Abstract
The bacterial stringent response is triggered by deficiencies of available nutrients and other environmental stresses. It is mediated by 5'-triphosphate-guanosine-3'-diphosphate and 5'-diphosphate-guanosine-3'-diphosphate (collectively (p)ppGpp) and generates global changes in gene expression and metabolism that enable bacteria to adapt to and survive these challenges. Borrelia burgdorferi encounters multiple stressors in its cycling between ticks and mammals that could trigger the stringent response. We have previously shown that the B. burgdorferi stringent response is mediated by a single enzyme, RelBbu, with both (p)ppGpp synthase and hydrolase activities, and that a B. burgdorferi 297 relBbu null deletion mutant was defective in adapting to stationary phase, incapable of down-regulating synthesis of rRNA and could not infect mice. We have now used this deletion mutant and microarray analysis to identify genes comprising the rel regulon in B. burgdorferi cultured at 34°C, and found that transcription of genes involved in glycerol metabolism is induced by rel Bbu. Culture of the wild type parental strain, the rel Bbu deletion mutant and its complemented derivative at 34°C and 25°C in media containing glucose or glycerol as principal carbon sources revealed a growth defect in the mutant, most evident at the lower temperature. Transcriptional analysis of the glp operon for glycerol uptake and metabolism in these three strains confirmed that rel Bbu was necessary and sufficient to increase transcription of this operon in the presence of glycerol at both temperatures. These results confirm and extend previous findings regarding the stringent response in B. burgdorferi. They also demonstrate that the stringent response regulates glycerol metabolism in this organism and is likely crucial for its optimal growth in ticks.
Introduction
Nutritional exigencies and other environmental challenges are met by bacteria through the use of global regulatory pathways [1–4]. The comprehensive shifts in gene expression induced by these regulatory pathways at the transcriptional and post-transcriptional level permit the rapid and dramatic modulation of bacterial growth and metabolism needed to adapt to these challenges. One of these global regulatory responses, the stringent response, is conserved in virtually all bacteria. It was originally described in Escherichia coli in association with amino acid scarcity, but has subsequently been shown to be triggered by many other environmental stressors including insufficiencies of iron, glucose and fatty acids [1–4]. It is mediated by the nucleotide alarmones guanosine-3'-diphosphate-5'-diphosphate and guanosine-3'-diphosphate-5'-triphosphate (collectively (p)ppGpp), each with a similar but distinct regulatory potential [1,2,5].
In E. coli, cytosolic levels of (p)ppGpp are regulated by RelA, a synthase, and SpoT, an enzyme with both synthase and hydrolase activities [1,4]. In other bacteria including B. burgdorferi, a single gene, rel or rsh (rel/spo homolog), encodes an enzyme with both synthase and hydrolase activities [6–8]. In those bacteria with a single rel/rsh ortholog, the N-terminal domain is responsible for the dual enzymatic activity while the C-terminal domain contains potential regulatory elements. Cytosolic levels of (p)ppGpp may also be controlled by other small GTPases [9]. Triggering of the stringent response by uncharged tRNA and activation and diffusion of ribosomal-bound RelA generates (p)ppGpp and leads to global changes in gene expression and intermediary metabolism [1–4]. These include decreased synthesis of stable rRNA and tRNA, proteolysis of ribosomal proteins, increased synthesis of amino acids, inhibition of motility, activation of rpoN-rpoS regulons and changes in carbon source utilization [1–3,10–13]. The net result is a shift to a slow- or non-growing state. Once stresses triggering the stringent response are removed, the short half-life of RelA/Rel and (p)ppGpp facilitates renewed synthesis of macromolecules and resumption of growth [1–4]. The ability of (p)ppGpp to shift transcription depends on its interaction with RNA polymerase, directing transcription from σ70 promoters to alternative promoters [1–3,13,14] often in synergy with the small regulator, DksA [11,12,15–17], as well as interactions with other proteins and regulatory RNAs [18]. In E. coli, relA expression is also regulated by the carbon storage regulator CsrA [19].
The stringent response is involved in bacterial virulence at multiple levels, having been shown to facilitate survival of extracellular pathogens in the host, transmissibility and persistence of a variety of intracellular pathogens [18,20,21], production of toxins [22], and host-vector cycling of vector-transmitted pathogens [23,24]. It also appears to be involved in the development of antimicrobial tolerance in bacteria by increasing the number of persister cells in culture [25–27]. Mutants of pathogens unable to produce (p)ppGpp are generally attenuated and have been proposed as live vaccines [24,28,29], while compounds able to block the production of (p)ppGpp may have therapeutic potential [30].
The life cycle of Borrelia burgdorferi sensu lato depends on its survival in several tissues and organs of ixodid tick vectors and mammalian reservoirs where it is exposed to challenging, variable and rapidly shifting availability of a range of nutrients [31,32]. The two component system-triggered regulatory pathway composed of Hk1/Rrp1/c-di-GMP increases borrelial survival during the larval and nymphal blood meals [33,34]. This pathway stimulates B. burgdorferi glycerol metabolism and together with genes of the glp operon is crucial for maximum fitness of the bacterium under these conditions [35,36]. Initial survival in mammalian reservoirs, in contrast, predominantly involves the Rrp2/RpoN(BosR)/RpoS cascade [33,34]. Linkage of other potential global regulators to these regulatory loops in Borrelia spp. (e.g., the stringent response, the carbon storage regulator protein CsrA) is still being characterized [37–43].
B. burgdorferi B31 contains a single chromosomal rel gene, rel Bbu (BB0198, nt195693–197696), orthologous to E. coli relA and spoT [38,44,45]. Cloned rel Bbu transcribed from its own promoter produced rel Bbu mRNA and RelBbu protein [37], was the only source of (p)ppGpp production in B. burgdorferi [37,38], and could complement an E. coli relA/spoT double null mutant [37]. The stringent response in B. burgdorferi was ameliorated during in vitro growth in the presence of tick cells and in engorged ticks [37,44]. Expression of rel Bbu mRNA increased under in vitro conditions that presumably simulate the unfed tick state [46]. Amounts of RelBbu were higher in B. burgdorferi growing in dialysis membrane chambers in vivo than in organisms growing in vitro despite similar levels of rel Bbu mRNA under these two conditions [37]. A B. burgdorferi rel Bbu null mutant (Δrel Bbu) did not down-regulate synthesis of rRNA in stationary phase, a phenotype resembling that of a “relaxed” E. coli mutant, and was not infectious in mice [38,47].
In order to expand our understanding of the role of rel Bbu and the stringent response in borrelial gene regulation and carbon source utilization, we have used B. burgdorferi microarrays to compare the global transcriptome in wild type B. burgdorferi 297 and its Δrel Bbu derivative during in vitro growth, and found the mutant to have a substantially altered transcriptome. Additional analyses confirmed that genes involved in glycerol utilization and metabolism are modulated by (p)ppGpp under these conditions [35,36].
Results
Effect of deletion of rel Bbu on B. burgdorferi gene expression during exponential and stationary growth phases
The global alarmone nature of (p)ppGpp indicated that microarray analysis of wild type and Δrel Bbu strains would be useful for examining the role of RelBbu and thus of (p)ppGpp in B. burgdorferi 297. From the multiplicity of growth conditions that could be chosen for these studies, we focused on the role of RelBbu during in vitro growth at 34°C under two growth phases, exponential and stationary. Genes were considered to have altered expression between wild type and mutant strains if transcript levels differed by at least 2-fold at P ≤ 0.02. Increased expression of genes in the mutant relative to wild type implies (p)ppGpp-mediated repression of these genes in the wild type, whereas decreased expression is consistent with (p)ppGpp-mediated induction in the wild type. During the exponential phase of growth, 38 genes exhibited increased expression and 37 genes exhibited decreased expression in the mutant as compared to the parental B. burgdorferi wild type strain (S1 Table). The Δrel Bbu phenotype was more evident during stationary phase: 174 genes showed increased expression and 103 showed decreased expression in comparison to the wild type strain (S2 Table). Nineteen of 38 genes with higher expression in the exponential phase in B. burgdorferi Δrel Bbu were also elevated in stationary phase; 25 of 37 genes with lower expression in exponential phase in the mutant were also lower in stationary phase. These findings indicate that RelBbu regulates expression of these genes in a similar manner in both exponential and stationary growth phases (S1 Table, S2 Table), and are also consistent with the production of (p)ppGpp during both growth phases in vitro [48,49]
For validation of the microarray results, 16 genes (11 chromosomal, 5 plasmid) were selected for analysis by quantitative reverse transcription real-time PCR (RT-PCR) (Fig. 1). These genes were chosen on the basis of assigned functions: transcriptional regulators (dksA, rpoD); metabolism/transporters (glpK, glpD, mvaA, apt, oppA-3, oppF, potC, potA, chbC); cell envelope/membrane proteins (bmpD, BBA03, BBA74, BBB07, blyA). Expression levels of mRNA determined by quantitative RT-PCR were consistent with results from microarrays for 10 of 16 genes in exponentially growing cells and for 15 of 16 genes in stationary phase cells (Fig. 1). They were discordant for potA, mvaA, rpoD, apt, BBB07, and blyA whose expression in the exponential phase was elevated in the mutant by RT-PCR but not by microarray analysis, and for oppF whose expression in the stationary phase was decreased in the mutant by microarray but not by RT-PCR analysis (Table 1, S1 Table, S2 Table, Fig. 1).
Fig 1. Transcriptional analysis by qRT-PCR of selected genes in B. burgdorferi 297 Δrel Bbu relative to expression of these genes in the wild type parental strain during (A) exponential and (B) stationary phases of growth in BSK-H at 34°C.
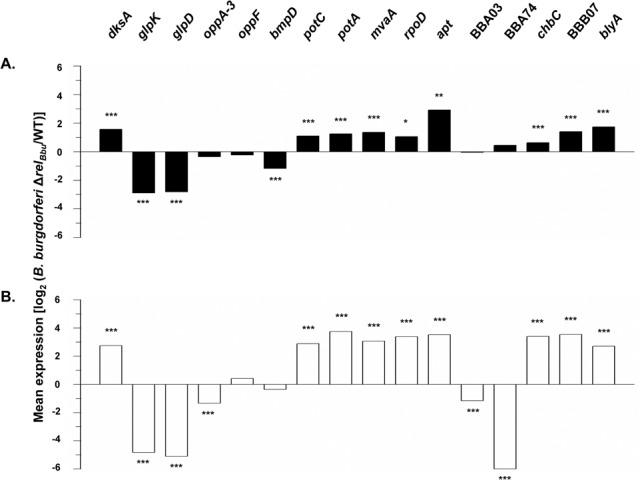
*, P < 0.02; **, P < 0.005; ***, P < 0.001. Genes selected either had an assigned function and were regulated in operons, were involved in various aspects of cellular metabolism, were regulatory in nature or were localized to the cell envelope. Increased expression of genes in the mutant relative to wild type is consistent with rel-mediated repression of these genes in the wild type, while decreased expression is consistent with rel-mediated induction in the wild type. RT-PCR data were discordant with microarrays for potA, mvaA, rpoD, apt, BBB07, and blyA in the exponential phase of growth, and for oppF in the stationary phase of growth. See Materials and Methods for details of transcriptional analyses.
Table 1. Modulated selected genes with annotated function in B. burgdorferi 297 Δrel Bbu during growth in vitro at 34°C a .
| Exponential phase | Stationary phase | ||||
|---|---|---|---|---|---|
| Gene | Description | Mean expression (log2 [ΔrelBbu/WT]) | P | Mean expression (log2 [ΔrelBbu/WT]) | P |
| Transcriptional regulators | |||||
| BB0168 | dnaK suppressor (dksA) | 1.31 | 0.002 | 3.26 | <0.001 |
| BB0419 | response regulatory protein (rrp-1) | 1.42 | 0.001 | ||
| BB0420 | sensory transduction histidine kinase/response regulator (hk1) | 2.65 | 0.001 | 2.39 | <0.001 |
| BB0712 | RNA polymerase σ70 factor (rpoD) | 1.05 | 0.013 | 2.19 | <0.001 |
| DNA synthesis/repair | |||||
| BB0022 | Holliday junction DNA helicase (ruvB) | 2.58 | <0.001 | ||
| BB0344 | DNA helicase (uvrD) | 2.79 | 0.006 | ||
| BB0438 | DNA polymerase III, β subunit (dnaN) | 2.41 | 0.006 | 1.56 | <0.001 |
| BB0552 | DNA ligase (lig) | 2.64 | 0.010 | ||
| BB0579 | DNA polymerase III, α subunit (dnaE) | 4.95 | 0.001 | ||
| BB0710 | DNA primase (dnaG), authentic frameshift | 1.26 | <0.001 | 1.33 | <0.001 |
| BB0836 | excinuclease ABC, B subunit (uvrB) | 2.35 | <0.001 | ||
| BB0837 | excinuclease ABC, A subunit (uvrA) | 1.04 | 0.003 | ||
| Cell division | |||||
| BB0177 | glucose inhibited division protein B (gidB) | 3.81 | 0.007 | ||
| BB0178 | glucose inhibited division protein A (gidA) | 3.07 | 0.005 | ||
| BB0302 | cell division protein (ftsW) | 2.96 | 0.001 | ||
| BB0781 | GTP-binding protein (obg) | 1.03 | 0.004 | 1.95 | <0.001 |
| BB0789 | cell division protein (ftsH) | 1.01 | <0.001 | ||
| Protein synthesis | |||||
| BB0229 | ribosomal protein L31 (rpmE) | 1.54 | <0.001 | ||
| BB0251 | leucyl-tRNA synthetase (leuS) | 3.41 | <0.001 | ||
| BB0514 | phenylalanyl-tRNA synthetase, β subunit (pheT) | 1.63 | 0.009 | ||
| BB0615 | ribosomal protein S4 (rpsD) | 2.87 | <0.001 | ||
| BB0691 | translation elongation factor G (fus-2) | 0.96 | 0.001 | ||
| BB0778 | ribosomal protein L21 (rplU) | 1.51 | 0.002 | ||
| BB0780 | ribosomal protein L27 (rpmA) | 1.03 | <0.001 | ||
| Motility/chemotaxis | |||||
| BB0147 | flagellar filament 41 kDa core protein (flaB) | -1.31 | 0.001 | ||
| BB0181 | flagellar hook-associated protein (flgK) | -1.14 | 0.010 | ||
| BB0271 | flagellar biosynthesis protein (flhA) | 1.95 | 0.002 | ||
| BB0578 | methyl-accepting chemotaxis protein (mcp-1) | 3.06 | 0.012 | ||
| BB0668 | flagellar filament outer layer protein (flaA) | -1.13 | <0.001 | ||
| BB0670 | purine-binding chemotaxis protein (cheW-3) | 2.03 | 0.011 | ||
| BB0775 | flagellar hook-basal body complex protein (flhO) | 1.03 | <0.001 | ||
| Cell envelope | |||||
| BB0382d | basic membrane protein B (bmpB) | -1.02 | 0.012 | -1.59 | <0.001 |
| BB0383 | basic membrane protein A (bmpA) | -1.15 | 0.016 | ||
| BB0385 | basic membrane protein D (bmpD) | -2.93 | 0.007 | ||
| BBA15 | outer surface protein A (ospA) | -1.81 | 0.001 | ||
| BBA16 | outer surface protein B (ospB) | -1.54 | 0.002 | ||
| BBA60 | surface lipoprotein P27 | -5.07 | <0.001 | ||
| BBA74 | membrane-associated periplasmic protein | -2.03 | <0.001 | ||
| BBB07 | α3β1 integrin-binding protein | 1.42 | <0.001 | ||
| BBJ41 | antigen P35, putative | -8.62 | <0.001 | ||
| BBM23 b | holin (blyA) | 1.74 | <0.001 | 1.43 | <0.001 |
| BBN24 c | holin (blyB) | 2.99 | <0.001 | 1.17 | <0.001 |
| Central metabolism/carbon source transporters | |||||
| BB0240 | glycerol uptake facilitator (glpF) | -4.54 | <0.001 | -4.76 | <0.001 |
| BB0241 | glycerol kinase (glpK) | -8.27 | <0.001 | -5.47 | <0.001 |
| BB0243 | glycerol-3-phosphate dehydrogenase (glpD) | -6.52 | <0.001 | -3.79 | <0.001 |
| BB0328 | oligopeptide ABC transporter, periplasmic oligopeptide-binding protein (oppA-1) | -1.16 | 0.001 | -2.65 | 0.002 |
| BB0329 | oligopeptide ABC transporter, periplasmic oligopeptide-binding protein (oppA-2) | -1.33 | <0.001 | ||
| BB0330 | oligopeptide ABC transporter, periplasmic oligopeptide-binding protein (oppA-3) | -4.07 | 0.015 | ||
| BB0334 | oligopeptide ABC transporter, ATP-binding protein (oppD) | -3.25 | 0.006 | -1.21 | 0.002 |
| BB0335 | oligopeptide ABC transporter, ATP-binding protein (oppF) | -1.51 | <0.001 | ||
| BBB04 | chitobiose transporter protein (chbC) | 1.83 | 0.004 | 2.11 | <0.001 |
| BBB05 | chitobiose transporter protein (chbA) | 4.05 | 0.013 | 5.07 | <0.001 |
| BBB06 | chitobiose transporter protein (chbB) | 5.02 | <0.001 | ||
| BB0683 | 3-hydroxy-3-methylglutaryl-CoA synthase (hmgs) | 1.23 | 0.010 | 1.65 | <0.001 |
| BB0685 | 3-hydroxy-3-methylglutaryl-CoA reductase (mvaA) | 1.36 | <0.001 | 4.39 | <0.001 |
a. Transcriptional analysis from microarrays (regular font) or RT-PCR (boldface). Where data from RT-PCR is shown, microarrays showed no significant difference in gene expression between B. burgdorferi 297 Δrel Bbu and wild type.
b. Expression values for blyA orthologs BBM23, BBP23, BBR23 that showed increased expression in stationary phase and BBN23, BBR23 and BBS23 that showed increased expression in exponential phase were considered as a single transcript because they are 100% identical in sequence.
c. Expression values for blyB orthologs BBN24, BBR24, BBS24 that showed increased expression in stationary phase and BBN24, BBR24, and BBS23 that showed increased expression in exponential phase were considered as a single transcript because they are 100% identical in sequence.
B. burgdorferi RelBbu regulon in exponential and stationary growth phases
More than 50% of the genes showing changes in transcriptional levels by microarray analysis (S1 Table, S2 Table) are annotated as hypothetical proteins and have no predicted biological role. This made functional analysis difficult. Genes with annotated functions in the B. burgdorferi RelBbu regulon included those encoding transcriptional regulators or proteins involved in DNA synthesis and repair, cell division, protein synthesis, motility and chemotaxis, cell envelope synthesis, central metabolism and carbon source transport (Table 1, Fig. 1). These are all functions regulated by the stringent response in E. coli and other bacteria [1,12,50]
Several important transcriptional regulators were induced as a result of rel Bbu deletion. The stringent response regulator dksA (BB0168) and σ70 (rpoD, BB0712) were upregulated in the Δrel Bbu mutant in both exponential and stationary growth phases (Table 1, Fig. 1). Genes coding for the two-component regulatory system 1 were also upregulated in the Δrel Bbu mutant. Expression of rrp1 (BB0419) increased in stationary phase while expression of sensory histidine kinase hk1 (BB0420) increased in both growth phases (Table 1), indicating that expression of these genes is repressed in the presence of (p)ppGpp. Neither rpoN nor rpoS was modulated by the stringent response in B. burgdorferi. This lack of effect of the B. burgdorferi stringent response on RpoS and OspC expression had been previously reported [38].
The rel mutant exhibited altered expression for genes encoding proteins involved in central metabolism and transport of carbon sources. Transcripts for two of the three subunits of the chitobiose transporter, chbC (BBB04) and chbA (BBB05), were elevated in the mutant during exponential phase and all three subunits (including chbB, BBB06) were elevated during stationary phase (Table 1, Fig. 1). In contrast, expression of the glp operon (BB0240-BB0243), encoding proteins involved in glycerol transport and metabolism were diminished in the rel Bbu mutant (Table 1, Fig. 1). Genes encoding components of the oligopeptide ABC transporter, oppA (BB0328) and oppD (BB0334) were decreased in the rel mutant during the exponential phase, while many more of the genes of this transporter (BB0328-BB330, BB0334, BB0335) decreased in the mutant during stationary phase (Table 1, S1 Table, S2 Table). Expression of HMG-CoA synthase (hmgs, BB0683) was increased in the rel mutant during both exponential and stationary phase and HMG-CoA reductase (mvaA, BB0685) expression was increased during stationary phase (Table 1, Fig. 1), indicating that (p)ppGpp repression of the mevalonate pathway is part of the stringent response in B. burgdorferi [51].
Effect of glucose and glycerol on growth of B. burgdorferi rel Bbu mutant and derivatives
Microarray analysis strongly suggested that the lack of (p)ppGpp in the rel Bbu deletion mutant reduced expression of the glp operon. Because glycerol and glucose are among the limited number of carbohydrates that can support B. burgdorferi growth [52–54], we investigated the ability of B. burgdorferi and its rel deletion mutant to utilize glycerol in BSK-Lite medium containing glycerol as the principal carbon source. The B. burgdorferi 297 parental wild type strain reached significantly higher cell concentrations during stationary phase than did the Δrel Bbu derivative during growth at either 25°C or 34°C in BSK-Lite containing glucose as the principal carbon source (P < 0.001, one-way ANOVA, Bonferroni post-test) (Fig. 2A and 2C). This impairment was partially reversed in the complemented strain (P < 0.001) (Fig. 2). These data are in agreement with earlier studies using BSK-H media containing glucose as the principal carbon source [38].
Fig 2. Growth of wild type B. burgdorferi 297 (solid circles), Δrel Bbu (open circles), or Δrel Bbu complemented with pKFSS1-Δrel Bbu (solid triangles) in BSK-Lite containing either glucose or glycerol as principal carbon sources.
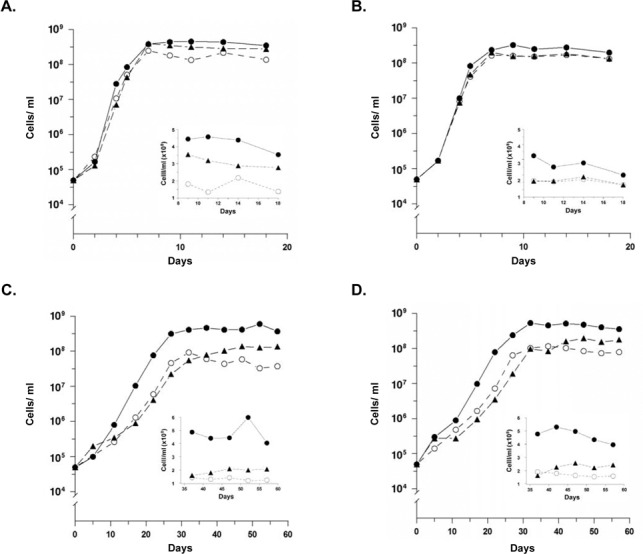
Insets show enlargement of cell concentrations for clarity. Glucose-containing medium, 34°C (A). Differences between stationary phase cell concentrations of wild type and Δrel Bbu mutant are significant as are differences in stationary phase concentrations between the Δrel Bbu mutant and its complemented derivative (P < 0.001, one-way ANOVA, Bonferroni post-test). Glycerol-containing medium, 34°C (B). Differences between stationary phase cell concentrations of wild type and Δrel Bbu mutant are significant (P < 0.001, one-way ANOVA, Bonferroni post-test). Differences in stationary phase concentrations between the Δrel Bbu mutant and its complemented derivative are not significant (P > 0.05, one-way ANOVA, Bonferroni post-test). Glucose-containing medium, 25°C (C). Differences between stationary phase cell concentrations of wild type and Δrel Bbu mutant are significant as are differences in stationary phase concentrations between the Δrel Bbu mutant and its complemented derivative (P < 0.001, one-way ANOVA, Bonferroni post-test). Glycerol-containing medium, 25°C (D). Differences between stationary phase cell concentrations of wild type and Δrel Bbu mutant are significant (P < 0.001, one-way ANOVA, Bonferroni post-test) as are differences in stationary phase concentrations between the Δrel Bbu mutant and its complemented derivative (P < 0.05, one-way ANOVA, Bonferroni post-test).
During growth at 34°C in BSK-Lite with glycerol as the principal carbon source, the wild type strain reached significantly higher cell concentrations in the stationary phase than the Δrel Bbu derivative and was not affected by complementation with the extrachromosomally-located gene (P < 0.01, one-way ANOVA, Bonferroni post-test) (Fig. 2B). During growth at 25°C in BSK-Lite containing glycerol, the wild type strain also reached a significantly higher cell density in the stationary phase than the Δrel Bbu mutant (P < 0.001, one-way ANOVA, Bonferroni post-test). The difference in cell concentrations between wild type B. burgdorferi and the rel Bbu mutant was more obvious at 25°C than at 34°C with either glucose or glycerol suggesting a more important role for the stringent response in metabolic regulation at 25°C than at 34°C.
Transcriptional analysis of rel Bbu and the glp operon in B. burgdorferi rel Bbu mutant and derivatives
The impairment of growth experienced by the Δrel Bbu mutant, coupled with the transcriptome data indicating reduced expression of the glp operon in the mutant, suggested that the stringent response might play a role in glycerol utilization in B. burgdorferi, particularly at 25°C. Transcriptional analysis in temperature-shifted organisms during logarithmic growth of wild type and Δrel Bbu mutant strains at 34°C and 25°C confirmed that rel Bbu was not expressed in the deletion mutant and that expression of rel Bbu at 34°C was greater in wild type cells growing on glycerol than in cells growing on glucose (Fig. 3). It also confirmed the role of (p)ppGpp in modifying transcription of glycerol metabolism genes. Transcript levels for the glycerol uptake facilitator (glpF, BB0240) and glycerol-3-phosphate dehydrogenase (glpD, BB0243), the first and the last genes in the glp operon, were substantially higher in wild type B. burgdorferi and in complemented Δrel Bbu mutant grown in media with glycerol as principal carbon source than in cells grown at 34°C in media with glucose as principal carbon source (Fig. 4A, 4B). Under both conditions, transcription of these genes was considerably lower in the Δrel Bbu mutant compared to the wild type or complemented strains (Fig. 4A, 4B). At 25°C, there was essentially no transcription of glpD and glpF in wild type or mutant B. burgdorferi grown in medium with glucose as principal carbon source (Fig. 4C). In contrast, these genes were actively transcribed in wild type and rel Bbu complemented B. burgdorferi strains, but not in the Δrel Bbu mutant during growth in medium with glycerol as principal carbon source at this temperature (Fig. 4D). These changes in transcriptional levels of glp operon genes are expected to correlate with changes in levels of glp gene products, as we previously determined with glpD [36] and data not shown.
Fig 3. Transcriptional analysis of rel Bbu in wild type B. burgdorferi 297 (solid bars), Δrel Bbu (open bars), and Δrel Bbu complemented with pKFSS1-Δrel Bbu (grey bars) during logarithmic growth in BSK-Lite medium containing glucose or glycerol as principal carbon sources.
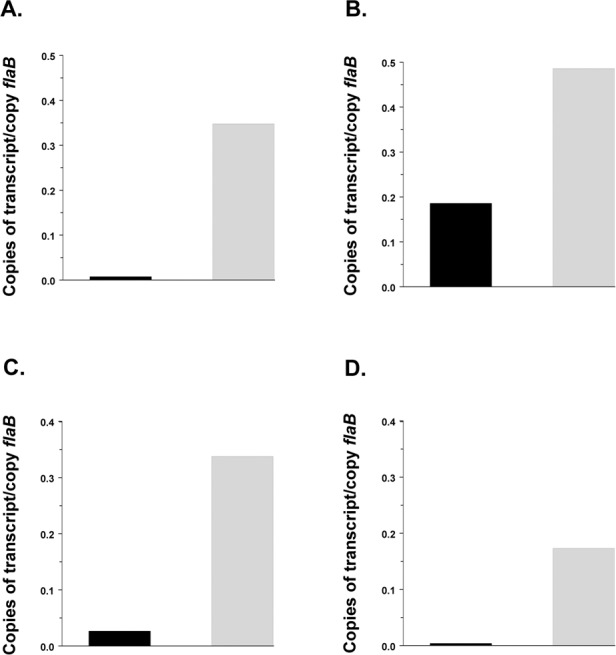
No bar is visible for the Δrel Bbu mutant because its expression level was zero. Glucose-containing medium, 34°C (A). Glycerol-containing medium, 34°C (B). Glucose-containing medium, 25°C (C). Glycerol-containing medium, 25°C (D).
Fig 4. Transcriptional analysis of glp operon genes in wild type B. burgdorferi 297 (solid bars), Δrel Bbu (open bars), and Δrel Bbu complemented with pKFSS1-Δrel Bbu (grey bars) during late logarithmic growth phase in BSK-Lite medium containing glucose or glycerol as principal carbon sources.
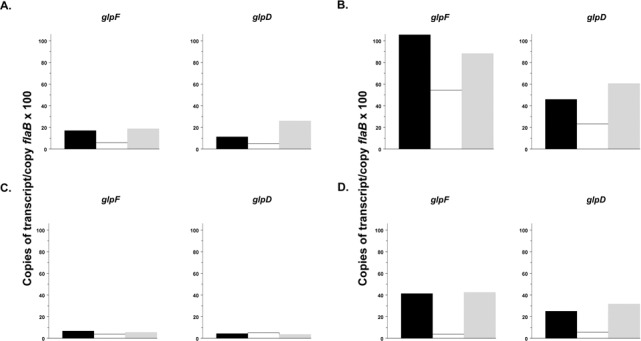
Glucose-containing medium, 34°C (A). Glycerol-containing medium, 34°C (B). Glucose-containing medium, 25°C (C). Glycerol-containing medium, 25°C (D).
Co-regulation of gene expression by stringent response and other global regulators in B. burgdorferi
To determine if the stringent response regulates glycerol uptake in B. burgdorferi indirectly through transcriptional cross talk with other global regulators, transcript levels of rpoS, rpoN, bosR, rrp-1 and rrp-2 were determined in wild type B. burgdorferi and its rel Bbu derivatives during temperature-shifted exponential growth at 25°C and 34°C in media with glucose or glycerol as principal carbon sources. There were no significant differences in transcription of any of these genes between the wild type and the rel Bbu mutant during exponential growth in either medium at either temperature (data not shown), similar to results from microarrays derived from the non-temperature-shifted exponential growth of B. burgdorferi at 34°C (Table 1, S1 Table).
Discussion
The (p)ppGpp-mediated stringent response is a broadly conserved global regulatory response in bacteria that acts to preserve critical amounts of intracellular molecules in response to nutritional scarcity and/or other environmental stressors [1–3,5,48,50,55] such as those faced by B. burgdorferi in the distinctive vector and mammalian host environments it encounters in the course of its life cycle. We previously demonstrated that the B. burgdorferi stringent response could control growth, rRNA accumulation and virulence [38,47]. Microarray analysis validated by direct examination of gene expression of selected genes has shown that deletion of rel Bbu and the subsequent lack of (p)ppGpp affects transcription of many genes in B. burgdorferi (75 during the exponential growth phase, 277 during stationary phase), and have confirmed production of this alarmone during both exponential and stationary growth phases [49]. The greater effects of (p)ppGpp during stationary phase were to be expected on the basis of our previous work showing that the lack of synthesis of (p)ppGpp preferentially affected borrelial growth and rRNA accumulation during transition from exponential growth to stationary phase and in the stationary phase itself [38,47]. The subsequent more detailed characterization of genes coding for glycerol transport and metabolism in the present study confirmed the regulation of glycerol metabolism in B. burgdorferi by the stringent response, a regulation likely crucial for its optimal growth in ticks.
Genes whose transcription was affected by deletion of rel Bbu included those associated with DNA synthesis and repair, protein synthesis, cell motility and chemotaxis, cell wall synthesis and carbohydrate intermediary metabolism (Table 1, S1 Table, S2 Table). If genes with greater expression in the rel Bbu deletion mutant than in the parental wild type are assumed to be repressed by (p)ppGpp (or other regulatory factors whose expression requires the presence of (p)ppGpp) and those with reduced expression in the rel Bbu mutant are assumed to be induced in the presence of (p)ppGpp (or other regulatory factors whose synthesis is stimulated by (p)ppGpp), the stringent response in B. burgdorferi shares many similarities with this response in other bacteria [2,3,5,6,48,55–59]. The stringent response classically depresses motility and chemotaxis to save energy, and together with DksA, acts especially in stationary phase [11,12]. Modulation of borrelial motility by the stringent response may be particularly relevant in ticks where B. burgdorferi undergo temporal and spatial starvation and must migrate from the gut to the salivary glands to complete the enzootic cycle [60,61].
Despite these similarities, the stringent response in B. burgdorferi is not identical to that of other bacteria (Table 1, S1 Table, S2 Table). This is not surprising, given the phylogenetic distance between spirochetes and, for example, gammaproteobacteria such as E. coli [62]. While the stringent response in E. coli modulates expression of rpoN and rpoS [10,11,63], the stringent response in B. burgdorferi does not. Interestingly, comparison of microarray studies of null mutants of rel/(p)ppGpp (this study) and rpoS [64] uncovered 17 genes that were modulated in both studies (Table 2). Fifteen of these 17 genes were repressed by (p)ppGpp only in the stationary phase, while all 17 were activated by RpoS. This might suggest that the effects of (p)ppGpp and RpoS on gene expression in B. burgdorferi during in vitro growth are independent of, or counterbalance, each other. Unfortunately, comparison between the in vitro stringent response regulon in cells at stationary phase and the RpoS regulon in exponentially growing cells may not be relevant, particularly since both microarrays and quantitative RT-PCR found no significant differences in transcript levels between these and other global regulators in B. burgdorferi wild type and rel Bbu mutant during exponential growth at 34°C in vitro. While several genes induced by (p)ppGpp (the glp operon, BB365, ospA/B, BBA62, BBA69, BBA74, BBD18, BBJ41) during stationary phase in vitro were repressed by RpoS under mammalian host-like conditions [64], the difference between these two culture conditions again makes evaluation of any potential interplay between these two transcriptional regulators uncertain.
Table 2. Genes modulated during growth in vitro in B. burgdorferi Δrel Bbu (S2 Table) and B. burgdorferi ΔrpoS [64].
| Mean expression | |||
|---|---|---|---|
| Gene | Description | log2 (Δrel Bbu/WT) | log2 (ΔrpoS Bbu/WT) |
| BB0670 | purine-binding chemotaxis protein (cheW-3) | 2.03 | -2.56 |
| BB0782 | nicotinate (nicotinamide) nucleotide adenylyltransferase | 2.99 | -2.88 |
| BBA12 | conserved hypothetical protein | 2.81 | -2.73 |
| BBA60 | surface lipoprotein P27 | -5.07 | -2.58 |
| BBM01 | hypothetical protein | 3.57 | -3.05 |
| BBM08 | conserved hypothetical protein | 3.04 | -2.76 |
| BBN01 | hypothetical protein | 3.82 | -2.85 |
| BBN29 | hypothetical protein, paralogous family 161, authentic point mutation | 3.07 | -4.62 |
| BBO03 | hypothetical protein | 3.91 | -2.59 |
| BBO04 | hypothetical protein | 3.08 | -2.85 |
| BBO29 | hypothetical protein | 2.54 | -3.30 |
| BBP21 | conserved hypothetical protein | 2.14 | -2.81 |
| BBP25 | conserved hypothetical protein | 4.83 | -3.28 |
| BBP28 | Lipoprotein | -3.07 | -3.72 |
| BBR02 | hypothetical protein, paralogous family 147, authentic frameshift | 4.36 | -3.31 |
| BBR29 | conserved hypothetical protein | 2.83 | -7.17 |
| BBS01 | hypothetical protein | 4.22 | -3.14 |
Additional differences between the stringent response in B. burgdorferi and that of other bacteria result from genes modulated by the stringent response that are unique to this bacterium (e.g., genes for antigenic surface proteins, bly genes) [52,65,66]. Regulation of these genes by the stringent response could influence the composition of the B. burgdorferi cell envelope to provide a surface better able to endure variable environments such as those encountered in the tick [34]. The ability of the stringent response to modulate peptidoglycan synthesis through its modulation of the mevalonate pathway could also result in antigenic alterations and round forms [67]. Existence of these changes could suggest that niche nutritional signals can be coupled to cell surface antigenic composition and functionality to develop interactions that favor B. burgdorferi residence [34,60,61].
Stringent response-mediated regulation of genes involved in nutrient transport and metabolism is common in E. coli and many other bacteria [1,5]. B. burgdorferi lacks amino acid biosynthetic pathways [52] and is dependent on oligopeptide uptake to satisfy its requirement for amino acids. Transcriptional modulation of the oligopeptide transporter system by the stringent response observed in the present study expands the role of (p)ppGpp to this essential borrelial function [68,69]. Expression of the glp operon (Table 1) was inhibited in the rel mutant in both exponential and stationary growth phases, suggesting that transcription of this operon is induced by (p)ppGpp. In contrast, transcription of genes encoding the three components of the chitobiose phosphotransferase system transporter located on plasmid cp26 (Table 1) was elevated in the rel mutant in the exponential and stationary phase of growth, suggesting that their transcription is directly or indirectly repressed by (p)ppGpp. Alterations in expression of genes involved in utilization of carbon sources such as glycerol and chitobiose strongly suggest that the borrelial stringent response plays an important role in modifying synthesis of macromolecules, central metabolism and carbon utilization in ticks [2,50,55].
Previous reports of regulation of glp operon expression by RpoS and Rrp1 [35,64], and the essential role of glycerol uptake and metabolism for maximum B. burgdorferi fitness in ticks [35,36] prompted a more detailed examination of the regulation of this operon by the stringent response and (p)ppGpp. Decreased ability of the rel Bbu mutant to grow in medium with glycerol as the principal carbon source (Fig. 2B, 2D) and repression of transcription of glycerol metabolism genes at 25°C compared to 34°C during exponential growth (Fig. 4B, 4D) is similar to a phenotype previously described for a B. burgdorferi rrp1 mutant that cannot synthesize c-di-GMP [35]. This could suggest that the stringent response controls glycerol uptake by down-modulating Rrp1 in B. burgdorferi at 25°C in ticks. Because (p)ppGpp repressed rrp1 in stationary phase and induced expression of the glp operon in both exponential and stationary phases at 34°C (Table 1), one would expect to see increased c-di-GMP and increased transcription of glycerol metabolism genes in the rel Bbu mutant rather than the observed decrease. This could suggest that despite down-modulation of Rrp1 expression by (p)ppGpp, sufficient Rrp1 remains to allow production of c-di-GMP. This might indicate that c-di-GMP and (p)ppGpp act synergistically to increase expression of the glp operon in both exponential and stationary phases (Table 1), and are both necessary for development of optimal regulatory activity. Alternatively, synthesis of c-di-GMP by Rrp1 might be dependent on the presence of (p)ppGpp [70]. In any case, these data suggest that (p)ppGpp adds an additional layer of control to the regulation of glp operon expression by Rrp1 [35]. The precise mechanism by which (p)ppGpp and c-di-GMP act to induce glp operon expression, and the inter-relationships of rel Bbu and rrp1 in modulating this expression, clearly warrant further study.
The greater expression of the glp genes at 34°C contradicts previous results where these genes were better expressed at 25°C than at 34°C [36]. Although there was little rel Bbu transcript in the wild type at 25°C, it remains unclear how much RelBbu activity is required for expression of glycerol utilization genes. The apparent discrepancy between the low expression of rel Bbu and the glp genes at 25°C with strong growth of wild type B. burgdorferi 297 and its complemented derivative at this temperature could indicate that this amount of rel Bbu expression produces adequate (p)ppGpp to maintain sufficient expression of glpD and glpF for growth under these conditions [49]. It could also result from strain differences, since different B. burgdorferi strains have been shown to exhibit differences in the stringent response [37,38].
Expression of the genes encoding components of the chitobiose PTS was elevated in the rel Bbu mutant, implying that (p)ppGpp inhibits chitobiose uptake. In contrast to the glp operon which is regulated by RpoS, Rrp1 and BosR, chbCAB transporter genes are not members of any of these regulons [35,64,71]. However, a recent study did suggest that chbC expression was significantly lower in an Rrp1 mutant not expressing RpoS or BosR [72]. While this implies that either RpoS or BosR are required for minimal expression of chbC and is consistent with the findings of Rhodes et al. [73], it is not consistent with those of Pappas et al [36]. These latter workers found that chbC transcripts were significantly higher in ticks than in mouse joints [36]. Although a chbC mutant could not utilize chitobiose, it could successfully complete the mouse-tick infectious cycle, suggesting that chitobiose utilization is dispensable for this latter process [74]. Resolution of these apparently contradictory findings and the possible role of (p)ppGpp in modulating chitobiose uptake will require further research.
The relationship between glycerol and chitobiose utilization is complex. Modulation of genes involved in utilization of glycerol and chitobiose by global regulators is likely to be relevant for survival of B. burgdorferi in ticks [35,36,72–75]. The ability of B. burgdorferi to utilize glycerol in unfed nymphs may in fact be essential for effectively surviving the non-replicative quiescent state [76]. In other bacteria, the stringent response and glycerol metabolism are associated with establishing and maintaining the persistent state [76,77]. The data presented here could suggest that a similar situation is operative in B. burgdorferi.
Although both the glp operon and chitobiose transporter genes are expressed at significantly higher levels in ticks than in mammals [36], the regulatory mechanisms controlling utilization of the two carbohydrates are distinct. Repression of glp operon expression during the mammalian phase is RpoS-dependent, but this has not been definitively demonstrated for chbC. Elevation of transcripts for all components of the chiotobiose transporter in the rel Bbu mutant suggests that (p)ppGpp might be the regulatory molecule responsible for expression of the chitobiose transport genes. Further studies of B. burgdorferi growing under in vivo conditions throughout the enzootic cycle will be necessary to clarify the potential contribution of rel and (p)ppGpp to the regulation of glycerol and chitobiose utilization.
It was previously suggested that B. burgdorferi glp mutants survived in flat and feeding nymphs because they had access to chitobiose that became available during dissolution of the peritrophic matrix and molting [36]. It could be hypothesized that the stringent response in B. burgdorferi links nutritional stress with sequential utilization of hexoses, glycerol and chitobiose [1,50,53,55]. The role played by (p)ppGpp in this sequential utilization during shifts from exponential phase in feeding larvae to stationary phase in flat nymphs and again to exponential growth in feeding nymphs is consonant with the role this alarmone plays in other bacteria, and is comparable to diauxic growth in E. coli facing similar nutritional challenges [1,50,53,55].
There are several possible mechanisms by which (p)ppGpp could regulate gene expression in B. burgdorferi. It could directly affect transcription, or act indirectly by changing levels of other transcriptional regulators [2,3,19,48] or by modifying protein function by direct binding [78]. For example, we found that mRNA expression of two transcriptional regulators, DksA and Rrp1 was increased in the rel Bbu deletion mutant relative to wild type (Table 1). While this is consistent with repression of expression of these genes by (p)ppGpp, it could also result from a mechanism where the absence of one of these regulators was compensated by overproduction of another [10,11,79]. In E. coli, an increase in DksA can compensate for the lack of (p)ppGpp and can act synergistically, antagonistically and independently of (p)ppGpp to regulate mRNA transcription [80]. In other bacteria, DksA is required for negative regulation of rRNA and ribosomal protein expression and positive regulation of maintenance and stress-resistance promoters during the stringent response [17,81,82]. In the present case, our results suggest that DksA may act synergistically with (p)ppGpp in B. burgdorferi; further work is needed to determine whether there is a total or partial coincidence of the Rel and DskA regulons in this organism. (p)ppGpp does not seem to be involved in regulation of Rrp2, RpoN and BosR in B. burgdorferi since transcript amounts for the genes encoding these regulators were not affected by the deletion of rel Bbu.
Stringent response regulation of glycerol and chitobiose gene expression might be mediated directly at the level of transcription by interactions of (p)ppGpp and DksA with the RNA polymerase complex at the glycerol and chitobiose gene promoters [1,14,83]. That there were no differences in transcriptional levels of rpoS, rpoN, bosR, and rrp2 between the rel Bbu mutant and wild type B. burgdorferi under any experimental conditions examined suggests a direct interaction of (p)ppGpp with the RNA polymerase complexes at the glp and chitobiose operon promoters or interactions with still uncharacterized regulators [1,14,48,50,83]. The B. burgdorferi genome does not encode regulators such as SlyA and PigR that are directly involved in transcriptional regulation mediated by (p)ppGpp in other bacteria, but the role of DksA as a global regulator in B. burgdorferi has not yet been defined [1]. In E. coli, the CsrA and stringent response regulatory pathways have been shown to be linked [19]. The recently characterized B. burgdorferi CsrA and PlzA homologues could therefore be potential regulators responsible for linking the stringent response to Hk2-Rrp2/BosR-RpoN/RpoS and the Hk1-Rrp1-c-di-GMP cascades in their ability to regulate glycerol and chitobiose metabolism [19,39,43,84]. However, more recent work suggests that CsrA is not involved in regulation of the RpoN-RpoS axis in B. burgdorferi [85].
In summary, we have established the stringent response/(p)ppGpp regulon in B. burgdorferi during exponential and stationary phase growth in vitro. We show that (p)ppGpp stimulates induction of the glp operon and repression of chitobiose transporter gene expression. A limitation of the present findings is that they are based on expression analysis of B. burgdorferi during in vitro culture. Additional studies will be necessary to examine the nature of the stringent response in vivo in both ticks and mammals. Possible differences in the stringent response in B. burgdorferi strains with differing virulence potential may also warrant investigation.
Materials and Methods
B. burgdorferi strains and culture conditions
B. burgdorferi 297 (clone BbAH130) was kindly provided by Dr. M. V. Norgard, University of Texas Southwestern Medical Center. This strain was the parental strain for the B. burgdorferi Δrel Bbu deletion mutant and its complemented derivative, B. burgdorferi Δrel Bbu pKFSS1-rel Bbu [38]. B. burgdorferi strains were maintained at 34°C in Barbour-Stoenner-Kelley (BSK)-H (Sigma-Aldrich, St. Louis, MO) supplemented with 6% heat-inactivated rabbit serum (Sigma). B. burgdorferi Δrel Bbu was grown in the presence of 400 μg/ml of kanamycin (Sigma), B. burgdorferi Δrel Bbu pKFSS1-rel Bbu was grown in the presence of 400 μg/ml of kanamycin and 50 μg/ml of streptomycin (Sigma).
For microarray experiments, strains were grown continuously at 34°C, and were not temperature shifted [46]. For growth experiments and expression analysis of rel Bbu and glp genes in modified growth medium with different carbon sources at different temperatures, B. burgdorferi 297 and Δrel Bbu derivatives were temperature shifted. The cells for each strain were grown to late log phase (5–10 x 107 cells/ml) in BSK-II medium [86] at 25°C, and then diluted to a final concentration of 5 x 104 in 40 ml of BSK-Lite with N-acetylglucosamine [36,53] and either 0.4% glucose or 0.4% glycerol as principal carbon source. Quadruplicate 5 ml aliquots of each strain with each principal carbon source were incubated at 25°C or 34°C for up to 60 days, or until one week after stationary phase was reached. Technical replicate tubes were counted daily (for cultures incubated at 34°C) or every two days (for cultures incubated at 25°C) by dark field microscopy.
Microarray analysis
B. burgdorferi parental and Δrel Bbu cells were collected from duplicate cultures during exponential growth (5 x 107 cells/ml for wild type, 2 x 107 cells/ml for Δrel Bbu) and stationary phase (4x108 cells/ml for wild type, 8x107 cells/ml for Δrel Bbu) at 34°C. RNA was isolated using TRIzol (Invitrogen Life Technologies, Carlsbad, CA), treated with RQ1 RNase-free DNase (Promega Corporation, Madison, WI) and fluorescently labeled with Cy3 or Cy5 dye by reverse transcription. Microarray hybridizations were performed as described [64] with cDNA prepared from cells for each growth phase with two biological replicates and with two technical replicates (dye swap). Data acquired using GenePix software were transferred to Microsoft Excel for background subtraction and normalization [64]. Significance of differential expression was determined by two-tailed, unpaired Student t test at P<0.02 and fold-comparison >2. Genes located on plasmids of B. burgdorferi B31 that are printed on microarray slides, but are absent in B. burgdorferi 297 [87] were removed from the final output. Microarray data have been submitted under ArrayExpress (https://www.ebi.ac.uk/arrayexpress), accession number E-MTAB-3029.
Reverse transcription and real-time PCR
Two reverse transcription real-time PCR protocols were used in this work: one to validate the results of the microarrays of B. burgdorferi strains grown in exponential and stationary phases and the other to determine gene expression in B. burgdorferi strains growing in BSK-Lite.
To validate microarray results, cDNA was synthesized using 1 μg of total B. burgdorferi RNA isolated for microarray analysis, the primers listed in Table 2, and AMV reverse transcriptase (Promega) following the manufacturer’s instructions. The resulting cDNA was quantified by real-time PCR with primers specific for each gene (S3 Table) using the LightCycler Master SYBR Green I Mixture (Roche) and a LightCycler Real-time PCR instrument (Roche). To compare mRNA levels, PCR reactions were performed with both biological replicates used for microarray analysis. Each experimental sample was analyzed in duplicate. Genomic DNA from 103–106 cells of the corresponding B. burgdorferi strain was used as a standard to estimate the amount of cDNA of genes studied in each real-time PCR. Transcript amounts for each gene were normalized to cDNA of constitutively expressed flaB. Results are reported as log2 mean expression of relevant genes in B. burgdorferi Δrel Bbu relative to their expression in the parental wild type strain.
To determine gene expression in B. burgdorferi strains growing in BSK-Lite, RNA was isolated from late log phase cells growing at 34°C and 25°C using TRIzol and treated twice with the Ambion DNA free kit (Ambion, Austin, TX) according to the manufactuer’s instructions to remove DNA. cDNA was synthesized using 2 μg of purified RNA, random hexamer primers (Promega), and AMV reverse transcriptase enzyme (Promega). Real-time PCR reactions were performed as previously described [88] using primers listed in S3 Table. Copy number for flaB was determined for each biological sample and copy number for each gene was then normalized to copies of flaB.
Data analysis
Statistical analysis of growth curves was performed on log2 transformed data using one-way ANOVA. Significance was defined as P ≤ 0.05. Data from microarrays were analyzed as previously described [64].
Supporting Information
(DOC)
(DOC)
(DOC)
Data Availability
All relevant data are within the paper and its Supporting Information files. Microarray data have been submitted under ArrayExpress (https://www.ebi.ac.uk/arrayexpress), accession number E-MTAB-3029.
Funding Statement
This work was supported by grant 5R01 AI48856-07 from National Institutes of Health http://www.nih.gov/ to FCC and by grant 5R01 AI45801 from National Institutes of Health to IS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Potrykus K, Cashel M (2008) (p)ppGpp: still magical? Annu Rev Microbiol 62: 35–51. 10.1146/annurev.micro.62.081307.162903 [DOI] [PubMed] [Google Scholar]
- 2. Traxler MF, Summers SM, Nguyen HT, Zacharia VM, Hightower GA et al. (2008) The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli . Mol Microbiol 68: 1128–1148. 10.1111/j.1365-2958.2008.06229.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Traxler MF, Zacharia VM, Marquardt S, Summers SM, Nguyen HT et al. (2011) Discretely calibrated regulatory loops controlled by ppGpp partition gene induction across the 'feast to famine' gradient in Escherichia coli . Mol Microbiol 79: 830–845. 10.1111/j.1365-2958.2010.07498.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. English BP, Hauryliuk V, Sanamrad A, Tankov S, Dekker NH et al. (2011) Single-molecule investigations of the stringent response machinery in living bacterial cells. Proc Natl Acad Sci U S A 108: 365–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boutte CC, Crosson S (2013) Bacterial lifestyle shapes stringent response activation. Trends Microbiol 21: 174–180. 10.1016/j.tim.2013.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Geiger T, Goerke C, Fritz M, Schafer T, Ohlsen K et al. (2010) Role of the (p)ppGpp synthase RSH, a RelA/SpoT homolog, in stringent response and virulence of Staphylococcus aureus . Infect Immun 78: 1873–1883. 10.1128/IAI.01439-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Atkinson GC, Tenson T, Hauryliuk V (2011) The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS One 6: e23479 10.1371/journal.pone.0023479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. He P, Deng C, Liu B, Zeng L, Zhao W et al. (2013) Characterization of a bifunctional enzyme with (p)ppGpp-hydrolase/synthase activity in Leptospira interrogans . FEMS Microbiol Lett 348: 133–142. 10.1111/1574-6968.12279 [DOI] [PubMed] [Google Scholar]
- 9. Persky NS, Ferullo DJ, Cooper DL, Moore HR, Lovett ST (2009) The ObgE/CgtA GTPase influences the stringent response to amino acid starvation in Escherichia coli . Mol Microbiol 73: 253–266. 10.1111/j.1365-2958.2009.06767.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bernardo LM, Johansson LU, Skarfstad E, Shingler V (2009) σ54-promoter discrimination and regulation by ppGpp and DksA. J Biol Chem 284: 828–838. 10.1074/jbc.M807707200 [DOI] [PubMed] [Google Scholar]
- 11. Brown L, Gentry D, Elliott T, Cashel M (2002) DksA affects ppGpp induction of RpoS at a translational level. J Bacteriol 184: 4455–4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lemke JJ, Durfee T, Gourse RL (2009) DksA and ppGpp directly regulate transcription of the Escherichia coli flagellar cascade. Mol Microbiol 74: 1368–1379. 10.1111/j.1365-2958.2009.06939.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sharma UK, Chatterji D (2010) Transcriptional switching in Escherichia coli during stress and starvation by modulation of σ70 activity. FEMS Microbiol Rev 34: 646–657. 10.1111/j.1574-6976.2010.00223.x [DOI] [PubMed] [Google Scholar]
- 14. My L, Rekoske B, Lemke JJ, Viala JP, Gourse RL et al. (2013) Transcription of the Escherichia coli fatty acid synthesis operon fabHDG is directly activated by FadR and inhibited by ppGpp. J Bacteriol 195: 3784–3795. 10.1128/JB.00384-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Perederina A, Svetlov V, Vassylyeva MN, Tahirov TH, Yokoyama S et al. (2004) Regulation through the secondary channel—structural framework for ppGpp-DksA synergism during transcription. Cell 118: 297–309. [DOI] [PubMed] [Google Scholar]
- 16. Lemke JJ, Sanchez-Vazquez P, Burgos HL, Hedberg G, Ross W et al. (2011) Direct regulation of Escherichia coli ribosomal protein promoters by the transcription factors ppGpp and DksA. Proc Natl Acad Sci U S A 108: 5712–5717. 10.1073/pnas.1019383108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gummesson B, Lovmar M, Nystrom T (2013) A proximal promoter element required for positive transcriptional control by guanosine tetraphosphate and DksA protein during the stringent response. J Biol Chem 288: 21055–21064. 10.1074/jbc.M113.479998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dalebroux ZD, Swanson MS (2012) ppGpp: magic beyond RNA polymerase. Nat Rev Microbiol 10: 203–212. 10.1038/nrmicro2720 [DOI] [PubMed] [Google Scholar]
- 19. Edwards AN, Patterson-Fortin LM, Vakulskas CA, Mercante JW, Potrykus K et al. (2011) Circuitry linking the Csr and stringent response global regulatory systems. Mol Microbiol 80: 1561–1580. 10.1111/j.1365-2958.2011.07663.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dalebroux ZD, Svensson SL, Gaynor EC, Swanson MS (2010) ppGpp conjures bacterial virulence. Microbiol Mol Biol Rev 74: 171–199. 10.1128/MMBR.00046-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Klinkenberg LG, Lee JH, Bishai WR, Karakousis PC (2010) The stringent response is required for full virulence of Mycobacterium tuberculosis in guinea pigs. J Infect Dis 202: 1397–1404. 10.1086/656524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pal RR, Bag S, Dasgupta S, Das B, Bhadra RK (2012) Functional characterization of the stringent response regulatory gene dksA of Vibrio cholerae and its role in modulation of virulence phenotypes. J Bacteriol 194: 5638–5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Charity JC, Blalock LT, Costante-Hamm MM, Kasper DL, Dove SL (2009) Small molecule control of virulence gene expression in Francisella tularensis . PLoS Pathog 5: e1000641 10.1371/journal.ppat.1000641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sun W, Roland KL, Branger CG, Kuang X, Curtiss R III (2009) The role of relA and spoT in Yersinia pestis KIM5 pathogenicity. PLoS One 4: e6720 10.1371/journal.pone.0006720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mwangi MM, Kim C, Chung M, Tsai J, Vijayadamodar G et al. (2013) Whole-genome sequencing reveals a link between β-lactam resistance and synthetases of the alarmone (p)ppGpp in Staphylococcus aureus . Microb Drug Resist 19: 153–159. 10.1089/mdr.2013.0053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Geiger T, Kastle B, Gratani FL, Goerke C, Wolz C (2014) Two small (p)ppGpp synthases in Staphylococcus aureus mediate tolerance against cell envelope stress conditions. J Bacteriol 196: 894–902. 10.1128/JB.01201-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Amato SM, Brynildsen MP (2014) Nutrient transitions are a source of persisters in Escherichia coli biofilms. PLoS One 9: e93110 10.1371/journal.pone.0093110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Na HS, Kim HJ, Lee HC, Hong Y, Rhee JH et al. (2006) Immune response induced by Salmonella typhimurium defective in ppGpp synthesis. Vaccine 24: 2027–2034. [DOI] [PubMed] [Google Scholar]
- 29. Park SI, Jeong JH, Choy HE, Rhee JH, Na HS et al. (2010) Immune response induced by ppGpp-defective Salmonella enterica serovar Gallinarum in chickens. J Microbiol 48: 674–681. 10.1007/s12275-010-0179-6 [DOI] [PubMed] [Google Scholar]
- 30. Wexselblatt E, Oppenheimer-Shaanan Y, Kaspy I, London N, Schueler-Furman O et al. (2012) Relacin, a novel antibacterial agent targeting the stringent response. PLoS Pathog 8: e1002925 10.1371/journal.ppat.1002925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Godfrey HP, Bugrysheva JV, Cabello FC (2002) The role of the stringent response in the pathogenesis of bacterial infections. Trends Microbiol 10: 349–351. [DOI] [PubMed] [Google Scholar]
- 32. Samuels DS (2011) Gene regulation in Borrelia burgdorferi . Annu Rev Microbiol 65: 479–499. 10.1146/annurev.micro.112408.134040 [DOI] [PubMed] [Google Scholar]
- 33. Skare JT, Carroll JA, Yang XF, Samuels DS, Akins DR (2010) Gene regulation, transcriptomics and proteomics. In: Samuels DS, Radolf JD, editors. Borrelia. Molecular biology, host interaction and pathogenesis. Norfold, UK: Caister Academic Press; pp. 67–101. [Google Scholar]
- 34. Radolf JD, Caimano MJ, Stevenson B, Hu LT (2012) Of ticks, mice and men: understanding the dual-host lifestyle of Lyme disease spirochaetes. Nat Rev Microbiol 10: 87–99. 10.1038/nrmicro2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. He M, Ouyang Z, Troxell B, Xu H, Moh A et al. (2011) Cyclic di-GMP is essential for the survival of the lyme disease spirochete in ticks. PLoS Pathog 7: e1002133 10.1371/journal.ppat.1002133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pappas CJ, Iyer R, Petzke MM, Caimano MJ, Radolf JD et al. (2011) Borrelia burgdorferi requires glycerol for maximum fitness during the tick phase of the enzootic cycle. PLoS Pathog 7: e1002102 10.1371/journal.ppat.1002102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bugrysheva JV, Dobrikova EY, Sartakova ML, Caimano MJ, Daniels TJ et al. (2003) Characterization of the stringent response and rel Bbu expression in Borrelia burgdorferi . J Bacteriol 185: 957–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bugrysheva JV, Bryksin AV, Godfrey HP, Cabello FC (2005) Borrelia burgdorferi rel is responsible for generation of guanosine-3'-diphosphate-5'-triphosphate and growth control. Infect Immun 73: 4972–4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Karna SL, Sanjuan E, Esteve-Gassent MD, Miller CL, Maruskova M et al. (2011) CsrA modulates levels of lipoproteins and key regulators of gene expression critical for pathogenic mechanisms of Borrelia burgdorferi . Infect Immun 79: 732–744. 10.1128/IAI.00882-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Karna SL, Prabhu RG, Lin YH, Miller CL, Seshu J (2013) Contributions of environmental signals and conserved residues to the functions of carbon storage regulator A of Borrelia burgdorferi . Infect Immun 81: 2972–2985. 10.1128/IAI.00494-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ouyang Z, Deka RK, Norgard MV (2011) BosR (BB0647) controls the RpoN-RpoS regulatory pathway and virulence expression in Borrelia burgdorferi by a novel DNA-binding mechanism. PLoS Pathog 7: e1001272 10.1371/journal.ppat.1001272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sze CW, Morado DR, Liu J, Charon NW, Xu H et al. (2011) Carbon storage regulator A (CsrABb) is a repressor of Borrelia burgdorferi flagellin protein FlaB. Mol Microbiol 82: 851–864. 10.1111/j.1365-2958.2011.07853.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sze CW, Li C (2011) Inactivation of bb0184, which encodes carbon storage regulator A, represses the infectivity of Borrelia burgdorferi . Infect Immun 79: 1270–1279. 10.1128/IAI.00871-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bugrysheva JV, Dobrikova EY, Godfrey HP, Sartakova ML, Cabello FC (2002) Modulation of Borrelia burgdorferi stringent response and gene expression during extracellular growth with tick cells. Infect Immun 70: 3061–3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Concepcion MB, Nelson DR (2003) Expression of spoT in Borrelia burgdorferi during serum starvation. J Bacteriol 185: 444–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Revel AT, Talaat AM, Norgard MV (2002) DNA microarray analysis of differential gene expression in Borrelia burgdorferi, the Lyme disease spirochete. Proc Natl Acad Sci U S A 99: 1562–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bugrysheva JV, Godfrey HP, Schwartz I, Cabello FC (2011) Patterns and regulation of ribosomal RNA transcription in Borrelia burgdorferi . BMC Microbiol 11: 17 10.1186/1471-2180-11-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Durfee T, Hansen AM, Zhi H, Blattner FR, Jin DJ (2008) Transcription profiling of the stringent response in Escherichia coli . J Bacteriol 190: 1084–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Balsalobre C (2011) Concentration matters!! ppGpp, from a whispering to a strident alarmone. Mol Microbiol 79: 827–829. 10.1111/j.1365-2958.2010.07521.x [DOI] [PubMed] [Google Scholar]
- 50. Liu M, Durfee T, Cabrera JE, Zhao K, Jin DJ et al. (2005) Global transcriptional programs reveal a carbon source foraging strategy by Escherichia coli . J Biol Chem 280: 15921–15927. [DOI] [PubMed] [Google Scholar]
- 51. Van Laar TA, Lin YH, Miller CL, Karna SL, Chambers JP et al. (2012) Effect of levels of acetate on the mevalonate pathway of Borrelia burgdorferi . PLoS One 7: e38171 10.1371/journal.pone.0038171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R et al. (1997) Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi . Nature 390: 580–586. [DOI] [PubMed] [Google Scholar]
- 53. von Lackum K., Stevenson B (2005) Carbohydrate utilization by the Lyme borreliosis spirochete, Borrelia burgdorferi . FEMS Microbiol Lett 243: 173–179. [DOI] [PubMed] [Google Scholar]
- 54. Hoon-Hanks LL, Morton EA, Lybecker MC, Battisti JM, Samuels DS et al. (2012) Borrelia burgdorferi malQ mutants utilize disaccharides and traverse the enzootic cycle. FEMS Immunol Med Microbiol 66: 157–165. 10.1111/j.1574-695X.2012.00996.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Traxler MF, Chang DE, Conway T (2006) Guanosine 3',5'-bispyrophosphate coordinates global gene expression during glucose-lactose diauxie in Escherichia coli . Proc Natl Acad Sci U S A 103: 2374–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Potrykus K, Murphy H, Philippe N, Cashel M (2011) ppGpp is the major source of growth rate control in E. coli . Environ Microbiol 13: 563–575. 10.1111/j.1462-2920.2010.02357.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mechold U, Malke H (1997) Characterization of the stringent and relaxed responses of Streptococcus equisimilis . J Bacteriol 179: 2658–2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kazmierczak KM, Wayne KJ, Rechtsteiner A, Winkler ME (2009) Roles of rel Spn in stringent response, global regulation and virulence of serotype 2 Streptococcus pneumoniae D39. Mol Microbiol 72: 590–611. 10.1111/j.1365-2958.2009.06669.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gaca AO, Abranches J, Kajfasz JK, Lemos JA (2012) Global transcriptional analysis of the stringent response in Enterococcus faecalis . Microbiology 158: 1994–2004. 10.1099/mic.0.060236-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Dunham-Ems SM, Caimano MJ, Pal U, Wolgemuth CW, Eggers CH et al. (2009) Live imaging reveals a biphasic mode of dissemination of Borrelia burgdorferi within ticks. J Clin Invest 119: 3652–3665. 10.1172/JCI39401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sultan SZ, Manne A, Stewart PE, Bestor A, Rosa PA et al. (2013) Motility is crucial for the infectious life cycle of Borrelia burgdorferi . Infect Immun 81: 2012–2021. 10.1128/IAI.01228-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gupta RS (2000) The phylogeny of proteobacteria: relationships to other eubacterial phyla and eukaryotes. FEMS Microbiol Rev 24: 367–402. [DOI] [PubMed] [Google Scholar]
- 63. Kvint K, Farewell A, Nystrom T (2000) RpoS-dependent promoters require guanosine tetraphosphate for induction even in the presence of high levels of σS . J Biol Chem 275: 14795–14798. [DOI] [PubMed] [Google Scholar]
- 64. Caimano MJ, Iyer R, Eggers CH, Gonzalez C, Morton EA et al. (2007) Analysis of the RpoS regulon in Borrelia burgdorferi in response to mammalian host signals provides insight into RpoS function during the enzootic cycle. Mol Microbiol 65: 1193–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Casjens S, van VR, Tilly K, Rosa PA, Stevenson B (1997) Homology throughout the multiple 32-kilobase circular plasmids present in Lyme disease spirochetes. J Bacteriol 179: 217–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Damman CJ, Eggers CH, Samuels DS, Oliver DB (2000) Characterization of Borrelia burgdorferi BlyA and BlyB proteins: a prophage-encoded holin-like system. J Bacteriol 182: 6791–6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Dunham-Ems SM, Caimano MJ, Eggers CH, Radolf JD (2012) Borrelia burgdorferi requires the alternative sigma factor RpoS for dissemination within the vector during tick-to-mammal transmission. PLoS Pathog 8: e1002532 10.1371/journal.ppat.1002532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lin B, Short SA, Eskildsen M, Klempner MS, Hu LT (2001) Functional testing of putative oligopeptide permease (Opp) proteins of Borrelia burgdorferi: a complementation model in opp - Escherichia coli . Biochim Biophys Acta 1499: 222–231. [DOI] [PubMed] [Google Scholar]
- 69. Wang XG, Kidder JM, Scagliotti JP, Klempner MS, Noring R et al. (2004) Analysis of differences in the functional properties of the substrate binding proteins of the Borrelia burgdorferi oligopeptide permease (Opp) operon. J Bacteriol 186: 51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Novak EA, Sultan SZ, Motaleb MA (2014) The cyclic-di-GMP signaling pathway in the Lyme disease spirochete, Borrelia burgdorferi . Front Cell Infect Microbiol 4:56 eCollection;%2014.: 56. 10.3389/fcimb.2014.00056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ouyang Z, Kumar M, Kariu T, Haq S, Goldberg M et al. (2009) BosR (BB0647) governs virulence expression in Borrelia burgdorferi . Mol Microbiol 74: 1331–1343. 10.1111/j.1365-2958.2009.06945.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Sze CW, Smith A, Choi YH, Yang X, Pal U et al. (2013) Study of the response regulator Rrp1 reveals its regulatory role in chitobiose utilization and virulence of Borrelia burgdorferi . Infect Immun 81: 1775–1787. 10.1128/IAI.00050-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Rhodes RG, Coy W, Nelson DR (2009) Chitobiose utilization in Borrelia burgdorferi is dually regulated by RpoD and RpoS. BMC Microbiol 9: 108 10.1186/1471-2180-9-108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Tilly K, Grimm D, Bueschel DM, Krum JG, Rosa P (2004) Infectious cycle analysis of a Borrelia burgdorferi mutant defective in transport of chitobiose, a tick cuticle component. Vector Borne Zoonotic Dis 4: 159–168. [DOI] [PubMed] [Google Scholar]
- 75. Tilly K, Elias AF, Errett J, Fischer E, Iyer R et al. (2001) Genetics and regulation of chitobiose utilization in Borrelia burgdorferi . J Bacteriol 183: 5544–5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Amato SM, Fazen CH, Henry TC, Mok WW, Orman MA et al. (2014) The role of metabolism in bacterial persistence. Front Microbiol 5: 70 10.3389/fmicb.2014.00070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Helaine S, Cheverton AM, Watson KG, Faure LM, Matthews SA et al. (2014) Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science 343: 204–208. 10.1126/science.1244705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kanjee U, Ogata K, Houry WA (2012) Direct binding targets of the stringent response alarmone (p)ppGpp. Mol Microbiol 85: 1029–1043. 10.1111/j.1365-2958.2012.08177.x [DOI] [PubMed] [Google Scholar]
- 79. Chandrangsu P, Lemke JJ, Gourse RL (2011) The dksA promoter is negatively feedback regulated by DksA and ppGpp. Mol Microbiol 80: 1337–1348. 10.1111/j.1365-2958.2011.07649.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Magnusson LU, Gummesson B, Joksimovic P, Farewell A, Nyström T (2007) Identical, independent, and opposing roles of ppGpp and DksA in Escherichia coli . J Bacteriol 189: 5193–5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kolmsee T, Delic D, Agyenim T, Calles C, Wagner R (2011) Differential stringent control of Escherichia coli rRNA promoters: effects of ppGpp, DksA and the initiating nucleotides. Microbiology 157: 2871–2879. 10.1099/mic.0.052357-0 [DOI] [PubMed] [Google Scholar]
- 82. Lennon CW, Ross W, Martin-Tumasz S, Toulokhonov I, Vrentas CE et al. (2012) Direct interactions between the coiled-coil tip of DksA and the trigger loop of RNA polymerase mediate transcriptional regulation. Genes Dev 26: 2634–2646. 10.1101/gad.204693.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Roberts JW (2009) Promoter-specific control of E. coli RNA polymerase by ppGpp and a general transcription factor. Genes Dev 23: 143–146. 10.1101/gad.1770509 [DOI] [PubMed] [Google Scholar]
- 84. He M, Zhang JJ, Ye M, Lou Y, Yang XF (2014) Cyclic Di-GMP receptor PlzA controls virulence gene expression through RpoS in Borrelia burgdorferi . Infect Immun 82: 445–452. 10.1128/IAI.01238-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Ouyang Z, Zhou J, Norgard MV (2014) CsrA (BB0184) is not involved in the activation of the RpoN-RpoS regulatory pathway in Borrelia burgdorferi . Infect Immun 82: 1511–1522. 10.1128/IAI.01555-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Barbour AG (1984) Isolation and cultivation of Lyme disease spirochetes. Yale J Biol Med 57: 521–525. [PMC free article] [PubMed] [Google Scholar]
- 87. Casjens SR, Mongodin EF, Qiu WG, Luft BJ, Schutzer SE et al. (2012) Genome stability of Lyme disease spirochetes: comparative genomics of Borrelia burgdorferi plasmids. PLoS One 7: e33280 10.1371/journal.pone.0033280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Iyer R, Mukherjee P, Wang K, Simons J, Wormser GP et al. (2013) Detection of Borrelia burgdorferi nucleic acids after antibiotic treatment does not confirm viability. J Clin Microbiol 51: 857–862. 10.1128/JCM.02785-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
(DOC)
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. Microarray data have been submitted under ArrayExpress (https://www.ebi.ac.uk/arrayexpress), accession number E-MTAB-3029.


