Abstract
Accurate and high throughput cell sorting is a critical enabling technology in molecular and cellular biology, biotechnology, and medicine. While conventional methods can provide high efficiency sorting in short timescales, advances in microfluidics have enabled the realization of miniaturized devices offering similar capabilities that exploit a variety of physical principles. We classify these technologies as either active or passive. Active systems generally use external fields (e.g., acoustic, electric, magnetic, and optical) to impose forces to displace cells for sorting, whereas passive systems use inertial forces, filters, and adhesion mechanisms to purify cell populations. Cell sorting on microchips provides numerous advantages over conventional methods by reducing the size of necessary equipment, eliminating potentially biohazardous aerosols, and simplifying the complex protocols commonly associated with cell sorting. Additionally, microchip devices are well suited for parallelization, enabling complete lab-on-a-chip devices for cellular isolation, analysis, and experimental processing. In this review, we examine the breadth of microfluidic cell sorting technologies, while focusing on those that offer the greatest potential for translation into clinical and industrial practice and that offer multiple, useful functions. We organize these sorting technologies by the type of cell preparation required (i.e., fluorescent label-based sorting, bead-based sorting, and label-free sorting) as well as by the physical principles underlying each sorting mechanism.
Keywords: microfluidic, cell sorting, flow cytometry, lab on a chip, rare cell isolation
Introduction
Isolating and sorting cells from complex, heterogeneous mixtures represents a critical task in many areas of biology, biotechnology, and medicine. Cell sorting is often used to enrich or purify cell samples into well-defined populations to enhance efficiency in research and development applications. Cell sorting also serves as the first step in many diagnostic and therapeutic practices, such as the enrichment of hematopoietic stem cells for autologous patient treatments.1 The need to sort cells is rapidly expanding toward the isolation of rarer target cell populations, including the enrichment of circulating tumor cells (CTCs), hematopoietic stem cells (HSCs), and circulating fetal cells (CFCs) from blood.2–5 Meanwhile, the growing interest in theranostics and personalized medicine, in which treatments are tailored to the prognoses of patients, is further driving the demand for rapid and high performance cell sorting.6
The first commercial cell sorter, which exploits a technique broadly known as fluorescence-activated cell sorting (FACS), was invented in 1969 by Herzenberg et al.7 Soon after its seminal introduction, FACS was optimized into a translatable technology and later propelled through multiple evolutions that have now firmly established it as the benchmark for modern cell sorting devices.8, 9 Now, FACS technologies are automated, robust, and capable of exceptional specificity when using multiple morphological and fluorescent cell signatures (e.g., cell surface labels, cell size, and granularity). These systems are also capable of multiplexed detections, analyses, and sorting speeds up to 50,000 cells per second.10, 11 Similarly, magnetic-activated cell sorting devices, which generally operate by separating cells with magnetic labels from unlabeled cells in a column via a permanent magnet, are widely used due to their rapid, batchwise processing.12
The immense contributions of current commercial cell sorting platforms are tempered by several significant and persistent limitations, however, including: limited sample throughput and processing speeds that would make processing clinical-scale samples (>500 million cells) unfeasible, high operating pressures that could result in a loss of function or viability, bulky instrumentation that occupy large bench footprints, technical expertise necessary for operating complex machinery, and increased risk of sample contamination and safety concerns due to the sorting of aerosolized samples. These limitations, as well as high unit and sample processing costs, must be overcome to enable more efficient clinical application and commercialization. Consequently, the next generation of cell sorting devices must meet higher standards for performance, versatility, and convenience, including: (i) faster sorting rates, (ii) equal or improved accuracies, (iii) ability to process native biological fluids, (iv) ability to process diverse cell types, (v) enhanced capabilities for multiplexed sorting, (vi) simpler operating procedures enabling fully automated systems, (vii) reduced biohazard risk by eliminating aerosols, (viii) reduced cost, and (ix) reduced size for operational convenience and portability.
To address these needs, researchers are actively looking toward microfluidic devices as the platform for the next generation translatable cell sorter. Microchip devices are a proven technology for cellular handling as they can offer precise spatial and temporal control in a greatly miniaturized platform.11, 13 These devices can be easily made using standard microfabrication tools, which lowers cost and simplifies commercialization efforts. In addition, microfluidics can be used to detect, focus, mix, count, lyse, and analyze individual cells on an integrated platform for complete lab-on-a-chip applications.14–19
This review will survey recent developments in microchip cell sorting by organizing each technology into one of three principal categories based on its primary cell recognition modality: (i) fluorescent label-based, (ii) bead-based, and (iii) label-free cell sorting. Within each category, several subsections are provided to further categorize each technology by the physical principles governing the sorting process. We emphasize more recent technologies, especially those that integrate multiple functions on the same device toward a fully integrated point-of-use device.
Fluorescent Label-Based Cell Sorting
Fluorescent label-based cell sorting relies on fluorescent probes or stains to identify cells by type. In traditional FACS, fluorescently-labeled cells organized in a laminar flow stream encounter a focused laser beam that scatters into a detector. The fluorescent signal is then analyzed to assign each cell a type for discrete sorting, whereby in the case of FACS, each cell is encapsulated into an aerosol droplet that is charged and electrostatically sorted.9 To circumvent the need to form of aerosol droplets, many research groups have used fluorescent labels to identify cells in the microfluidic regime for sorting by a variety of mechanisms, as described in detail in this section. Similar to FACS devices, these technologies generally operate by ordering cells in flow streams for: (i) serial interrogation by laser light, (ii) real-time classification, and (iii) rapid, command-driven sorting. Since each cell is processed discretely, fluorescent label-based approaches are often associated with high efficiencies. Further, immunostaining assays are ubiquitous, reliable, and require less preparation time than bead-based labeling, which can help reduce experimental error. These advantages have made fluorescent label-based technologies the mainstays of modern cell sorting technologies and a viable option for many microchip cell sorting devices.
A. Electrokinetic Mechanisms
Electrokinetics describes a family of effects stemming from an applied electric field that results in the migration of particles or cells.20, 21 In addition to charging aerosol droplets for electrostatic sorting (as in FACS), electrokinetic forces can be used to directly displace cells, or cell-containing droplets, in fluids. For the purposes of this review, electrokinetic manipulations are divided into three categories: electrophoresis, dielectrophoresis, and electroosmotic flow. While these mechanisms are phenomenologically distinct, the forces they exert on cells are well-suited for sorting within the length scales of microfluidic devices.
Electrophoresis
Electrophoresis refers to the movement of suspended particles toward an oppositely charged electrode in direct current (DC). Since most cells possess a slight negative charge due to a locus of chemical groups on their surface, they migrate toward the positive electrode during electrophoresis, and the electrophoretic force exerted on that cell is proportional to its charge.20 Takahashi et al. applied electrophoresis to sort cells in a microchip in which an upstream fluorescence detector identified labeled cells for rapid electrostatic sorting downstream.22 Yao et al. developed a similar device based on gravity that operated in an upright orientation to process cells without convective flow.23 A more recent example by Guo et al. showed electrophoretic sorting with much higher throughputs by sorting water-in-oil droplets under continuous flow.24 In this system, prefocused cells were encapsulated into droplets such that droplets containing single cells were sorted from droplets containing no cells or multiple cells.
Dielectrophoresis
In contrast to electrophoresis, where cells move in a uniform electric field due to their surface charge, dielectrophoresis (DEP) refers to the movement of cells in a non-uniform electric field due to their polarizability. For movement in response to a dielectrophoretic force, cells do not need to possess a surface charge because, unlike a DC field, an alternating current (AC) is capable of polarizing the cell (i.e., inducing a dipole moment across the cell).20 Once exposed to an AC field, cells migrate either toward or away from the region of strongest field intensity depending on the electrical permeability of the cell and the fluid. Cells with a higher permeability than the fluid are attracted toward the field maxima, which is known as positive DEP (pDEP).25 The opposite is true for negative DEP (nDEP), which is often preferable to minimize deleterious effects on cell function or viability that might occur due to high field strengths. The magnitude of the dielectrophoretic force is dependent on the size and properties of the cell, fluid, and the parameters of the electric field, which is useful for sorting cells by size and dielectric properties.20 Wang et al. developed a system using lateral nDEP, whereby a set of interdigitated electrodes aligned along both sides of a microfluidic channel provides repulsive forces to organize cells by precise distances from the microfluidic channel walls, to enable enhanced on-chip cytometry and sorting across five channels.26
In contrast to directly sorting cells in a buffered suspension, several groups have developed systems to encapsulate single cells into emulsified droplets for sorting using DEP, thus enabling continuous genomic and proteomic analyses downstream.27–29 Unlike FACS, which can generate potentially biohazardous aerosols, water-in-oil droplets provide a safe and rapid way to analyze individual cells post-sorting. Baret et al. applied DEP in a fluorescence-activated droplet sorter to separate up to 2,000 cells/sec.27 Agresti et al. used emulsions to generate picoliter-volume reaction vessels for detecting new variants of molecular enzymes and dielectrophoretic sorting.28 Mazutis et al. showed that cells compartmentalized into emulsions with beads coated with capture antibodies can be used to analyze the secretion of antibodies from cells for downstream sorting using DEP (Fig. 1).29 These advances hold promise for creating the next generation of cell sorting devices that could also enable clinical detection, analysis, and diagnosis using a single microchip.
Fig. 1.
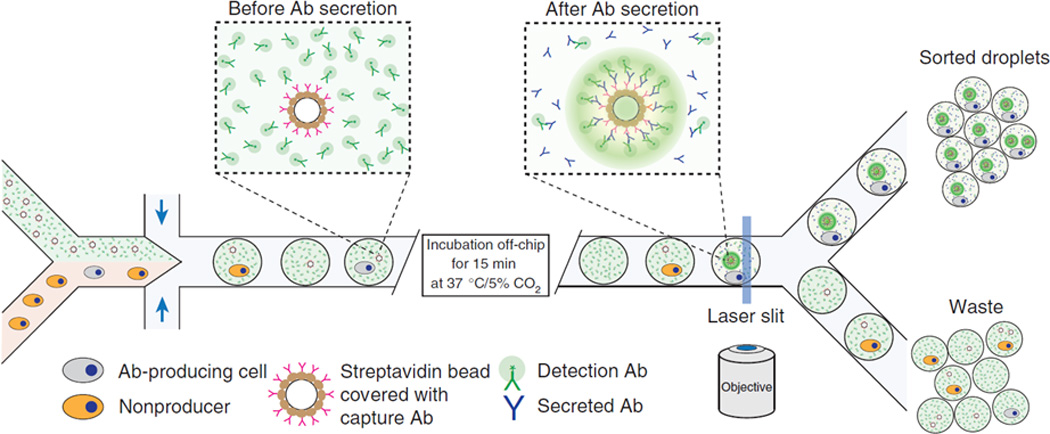
Droplet-based microreactor and cell sorter using DEP. Cells expressing and secreting a target antibody (depicted in grey) and cells not expressing a target antibody (depicted in orange) are encapsulated in droplets with a fluorescent detection antibody, incubated off-chip to permit the production of secretion antibodies, and sorted according to the increased fluorescence signal from the localized packing of detection antibodies on the surface of the bead covered with capture and secretion antibodies. Reprinted with permission from Mazutis et al29 Copyright 2013 Nature Publishing Group.
Electroosmotic Flow
Unlike the electrically driven migration of cells within a stationary fluid (such as in electrophoresis and DEP), electroosmotic flow refers to the movement of a fluid due to the electrically induced migration of solvated ions, thereby transporting cells suspended within the fluid (Fig. 2).30 This principle was applied to sort fluorescent from non-fluorescent cells in microfabricated FACS devices using DC electroosmosis.31–33 While effective, a major disadvantage of DC electric fields for sorting cells is that Faradaic reactions (e.g., electrolysis of water) generally occur at the anode and cathode to maintain a constant electric field, which can generate bubbles and harmful compounds, such as hydrogen peroxide, that may adversely affect cellular viability and solution pH if not carefully monitored and regulated.34 As such, Puttaswamy et al. coupled nDEP with AC electroosmosis to mitigate these effects while retaining the ability to focus, transport, and sort cells.35
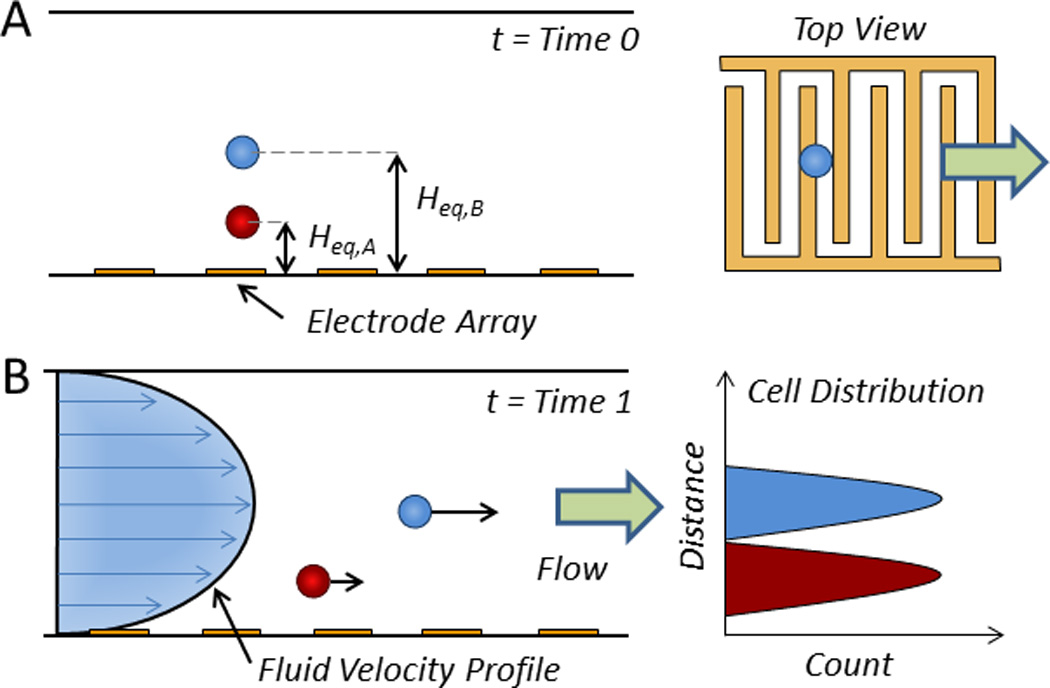
Fig. 2.
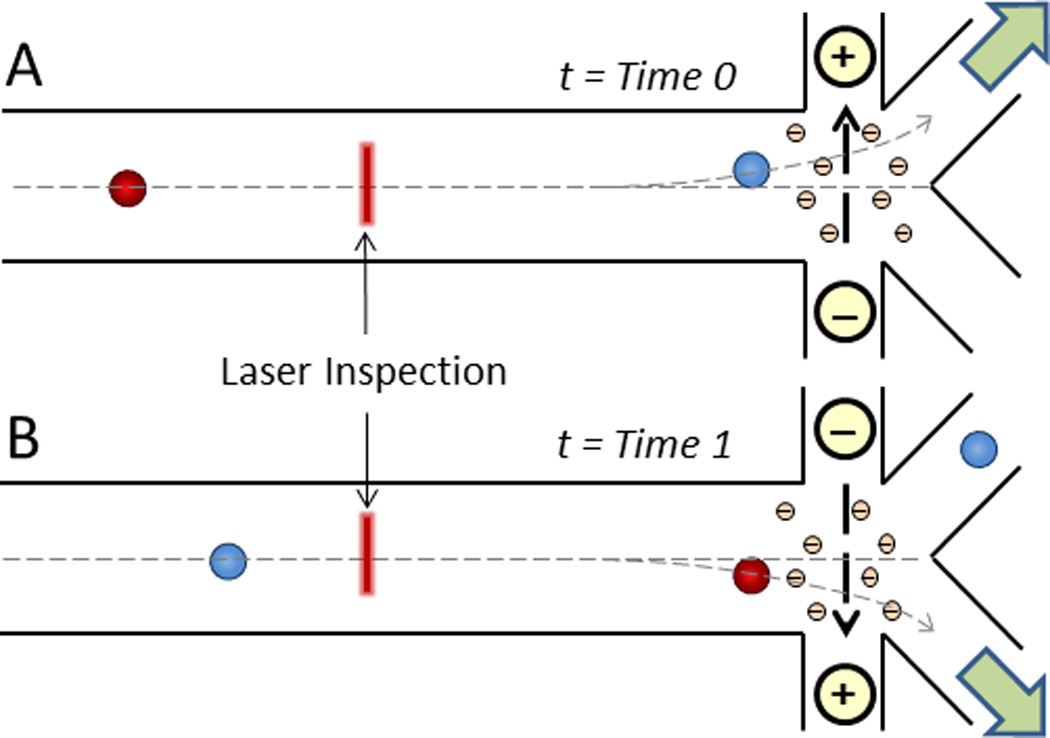
Direct current (DC) electroosmotic cell sorting. Following laser inspection and cell identification, solvated negative ions in the counterionic layer along the positively charged microchannel floor migrate to the oppositely charged electrode, thereby dragging the surrounding liquid for cell transport30 to either A) the first outlet (t = Time 0) or B) the second outlet (t = Time 1).31
B. Acoustophoresis
Acoustophoresis refers to the movement of an object in response to an acoustic pressure wave. Recently, acoustic microfluidic (i.e., acoustofluidic) technologies have provided many new areas of development within analytical flow cytometry, including the sorting of cells.36–38 Acoustic forces are amenable to cell handling as they can provide rapid and precise spatial control in microchips without affecting cellular viability.25, 39–41 In this context, acoustic waves can be divided into three categories: bulk standing waves,42 standing surface acoustic waves (SSAWs),43 and traveling waves.44
Acoustic standing waves form through the disturbance of a medium (e.g., fluid or a surface) by pressure waves of equal magnitude and frequency traveling in opposite directions that result in a single stationary wave containing fixed regions (i.e., nodes) that exhibit a lack of pressure fluctuation and alternating regions (i.e., antinodes) that exhibit alternating pressure maxima and minima. These contrasting regions can provide precise spatiotemporal manipulation of individual cells, as discussed below.
Bulk Acoustic Standing Waves
Bulk acoustic standing waves occur when a microfluidic channel is excited by ultrasound to a resonance mode in which the applied wavelength matches the spatial dimensions of the microfluidic channel. Here, suspended particles with a radius, a, experience radiation forces proportional to the square of the pressure amplitude, p0:
| (1) |
where λ, β, and ρ denote the ultrasonic wavelength, compressibility, density, and the subscripts c and f denote the cell and fluid, respectively.45, 46 The magnitude of this force is strongly dependent on the volume of the cell, and the direction of this force is determined by the acoustic contrast factor, ϕ, which depends on the densities and compressibilities of the cell and the fluid. If the ϕ is positive, cells will travel towards the node(s) of an acoustic standing wave (Fig. 3A), whereas, if the ϕ is negative, cells (or particles) will travel towards the antinodes (Fig. 3B). Most cells in physiological buffers display a positive ϕ due to their relatively higher densities and thus can be precisely and rapidly focused in microfluidic channels without additional diluent buffers (i.e., sheath fluids).47, 48 Bulk acoustic waves were first used to sort cells by Johansson et al., in which fluorescently labeled cells detected by a camera triggered an ultrasound transducer that directed that cell from its initial streamline toward the pressure node, thereby modifying its trajectory to the target outlet.42 More recently, Jakobsson et al. demonstrated an acoustofluidic device to focus cells to one side of a microfluidic channel for fluorescence detection, real-time classification, and acoustophoretic sorting.49
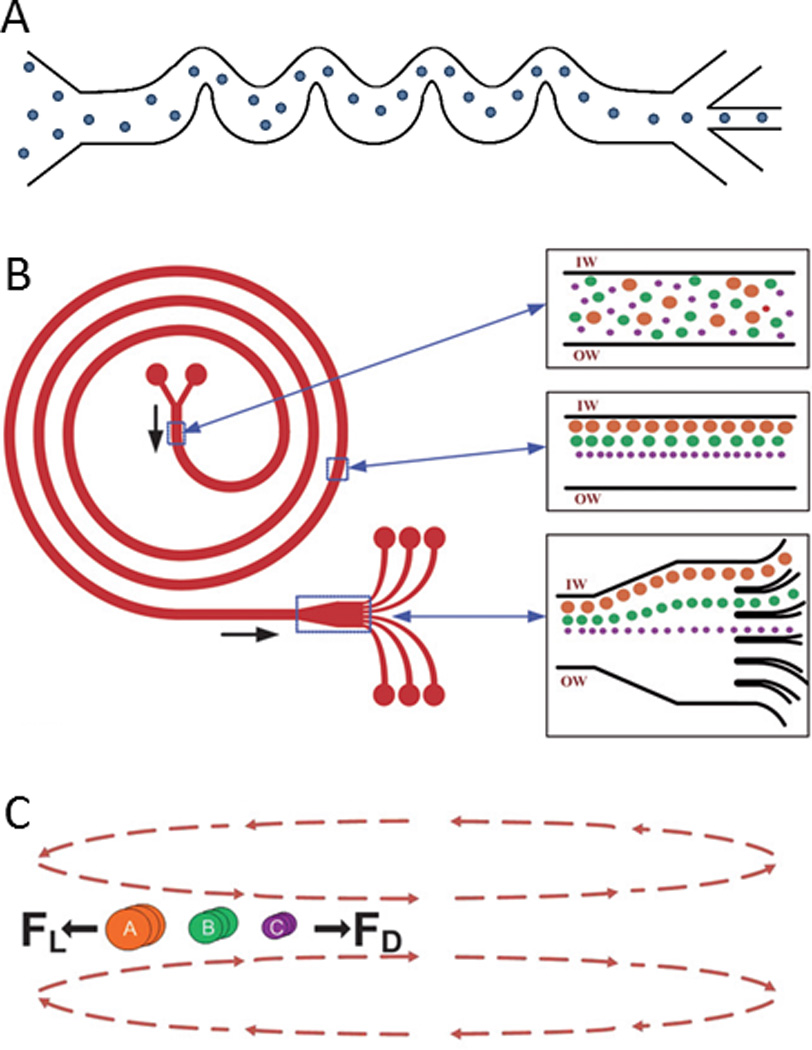
Fig. 3.
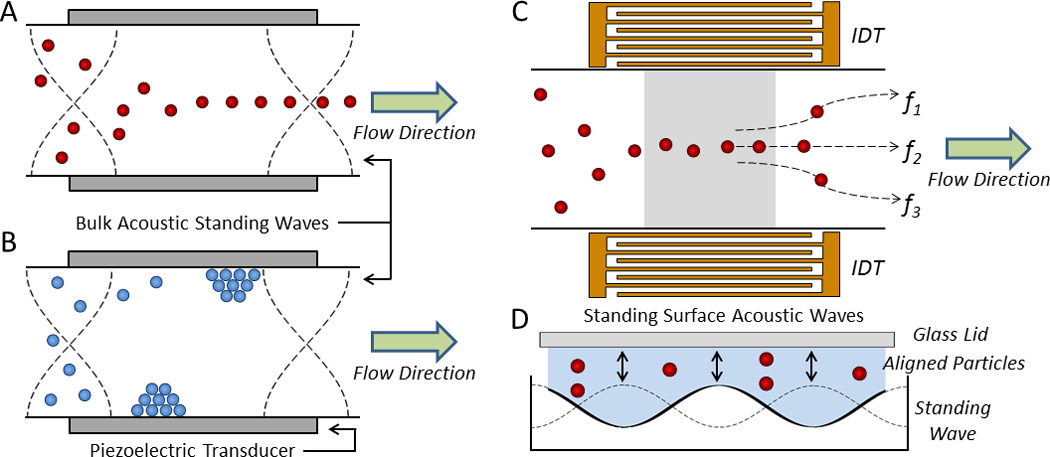
Acoustofluidic manipulation of cells and particles. In a bulk acoustic standing wave, objects with a A) positive and B) negative ϕ migrate to the pressure node(s) and antinodes, respectively.50 C) In a SSAW device, interdigital transducers (IDTs) focus cells along well-defined streamlines according to the driving frequencies of the IDTs (e.g., f1, f2, and f3) for sorting across multiple outlets. D) The cross section of a SSAW device containing four pressure nodes.43
Standing Surface Acoustic Waves
In contrast to bulk acoustic standing waves, SSAW devices form a standing wave along the floor of the microfluidic channel using interdigital transducers (IDTs), providing the mechanical perturbations necessary to position cells along well-defined flow streams in the fluid above (Fig. 3C,D).51 SSAW devices show particular promise for fluorescent label-based cell sorting since a single device can provide a large range of frequencies for dexterous spatial control of single cells and, in turn, multiple channels for sorting.52, 53 These devices have efficiently sorted cells in buffer as well as in water-in-oil droplets across five fluidic channels.54, 55 Ding et al. further showed that SSAW devices can function as acoustic tweezers to manipulate the spatial orientation and patterning of cells and whole organisms such as C. elegans.56
Traveling Acoustic Waves
While most acoustically actuated microfluidic sorting devices developed thus far employ bulk standing waves or SSAWs, traveling acoustic waves have also been used to sort cells in the microfluidic regime. Cho et al. devised a system where a transducer actuates to deflect target cells into the side outlets at rates above 1,000 cells/sec.44 Franke et al. exploited acoustic streaming forces generated from non-standing surface acoustic waves to redirect fluid flow from one outlet to an adjacent outlet.57 More recently, Schmid et al. developed a device that produces a non-standing surface acoustic wave that effectively creates a traveling wave that emanates from the surface to the microfluidic cavity to continuously deflect cells or droplet-containing cells at rates above 1,000 cells/sec.58
C. Optical Manipulations
In addition to forces from pressure waves or electric fields, radiation forces produced by a highly focused optical beam have been used for cell manipulation.59 Optical forces are considered advantageous due to the preservation of cell function, precise spatial control in three dimensions, and manipulation of very small objects (e.g., on the atomic or molecular scale).60 A focused laser beam can trap cells due to a mismatch in the refractive index between the cell and its surrounding fluid, producing optical scattering and gradient forces. Scattering forces tend to push objects away from the source of light, whereas gradient forces attract objects to the point of highest intensity produced by the light (i.e., the focusing maxima) (Fig. 4A). Similar to pDEP, objects travel toward the beam maxima and become optically trapped when the gradient forces overcome scattering forces, producing so-called optical tweezers. Since its inception, this technique has ushered many new types of analyses for single cells, particles, and molecules.61
Fig. 4.
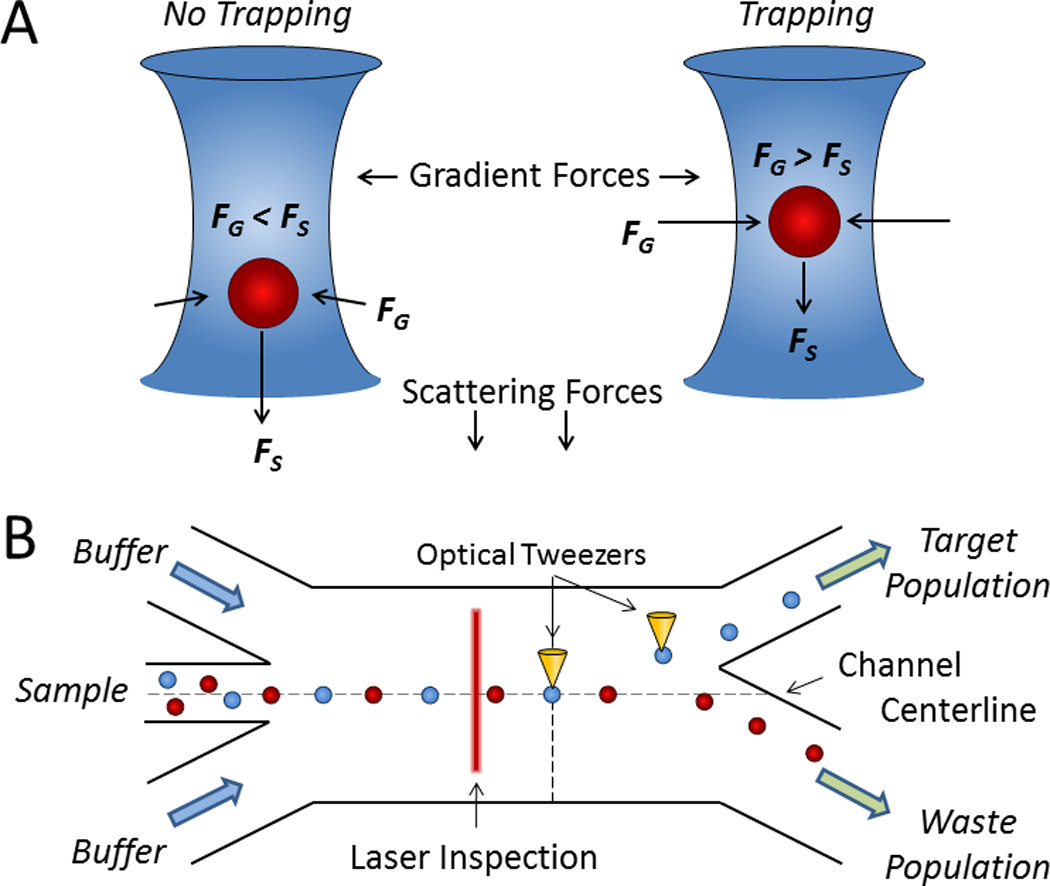
Cell sorting by optical force switching. A) When scattering forces (FS) exceed gradient forces (FG) from a focused laser beam, cells are deflected (left); however, when FG exceeds FS, cells are optically trapped (right).61 B) A hydrodynamically focused stream of cells is aligned toward the waste outlet whereupon cells of interest detected by laser inspection are captured and displaced by optical tweezers for sorting.68
Optics were first used to levitate and transport individual particles in a fluid by Ashkin in 1970.62 Eventually, optical radiation forces were applied to cell sorting by propelling single cells through a microfluidic channel, whereby the long traversal times generally resulted in low throughputs.63 However, recent innovations in miniaturized optical devices have reinvigorated the use of optics as switches, which have made optics-based methods more feasible for fluorescent label-based cell sorting.64, 65 Several groups have used focused optical beams to impose switchable radiation forces on a continuous stream of focused cells for on-chip sorting (Fig. 4B).65–68 Recently, Wu et al. developed a pulsed laser fluorescence-based cell sorter that rapidly forms a stationary bubble due to localized heating to controllably deflect target cells at 20,000 cells/sec.69, 70
D. Mechanical Systems
Instead of using external fields, mechanical sorting systems use discrete mechanical forces to sort cells. Krüger et al. developed an external rotary valve to regulate the throughput within a multi-inlet disposable microfluidic chip by hydrodynamic flow switching for cell counting and sorting.71 Fu et al. devised a similar device to sort E. coli that also includes a trapping component for enhanced analysis.72 This design was later simplified to hydrodynamically focus and isolate latex beads from red blood cells.73 Chen et al. used a similar approach that employed negative pressure gradients to draw fluid from chips and switch the flow of cells to various outlets using gating valves.74
Some groups have developed on-chip flow switching mechanisms instead of relying on external valves to direct fluid flow. For example, Ho et al. designed a micromachined T-switch actuated by electrolytically-produced bubbles to direct the flow of individual cells for binary sorting (Fig. 5).75 And Shirasaki et al. demonstrated cell sorting by using laser light to rapidly heat thermoreversible gelation polymers that controllably actuate the opening and closing of a microchannel.76
Fig. 5.
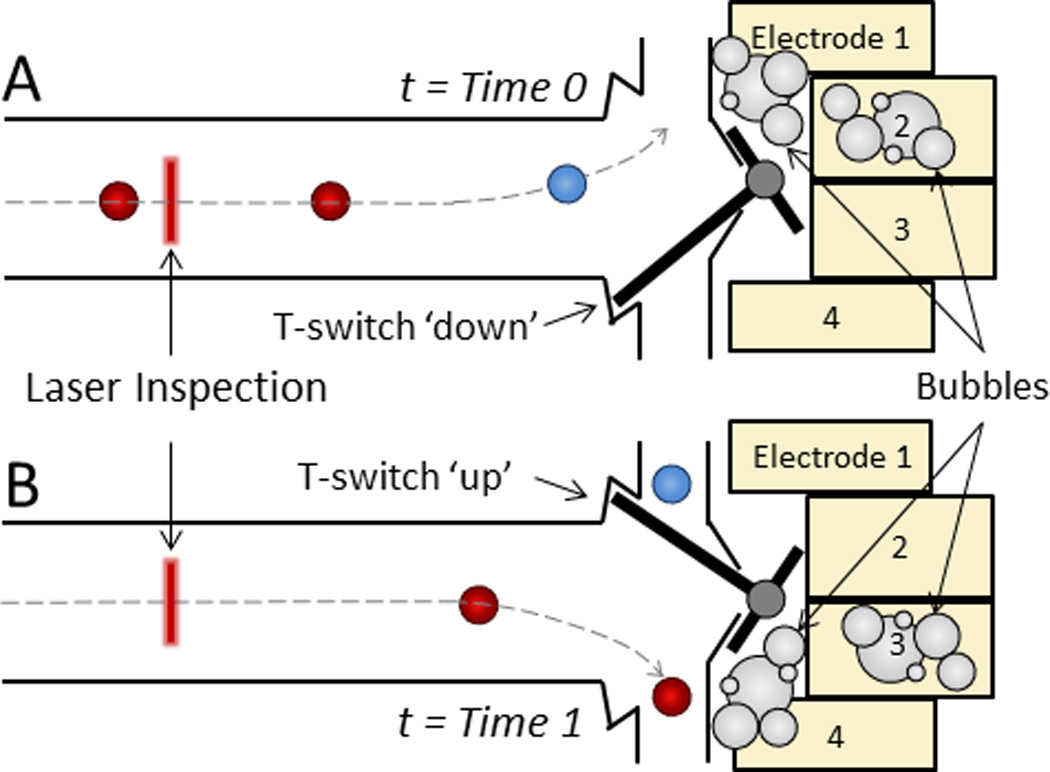
Electromechanical T-switch for cell sorting. A microfabricated cantilever beam reversibly shifts from A) a ‘down’ position (t = Time 0) to B) an ‘up’ position (t = Time 1) when a corresponding pair of electrodes is activated to generate bubbles via isothermal electrolysis, which in turn exerts a mechanical force on the T-switch to redirect fluid flow.75
Bead-Based Cell Sorting
While fluorescent label-based cell sorting offers several advantages, the inconvenience of serial detections, discrete sorting, and the inability of the fluorescent labels to directly contribute to the sorting process has led to the development of other effective techniques for rapid cell sorting, including bead-based and label-free cell sorting systems. Bead-based cell sorting systems depend on particles of a specific material, size, and surface-binding capacity to capture target cells, or sometimes non-target cells, for sorting in an external field. The attachment of beads allows bound complexes to experience a force that is different from unbound cells. Consequently, bead-based cell sorting does not require the serial interrogation of cells and can manipulate groups of cells simultaneously. This affords bead-based systems the ability to sort cells at potentially faster rates and without large volumes of diluent. Additionally, bead-based assays share the immunospecificity of fluorescent label-based sorting, providing the potential for highly accurate targeting and reproducible results. The first commercial bead-based cell sorting system was a benchtop magnetic cell sorter described by Miltenyi et al. in 1989.12 This method of cell isolation was soon miniaturized to the microfluidic domain to sort cells with magnetic and other types of particulate labels.
A. Magnetophoresis
Cell sorting by magnetophoresis (MAP) is facilitated by the use of permanent magnets or electromagnetic coils to exert forces on cells labeled with magnetic particles, magnetically responsive cells, or cells suspended in ferrofluid. In the case of magnetic bead-based sorting, cell surface markers are labeled with particles (typically nanoscopic) containing iron. Cell sorting by MAP is usually performed to either debulk cell populations or to isolate rare cells from native biofluids;77 and similar to acoustophoretic or dielectrophoretic forces, magnetophoretic forces are highly dependent on size of the magnetic moiety.78
The concept for magnetic cell sorting developed by Miltenyi was quickly miniaturized into a microchip device to sort magnetically-labeled cells from unlabeled cells under free-flow MAP.79–81 Using similar principles, Adams et al. developed a multiplexed cell sorting device using MAP where one cell type labeled with large magnetic beads was separated due to the larger magnetic force acting upon it, and a second cell type labeled with small magnetic beads was then separated from unlabeled cells.82 Similarly, Carr et al. developed a multiplexed bead sorter that sorts magnetic beads across 25 output fractions based on the magnitude of their magnetic moments.83
Creating reliable technologies for isolating rare cells such as CTCs, HSCs, and CFCs remains a major challenge in research and medicine. In particular, CTC isolation is a daunting task due to their low abundance in peripheral blood and their “stealthy” behavior during epithelial-mesenchymal transitions.2, 5, 84 Recently, however, significant progress has been made in isolating CTCs via MAP. Hoshino et al. devised a system can capture CTCs from blood cells in ratios as low 1:109 (Fig. 6A).85 In their system, CTCs were labeled with antibodies against epithelial cell adhesion molecule (EpCAM) that were conjugated to iron oxide nanoparticles, separated, and then stained with anti-cytokeratin, DAPI, and anti-CD45 to confirm the captured cells originated from tumors. Numerical simulations later showed that an inverted system (i.e., where the magnetic field is oriented opposite to gravity) can help improve the fidelity of rare cell isolation.86 Darabi et al. developed a similar device with a micropatterned magnetic grid for isolating lymphocytes.87 Kang et al. developed a microchip device with small parallel chambers perpendicular to the main channel to isolate and retain magnetically labeled CTCs from blood for on-chip cell culture.88 For more precise control over the deposition of cells, our group has recently shown that the magnetic labels can be used to isolate cells from a population and organize those cells into well-defined compartments across a large magnetographic array for rapid enumeration, cell pairing, and single-cell analysis.89
Fig. 6.
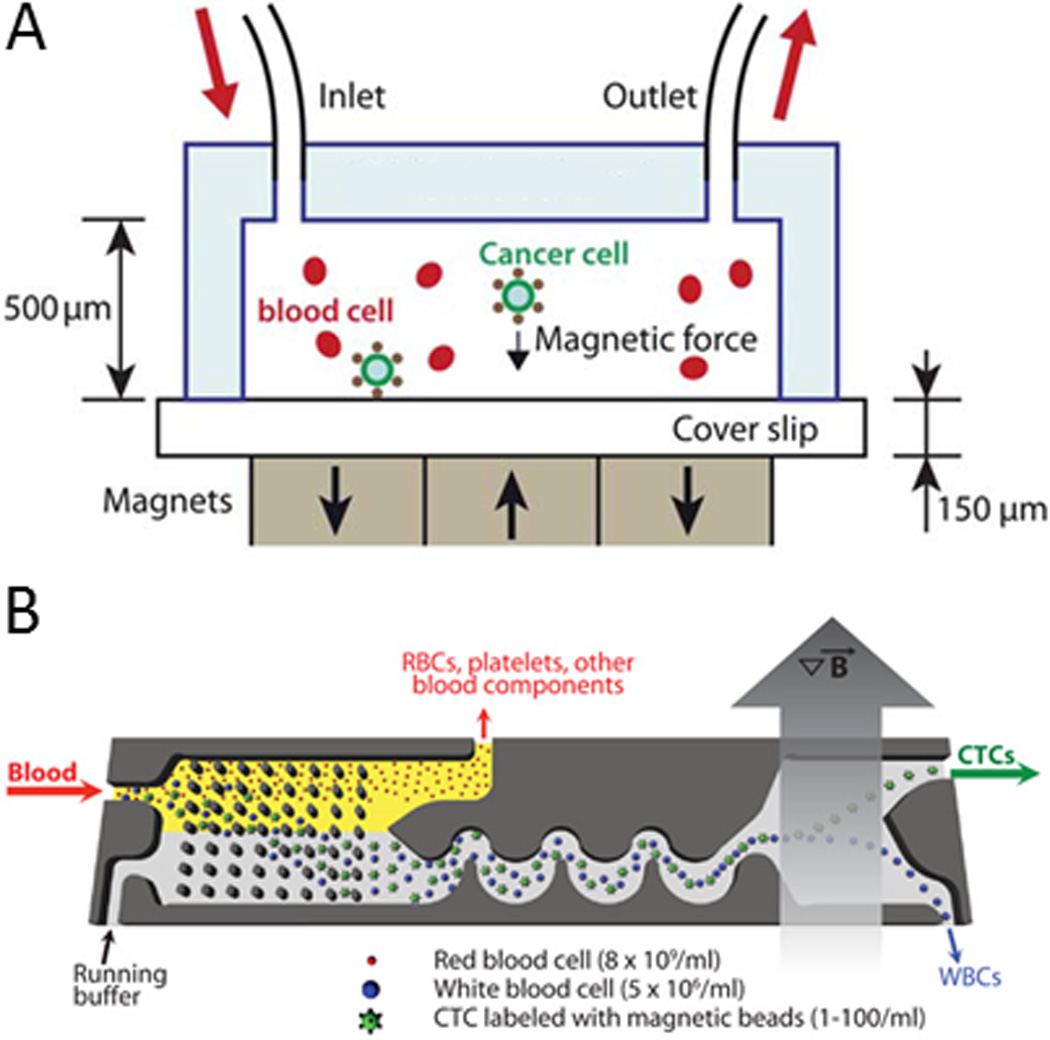
Magnetic isolation of CTCs in microfluidic devices. A) Magnetic beads conjugated with anti-EpCAM are shown to capture and isolate CTCs under free flow in a magnetic field. Reprinted with permission from Hoshino et al85 Copyright 2011 Royal Society of Chemistry. B) Magnetically labeled CTCs and leukocytes are filtered from blood via deterministic lateral displacement (see Fig. 12), focused via inertial focusing (see Fig. 10A), and sorted in a magnetic field. Reprinted with permission from Ozkumar et al90 Copyright 2013 Science Translational Medicine.
Chen et al. recently developed a device that can capture magnetically-labeled cells for sequential encapsulation into aqueous picoliter vessels containing reagents for single cell analysis.91 Researchers from the Toner lab recently developed the innovative CTC-iChip that contains three modules on a single microchip to separate CTCs from peripheral blood for diagnostic analysis: (1) deterministic lateral displacement (a method described below) to debulk nucleated cells (e.g., leukocytes and CTCs) from erythrocytes and platelets, (2) inertial focusing to align cells into a focused streamline (a method described in detail below), and (3) an external magnetic field to separate magnetically labeled CTCs from leukocytes (or magnetically labeled leukocytes from CTCs) (Fig. 6B).90, 92 These bead-based technologies show great promise for isolating tumor cells from unmodified biological fluids on a single chip with minimal processing steps for rapid analysis and diagnosis.
B. Acoustophoresis
Elastomeric beads (or particles) show promise in acoustofluidic systems due to their unique focusing behaviors. Unlike cells, elastomeric particles focus along the antinodes of an acoustic standing wave due their relatively low density and bulk modulus compared to aqueous fluids (i.e., ϕ < 0; Eqn. 1).93, 94 Using silicone elastomeric particles synthesized from nucleation and growth techniques, we have developed a system where biofunctionalized elastomeric particles decorated with streptavidin can capture cells displaying biotinylated antibodies and confine those cells to the antinodes of an acoustic standing wave (Fig. 7).50, 95 This method of cell confinement can enable rapid, continuous, and discriminate sorting in which unlabeled cells focus at the acoustic node(s) and labeled cells focus at the acoustic antinodes for downstream collection.93 Also, Lenshof et al. demonstrated that target cells labeled with dense (ϕ > 0) microbeads could be separated from same-sized non-target cells by virtue of their disparate rate of acoustophoretic migration.96
Fig. 7.
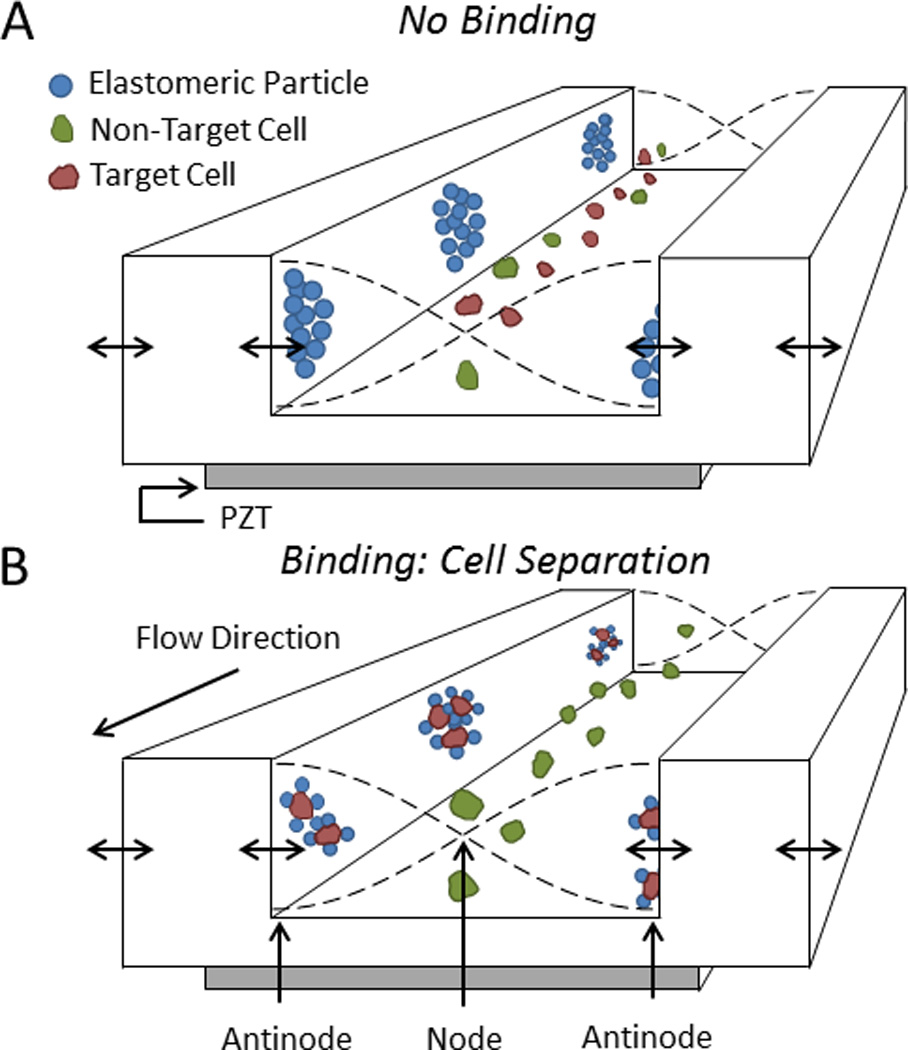
Acoustic separation of cells using elastomeric particles. A) Elastomeric particles and cells focus to the antinodes and node(s) of an acoustic standing wave, respectively. B) When elastomeric particles bind to target cells, those complexes displace to the pressure antinodes for separation from non-target cells. Adapted from Shields IV et al50 2014 American Chemical Society.
C. Electrokinetic Mechanisms
Electric forces exerted on cells and particles scale according to their size, thereby enabling the possibility of non-sequential sorting.20 However, since many cell populations exhibit similar size morphologies, Hu et al. used beads to bind to target cells such that the bound complexes experience greater dielectrophoretic forces in alternating current and thus separate from non-labeled cells.97 In this system, cells labeled with beads traveled farther than cells without beads toward a separate outlet. This system was modified to sort three types of bacteria in a single pass using polystyrene beads of two distinct sizes.98 Later, a magnetic trapping component was added to isolate magnetically-labeled cells from the bulk population in an integrated dielectrophoretic-magnetic activated cell sorter (Fig. 8).99 In addition, Cheng et al. fabricated a DEP device that can filter, focus, sort, and trap cells on a single integrated microchip.100 Their sorting mechanism used angled electrodes and nDEP to deflect bacteria into individual channels and surface enhanced Raman scattering to characterize the samples.
Fig. 8.
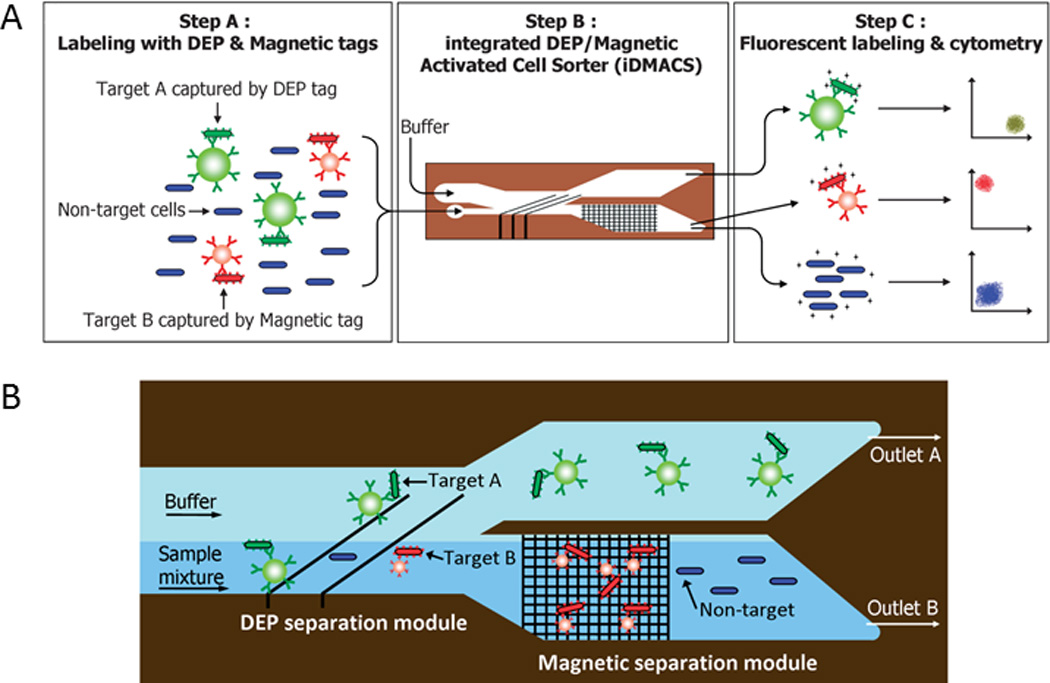
Multi-target cell sorting using DEP and magnetic trapping. A) Target Cell A is captured with polystyrene beads depicted in green and Target Cell B is captured with superparamagnetic beads depicted in red. B) Target Cell A is removed from non-target cells (depicted in blue) by dielectrophoretic forces and Target Cell B is removed from non-target cells by magnetic trapping. Reprinted with permission from Kim et al99 Copyright 2009 Royal Society of Chemistry.
Label-Free Cell Sorting
Label-free cell sorting in microfluidic devices is perhaps the most studied of the three approaches as it encompasses not only active systems (i.e., systems that rely on the use of external fields cells for sorting), but also passive systems, which include mechanisms such as inertial flow, on-chip filtration, and immobilization. In these cases, instead of solely relying on surface markers for labeling cells with fluorophores or beads, label-free sorting relies on the physical differences in the properties of cells such as size, shape, density, elasticity, polarizability, and magnetic susceptibility. The earliest automated cell sorter did not require any labels as it sorted cells by volume using an impedance technique based on the Coulter principle.101 This technique was developed in 1965 by Fulwyler and is widely regarded as the forerunner of modern flow cytometers and FACS devices.102 These key concepts have since been expanded to sort cells in the microfluidic regime. Of the sorting assays described, label-free sorting generally requires the least amount of preparation, making it a highly attractive option for cell sorting.
A. Acoustophoresis
Bulk Acoustic Standing Waves
Unlike the previously described acoustofluidic systems, label-free acoustofluidic systems generally sort cells based on differences in their size. This method of cell sorting relies on the initial placement of cells in streamlines away from the pressure node(s) for their rapid displacement to the node(s) upon actuation of the device (Fig. 3(A)), thereby separating cells of disparate size or acoustic properties. This concept was first used by Petersson et al. to purify erythrocytes from a mixture of lipid particles.103, 104 In a later study, Petersson et al. used hydrodynamic forces to direct particles and cells to the side walls of an acoustofluidic device (i.e., corresponding to the pressure antinodes), whereupon they migrated toward the node (i.e., the channel center) of the acoustic standing wave at rates proportional to their volume.105 Similar to DEP and MAP, larger cells experience greater forces in an acoustic standing wave, and thus respond faster to the radiation forces (Eqn. 1), directing larger cells to the center outlet and smaller cells to the side outlets. This concept was later used for debulking platelets from peripheral blood progenitor cells by Dykes et al.,106 for enriching prostate cancer cells from leukocytes by Augustsson et al.,107 and for sorting viable from non-viable cells by Yang et al.108 Using a thin wall in an acoustic resonating chip, Fong et al. created an H-filter device that can form bulk acoustic standing waves with the node offset from the center of a microfluidic channel for a new method of acoustic cell sorting.109
Standing Surface Acoustic Waves
While the majority of SSAW devices use fluorescent tags to sequentially identify and sort cells, Nam et al. developed a SSAW device to separate platelets from whole blood.110 In their system, blood was injected into the device and sheath fluid focused the cells into a confined band. A separation then followed in which larger cells (e.g., leukocytes and erythrocytes) experienced greater acoustic radiation forces causing them to migrate farther then the surrounding platelets. More recently, researchers from the Huang group have used SSAW devices to sort cells by compressibility111 and wash cells by separating leukocytes from lysed red blood cells.112
B. Electrokinetics
Dielectrophoresis
Label-free cell sorting devices can employ DEP to separate cells based on their intrinsic dielectric properties (i.e., polarizability). Huang et al. first demonstrated this concept by sorting five cell lines with a microelectronic array.113 Also, Cummings et al. designed a microchip containing arrays of insulating posts where the resulting nonuniform electric field across the posts exerted different magnitudes of force on cells as they passed.114 Later, this principle was used to generate nonuniform electric fields near insulating ridges on a microchip to filter and concentrate cells.115 Vahey et al. introduced an isodielectric separation system for sorting cells without labels.116 In this system, cells experience dielectrophoretic forces that displace them to their natural equilibrium point across a conductivity gradient to where their net polarization vanishes (i.e., isodielectric point). With this principle, cells with different conductivities displace by discrete distances along that gradient for efficient on-chip sorting.
An interesting extension of dielectric cell sorting uses field-flow fractionation (FFF), which includes a family of approaches (e.g., gravitational, centrifugal, thermal, magnetic, and electrical) to position cells by precise distances from the microchannel floor and thus to distinct regions of the parabolic flow profile for separation by flow.117–119 Vykoukal et al. first developed a DEP FFF system to isolate stem cells from enzyme-digested adipose tissue.120 This system contained patterned microelectrodes along the bottom of a microfluidic channel that exerted dielectric forces in the opposite direction of gravitational forces, which enabled each cell type to reach an equilibrium height based on its surface charge for rapid sorting under flow (Fig. 9).120 Cui et al. demonstrated that a pulsed dielectric cell sorter could utilize nDEP to trap cells upstream for subsequent elution by size.121 When the AC field was activated, cells became indiscriminately trapped to the ceiling of the microchannel via nDEP. Then after the field was disengaged, larger cells eluted from the microchannel faster than the smaller cells due to larger fluid forces. Then after reactivating the field, smaller cells were dielectrically trapped again whereas the larger cells escaped the electrodes.
Fig. 9.
Sorting by nDEP-assisted field-flow fractionation. A) An array of electrodes displaces cells to equilibrium positions above the floor of a microfluidic channel according to their type (t = Time 0) and B) sorts those cells by propelling them through the channel at rates according to their distance from the wall (t = Time 1).120
Some groups have coupled hydrodynamic lift forces, which act on cells exposed to a shear flow,122 with DEP to sort cells by size or electric permeability. Doh and Cho developed a microchip to separate viable from non-viable cells due to their dissimilar polarizabilities, resulting in pDEP and nDEP, respectively.123 Moon et al. and Shim et al. used hydrodynamic lift coupled with DEP to sort breast cancer cells and CTCs without labels, respectively.124, 125 Kim et al. sorted cells based on their different sizes during the cell cycle using a DEP fractionation technique.126 Hydrodynamic forces were used to concentrate cells to one side of the microfluidic channel whereupon cells encountered an AC field that displaced the cells across the channel by distances according to their size and dielectric properties. This technique was later used to separate viable from nonviable yeast cells.127 An interesting dual function microchip was developed by Sun et al. that could simultaneously sort cells using DEP and size cells using the Coulter Principle via an integrated transistor.128
C. Magnetophoresis
While the majority of magnetic cell sorting devices require nanoparticles to capture and isolate cells (Fig. 6), erythrocytes can be sorted from blood in a similar fashion solely due to their natural iron content in methemoglobin.129, 130 This label-free method of cell sorting has been used for debulking peripheral blood by removing erythrocytes from leukocytes, platelets, lipids, and plasma. Isolating erythrocytes from blood using external magnetic fields was first demonstrated in 1975 by Melville et al.131 and was later refined by Zborowski et al. using cell tracking velocimetry132 and by Han et al. embedding a ferromagnetic wire inside of a microdevice.133–135 This method of magnetic manipulation from embedded ferromagnetic microwires was more fully developed later by others.136–138 Furlani presented a method for separating red blood cells from white blood cells in plasma by embedding soft-magnetic elements in an array along a microfluidic channel.139
Since these magnetic, label-free approaches are only suitable for blood cells with natural iron content, several groups have employed nontraditional means for continuously sorting other types of cells in magnetic fields. Roberts et al. created a device that could sort macrophages from monocytes due to their dissimilar internalization rate of iron nanoparticles.140 Since macrophages can endocytose more iron content per cell than monocytes, macrophages can undergo greater loadings of magnetic material and thus experience greater MAP forces in an external field.
Some groups have sorted diamagnetic (i.e., unlabeled) cells by suspending them in ferrofluids, which can separate cells due to differences in their size, shape, or deformability.141 Ferrofluids are liquids that are strongly magnetizable (e.g., water containing iron oxide nanoparticles) and can thus transport diamagnetic particles in a magnetic field. Zeng et al. developed a system where an upstream magnet focused cells suspended in ferrofluid into a single stream, and a downstream magnet displaced those cells according to their size.142 Similarly, Shen et al. used a paramagnetic solution (i.e., of paramagnetic ions) to exert repulsive forces to sort cells by size.143
D. Optics
Similar to the optical switching mechanisms described previously, Hoi et al. created a layered device with perpendicular flow channels on top of one another.144 In this system, a microscope detects unlabeled cells as they pass through the first channel, which triggers a pulsed laser to rapidly force cells into the second channel at their junction. Lau et al. used laser tweezers for an automated Raman-activated cell sorter that functions without fluorescent labels.145 While the majority of optical systems employ one focused laser beam, MacDonald et al. used a body-centered tetragonal optical lattice to fractionate a two-component cell mixture in a continuous flow microfluidic device.146
A major limitation of optical tweezers is their inherently slow processing that stymies the potential for high throughput sorting. Optoelectronic tweezers, however, refer to a class of tools that employ optics and electronics by projecting optical images on a photosensitive substrate. The projected images effectively form transient electrodes on the bottom surface to direct an AC field from a conductive, transparent lid for the precise spatial arrangement of micro- and nanoparticles.147 Optoelectronic tweezers can thus provide a low-power means to trap, transport, and sort cells without labels.147 And while optical tweezers provide high-resolution manipulations, they can also require optical intensities reportedly 100,000 times greater than optoelectronic tweezers while only providing small regions for cellular manipulation.148 In an early example, Chiou et al. used optoelectronic tweezers to pattern cells into a large, well-defined array.148 Now, optoelectronic tweezers are used to sort cells based on size due to the differing induced velocities in response to applied dielectrophoretic forces149, 150 as well as viable cells from non-viable cells due to differences in membrane potentials.151 These devices have been coupled with electrowetting-on-dielectrics mechanisms to enable multiple delivery rounds of drugs or nutrients to cells followed by electrically-driven waste removal.152 Optoelectronic tweezers have been integrated into microfluidic devices for single-cell preparation and analysis, enabling the high precision benefits of optics devices with higher throughputs.153
E. Passive Cell Sorting
The technologies described thus far involve active methods for cell separation; however, the remaining systems involve passive approaches to separate, isolate, or enrich cell populations.154 Passive systems consist of a variety of methods that do not rely on fluorescent labels or beads. Instead, these methods rely on the inherent differences in cellular morphology between cell groups (e.g., size, shape, compressibility, and density) and can sort cells using inertial forces, hydrodynamic spreading, deterministic lateral displacement, filtration, transient cellular adhesion, and cellular immobilization.155
Inertial Focusing in Curved Channels
Inertial forces can result in the induced migration of cells or particles across streamlines in laminar flow streams. Typically, inertial forces emanate from boundary effects of fluid flow adjacent to the walls of a microfluidic channel, causing lift. Inertial focusing in curved channels refers to a subset of distinct phenomenological techniques for cell fractionation, which includes the use of serpentine or Archimedean spiral patterns for cell ordering and sorting.156 Di Carlo et al. demonstrated that cells could be differentially focused and sorted based on size under laminar flow using a serpentine pattern (Fig. 10A).157 A major benefit of this system is its high throughput (e.g., 1.5 mL/min) without sheath flow or sequential cell manipulation, which is useful for processing native biological fluids and flow cytometry.158, 159 As previously discussed, an inertial flow microfluidic chip with a deterministic lateral displacement module was incorporated into a magnetic-label based system for CTC isolation (Fig. 6B).90
Fig. 10.
Inertial microfluidics for cell sorting in curved microchannels. A) A serpentine microfluidic channel can focus cells into a single streamline. B) A spiral microfluidic channel can sort cells by size (IW and OW indicate the inner wall and outer wall, respectively). C) The cross section of the microfluidic channel showing the balance between lift forces (FL) and Dean drag forces (FD). Reprinted with permission from Kuntaegowdanahalli et al161 Copyright 2009 Royal Society of Chemistry.
Similarly, Russom et al. created a system with a spiral channel to apply centrifugal forces to focus and sort cells.160 As in the case of serpentine patterns, cells can be sorted in a spiral microchannel due to their differential focusing for rapid separation upon branching into multiple channels (e.g., six, Fig. 10B,C).161 Recently, this system was used by Hou et al. to retrieve CTCs from blood using an approach named Dean flow fractionation162 and by Nivedita et al. to separate blood cells.163 Guan et al. and Warkiani et al. demonstrated that a channel with a trapezoidal cross section could further enhance the performance of a spiral microfluidic device to focus and sort cells.164, 165
Pinched Flow and Hydrodynamic Spreading
In addition to curved channels, inertial forces can play a critical role in straight channels.156 For example, Parichehreh et al. were able to enrich nucleated cell populations in blood using inertial forces in straight microfluidic channels of large aspect ratios.166 Similarly, pinched flow fractionation and hydrodynamic spreading are two examples where inertial forces play a role in the ordering of cells for the subsequent sorting. Pinched flow fractionation occurs when a flow stream of cells is pinched by a narrow channel cross section such that cells are constrained and aligned against a side wall and subsequently separate by size once the channel broadens due to the laminar flow profile (Fig. 11).167 This alignment effect is typically enhanced by sheath fluid, which pushes cells against a wall such that the center of the larger cells are farther from the wall surface than the center of smaller cells, thus giving cells of different sizes slightly different flow trajectory upon broadening of the channel. This method for sorting has been expanded by the addition of multiple asymmetric outlets for better hydrodynamic control168 as well as by the spatial reorientation of the microfluidic device for gravitationally enhanced separation between cell populations of different size and mass.169
Fig. 11.
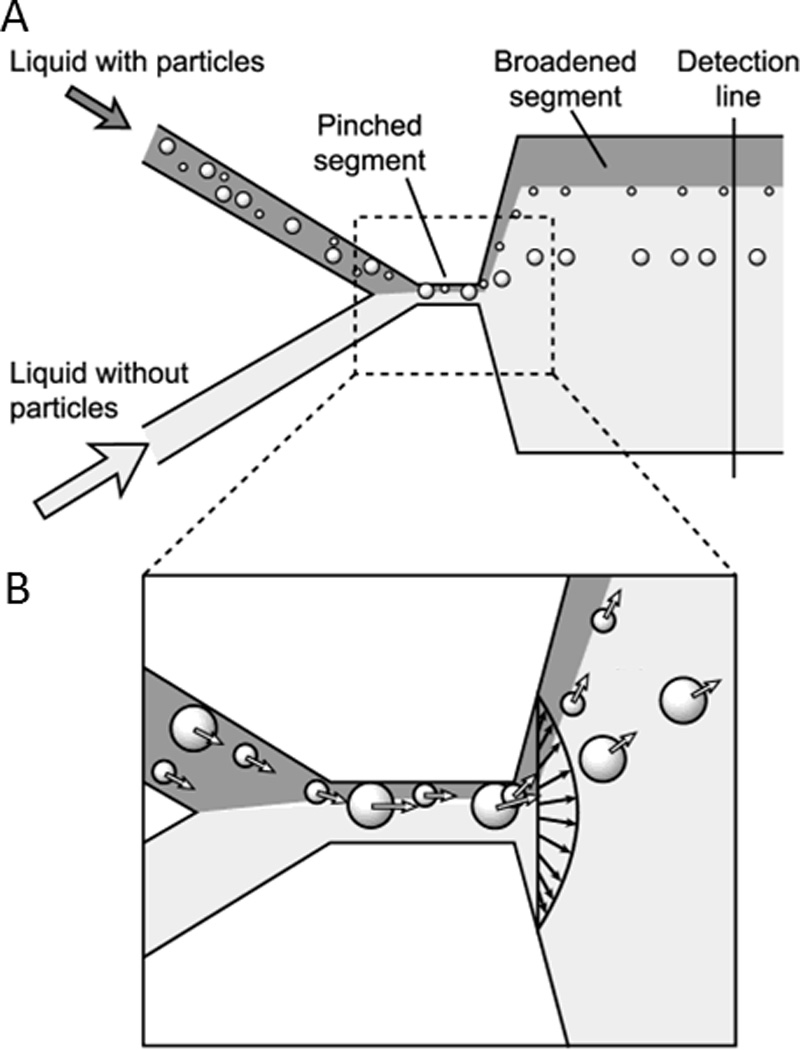
Cell sorting by pinched flow fractionation. A) In the pinched segment, cells are first pushed against the wall, and then separated by size upon broadening of the microfluidic channel. B) Cells are aligned in the pinched segment of the channel and follow separate streamlines for sorting by size after exiting the pinched segment. Reprinted with permission from Yamada et al167 Copyright 2004 American Chemical Society.
Similar to pinched flow fractionation, some groups have used hydrodynamic techniques to direct cells against one wall of a microfluidic channel without a pinched segment for sorting upon broadening of the channel in a method called hydrodynamic spreading.170, 171 Wu et al. enhanced this effect by coupling hydrodynamic spreading with electroosmosis to sort E. coli and yeast cells.172 Instead of using hydrodynamics or pinched flow to align cells against one wall of a microfluidic channel, Mach et al. used inertial forces in a straight channel with a large aspect ratio cross section (i.e., height > width) to separate pathogenic bacteria from blood upon hydrodynamic spreading.173 Microfluidic channels with large aspect ratio cross sections can localize to specific regions due to inertial lift and shear forces, which are readily influenced by channel geometry, particle size, and flow properties changes in the Reynolds number for sorting upon hydrodynamic spreading.174 This concept was exploited to isolate rare tumor cells spiked in blood175 and sort cells by their deformability.176 Instead of using pinched flow or inertial forces, Geislinger et al. devised a hydrodynamic lift system enhanced by hydrodynamic spreading to separate CTC and red blood cells.177, 178
Deterministic Lateral Displacement
Given the standard fabrication methods used to create microfluidic chips, micropatterns within a microfluidic channel (e.g., posts, ridges, groves, ratchets) provide a convenient way to spatially manipulate cells. Deterministic lateral displacement is an important example, where cells navigate through an array of posts for sorting by size. In these systems, control over sorting is given by the design of the array features such that cells smaller than a critical radius (a < Rc) move with the convective flow and cells larger than a critical radius (a > Rc) move in a direction dictated by the arrays (Fig. 12).179 Using a periodic array of microposts, in which each row of posts is offset by a certain periodic distance, Huang et al. showed that smaller cells could more easily traverse between obstacles than larger cells, enabling their separation.180 The critical sizes of cells and microposts were carefully evaluated by Inglis et al.181 and have been used to fractionate undiluted whole blood samples as well as isolate cancer cells.182–184 Later, Beech et al. showed that deterministic lateral displacement was useful for sorting cells by not only size, but by shape and deformability.179
Fig. 12.
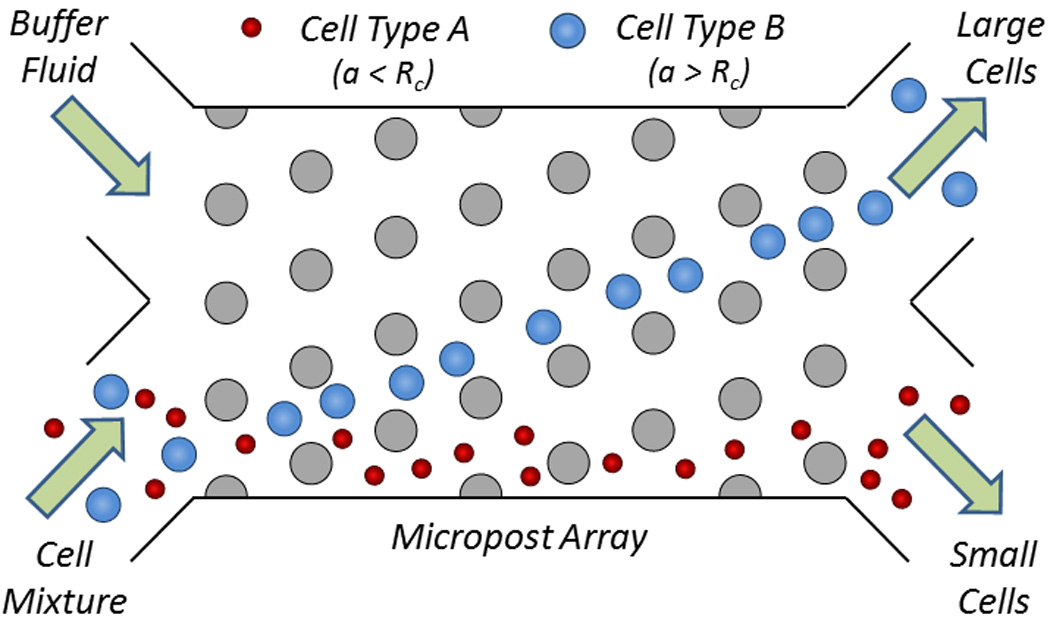
Cell sorting by deterministic lateral displacement. Large cells (depicted in blue) migrate away from the small cells (depicted in red) in the initial streamline due to the engineered size and spacing of the microposts in the microfluidic channel.181
Hydrophoretic Filtration
Ridge-induced hydrophoretic filtration relies on the formation of a lateral pressure gradient within a microfluidic channel due to flow-altering micropatterns. A successive array of slanted obstacles on the microchannel floor and ceiling induces a pressure gradient across the width of the channel to focus cells to precise locations within the generated local pressure field according to type and then separates those cells.185 Using this phenomenon, Choi et al. developed a device that exploits differences in both the size and deformability of cells for passive sorting.186
Similarly, a device with successive ridges along the channel ceiling has been shown to focus, guide, and sort cells (Fig. 13).187 This device, previously called a microvortex manipulator, contains an array of herringbone shaped slanted grooves in the ceiling of a microfluidic channel that passively focuses cells to defined regions of a microfluidic channel and sorts cells based on their density.188 Using a different style of microvortex, Sollier et al. sorted CTCs using a sudden expansion (i.e., by introducing sharp corners perpendicular to the flow direction) in a microfluidic channel to create predictable eddy flows to capture CTCs from blood.189 A similar microfluidic channel design with multiple orifices has been used to sort CTCs from white blood cells.190
Fig. 13.
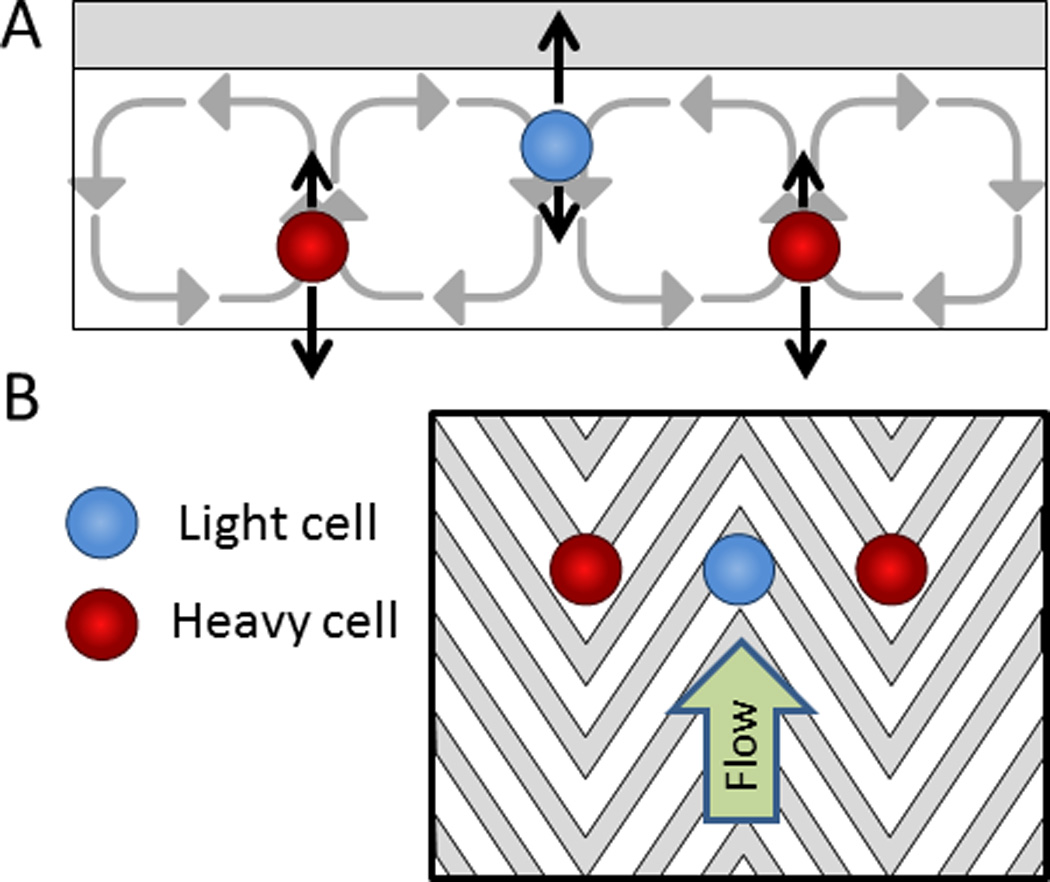
Hydrophoretic cell focusing and sorting. A) A simplified free-body diagram of cells with a low density (shown in blue) separating from cells with a high density (shown in red). The black arrows pointing upwards represent buoyancy forces and the black arrows pointing downwards represent settling forces. B) A top view of the microfluidic channel with herringbone grooves in the ceiling to guide the focusing and separation of cells by density.187
Size Exclusion Filtration
Instead of engineering the size and pattern of uniformly spaced posts to sort cells, size exclusion filtration refers to the use of posts with tiered (i.e., decreasing) spacing as a function of distance for sorting cells in a non-binary fashion. Size exclusion filters consist of a series of linear arrays of pillars that selectively group cells by size and shape. Mohamed et al. developed a size exclusion filter to isolate CFCs from maternal blood.191 Later, McFaul et al. and Preira et al. showed how structural ratchets can be used to sort cells based on size as well as deformability when used with oscillatory applied pressures (Fig. 14).192, 193
Fig. 14.
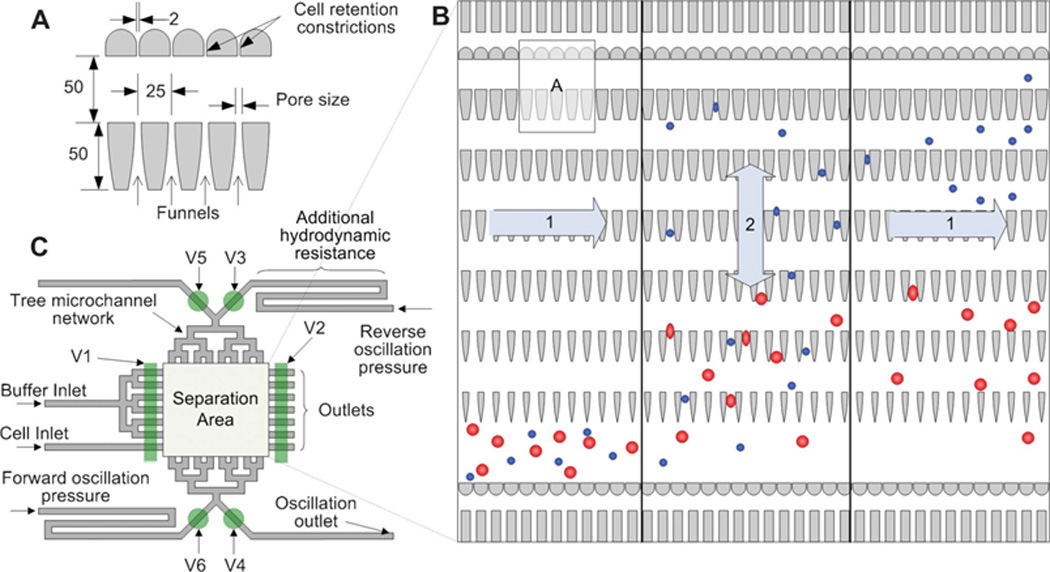
Micropatterned ratchets for isolating cells by size and deformability. A) A design of microratchet funnels for fractionating cell populations (dimensions in µm). B) The operation area of the device whereby cells (1, left) enter the chip, (2) reversibly flow through the ratchets for separation by size, and (1, right) exit the chip. C) Schematic of a size exclusion filtration device with various inlets, outlets, and valves (V1-V6). Reprinted with permission from McFaul et al192 Copyright 2012 Royal Society of Chemistry.
Cross-Flow Filtration
The filtration of cell-containing fluids is one of the earliest methods used to fractionate cell populations. Examples of these filters include weir filters, which contain large barriers to trap large cells (Fig. 15A); pillar filters, which contain a row of same-sized microposts to trap large cells (Fig. 15B); and membrane filters, which contain an array of pores on the floor or ceiling to trap large cells.194 A major limitation of these filters is that they often clog by trapping larger cells or debris. Recently, however, Earhart et al. devised a membrane filter that could reduce this clogging by using larger pore sizes and incorporating a magnetic component to capture magnetically labeled cells (e.g., CTCs) at the edges of the pores, thereby allowing unlabeled cells to easily pass through the filter.195
Fig. 15.
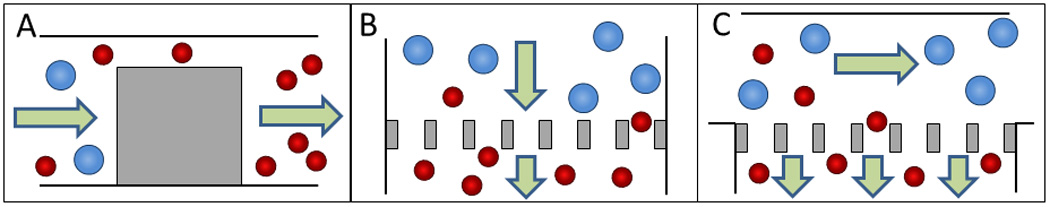
Microfluidic filtration mechanisms. Schematic of a A) weir filter, B) pillar filter, and C) cross-flow filter to separate smaller cells (depicted in red) from larger cells (depicted in blue).194
In contrast, cross-flow filtration, sometimes referred to as tangential flow filtration, uses an array of lateral slits aligned in the direction of flow to fractionate cell populations by size (Fig. 15C). This method of filtration is a major advancement over early types of filters such as weir, pillar, and membrane because of their decreased likelihood for clogging because the behave more like a sieve than a dead-end filter.196 Several cross-flow filter designs have been developed for size-based sorting applications such as the separation of white blood cells from whole blood, plasma from whole blood, and myocytes from non-myocytes.196–200 Using a cross-flow filter, Chen et al. developed an integrated system for sorting and lysing cells on a single chip with capabilities for DNA purification that can yield amounts comparable to commercial centrifugation methods.201
Hydrodynamic Filtration
The last type of microfluidic filtration is hydrodynamic filtration. Here, aligned cells are separated by multiple branched outlets, whereby the fluid draining from the outlets pulls cells from the walls of the main channel at rates that scale according to their size (Fig. 16).202 Smaller cells exit the proximal outlets because their center is closer to the wall of the microfluidic channel, enabling their controlled shunting from larger cells (Fig. 16B–C).Yamada et al. first showed a hydrodynamic filtration device and its use for the selective enrichment of leukocytes and liver cells.202–204 Later, Mizuno et al. combined hydrodynamic filtration to first separate lymphocytes by size, and second to separate lymphocytes by surface marker expression using MAP for multiplexed sorting.205 After cells are filtered, the magnetic field pulls cells with more magnetic content (due to their higher expression levels) toward the outlet closer to the magnet, whereas cells with lower expression levels exit the outlet farthest from the magnet.
Fig. 16.
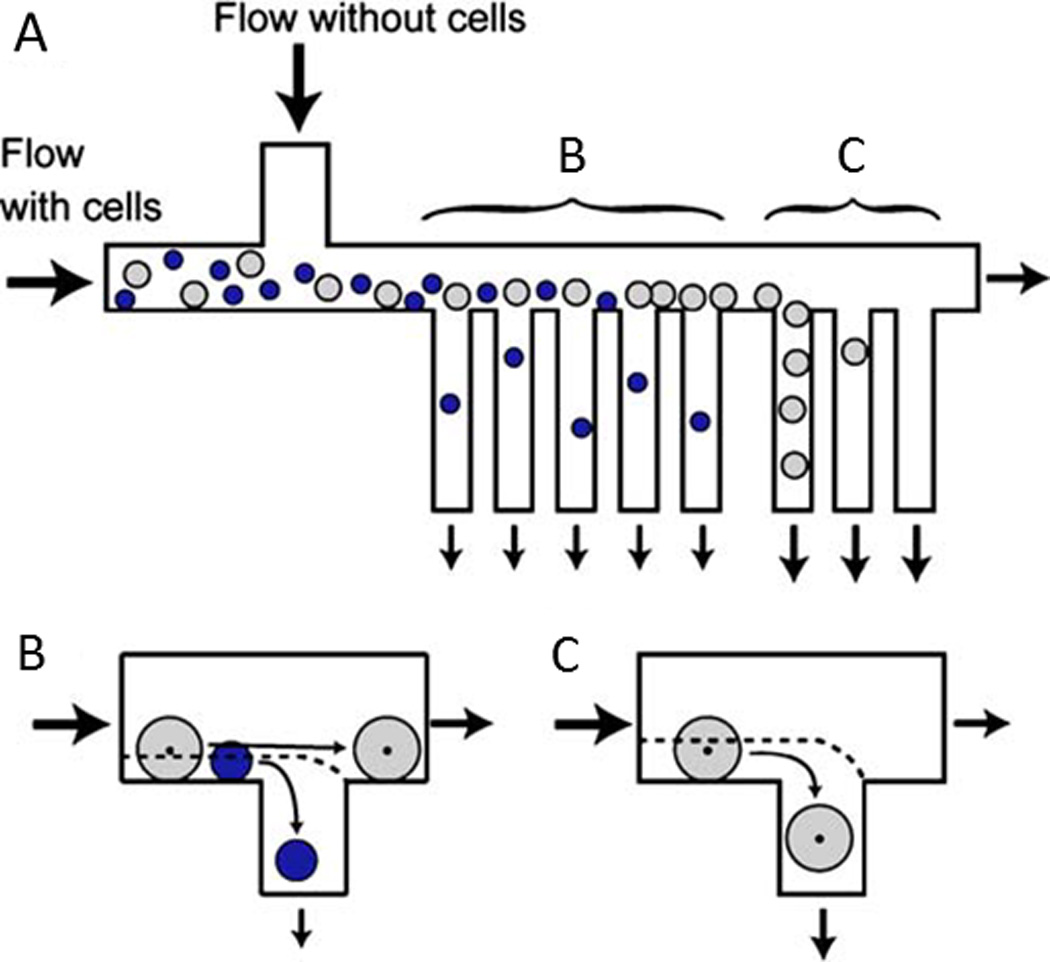
Cell sorting by hydrodynamic filtration. A) Cells are injected into the microfluidic device and are pushed toward the outlets. B) Small cells exit out of the proximal branches whereas C) large cells exit out of the distal branches. Reprinted with permission from Yamada et al202 Copyright 2007 Springer.
Transient Cellular Adhesion
Instead of relying in differences in the size, deformability, and membrane polarizability of cells, some groups are examining the impermanent adhesion of cells to a surface to enable a unique class of microfluidic cell sorting methods.206 Lee et al. showed that a flat surface containing a striated pattern of P-selectin could induce the transient cellular adhesion and lateral displacement of HL60 cells due to high affinity between the PSGL-1 ligand and P-selectin.207 Bose et al. showed this system could isolate neutrophils from blood with high enrichment performances without significant cellular agitation.208
A similar technique, deterministic cell rolling, is used to sort cells based on their surface interactions with a microfluidic channel. Choi et al. showed that a series of successive ridges along the floor of a microfluidic channel can induce repeated cellular collisions for sorting.209 In this device, the surface is modified with P-selectin to elicit a brief interaction with target cells and push those cells toward one side of the microfluidic channel (i.e., the gutter side). Non-target cells flow above the ridges toward the other side of the microfluidic channel (i.e., the focusing side) (Fig. 17).209, 210
Fig. 17.
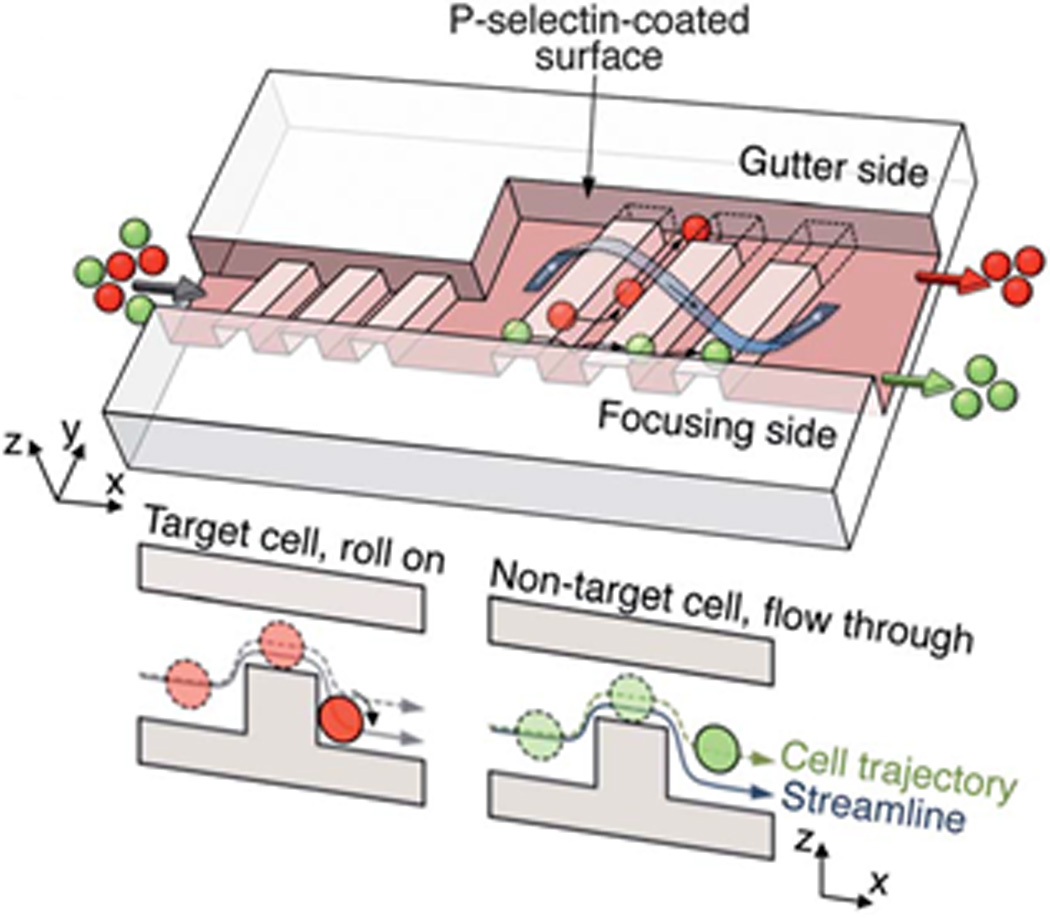
Cell sorting by deterministic cell rolling. Target cells (red) interact with the surface, roll across the ridges, and laterally displace toward the gutter side whereas non-target cells (green) flow over the ridges, not interacting with the surface, and exit on the focusing side. Reprinted with permission from Choi et al209 Copyright 2012 Royal Society of Chemistry.
Cellular Immobilization
Instead of relying on transient interactions between cells and a functionalized surface for sorting, which can suffer from limited throughputs due to the reliance of low flow rates, many groups are investigating the non-transient capture, or immobilization, of cells on surfaces or in columns. One of the earliest approaches for fractionating cell populations by immobilization was through affinity chromatography, which sorts by the high affinity interactions between cell-surface receptors and immobilized ligands on the chromatography matrix.211, 212 This technique is capable of yielding results that do not depend on the size, density, or charge of the cells, and can be performed in various types of matrices.213
Many of the more recent cell immobilization devices use antibodies for cell-specific isolation, often from whole blood.214–216 For example, Hyun et al. used a microfluidic device coated with antibodies for the enrichment of CTCs.217 Instead of using antibodies, Chen et al. developed a nanoroughened surface to selectively capture CTCs based on their differential adhesion preference compared to non-CTCs.218 Similarly, Singh et al. developed a shear-based system to isolate human pluripotent stem cells where intact colonies were rapidly isolated based on their adhesion strength to the microfluidic channel floor.219 These techniques show promise for isolating rare cells such as CTCs due to their ability to process large sample volumes and recover rare cells from liquid biopsies (e.g., less than 1 cell in 1 mL of blood) without relying on surface markers (e.g., EpCAM), which are often downregulated during the mesenchymal-epithelial transition.2, 220
The CTC-chip, which is the predecessor of the previously mentioned CTC-iChip, uses microposts conjugated with anti-EpCAM to immobilize CTCs.221 In a similar system, antibodies conjugated to a pitched substrate are able to capture specific cells for rapid detection assays.222 Yu et al. developed a magneto-controllable device where antibody-conjugated iron nanoparticles are magnetically localized around the circumference of microposts within a microfluidic cavity, whereupon cancer cells expressing the target antigen are captured for rapid release after disengaging the magnetic field.223 Saliba et al. designed a system using a similar concept, which employs the self-assembly of antibody-labeled magnetic microbeads to form hexagonally spaced pillars under an applied magnetic field (Fig. 18).224 The self-assembled columns specifically capture and retain tumor cells that can be recovered for analysis after disengaging the magnetic field. Yoon et al. immobilized CTCs from blood using functionalized graphene oxide nanosheets.225 Finally, Mittal et al. functionalized a fluid-permeable surface with antibodies for the selective capture of CTCs.226
Fig. 18.
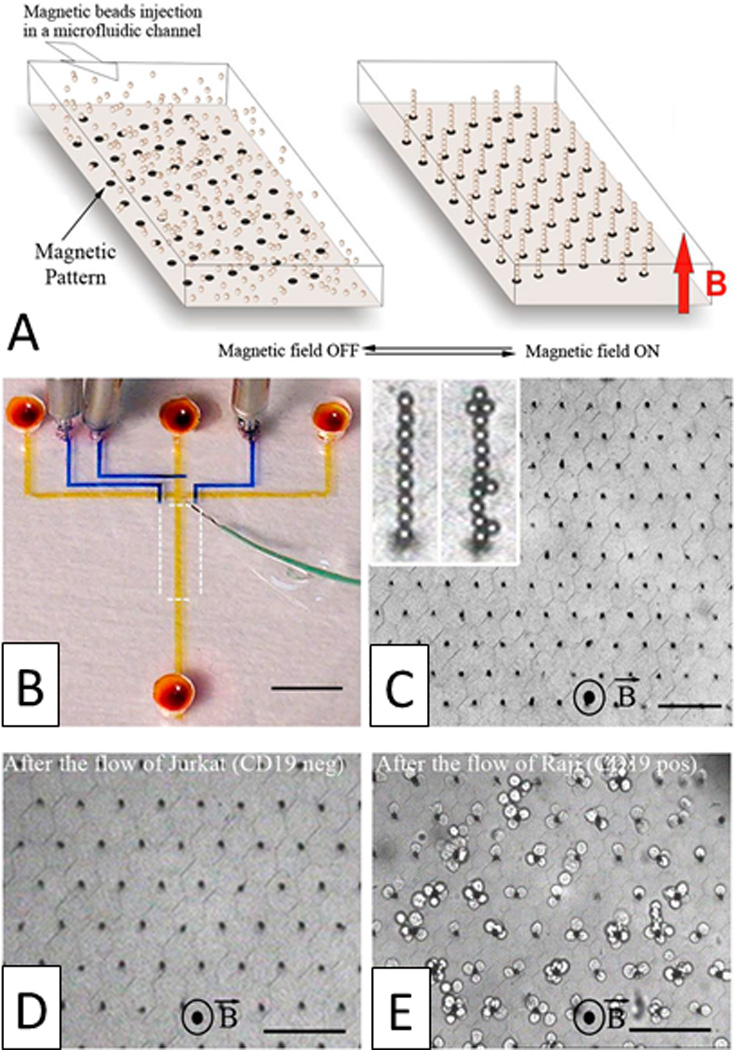
Magnetic self-assembly of biofunctional magnetic beads for isolating rare cells. A) Schematic of a hexagonal array of magnetic ink (left) can guide the self-assembly of magnetic beads conjugated with anti-CD19 mAb in the presence of a vertical magnetic field (right). B) Photograph of the microfluidic device. Optical micrographs of the columns after C) the assembly of magnetic beads, D) the passage of 1,000 Jurkat cells (CD19 negative), and E) the passage of 400 Raji cells (CD19 positive) (scale bar: 80 µm). Reprinted with permission from Saliba et al224 Copyright 2010 Proceedings of the National Academy of Sciences of the USA.
While antibodies are traditionally used to capture cells for sorting, aptamers also show promise for the high affinity capture of cells. Xu et al. developed an aptamer-based microfluidic system to passively capture CTCs, thereby enriching rare cells 135-fold in a single pass.227 Similarly, Sheng et al. used microposts conjugated with aptamers for the isolation of CTCs228 and Zhao et al. developed a biosensing network containing DNA tentacles functionalized with aptamers along the terminal end for capturing cells.229
Conclusion
Advances in cell biology, cell-based diagnostics, and cellular therapies, along with the promise of point-of-care and personalized medicine, have increased the need for cell sorting devices in both basic research and in clinical applications that are safer, more efficient, and easier to use than current technologies. To address this critical need, researchers are looking toward microfluidics due to their accessible fabrication, low reagent consumption, and small footprints which offer significantly scaled-down sizes with design features that are commensurate with single cells.230 Further, microfluidic devices can offer improved safety over traditional cell sorting technologies by eliminating potentially biohazardous aerosols.
Many of the microfluidic technologies described in this review are still in the prototype or proof-of-concept stage; few have demonstrated the clinical performance of a fully integrated and validated system. In order to reach their full potential, these technologies still require substantial commercial investment for standardization, manufacturability, and repeatable performance. As such, microfluidic technologies still suffer from a few key limitations that must be overcome. For example, many of these devices suffer from low throughput due to single-channel or single-orifice designs. Advances in parallelization and other innovative scale-up approaches could help to overcome this barrier. This would prove particularly valuable for diagnostic applications that target rare cell populations from liquid biopsies (e.g., such as CTCs from blood) or multiplexed cell sorting from large sample volumes. Secondly, the limited lifespan of microfluidic chips, which are typically shortened due to blocking or clogging, is another barrier to routine research or clinical use. The commercialization of cell sorting technologies may look to disposable chips to overcome these short lifetimes and enable a sterile, user-friendly operation. Finally, many microfluidic cell sorting systems still require complex sample preparations that reduce ease-of-use and can compromise sorting accuracies due to user variations. Therefore, a major challenge remains to build microfluidic devices that require minimal sample preparation and operational training.
The future of microfluidic cell sorting is promising. With the large variety of cell sorting devices (e.g., from active systems that use acoustic, electric, magnetic, or optical forces to passive systems that use inertial effects, filters, or immobilization procedures), microfluidic cell sorting technologies can offer speeds and accuracies in a number of ways that rival current commercial devices in a platform that is more efficient, less cumbersome, and offers a more straightforward standard operating procedure. Unlike most commercial cell sorting machines, microfluidic devices are highly modular, thereby offering the capacity to perform multiple functions (such as mixing, counting, and lysing analyzing single cells) in a single compact device. This ability positions microfluidic technologies at the forefront of the next generation of cell handling devices by providing the capability of performing high-level, post-sorting analyses such as biochemical, secretion, and cell culture assays for true lab-on-a-chip devices.
Acknowledgements
This work was supported by the National Science Foundation’s (NSF’s) Research Triangle Materials Research Science and Engineering Center (MRSEC, DMR-1121107), the National Institutes of Health (R21GM111584), and a NSF Graduate Research Fellowship (GRF-1106401) to C.W.S. The authors have no conflicts of interest.
References
- 1.Tomlinson MJ, Tomlinson S, Yang XB, Kirkham J. Journal of tissue engineering. 2013;4:2041731412472690. doi: 10.1177/2041731412472690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong AJ, Marengo MS, Oltean S, Kemeny G, Bitting RL, Turnbull JD, Herold CI, Marcom PK, George DJ, Garcia-Blanco MA. Molecular cancer research : MCR. 2011;9:997–1007. doi: 10.1158/1541-7786.MCR-10-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wognum AW, Eaves AC, Thomas TE. Archives of medical research. 2003;34:461–475. doi: 10.1016/j.arcmed.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 4.Bischoff F, Marquez-Do D, Martinez D, Dang D, Horne C, Lewis D, Simpson L. Clin Genet. 2003;63:483–489. doi: 10.1034/j.1399-0004.2003.00087.x. [DOI] [PubMed] [Google Scholar]
- 5.Chen Y, Li P, Huang PH, Xie Y, Mai JD, Wang L, Nguyen NT, Huang TJ. Lab Chip. 2014;14:626–645. doi: 10.1039/c3lc90136j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chin L, Andersen JN, Futreal PA. Nature medicine. 2011;17:297–303. doi: 10.1038/nm.2323. [DOI] [PubMed] [Google Scholar]
- 7.Hulett HR, Bonner WA, Barrett J, Herzenberg LA. Science. 1969;166:747–749. [PubMed] [Google Scholar]
- 8.Herzenberg LA, Parks D, Sahaf B, Perez O, Roederer M, Herzenberg LA. Clinical chemistry. 2002;48:1819–1827. [PubMed] [Google Scholar]
- 9.Bonner WA, Hulett HR, Sweet RG, Herzenberg LA. Review of Scientific Instruments. 1972;43:404–409. doi: 10.1063/1.1685647. [DOI] [PubMed] [Google Scholar]
- 10.Gossett DR, Weaver WM, Mach AJ, Hur SC, Tse HT, Lee W, Amini H, Di Carlo D. Analytical and bioanalytical chemistry. 2010;397:3249–3267. doi: 10.1007/s00216-010-3721-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Piyasena ME, Graves SW. Lab on a Chip. 2014;14:1044–1059. doi: 10.1039/c3lc51152a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miltenyi S, Muller W, Weichel W, Radbruch A. Cytometry. 1990;11:231–238. doi: 10.1002/cyto.990110203. [DOI] [PubMed] [Google Scholar]
- 13.El-Ali J, Sorger PK, Jensen KF. Nature. 2006;442:403–411. doi: 10.1038/nature05063. [DOI] [PubMed] [Google Scholar]
- 14.Zhao M, Schiro PG, Kuo JS, Koehler KM, Sabath DE, Popov V, Feng Q, Chiu DT. Analytical chemistry. 2013;85:2465–2471. doi: 10.1021/ac400193b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang S, Undar A, Zahn JD. Lab Chip. 2006;6:871–880. doi: 10.1039/b516401j. [DOI] [PubMed] [Google Scholar]
- 16.Mernier G, Piacentini N, Braschler T, Demierre N, Renaud P. Lab Chip. 2010;10:2077–2082. doi: 10.1039/c000977f. [DOI] [PubMed] [Google Scholar]
- 17.Sheng W, Ogunwobi OO, Chen T, Zhang J, George TJ, Liu C, Fan ZH. Lab Chip. 2014;14:89–98. doi: 10.1039/c3lc51017d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nan L, Jiang Z, Wei X. Lab Chip. 2014;14:1060–1073. doi: 10.1039/c3lc51133b. [DOI] [PubMed] [Google Scholar]
- 19.Nilsson J, Evander M, Hammarstrom B, Laurell T. Analytica chimica acta. 2009;649:141–157. doi: 10.1016/j.aca.2009.07.017. [DOI] [PubMed] [Google Scholar]
- 20.Voldman J. Annual review of biomedical engineering. 2006;8:425–454. doi: 10.1146/annurev.bioeng.8.061505.095739. [DOI] [PubMed] [Google Scholar]
- 21.Gonzalez CF, Remcho VT. Journal of Chromatography A. 2005;1079:59–68. doi: 10.1016/j.chroma.2005.03.070. [DOI] [PubMed] [Google Scholar]
- 22.Takahashi K, Hattori A, Suzuki I, Ichiki T, Yasuda K. Journal of nanobiotechnology. 2004:2. doi: 10.1186/1477-3155-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yao B, Luo G, Feng X, Wang W, Chen L, Wang Y. Lab on a Chip. 2004;4:603–607. doi: 10.1039/b408422e. [DOI] [PubMed] [Google Scholar]
- 24.Guo F, Ji X-H, Liu K, He R-X, Zhao L-B, Guo Z-X, Liu W, Guo S-S, Zhao X-Z. Applied Physics Letters. 2010;96:193701. [Google Scholar]
- 25.Lenshof A, Laurell T. Chemical Society reviews. 2010;39:1203–1217. doi: 10.1039/b915999c. [DOI] [PubMed] [Google Scholar]
- 26.Wang L, Flanagan LA, Monuki E, Jeon NL, Lee AP. Lab Chip. 2007;7:1114–1120. doi: 10.1039/b705386j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baret JC, Miller OJ, Taly V, Ryckelynck M, El-Harrak A, Frenz L, Rick C, Samuels ML, Hutchison JB, Agresti JJ, Link DR, Weitz DA, Griffiths AD. Lab Chip. 2009;9:1850–1858. doi: 10.1039/b902504a. [DOI] [PubMed] [Google Scholar]
- 28.Agresti JJ, Antipov E, Abate AR, Ahn K, Rowat AC, Baret JC, Marquez M, Klibanov AM, Griffiths AD, Weitz DA. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:4004–4009. doi: 10.1073/pnas.0910781107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mazutis L, Gilbert J, Ung WL, Weitz DA, Griffiths AD, Heyman JA. Nature protocols. 2013;8:870–891. doi: 10.1038/nprot.2013.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Velev OD, Bhatt KH. Soft Matter. 2006;2:738. doi: 10.1039/b605052b. [DOI] [PubMed] [Google Scholar]
- 31.Fu AY, Spence C, Scherer A, Arnold FH, Quake SR. Nature Biotechnology. 1999;17:1109–1111. doi: 10.1038/15095. [DOI] [PubMed] [Google Scholar]
- 32.Dittrich PS, Schwille P. Analytical chemistry. 2003;75:5767–5774. doi: 10.1021/ac034568c. [DOI] [PubMed] [Google Scholar]
- 33.Sun Y, Lim CS, Liu AQ, Ayi TC, Yap PH. Sensors and Actuators A: Physical. 2007;133:340–348. [Google Scholar]
- 34.Erlandsson PG, Robinson ND. Electrophoresis. 2011;32:784–790. doi: 10.1002/elps.201000617. [DOI] [PubMed] [Google Scholar]
- 35.Valagerahally Puttaswamy S, Sivashankar S, Yeh C-H, Chen R-J, Liu CH. Microelectronic Engineering. 2010;87:2582–2591. [Google Scholar]
- 36.Ward M, Turner P, DeJohn M, Kaduchak G. 2009 [Google Scholar]
- 37.Austin Suthanthiraraj PP, Piyasena ME, Woods TA, Naivar MA, Lomicronpez GP, Graves SW. Methods. 2012;57:259–271. doi: 10.1016/j.ymeth.2012.02.013. [DOI] [PubMed] [Google Scholar]
- 38.Kaduchak G, Ward MD. 2013 [Google Scholar]
- 39.Lenshof A, Magnusson C, Laurell T. Lab Chip. 2012;12:1210–1223. doi: 10.1039/c2lc21256k. [DOI] [PubMed] [Google Scholar]
- 40.Burguillos MA, Magnusson C, Nordin M, Lenshof A, Augustsson P, Hansson MJ, Elmer E, Lilja H, Brundin P, Laurell T, Deierborg T. PloS one. 2013;8:e64233. doi: 10.1371/journal.pone.0064233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laurell T, Petersson F, Nilsson A. Chemical Society reviews. 2007;36:492–506. doi: 10.1039/b601326k. [DOI] [PubMed] [Google Scholar]
- 42.Johansson L, Nikolajeff F, Johansson S, Thorslund S. Analytical chemistry. 2009;81:5188–5196. doi: 10.1021/ac802681r. [DOI] [PubMed] [Google Scholar]
- 43.Ding X, Li P, Lin SC, Stratton ZS, Nama N, Guo F, Slotcavage D, Mao X, Shi J, Costanzo F, Huang TJ. Lab Chip. 2013;13:3626–3649. doi: 10.1039/c3lc50361e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cho SH, Chen CH, Tsai FS, Godin JM, Lo YH. Lab Chip. 2010;10:1567–1573. doi: 10.1039/c000136h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gor’kov LP. Soviet Physics Doklady. 1962;6:773–775. [Google Scholar]
- 46.Bruus H. Lab Chip. 2012;12:1014–1021. doi: 10.1039/c2lc21068a. [DOI] [PubMed] [Google Scholar]
- 47.Augustsson P, Laurell T. Lab Chip. 2012;12:1742–1752. doi: 10.1039/c2lc40200a. [DOI] [PubMed] [Google Scholar]
- 48.Goddard G, Martin JC, Graves SW, Kaduchak G. Cytometry. Part A : the journal of the International Society for Analytical Cytology. 2006;69:66–74. doi: 10.1002/cyto.a.20205. [DOI] [PubMed] [Google Scholar]
- 49.Jakobsson O, Grenvall C, Nordin M, Evander M, Laurell T. Lab Chip. 2014;14:1943–1950. doi: 10.1039/c3lc51408k. [DOI] [PubMed] [Google Scholar]
- 50.Shields CW, IV, Johnson LM, Gao L, López GP. Langmuir. 2014;30:3923–3927. doi: 10.1021/la404677w. [DOI] [PubMed] [Google Scholar]
- 51.Shi J, Huang H, Stratton Z, Huang Y, Huang TJ. Lab Chip. 2009;9:3354–3359. doi: 10.1039/b915113c. [DOI] [PubMed] [Google Scholar]
- 52.Wang Z, Zhe J. Lab Chip. 2011;11:1280–1285. doi: 10.1039/c0lc00527d. [DOI] [PubMed] [Google Scholar]
- 53.Lin SC, Mao X, Huang TJ. Lab Chip. 2012;12:2766–2770. doi: 10.1039/c2lc90076a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ding X, Lin SC, Lapsley MI, Li S, Guo X, Chan CY, Chiang IK, Wang L, McCoy JP, Huang TJ. Lab Chip. 2012;12:4228–4231. doi: 10.1039/c2lc40751e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li S, Ding X, Guo F, Chen Y, Lapsley MI, Lin SC, Wang L, McCoy JP, Cameron CE, Huang TJ. Analytical chemistry. 2013;85:5468–5474. doi: 10.1021/ac400548d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ding X, Lin SCS, Kiraly B, Yue H, Li S, Chiang IK, Shi J, Benkovic SJ, Huang TJ. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:11105–11109. doi: 10.1073/pnas.1209288109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Franke T, Braunmuller S, Schmid L, Wixforth A, Weitz DA. Lab Chip. 2010;10:789–794. doi: 10.1039/b915522h. [DOI] [PubMed] [Google Scholar]
- 58.Schmid L, Weitz DA, Franke T. Lab Chip. 2014;5:3710–3718. doi: 10.1039/c4lc00588k. [DOI] [PubMed] [Google Scholar]
- 59.Jonas A, Zemanek P. Electrophoresis. 2008;29:4813–4851. doi: 10.1002/elps.200800484. [DOI] [PubMed] [Google Scholar]
- 60.Neuman KC, Nagy A. Nature methods. 2008;5:491–505. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moffitt JR, Chemla YR, Smith SB, Bustamante C. Annual review of biochemistry. 2008;77:205–228. doi: 10.1146/annurev.biochem.77.043007.090225. [DOI] [PubMed] [Google Scholar]
- 62.Ashkin A. Physical Review Letters. 1970;24:156–159. [Google Scholar]
- 63.Buican T, Smyth M, Crissman H, Salzman G, Stewart C, Martin J. Applied Optics. 1987;26:5311–5316. doi: 10.1364/AO.26.005311. [DOI] [PubMed] [Google Scholar]
- 64.Applegate RW, Jr, Squier J, Vestad T, Oakey J, Marr DW. Optics Express. 2004;12:4390–4398. doi: 10.1364/opex.12.004390. [DOI] [PubMed] [Google Scholar]
- 65.Wang MM, Tu E, Raymond DE, Yang JM, Zhang H, Hagen N, Dees B, Mercer EM, Forster AH, Kariv I, Marchand PJ, Butler WF. Nat Biotechnol. 2005;23:83–87. doi: 10.1038/nbt1050. [DOI] [PubMed] [Google Scholar]
- 66.Applegate RW, Jr, Squier J, Vestad T, Oakey J, Marr DW, Bado P, Dugan MA, Said AA. Lab Chip. 2006;6:422–426. doi: 10.1039/b512576f. [DOI] [PubMed] [Google Scholar]
- 67.Perroud TD, Kalser JN, Sy JC, Lane TW, Branda CS, Singh AK, Patel KD. Analytical Chemistry. 2008;90:6365–6372. doi: 10.1021/ac8007779. [DOI] [PubMed] [Google Scholar]
- 68.Wang X, Chen S, Kong M, Wang Z, Costa KD, Li RA, Sun D. Lab Chip. 2011;11:3656–3662. doi: 10.1039/c1lc20653b. [DOI] [PubMed] [Google Scholar]
- 69.Wu TH, Chen Y, Park SY, Hong J, Teslaa T, Zhong JF, Di Carlo D, Teitell MA, Chiou PY. Lab Chip. 2012;12:1378–1383. doi: 10.1039/c2lc21084c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen Y, Chung AJ, Wu TH, Teitell MA, Di Carlo D, Chiou PY. Small. 2014;10:1746–1751. doi: 10.1002/smll.201302885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kruger J, Singh K, O’Neill A, Jackson C, Morrison A, O’Brien P. Journal of Micromechanics and Microengineering. 2002;12:486–494. [Google Scholar]
- 72.Fu AY, Chou HP, Spence C, Arnold FH, Quake SR. Analytical Chemistry. 2002;74:2451–2457. doi: 10.1021/ac0255330. [DOI] [PubMed] [Google Scholar]
- 73.Wolff A, Perch-Nielsen IR, Larsen UD, Friis P, Goranovic G, Poulsen CR, Kutter JP, Telleman P. Lab Chip. 2003;3:22–27. doi: 10.1039/b209333b. [DOI] [PubMed] [Google Scholar]
- 74.Chen P, Feng X, Hu R, Sun J, Du W, Liu BF. Analytica chimica acta. 2010;663:1–6. doi: 10.1016/j.aca.2010.01.046. [DOI] [PubMed] [Google Scholar]
- 75.Ho CT, Lin RZ, Chang HY, Liu CH. Lab Chip. 2005;5:1248–1258. doi: 10.1039/b507575k. [DOI] [PubMed] [Google Scholar]
- 76.Shirasaki Y, Tanaka J, Makazu H, Tashiro K, Shoji S, Tsukita S, Funatsu T. Analytical chemistry. 2008;78:695–701. doi: 10.1021/ac0511041. [DOI] [PubMed] [Google Scholar]
- 77.Zborowski M, Chalmers JJ. Analytical chemistry. 2011;83:8050–8056. doi: 10.1021/ac200550d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gao Y, Jian YC, Zhang LF, Huang JP. J. Phys. Chem. C. 2007;111:10785–10791. [Google Scholar]
- 79.Pamme N, Wilhelm C. Lab Chip. 2006;6:974–980. doi: 10.1039/b604542a. [DOI] [PubMed] [Google Scholar]
- 80.Xia N, Hunt TP, Mayers BT, Alsberg E, Whitesides GM, Westervelt RM, Ingber DE. Biomedical microdevices. 2006;8:299–308. doi: 10.1007/s10544-006-0033-0. [DOI] [PubMed] [Google Scholar]
- 81.Lai JJ, Nelson KE, Nash MA, Hoffman AS, Yager P, Stayton PS. Lab Chip. 2009;9:1997–2002. doi: 10.1039/b817754f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Adams JD, Kim U, Soh HT. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:18165–18170. doi: 10.1073/pnas.0809795105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Carr C, Espy M, Nath P, Martin SL, Ward MD, Martin J. J Magn Magn Mater. 2009;321:1440–1445. doi: 10.1016/j.jmmm.2009.02.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yu M, Stott S, Toner M, Maheswaran S, Haber DA. The Journal of cell biology. 2011;192:373–382. doi: 10.1083/jcb.201010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hoshino K, Huang YY, Lane N, Huebschman M, Uhr JW, Frenkel EP, Zhang X. Lab Chip. 2011;11:3449–3457. doi: 10.1039/c1lc20270g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hoshino K, Chen P, Huang YY, Zhang X. Analytical chemistry. 2012;84:4292–4299. doi: 10.1021/ac2032386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Darabi J, Guo C. Biomicrofluidics. 2013;7:54106. doi: 10.1063/1.4821628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kang JH, Krause S, Tobin H, Mammoto A, Kanapathipillai M, Ingber DE. Lab Chip. 2012;12:2175–2181. doi: 10.1039/c2lc40072c. [DOI] [PubMed] [Google Scholar]
- 89.Shields CW, IV, Livingston CE, Yellen BB, López GP, Murdoch DM. Biomicrofluidics. 2014;8:041101. doi: 10.1063/1.4885840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ozkumur E, Shah AM, Ciciliano JC, Emmink BL, Miyamoto DT, Brachtel E, Yu M, Chen PI, Morgan B, Trautwein J, Kimura A, Sengupta S, Stott SL, Karabacak NM, Barber TA, Walsh JR, Smith K, Spuhler PS, Sullivan JP, Lee RJ, Ting DT, Luo X, Shaw AT, Bardia A, Sequist LV, Louis DN, Maheswaran S, Kapur R, Haber DA, Toner M. Science translational medicine. 2013;5:179ra147. doi: 10.1126/scitranslmed.3005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen A, Byvank T, Chang WJ, Bharde A, Vieira G, Miller BL, Chalmers JJ, Bashir R, Sooryakumar R. Lab Chip. 2013;13:1172–1181. doi: 10.1039/c2lc41201b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Karabacak NM, Spuhler PS, Fachin F, Lim EJ, Pai V, Ozkumur E, Martel JM, Kojic N, Smith K, Chen PI, Yang J, Hwang H, Morgan B, Trautwein J, Barber TA, Stott SL, Maheswaran S, Kapur R, Haber DA, Toner M. Nature protocols. 2014;9:694–710. doi: 10.1038/nprot.2014.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cushing KW, Piyasena ME, Carroll NJ, Maestas GC, Lopez BA, Edwards BS, Graves SW, Lopez GP. Analytical chemistry. 2013;85:2208–2215. doi: 10.1021/ac3029344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Johnson LM, Gao L, Shields CW, IV, Smith M, Efimenko K, Cushing K, Genzer J, Lopez GP. Journal of nanobiotechnology. 2013;11:22. doi: 10.1186/1477-3155-11-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shields CW, IV, Sun D, Johnson KA, Duval KA, Rodriguez AV, Gao L, Dayton PA, Lopez GP. Angew Chem Int Ed Engl. 2014;53:8070–8073. doi: 10.1002/anie.201402471. [DOI] [PubMed] [Google Scholar]
- 96.Lenshof A, Jamal A, Dykes J, Urbansky A, Åstrand-Grundström I, Laurell T, Scheding S. Cytometry Part A. 2014;85:933–941. doi: 10.1002/cyto.a.22507. [DOI] [PubMed] [Google Scholar]
- 97.Hu X, Bessette PH, Qian J, Meinhart CD, Daugherty PS, Soh HT. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15757–15761. doi: 10.1073/pnas.0507719102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kim U, Qian J, Kenrick SA, Daugherty PS, Soh HT. Analytical chemistry. 2008;80:8656–8661. doi: 10.1021/ac8015938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kim U, Soh HT. Lab Chip. 2009;9:2313–2318. doi: 10.1039/b903950c. [DOI] [PubMed] [Google Scholar]
- 100.Cheng IF, Chang H-C, Hou D, Chang H-C. Biomicrofluidics. 2007;1:021503. [Google Scholar]
- 101.Coulter W. Proc Natl Electron Conf. 1957;12:1034–1040. [Google Scholar]
- 102.Fulwyler MJ. Science. 1965;150:910–911. doi: 10.1126/science.150.3698.910. [DOI] [PubMed] [Google Scholar]
- 103.Petersson F, Nilsson A, Holm C, Jonsson H, Laurell T. The Analyst. 2004;129:938–943. doi: 10.1039/b409139f. [DOI] [PubMed] [Google Scholar]
- 104.Petersson F, Nilsson A, Holm C, Jonsson H, Laurell T. Lab Chip. 2005;5:20–22. doi: 10.1039/b405748c. [DOI] [PubMed] [Google Scholar]
- 105.Petersson F, Aberg L, Sward-Nilsson AM, Laurell T. Analytical Chemistry. 2007;79:5117–5123. doi: 10.1021/ac070444e. [DOI] [PubMed] [Google Scholar]
- 106.Dykes J, Lenshof A, Astrand-Grundstrom I, Laurell T, Scheding S. PloS one. 2011;6:e23074. doi: 10.1371/journal.pone.0023074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Augustsson P, Magnusson C, Nordin M, Lilja H, Laurell T. Anal. Chem. 2012;84:7954–7962. doi: 10.1021/ac301723s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yang AH, Soh HT. Analytical chemistry. 2012;84:10756–10762. doi: 10.1021/ac3026674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fong EJ, Johnston AC, Notton T, Jung SY, Rose KA, Weinberger LS, Shusteff M. The Analyst. 2014;139:1192–1200. doi: 10.1039/c4an00034j. [DOI] [PubMed] [Google Scholar]
- 110.Nam J, Lim H, Kim D, Shin S. Lab Chip. 2011;11:3361–3364. doi: 10.1039/c1lc20346k. [DOI] [PubMed] [Google Scholar]
- 111.Ding X, Peng Z, Lin SC, Geri M, Li S, Li P, Chen Y, Dao M, Suresh S, Huang TJ. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:12992–12997. doi: 10.1073/pnas.1413325111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li S, Ding X, Mao Z, Chen Y, Nama N, Guo F, Li P, Wang L, Cameron CE, Huang TJ. Lab Chip. 2014;15:331–338. doi: 10.1039/c4lc00903g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Huang Y, Joo S, Duhon M, Heller M, Wallace B, Xu X. Analytical chemistry. 2002;74:3362–3371. doi: 10.1021/ac011273v. [DOI] [PubMed] [Google Scholar]
- 114.Cummings EB, Singh AK. Analytical Chemistry. 2003;75:4724–4731. doi: 10.1021/ac0340612. [DOI] [PubMed] [Google Scholar]
- 115.Barrett LM, Skulan AJ, Singh AK, Cummings EB, Fiechtner GJ. Analytical chemistry. 2005;77:6798–6804. doi: 10.1021/ac0507791. [DOI] [PubMed] [Google Scholar]
- 116.Vahey MD, Voldman J. Analytical Chemistry. 2008;80:3135–3143. doi: 10.1021/ac7020568. [DOI] [PubMed] [Google Scholar]
- 117.Roda B, Zattoni A, Reschiglian P, Moon MH, Mirasoli M, Michelini E, Roda A. Analytica chimica acta. 2009;635:132–143. doi: 10.1016/j.aca.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 118.Giddings JC. Science. 1993;260:1456–1465. doi: 10.1126/science.8502990. [DOI] [PubMed] [Google Scholar]
- 119.Giddings JC. Separation Science and Technology. 1985;20:749–768. [Google Scholar]
- 120.Vykoukal J, Vykoukal DM, Freyberg S, Alt EU, Gascoyne PR. Lab Chip. 2008;8:1386–1393. doi: 10.1039/b717043b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cui HH, Voldman J, He XF, Lim KM. Lab Chip. 2009;9:2306–2312. doi: 10.1039/b906202e. [DOI] [PubMed] [Google Scholar]
- 122.Karimi A, Yazdi S, Ardekani AM. Biomicrofluidics. 2013;7:21501. doi: 10.1063/1.4799787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Doh I, Cho Y-H. Sensors and Actuators A: Physical. 2005;121:59–65. [Google Scholar]
- 124.Moon HS, Kwon K, Kim SI, Han H, Sohn J, Lee S, Jung HI. Lab Chip. 2011;11:1118–1125. doi: 10.1039/c0lc00345j. [DOI] [PubMed] [Google Scholar]
- 125.Shim S, Stemke-Hale K, Tsimberidou AM, Noshari J, Anderson TE, Gascoyne PR. Biomicrofluidics. 2013;7:11807. doi: 10.1063/1.4774304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kim U, Shu CW, Dane KY, Daugherty PS, Wang JY, Soh HT. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:20708–20712. doi: 10.1073/pnas.0708760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Valero A, Braschler T, Demierre N, Renaud P. Biomicrofluidics. 2010;4:022807. doi: 10.1063/1.3430542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sun J, Gao Y, Isaacs RJ, Boelte KC, Lin PC, Boczko EM, Li D. Analytical chemistry. 2012;84:2017–2024. doi: 10.1021/ac203212g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Toner M, Irimia D. Annual review of biomedical engineering. 2005;7:77–103. doi: 10.1146/annurev.bioeng.7.011205.135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hou HW, Bhagat AAS, Lee WC, Huang S, Han J, Lim CT. Micromachines. 2011;2:319–343. [Google Scholar]
- 131.Melville D, Paul F, Roath S. Nature. 1975;255:706. [Google Scholar]
- 132.Zborowski M, Ostera GR, Moore LR, Miliron S, Chalmers JJ, Schechter AN. Biophysical Journal. 2003;84:2638–2645. doi: 10.1016/S0006-3495(03)75069-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Han K-H, Bruno Frazier A. Journal of Applied Physics. 2004;96:5797. [Google Scholar]
- 134.Han K-H, Frazier AB. Journal of Microelectromechanical Systems. 2005;14:1422–1431. [Google Scholar]
- 135.Han KH, Frazier AB. Lab Chip. 2006;6:265–273. doi: 10.1039/b514539b. [DOI] [PubMed] [Google Scholar]
- 136.Jung J, Han K-H. Applied Physics Letters. 2008;93:223902. [Google Scholar]
- 137.Qu BY, Wu ZY, Fang F, Bai ZM, Yang DZ, Xu SK. Analytical and bioanalytical chemistry. 2008;392:1317–1324. doi: 10.1007/s00216-008-2382-4. [DOI] [PubMed] [Google Scholar]
- 138.Seo H-K, Kim Y-H, Kim H-O, Kim Y-J. Journal of Micromechanics and Microengineering. 2010;20:095019. [Google Scholar]
- 139.Furlani EP. Journal of Physics D: Applied Physics. 2007;40:1313–1319. [Google Scholar]
- 140.Robert D, Pamme N, Conjeaud H, Gazeau F, Iles A, Wilhelm C. Lab Chip. 2011;11:1902–1910. doi: 10.1039/c0lc00656d. [DOI] [PubMed] [Google Scholar]
- 141.Zhu T, Cheng R, Lee SA, Rajaraman E, Eiteman MA, Querec TD, Unger ER, Mao L. Microfluidics and Nanofluidics. 2012;13:645–654. doi: 10.1007/s10404-012-1004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zeng J, Deng Y, Vedantam P, Tzeng T-R, Xuan X. Journal of Magnetism and Magnetic Materials. 2013;346:118–123. [Google Scholar]
- 143.Shen F, Hwang H, Hahn YK, Park JK. Analytical chemistry. 2012;84:3075–3081. doi: 10.1021/ac201505j. [DOI] [PubMed] [Google Scholar]
- 144.Hoi SK, Udalagama C, Sow CH, Watt F, Bettiol AA. Applied Physics B. 2009;97:859–865. [Google Scholar]
- 145.Lau AY, Lee LP, Chan JW. Lab Chip. 2008;8:1116–1120. doi: 10.1039/b803598a. [DOI] [PubMed] [Google Scholar]
- 146.MacDonald MP, Spalding GC, Dholakia K. Nature. 2003;426:421–424. doi: 10.1038/nature02144. [DOI] [PubMed] [Google Scholar]
- 147.Wu MC. Nature Photonics. 2011;5:322–324. [Google Scholar]
- 148.Chiou PY, Ohta AT, Wu MC. Nature. 2005;436:370–372. doi: 10.1038/nature03831. [DOI] [PubMed] [Google Scholar]
- 149.Huang SB, Chen J, Wang J, Yang CL, Wu MH. International Journal of Electrochemical Science. 2012;7:12656–12667. [Google Scholar]
- 150.Huang SB, Wu MH, Lin YH, Hsieh CH, Yang CL, Lin HC, Tseng CP, Lee GB. Lab Chip. 2013;13:1371–1383. doi: 10.1039/c3lc41256c. [DOI] [PubMed] [Google Scholar]
- 151.Ohta AT, Chiou PY, Phan HL, Sherwood SW, Yang JM, Lau ANK, Hsu HY, Jamshidi A, Wu MC. Journal of Selected Topics in Quantum Electronics - IEEE. 2007;13:235–243. [Google Scholar]
- 152.Shah GJ, Ohta AT, Chiou EPY, Wu MC, Kim CJ. Lab on a chip. 2009;9:1732–1739. doi: 10.1039/b821508a. [DOI] [PubMed] [Google Scholar]
- 153.Huang KW, Wu YC, Lee JA, Chiou PY. Lab Chip. 2013;13:3721–3727. doi: 10.1039/c3lc50607j. [DOI] [PubMed] [Google Scholar]
- 154.Sollier E, Rostaing H, Pouteau P, Fouillet Y, Achard J-L. Sensors and Actuators B: Chemical. 2009;141:617–624. [Google Scholar]
- 155.Tsutsui H, Ho CM. Mechanics research communications. 2009;36:92–103. doi: 10.1016/j.mechrescom.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Di Carlo D. Lab Chip. 2009;9:3038–3046. doi: 10.1039/b912547g. [DOI] [PubMed] [Google Scholar]
- 157.Di Carlo D, Irimia D, Tompkins RG, Toner M. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:18892–18897. doi: 10.1073/pnas.0704958104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Di Carlo D, Edd JF, Irimia D, Tompkins RG, Toner M. Analytical Chemistry. 2008;80:2204–2211. doi: 10.1021/ac702283m. [DOI] [PubMed] [Google Scholar]
- 159.Oakey J, Applegate RW, Jr, Arellano E, Di Carlo D, Graves SW, Toner M. Analytical chemistry. 2010;82:3862–3867. doi: 10.1021/ac100387b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Russom A, Gupta AK, Nagrath S, Di Carlo D, Edd JF, Toner M. New J Phys. 2009;11:75025. doi: 10.1088/1367-2630/11/7/075025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kuntaegowdanahalli SS, Bhagat AA, Kumar G, Papautsky I. Lab Chip. 2009;9:2973–2980. doi: 10.1039/b908271a. [DOI] [PubMed] [Google Scholar]
- 162.Hou HW, Warkiani ME, Khoo BL, Li ZR, Soo RA, Tan DS-W, Lim W-T, Han J, Bhagat AAS, Lim CT. Scientific Reports. 2013;3:1–8. doi: 10.1038/srep01259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Nivedita N, Papautsky I. Biomicrofluidics. 2013;7:54101. doi: 10.1063/1.4819275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Guan G, Wu L, Bhagat AA, Li Z, Chen PC, Chao S, Ong CJ, Han J. Sci Rep. 2013;3:1475. doi: 10.1038/srep01475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Warkiani ME, Guan G, Luan KB, Lee WC, Bhagat AA, Chaudhuri PK, Tan DS, Lim WT, Lee SC, Chen PC, Lim CT, Han J. Lab Chip. 2014;14:128–137. doi: 10.1039/c3lc50617g. [DOI] [PubMed] [Google Scholar]
- 166.Parichehreh V, Medepallai K, Babbarwal K, Sethu P. Lab Chip. 2013;13:892–900. doi: 10.1039/c2lc40663b. [DOI] [PubMed] [Google Scholar]
- 167.Yamada M, Nakashima M, Seki M. Analytical Chemistry. 2004;76:5465–5471. doi: 10.1021/ac049863r. [DOI] [PubMed] [Google Scholar]
- 168.Takagi J, Yamada M, Yasuda M, Seki M. Lab Chip. 2005;5:778–784. doi: 10.1039/b501885d. [DOI] [PubMed] [Google Scholar]
- 169.Huh D, Bahng JH, Ling Y, Wei H, Kripfgans OD, Fowlkes JB, Grotberg JB, Takayama S. Analytical Chemistry. 2007;79:1369–1376. doi: 10.1021/ac061542n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zhang X, Cooper JM, Monaghan PB, Haswell SJ. Lab Chip. 2006;6:561–566. doi: 10.1039/b515272k. [DOI] [PubMed] [Google Scholar]
- 171.Wu Z, Willing B, Bjerketorp J, Jansson JK, Hjort K. Lab on a chip. 2009;9:1193–1199. doi: 10.1039/b817611f. [DOI] [PubMed] [Google Scholar]
- 172.Wu Z, Liu AQ, Hjort K. Journal of Micromechanics and Microengineering. 2007;17:1992–1999. [Google Scholar]
- 173.Mach AJ, Di Carlo D. Biotechnology and bioengineering. 2010;107:302–311. doi: 10.1002/bit.22833. [DOI] [PubMed] [Google Scholar]
- 174.Bhagat AAS, Kuntaegowdanahalli SS, Papautsky I. Physics of Fluids. 2008;20:101702. [Google Scholar]
- 175.Zhou J, Giridhar PV, Kasper S, Papautsky I. Lab Chip. 2013;13:1919–1929. doi: 10.1039/c3lc50101a. [DOI] [PubMed] [Google Scholar]
- 176.Hur SC, Henderson-MacLennan NK, McCabe ER, Di Carlo D. Lab Chip. 2011;11:912–920. doi: 10.1039/c0lc00595a. [DOI] [PubMed] [Google Scholar]
- 177.Geislinger TM, Franke T. Biomicrofluidics. 2013;7:44120. doi: 10.1063/1.4818907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Geislinger TM, Eggart B, Braunmuüller S, Schmid L, Franke T. Applied Physics Letters. 2012;100:183701. [Google Scholar]
- 179.Beech JP, Holm SH, Adolfsson K, Tegenfeldt JO. Lab Chip. 2012;12:1048–1051. doi: 10.1039/c2lc21083e. [DOI] [PubMed] [Google Scholar]
- 180.Huang LR, Cox EC, Austin RH, Sturm JC. Science. 2004;304:987–990. doi: 10.1126/science.1094567. [DOI] [PubMed] [Google Scholar]
- 181.Inglis DW, Davis JA, Austin RH, Sturm JC. Lab Chip. 2006;6:655–658. doi: 10.1039/b515371a. [DOI] [PubMed] [Google Scholar]
- 182.Davis JA, Inglis DW, Morton KJ, Lawrence DA, Huang LR, Chou SY, Sturm JC, Austin RH. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:14779–14784. doi: 10.1073/pnas.0605967103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Huang R, Barber TA, Schmidt MA, Tompkins RG, Toner M, Bianchi DW, Kapur R, Flejter WL. Prenatal diagnosis. 2008;28:892–899. doi: 10.1002/pd.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Liu Z, Huang F, Du J, Shu W, Feng H, Xu X, Chen Y. Biomicrofluidics. 2013;7:11801. doi: 10.1063/1.4774308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Choi S, Park JK. Lab Chip. 2007;7:890–897. doi: 10.1039/b701227f. [DOI] [PubMed] [Google Scholar]
- 186.Choi S, Song S, Choi C, Park JK. Lab Chip. 2007;7:1532–1538. doi: 10.1039/b705203k. [DOI] [PubMed] [Google Scholar]
- 187.Hsu CH, Di Carlo D, Chen C, Irimia D, Toner M. Lab Chip. 2008;8:2128–2134. doi: 10.1039/b813434k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Stott SL, Hsu CH, Tsukrov DI, Yu M, Miyamoto DT, Waltman BA, Rothenberg SM, Shah AM, Smas ME, Korir GK, Floyd FP, Jr, Gilman AJ, Lord JB, Winokur D, Springer S, Irimia D, Nagrath S, Sequist LV, Lee RJ, Isselbacher KJ, Maheswaran S, Haber DA, Toner M. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:18392–18397. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Sollier E, Go DE, Che J, Gossett DR, O'Byrne S, Weaver WM, Kummer N, Rettig M, Goldman J, Nickols N, McCloskey S, Kulkarni RP, Di Carlo D. Lab Chip. 2014;14:63–77. doi: 10.1039/c3lc50689d. [DOI] [PubMed] [Google Scholar]
- 190.Hyun KA, Kwon K, Han H, Kim SI, Jung HI. Biosensors & bioelectronics. 2013;40:206–212. doi: 10.1016/j.bios.2012.07.021. [DOI] [PubMed] [Google Scholar]
- 191.Mohamed H, Turner JN, Caggana M. Journal of chromatography. A. 2007;1162:187–192. doi: 10.1016/j.chroma.2007.06.025. [DOI] [PubMed] [Google Scholar]
- 192.McFaul SM, Lin BK, Ma H. Lab Chip. 2012;12:2369–2376. doi: 10.1039/c2lc21045b. [DOI] [PubMed] [Google Scholar]
- 193.Preira P, Grandne V, Forel JM, Gabriele S, Camara M, Theodoly O. Lab Chip. 2013;13:161–170. doi: 10.1039/c2lc40847c. [DOI] [PubMed] [Google Scholar]
- 194.Ji HM, Samper V, Chen Y, Heng CK, Lim TM, Yobas L. Biomedical microdevices. 2008;10:251–257. doi: 10.1007/s10544-007-9131-x. [DOI] [PubMed] [Google Scholar]
- 195.Earhart CM, Hughes CE, Gaster RS, Ooi CC, Wilson RJ, Zhou LY, Humke EW, Xu L, Wong DJ, Willingham SB, Schwartz EJ, Weissman IL, Jeffrey SS, Neal JW, Rohatgi R, Wakelee HA, Wang SX. Lab Chip. 2014;14:78–88. doi: 10.1039/c3lc50580d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Murthy SK, Sethu P, Vunjak-Novakovic G, Toner M, Radisic M. Biomedical microdevices. 2006;8:231–237. doi: 10.1007/s10544-006-8169-5. [DOI] [PubMed] [Google Scholar]
- 197.Sethu P, Sin A, Toner M. Lab Chip. 2006;6:83–89. doi: 10.1039/b512049g. [DOI] [PubMed] [Google Scholar]
- 198.VanDelinder V, Groisman A. Analytical Chemistry. 2006;78:3765–3771. doi: 10.1021/ac060042r. [DOI] [PubMed] [Google Scholar]
- 199.VanDelinder V, Groisman A. Analytical Chemistry. 2007;79:2023–2030. doi: 10.1021/ac061659b. [DOI] [PubMed] [Google Scholar]
- 200.Chen X, Cui D, Liu C, Li H. Sensors and Actuators B: Chemical. 2008;130:216–221. [Google Scholar]
- 201.Chen X, Cui D, Liu C, Li H, Chen J. Analytica chimica acta. 2007;584:237–243. doi: 10.1016/j.aca.2006.11.057. [DOI] [PubMed] [Google Scholar]
- 202.Yamada M, Kano K, Tsuda Y, Kobayashi J, Yamato M, Seki M, Okano T. Biomedical microdevices. 2007;9:637–645. doi: 10.1007/s10544-007-9055-5. [DOI] [PubMed] [Google Scholar]
- 203.Yamada M, Seki M. Lab Chip. 2005;5:1233–1239. doi: 10.1039/b509386d. [DOI] [PubMed] [Google Scholar]
- 204.Yamada M, Seki M. Analytical chemistry. 2006;78:1357–1362. doi: 10.1021/ac0520083. [DOI] [PubMed] [Google Scholar]
- 205.Mizuno M, Yamada M, Mitamura R, Ike K, Toyama K, Seki M. Analytical chemistry. 2013;85:7666–7673. doi: 10.1021/ac303336f. [DOI] [PubMed] [Google Scholar]
- 206.Didar TF, Tabrizian M. Lab Chip. 2010;10:3043–3053. doi: 10.1039/c0lc00130a. [DOI] [PubMed] [Google Scholar]
- 207.Lee CH, Bose S, Van Vliet KJ, Karp JM, Karnik R. Langmuir. 2011;27:240–249. doi: 10.1021/la102871m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Bose S, Singh R, Hanewich-Hollatz M, Shen C, Lee CH, Dorfman DM, Karp JM, Karnik R. Sci Rep. 2013;3:2329. doi: 10.1038/srep02329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Choi S, Karp JM, Karnik R. Lab Chip. 2012;12:1427–1430. doi: 10.1039/c2lc21225k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Choi S, Levy O, Coelho MB, Cabral JM, Karp JM, Karnik R. Lab Chip. 2014;14:161–166. doi: 10.1039/c3lc50923k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Baran MM, Allen DM, Russell SR, Scheetz II ME, JMonthony JF. Journal of Immunological Methods. 1982;53:321–334. doi: 10.1016/0022-1759(82)90179-x. [DOI] [PubMed] [Google Scholar]
- 212.Hertz CM, Graves DJ, Lauffenburger DA, Serota FT. Biotechnology and Bioengineering. 1984;27:603–612. doi: 10.1002/bit.260270509. [DOI] [PubMed] [Google Scholar]
- 213.Kumar A, Srivastava A. Nature protocols. 2010;5:1737–1747. doi: 10.1038/nprot.2010.135. [DOI] [PubMed] [Google Scholar]
- 214.Cheng X, Irimia D, Dixon M, Sekine K, Demirci U, Zamir L, Tompkins RG, Rodriguez W, Toner M. Lab Chip. 2007;7:170–178. doi: 10.1039/b612966h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Pullagurla SR, Witek MA, Jackson JM, Lindell MA, Hupert ML, Nesterova IV, Baird AE, Soper SA. Analytical chemistry. 2014;86:4058–4065. doi: 10.1021/ac5007766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Li P, Gao Y, Pappas D. Analytical chemistry. 2012;84:8140–8148. doi: 10.1021/ac302002a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Hyun KA, Lee TY, Jung HI. Analytical chemistry. 2013;85:4439–4445. doi: 10.1021/ac3037766. [DOI] [PubMed] [Google Scholar]
- 218.Chen W, Weng S, Zhang F, Allen S, Li X, Bao L, Lam RHW, Macoska JA, Merajver SD, Fu J. ACS Nano. 2013;7:566–575. doi: 10.1021/nn304719q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Singh A, Suri S, Lee T, Chilton JM, Cooke MT, Chen W, Fu J, Stice SL, Lu H, McDevitt TC, Garcia AJ. Nature methods. 2013;10:438–444. doi: 10.1038/nmeth.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Li P, Stratton ZS, Dao M, Ritz J, Huang TJ. Lab Chip. 2013;13:602–609. doi: 10.1039/c2lc90148j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Mahara A, Yamaoka T. Biotechnology progress. 2010;26:441–447. doi: 10.1002/btpr.354. [DOI] [PubMed] [Google Scholar]
- 223.Yu X, He R, Li S, Cai B, Zhao L, Liao L, Liu W, Zeng Q, Wang H, Guo SS, Zhao XZ. Small. 2013;9:3895–3901. doi: 10.1002/smll.201300169. [DOI] [PubMed] [Google Scholar]
- 224.Saliba AE, Saias L, Psychari E, Minc N, Simon D, Bidard FC, Mathiot C, Pierga JY, Fraisier V, Salamero J, Saada V, Farace F, Vielh P, Malaquin L, Viovy JL. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14524–14529. doi: 10.1073/pnas.1001515107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Yoon HJ, Kim TH, Zhang Z, Azizi E, Pham TM, Paoletti C, Lin J, Ramnath N, Wicha MS, Hayes DF, Simeone DM, Nagrath S. Nature nanotechnology. 2013;8:735–741. doi: 10.1038/nnano.2013.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Mittal S, Wong IY, Deen WM, Toner M. Biophysical journal. 2012;102:721–730. doi: 10.1016/j.bpj.2011.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Xu Y, Phillips JA, Yan J, Li Q, Fan ZH, Tan W. Analytical chemistry. 2009;81:7436–7442. doi: 10.1021/ac9012072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Sheng W, Chen T, Kamath R, Xiong X, Tan W, Fan ZH. Analytical chemistry. 2012;84:4199–4206. doi: 10.1021/ac3005633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Zhao W, Cui CH, Bose S, Guo D, Shen C, Wong WP, Halvorsen K, Farokhzad OC, Teo GS, Phillips JA, Dorfman DM, Karnik R, Karp JM. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:19626–19631. doi: 10.1073/pnas.1211234109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Sackmann EK, Fulton AL, Beebe DJ. Nature. 2014;507:181–189. doi: 10.1038/nature13118. [DOI] [PubMed] [Google Scholar]


