Abstract
In the present study, an artificial spider silk gene, 6mer, derived from the consensus sequence of Nephila clavipes dragline silk gene, was fused with different silica-binding peptides (SiBPs), A1, A3 and R5, to study the impact of the fusion protein sequence chemistry on silica formation and the ability to generate a silk–silica composite in two different bioinspired silicification systems: solution–solution and solution– solid. Condensed silica nanoscale particles (600–800 nm) were formed in the presence of the recombinant silk and chimeras, which were smaller than those formed by 15mer-SiBP chimeras [1], revealing that the molecular weight of the silk domain correlated to the sizes of the condensed silica particles in the solution system. In addition, the chimeras (6mer-A1/A3/R5) produced smaller condensed silica particles than the control (6mer), revealing that the silica particle size formed in the solution system is controlled by the size of protein assemblies in solution. In the solution–solid interface system, silicification reactions were performed on the surface of films fabricated from the recombinant silk proteins and chimeras and then treated to induce β-sheet formation. A higher density of condensed silica formed on the films containing the lowest β-sheet content while the films with the highest β-sheet content precipitated the lowest density of silica, revealing an inverse correlation between the β-sheet secondary structure and the silica content formed on the films. Intriguingly, the 6mer-A3 showed the highest rate of silica condensation but the lowest density of silica deposition on the films, compared with 6mer-A1 and -R5, revealing antagonistic crosstalk between the silk and the SiBP domains in terms of protein assembly. These findings offer a path forward in the tailoring of biopolymer–silica composites for biomaterial related needs.
Keywords: Spider silk, Silica, Silicification, Fusion proteins
1. Introduction
Biomaterials have been designed and implemented for replacing damaged or diseased tissues or organs. Silk proteins from spider or silkworm silk have been fabricated into various material formats to address this need [2–6]. Key features for these types of protein-based biomaterials include the chemistry, structure and morphology of the materials which can provide good compatibilities to cells and tissues. Versatile and designer features are useful in these types of biomaterials as they are genetically encoded proteins from which selective and targeted changes in sequence chemistry can be incorporated with tight control. Recombinant spider silk proteins are usually produced by bacterial expression as native sources of spider silk are low in yield and suffer from purification challenges. Recombinant spider silk protein has the advantage of the option to be fused with new functional protein domains to generate novel functional polymers [7–11].
Biosilica is found in a wide array of organisms, ranging from single-celled diatoms to plants and sponges, forming various material morphologies that confer protection or mechanical support [12]. Biosilicification has provided important inspiration for the design of novel biomaterials for tissue engineering [2,11] and biosensors [13]. Silaffin, a peptide implicated in the biogenesis and deposition of silica in diatoms and sea snails, is of interest for the synthesis of materials with specific properties because of its ability to promote silica formation in vitro [14,15]. A short silica-binding peptide (SiBP), R5, which is derived from the silaffin protein of the diatom, Cylindrotheca fusiformis [14], was fused with glucose oxidase [13] and protein polymers [11,16] to precipitate silica in vitro, forming novel materials with different properties. Our previous work described a chimera, consisting of the spider dragline silk polymer and R5, resulting in the precipitation of nanoscale silica particles [11]. Further, the silk–silica composite morphology and structure were regulated by controlling processing conditions to produce films which promoted osteogenic differentiation of mesenchymal stem cells (hMSCs), indicating utility for osteogenic tissue engineering [2,11]. Other than R5, fragments of the R5 sequence revealed an 11 amino acid long fragment, A1, that had a similar effect as R5 in the precipitation of silica in vitro [17]. Aside from these native sequences above, in vitro screening by phage displays isolated peptides showing binding activity to silica surfaces, where the peptide A3 (or Si4-1), had the highest activity for binding silica compared with other peptide sequences [18].
Recently, an artificial silk polymer, 15mer ((SGRGGLGGQGAGAAAAAGGAGQGGYGGLGSQGT)15, 39 kDa), derived from the consensus repeat of Nephila clavipes dragline silk protein [19,20] was fused with the SiBPs (A1, A3 and R5) and silicification was performed at a series of pHs [1], demonstrating that silica condensation rates and silk–silica composite morphology correlated with colloidal instability of the chimeric proteins which was controlled by pH. In addition, another SiBP Pep1 (KSLSRHDHIHHH), also isolated from phage display, was chemically fused to spider silk polymers, 6mer and 15mer [21], and these polymers showed increased silica condensation compared with the control, silk protein alone. These studies illustrate the potential for utilizing silica precipitating domains in combination with a bulk biomaterial, silk protein, to generate new and interesting biomaterials.
The previously listed studies were conducted primarily in solution, and a few studies have reported diversity of morphology or capacity of bio-silicification on solid-state materials, such as would be potentially valuable for hard tissue regeneration. In the present study, the artificial spider silk polymer, 6mer ((SGRGGLGGQGAGAAAAAGGAGQGGYGGLGSQGT)6, 16 kDa) [10], was utilized to produce three different chimeras by fusing SiBPs A1, A3 and R5, respectively, to the C-terminus, to investigate the effect of molecular weight (MW) of silk domain in the fusion protein on the formation of silk–silica composite particles at the solution–solution interface. In addition, recombinant silk protein and chimeras were fabricated into solid-state materials to investigate capacity and morphology of silica condensation on their surfaces.
2. Materials and methods
2.1. Constructs of recombinant silk protein or chimeras
The vector pET30L carrying the artificial silk protein 6mer (SGRGGLGGQGAGAAAAAGGAGQGGYGGLGSQGT)6 (pET30L-6mer) derived from N. clavipes dragline silk protein was constructed previously [10,22]. To prepare the chimeras with SiBPs fused at the C-terminus of the 6mer, pET30L-6mer was digested by SpeI and then treated by antarctic phosphatase (NEB; MA) to prevent self-ligation. The nucleotide sequences of SiBPs, A1 (SGSKGSKRRIL), A3 (MSPHPHPRHHHT) and R5 (SSKKSGSYSGSKGSKRRIL) were designed with restriction endonuclease sites NheI and SpeI flanked at 5′ and 3′ terminals, respectively. Codons were optimized for expression in Escherichia coli strain Bl21(DE3) by using the on-line tool OPTIMIZER [23] and were synthesized commercially (Invitrogen, New York) (Table S.1). The synthesized nucleotides were annealed to generate double strands and then ligated to generate the constructs pET30L-6mer-A1/A3/R5.
2.2. Expression and purification
The silk protein and the chimeras were expressed in E. coli strain Bl21 Star (DE3) (Invitrogen, New York). Cells were cultivated at 37 °C in lysogeny broth (LB) medium with 50 μg ml–1 Kanamycin until an optical density of between 0.6 and 0.8 at 600 nm was reached. Then isopropyl β-d-1-thiogalactopyranoside (Sigma-Aldrich, MO) was added to give a final concentration of 1 mM to induce protein expression. Cells were harvested by centrifugation for 20 min at 8000 rpm after 5 h of expression. For large-scale expression a Bioflo 3000 (New Brunswick Scientific, NJ) fermentor was used. The pellet was resuspended in denaturing buffer (100 mM NaH2PO4, 10 mM Tris–HCl, 8 M urea) and stirred overnight at 4 °C. Recombinant silk protein and chimeras were purified by Ni-NTA affinity chromatography. The recombinant silk protein and chimeras eluted from the Ni-NTA resin were dialyzed against 100 mM phosphate buffer (pH 5.5) for 1 day followed by distilled water for 3 days using cellulose ester snake skin tubing with MWCO 10,000 Da (Thermo Scientific, MA). The precipitates were removed by filtration using 5.0 μm sterile syringe filters (Millipore, Billerica, MA). Finally, the dialyzed proteins were lyophilized. Protein identity and purity were confirmed by SDS-PAGE (Invitrogen, New York) and matrix-assisted laser desorption/ionization–time of flight (MALDI-TOF) mass spectrometry (Tufts core facility, Boston, MA).
2.3. Dynamic light scattering and zeta potential measurements
Dynamic light scattering and zeta potential measurements were used to measure the average particle diameter and charge on the proteins over the pH range 3–9. Solutions of the chimeric proteins were made in aqueous 0.1 M citric acid solutions to a concentration of 1 mg ml–1 and then filtered through a 450 nm membrane to remove any undispersed aggregates. Measurements were made using a Malvern nanoS Zetasizer where dispersed phase properties of latex and a dispersion phase of 0.1 M tris buffer were chosen for a compatible refractive index, viscosity and dielectric constant. Five separate measurements were made at each pH for particle size and charge analysis and the results were averaged (any outliers were removed where necessary but not more than one allowed per data set). The pH was adjusted between analyses for subsequent measurement by the addition of measured amounts of 1.0 M bis-tris propane, and the measurements were repeated to give size and zeta potential data over a pH range 3–9.
2.4. Bioinspired silicification in solution
The silicification reactions were performed according to our prior methods [1,21]. The lyophilized 6mer and chimera proteins were dissolved in double distilled water and then filtered with 5.0 μm syringe filters (Millipore, MA) to remove insoluble precipitates. The protein concentration was measured by bicinchoninic acid assay (Thermo Scientific, MA). 174 μl of 0.5 mg ml–1 protein solutions was buffered to pH 7.0 by adding 13 μl of 1 M bis-tris propane and 7 μl of 1 M citric acid buffers. 6 μl of prehydrolyzed tetraethoxysilane (TEOS) solution (prepared by mixing 2.23 ml of TEOS (Sigma-Aldrich, MO) with 7.76 ml of 50% ethanol/water solution, 100 μl of 1 M HCl and standing at room temperature for 15 min) was added and mixed thoroughly. The condensed and aggregated silk–silica precipitates were collected after 1 h by centrifugation at 13,000 rpm for 5 min and washed twice with water and finally freeze-dried.
2.5. Turbidity assay
The process of silica precipitation in solution at pH 7.0 was monitored by turbidity at 595 nm [1] with a plate reader Spectra-Max M2 (Molecular Device, CA) from three independent experiments.
2.6. Preparation of silk protein and chimera films
The lyophilized recombinant proteins were dissolved in hexafluoroisopropanol (HFIP) (Sigma-Aldrich, MO) at a concentration of 2.5% (w/v). 30 μl of the protein–HFIP solution was pipetted onto polydimethylsiloxane (PDMS) substrates (6 mm in diameter) and air-dried in a fume hood. To induce β-sheet formation, the films were then treated by water vapor annealing, using an Isotemp vacuum oven (Fisher Scientific, NH) and 70% methanol, respectively. For water vapor annealing, the films were treated at 50 °C and 70 °C, respectively, and at 25 inches of mercury (inHg) for 2 h [24]. For methanol treatment, the films were immersed in 70% methanol overnight and then air-dried [11].
2.7. Silicification reaction on films
A total of 1 ml of 30 mM prehydrolyzed TEOS in 100 mM bis-tris propane/citric acid buffer at pH 7.0 was added into each well of a 24-well plate to cover the film formed on the PDMS substrate for one hour at room temperature. The films were then washed two times with distilled water and left to dry overnight in the fume hood.
2.8. Infrared spectroscopy
Fourier transform infrared spectroscopy (FTIR) was carried out using a FT/IR-6200 (JASCO, OK) to investigate protein conformation. Absorbance spectra were collected from 4000 to 600 cm −1 by averaging 32 scans at a resolution of 4.0 cm −1 and the background of PDMS was subtracted. Fourier self-deconvolution was performed with OPUS software (Bruker, MA) with Lorentzian peak profile (half-bandwidth of 25 cm −1 and a noise reduction factor of 0.3). Ten peaks were selected with a Gaussian algorithm across the amide I absorption band (1720–1580 cm −1) corresponding to β-turns (1668–1696 cm −1), α-helix (1658–1667 cm −1), random coil (1630–1657 cm −1) and β sheet (1618–1629 cm −1 and 1697–1703 cm −1) [25,26].
2.9. Scanning electron microscopy (SEM) and energy-dispersive X-ray spectroscopy (EDX)
Samples were coated with gold and then observed using a Carl Zeiss (Carl Zeiss SMT, Germany) Ultra 55 field emission scanning electron microscope (FESEM) at an accelerated voltage of 5 kV. Particle size and the diameter standard deviation were calculated by averaging the diameter of at least 100 particles. EDX was performed using Zeiss EVO 55 Environmental SEM coupled with an EDAX EDX system (Harvard University Center for Nanoscale Systems).
2.10. Statistical analysis
All quantitative analyses were performed at least in three repeats and average size of particles was determined by measurement of more than 100 particles. Results are presented based on averages and standard deviations as error bars. The significant difference between two samples was determined by p-value calculated using Student's t-test.
3. Results
3.1. Biosynthesis of silk protein and chimeras
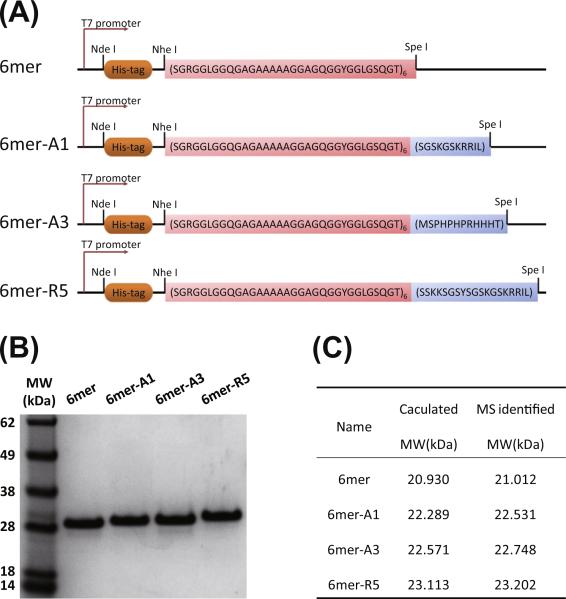
The strategy for biosynthesis of silk protein and the chimeras was adopted as described previously [10,22]. The constructs of the 6mer or the chimeras (6mer-A1, A3 or R5) are illustrated in Fig. 1A. NheI-SpeI digestion and DNA sequencing confirmed the inserts. SDS-PAGE indicated both expression and purification of the 6mer and the chimeras were successful, and no obvious impurities were detected on the gels (Fig. 1B). The MW of each protein identified by mass spectroscopy matched the target calculation (Fig. 1C), indicating the expressed and the purified proteins were correctly generated. In the case of the 6mer, the molecular weight was above 28 kDa on the gels, but was identified at 21–23 kDa by mass spectroscopy. This discrepancy has been reported previously for repetitive silk-based copolymers and likely reflects the hydro-phobic nature and repetitive features in the proteins [10,27,28].
Fig. 1.
Biosynthesis of 6mer and chimeras: (A) scheme of constructs of 6mer and 6mer-SiBP chimeras; (B) SDS-PAGE of purified recombinant proteins; (C) molecular weight identified by MALDI-TOF mass spectrometry.
3.2. Bioinspired silicification catalyzed by silk protein and chimeras in solution
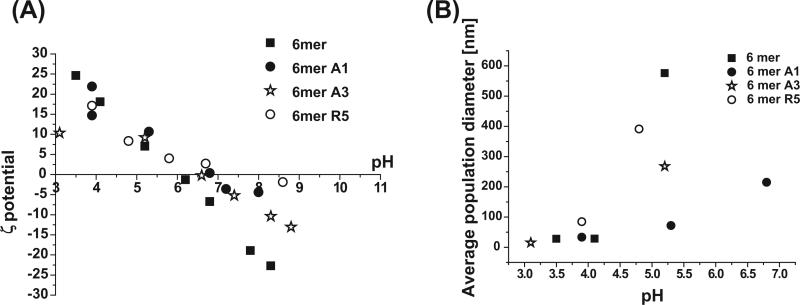
Silicification reactions were performed in solution in order to observe the morphology of the silk–silica composite structures formed. The colloidal instability of silk protein and chimeras in solution, controlled by pH, accelerates the condensation of silica and promotes the formation of spherical particles [1]. The silicification reaction was performed at pH 7.0 in the present study as the 6mer and the chimeras showed the greatest colloidal instability at near neutral pH (Fig. 2A). In addition, population diameter measurements of the silk protein and the chimeras by DLS showed increasing particle sizes with increasing pH from 3 to 5.5, with the 6mer aggregating into the largest particles with a diameter of ~600 nm (Fig. 2B). At pH 7 only the 6mer-A1 chimera produced a stable particle population. The other chimeras and the 6mer showed aggregation and precipitation with apparent high diameters and high polydispersity at this pH and above.
Fig. 2.
Relationship between pH and surface charge (A) and average particle size (B) of 6mer and 6mer-SiBP chimeras.
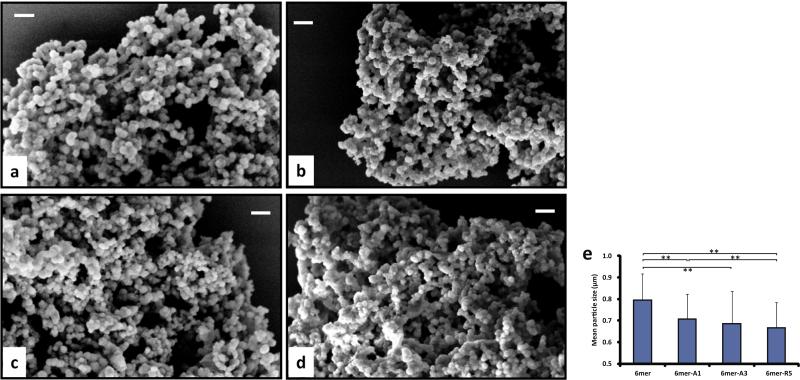
The morphology of the silk–silica composite structures formed is depicted in Fig. 3. Spherical particles were observed for the 6mer and each of chimeras and these particles formed interconnected dense networks. However, the spherical particles formed in the presence of either the 6mer or each of the chimeras were heterogenous in size (Fig. 3a–e). The 6mer produced particles of 790 ± 120 nm in diameter, while the chimera protein 6mer-A1, -A3 and -R5 produced particles of 710 ± 110, 690 ± 150 and 670 ± 120 nm in diameter, respectively, which were all a little bit smaller than those of silk protein alone, the 6mer.
Fig. 3.
Morphology (a–d) and diameter (e) of condensed silk–silica composites generated by 6mer and 6mer-SiBP chimeras in solution-solution system: (a) 6mer, (b) 6mer-A1, (c) 6mer-A3, (d) 6mer-R5. Scale bar = 2 μm. **p < 0.01.
3.3. Rate of silica condensation accelerated by silk and chimeras in solution
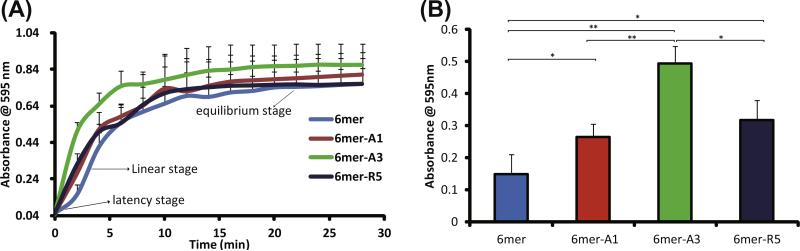
The rate of silica condensation accelerated by silk protein and chimeras in solution was investigated by change in turbidity (Fig. 4). The silk protein- and chimeras-accelerated silica condensation reached equilibrium after ~30 min (Fig. 4A). Silica condensation was accelerated by the 6mer due to the charged amino acid residues in the sequences (arginine and His-tag). However, the rate of reaction accelerated by the 6mer control, as expected, was lower than each of the chimeras. A delay at 2 min (latent stage) for the reaction with the 6mer was observed before the reaction proceeded rapidly (linear stage), while no latent stage was observed for the chimeras, indicating the SiBPs fused at the C-terminus of the 6mer increased activity of silica condensation. The 6mer-A3 showed the greatest acceleration of silica condensation, and there was no significant difference between the 6mer-A1 and 6mer-R5 (Fig. 4B). The chimera carrying the SiBP A3 showed the highest acceleration of silica condensation, consistent with a previous report describing silica formation using 15mer chimeras [1].
Fig. 4.
Rate of silica condensation induced by 6mer and 6mer-SiBP chimeras in solution–solution system: (A) time-dependent turbidity in silicification reaction; (B) turbidity at 2 min.
3.4. Bioinspired silicification on films
In order to study silica condensation on surfaces, the recombinant spider silk protein and the chimeras were fabricated into films using methods described previously [11]. In the films, shifts from random coil to β-sheet were induced to prevent dissolution of films by treating with water vapor at 50 °C and 70 °C, as well as 70% methanol. Secondary structure conformation of the protein in the films was investigated by FTIR. The 6mer or each of the chimeras showed strong peaks in the amide I (1690–1600 cm −1) and the amide II (1575–1480 cm −1) regions (Fig. S.1). For the untreated films, the 6mer and the chimeras only showed peaks at 1649 cm −1 in the amide I region, indicating the presence of random coils. After self-assembly, a shift in the main peak from 1649 cm −1 to 1624 cm −1 was observed for the silk protein and each of the chimeras indicating the formation of β-sheets.
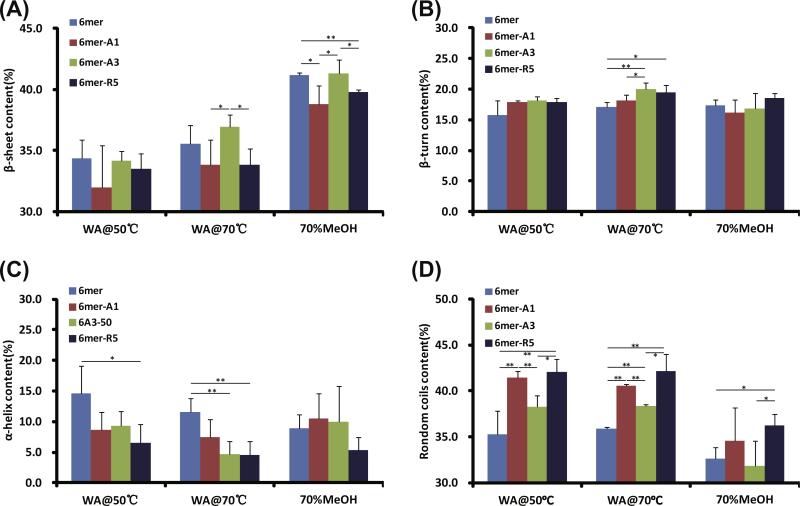
The FTIR spectra were deconvoluted and fitted with peaks assigned to β-sheet, β-turn, α-helix and random coils (Fig. 5 and Table S.2). Overall, more than 30% β-sheet was induced in the films for the 6mer or each of the chimeras by annealing. The lowest β-sheet content in the films was induced by water vapor annealing at 50 °C, and a medium content of β-sheet content at 70 °C, respectively. As expected, the highest β-sheet content was from treatment with 70% methanol. A pattern for each treatment was observed such that the chimeras 6mer-A1 and 6mer-R5 showed a lower content of β-sheet and a correspondingly higher content of random coils as opposed to the 6mer and 6mer-A3 chimeras (Fig. 5A and D). Lower α-helix content was induced in the chimera films than in the 6mer film by water vapor annealing (Fig. 5C).
Fig. 5.
Conformation and content of secondary structures of proteins in films after various treatments. Films fabricated from 6mer or each of chimeras were treated by water vapor annealing at 50 °C (WA@50 °C) and at 70 °C (WA@70 °C) as well as by 70% methanol treatment (70%MeOH). *0.01 < p < 0.05; **p < 0.01.
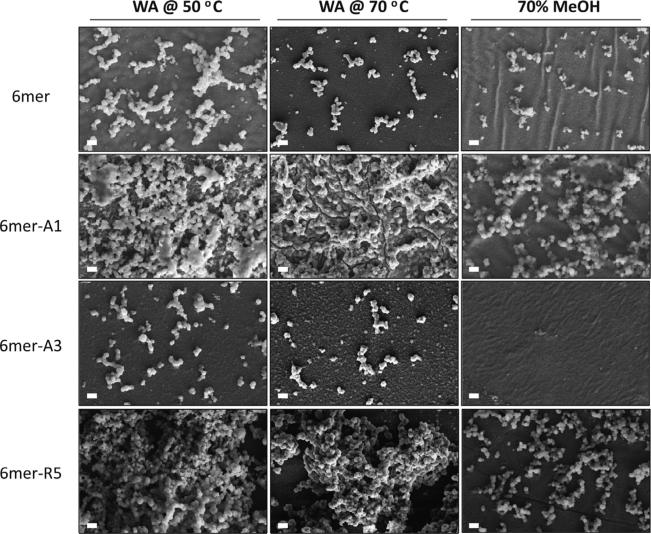
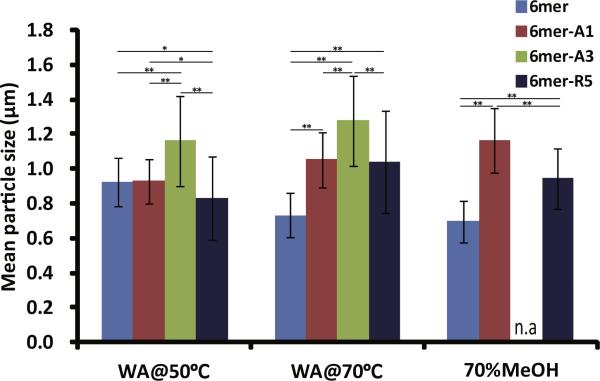
Silicification reactions were performed on the films after β-sheet induction by the various treatments (Figs. 6 and 7). A high density of condensed silica particles interconnected into three-dimensional networks was observed on the surface of 6mer-R5 films which were treated by water vapor annealing methods, while a low density of silica particles was formed on the film treated by 70% methanol. Although a lower density of silica particles were formed on the 6mer-A1 films than on the 6mer-R5 films, condensed silica particles were fused together on the 6mer-A1 films when the films were treated by water vapor annealing or methanol annealing. A low density of silica particles was observed on the 6mer-A3 films treated by water vapor annealing, but no condensed silica particles were observed on the 6mer-A3 films treated by 70% methanol. The sizes of condensed silica particles formed on the films are shown in Fig. 7. The size of particles formed on the 6mer films was less than 1 μm average diameter and comparatively smaller than that on the chimera films when treated by water vapor annealing at 70 °C and by 70% methanol. For silk chimera films, 6mer-A3 produced the largest particles which were maximally 1.4 μm in diameter on the film treated by water vapor annealing at 70 °C, while 6mer-A1 and 6mer-R5 generated particles with the size ~1.0 lm on the films treated by water vapor annealing as well as 70% methanol.
Fig. 6.
Morphology of condensed silica structures on films after various treatments. Films fabricated from 6mer or each of chimeras were treated by water vapor annealing at 50 °C (WA@50 °C) and at 70 °C (WA@70 °C) as well as by 70% methanol treatment (70%MeOH). Scale bar = 2 μm.
Fig. 7.
Size of condensed silica particles formed on films which were treated by water vapor annealing at 50 °C (WA@50 °C) and at 70 °C (WA@70 °C) as well as by 70% methanol treatment (70%MeOH). *0.001 < p < 0.01; **pxs < 0.001.
Besides those obviously observed particles on the films, many nanoscale features (50–100 nm in diameter) were observed especially on the 6mer and 6mer-A3 films treated by water vapor annealing at 70 °C (Fig. 6). To confirm these nanoscale features were formed from silica condensation, elemental mapping of samples was performed by EDX (Fig. 8). On the 6mer and the 6mer-A3 films, silicon was detected anywhere on the films besides the position of silica particles, suggesting silica bonding to the films, although no obvious silica particles formed. It indicated those nanoscale features were made of condensed silica and likely nas-cent silica particles. In addition, the ratio of carbon (C) to silicon (Si) was 0.28 on both the 6mer-A1 and -R5 films, 1.2 on the 6mer and 1.6 on the 6mer-A3 films, indicating a higher density of condensed silica on 6mer-A1 and -R5 films than on 6mer (control) or 6mer-A3 films, consistent with the SEM observations (Fig. 6).
Fig. 8.
Elemental analysis of structures on the films treated by water vapor annealing at 70 °C. Elemental maps are shown for carbon (C), oxygen (O) and silicon (Si). Insets are EDX spectra for samples. White arrows indicate the position of particles identified by the elemental mapping for 6mer and 6mer-A3.
4. Discussion
Control of silica condensation was investigated by performing silicification reactions with a series of genetically engineered 6mer and 6mer-SiBP fusion proteins at two different interfaces: solution–solution and solution–solid. In the solution–solution studies, silk protein and silk–SiBP fusion proteins interacted with prehydrolyzed TEOS to accelerate silica condensation. The correlation between protein colloidal instability and silica condensation has been discussed previously [1]. Particles were produced with mean diameters ranging from 600 to 800 nm by either the 6mer or each of the fusion proteins (Fig. 3e), which was consistent with the previously reported finding that particles formed from the fusion protein 6mer-pep1 were 690 ± 130 nm in diameter [21]. However, particles produced in the presence of the 15mer or 15mer-SiBP fusion proteins at neutral pH conditions ranged from 1.0 to 2.0 lm in diameter [1,11,21,29,30]. Furthermore, the particles generated from the 6mer-SiBP chimeras were smaller than those generated from the 6mer alone (Fig. 3e). The same phenomenon was observed for particles formed in the presence of the chimera 6mer-pep1, which were smaller than those prepared in the presence of the 6mer alone [21]. One mechanism to explain in part the above phenomenon is that silk protein or silk–SiBP chimeras formed colloidal species in solution and acted as templates to precipitate silica [1,31] . This is likely to be due to subtle differences in charge state at pH 7. The chimeras all have additional side chain chargeable groups likely to impose a more positively charged surface. The increased charge density is then offset by a reduction in particle size to increase surface area and hence reduce the charge density, resulting in a lower particle size at equilibrium. Overall, there is an interplay between the hydrophobic effect driving complete phase separation and the surface charge favoring dissolution.
In addition, silica condensation is also influenced by a charge relay effect which controls the catalytic activity of the SiBPs to accelerate silica condensation [31,32]. Charged groups in the SiBPs may interact differently so that the SiBPs present different capacities to accelerate silica condensation, allowing the silk–SiBP chimeras to generate a diversity of size of condensed silica particles. Thus, a potential mechanism to control the size of silk–silica particles at the nano- and micro-scale can be pursued by the sizes of the silk domains as well as the charges of silica-binding domains generated by sequence chemistry.
In the solution–solid interface system, silica condensation was carried out on the surface of the films of the recombinant 6mer and 6mer-SiBP fusion proteins. Before conducting the silica condensation reaction, the films were treated by water vapor annealing at 50 °C and 70 °C as well as by 70% methanol. As expected, the lowest β-sheet content formed in the films treated at 50 °C and highest β-sheet content in the films treated by 70% methanol (Fig. 5A) [24]. Intriguingly, the films treated by water vapor annealing at 50 °C generated highest while those treated 70% methanol generated lowest density of condensed silica particles on the either the 6mer or each of the chimeras (Fig. 6). This phenomenon may be correlated to β-sheet content in the films, which form due to the hydrophobic polyalanine domains [33], perhaps shielding the silica-binding domains from the condensation reactions.
The 6mer-A1/-R5 films produced a higher density of condensed silica particles than the 6mer or the 6mer-A3 films which were treated with the same conditions (Fig. 6). However, the 6mer-A3 showed the greater rate of inducing silica precipitation than 6mer-A1 and -R5 in the solution–solution interface system (Fig. 4). This phenomenon may be related to crosstalk between the silk domain and the SiBP domains. In the process of annealing the films, the silk protein self-assembled into energy-stable conformations, β-sheet crystals [24,25]. The silica binding peptides A1 and R5 are able to precipitate silica efficiently because of the presence of the motif “RRIL”, which confers active silica precipitating assemblies [17]. It appeared that the introduction of A1 and R5, rather than A3, to the C-terminus of silk domain affected β-sheet formation in the chimera proteins (Fig. 5A). In the solution, random coil was the major structure and no β-sheet formed from the silk domains [9]. Therefore, assembly of A1 and R5, rather than A3, may antagonize β-sheet formation driven by the silk domain in the films, accounting for the fact that that 6mer-A3 showed the highest rate of silica precipitation at the solution–solution interface with the lowest density of silica deposition at the solution–solid interface.
These observations suggest that morphology of condensed silica structures and the rate of silica condensation on the surface of solid silk–SiBP materials can be tuned by fusing SiBPs with different capacities of self-assembly as well as by controlling the post-fabrication processing of the materials.
5. Conclusions
Bioinspired silicification was performed with genetically engineered silk protein and silk–SiBP fusion proteins using both solution–solution and solution–solid interfaces. Nanoparticles were produced in the solution studies and particles formed by fusion proteins were smaller than when the molecular weight of the silk domain was smaller, suggesting that molecular weight plays a role in controlling the size of silk–silica particles. This feature impacts silica degradation and may influence cell growth and differentiation in bone regeneration [29,30]. In solution–solid interface systems, silica particles were formed on the films and 6mer-A1/-R5 films supported a higher silica precipitation activity. This feature was at least in part related to the β-sheet content of the films, further offering control of silica condensation rate and precipitation activity. Different morphologies of condensed silica structures were observed on the 6mer-A1 films because the silica particles appeared to be incorporated into the films, suggesting that there was a crosstalk between silk domain and SiBPs that led to control of the morphology of the condensed silica structures which may influence interfacial bonding between materials and cells or tissues in bone regeneration. Thus, tuning bioinspired silicification with artificial macromolecular-SiBP chimeras by controlling the molecular weight of the silk domain and the nature of the SiBP sequences, along with post treatment processes to impact secondary structure of the proteins, can play roles in generating tuned biopolymer–silica composites for use in biomedical engineering.
Supplementary Material
Acknowledgements
We thank the NIH (R01 DE017207) and Natural Science Foundation of China (NSFC, project no. 51273138) for support of this research.
Footnotes
Appendix A. Figures with essential colour discrimination
Certain figures in this article, particularly Figs. 1, 3–5, 7 and 8 is difficult to interpret in black and white. The full colour images can be found in the on-line version, at http://dx.doi.org/10.1016/j.actbio.2014.10.040.
Appendix B. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.actbio.2014.10.040.
References
- 1.Belton DJ, Mieszawska AJ, Currie HA, Kaplan DL, Perry CC. Silk–silica composites from genetically engineered chimeric proteins: materials properties correlate with silica condensation rate and colloidal stability of the proteins in aqueous solution. Langmuir. 2012;28:4373–81. doi: 10.1021/la205084z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mieszawska AJ, Nadkarni LD, Perry CC, Kaplan DL. Nanoscale control of silica particle formation via silk–silica fusion proteins for bone regeneration. Chem Mater. 2010;22:5780–5. doi: 10.1021/cm101940u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tang-Schomer MD, White JD, Tien LW, Schmitt LI, Valentin TM, Graziano DJ, et al. Bioengineered functional brain-like cortical tissue. Proc Natl Acad Sci USA. 2014 doi: 10.1073/pnas.1324214111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wray LS, Tsioris K, Gi ES, Omenetto FG, Kaplan DL. Slowly degradable porous silk microfabricated scaffolds for vascularized tissue formation. Adv Funct Mater. 2013;23:3404–12. doi: 10.1002/adfm.201202926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu J, Rnjak-Kovacina J, Du Y, Funderburgh ML, Kaplan DL, Funderburgh JL. Corneal stromal bioequivalents secreted on patterned silk substrates. Biomaterials. 2014;35:3744–55. doi: 10.1016/j.biomaterials.2013.12.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xia X, Wang M, Lin Y, Xu Q, Kaplan DL. Hydrophobic drug-triggered self-assembly of nanoparticles from silk–elastin-like protein polymers for drug delivery. Biomacromolecules. 2014 doi: 10.1021/bm4017594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Currie HA, Deschaume O, Naik RR, Perry CC, Kaplan DL. Genetically engineered chimeric silk–silver binding proteins. Adv Funct Mater. 2011;21:2889–95. doi: 10.1002/adfm.201100249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gomes SC, Leonor IB, Mano JF, Reis RL, Kaplan DL. Antimicrobial functionalized genetically engineered spider silk. Biomaterials. 2011;32:4255–66. doi: 10.1016/j.biomaterials.2011.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang J, Wong C, George A, Kaplan DL. The effect of genetically engineered spider silk–dentin matrix protein 1 chimeric protein on hydroxyapatite nucleation. Biomaterials. 2007;28:2358–67. doi: 10.1016/j.biomaterials.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 10.Numata K, Subramanian B, Currie HA, Kaplan DL. Bioengineered silk protein-based gene delivery systems. Biomaterials. 2009;30:5775–84. doi: 10.1016/j.biomaterials.2009.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.WongPoFoo C, Patwardhan SV, Belton DJ, Kitchel B, Anastasiades D, Huang J, et al. Novel nanocomposites from spider silk–silica fusion (chimeric) proteins. Proc Natl Acad Sci USA. 2006;103:9428–33. doi: 10.1073/pnas.0601096103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schroder HC, Wang XH, Tremel W, Ushijima H, Muller WEG. Biofabrication of biosilica-glass by living organisms. Nat Prod Rep. 2008;25:455–74. doi: 10.1039/b612515h. [DOI] [PubMed] [Google Scholar]
- 13.Choi O, Kim BC, An JH, Min K, Kim YH, Um Y, et al. A biosensor based on the self-entrapment of glucose oxidase within biomimetic silica nanoparticles induced by a fusion enzyme. Enzyme Microb Technol. 2011;49:441–5. doi: 10.1016/j.enzmictec.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 14.Kroger N, Deutzmann R, Sumper M. Polycationic peptides from diatom biosilica that direct silica nanosphere formation. Science. 1999;286:1129–32. doi: 10.1126/science.286.5442.1129. [DOI] [PubMed] [Google Scholar]
- 15.Kroger N, Deutzmann R, Sumper M. Silica-precipitating peptides from diatoms. The chemical structure of silaffin-A from Cylindrotheca fusiformis. J Biol Chem. 2001;276:26066–70. doi: 10.1074/jbc.M102093200. [DOI] [PubMed] [Google Scholar]
- 16.Marner WD, 2nd, Shaikh AS, Muller SJ, Keasling JD. Morphology of artificial silica matrices formed via autosilification of a silaffin/protein polymer chimera. Biomacromolecules. 2008;9:1–5. doi: 10.1021/bm701131x. [DOI] [PubMed] [Google Scholar]
- 17.Knecht MR, Wright DW. Functional analysis of the biomimetic silica precipitating activity of the R5 peptide from Cylindrotheca fusiformis. Chem Commun. 2003:3038–9. doi: 10.1039/b309074d. [DOI] [PubMed] [Google Scholar]
- 18.Naik RR, Brott LL, Clarson SJ, Stone MO. Silica-precipitating peptides isolated from a combinatorial phage display peptide library. J Nanosci Nanotechnol. 2002;2:95–100. doi: 10.1166/jnn.2002.074. [DOI] [PubMed] [Google Scholar]
- 19.Bini E, Foo CW, Huang J, Karageorgiou V, Kitchel B, Kaplan DL. RGD-functionalized bioengineered spider dragline silk biomaterial. Biomacromolecules. 2006;7:3139–45. doi: 10.1021/bm0607877. [DOI] [PubMed] [Google Scholar]
- 20.Prince JT, McGrath KP, DiGirolamo CM, Kaplan DL. Construction, cloning, and expression of synthetic genes encoding spider dragline silk. Biochemistry. 1995;34:10879–85. doi: 10.1021/bi00034a022. [DOI] [PubMed] [Google Scholar]
- 21.Canabady-Rochelle LL, Belton DJ, Deschaume O, Currie HA, Kaplan DL, Perry CC. Bioinspired silicification of silica-binding peptide–silk protein chimeras: comparison of chemically and genetically produced proteins. Biomacromolecules. 2012;13:683–90. doi: 10.1021/bm201555c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rabotyagova OS, Cebe P, Kaplan DL. Self-assembly of genetically engineered spider silk block copolymers. Biomacromolecules. 2009;10:229–36. doi: 10.1021/bm800930x. [DOI] [PubMed] [Google Scholar]
- 23.Puigbo P, Guzman E, Romeu A, Garcia-Vallve S. OPTIMIZER: a web server for optimizing the codon usage of DNA sequences. Nucleic Acids Res. 2007;35:W126–31. doi: 10.1093/nar/gkm219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu X, Shmelev K, Sun L, Gil ES, Park SH, Cebe P, et al. Regulation of silk material structure by temperature-controlled water vapor annealing. Biomacromolecules. 2011;12:1686–96. doi: 10.1021/bm200062a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu X, Kaplan D, Cebe P. Determining beta-sheet crystallinity in fibrous proteins by thermal analysis and infrared spectroscopy. Macromolecules. 2006;39:6161–70. [Google Scholar]
- 26.Huang WW, Krishnaji S, Hu X, Kaplan D, Cebe P. Heat capacity of spider silk-like block copolymers. Macromolecules. 2011;44:5299–309. doi: 10.1021/ma200563t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gomes S, Leonor IB, Mano JF, Reis RL, Kaplan DL. Spider silk–bone sialoprotein fusion proteins for bone tissue engineering. Soft Matter. 2011;7:4964–73. [Google Scholar]
- 28.Xia XX, Xu Q, Hu X, Qin G, Kaplan DL. Tunable self-assembly of genetically engineered silk–elastin-like protein polymers. Biomacromolecules. 2011;12:3844–50. doi: 10.1021/bm201165h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin W, Huang Y-w, Zhou X-D, Ma Y. In vitro toxicity of silica nanoparticles in human lung cancer cells. Toxicol Appl Pharmacol. 2006;217:252–9. doi: 10.1016/j.taap.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 30.Mieszawska AJ, Fourligas N, Georgakoudi I, Ouhib NM, Belton DJ, Perry CC, et al. Osteoinductive silk–silica composite biomaterials for bone regeneration. Biomaterials. 2010;31:8902–10. doi: 10.1016/j.biomaterials.2010.07.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Belton DJ, Patwardhan SV, Annenkov VV, Danilovtseva EN, Perry CC. From biosilicification to tailored materials: optimizing hydrophobic domains and resistance to protonation of polyamines. Proc Natl Acad Sci USA. 2008;105:5963–8. doi: 10.1073/pnas.0710809105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuno T, Nonoyama T, Hirao K, Kato K. Influence of the charge relay effect on the silanol condensation reaction as a model for silica biomineralization. Langmuir. 2011;27:13154–8. doi: 10.1021/la202576v. [DOI] [PubMed] [Google Scholar]
- 33.Rabotyagova OS, Cebe P, Kaplan DL. Role of polyalanine domains in beta-sheet formation in spider silk block copolymers. Macromol Biosci. 2010;10:49–59. doi: 10.1002/mabi.200900203. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.