, the whole brain jackknife sensitivity analysis indicated a CMRgl reduction in the right middle frontal gyrus, which was highly replicable because they were consistent through all the combinations of the four studies.
Table 5.
Regional differences of CMRgl between APOE ε4+ and APOE ε4−
| Maximum | |||||
|---|---|---|---|---|---|
| Region | Talairach coordinates x, y, z |
SDM value |
Uncorrected p value |
Cluster breakdown (number of voxel) |
Jackknife sensitivity analysis (combination of studies detecting the differences) |
| Right middle frontal gyrus, Brodmann area 9 | 40, 36, 34 | 4.412 | 0.000012845 | 276 | 4 out of 4 |
| Right cingulate gyrus, Brodmann area 24 | 6, 8, 26 | 3.200 | 0.001856134 | 76 | 4 out of 4 |
| Left precuneus, Brodmann area 31 | −14, −44, 42 | 2.605 | 0.002793834 | 18 | 4 out of 4 |
| Left cingulate gyrus, Brodmann area 23 | −2, −34, 26 | 2.330 | 0.003493899 | 28 | 4 out of 4 |
Regions identified by meta-analysis of coordinates from four studies (voxelwise p<0.005).
APOE, the apolipoprotein E gene; CMRgl, cerebral metabolic rates for glucose; SDM, signed differential mapping.
DISCUSSION
The main purposes of our study were to analyse the published literature and to attempt to clarify the role of the APOE ε4 genotype. In this systematic review and meta-analysis comprising 14 studies with 1628 participants, the APOE ε4+ (including APOE ε4ε4) genotypes were associated with atrophic hippocampus, increased amyloid deposition and decreased CMRgl, especially in the right middle frontal gyrus.
One previous meta-analysis had examined the hippocampal volume and asymmetry in patients with MCI and AD.2 In their asymmetry analysis, a left-less-than-right pattern is found consistently in all three groups (MCI, AD and controls). However, previous reviews did not examine the association of APOE ε4 with the hippocampal atrophy. Mechanisms underlying the associations of APOE genotypes with hippocampal atrophy are unclear. Our meta-analyses reported more severe hippocampal involvement in patients with the APOE ε4 allele, which is compatible with the results of neuropathological and neurochemical studies.28,29 An association between the APOE ε4 genotype with densities of senile plaques and neurofibrillary tangles has been demonstrated in patients with AD.29 It is reported that densities of senile plaques and neurofibrillary tangles were related to the APOE ε4 allele only in the hippocampus, but there is no significant correlation between the ε4 allele frequency and density of senile plaques or neurofibrillary tangles in the neocortex (frontal, temporal and parietal lobes). Besides, evidence from cellular research and animal research demonstrates the detrimental effect of APOE ε4, reduced effectiveness in clearance of Aβ and modulation of τ phosphorylation in the hippocampus.30–33 To date, a few studies have tried to investigate the APOE ε4 effect on the hippocampal atrophy at the subregional level,34,35 however, these studies failed to detect an effect in regions not involved by the pathology. The result of the present meta-analysis supports previous investigations in showing that the regions most affected by AD pathology are more atrophic in carriers of the APOE ε4 allele. Whether the hippocampal atrophy is primarily due to the neuronal cell loss in the hippocampus or secondary to the degeneration of extra hippocampal fibres converging thereon remains to be determined. More and more studies on the association between APOE ε4 carrier status and hippocampal volume have become available, the exact mechanism will be worked out soon.
To our knowledge, no meta-analysis has to date evaluated the relationship between APOE ε4 polymorphism and PET-defined amyloid deposition via brain [18F] FDG uptake ratio (SUVR) or [11C] PiB uptake. We found a statistically significant association between APOE ε4 and PET-amyloid tracers, suggesting its potential effects on cortical amyloid burden. In line with our results, many of those studies that did not meet the strict inclusion criteria for the meta-analysis also found a positive association between the amyloid deposition via PET-amyloid tracers and AD. Grimmer et al27 reported that the cerebral [11C] PiB uptake ratio increased significantly in patients with AD during a relatively short interval of its clinical course, which was gene-dose-dependent to the number of APOE ε4 alleles. Another paper also identified the significant association between [11C] PiB uptake and the APOE ε4 allele in bilateral temporoparietal and frontal cortices.26 These studies were not directly comparable within our meta-analyses because of a lack of the associated original data or because they used a different method of calculation. In contrast, one of the included studies reported increased amyloid pathology in the frontal cortex in APOE ε4−patients.20 It seems rather paradoxical that a lack of the major genetic risk factor for AD is associated with increased pathological burden. It was elucidated that such inconsistent results might be related to confounding factors interfering with different assay protocols and demographic characteristics.
Amyloid plaques and τ neurofibrillary tangles, the pathological hallmarks of AD, begin accumulating in the healthy human brain decades before clinical dementia symptoms can be detected. In our analyses, a carrier of APOE ε4 was associated with increased amyloid load. It was hypothesised that APOE may function as an Aβ-binding protein that induces a pathological β sheet conformational change in Aβ,36 due to the strong association between APOE and Aβ in the brain. Initial histopathological studies investigating the relationship between amyloid plaques and APOE isoforms had demonstrated a positive correlation between plaque density and APOE ε4 allele dose.37 To explain the difference in amyloid plaque load between APOE ε4+ and APOE ε4− patients, it is speculated that the Aβ deposition may have been ongoing for a longer time in APOE ε4+ patients.20 This hypothesis was supported by data from studies in epidemiology and neuropathology, which demonstrated earlier onset of disease, or higher cerebral amyloid load in younger APOE ε4+ subjects with AD.38 Meanwhile, the second possible explanation may be found in a higher speed of amyloid accumulation in APOE ε4+ subjects over time during the course of disease. Additional studies are needed to explore this relationship further, including analyses in subgroups divided by diagnose from a more homogeneous population.
VBM results, which pooled VBM studies for a meta-analysis of CMRgl differences between APOE ε4+ and APOE ε4− subjects, also revealed robust cerebral glucose metabolite changes in the brain, especially the right middle frontal gyrus. Our analysis showed statistically significant reductions in metabolism in APOE ε4 carriers compared with non-carriers. This result remained largely unchanged when jackknife sensitivity analysis was performed. Thus, our results were robust and highly replicable. Subjects included in our meta-analysis for FDG-PET were mostly cognitive normal subjects at increased genetic risk for AD. This may be supported by the possibility that there is brain metabolic decrease in APOE carriers before the occurrence of AD. This voxelwise approach revealed a change in the medial surface, which is part of the default mode network. This network becomes disconnected in AD39 and was reported with evidence of Aβ deposition in normal older people.40 However, there was no obvious explanation as to why the right middle frontal gyrus was more affected in APOE ε4 carriers.
This is the largest systematic review and meta-analysis of the association of the APOE ε4 polymorphism with three neuroimaging markers of AD. In contrast with previous reviews, we have also included voxel-based meta-analysis.
There are several limitations in our meta-analyses. One limitation is publication bias, because some relevant studies may not have been included in the MEDLINE database and were inevitably missed in this study. Second, subjects included were of three diagnostic categories, MCI, AD and healthy subjects. AD was recognised with the pathological change in hippocampal atrophy, amyloid deposition and cerebral hypometabolism; MCI was recognised as the closest to AD. In our meta-analyses, the number of subgroups divided by subjects was not sufficient to conduct a meta-analysis. Third, we were limited by the fact that most studies provided effect estimates for APOE ε4+ vs APOE ε4− only, with varying reference groups. Many studies have used APOE ε4ε4, ε3ε3 and other genotypes as reference groups, rather than APOE ε4+ and ε4−, which may have reduced our power to detect association. Fourth, the detection of hippocampal volume could be correlated with magnetic strength and some studies included in this meta-analysis were performed using MRI scanners <1.5 T; the analysis for hippocampal volume is different in different laboratories. This may have led to inaccuracy in hippocampal volume. Fifth, the FDG-PET meta-analysis was based on the pooling of stereotactic coordinates with statistically significant differences rather than on raw data from the included studies and this may lead to less accurate results. However, this is the same in all meta-analytical approaches. Finally, as in all systematic reviews and meta-analyses of published data, some degree of publication bias can’t be excluded. Thus, confirmation of our findings could be obtained in the future by meta-analyses with large consortia using more harmonised phenotypical criteria and analytical models.
In summary, these multiple meta-analyses not only identify the APOE ε4 allele to be associated with atrophic hippocampal volume, increased amyloid deposition and hypometabolism in the brain, but also corroborate that the APOE gene is correlated with neuron loss and deposition of amyloid plaques, which are characterised neuropathological features of AD. Future studies examining the association between neuropathology of AD and APOE genotypes could also help in understanding the pathophysiological role of APOE in AD. Finally, we suggest that the potential of PET and MRI has not been fully explored and future studies could contribute to standardising the neuroimaging techniques for wider applications.
Footnotes
Contributors J-TY designed the analysis. C-CT, X-FM, CW and P-RH collected and abstracted the data. YL and H-FW carried out the statistical analysis. YL and J-TY drafted the manuscript. SLR and AJS provided some original data. All authors analysed and interpreted the data and critically revised the manuscript for important intellectual content. The contents of this study are solely the responsibility of the authors and do not necessarily represent the official view of their institutions or any other party. J-TY and LT have full access to all of the data and take full responsibility for the data, the analyses, and interpretation. All authors reviewed and approved the final report.
Competing interests None.
Provenance and peer review Not commissioned; externally peer reviewed.
REFERENCES
- 1.Petersen RC, Jack CR, Jr, Xu YC, et al. Memory and MRI-based hippocampal volumes in aging and AD. Neurology. 2000;54:581–587. doi: 10.1212/wnl.54.3.581. [DOI] [PubMed] [Google Scholar]
- 2.Shi F, Liu B, Zhou Y, et al. Hippocampal volume and asymmetry in mild cognitive impairment and Alzheimer’s disease: meta-analyses of MRI studies. Hippocampus. 2009;19:1055–1064. doi: 10.1002/hipo.20573. [DOI] [PubMed] [Google Scholar]
- 3.Geroldi C, Laakso MP, DeCarli C, et al. Apolipoprotein E genotype and hippocampal asymmetry in Alzheimer’s disease: a volumetric MRI study. J Neurol Neurosurg Psychiatry. 2000;68:93–96. doi: 10.1136/jnnp.68.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reiman EM, Uecker A, Caselli RJ, et al. Hippocampal volumes in cognitively normal persons at genetic risk for Alzheimer’s disease. Ann Neurol. 1998;44:288–291. doi: 10.1002/ana.410440226. [DOI] [PubMed] [Google Scholar]
- 5.Klunk WE, Engler H, Nordberg A, et al. Imaging brain amyloid in Alzheimer’s disease with Pittsburgh Compound-B. Ann Neurol. 2004;55:306–319. doi: 10.1002/ana.20009. [DOI] [PubMed] [Google Scholar]
- 6.Hashimoto M, Yasuda M, Tanimukai S, et al. Apolipoprotein E epsilon 4 and the pattern of regional brain atrophy in Alzheimer’s disease. Neurology. 2001;57:1461–1466. doi: 10.1212/wnl.57.8.1461. [DOI] [PubMed] [Google Scholar]
- 7.Radua J, Mataix-Cols D, Phillips ML, et al. A new meta-analytic method for neuroimaging studies that combines reported peak coordinates and statistical parametric maps. Eur Psychiatry. 2012;27:605–611. doi: 10.1016/j.eurpsy.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 8.Risacher SL, Kim S, Shen L, et al. The role of apolipoprotein E (APOE) genotype in early mild cognitive impairment (E-MCI) Front Aging Neurosci. 2013;5:11. doi: 10.3389/fnagi.2013.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song F, Sheldon TA, Sutton AJ, et al. Methods for exploring heterogeneity in meta-analysis. Eval Health Prof. 2001;24:126–151. doi: 10.1177/016327870102400203. [DOI] [PubMed] [Google Scholar]
- 10.Egger M, Smith GD, Phillips AN. Meta-analysis: principles and procedures. BMJ. 1997;315:1533–1537. doi: 10.1136/bmj.315.7121.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bora E, Fornito A, Radua J, et al. Neuroanatomical abnormalities in schizophrenia: a multimodal voxelwise meta-analysis and meta-regression analysis. Schizophr Res. 2011;127:46–57. doi: 10.1016/j.schres.2010.12.020. [DOI] [PubMed] [Google Scholar]
- 12.Hart H, Radua J, Mataix-Cols D, et al. Meta-analysis of fMRI studies of timing in attention-deficit hyperactivity disorder (ADHD) Neurosci Biobehav Rev. 2012;36:2248–2256. doi: 10.1016/j.neubiorev.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 13.Jacobs HI, Radua J, Luckmann HC, et al. Meta-analysis of functional network alterations in Alzheimer’s disease: toward a network biomarker. Neurosci Biobehav Rev. 2013;37:753–765. doi: 10.1016/j.neubiorev.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 14.Shi J, Lepore N, Gutman BA, et al. Genetic influence of apolipoprotein E4 genotype on hippocampal morphometry: an N=725 surface-based Alzheimer’s disease neuroimaging initiative study. Hum Brain Mapp. 2014 doi: 10.1002/hbm.22447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mori E, Lee K, Yasuda M, et al. Accelerated hippocampal atrophy in Alzheimer’s disease with apolipoprotein E epsilon4 allele. Ann Neurol. 2002;51:209–214. doi: 10.1002/ana.10093. [DOI] [PubMed] [Google Scholar]
- 16.Pievani M, Galluzzi S, Thompson PM, et al. APOE4 is associated with greater atrophy of the hippocampal formation in Alzheimer’s disease. NeuroImage. 2011;55:909–919. doi: 10.1016/j.neuroimage.2010.12.081. [DOI] [PubMed] [Google Scholar]
- 17.Protas HD, Chen K, Langbaum JB, et al. Posterior cingulate glucose metabolism, hippocampal glucose metabolism, and hippocampal volume in cognitively normal, late-middle-aged persons at 3 levels of genetic risk for Alzheimer disease. JAMA Neurol. 2013;70:320–325. doi: 10.1001/2013.jamaneurol.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rowe CC, Ellis KA, Rimajova M, et al. Amyloid imaging results from the Australian Imaging, Biomarkers and Lifestyle (AIBL) study of aging. Neurobiol Aging. 2010;31:1275–1283. doi: 10.1016/j.neurobiolaging.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 19.Fleisher AS, Chen K, Liu X, et al. Apolipoprotein E epsilon4 and age effects on florbetapir positron emission tomography in healthy aging and Alzheimer disease. Neurobiol Aging. 2013;34:1–12. doi: 10.1016/j.neurobiolaging.2012.04.017. [DOI] [PubMed] [Google Scholar]
- 20.Ossenkoppele R, van der Flier WM, Zwan MD, et al. Differential effect of APOE genotype on amyloid load and glucose metabolism in AD dementia. Neurology. 2013;80:359–365. doi: 10.1212/WNL.0b013e31827f0889. [DOI] [PubMed] [Google Scholar]
- 21.Yi D, Lee DY, Sohn BK, et al. Beta-Amyloid Associated Differential Effects of APOE epsilon4 on Brain Metabolism in Cognitively Normal Elderly. Am J Geriatr Psychiatry. 2014 doi: 10.1016/j.jagp.2013.12.173. [DOI] [PubMed] [Google Scholar]
- 22.Jagust WJ, Landau SM. Apolipoprotein E, not fibrillar beta-amyloid, reduces cerebral glucose metabolism in normal aging. J Neurosci. 2012;32:18227–18233. doi: 10.1523/JNEUROSCI.3266-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langbaum JB, Chen K, Caselli RJ, et al. Hypometabolism in Alzheimer-affected brain regions in cognitively healthy Latino individuals carrying the apolipoprotein E epsilon4 allele. Arch Neurol. 2010;67:462–468. doi: 10.1001/archneurol.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reiman EM, Chen K, Alexander GE, et al. Functional brain abnormalities in young adults at genetic risk for late-onset Alzheimer’s dementia. Proc Natl Acad Sci. 2003;101:284–289. doi: 10.1073/pnas.2635903100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mosconi L, Sorbi S, Nacmias B, et al. Age and ApoE genotype interaction in Alzheimer’s disease: an FDG-PET study. Psychiatry Res. 2004;130:141–151. doi: 10.1016/j.pscychresns.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 26.Drzezga A, Grimmer T, Henriksen G, et al. Effect of APOE genotype on amyloid plaque load and gray matter volume in Alzheimer disease. Neurology. 2009;72:1487–1494. doi: 10.1212/WNL.0b013e3181a2e8d0. [DOI] [PubMed] [Google Scholar]
- 27.Grimmer T, Tholen S, Yousefi BH, et al. Progression of cerebral amyloid load is associated with the apolipoprotein E epsilon4 genotype in Alzheimer’s disease. Biol Psychiatry. 2010;68:879–884. doi: 10.1016/j.biopsych.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Allen SJ, MacGowan SH, Tyler S, et al. Reduced cholinergic function in normal and Alzheimer’s disease brain is associated with apolipoprotein E4 genotype. Neurosci Lett. 1997;239:33–36. doi: 10.1016/s0304-3940(97)00872-0. [DOI] [PubMed] [Google Scholar]
- 29.Berg L, McKeel DW, Jr, Miller JP, et al. Clinicopathologic studies in cognitively healthy aging and Alzheimer’s disease: relation of histologic markers to dementia severity, age, sex, and apolipoprotein E genotype. Arch Neurol. 1998;55:326–335. doi: 10.1001/archneur.55.3.326. [DOI] [PubMed] [Google Scholar]
- 30.Deane R, Sagare A, Hamm K, et al. apoE isoform-specific disruption of amyloid beta peptide clearance from mouse brain. J Clin Invest. 2008;118:4002–4013. doi: 10.1172/JCI36663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mahley RW. Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science. 1988;240:622–630. doi: 10.1126/science.3283935. [DOI] [PubMed] [Google Scholar]
- 32.Poirier J, Davignon J, Bouthillier D, et al. Apolipoprotein E polymorphism and Alzheimer’s disease. Lancet. 1993;342:697–699. doi: 10.1016/0140-6736(93)91705-q. [DOI] [PubMed] [Google Scholar]
- 33.Ye S, Huang Y, Mullendorff K, et al. Apolipoprotein (apo) E4 enhances amyloid beta peptide production in cultured neuronal cells: apoE structure as a potential therapeutic target. Proc Natl Acad Sci USA. 2005;102:18700–18705. doi: 10.1073/pnas.0508693102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morra JH, Tu Z, Apostolova LG, et al. Automated mapping of hippocampal atrophy in 1-year repeat MRI data from 490 subjects with Alzheimer’s disease, mild cognitive impairment, and elderly controls. NeuroImage. 2009;45:S3–S15. doi: 10.1016/j.neuroimage.2008.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mueller SG, Weiner MW. Selective effect of age, Apo e4, and Alzheimer’s disease on hippocampal subfields. Hippocampus. 2009;19:558–564. doi: 10.1002/hipo.20614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Naslund J, Thyberg J, Tjernberg LO, et al. Characterization of stable complexes involving apolipoprotein E and the amyloid beta peptide in Alzheimer’s disease brain. Neuron. 1995;15:219–228. doi: 10.1016/0896-6273(95)90079-9. [DOI] [PubMed] [Google Scholar]
- 37.Rebeck GW, Reiter JS, Strickland DK, et al. Apolipoprotein E in sporadic Alzheimer’s disease: allelic variation and receptor interactions. Neuron. 1993;11:575–580. doi: 10.1016/0896-6273(93)90070-8. [DOI] [PubMed] [Google Scholar]
- 38.Ashford JW. APOE genotype effects on Alzheimer’s disease onset and epidemiology. J Mol Neurosci. 2004;23:157–165. doi: 10.1385/JMN:23:3:157. [DOI] [PubMed] [Google Scholar]
- 39.Greicius MD, Srivastava G, Reiss AL, et al. Default-mode network activity distinguishes Alzheimer’s disease from healthy aging: evidence from functional MRI. Proc Natl Acad Sci USA. 2004;101:4637–4642. doi: 10.1073/pnas.0308627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mormino EC, Smiljic A, Hayenga AO, et al. Relationships between beta-amyloid and functional connectivity in different components of the default mode network in aging. Cereb Cortex. 2011;21:2399–2407. doi: 10.1093/cercor/bhr025. [DOI] [PMC free article] [PubMed] [Google Scholar]