Abstract
8-oxoguanine DNA glycosylase (Oggl) repairs 8-oxo-7,8-dihydroxyguanine (8-oxoG), one of the most abundant DNA adducts caused by oxidative stress. In the mitochondria, Oggl is thought to prevent activation of the intrinsic apoptotic pathway in response to oxidative stress by augmenting DNA repair. However, the predominance of the β-Oggl isoform, which lacks 8-oxoG DNA glycosylase activity, suggests that mitochondrial Oggl functions in a role independent of DNA repair. We report here that overexpression of mitochondria-targeted human α-hOggl (mt-hOggl) in human lung adenocarcinoma cells with some alveolar epithelial cell characteristics (A549 cells) prevents oxidant-induced mitochondrial dysfunction and apoptosis by preserving mitochondrial aconitase. Importantly, mitochondrial α-hOggl mutants lacking 8-oxoG DNA repair activity were as effective as wild-type mt-hOggl in preventing oxidant-induced caspase-9 activation, reductions in mitochondrial aconitase and apoptosis suggesting that the protective effects of mt-hOgg 1 occur independent of DNA repair. Notably, wild-type and mutant mt-hOggl co-precipitate with mitochondrial aconitase. Furthermore, overexpression of mitochondrial aconitase abolishes oxidant-induced apoptosis whereas hOggl silencing using shRNA reduces mitochondrial aconitase and augments apoptosis. These findings suggest a novel mechanism that mt-hOggl acts as a mitochondrial aconitase chaperone protein to prevent oxidant-mediated mitochondrial dysfunction and apoptosis that might be important in the molecular events underlying oxidant-induced toxicity.
Keywords: DNA repair, aconitase, Oggl, free radicals, asbestos, mitochondria
Introduction
Reactive oxygen species (ROS) produced under physiologic conditions mediate various cellular signaling pathways but higher levels of ROS can induce oxidative DNA damage and apoptosis that contribute to tumorigenesis, aging, and degenerative diseases (1–3). As compared to nuclear DNA, mitochondrial DNA (mtDNA) is far more susceptible to ROS mediated damage because of its proximity to the electron transport chain and the relatively limited mitochondrial DNA repair capacity (1,4). One of the most abundant DNA adducts caused by oxidative stress, 8-oxo-7,8-dihydroxyguanine (8-oxoG), is causally associated with several cancers and neurodegenerative diseases because it induces mutations by preferentially mispairing with adenosine during replication (5,6). In mitochondria, the base excision repair pathway is primarily responsible for removing 8-oxoG from DNA (1). In humans, 8-oxoG is repaired by the 8-oxoguanine DNA glycosylase (hOggl), an enzyme that recognizes and hydrolyzes the aberrant base from the DNA backbone (7–9). Mutations or polymorphisms of the hOGGl gene, which is located on chromosome 3p26.2, increase the risk of various malignancies, including lung cancer (1,10–13). Alternative splicing of the hOGGl transcript results in two major isoforms: αOggl and βOggl. Mitochondria-targeted α-hOggl (mt-hOggl) overexpression blocks mitochondria-regulated apoptosis caused by oxidative stress in part by augmenting mtDNA repair mechanisms (14–17). Curiously, the mitochondrial levels of βOggl are 20-fold greater than αOggl, yet βOggl lacks 8-oxoG DNA glycosylase activity (18). The predominance of βOggl in the mitochondria suggests that hOggl may function in a role independent of DNA repair.
In this study, we explored the interaction between Oggl and mitochondrial aconitase, an iron-sulfur-containing tricarboxycylic acid (TCA) cycle enzyme that is vulnerable to oxidative inactivation and has been implicated as a mitochondrial redox sensor (19). Aconitase, which is crucial for maintaining mtDNA in yeast (20), co-precipitates with frataxin, an iron chaperone protein that blocks aconitase oxidative inactivation and/or potentiates its reactivation (21). We demonstrate that mt-hOggl overexpression prevents oxidant-induced mitochondrial dysfunction and apoptosis by blocking the decrease in mitochondrial aconitase. This function of mt-hOggl is independent of its DNA repair function as mt-hOggl mutants that lack 8-oxoG DNA glycosylase activity were as effective as wild-type mt-hOgg 1 in preventing oxidant-induced mitochondrial dysfunction, the decrease in aconitase activity and DNA fragmentation. We show that mt-hOggl co-precipitates with mitochondrial aconitase and that this blocks oxidant-induced decreases in mitochondrial aconitase protein expression and activity. Our results suggest that mitochondrial Oggl may act as a chaperone protein of aconitase to prevent oxidant-induced degradation of aconitase and mitochondria-regulated apoptosis. These data provide a novel mechanism linking oxidant-mediated DNA damage, aconitase and apoptosis.
Experimental Procedures
Reagents
All reagents were purchased from Sigma Inc (St Louis, MO) unless otherwise noted. Amosite asbestos fibers used in these experiments were Union International Centere le Cancer reference standard samples kindly supplied by Drs. V. Timbrell (22) and Andy Ghio (Environmental Protection Agency). These amphibole fibers are 70% respirable (length between 2–5 µm) while the remainder are > 5 µm in length. Stock solutions (5 mg/ml) were prepared in Hanks balanced salt solution (HBSS) with calcium, magnesium, and 15 mM HEPES. All suspensions were autoclaved and stored at 4°C. Samples were warmed to 37°C and vigorously vortexed prior to usage to ensure a uniform suspension and used as previously described (23–26).
Cell Culture
A549 cells, human lung adenocarcinoma cells with some features of alveolar epithelial type II (AT2) cells and a wild-type p53, were obtained from the American Type Culture Collection (ATCC, Rockville, MD). In general, A549 cells were plated in 6 well plates at a seeding density of 1 × 106 and grown to confluence over 24 h in Dulbeco's modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) while in a humidified 37°C incubator containing 5% CO2. To limit cell proliferation, the media was changed to DMEM with 0.5% FBS for an additional 24 h before adding either an endogenous (H2O2) or exogenous (asbestos) oxidative stress for an additional 24h in DMEM without serum.
Transient Transfection Protocols
Mitochondrial overexpression of the α-isoform of the hOGGl gene (kind gift from Dr. Glenn Wilson described elsewhere [14,27]) in A549 cells was accomplished by transient transfection using either recombinant Adenovirus (16) or by lipofectamine according to established protocols. Recombinant Adenoviruses used in these studies, which have been described by others elsewhere (16), consisted of an empty vector Ad.889 lacking the first cistron as a control for non-specific viral effects, and Ad.979 that encodes the α-isoform of the hOGGl gene using a CMV promoter and a mitochondrial targeting sequence for human manganese superoxide dismutase (SOD2) and modified to incorporate a stop codon. Nearly confluent A549 cells were transfected with Ad.889 or Ad.979 with viral particle concentrations corresponding to multiplicity of infections (MOI) of six (6×), ten (lO×), fifteen (15×) or twenty (20×) viral particles per A549 cell. The cells were studied 48 h after transfection. The optimal mt-hOggl protein expression was noted with a MOI of lO× and that was utilized for all subsequent experiments.
In separate experiments, wild-type and mutant α-Oggl plasmid constructs previously described with the modifications detailed below (18) were transiently transfected into A549 cells using lipofectamine 2000 (Invitrogen) according to the manufacture’s recommendations. Briefly, A549 cells (1 million cells in 2 ml of DMEM without antibiotics) were placed in each well of a 6 well plate for 24 h in a humidified 37°C incubator containing 5% CO2 so that the cells were ~95% confluent at the time of transfection the next day. For each transfection, complexes were prepared in a Opti-MEM® (Invitrogen) using 4 µg of plasmid DNA and 10 µl of Lipofectamine™ 2000, incubated for 20 min at room temperature, and then incubated at 37°C in a CO2 incubator for 4–6 h before the medium was changed to DMEM with 10% FBS. After incubation for 48 h, the cells were tested for oxidant-induced caspase-9 activation, mt-aconitase activity and DNA fragmentation.
Western Analysis
Cell lysates were collected and immunoblotting were performed as described (26). For localization studies, we separated the total cellular protein into the mitochondrial and the cytosolic fractions using an Apoalert fractionation kit (BD Biosciences Clontech, Palo Alto, CA) as described (26). The antibodies for Western blotting included polyclonal antibodies directed against hOgg 1 (Novus Biological, Littleton, CO), mitochondrial aconitase (kind gift of Dr L. Sweda; 21), cytochrome oxidase IV (1:1000; COX IV; Cell Signaling Technology) and beta tubulin (1:1000; Santa Cruz Biotechnologies; Santa Cruz, CA). The protein bands were visualized by enhanced chemiluminescence reaction (Amersham Biosciences, Indianapolis, IN) and quantified by densitometry using Eagle Eye software (Stratagene, La Jolla, CA).
8-Oxo-Guanine Incision Assay
A549 cell 8-oxoG incision was performed using mitochondrial extracts as previously described (18). Briefly, empty vector control and mt-hOggl overexpressing A549 cells were treated with asbestos 25 ug/cm2 for 24 h and mitochondrial protein was extracted as described (26). 8-oxoG-containing 21-mer with the sequence5’-CAGCCAATCAGTXCACCATTC-3’ (X: 8-oxoG), and its complementary strand were chemically synthesized (The Midland Certified Reagent Co., Midland, TX), 3’-end-labeled using terminal transferase and [32P]ddATP (Amersham Biosciences, 3000 Ci/mmol), and annealed with its complementary oligonucleotide to form the duplex DNA that was used as the assay substrate. The duplex substrate DNA (20 pmol) was incubated with the mitochondrial extracts (25 µg of protein) at 37°C for 1 h in 1 ml of reaction mixture (50 mM Tris-HCl, 50 mM KC1, and 1 mM EDTA, pH 7.5). As a negative control, substrate DNA that was not exposed to mitochondrial extracts was subjected to the same procedure. The reaction was terminated by heating at 90°C for 3 min. Reaction products were electrophoresed on 20% denaturing (7 M urea) polyacrylamide gels (DNA sequencing gel). After electrophoresis, the gels were wrapped in Saran Wrap and the amounts of the cleaved substrate were determined by autoradiography and Quantity One software (Bio-Rad Laboratories). 8-oxoG incision activity was calculated from the ratio of the radioactivity in the band corresponding to the damage-specific cleavage product (13-mer) divided by the total radioactivity in the lane (13mer plus 21mer) and expressed as fold control levels.
Mitochondrial and Apoptosis Assays
AEC AΔΨm and caspase-9 activation were assessed as we have described (24–26). Briefly, AEC ΔΨm was based upon the percentage difference in the ratio of tetramethylrhodamine ethyl ester (TMRE) and MitoTracker green fluorescence corrected for the background fluorescence. Caspase-9 activity was assessed by a commercially available fluorometric assay kit (Roche Diagnostics, Indianapolis, IN). Apoptosis was assessed using a histone-associated DNA fragmentation (mono and oligo nucleosomes) ELISA assay (Roche diagnostics, Indianapolis, IN).
Measurement of ROS production by Dichlorofluorescence
A549 cell ROS production was determined using 2', 7’-dichlorodihydrofluorescein diacetate (DCFH-DA; Molecular Probes, Eugene, OR) as described (25). Briefly, after the cells were transfected with mt-hOggl or empty vector controls for 48 h, they were treated for various times (1 h or 24 h) with control media, asbestos (5 or 25 µg/cm2), or Antimycin A (10 nM), washed with PBS and then loaded for 30 min with DCFH-DA (10 µM) in MEM without phenol red. The acetoxymethyl group on DCFH-DA is cleaved by nonspecific esterases within the cell, resulting in a nonfluorescent charged molecule that does not cross the cell membrane. Intracellular ROS irreversibly oxidize the DCFH-DA to dichlorofluorescein (DCF), which is a fluorescent product. After treatment, the media were removed, the cells were lysed, centrifuged to eliminate debris, and the fluorescence in the supernatant was assessed using a fluorometer (excitation 500 nm/emission 530 nm). The data are expressed as relative fluorescence units (fold control).
Measurement of ROS production by targeted-redox sensors
We also used reduction-oxidation sensitive green fluorescent protein (roGFP) targeted to the mitochondria or cytosol that enables real time visualization of the oxidative state of the indicator by displaying rapid and reversible ratiometric changes in fluorescence in response to changes in redox potential (28,29). Briefly, A549 and mt-hOggl overexpressed cells were infected with an adenovirus encoding roGFP with or without a mitochondrial localization sequence. Forty-eight hours after infection, the cells were exposed to vehicle or asbestos for 24h. The cells were then removed from the plate with trypsin and equal aliquots of the resulting suspension were transferred to tubes containing media alone or media containing lmM DTT or lmM t-butyl-hydroperoxide (t-BOOH). After 10 min, the ratio of fluorescence (em=535nm) at ex=400 and 490 nm was measured in 5,000 cells per condition by flow cytometry using a DakoCytomation MoFlo high speed multilaser droplet cell sorter, which allows for high speed analysis of a single cell at multiple excitation wavelengths. The oxidation state of the probe was calculated as the completely reduced ratio (DTT) less the untreated value divided by the difference in the ratio observed with DTT and t-BOOH. As a control, cells not expressing Ro-GFP were grown under identical conditions and subjected to the same procedure.
Aconitase Activity
Aconitase activity was measured by a commercially available spectrophotometric assay (OxisResearch, Portland, OR) that measures the formation of NADPH at 340 nm in a coupled assay with citrate as the substrate of aconitase and the isocitrate formed is converted to alpha-ketoglutarate by NADP+-dependent isocitrate dehydrogenase (19). The data were expressed as RU / µg protein and expressed as fold-control levels.
Aconitase Immunopurification
Mitochondrial extracts (150 µg in 100 µ1) prepared as previously described (26) were incubated with 10 µ1 of anti-hOggl at 4°C for 1 h. The mixture was then treated with protein G-Sepharose, incubated for 16 h (4°C), centrifuged at l000g for 5 min, the beads were washed three times with PBS, pH 7.6 containing 1% NP-40 and resuspended in gel-loading buffer containing SDS but without reductant. The membrane was then probed by Western blot analysis with antibodies directed for mitochondrial aconitase, IgG or hOggl.
Mitochondrial-targeted Wild-type α-Oggl and α-Oggl Mutant Plasmid Constructs
To determine whether 8oxoG DNA glycocylase activity is required to mediate the protective effects of mt-hOggl, we utilized wild-typeα-Oggl and mutantα-Oggl plasmid constructs with specific amino acid substitutions in theα-0 helix ofαOggl to that of theβOggl isoform that lacks 8-oxoG DNA repair (V317G lacks 8-oxoG binding and has decreased 8-oxoG incision activity; longα/β-317–323 completely lacks both 8-oxoG binding and incision activity) as previously described (18) with the modification of its vector backbone from the pQE vector to pcDNA3.1 (+) (Invitrogen), the addition of 5’ mitochondrial localizing sequence from the aminoterminal region of subunit IV of the cytochrome c oxidase gene, partial deletion of the nuclear localizing sequence (NLS, 335–342; ΔNLS 329–344) from the 3’ terminus and 3’ addition of myc tag for all the constructs. These constructs were verified by DNA sequencing. Mitochondrial localization of the proteins was confirmed by immunostaining using C-Myc mouse monoclonal antibodies (Santa Cruz Biotechnology). The plasmids were transiently transfected into A549 cells using lipofectamine 2000 (Invitrogen) according to the manufacture’s recommendations.
To determine whether mitochondrial aconitase co-immunoprecipitates with overexpressed wild-type α-Oggl and mutant longaα/β317–323-Oggl, mitochondrial extracts (150 µg/µl) from A549 cells overexpressing either wild-typeα-Oggl or mutant longα/β317–323-Oggl were obtained and then incubated with 10 µ1 of antimyc (Abeam Inc, Cambridge, MA) at 4°C for 1 h. Immunoprecipitation was performed using the Seize X protein A immunoprecipitation kit (Pierce Biotechnology Inc, Rockford, IL) according to the manufacture’s recommendation and prepared for Western blot analysis as described above using antibodies directed against mitochondrial aconitase, hOggl, or COX IV.
Overexpression of Aconitase
An aconitase 2 construct (Invitrogen; full length clone CS0DL005YJ10 in pCMV-SPORT6) was transiently transfected into A549 cells using lipofectamine 2000 (Invitrogen) as described above. After incubation for 48 h, the cells were tested for mitochondrial aconitase transgene expression (activity and protein expression) as well as caspase-9 activity and DNA fragmentation in the presence and absence of asbestos.
Transient RNA Interference Transfection
Small double-stranded shRNAs that silenced hOGGl and scrambled shRNA controls were purchased from Invitrogen. All experiments were performed with the shRNA sense, 5'UCC AAG GUG UGC GAC UGC UGC GAC A3' and antisense, 5'-UGU CGC AGC AGU CGC ACA CCU UGG A3' and a scrambled dsRNA negative control HI GC (Invitrogen). Briefly, A549 cells were seeded at 8 × 105/6-well dish or 1 × 107 /150 mm dish 24 h before transfection using 5 µ1 or 180 µ1 of lipofectamine 2000, respectively and a total of 100 pmol (6-well) and 1000 pmol (150 mm Dish) of RNA. The cells were harvested 1 d later by trypsinization and used for isolation of RNA and determination of protein concentration.
Real-time RT-PCR
Total RNA was isolated from transfected cells using Trizol Reagent (Invitrogen) and cDNA was prepared using 0.5 µg of oligo-d(T) primers and the Superscript III RNase H reverse transcriptase (Invitrogen) according to the manufacturer's protocol. Real time RT-PCR was used to verify the differential expression of selected human genes (OGGl and actin). Briefly, specific target primers and probe sets (Taqman Assays, Applied Biosystems), their corresponding probes, and template RNA were combined with TaqMan mastermix solution (Universal PCR Mastermix, Applied Biosystems) according to the manufacturer’s protocol. Amplified target sequences were detected with the ABI Prism 7300 sequence detector (Applied Biosystems). All reactions were run in triplicate and OGGl gene expression values were expressed as fold-control normalized to actin.
Statistical analysis
The results of each experimental condition were determined from the mean of triplicate trials. Data were expressed as the means ± SEM (n=6 unless otherwise stated). A two-tailed Student's t-test was used to assess the significance of differences between two groups. Analysis of variance was used when comparing more than two groups; differences between two groups within the set were analyzed by a Fisher's protected least significant differences test. Probability values <0.05 were considered significant.
Results
Mitochondrial targeted hOggl blocks oxidant-induced mitochondrial dysfunction and apoptosis
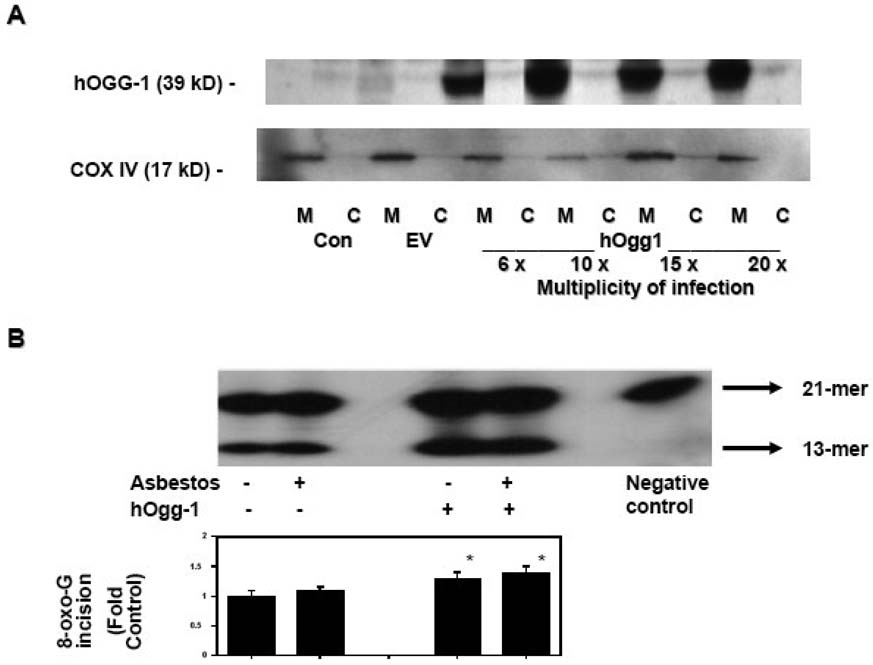
To investigate the mechanism by which mt-hOggl prevents ROS-induced apoptosis, we transiently overexpressed mt-hOggl in A549 cells using an adenoviral vector containing the α isoform of the hOGGl gene driven by the CMV promoter and containing an appended mitochondrial targeting sequence from human SOD2 as previously described (16). We used human A549 cells, which are derived from tumors of AT2 cells, because they facilitate molecular studies that are not feasible with primary isolated AT2 cells. We previously showed that A549 cells, similar to rat AT2 cells, undergo ROS (H2O2 and amosite asbestos)-induced p53-dependent transcription that mediates mitochondria-regulated apoptosis (23–26). In accord with studies in other cell types (16), transient transfection of A549 cells increased the levels of mitochondrial targeted hOggl protein as compared to controls (nontransfected or empty vector-treated) (Fig 1A). We detected minimal mitochondrial hOggl protein expression in control, nontransfected (M-Con lane) or empty vector-transfected (M-EV lane) A549 cells but noted more than a ten-fold increase in mitochondrial hOggl protein expression after adenoviral transient transfection (M-hOggl 6×-, 10×-, 15×-, and 20×-viral particles per A549 cell lanes) despite similar mitochondrial protein loading as assessed by COX IV expression. Consistent with previous reports showing that overexpression of mt-hOggl promotes mt-DNA repair after oxidative stress (14–17), we confirmed that mitochondria obtained from mt-hOggl overexpressing A549 cells increase 8-oxoG DNA repair activity as compared to mitochondria from empty vector-treated cells controls (~40% increase; p < .05 v. control; n=3) (Fig 1B). Although asbestos can augment 8-oxoG DNA repair by hOggl, mitochondrial 8-oxoG DNA repair has not been investigated (30). As shown in Figure 1B, asbestos did not augment mitochondrial 8-oxoG DNA repair as compared to empty vector treated cells nor did it alter the increased activity noted in mt-hOggl overexpressing cells.
Figure 1. Mitochondria-targeted hOggl augments hOggl mitochondrial protein and 8-oxoG DNA glycosylase activity in A549 cells.
(A) Mt-hOggl was transiently overexpressed using an adenovirus containing the α isoform of the hOGGl gene with a mitochondrial targeting sequence. A representative Western blot analysis of hOggl protein expression in mitochondrial (M) and cytosolic (C) fractions of control (con), empty vector (ev), and hOggl overexpression in A549 cells exposed to various multiplicity of infection (MOI) for 48 h. A MOI of lOx was used for all subsequent adenoviral transient transfection experiments. The Western blot is representative of three separate experiments. (B): Mitochondria-targeted hOggl augments 8-oxoG DNA glycosylase activity. A549 cells were transiently transfected with empty vector control (lanes 1 and 2) or mt-hOggl (lanes 3 and 4) as described in the Experimental Procedures and then treated with control media or asbestos (25 µg/cm2) for 24 h. 8-oxoG incision activity was calculated as described in the Experimental Procedures and expressed as fold control levels. (* p < .05 v. empty vector controls; n=3).
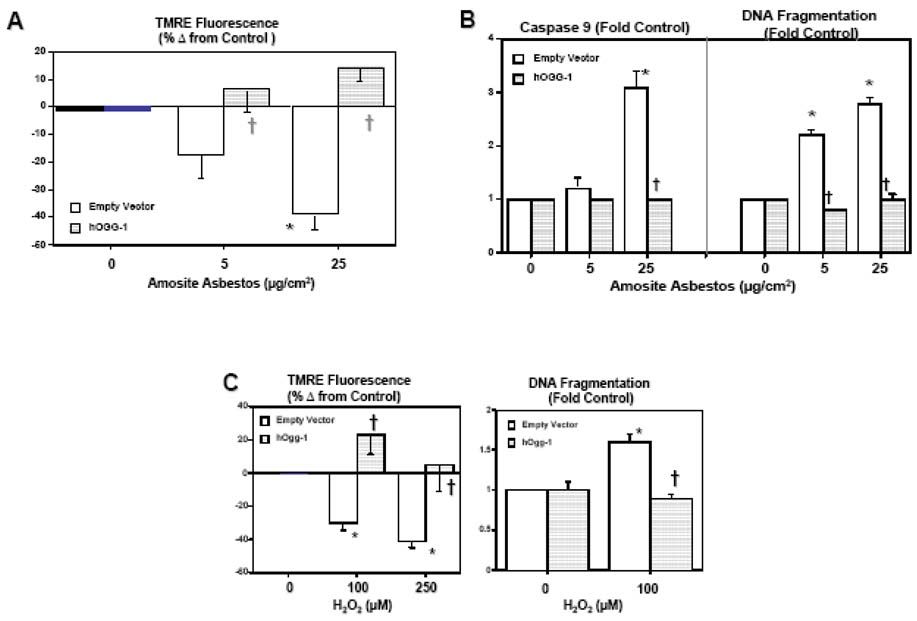
Empty vector-treated A549 cells exposed to H2O2 or amosite asbestos for 24 h demonstrated mitochondrial dysfunction, as assessed by a loss of mitochondrial membrane potential (ΔΨm), activation of caspase-9, and apoptosis, as assessed by DNA fragmentation (Fig 2). These findings with empty vector-treated cells were comparable to our earlier studies using A549 and rat AT2 cells (23–25). Furthermore, we have shown that the levels of H2O2- or asbestos-induced A549 cell apoptosis assessed by DNA fragmentation parallel the levels of apoptosis assessed by other measures including TUNEL staining, nuclear morphology, annexin V staining and caspase-3 activation (23–26). Notably, A549 cells overexpressing mt-hOggl completely blocked H2O2- or asbestos-induced mitochondrial dysfunction and DNA fragmentation (Fig 2). Asbestos (25 µg/cm2) increased TMRE fluorescence by ~10% (Figure 2A) but this difference did not reach statistical significance as compared to untreated mt-hOggl overexpressing A549 cells and we did not detect evidence of asbestos-induced caspase 9 activation or DNA fragmentation in mt-hOggl overexpressing cells (Fig 2).
Figure 2. Mitochondria-targeted hOggl blocks oxidant-induced A549 cell mitochondrial dysfunction and apoptosis.
(A) Mt-hOgg-1 overexpression prevents amosite asbestos-induced ΔΨm as assessed using a tetramethylrhodamine ethyl ester (TMRE) fluorometric technique. (B): Mt-hOggl overexpression abolishes asbestos-induced caspase-9 activation and DNA nucleosomal fragmentation.(C): Mt-hOggl overexpression blocks H2O2-induced ΔΨm (TMRE fluorescence) and DNA fragmentation. * p < .05 v. no asbestos or H2O2; † p < .05 v. empty vector cells exposed to the same dose of asbestos or H2O2.
Mitochondrial targeted hOggl does not prevent ROS production but does block oxidant-induced decreases in mitochondrial aconitase activity and protein expression
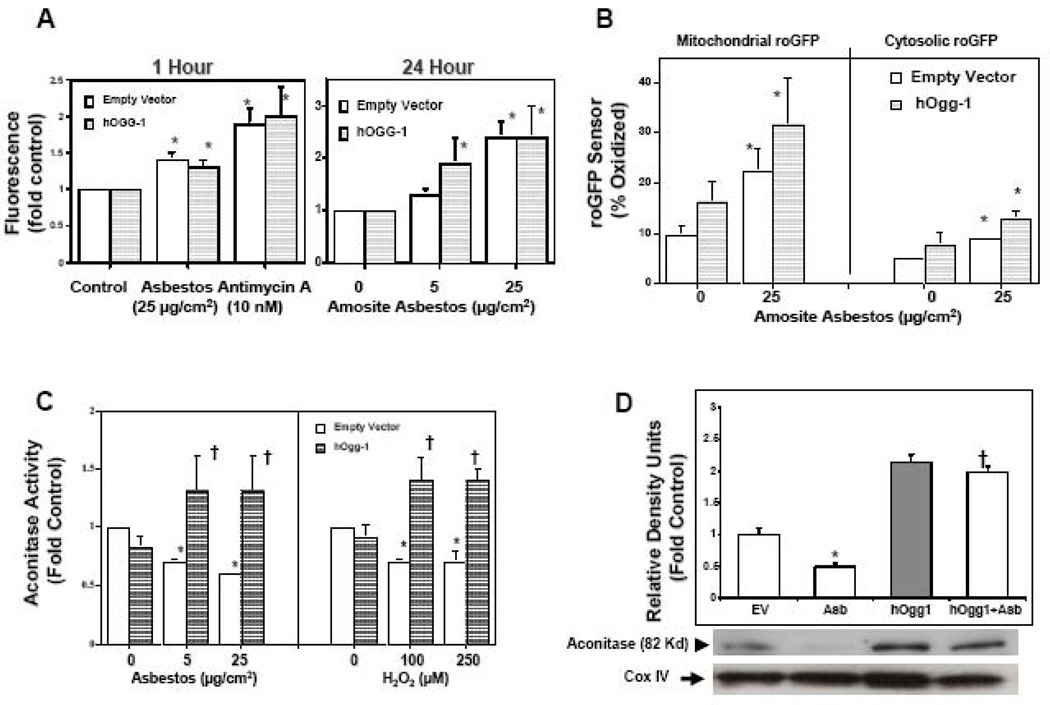
To determine whether mt-hOggl decreases the levels of oxidative stress, we used a 2’,7’-dichlorodihydro-fluorescein diacetate assay as described (24). We found that mt-hOggl did not alter the levels of oxidative stress caused by either asbestos or Antimycin A, the latter of which induces mitochondrial ROS production by blocking complex III in the respiratory chain (Fig 3A). To specifically assess mitochondrial oxidant stress, we used an adenovirally expressed roGFP targeted to the mitochondrial matrix or cytosol that enables real-time visualization of the oxidation status in the subcellular compartment where it is expressed (28,29). Asbestos increased ROS production in the mitochondria and, to a lesser extent, the cytosol (Fig 3B). The targeted roGFP sensors revealed more asbestos-induced oxidant stress in the mitochondria as compared to cytosol (22% v. 8% oxidized, respectively), which is in accord with our prior report implicating mitochondrial ROS in mediating asbestos-induced AT2 cell apoptosis (25). Similar to the DCF data (Fig 3A), mt-hOggl overexpression did not attenuate asbestos-induced oxidative stress (Fig 3B). Thus, we could not detect any attenuation of oxidant stress by mt-hOggl that would account for its protective effect.
Figure 3. Mitochondria-targeted hOggl does not alter oxidant-induced ROS production but does prevent reductions in mitochondrial aconitase.
(A) Mt-hOggl overexpression does not alter amosite asbestos- or Antimycin A-induced ROS production as assessed by a dichlorofluorescein (DCF) assay. (B) Mt-hOggl overexpression does not block asbestos-induced ROS production as assessed by a roGFP sensor targeted to the mitochondria or cytosol. (C) As expected, mitochondrial aconitase activity is reduced by oxidative stress (24 h exposure to amosite asbestos or H2O2). Notably, mt-hOggl overexpression completely preserves mitochondrial aconitase activity in the setting of oxidative stress. (D) Amosite asbestos decreases mitochondrial aconitase protein expression and mt-hOggl overexpression blocks this. The levels of mitochondrial aconitase expression from three experiments are shown in a densitometric analysis. Expression of cytochrome oxidase IV (COX IV) was used to confirm the presence of mitochondrial protein and comparable loading. * p < .05 v. empty vector controls not exposed to H2O2, Antimycin A, or asbestos; † p < .05 v. empty vector cells exposed to the same dose of H2O2 or asbestos.
To determine whether the protective effects of mt-hOggl are mediated by preserving mitochondrial aconitase against oxidant-induced degradation (31), we assessed aconitase activity and protein expression. In accordance with the rapid onset of H2O2-induced decreases in aconitase activity in cardiac mitochondria (19), we noted that a 30 min exposure to either 100 µM of H2O2 or 25 µg/cm2 of asbestos reduced mitochondrial aconitase activity in A549 cells (fold control: 0.80 ± 0.10 and 0.80 ± 0.03, respectively; n = 6; p < 0.05 v. control) and these changes persisted at 24 h (Fig 3C). Notably, mt-hOggl overexpression completely blocked oxidant-induced decreases in mitochondrial aconitase activity (Fig 3C). Similar to mitochondrial aconitase activity, asbestos decreased mitochondrial aconitase protein levels and mt-hOggl overexpression abolished this effect (Fig 3D). Collectively, these data show that mt-hOggl overexpression preserves mitochondrial aconitase activity and protein expression in the setting of oxidative stress from either H2O2 or asbestos.
Mitochondrial hOggl prevents oxidant-induced toxicity independent of its 8-oxoG DNA repair activity
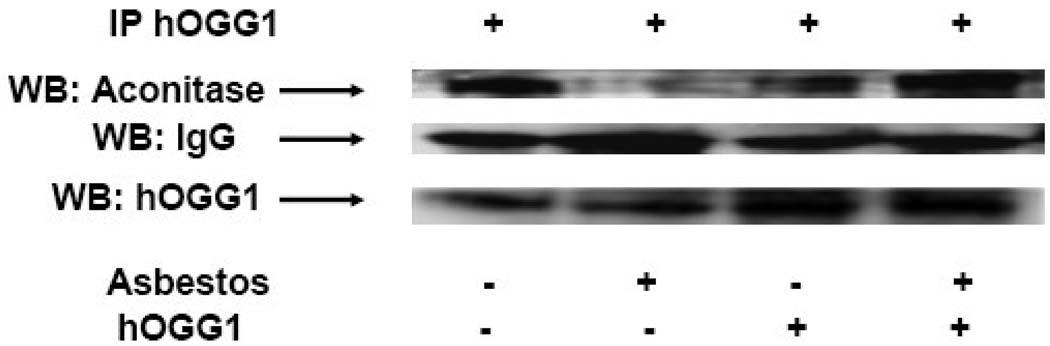
Because human mitochondria contain ~20-fold more of the βOggl isoform that lacks 8-oxoG DNA glycosylase activity (18), we reasoned that mt-hOggl might prevent oxidant-induced aconitase inactivation by acting as a molecular chaperone similar to frataxin (21). Although stable protein-protein interactions between various base excision repair members have not been demonstrated, hOggl does interact with select proteins such as XRCC1, a scaffolding protein, and protein kinases (e.g. Cdk4 and C-Abl) (32,33). Notably, mitochondrial aconitase co-immunoprecipitated with endogenous mitochondrial hOggl (Fig 4A; lane 1) and exposure to asbestos decreased this association (Fig 4A; 2nd lane). Further, mitochondria-targeted hOggl overexpressing A549 cells also produced a protein that co-immunoprecipitated with mitochondrial aconitase (Fig 4A; lane 3) and completely blocked asbestos-induced decreases in mitochondrial aconitase (Fig 4A; 2nd and 4th lanes). The changes in mitochondrial aconitase co-immunoprecipitation with hOggl parallel our earlier findings with aconitase activity and protein expression (Fig 3C and D).
Figure 4. Mitochondrial hOggl co-precipitates with mitochondrial aconitase.
Immunoprecipitation (IP) using a hOggl antibody was performed on mitochondria proteins obtained from control and mt-hOggl overexpressing A549 cells in the absence or presence of asbestos (25 µg/cm2) for 24 h as described in the Methods. Mitochondrial aconitase and loading controls (IgG and hOggl) were assessed by Western blotting (WB). The immunoprecipitation blots are representative of three independent experiments.
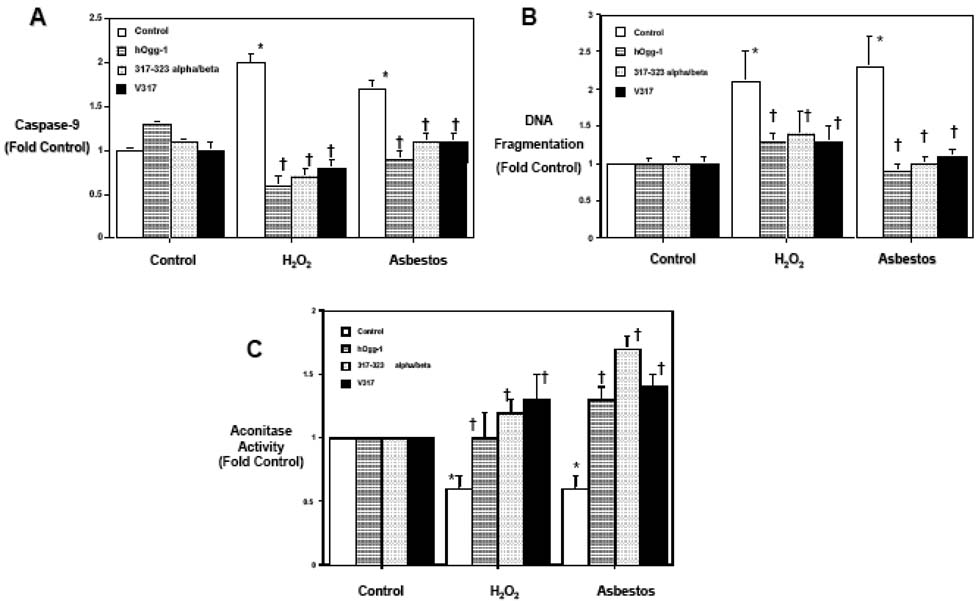
As βOggl lacks the C-terminal αO helix (aa 317–323) present in αOggl that is necessary for DNA binding and 8-oxoG DNA glycosylase activity (18), we reasoned that the protective effects of mitochondrial αOggl occur independent of 8oxoG DNA glycosylase activity by preserving mitochondrial aconitase. To address this possibility, we assessed the protective effects of two previously described mutant mt-Oggl proteins completely lacking in 8-oxoG DNA glycosylase activity (18). Specifically, the αO helix of the αOggl isoform was mutated by changing valine at amino acid 317 to the aliphatic amino acid glycine in the βOgg-l isoform (V317G) or by changing the entire 317–323 amino acid stretch of the αO helix of the αOggl isoform to the corresponding βOggl isoform (long α/β317–323) as characterized elsewhere (18). The wild-type and mutant Ogg 1 plasmid constructs were modified by placement in a mammalian expression vector, the addition of a mitochondrial localizing signal, partial deletion of the nuclear localizing sequence and the addition of a myc tag. After transient transfection with lipofectamine, we noted that either the V317G or the long α/β317–323 mt-hOggl mutants, similar to the wild-type mt-hOggl overexpression, completely blocked oxidant (H2O2 [100 µM for 24 h] or asbestos [25 µg/cm2 for 24h])-induced caspase-9 activation (Fig 5A), DNA fragmentation (Fig 5B), and fully preserved mitochondrial aconitase activity (Fig 5C). Taking advantage of the myc tag in our overexpressed mitochondrial wild-type and longα/β317–323-Oggl mutant constructs, we find that mitochondrial aconitase co-precipitates with both the wild-type and mutant hOggl (Fig 6). However, COX IV, a mitochondrial protein, does not co-precipitate with either construct but was present in all the mitochondrial protein extracts as expected prior to immunoprecipitation. These data demonstrate that mt-hOggl protects lung epithelial cells exposed to oxidative stress by a mechanism that does not require 8-oxoG DNA glycosylase activity but does involve its interaction with mitochondrial aconitase.
Figure 5. Mitochondrial hOggl prevents oxidant-induced toxicity independent of its 8-oxoG DNA repair activity.
Transient overexpression of empty vector control, wild-type mt-hOgg-1 as well as two previously described hOgg-1 mutant plasmid constructs lacking 8-oxoguanine DNA repair capacity (317–323 long alpha/beta and V317) were done by lipofectamine as described in the Methods (18). Mitochondrial localization was confirmed by C-Myc immunostaining. Similar to wild-type mt-hOgg-1, overexpression of mt-hOgg-1 mutants 317–323 long alpha/beta or V317 completely blocked oxidant induced caspase-9 activation (A), DNA fragmentation (B), and the decrease in mitochondrial aconitase activity (C) caused by H2O2 (100 µM) or asbestos (25 µg/cm2) for 24h. * p < .05 v. no H2O2 or asbestos; † p < .05 v. empty vector cells exposed to the same dose of 2O2 or asbestos.
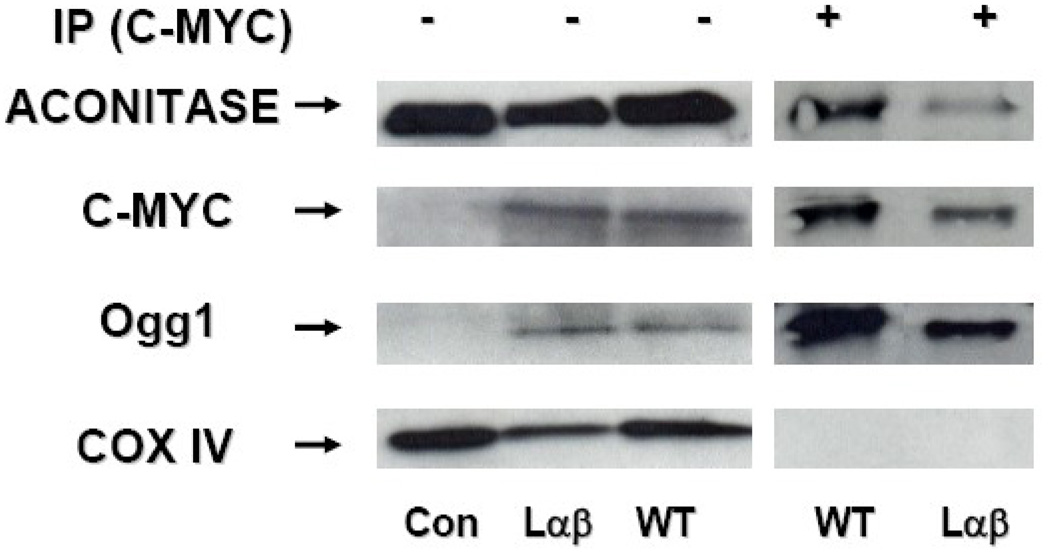
Figure 6. Mitochondrial wild-type and long alpha/beta 317–323 hOggl mutant co-precipitate with mitochondrial aconitase.
Mitochondrial proteins obtained from control (Con) and mt-hOggl (wild-type [WT] and long α/β 317–323 [Laβ]) overexpressing A549 cells were subjected to Western blot analysis before and after immunoprecipitation (IP) using antimyc performed as described in the Experimental Procedures. As expected, mitochondrial proteins probed before IP demonstrated hOgg 1 and c-myc only in the hOgg 1 overexpressing cells (either wild-type or Lαβ mutant hOggl) despite similar levels of mitochondrial aconitase and COX IV as control cells. After IP with c-myc, mitochondrial aconitase and hOggl, but not COX IV, co-precipitated with the myc constructs containing either wild-type or Lαβ mutant hOggl.
Mitochondria-targeted aconitase overexpression prevents oxidant-induced DNA fragmentation and caspase-9 activation
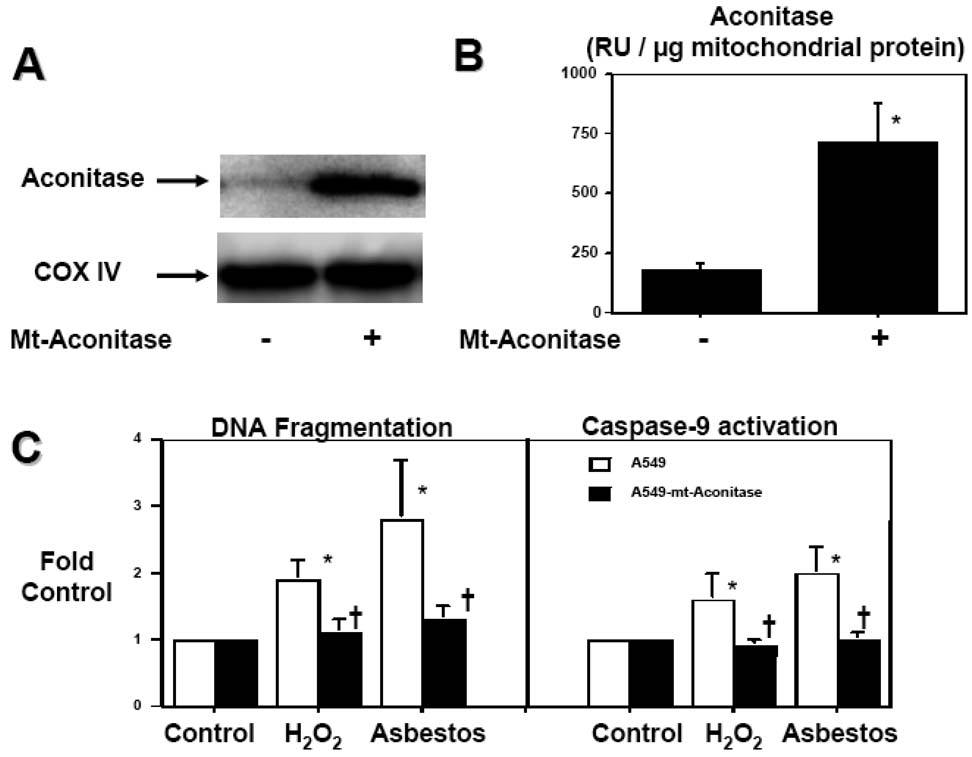
Because constitutively expressed mitochondrial aconitase promotes mtDNA stability in yeast (20), we tested whether mitochondrial aconitase overexpression could prevent oxidant-induced mitochondrial dysfunction and apoptosis in human A549 cells. We transiently overexpressed mitochondrial aconitase in A549 cells using an expression plasmid that encodes the cis-aconitase protein that functions in the mitochondria catalyzing citrate to isocitrate in the TCA cycle and is one of the mitochondrial matrix proteins that are preferentially degraded by Lon protease after oxidative modification (19,21,31). Compared to controls, overexpression of mitochondrial aconitase increased mt-aconitase protein expression (Fig 7A) and activity 4-fold (175±30 v. 714±167 RU / µg mitochondrial protein, respectively; n=3; Fig 7B). Notably, mt-aconitase overexpression blocked H2O2(100 µM)- and asbestos (25 µg/cm2)-induced DNA fragmentation and caspase-9 activation (Fig 7C).
Figure 7. Mitochondria-targeted aconitase overexpression prevents oxidant-induced A549 cell DNA fragmentation and caspase-9 activation.
(A) Mitochondria-targeted aconitase augments aconitase protein levels as assessed by Western blotting and (B) mitochondrial aconitase activity (n=3). (C) Mitochondria-targeted aconitase overexpression blocks oxidative stress-induced DNA fragmentation and caspase-9 activation caused by H2O2 (100 µM) or asbestos (25 µg/cm2) for 24h. * p < .05 v. no H2O2 or asbestos; † p < .05 v. empty vector cells exposed to the same dose of H2O2 or asbestos.
Our finding that mitochondrial aconitase overexpression blocks oxidant-induced mitochondrial dysfunction and apoptosis (Fig 7) and that mitochondrial aconitase co-precipitates with wild-type and mutant Oggl (Fig 4 and 6) suggests that Oggl functions as a molecular chaperone of aconitase protecting it from oxidative degradation by Lon protease (31). Support for this possibility comes from our finding that MG132 (250 µM), a protease inhibitor that blocks mitochondrial Lon protease activity (34), abolishes asbestos (25 µg/cm2)-induced reductions in mitochondrial aconitase activity after 24h (fold-control: Asbestos 0.6 ±0.1 (3) * and MG132 + Asbestos 1.4 ± 0.1 (3) †; * p < 0.05 v Control; † p < 0.05 v. Asbestos).
To address whether Oggl is necessary to prevent oxidant-induced changes in aconitase and apoptosis, hOggl mRNA levels were silenced using shRNA. Compared to scrambled control shRNA, shRNA against hOgglreduced hOggl mRNA expression by ~90% as assessed using real time PCR (Fig 8A). Notably, silencing of hOggl reduced mitochondrial aconitase levels by ~30% (Fig 8B) and enhanced oxidant-induced apoptosis (Fig 8C).
Figure 8. shRNA-mediated down-regulation of hOGGl mRNA reduces mitochondrial aconitase and augments oxidant-induced apoptosis.
Cells were transfected with mock, control shRNA or hOGGl shRNA for 24 h as delineated in the Experimental Procedures. As compared to mock shRNA controls, shRNA against hOGGl reduces OGG1 mRNA levels by ~90% (n=6) (A) and decreases mitochondrial aconitase levels by ~30% (n=3) (B). As compared to Mock shRNA-treated controls (Con), DNA fragmentation caused by asbestos (5–25 µg/cm2) or H2O2 (100 µM) for 24h was augmented by shRNA against hOGGl (n=6); data expressed as fold control (mock shRNA-treated) (C) * p < .05 v. Control; † p < .05 v. Mock shRNA.
Discussion
Mitochondrial oxidative DNA damage is implicated in the pathogenesis of malignancy, aging and degenerative disorders. The major findings in this study are that A549 cells overexpressing mt-hOggl block oxidant-induced ΔΨm, caspase-9 activation, and DNA fragmentation by preserving mitochondrial aconitase activity and protein levels. Further, we show that mt-hOggl co-precipitates with mitochondrial aconitase and that the protection conferred by mt-hOggl is independent of its 8-oxoG DNA glycosylase activity. Finally, we demonstrate that overexpression of mitochondrial aconitase abolishes oxidant-induced caspase-9 activation and DNA fragmentation.
Our findings with A549 lung cancer cells showing that mt-hOggl overexpression prevents oxidant-induced mitochondrial dysfunction and apoptosis are consistent with reports from several groups of investigators working in different model systems (14–17,35). In contrast, some investigators have shown enhanced cytotoxicity by chemotherapeutic agents after overexpression of mitochondrial hOggl type 2a in hepatocytes or N-methylpurine DNA glycosylase in human breast cancer cells (36,37). The explanation for these results is likely due to various factors including the cell type, specific DNA glycosylase overexpressed, the extent of oxidative mt-DNA damage, and the availability of down-stream base excision DNA repair enzymes to restore the apurinic/apyrimidinic (AP) sites generated by glycosylases (e.g. AP endonuclease, DNA polymerase, and DNA ligase).
An important observation in this study is that mt-hOggl overexpression blocks the decrease in mitochondrial aconitase activity and protein expression caused by oxidative stress (Fig 3C/D) without altering the levels of oxidative stress as assessed by both a fluorescence assay (Fig 3A) as well as with highly sensitive roGFP sensors targeted to either the mitochondria or cytosol (Fig 3B). Thus, unlike SOD2, a mitochondrial antioxidant that preserves mitochondrial aconitase activity and protein expression (38,39), we could not detect that mt-hOggl overexpression decreased the levels of oxidative stress. Further, that mt-hOggl co-precipitates with mitochondrial aconitase (both wild-type and long α/β 317–323 mutant) and that the overexpression of mt-hOggl prevents oxidant-induced reductions in aconitase whether or not mt-hOggl is capable of 8-oxoG DNA repair suggests a unique mitochondrial function of mt-hOgg 1 in preventing oxidative modifications of aconitase that might lead to its degradation by mitochondrial Lon protease (31). Mitochondrial hOggl may act as a molecular chaperone of aconitase in a manner similar to frataxin, a mitochondrial localized iron chaperone protein that blocks aconitase iron/sulfur disassembly by oxidative stress (21). Targeted depletion of hepatic frataxin in mice promotes tumor formation in livers that occurs in association with increased hepatocyte mitochondrial dysfunction, oxidative stress and apoptosis (40). However, unlike frataxin, we are unaware of any iron chelating properties of mt-hOggl and we did not detect any decrease in mitochondrial oxidative generation when mt-hOggl was overexpressed (Fig 3). Another possibility is that mt-hOggl blocks key oxidative modification sites on mitochondrial aconitase responsible for activating proteolytic degradation by mitochondrial Lon protease (19,31). Our finding that MG132, which is a protease inhibitor that blocks mitochondrial Lon protease activity (34), prevents asbestos-induced reductions in mitochondrial aconitase activity provides some support for this possibility. Although the sites of interaction between mt-hOggl and aconitase are unknown, one candidate involves the R229Q hOGGl mutation, which results in a protein that is unable to prevent oxidative stress-induced cell death when overexpressed in the mitochondria (41). Future studies are required to clarify these issues as well as to determine precisely how mt-hOggl interacts with aconitase to prevent oxidative modification that triggers aconitase degradation by Lon protease and whether other mtDNA repair proteins act similarly.
A novel finding in this study is that mt-hOgg 1 mutants lacking the key amino acids in the C-terminal αO helix necessary for 8-oxoG DNA repair prevent oxidant-induced mitochondrial dysfunction, decreases in mitochondrial aconitase and DNA fragmentation in a manner comparable to wild-type mt-hOggl (Fig 5). Because the mt-Oggl mutants used in this study were altered to the corresponding mitochondrial β0gg-l isoform (18), our data suggest that the βOggl isoform might function primarily to prevent oxidative modification of mitochondrial aconitase. The protective effect of hOggl mutants lacking DNA repair capacity on mitochondrial function and mitochondrial aconitase activity are in accord with the observation that 8-oxoG levels per se do not cause mitochondrial dysfunction (42). Several lines of evidence firmly implicate a novel interactive effect between mitochondrial hOggl and aconitase in preserving mitochondrial function and blocking oxidant-induced intrinsic apoptosis. First, overexpression of either mitochondrial hOggl (wild-type or mutants lacking DNA repair) or aconitase prevents mitochondria-regulated apoptosis caused by either asbestos or H2O2. Second, we showed that endogenous hOggl (Figure 4; lane 1), mitochondria-targeted wild-type hOggl (Figure 4; lanes 3 and 4) and mutant hOggl (Figure 6; Lαβ lane) co-precipitates with mitochondrial aconitase and that mitochondria-targeted hOggl overexpression prevents aconitase degradation caused by oxidative stress. Finally, hOggl underexpression reduces mitochondrial aconitase protein expression and augments apoptosis. Although the precise molecular mechanism(s) that account for mitochondrial protection are unclear, future studies are currently directed at determining whether aconitase stabilizes mtDNA in the setting of oxidative stress. A study in yeast showed that mitochondrial aconitase overexpression can rescue severe mtDNA instability in strains lacking Abf2p, a crucial mtDNA-binding protein, and that this occurs independent of aconitase catalytic activity (20). These investigators suggested that mitochondrial aconitase, a TCA cycle enzyme important for ATP production, might link cellular metabolism to mtDNA maintenance. As compared to untreated controls, mt-hOggl overexpressing cells have increased mitochondrial aconitase protein levels (Fig 3D) and augmented aconitase activity in the setting of oxidative stress (asbestos and H2O2). We detected negligible changes between untreated controls and mt-hOggl overexpressing cells in terms of mitochondrial membrane potential change (Fig 2), caspase-9 activation (Fig 2 and 5), aconitase activity (Fig 3C) and DNA fragmentation (Fig 2 and 5) that were statistically significant. Although the explanation accounting for these observations are not established, we speculate that this is in part due to the interactive effects reported herein that these two molecules co-precipitate and prevent oxidant-induced apoptosis by preserving mitochondrial function. Given that frataxin stabilizes mitochondrial aconitase (21) and has a role in regulating hepatocyte apoptosis (40), it will also be of interest understanding whether frataxin modulates oxidant-induced alveolar epithelial cell apoptosis, but such experiments are beyond the scope of the current paper.
We observed that silencing of hOgg 1 mRNA reduced mitochondrial aconitase levels and augmented oxidant-induced DNA fragmentation (Fig 8). Several recent studies have shown that hOggl underexpression affects oxidant-induced apoptosis. Youn and coworkers (43) showed that hOggl silencing using siRNA techniques modestly augments H2O2-induced apoptosis in human fibroblasts and lung carcinoma H1299 cells in a manner comparable to our findings with A549 cells. Similarly, hOGGl-deficient A549 cells established using a ribozyme gene targeting technique demonstrated enhanced DNA damage and reduced viability after exposure to cooking oil fumes (44). In contrast, oxidant-exposed murine embryonic fibroblasts deficient in both OGG1 and MYH (another mammalian enzyme involved in oxidative DNA repair) had no detectable apoptosis but did have increased G2/M cell cycle arrest (45). Our data support a role for Oggl in preventing oxidant-induced apoptosis in lung epithelial cells and extend the work of others by showing that Oggl acts in part by preserving mitochondrial aconitase.
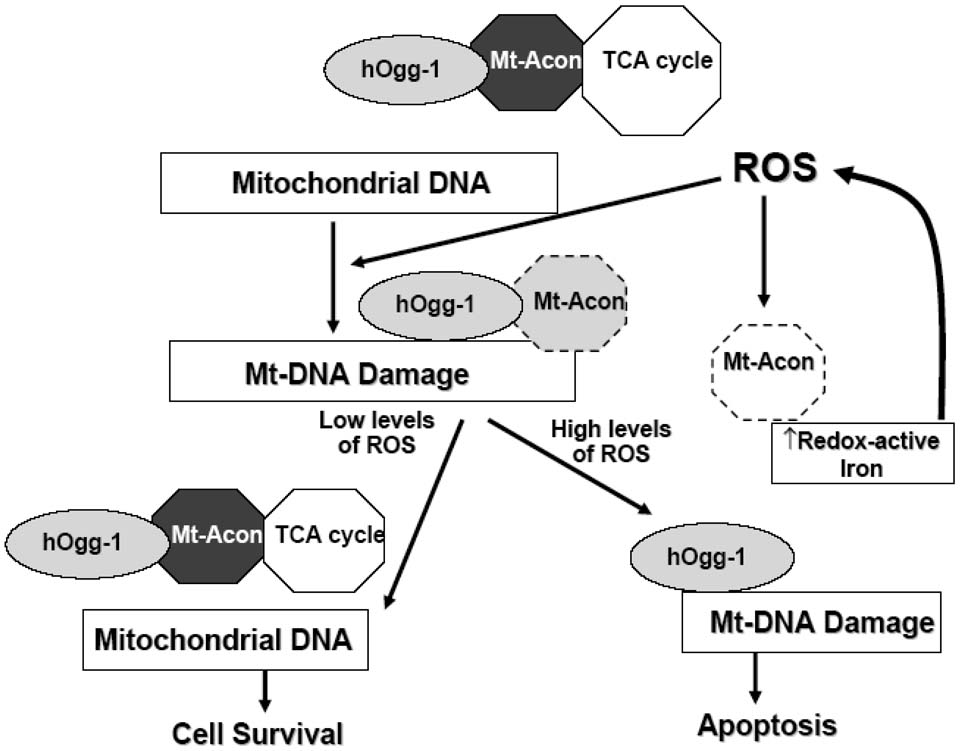
In conclusion, our data establish an innovative role for mt-hOggl protecting mitochondrial aconitase against oxidant-induced mitochondrial dysfunction and apoptosis. The protective effects of mt-hOggl occur independently of any antioxidant function and mitochondrial 8-oxoG DNA glycosylase activity. We demonstrate that mt-hOggl co-precipitates with mitochondrial aconitase and that over-expression of mitochondrial aconitase abolishes oxidant-induced lung epithelial cell apoptosis. A hypothetical model illustrating how these events might be coordinated is shown in Fig 9. Under physiologic conditions, mt-hOggl is associated with mitochondrial aconitase in the TCA cycle facilitating normal cellular metabolism by preventing oxidative modification of aconitase. Oxidative stress (e.g. H2O2 or asbestos fibers) induces mtDNA damage, such as 8-oxoG DNA adducts, which is a potent stimulus for the αhOggl isoform to bind mtDNA (46). Although the βOggl isoform does not bind mtDNA or repair 8-oxoG DNA adducts after oxidative stress (18), it might function to shield mitochondrial aconitase from oxidative inactivation, perhaps by working in conjunction with frataxin (21). In the setting of low levels of oxidative stress, the normal physiologic state is restored. However, in the setting of high levels of oxidative stress that overwhelm the antioxidant defenses and DNA repair capacity, mitochondrial aconitase is irreversibly altered leading to its breakdown, the release of redox-active iron from aconitase and apoptosis. This model concurs with a recent study showing that long-term iron overload results in mtDNA damage and subsequent reduction in the synthesis of respiratory chain subunits encoded by the mtDNA (47). The coupling of mt-hOggl to mitochondrial aconitase demonstrated herein represents a novel mechanism linking metabolism to mtDNA that may be important in the pathophysiologic events leading to oxidant-induced toxicity as seen in various tumors, degenerative disorders, cardiac diseases (e.g. hemochromatosis), respiratory disorders (e.g. asbestosis, pulmonary fibrosis) and aging.
Figure 9. Hypothetical model in which mt-hOggl and aconitase interact in the setting of oxidative stress to promote cell life or death.
Acknowledgements
The authors appreciate the kind gift of adenoviral vectors containing hOggl and empty vector controls provided by Dr. Glenn Wilson (University of South Alabama) and mitochondrial aconitase polyclonal antibody provided by Dr Luke Szweda (Case Western Reserve University). The authors acknowledge the technical efforts provided by Neil Bruce. This work was supported by a Merit Review grant from the Department of Veterans Affairs (D.W.K.), the National Institutes of Health Grants HL67835-01 (G.R.S.B.), and HL35440 (P.T.S).
Abbreviations
- Oggl
8-oxoguanine DNA glycosylase
- 8-oxoG
8-oxo-7,8-dihydroxyguanine
- mt-hOggl
mitochondria-targeted human α-hOggl
- ROS
reactive oxygen species
- mtDNA
mitochondrial DNA
- SOD2
manganese superoxide dismutase
- AT2
alveolar epithelial type II
- ΔΨm
mitochondrial membrane potential
- ro-GFP
redox-sensitive green fluorescent protein mutant
- TCA
tricarboxycylic acid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bohr VA, Stevnsner T, de Souza-Pinto NC. Mitochondrial DNA repair of oxidative damage in mammalian cells. Gene. 2002;286:127–134. doi: 10.1016/s0378-1119(01)00813-7. [DOI] [PubMed] [Google Scholar]
- 2.Van Houten B, Woshner V, Santos JH. Role of mitochondrial DNA in toxic responses to oxidative stress. DNA Repair. 2006;5:145–152. doi: 10.1016/j.dnarep.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 3.Kroemer G, Galluzzi L, Brenner C. Mitochondrial membrane permeabilization in cell death. Physiol. Rev. 2007;87:99–163. doi: 10.1152/physrev.00013.2006. [DOI] [PubMed] [Google Scholar]
- 4.Yakes FM, Van Houten B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc. Natl. Acad. Sci. U.S.A. 1997;94:514–519. doi: 10.1073/pnas.94.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richter C, Park JW, Ames BN. Normal oxidative damage to mitochondrial and nuclear DNA is extensive. Proc. Natl. Acad. Sci. U.S.A. 1988;85:6465–6467. doi: 10.1073/pnas.85.17.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dizdaroglu M, Jaruga P, Birincioglu M, Rodriguez H. Free radical-induced damage to DNA: mechanisms and measurements. Free Radic. Biol. Med. 2002;32:1102–1115. doi: 10.1016/s0891-5849(02)00826-2. [DOI] [PubMed] [Google Scholar]
- 7.Aburatani H, Hippo Y, Ishida T, Takashima R, Matsuba C, Kodama T, Takao M, Yasui A, Yamamoto K, Asano M. Cloning and characterization of mammalian 8-hydroxyguanine-specific DNA glycosylase/apurinic, apyrimidinic lyase, a functional mutM homologue. Cancer Res. 1997;57:2151–2156. [PubMed] [Google Scholar]
- 8.Radicella JP, Dherin C, Desmaze C, Fox MS, Boiteux S. Cloning and characterization of hOGGl, a human homologue of the OGGl gene of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 1997;94:8010–8015. doi: 10.1073/pnas.94.15.8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roldan-Arjona T, Wei YF, Carter KC, Klungland A, Anselmino C, Wang RP, Augustus M, Lindahl T. Molecular cloning and functional expression of a human cDNA encoding the antimutator enzyme 8-hydroxyguanine-DNA glycosylase Proc. Natl. Acad. Sci. U.S.A. 1997;94:8016–8020. doi: 10.1073/pnas.94.15.8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chevillard S, Radicella JP, Levalois C, Lebeau J, Poupon MF, Oudard S, Dutrillaux B, Boiteux S. Mutations in OGG1, a gene involved in the repair of oxidative DNA damage, are found in human lung and kidney tumors. Oncogene. 1998;16:3083–3086. doi: 10.1038/sj.onc.1202096. [DOI] [PubMed] [Google Scholar]
- 11.Fortini P, Pascucci B, Parlanti E, D'Errico M, Simonelli V, Dogliotti E. 8-Oxoguanine DNA damage: at the crossroad of alternative repair pathways. Mutat. Res. 2003;531:127–139. doi: 10.1016/j.mrfmmm.2003.07.004. [DOI] [PubMed] [Google Scholar]
- 12.Russo MT, De Luca G, Degan P, Parlanti E, Dogliotti E, Barnes DE, Lindahl T, Yang H, Miller JH, Bignami M. Accumulation of the oxidative base lesion 8-hydroxyguanine in DNA of tumor-prone mice defective in both the Myh and Oggl DNA glycosylases. Cancer Res. 2004;64:4411–4414. doi: 10.1158/0008-5472.CAN-04-0355. [DOI] [PubMed] [Google Scholar]
- 13.Mambo E, Chatterjee A, de Souza-Pinto NC, Mayard S, Hogue BA, Hoque MO, Dizdaroglu M, Bohr VA, Sidransky D. Oxidized guanine lesions and hOggl activity in lung cancer. Oncogene. 2005;24:4496–4508. doi: 10.1038/sj.onc.1208669. [DOI] [PubMed] [Google Scholar]
- 14.Dobson AW, Xu Y, Kelley MR, LeDoux SP, Wilson GL. Enhanced mitochondrial DNA repair and cellular survival after oxidative stress by targeting the human 8-oxoguanine glycosylase repair enzyme to mitochondria. J. Biol. Chem. 2000;275:37518–37523. doi: 10.1074/jbc.M000831200. [DOI] [PubMed] [Google Scholar]
- 15.Shukla A, Jung M, Stern M, Fukagawa NK, Taatjes DJ, Sawyer D, Van Houten B, Mossman BT. Asbestos induces mitochondrial DNA dmage and dysfunction linked to the development of apoptotic cell death. Am. J. Physiol. Lung Cell. Mol. Physiol. 2003;285:L1018–L1025. doi: 10.1152/ajplung.00038.2003. [DOI] [PubMed] [Google Scholar]
- 16.Ruchko M, Gorodnya O, LeDoux SP, Alexeyev MF, Al-Mehdi AB, Gillespie MN. Mitochondrial DNA damage triggers mitochondrial dysfunction and apoptosis in oxidant-challenged lung endothelial cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 2005;288:L530–L535. doi: 10.1152/ajplung.00255.2004. [DOI] [PubMed] [Google Scholar]
- 17.Rachek LI, Grishko VI, LeDoux SP, Wilson GL. Role of nitric oxide-induced mtDNA damage in mitochondrial dysfunction and apoptosis. Free Radic. Biol. Med. 2006;40:754–762. doi: 10.1016/j.freeradbiomed.2005.09.028. [DOI] [PubMed] [Google Scholar]
- 18.Hashiguchi K, Stuart JA, de Souza-Pinto NC, Bohr VA. The C-terminal αO helix of human Oggl is essential for 8-oxoguanine DNA glycosylase activity: the mitochondrial β-Oggl lacks this domain and does not have glycosylase activity. Nucleic Acids Res. 2004;32:5596–5608. doi: 10.1093/nar/gkh863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bulteau AL, Ikeda-Saito M, Szweda LI. Redox-dependent modulation of aconitase activity in intact mitochondria. Biochemistry. 2003;42:14846–14855. doi: 10.1021/bi0353979. [DOI] [PubMed] [Google Scholar]
- 20.Chen XJ, Wang X, Kaufman BA, Butow RA. Aconitase couples metabolic regulation to mitochondrial DNA maintenance. Science. 2005;307:714–717. doi: 10.1126/science.1106391. [DOI] [PubMed] [Google Scholar]
- 21.Bulteau AL, O'Neill HA, Kennedy MC, Ikeda-Saito M, Isaya G, Szweda LI. Frataxin acts as an iron chaperone protein to modulate mitochondrial aconitase activity. Science. 2004;305:242–245. doi: 10.1126/science.1098991. [DOI] [PubMed] [Google Scholar]
- 22.Timbrell V. In: Characteristics of International Union Against Cancer standard reference samples of asbestos. Shapiro HA, editor. Cape Town, South Africa: Oxford University Press; 1970. pp. 28–36. [Google Scholar]
- 23.Aljandali A, Pollack H, Yeldandi A, Li Y, Weitzman SA, Kamp DW. Asbestos-induced alveolar epithelial cell apoptosis: Role of iron-induced free radicals. J. Lab. Clin. Med. 2001;137:330–339. doi: 10.1067/mlc.2001.114826. [DOI] [PubMed] [Google Scholar]
- 24.Panduri V, Weitzman SA, Chandel N, Kamp DW. The mitochondria-regulated death pathway mediates asbestos-induced alveolar epithelial cell apoptosis. Am. J. Respir. Cell Mol. Biol. 2003;28:241–248. doi: 10.1165/rcmb.4903. [DOI] [PubMed] [Google Scholar]
- 25.Panduri V, Weitzman SA, Chandel NS, Kamp DW. Mitochondrial-derived free radicals mediate asbestos-induced alveolar epithelial cell apoptosis. Am. J. Physiol. Lung Cell. Mol. Physiol. 2004;286:L1220–L1227. doi: 10.1152/ajplung.00371.2003. [DOI] [PubMed] [Google Scholar]
- 26.Panduri V, Surapureddi S, Soberanes S, Weitzman SA, Chandel N, Kamp DW. p53 mediates amosite asbestos induced alveolar epithelial cell mitochondria-regulated apoptosis. Am. J. Respir. Cell Mol. Biol. 2006;34:443–452. doi: 10.1165/rcmb.2005-0352OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rachek LI, Grishko VI, Musiyenko SI, Kelley MR, LeDoux SP, Wilson GL. Conditional targeting of the DNA repair enzyme hOggl into mitochondria. J. Biol. Chem. 2002;277:44932–44937. doi: 10.1074/jbc.M208770200. [DOI] [PubMed] [Google Scholar]
- 28.Hanson GT, Aggeler R, Oglesbee D, Cannon M, Capaldi RA, Tsien RY, Remington SJ. Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. J. Biol. Chem. 2004;279:13044–13053. doi: 10.1074/jbc.M312846200. [DOI] [PubMed] [Google Scholar]
- 29.Dooley CT, Dore TM, Hanson GT, Jackson WC, Remington SJ, Tsien RY. Imaging dynamic redox changes in mammalian cells with green fluorescent protein indicators J. Biol. Chem. 2004;279:22284–22293. doi: 10.1074/jbc.M312847200. [DOI] [PubMed] [Google Scholar]
- 30.Kim H-N, Morimoto Y, Tsuda T, Ootsuyama Y, Hirohashi M, Hirano T, Tanaka I, Lim Y, Yun I-G, Kasai H. Changes in DNA 8-hydroxyguanine levels, 8-hydroxyguanine repair activity, and hOGGl and hMTHl mRNA expression in human lung alveolar epithelial cells induced by crocidolite asbestos. Carcinogenesis. 2001;22:265–269. doi: 10.1093/carcin/22.2.265. [DOI] [PubMed] [Google Scholar]
- 31.Bota DA, Davies KJA. Lon protease preferentially degrades oxidized mitochondrial aconitase by an ATP-stimulated mechanism. Nature Cell Biol. 2002;4:674–680. doi: 10.1038/ncb836. [DOI] [PubMed] [Google Scholar]
- 32.Marsin S, Vidal AE, Sossou M, Menissier-de Murcia J, Le Page F, Boiteux S, de Murcia G, Radicella JP. Role of XRCC1 in the coordination and stimulation of oxidative DNA damage repair initiated the DNA glycosylase hOGGl. J. Biol. Chem. 2003;278:44068–44074. doi: 10.1074/jbc.M306160200. [DOI] [PubMed] [Google Scholar]
- 33.Hu J, Imam SZ, Hashiguchi K, de Souza-Pinto NC, Bohr VA. Phosphorylation of human oxoguanine DNA glycosylase (a-OGGl) modulates its function. Nucleic Acids Res. 2005;33:3271–3282. doi: 10.1093/nar/gki636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Granot Z, Kobiler O, Melamed-Book N, Eimeri S, Bahat A, Lu B, Braun S, Maurizi MR, Suzuki CK, Oppenheim AB, Orly J. Turnover of mitochondrial steroidogenic acute regulatory (StAR) protein by lon protease: The unexpected effect of proteosome inhibitors. Mol Endocrinol. 2007;21:2164–2177. doi: 10.1210/me.2005-0458. [DOI] [PubMed] [Google Scholar]
- 35.Dobson AW, Grishko V, LeDoux S, Kelly MR, Wilson GL, Gillespie MN. Enhanced mtDNA repair capacity protects pulmonary artery endothelial cells from oxidant-mediated death. Am. J. Physiol. Lung Cell Mol. Physiol. 2002;283:L205–L210. doi: 10.1152/ajplung.00443.2001. [DOI] [PubMed] [Google Scholar]
- 36.Fishel ML, Seo YR, Smith ML, Kelly MR. Enhancing the DNA base excision repair pathway in the mitochondria; targeting and overexpressing iV-methylpurine DNA glycosylase in mitochondria leads to enhanced cell killing. Cancer Res. 2003;63:608–615. [PubMed] [Google Scholar]
- 37.Zhang H, Mizumachi T, Carcel-Trullols J, Li L, Naito A, Spencer HJ, Spring PM, Smoller BR, Watson AJ, Margison GP, Higuchi M, Fan C-Y. Targeting human 8-oxoguanine DNA glycosylase (hOGGl) to mitochondria enhances cisplatin cytotoxicity in hepatoma cells. Carcinogenesis. 2007;28:1629–1637. doi: 10.1093/carcin/bgm072. [DOI] [PubMed] [Google Scholar]
- 38.Gardner PR, Raineri I, Epstein LB, White CW. Superoxide radical and iron modulate aconitase activity in mammalian cells. J. Biol. Chem. 1995;270(13):339–405. doi: 10.1074/jbc.270.22.13399. [DOI] [PubMed] [Google Scholar]
- 39.Powell CS, Jackson RM. Mitochondrial complex I, aconitase, and succinate dehydrogenase during hypoxia-reoxygenation: modulation of enzyme activities by MnSOD. Am. J. Physiol. Lung Cell. Mol. Physiol. 2003;285:L189–L198. doi: 10.1152/ajplung.00253.2002. [DOI] [PubMed] [Google Scholar]
- 40.Thierbach R, Schultz TJ, Isken F, Voigt A, Mietzner B, Drewes G, et al. Targeted disruption of hepatic frataxin expression causes impaired mitochondrial function, decreased life span and tumor growth in mice. Human Mol. Genetics. 2005;14:3857–3864. doi: 10.1093/hmg/ddi410. [DOI] [PubMed] [Google Scholar]
- 41.Chatterjee A, Mambo E, Zhang Y, Deweese T, Sidransky D. Targeting of mutant hoggl in mammalian mitochondria and nucleus: effect on cellular survival upon oxidative stress. BMC Cancer. 2006;6:235–246. doi: 10.1186/1471-2407-6-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Souza-Pinto NC, Eide L, Hogue BA, Thybo T, Stevnsner T, Seeberg E, Klungland A, Bohr VA. Repair of 8-oxodeoxyguanosine lesions in mitochondrial DNA depends on the oxoguanine DNA glycosylase (OGG1) gene and 8-oxoguanine accumulates in mitochondrial DNA of OGG1-defective mice. Cancer Res. 2001;61:5378–5381. [PubMed] [Google Scholar]
- 43.Youn CK, Song PI, Kim MH, Kim JS, Hyun JW, Choi SJ, et al. Human 8-oxoguanine DNA glycosylase suppresses the oxidative stress-induced apoptosis through a p5 3-mediated signaling pathway in human fibroblasts. Mol Cancer Res. 2007;5:1083–1098. doi: 10.1158/1541-7786.MCR-06-0432. [DOI] [PubMed] [Google Scholar]
- 44.Wu M, Che W, Zhang Z. Enhanced sensitivity to DNA damage induced by cooking oil fumes in human OGG1 deficient cells. Environ MolMutagen. 2008;49:265–275. doi: 10.1002/em.20381. [DOI] [PubMed] [Google Scholar]
- 45.Xie Y, Yang H, Miller JH, Shih DM, Hicks GG, Xie J, Shiu P. Cells deficient in oxidative DNA damage repair genes Myh and Oggl are sensitive to oxidants with increased G2/M arrest and multinucleation. Carcinogenesis. 2008;29:722–728. doi: 10.1093/carcin/bgn033. [DOI] [PubMed] [Google Scholar]
- 46.Bruner SD, Norman DP, Verdine GL. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature. 2000;403:859–866. doi: 10.1038/35002510. [DOI] [PubMed] [Google Scholar]
- 47.Gao X, Campian JL, Qian M, Sun MQ, Eaton JW. Mitochondrial DNA damage in iron overload. J. Biol. Chem. 2009;284:4767–4775. doi: 10.1074/jbc.M806235200. [DOI] [PMC free article] [PubMed] [Google Scholar]