Abstract
Partial digestion of rat liver nuclei with staphylococcal nuclease results in the liberation of nucleo-protein complexes consisting of one or more upsilon bodies. By velocity centrifugation we have isolated the monomeric subunit in relatively pure form. We find that this subunit contains 185 base pairs of DNA and 240,000 daltons of protein, resulting in a protein to DNA ratio identical to that of unperturbed chromatin. The isolated monomeric particle is further susceptible to internal nuclease attack resulting in the solubilization of 46% of the monomeric DNA. Analysis of the resistant DNA reveals a complex but highly reproducible pattern of DNA fragments ranging from 160 to 60 base pairs in length. Analysis of the reassociation kinetics of the isolated subunit DNA reveals that most, if not all genomic sequences, are involved in this basic subunit structure. No special frequency class of DNA is absent from upsilon bodies. Furthermore, virtually all liver sequences transcribed into mRNA are present in upsilon body DNA. These results indicate that upsilon body formation may be random with respect to DNA sequence and suggest that the mere presence of upsilon bodies over a specific region of DNA is not sufficient to restrict its transcription.
Full text
PDF
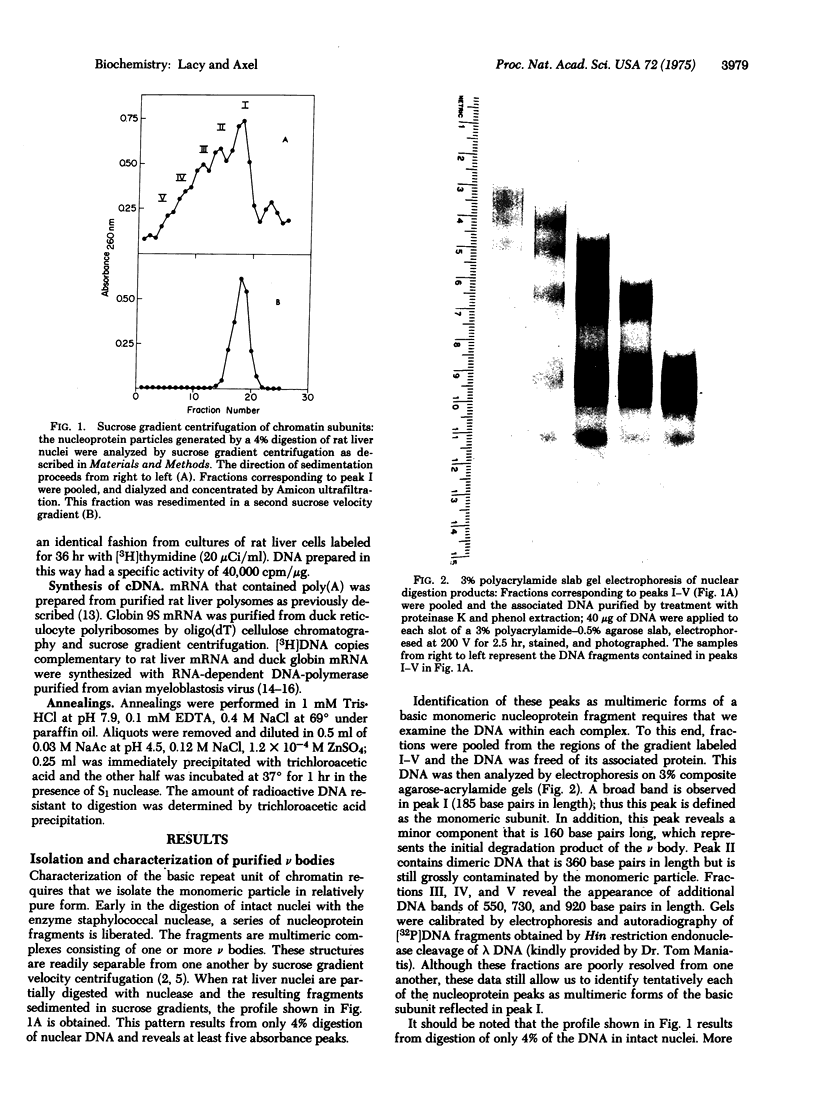
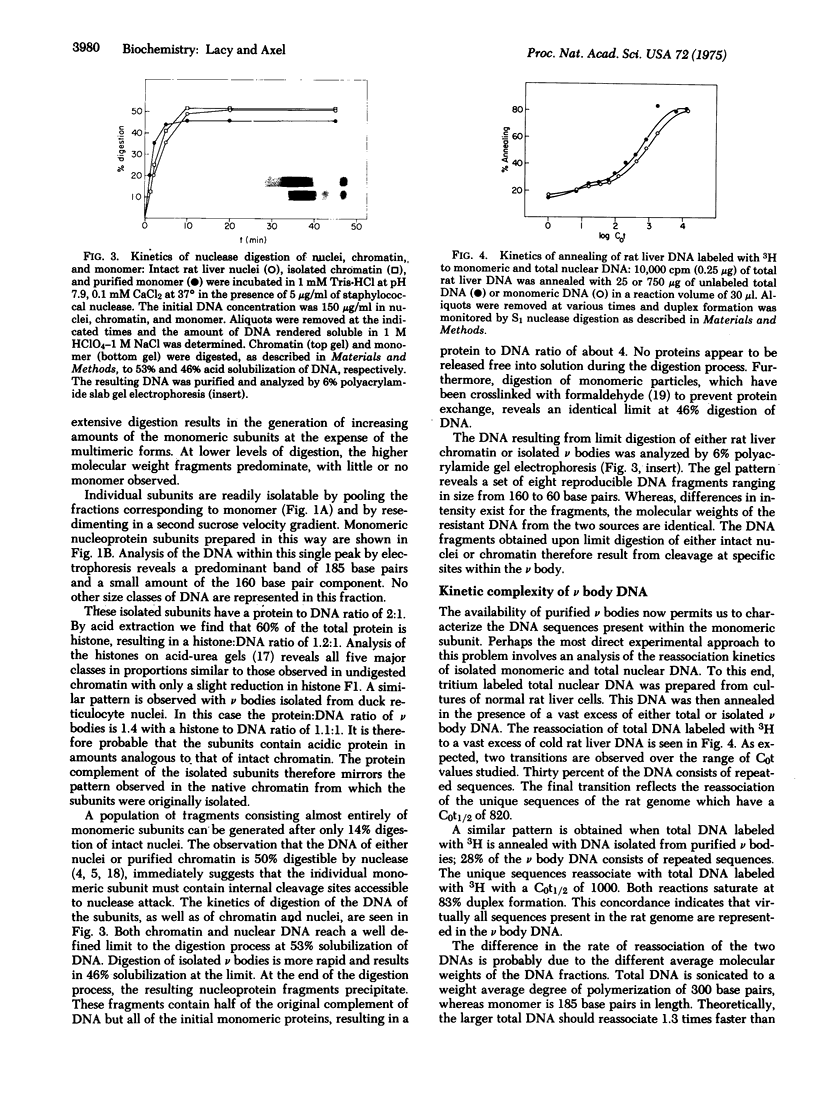
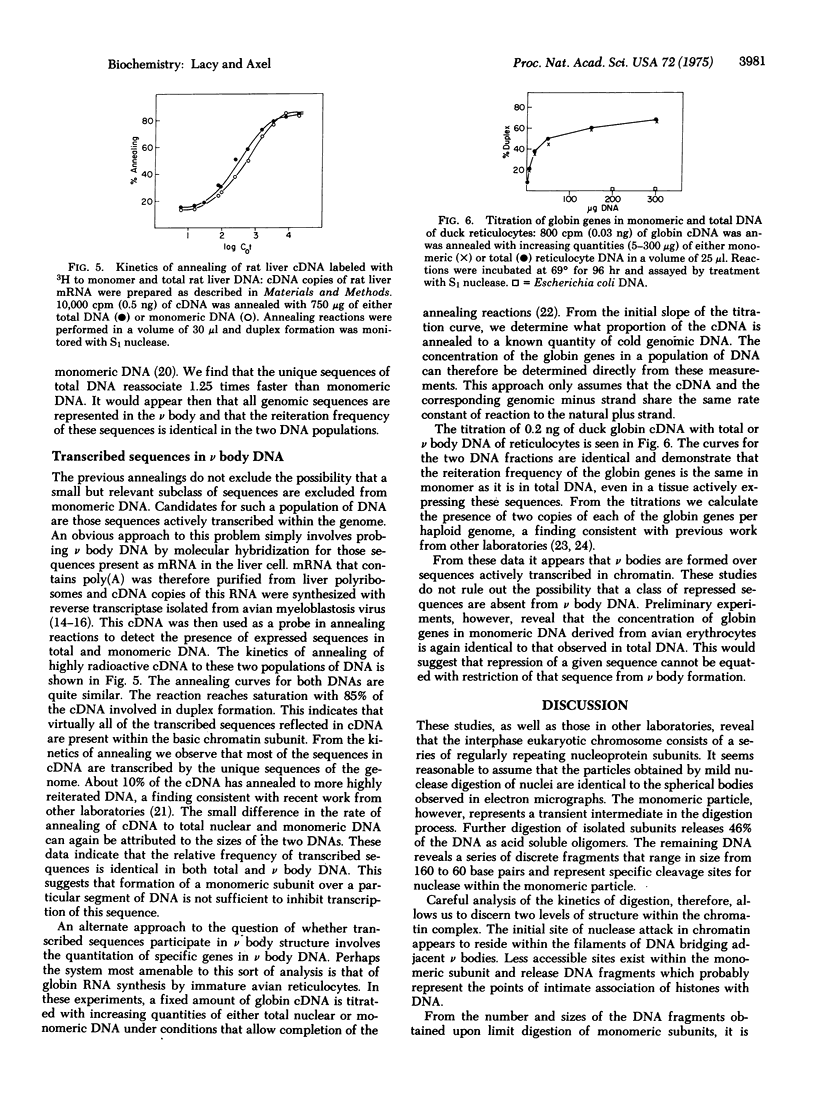

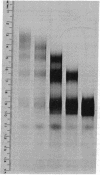
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Axel R., Cedar H., Felsenfeld G. Synthesis of globin ribonucleic acid from duck-reticulocyte chromatin in vitro. Proc Natl Acad Sci U S A. 1973 Jul;70(7):2029–2032. doi: 10.1073/pnas.70.7.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axel R., Cedar H., Felsenfield G. The structure of the globin genes in chromatin. Biochemistry. 1975 Jun 3;14(11):2489–2495. doi: 10.1021/bi00682a031. [DOI] [PubMed] [Google Scholar]
- Axel R. Cleavage of DNA in nuclei and chromatin with staphylococcal nuclease. Biochemistry. 1975 Jul;14(13):2921–2925. doi: 10.1021/bi00684a020. [DOI] [PubMed] [Google Scholar]
- Axel R., Melchior W., Jr, Sollner-Webb B., Felsenfeld G. Specific sites of interaction between histones and DNA in chromatin. Proc Natl Acad Sci U S A. 1974 Oct;71(10):4101–4105. doi: 10.1073/pnas.71.10.4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bank A., Terada M., Metafora S., Dow L., Marks P. A. In vitro synthesis of DNA components of human genes for globins. Nat New Biol. 1972 Feb 9;235(58):167–169. doi: 10.1038/newbio235167a0. [DOI] [PubMed] [Google Scholar]
- Bishop J. O., Freeman K. B. DNA sequences neighboring the duck hemoglobin genes. Cold Spring Harb Symp Quant Biol. 1974;38:707–716. doi: 10.1101/sqb.1974.038.01.076. [DOI] [PubMed] [Google Scholar]
- Bishop J. O., Morton J. G., Rosbash M., Richardson M. Three abundance classes in HeLa cell messenger RNA. Nature. 1974 Jul 19;250(463):199–204. doi: 10.1038/250199a0. [DOI] [PubMed] [Google Scholar]
- Brutlag D., Schlehuber C., Bonner J. Properties of formaldehyde-treated nucleohistone. Biochemistry. 1969 Aug;8(8):3214–3218. doi: 10.1021/bi00836a013. [DOI] [PubMed] [Google Scholar]
- Clark R. J., Felsenfeld G. Structure of chromatin. Nat New Biol. 1971 Jan 27;229(4):101–106. doi: 10.1038/newbio229101a0. [DOI] [PubMed] [Google Scholar]
- Hewish D. R., Burgoyne L. A. Chromatin sub-structure. The digestion of chromatin DNA at regularly spaced sites by a nuclear deoxyribonuclease. Biochem Biophys Res Commun. 1973 May 15;52(2):504–510. doi: 10.1016/0006-291x(73)90740-7. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D., Thomas J. O. Chromatin structure; oligomers of the histones. Science. 1974 May 24;184(4139):865–868. doi: 10.1126/science.184.4139.865. [DOI] [PubMed] [Google Scholar]
- Noll M. Subunit structure of chromatin. Nature. 1974 Sep 20;251(5472):249–251. doi: 10.1038/251249a0. [DOI] [PubMed] [Google Scholar]
- Olins A. L., Carlson R. D., Olins D. E. Visualization of chromatin substructure: upsilon bodies. J Cell Biol. 1975 Mar;64(3):528–537. doi: 10.1083/jcb.64.3.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudet P., Gross-Bellard M., Chambon P. Electron microscopic and biochemical evidence that chromatin structure is a repeating unit. Cell. 1975 Apr;4(4):281–300. doi: 10.1016/0092-8674(75)90149-x. [DOI] [PubMed] [Google Scholar]
- Packman S., Aviv H., Ross J., Leder P. A comparison of globin genes in duck reticulocytes and liver cells. Biochem Biophys Res Commun. 1972 Nov 1;49(3):813–819. doi: 10.1016/0006-291x(72)90483-4. [DOI] [PubMed] [Google Scholar]
- Panyim S., Chalkley R. The heterogeneity of histones. I. A quantitative analysis of calf histones in very long polyacrylamide gels. Biochemistry. 1969 Oct;8(10):3972–3979. doi: 10.1021/bi00838a013. [DOI] [PubMed] [Google Scholar]
- Peacock A. C., Dingman C. W. Resolution of multiple ribonucleic acid species by polyacrylamide gel electrophoresis. Biochemistry. 1967 Jun;6(6):1818–1827. doi: 10.1021/bi00858a033. [DOI] [PubMed] [Google Scholar]
- Ross J., Aviv H., Scolnick E., Leder P. In vitro synthesis of DNA complementary to purified rabbit globin mRNA (RNA-dependent DNA polymerase-reticulocyte-hemoglobin-density gradient centrifugation-oligo(dT) primer). Proc Natl Acad Sci U S A. 1972 Jan;69(1):264–268. doi: 10.1073/pnas.69.1.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahasrabuddhe C. G., Van Holde K. E. The effect of trypsin on nuclease-resistant chromatin fragments. J Biol Chem. 1974 Jan 10;249(1):152–156. [PubMed] [Google Scholar]
- Schutz G., Killewich L., Chen G., Feigelson P. Control of the mRNA for hepatic tryptophan oxygenase during hormonal and substrate induction. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1017–1020. doi: 10.1073/pnas.72.3.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma I. M., Temple G. F., Fan H., Baltimore D. In vitro synthesis of DNA complementary to rabbit reticulocyte 10S RNA. Nat New Biol. 1972 Feb 9;235(58):163–167. doi: 10.1038/newbio235163a0. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Van Lente F. Dissection of chromosome structure with trypsin and nucleases. Proc Natl Acad Sci U S A. 1974 Oct;71(10):4249–4253. doi: 10.1073/pnas.71.10.4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetmur J. G., Davidson N. Kinetics of renaturation of DNA. J Mol Biol. 1968 Feb 14;31(3):349–370. doi: 10.1016/0022-2836(68)90414-2. [DOI] [PubMed] [Google Scholar]
- Young B. D., Harrison P. R., Gilmour R. S., Birnie G. D., Hell A., Humphries S., Paul J. Kinetic studies of gene frequency. II. Complexity of globin complementary DNA and its hybridization characteristics. J Mol Biol. 1974 Apr 25;84(4):555–568. doi: 10.1016/0022-2836(74)90116-8. [DOI] [PubMed] [Google Scholar]