Abstract
Background
Prenatal alcohol exposure can kill developing neurons, leading to microencephaly and mental retardation. However, not all fetuses are equally vulnerable to alcohol’s neurotoxic effects. While some fetuses are severely affected and are ultimately diagnosed with fetal alcohol syndrome (FAS), others have no evidence of neuropathology and are behaviorally normal. These widely different outcomes among alcohol-exposed fetuses are likely due, in part, to genetic differences. Some fetuses possess genotypes that make them much more vulnerable than others to alcohol’s teratogenic effects. However, to date, only one gene has been identified whose mutation can worsen alcohol-induced behavioral deficits in an animal model of FAS. That gene is neuronal nitric oxide synthase (nNOS). The purpose of this study was to determine whether mutation of nNOS can likewise worsen alcohol-induced microencephaly and lead to permanent neuronal deficits.
Methods
Wild type and nNOS−/− mice received alcohol (0.0, 2.2, or 4.4 mg/g) daily over postnatal days (PD) 4–9. Beginning on PD 85, the mice underwent a series of behavioral tests, the results of which are reported in the companion paper. The brains were then weighed, and stereological cell counts were performed on the cerebral cortex and hippocampal formation, which are the brain regions that mediate the aforementioned behavioral tasks.
Results
Alcohol caused dose-dependent microencephaly, but only in the nNOS−/− mice and not in wild type mice. Alcohol-induced neuronal losses were more severe in the nNOS−/− mice than in the wild type mice in all of the brain regions examined, including the cerebral cortex, hippocampal CA3 subregion, hippocampal CA1 subregion, and dentate gyrus.
Conclusions
Targeted mutation of the nNOS gene increases the vulnerability of the developing brain to alcohol-induced growth restriction and neuronal losses. This increased neuropathology is associated with worsened behavioral dysfunction. The results demonstrate the critical importance of genotype in determining the outcome of developmental alcohol exposure.
Keywords: Cerebral cortex, hippocampus, dentate gyrus, stereology, FASD
Introduction
Alcohol can damage the developing brain in a variety of ways. Perhaps most important is neuronal death (Olney et al., 2000). Alcohol can kill developing neurons and lead to permanent deficits in neuronal number (Bonthius and West, 1991; Bonthius et al., 1992). These neuronal deficits contribute strongly to the microencephaly that commonly accompanies fetal alcohol spectrum disorder (FASD). Animal models have verified that alcohol is toxic to developing neurons and that the neuronal losses are highly correlated with behavioral deficits, including learning disorders, ataxia, and reduced seizure thresholds (Bonthius et al., 2001a,b; Goodlett et al., 1992; Thomas et al., 1998). Thus, neuronal death is a key pathogenic mechanism in FASD and is closely tied to both the structural and functional deficits induced by alcohol in the developing brain.
The presence, severity, and location of neuronal death differ markedly among children prenatally exposed to alcohol. Some have microencephaly, suggesting generalized and severe neuronal losses, while others have normal-sized brains with no evidence of neuronal deficits (Abel, 1995; Streissguth and Dehaene, 1993). In some children, neuronal losses are restricted to certain brain regions, such as the cerebellum or basal ganglia, while different brain regions are affected in others (Astley et al., 2009; Norman et al., 2009). These widely different patterns of neuropathology are probably due, at least in part, to genetic differences among the exposed fetuses (Warren and Li, 2005).
In the accompanying article, we demonstrated that homozygous mutation of one specific gene, neuronal nitric oxide synthase (nNOS), makes developing mice much more vulnerable to certain behavioral deficits following alcohol exposure (Karacay et al., 2014). These worsened behavioral deficits could be due to worsened neuronal losses. Indeed, we have demonstrated previously that neuronal losses induced by alcohol are greater in nNOS−/− mice than in wild type (Bonthius et al., 2002; 2006; de Licona et al., 2009). However, in all of these previous studies, neuronal numbers were determined acutely, in infant mice, immediately following the alcohol exposure. Because of background cell death (Hamburger, 1975) and the ability of some neuronal populations to proliferate in the postnatal period (Patten et al., 2013), the reductions in neuronal numbers in nNOS−/− mice observed in the neonates may not persist into adulthood. Thus, the goal of this study was to determine whether the worsened neuronal losses in nNOS−/− mice persist into adulthood. The results demonstrate for the first time that a specific genotype can predispose to permanently worsened neuroanatomical deficits in a model of FASD.
Materials and Methods
Animals
The mice used in this anatomical study were identical to those used in the behavioral study, as described in detail in the companion paper (Karacay et al., 2014). Briefly, the mouse strains were either homozygous null mutants for neuronal nitric oxide synthase (nNOS−/− mice) or their wild type control (F2 offspring of 129SVJ x C57B6 matings) (Huang et al., 1993). The Institutional Animal Care and Use Committee at the University of Iowa approved all of the procedures for this study.
Over postnatal days 4–9, the mice received intraperitoneal injections of alcohol (0.0, 2.2, or 4.4 mg/g/day), administered as 0%, 10%, and 20% (v/v) solutions, respectively (Bonthius et al., 2002). Males and females were included. Each treatment group for each genotype/sex combination consisted of 7–10 subjects (n = 7–10 per treatment/genotype/sex group). Beginning on PD 85, the animals underwent a series of behavior studies, including open field activity, the Morris water maze, and prepulse inhibition. Results of those behavioral studies are provided in the companion paper.
Tissue preparation and cutting of frozen sections
Following completion of the behavioral testing, the adult mice (PD 110–122) were weighed, anesthetized with pentobarbital, and perfused via the left cardiac ventricle with 0.9% NaCl, followed by 4% (w/v) paraformaldehyde fixative in 0.1 M phosphate buffer (pH=7.4). The brains were removed, weighed, and stored in fixative at 4°C for at least 24 hours. During processing, three of the brains were lost. The total number of brains included for each treatment group is shown in Table 1.
Table 1.
Treatment groups and number of subjects per group.
| Genotype | Alcohol dose (mg/g/day) |
Gender | n for brain weights and neuronal counts |
|---|---|---|---|
| Wild Type | 0.0 (injected control) | Male | 8 |
| Female | 7 | ||
| 2.2 | Male | 8 | |
| Female | 8 | ||
| 4.4 | Male | 8 | |
| Female | 7 | ||
| nNOS−/− | 0.0 | Male | 8 |
| Female | 7 | ||
| 2.2 | Male | 7 | |
| Female | 8 | ||
| 4.4 | Male | 8 | |
| Female | 7 |
The olfactory bulbs, cerebellum, and brain stem were removed. The forebrains were bisected at the sagittal midline. For cryoprotection, the right forebrains were placed in a 30% sucrose solution in phosphate buffer and gently rocked continuously on a rocker table at room temperature for 48 hours. The cryoprotected forebrains were positioned on the stage of a freezing microtome and cut exhaustively at a thickness of 40µm in the horizontal plane. All of the serial sections generated were saved in consecutive order with one section per well in 96-well tissue culture trays containing 4% paraformaldehyde fixative. The collected sections were stored at 4°C pending further processing.
Stereological Cell Counts
Stereological cell counts were conducted on neurons of the cerebral cortex, hippocampus, and dentate gyrus. These particular brain regions were chosen for study because they govern performance in the open field, the water maze, and prepulse inhibition (Vanderwolf et al., 1997; Morris et al., 1982; Swerdlow et al., 2001).
To quantify neurons of the hippocampal formation and cerebral cortex, we used the optical disector method of stereology. The technique of and rationale for the optical disector method has been described elsewhere (Gundersen et al., 1988; Bonthius et al., 1992; 2004b; West et al., 1988). The optical disector method requires determination of the volume containing the cells of interest, referred to as the “reference volume” (Vref), and the density (Nv) of the cells within that volume. The total number of neurons (N) is the product of the reference volume and the numerical density.
| (equation 1) |
To conduct the stereology, we utilized the frozen section method, previously described by our group (Bonthius et al., 2004b). Details of the methodology are provided in the Supplementary Materials.
The total number of cerebral cortex neurons, hippocampal CA1 pyramidal cells, hippocampal CA3 pyramidal cells, and granule cells of the dentate gyrus were stereologically quantified in animals of all three treatment groups and both genotypes. Methods utilized to identify the specific borders of the brain regions examined are provided in Figure 1 and described in detail in the Supplementary Materials.
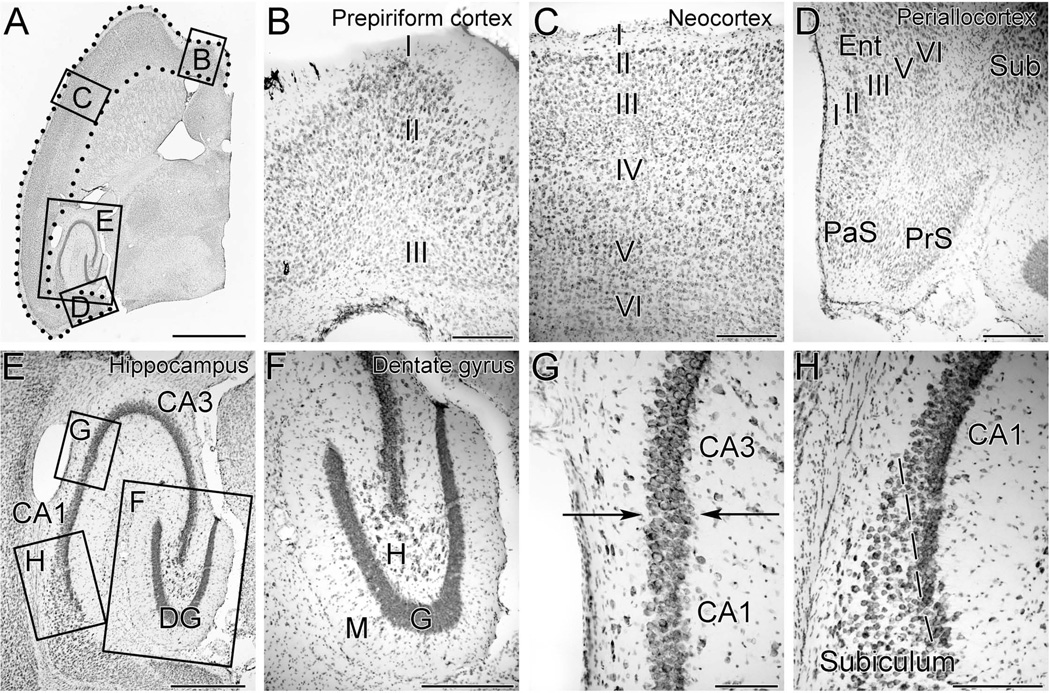
Figure 1. Photomicrographs of a Nissl-stained 40-µm-thick horizontal section of mouse forebrain at a mid-temporal plane demonstrating the boundaries of brain regions examined in this study.
A. The cerebral cortex included all cortical tissue extending from the interhemispheric fissure anteriorly to the subiculum posteriorly and is demarcated by the region within the dotted line.
B. Higher magnification view of the box B in panel A demonstrates the prepiriform cortex, a three-layered cortex, located anteriorly within the cerebral cortex.
C. Higher magnification view of the box C in panel A demonstrates the neocortex, which consists of six distinct cellular layers and constitutes the bulk of the mouse cerebral cortex.
D. Higher magnification view of the box D in panel A demonstrates the periallocortex, a five-layered cortical structure that includes the entorhinal cortex (Ent). The parasubiculum (PaS) and presubiculum (PrS) were also included in the measurements of cerebral cortex, while the more anterior subiculum (Sub) was not.
E. Higher magnification view of the box E in panel A demonstrates the subregions of the hippocampal formation, including the dentate gyrus (DG), CA3 pyramidal cells and CA1 pyramidal cells.
F. Higher magnification of box F in panel E demonstrates that the granule cells of the dentate gyrus (G) can be easily distinguished from cells of the molecular layer (M), and hilus (H).
G. Higher magnification of box G in panel E demonstrates the border between CA3 and CA1 (arrows). CA3 pyramidal cells have larger cell bodies and a lower packing density than the pyramidal cells of CA1.
H. Higher magnification of box H in panel E demonstrates the border (dashed line) between CA1 and the subiculum. The CA1 pyramidal cells abut one another and have a higher packing density than do the cells of the subiculum.
Scale bars represent 2 mm in A, 200 um in B-D, 500um in E, 300 um in F, 100 um in G, and 200 um in H.
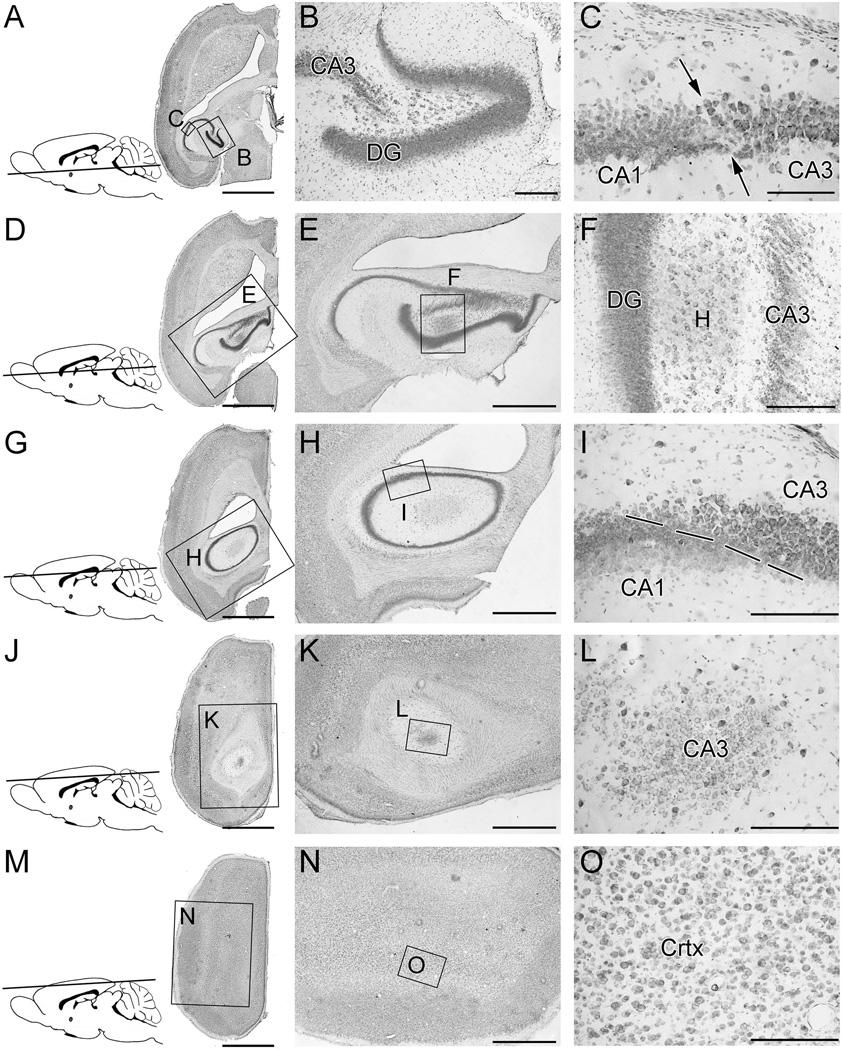
Quantification of cortical and hippocampal neurons required identification of brain region boundaries in the horizontal plane along the entire ventral-dorsal axis. When examining horizontal brain sections, as one progresses from ventral planes to dorsal planes, the shape and position of the hippocampal formation and of its subfields change considerably (Figure 2). Subfields of the hippocampal formation occupy “classical” locations and spatial relationships in the ventral regions, but not in the more dorsal regions. This can make identification of the boundaries of CA1, CA3, and the dentate gyrus challenging in the dorsal locations. However, this challenge was easily overcome by (a) utilizing adjacent sections to sequentially trace the location of the subfields and by (b) examining the sections at higher power, where the different cell sizes and packing densities provided clear guidance regarding the location of subfield boundaries.
Figure 2. Identification of brain region boundaries at various ventral-dorsal levels.
These are Nissl-stained 40-µm-thick horizontal sections of an adult mouse brain. The inset figure in each row shows the approximate horizontal plane through the brain from which the section in that row was derived.
A–C: At this level, which is at a slightly more dorsal level than that shown in figure 1, the hippocampal formation still has a “classical arrangement.” However, the width of the dentate gyrus has expanded. The border between CA1 and CA3 can be distinguished by the different cell soma sizes and packing densities.
D-F: At this level, the shape and location of the hippocampal formation have changed substantially, relative to those observed at more ventral levels. CA3 remains separated from the dentate gyrus by the hilus (H).
G–I: At this level, dentate gyrus granule cells are no longer present. The cytoarchitectural arrangement of the CA1 and CA3 subregions is an oval. The boundary between CA1 and CA3 is readily recognizable in the higher power photomicrograph (I) by differences in soma size and packing density.
J-L: At this level, the section passes tangentially through the dorsal-most portion of CA3. Neither CA1 nor dentate granule cells are present at this level.
M-O: This section passes exclusively through cerebral cortex. No component of the hippocampal formation is present.
Scale bars represent 2 mm in A, D, G, J, and M; 1 mm in E, H, K, and N; 200 um in B, F, L, I and O; 100 um in C
Throughout this study, systematic random sampling was used at all levels of sampling (Sterio, 1984). Table 2 shows the specific sampling parameters utilized for quantifying neurons of the cerebral cortex and hippocampal formation. Further details of the ways in which systematic random sampling was applied in these studies are supplied in the Supplementary Materials.
Table 2.
Stereology sampling scheme used with the optical disector method to quantify neurons of the cerebral cortex and hippocampal formation.
| Cerebral cortex | CA3 pyramidal cells |
CA1 pyramidal cells |
Dentate granule cells |
|
|---|---|---|---|---|
| Disector height | 12 µm | 12 µm | 12 µm | 12 µm |
| Counting frame area | 25 µm x 25 µm | 18 µm x 18 µm | 16 µm x 16 µm | 10 µm x 10 µm |
| Raster pattern sampling step | 1/238 | 1/18 | 1/15 | 1/40 |
| x,y step | 148,750 µm2 | 5832 µm2 | 3840 µm2 | 4000 µm2 |
| Section sampling fraction for Vref | 1/4 | 1/4 | 1/4 | 1/4 |
| Section sampling fraction for Nv | 1/24 | 1/12 | 1/12 | 1/12 |
Statistical analyses
All statistical analyses were conducted with SPSS statistical software. Total brain weight was analyzed by analysis of variance (ANOVA) in which genotype, alcohol treatment, and sex were the fixed factors.
To assess the effect of alcohol on cell survival, neuronal numbers for each of the individual cell populations (CA1 pyramidal cells, CA3 pyramidal cells, dentate gyrus granule cells, and cerebrocortical neurons) were analyzed together by three-way MANOVA in which genotype, alcohol treatment group, and sex were the fixed (between subjects) factors, while the dependent variables were the total number of cells in each of the four brain regions. The MANOVA, in which all cell populations were analyzed together, was followed by ANOVAs, in which each cell population was analyzed separately.
The effect of alcohol on neuronal number for each of the four cell populations was also expressed as a percent reduction from control. For the alcohol-exposed animals, this percent reduction from control is a direct measure of alcohol-induced cell loss. The percent reduction was calculated by subtracting the number of surviving neurons in each brain region in each alcohol-exposed mouse from the corresponding mean control value of the same brain region, then dividing by the mean control value and multiplying by 100. These alcohol-induced percent cell losses for all four of the cell populations were analyzed together by three-way MANOVA, with genotype, treatment group (2.2 and 4.4 mg/g), and sex as the fixed factors and with percent reduction in each cell population as the dependent variables. A significant main effect of genotype in this analysis would indicate that alcohol’s effect on behavior depended on genotype.
For all of the post-hoc analyses in this study, specific between-group comparisons were conducted with Bonferroni adjustments for multiple comparisons.
The results of the behavior studies are described in the companion paper (Karacay et al., 2014). To determine the relationships among cell numbers and behavioral performances, correlational analyses were conducted between each of the quantified cell populations (cerebral cortex, CA1, CA3, and dentate gyrus granule cells) and the most relevant overall measure of performance on each behavioral test (total line crossings for the open field, percent time in the target quadrant for the water maze, and percent inhibition at 16 dB on the PPI test). For these correlational analyses, Pearson correlation coefficients were calculated for each genotype (wild type and nNOS−/− separately) and for all mice in the study (wild type and nNOS−/− combined). Correlations were considered significant at the 0.05 level and below.
Results
Alcohol caused more severe microencephaly in nNOS−/− mice than in wild type
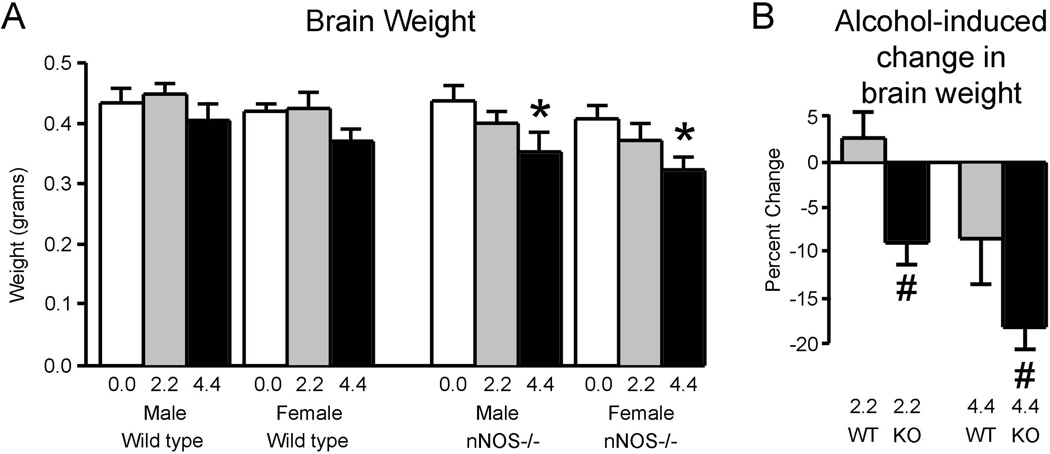
As shown in Figure 3A, brain weights were affected by sex, genotype, and alcohol exposure. For both genotypes, female brains were modestly but consistently smaller than male brains. This led to a significant effect of sex [F(1, 79) = 29.3; p<0.001]. The brains of nNOS−/− mice were slightly, but consistently, smaller than wild type, which led to a significant effect of genotype in the analysis [F(1, 79) = 39.7; p<0.001]. Alcohol reduced brain weights in a dose-dependent fashion for both genotypes and both sexes. This led a significant effect of treatment [F(2, 79) = 77.6; p<0.001]. However, the effect of alcohol on brain growth was more pronounced in the nNOS−/− mice than in wild type, as confirmed by a significant genotype x treatment interaction [F(2, 79) = 14.0; p<0.001]. Post hoc analyses revealed that statistically significant effects of alcohol on brain weight occurred only the nNOS−/− mice and only at the high alcohol dose.
Figure 3. Alcohol caused more severe microencephaly in nNOS−/− mice than in wild type mice.
A. Brain weights of adult male and female mice of the wild type and nNOS−/− strains following exposure to various doses of alcohol during development. Alcohol led to significant dose-dependent reductions in brain weight, but only in the nNOS−/− mice. Post-hoc statistical comparisons for this data set were conducted by comparing animals of the same genotype and sex.
B. Alcohol-induced brain weight reductions were significantly more severe in the nNOS−/− mice than in wild type at both the low (2.2 mg/g) and high (4.4 mg/g) alcohol doses. Because there were no sex differences in alcohol-induced brain weight reductions, data for the two sexes are combined.
* Significantly different from the no alcohol group of the same sex and genotype (p<0.05).
# Significantly different from the wild type group administered the same dose of alcohol (p<0.05).
Figure 3B depicts percent reductions in brain weight (for the combined males and females in each group) induced by alcohol and demonstrates that the nNOS−/− mice were more affected than wild type. This greater effect in the mutant mice was confirmed by a significant effect of genotype in the ANOVA of alcohol-induced changes in brain weight [F(1,53) = 93.9; p<0.001]. Among mice receiving either the 2.2 or 4.4 mg/g/day alcohol doses, the percent reductions in brain weight induced by alcohol were significantly greater in the nNOS−/− mice than in wild type. Thus, alcohol can permanently restrict brain growth in mice, and absence of the nNOS gene worsens this microencephaly.
Alcohol caused greater neuronal losses in nNOS−/− mice than in wild type
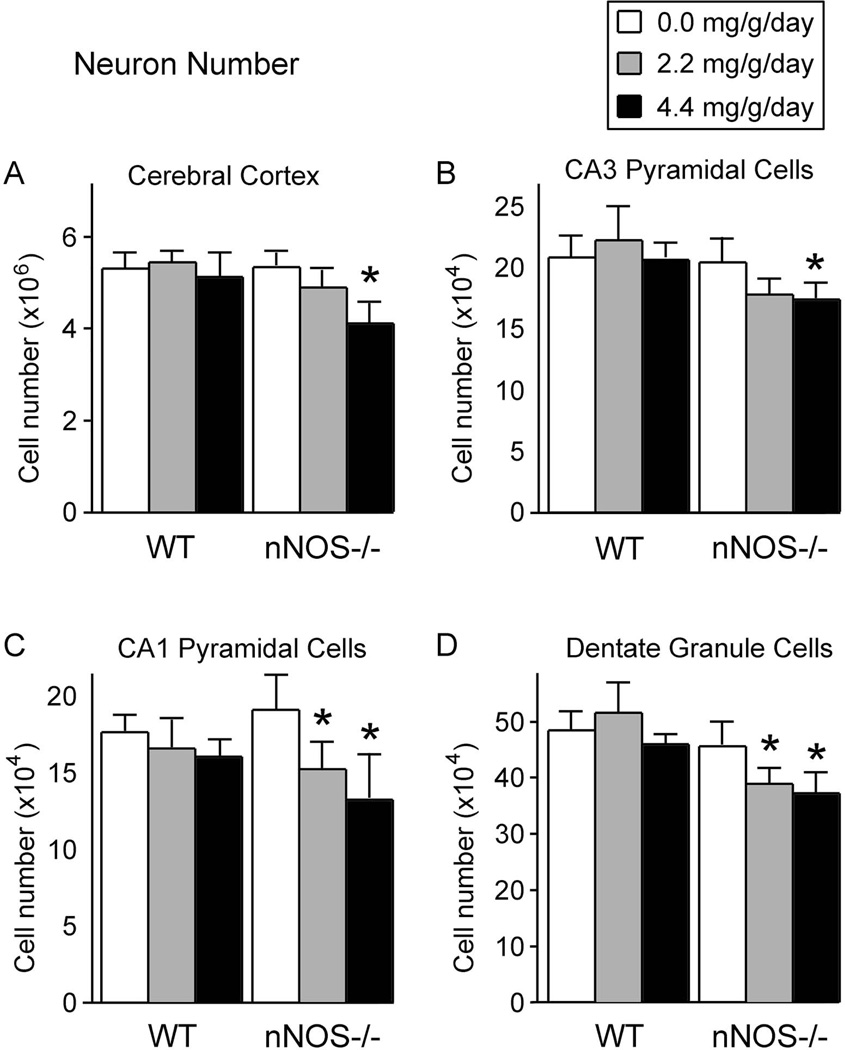
Figure 4 shows the total number of neurons within the cerebral cortex and subfields of the hippocampal formation in wild type and nNOS−/− mice. Because the analysis showed no significant main or interactive effects of sex on any of the cell populations, data from the two sexes were merged. The MANOVA, in which all neuronal populations (cortex, CA1, CA3, and dentate gyrus) were analyzed together, showed a significant genotype x treatment interaction [F(8, 152.0) = 3.868; p<0.001], indicating that the effect of alcohol on neuronal number depended on genotype. Follow-up ANOVAs in which each of the four cell populations was considered separately confirmed a significant genotype x treatment interaction for the cerebral cortex [F(2, 79) = 4.309; p<0.05], CA3 pyramidal cells [F(2, 79) = 4.649; p<0.05], CA1 pyramidal cells [F(2, 79) = 6.460; p<0.005], and dentate gyrus granule cells [F(2, 79) = 6.027; p<0.005].
Figure 4. The effect of alcohol on the total number of neurons within the cerebral cortex and subregions of the hippocampal formation in wild type and nNOS−/− mice.
In all four brain regions, in the absence of alcohol, wild type mice and nNOS−/− mice had equivalent numbers of neurons. Thus, absence of nNOS alone had no effect on neuron number. In the cerebral cortex (A) and in the CA3 hippocampal subregion (B), alcohol induced dose-dependent cell losses, but the losses were significant only in the nNOS−/− mice and only at the high alcohol dose. In the CA1 hippocampal subregion (C) and in the dentate gyrus (D), alcohol induced dose-dependent cell losses in the nNOS−/− mice only, and the reductions were significant at both the low and high alcohol doses.
All measures represent means. Error bars represent standard error of the mean.
* Significantly different from the no alcohol group of the same genotype (p<0.05).
As shown in Figure 4A, in the absence of alcohol, nNOS−/− mice and wild type mice had equivalent numbers of cerebral cortex neurons. Thus, absence of the nNOS gene alone did not induce any change in neuronal number within the cerebral cortex. Alcohol did affect the number of cerebral cortical neurons. However, the patterns in which alcohol affected cortical neuron number depended on genotype and dose. In particular, the low dose of alcohol (2.2 mg/g) did not statistically affect neuronal number in either genotype. The high dose of alcohol (4.4 mg/g) had no effect in wild type mice, but significantly reduced cortical neuron number in the nNOS−/− mice. This more severe effect of alcohol in the mutant mice than in wild type was verified by a significant genotype x treatment interaction [F(2, 79) = 4.309; p<0.05].
The greater vulnerability of the nNOS−/− mice to alcohol-induced loss of cortical neurons was further verified in the analysis of percent cell reductions (figure 5A). Both the 2.2 and 4.4 mg/g/day alcohol doses led to significantly greater cell losses in the nNOS−/− mice than in the wild type mice. At the high dose of alcohol (4.4 mg/g/day), neuronal losses in the cerebral cortex of nNOS−/− mice were three times greater than in wild type. These same patterns of neuronal vulnerability to alcohol in the cerebral cortex were also observed in CA3 pyramidal cells (figures 4B and 5B).
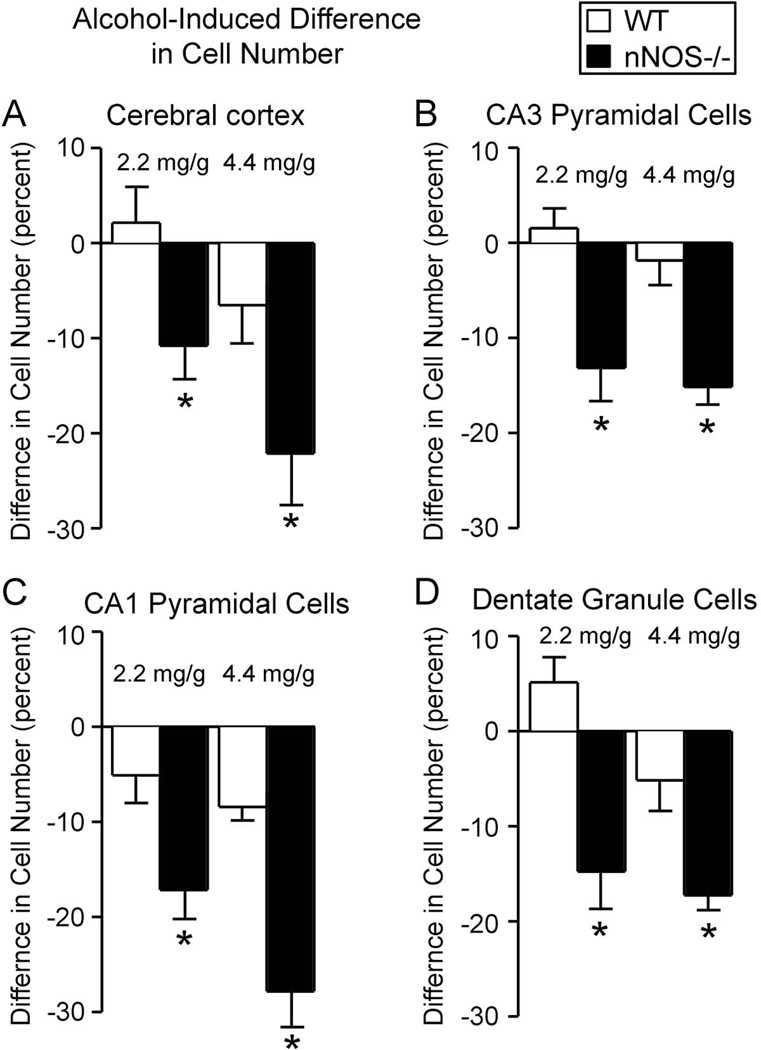
Figure 5. Alcohol induced greater cell losses in nNOS−/− mice than in wild type mice.
Shown here are the percent changes in cell number induced by alcohol in the cerebral cortex (A), CA3 hippocampal subregion (B), CA1 hippocampal subregion (C), and dentate gyrus (D). In all of the regions examined, and at both alcohol doses, alcohol-induced neuronal losses were significantly more severe in nNOS−/− mice than in wild type mice. Thus, absence of nNOS gene expression enhanced the vulnerability of forebrain neurons to alcohol-induced cell loss.
* Significantly different from the wild type group administered the same dose of alcohol (p<0.05).
The greater vulnerability to alcohol-induced cell loss of the nNOS−/− mice was even more apparent in the CA1 pyramidal cells and dentate gyrus granule cells (Figures 4C and 4D). In particular, for these neuronal populations, both the low dose and the high dose of alcohol significantly reduced the number of surviving neurons in the nNOS−/− mice, while neither dose had any significant effect in the wild type mice. Furthermore, the percent cell losses in both CA1 and the dentate gyrus were significantly greater in the nNOS−/− mice than in wild type at both alcohol concentrations (Figure 5C and 5D). In all four cell populations, the lower dose of alcohol (2.2 mg/g/day) led to more severe cell losses in the nNOS−/− mice than did the high alcohol dose in the wild type mice. Thus, absence of nNOS gene function substantially worsened alcohol-induced cell losses in multiple brain regions.
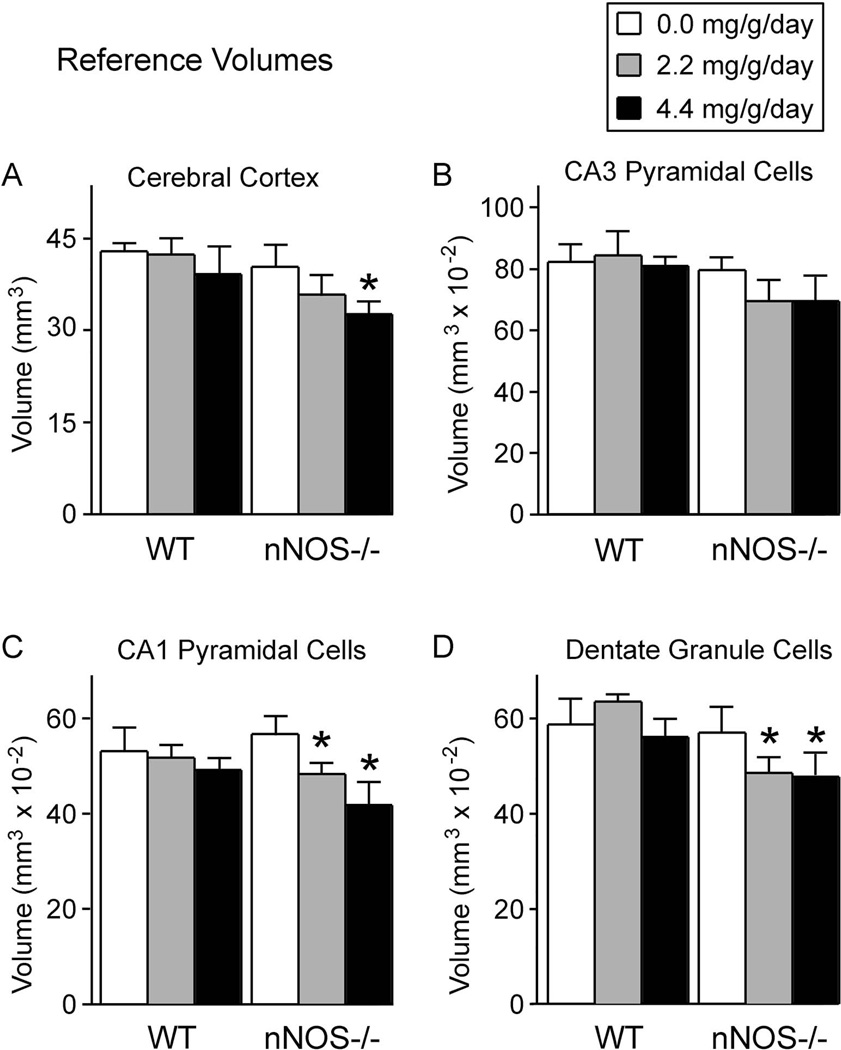
The reductions in cell number were not reflected by changes in cell densities. Neither genotype nor alcohol had any effect on cell densities of any of the four brain regions examined (Table 3). Instead, the reductions in cell number were reflected by reductions of the volumes in which the cells resided (the reference volumes). As shown in figure 6, alcohol significantly reduced the reference volumes of the cerebral cortex, CA1 pyramidal cells, and dentate gyrus. However, these alcohol-induced reductions in reference volume occurred only in the nNOS−/− mice, and not in the wild type mice.
Table 3.
Cellular densities (Nv) (x10,000) of neurons within the cerebral cortex and hippocampal formation*.
| Brain Region |
Wild type | nNOS−/− | ||||
|---|---|---|---|---|---|---|
| 0.0 mg/g/day | 2.2 mg/g/day | 4.4 mg/g/day | 0.0 mg/g/day | 2.2 mg/g/day | 4.4 mg/g/day | |
| Cerebral Cortex | 12.92 ± 0.34 | 13.21± 0.29 | 13.21 ± 0.18 | 13.30 ± 0.22 | 13.39 ± 0.14 | 12.94 ± 0.27 |
| CA3 Pyramidal Cells | 25.73 ± 0.34 | 26.00 ± 0.27 | 25.86 ± 0.64 | 25.74 ± 0.26 | 25.48 ± 0.76 | 25.08 ± 0.39 |
| CA1 Pyramidal Cells | 32.42 ± 0.53 | 32.67 ± 0.74 | 32.72 ± 0.31 | 32.09 ± 0.69 | 32.35 ± 0.51 | 31.84 ±0.45 |
| Dentate Gyrus Granule Cells | 82.52 ± 1.13 | 80.94 ± 1.88 | 83.33 ± 0.86 | 81.00 ± 1.87 | 81.93 ± 1.31 | 80.79 ± 1.16 |
Densities are expressed as cells per cubic mm. Numbers are means ± SEM. Cell densities differed among brain regions, but there were no significant differences in cell densities between the two genotypes or among the alcohol treatment groups.
Figure 6. Alcohol reduced reference volumes, but only in nNOS−/− mice.
Shown here are the volumes in which neurons were distributed (the reference volumes), determined by the Cavalieri method, for neurons of the cerebral cortex (A), CA3 hippocampal subregion (B), CA1 hippocampal subregion (C), and the dentate gyrus (D). In the absence of alcohol, reference volumes in the nNOS−/− mice were equivalent to those of wild type mice. Thus, absence of nNOS gene function alone did not affect the volumes of these forebrain regions. In contrast, in the presence of alcohol, reference volumes were reduced, but only in the nNOS−/− mice. Thus, the enhanced alcohol-induced neuronal losses in the nNOS−/− mice were reflected principally by reductions in reference volumes, rather than by reductions in cellular densities.
Behavior scores were highly correlated with neuron numbers for nNOS−/− mice, but not for wild type mice
In the companion paper, we describe the results of behavioral tests, including open field activity, Morris water maze, and paired pulse inhibition (Karacay et al., 2014). Because the behavioral tests and neuronal cell counts were conducted on the same animals in these studies, it was possible to assess the degree to which behavioral performances were correlated with neuronal number (Table 4). When all of the mice in the study (wild type and nNOS−/−) were considered together, strongly significant correlations emerged between all of the chosen behavioral measures and all of the cell types examined. When only the nNOS−/− mice were considered, most of these correlations remained significant. In contrast, when only the wild type mice were considered, almost all of the correlations became statistically insignificant. Thus, for the nNOS−/− mice, there were significant relationships between neuronal losses and behavioral deficits, and these relationships tended to be absent in wild type mice.
Table 4.
Correlation coefficients between behavioral results and neuron numbers1.
| Genotype | Behavioral test 2,3,4 |
Neuron number by Brain Region | |||
|---|---|---|---|---|---|
| Cerebral cortex neurons |
CA3 Pyramidal cells |
CA1 Pyramidal cells |
Dentate Gyrus neurons |
||
| Wild type | Open Field | NS | −0.322* | NS | NS |
| Morris maze | NS | NS | NS | NS | |
| PPI | NS | NS | NS | NS | |
| nNOS−/− | Open Field | −0.320* | NS | −0.360* | −0.346* |
| Morris maze | 0.444** | NS | 0.453** | NS | |
| PPI | 0.301* | NS | 0.310* | NS | |
| Wild type and nNOS−/− combined | Open Field | −0.370** | −0.394** | −0.350** | −0.418** |
| Morris maze | 0.370** | 0.279** | 0.346** | 0.293** | |
| PPI | 0.210* | 0.266* | 0.230* | 0.242* | |
Numbers are Pearson Correlation Coefficients.
For the Open Field Test, the total number of lines crossed was analyzed. Thus, the negative correlation coefficients imply that, as the number of neurons increased, the number of lines crossed decreased.
For the Morris Water Maze, the percent time spent in the target quadrant during the probe trial was analyzed. The positive correlation coefficients imply that, as the number of neurons increased, the time spent in the target quadrant increased.
For the paired pulse inhibition test (PPI), the percent inhibition at 16 dB was analyzed. Thus, the positive correlation coefficients imply that, as the number of neurons increased, the percent inhibition in response to the prepulse increased.
Significant at the 0.05 level.
Significant at the 0.01 level.
NS = not significant.
Discussion
This study demonstrated for the first time that homozygous mutation of a single gene (nNOS) can permanently worsen alcohol-induced brain injury. This finding is critically important because some human fetuses are much more vulnerable than others to alcohol’s neuroteratogenic effects (Abel, 1995). The results underline the importance of genetic differences in determining individual outcomes of prenatal alcohol exposure (Warren and Li, 2005).
This study of alcohol-exposed nNOS−/− mice, demonstrating worsened neuroanatomical deficits, is especially valuable when combined with the companion study, demonstrating worsened behavioral deficits in those same mice (Karacay et al., 2014). In the companion study, we found that nNOS−/− mice exposed to alcohol are substantially impaired in several behaviors, including the open field, water maze, and prepulse inhibition tests. In the present study, we found that these mice have corresponding dose-dependent neuronal losses in the cerebral cortex and hippocampal formation, which are the brain regions that mediate those behaviors (Vanderwolf et al., 1997; Morris et al., 1982; Swerdlow et al., 2001). Thus, a specific genotype can worsen the impact of developmental alcohol exposure on certain brain regions and on the behaviors governed by those brain regions.
Alcohol exposure in this study induced little neuronal loss in the wild type mice, suggesting that wild type mice are relatively insensitive to alcohol. This finding appears to contrast with reports from the Olney group, in which alcohol exposure during development induced substantial apoptosis (Young and Olney, 2005; Conti et al., 2009). However, methodologic differences between the studies are instructional. In Olney’s studies, larger doses of alcohol were administered, resulting in greater maximum blood alcohol concentrations. In addition, the region most markedly affected in Olney’s reports was the striatum, which was not examined in the present study. These differences underline the likely important interactions among blood alcohol concentration, brain region, and genotype in determining the outcome of fetal alcohol exposure.
The precise mechanism through which absence of nNOS increased the vulnerability of neurons to alcohol-induced death is unclear. However, whereas nNOS catalyzes the production of nitric oxide (NO), it is likely that the increased vulnerability was due to a lack of NO. Indeed, using in vitro systems, we have shown that NO donors can protect developing neurons against alcohol-induced death, while agents that inhibit NO production worsen alcohol-induced cellular losses (Bonthius et al., 2008; 2009). In addition, we have demonstrated that adenoviral vector-mediated delivery of the nNOS gene into cerebellar granule neurons protects them against alcohol toxicity, not only in nNOS−/−, but also in wild type granule neurons (Karacay et al., 2007). Thus, NO is neuroprotective against alcohol. NO produces this protective effect via a specific signaling pathway that utilizes a cyclic nucleotide and a protein kinase, the NO-cGMP-PKG pathway (Bonthius et al., 2004a). In the presence of alcohol, stimulation of this pathway enhances neuronal survival, while inhibition of the pathway induces neuronal death (Bonthius et al., 2004a).
One downstream target of the NO-cGMP-PKG pathway is NF-kB. We have recently shown that stimulation of the NO-cGMP-PKG pathway activates NF-kB, that activation of NF-kB protects neurons against alcohol toxicity, and that inhibition of NF-kB blocks the protective effects of the NO-cGMP-PKG pathway (Bonthius et al., 2008; 2009). Thus, the NO-cGMP-PKG pathway protects neurons against alcohol, at least in part, by activating NF-kB. A transcription factor, NF-kB can promote neuronal survival by altering gene expression (Beg and Baltimore, 1996). The gene or genes whose expression NF-kB alters to protect neurons against alcohol is unknown, but we hypothesize that at least one of the target genes is nerve growth factor (NGF). Both NO and NF-kB can up-regulate NGF (Heese et al, 1998; Xiong et al, 1999), and NGF can protect neurons against alcohol-induced death (Bonthius et al., 2003). Thus, nNOS may work via the NO-cGMP-PKG pathway and NF-kB to increase NGF expression as its neuroprotective mechanism against alcohol.
Several previous studies have shown that neuronal losses induced by alcohol are greater in nNOS−/− mice than in wild type (Bonthius et al., 2002; 2006; de Licona et al., 2009). However, in all of those studies, neuronal numbers were determined in infant mice, immediately following the alcohol exposure. Because of background cell death (Hamburger, 1975; Pennington et al., 1984) and the ability of some neuronal populations to proliferate in the postnatal period (Bayer, 1982), the reductions in neuronal numbers in nNOS−/− mice observed in the neonates may not last into adulthood. In contrast, this study was conducted after the mice reached adulthood and demonstrated that the worsened neuronal losses in nNOS−/− mice are permanent.
Human fetuses differ in their vulnerability to alcohol toxicity, and much of this variability is likely due to genetic differences. The present studies revealed that mice homozygous for a null mutation of nNOS have worsened microencephaly, neuronal loss, and behavioral dysfunction following alcohol exposure. This raises the question whether the increased vulnerability of some human fetuses could be due to nNOS mutations. Complete genetic deficiency of nNOS has not been described in humans. However, the human nNOS gene encodes 29 exons and includes several polymorphic regions (Hall et al., 1994). These polymorphisms of the nNOS gene influence susceptibility to several diseases, including asthma, epilepsy, infantile pyloric stenosis, and Parkinson’s disease (Chung et al., 1996; Grasemann et al., 1999; Levecque et al., 2003; Sayitoglu et al., 2006). Thus, it is possible that variants in the nNOS gene underlie or contribute to increased alcohol-vulnerability in many human fetuses.
This study identified for the first time a gene (nNOS) whose mutation worsens alcohol-induced brain injury. Through the use of homozygous knockout mice, two other genes have been identified whose mutation can also influence alcohol neuroteratogenicity. However, mutation of those genes protects the brain against alcohol toxicity. One of these genes is the pro-apoptotic BAX gene. Targeted deletion of BAX prevents alcohol-induced apoptosis of Purkinje cells (Young et al., 2003; Heaton et al., 2006). The second gene is tissue plasminogen activator (tPA), whose targeted deletion can prevent caspase-3-mediated neurodegeneration of several forebrain structures following developmental alcohol exposure (Noel et al., 2011). Another gene that influences the risk of fetal alcohol effects is pdgfra, whose mutation worsens alcohol-induced craniofacial defects in a zebrafish model (McCarthy et al., 2013). Taken together, these studies demonstrate that the effect of individual genes on the alcohol-exposed developing brain can range from strongly positive to strongly negative and suggest that the net genetic influence is likely a complex interplay of genes that worsen and ameliorate alcohol’s teratogenic effects
Supplementary Material
Acknowledgements
This work was funded by the John Martin Fund for Neuroanatomic Research and NIH grant 5R01AA021465-02 to DJB.
References
- Abel EL. An update on incidence of FAS: FAS is not an equal opportunity birth defect. Neurotoxicol Teratol. 1995;17:437–443. doi: 10.1016/0892-0362(95)00005-c. [DOI] [PubMed] [Google Scholar]
- Astley SJ, Aylward EH, Olson HC, Kerns K, Brooks A, Coggins TE, Davies J, Dorn S, Gendler B, Jirikowic T, Kraegel P, Maravilla K, Richards T. Magnetic resonance imaging outcomes from a comprehensive magnetic resonance study of children with fetal alcohol spectrum disorders. Alcohol Clin Exp Res. 2009;33:1671–1689. doi: 10.1111/j.1530-0277.2009.01004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer SA. Changes in the total number of dentate granule cells in juvenile and adult rats: a correlated volumetric and 3H-thymidine autoradiographic study. Exp Brain Res. 1982;46:315–323. doi: 10.1007/BF00238626. [DOI] [PubMed] [Google Scholar]
- Beg AA, Baltimore D. An essential role for NF-kappaB in preventing TNF-alpha-induced cell death. Science. 1996;274:782–784. doi: 10.1126/science.274.5288.782. [DOI] [PubMed] [Google Scholar]
- Bonthius DJ, West JR. Permanent neuronal deficits in rats exposed to alcohol during the brain growth spurt. Teratology. 1991;44:147–163. doi: 10.1002/tera.1420440203. [DOI] [PubMed] [Google Scholar]
- Bonthius DJ, Bonthius NE, Napper RMA, West JR. Early postnatal alcohol exposure acutely and permanently reduces the number of granule cells and mitral cells in the rat olfactory bulb: A stereological study. J Comp Neurol. 1992;324:557–566. doi: 10.1002/cne.903240408. [DOI] [PubMed] [Google Scholar]
- Bonthius DJ, Pantazis N, Karacay B, Bonthius NE, Taggard DA, Lothman EW. Alcohol exposure during the brain growth spurt promotes hippocampal seizures, kindling and spreading depression. Alcohol Clin Exp Res. 2001a;25:734–745. [PubMed] [Google Scholar]
- Bonthius DJ, Woodhouse J, Bonthius NE, Taggard DA, Lothman EW. Reduced seizure threshold and hippocampal cell loss in rats exposed to alcohol during the brain growth spurt. Alcohol Clin Exp Res. 2001b;25:70–82. [PubMed] [Google Scholar]
- Bonthius DJ, Tzouras G, Karacay B, Mahoney J, Hutton A, McKim R, Pantazis NJ. Deficiency of neuronal nitric oxide synthase (nNOS) worsens alcohol-induced microencephaly and neuronal loss in developing mice. Dev Brain Res. 2002;138:45–59. doi: 10.1016/s0165-3806(02)00458-3. [DOI] [PubMed] [Google Scholar]
- Bonthius DJ, Karacay B, Dai D, Pantazis NJ. FGF-2, NGF and IGF-1, but not BDNF, utilize a nitric oxide pathway to signal neurotrophic and neuroprotective effects against alcohol toxicity in cerebellar granule cell cultures. Dev Brain Res. 2003;140:15–28. doi: 10.1016/s0165-3806(02)00549-7. [DOI] [PubMed] [Google Scholar]
- Bonthius DJ, Karacay B, Dai D, Hutton A, Pantazis NJ. The NO-cGMP-PKG pathway plays an essential role in the acquisition of ethanol resistance by cerebellar granule neurons. Neurotoxicol Teratol. 2004a;26:47–57. doi: 10.1016/j.ntt.2003.08.004. [DOI] [PubMed] [Google Scholar]
- Bonthius DJ, McKim R, Koele L, Harb H, Karacay B, Mahoney J, Pantazis NJ. Use of frozen sections to determine neuronal number in the murine hippocampus and neocortex using the optical disector and optical fractionator. Brain Res Protocol. 2004b;14:45–57. doi: 10.1016/j.brainresprot.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Bonthius DJ, McKim RA, Koele L, Harb H, Hutton-Kehrberg A, Mahoney J, Karacay B. Severe alcohol-induced neuronal deficits in the hippocampus and neocortex of neonatal mice genetically deficient for neuronal nitric oxide synthase (nNOS) J Comp Neurol. 2006;499:290–305. doi: 10.1002/cne.21095. [DOI] [PubMed] [Google Scholar]
- Bonthius DJ, Bonthius NE, Li S, Karacay B. The protective effect of neuronal nitric oxide synthase (nNOS) against alcohol toxicity depends upon the NO-cGMP-PKG pathway and NF-kappaB. Neurotoxicology. 2008;29:1080–1091. doi: 10.1016/j.neuro.2008.08.007. [DOI] [PubMed] [Google Scholar]
- Bonthius DJ, Luong T, Bonthius NE, Karacay B. Nitric oxide utilizes NF-kB to signal its neuroprotective effect against alcohol toxicity. Neuropharmacology. 2009;56:716–731. doi: 10.1016/j.neuropharm.2008.12.006. [DOI] [PubMed] [Google Scholar]
- Chung E, Curtis D, Chen G, Marsden PA, Twells R, Xu W. Genetic evidence for the neuronal nitric oxide synthase gene (NOS1) as a susceptibility locus for infantile pyloric stenosis. Am J Hum Genet. 1996;58:363–370. [PMC free article] [PubMed] [Google Scholar]
- Conti A, Young C, Olney JW, Muglia LJ. Adenylyl cyclases types 1 and 8 promote pro-survival pathways after ethanol exposure in the neonatal brain. Neurobiol Dis. 2009;33:111–118. doi: 10.1016/j.nbd.2008.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Licona HK, Karacay B, Mahoney J, McDonald E, Luang T, Bonthius DJ. A single exposure to alcohol during brain development induces microencephaly and neuronal losses in genetically susceptible mice, but not in wild type mice. Neurotoxicology. 2009;30:459–470. doi: 10.1016/j.neuro.2009.01.010. [DOI] [PubMed] [Google Scholar]
- Goodlett CR, Bonthius DJ, Wasserman EA, West JR. In: An animal model of cental nervous system dysfunction associated with fetal alcohol exposure: behavioral and neuroanatomical correlates, in Learning and Memory: Behavioral and Biological Processes. Wasserman EA, Gormezano I, editors. Englewood, NJ: Lawrence Erlbaum; 1992. pp. 183–208. [Google Scholar]
- Grasemann H, Drazen JM, Deykin A, Israel E, De Sanctis GRT, Pillari A. Single tandem repeat polymorphisms in the neuronal nitric oxide synthase gene in different ethnic populations. Hum Hered. 1999;49:139–141. doi: 10.1159/000022861. [DOI] [PubMed] [Google Scholar]
- Gundersen HJG, Bagger P, Bendtsen TF, Evans SM, Korbo L. The new stereological tools: disector, fractionator, nucleator, and point sampled intercepts and their use in pathological research and diagnosis. APMIS. 1988;96:857–881. doi: 10.1111/j.1699-0463.1988.tb00954.x. [DOI] [PubMed] [Google Scholar]
- Hall AV, Antoniou H, Wang Y, Cheung AH, Arbus M, Olsen SL. Structural organization of the human neuronal nitric oxide synthase gene (NOS1) Biol Chem. 1994;269:33082–33090. [PubMed] [Google Scholar]
- Hamburger V. Cell death in the development of the lateral motor column of the chick embryo. J Comp Neurol. 1975;160:535–546. doi: 10.1002/cne.901600408. [DOI] [PubMed] [Google Scholar]
- Heaton MB, Paiva M, Madorsky I, Siler-Marsiglio K, Shaw G. Effect of bax deletion on ethanol sensitivity in the neonatal rat cerebellum. J Neurobiol. 2006;66:95–101. doi: 10.1002/neu.20208. [DOI] [PubMed] [Google Scholar]
- Heese K, Fiebich BL, Bauer J, Otten U. NF-kB modulates liposaccharide-induced microglial nerve growth factor expression. Glia. 1998;22:401–407. doi: 10.1002/(sici)1098-1136(199804)22:4<401::aid-glia9>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- Huang PL, Dawson TM, Bredt DS, Snyder SH, Fishman MC. Targeted disruption of the neuronal nitric oxide synthase gene. Cell. 1993;75:1273–1286. doi: 10.1016/0092-8674(93)90615-w. [DOI] [PubMed] [Google Scholar]
- Karacay B, Li G, Pantazis NJ, Bonthius DJ. Stimulation of the cAMP pathway protects cultured cerebellar granule neurons against alcohol-induced cell death by activating the neuronal nitric oxide synthase (nNOS) gene. Brain Res. 2007;1143:34–45. doi: 10.1016/j.brainres.2007.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karacay B, Bonthius NE, Plume J, Bonthius DJ. Genetic absence of nNOS worsens fetal alcohol effects in mice. I: Behavioral deficits. Alcohol Clin Exp Res. 2014 doi: 10.1111/acer.12616. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levecque C, Elbaz A, Clavel J, Richard F, Vidal JS, Amouyel P. Association between Parkinson’s disease and polymorphisms in the nNOS and iNOS genes in a community-based case control study. Hum Mol Genet. 2003;12:79–86. doi: 10.1093/hmg/ddg009. [DOI] [PubMed] [Google Scholar]
- McCarthy N, Wetherill L, Lovely CB, Swartz ME, Foroud TM, Eberhart JK. Pdgfra protects against ethanol-induced craniofacial defects in a zebrafish model of FASD. Development. 2013;140:3254–3265. doi: 10.1242/dev.094938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RG, Garrud J, Rawlins NP, O’Keefe J. Place navigation impaired in rats with hippocampal lesions. Nature. 1982;297:681–683. doi: 10.1038/297681a0. [DOI] [PubMed] [Google Scholar]
- Noel M, Norris EH, Strickland S. Tissue plasminogen activator is required for the development of fetal alcohol syndrome in mice. PNAS. 2011;108:5069–5074. doi: 10.1073/pnas.1017608108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman AL, Crocker N, Mattson SN, Riley EP. Neuroimaging and fetal alcohol spectrum disorders. Dev Disabil Res Rev. 2009;15:209–217. doi: 10.1002/ddrr.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olney JW, Ishimaru MJ, Bittigau P, Ikonomidou C. Ethanol-induced apoptotic neurodegeneration in the developing brain. Apoptosis. 2000;5:515–521. doi: 10.1023/a:1009685428847. [DOI] [PubMed] [Google Scholar]
- Patten AR, Moller DJ, Graham J, Gil-Mohapel J, Christie BR. Liquid diets reduce cell proliferation but not neurogenesis in the adult rat hippocampus. Neuroscience. 2013;254:173–184. doi: 10.1016/j.neuroscience.2013.09.024. [DOI] [PubMed] [Google Scholar]
- Pennington SN, Taylor WA, Cowan DH, Kalmus GW. A single dose of ethanol suppresses rat embryo development in vivo. Alcohol Clin.Exp Res. 1984;8:326–329. doi: 10.1111/j.1530-0277.1984.tb05521.x. [DOI] [PubMed] [Google Scholar]
- Sayitoglu MA, Satik S, Unlucerci Y, Ozbek U, Bekpinar S, Eskazan E, et al. Neuronal Nos (Nos 1) polymorphism in patients with epilepsy: a pilot study. J Neurol Sci (Turkish) 2006;23:20–25. [Google Scholar]
- Sterio DC. The unbiased estimation of number and sizes of arbitrary particles using the disector. J Microsc. 1984;134:127–136. doi: 10.1111/j.1365-2818.1984.tb02501.x. [DOI] [PubMed] [Google Scholar]
- Streissguth AP, Dehaene P. Fetal alcohol syndrome in twins of alcoholic mothers: concordance of diagnosis and IQ. Am J Med Genet. 1993;47:857–861. doi: 10.1002/ajmg.1320470612. [DOI] [PubMed] [Google Scholar]
- Swerdlow NR, Geyer MA, Braff DL. Neural circuit regulation of prepulse inhibition of startle in the rat: current knowledge and future challenges. Psychopharmacology. 2001;156:194–215. doi: 10.1007/s002130100799. [DOI] [PubMed] [Google Scholar]
- Thomas JD, Goodlett CR, West JR. Alcohol-induced Purkinje cell loss depends on developmental timing of alcohol exposure and correlates with motor performance. Brain Res Dev Brain Res. 1998;105:159–166. doi: 10.1016/s0165-3806(97)00164-8. [DOI] [PubMed] [Google Scholar]
- Vanderwolf CH, McLauchlin M, Dringenberg HC, Baker GB. Brain structures involved in the behavioral stimulant effect of central serotonin release. Brain Res. 1997;772:121–134. doi: 10.1016/s0006-8993(97)00870-6. [DOI] [PubMed] [Google Scholar]
- Xiong H, Yamada K, Jourki H, Kawamura M, Takei N, Han D. Regulation of nerve growth factor release by nitric oxide through cyclic GMP pathway in cortical glial cells. Mol Pharmacol. 1999;56:339–347. doi: 10.1124/mol.56.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren KR, Li TK. Genetic polymorphisms: impact on the risk for fetal alcohol spectrum disorders. Birth Defects Res A Clin Mol Teratol. 2005;73:195–203. doi: 10.1002/bdra.20125. [DOI] [PubMed] [Google Scholar]
- West MJ, Coleman PD, Flood DG. Estimating the number of granule cells in the dentate gyrus with the disector. Brain Res. 1988;448:167–172. doi: 10.1016/0006-8993(88)91114-6. [DOI] [PubMed] [Google Scholar]
- Young C, Klocke BJ, Tenkova T, Choi J, Labruyere J, Qin YQ, Holtzman DM, Roth KA, Olney JW. Ethanol-induced neuronal apoptosis in vivo requires BAX in the developing mouse brain. Cell Death Differentiation. 2003;10:1148–1155. doi: 10.1038/sj.cdd.4401277. [DOI] [PubMed] [Google Scholar]
- Young C, Olney JW. Neuroapoptosis in the infant mouse brain triggered by a transient small increase in blood alcohol concentration. Neurobiol Dis. 2005;22:548–554. doi: 10.1016/j.nbd.2005.12.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.