Abstract
Background
Although it is well known that drug pressure selects for drug-resistant parasites, the role of transmission reduction by insecticide-treated bed nets (ITNs) on drug resistance remains unclear. In this study, the drug resistance profile of current and previous first-line anti-malarials in Kenya was assessed within the context of drug policy change and scale-up of ITNs. National first-line treatment changed from chloroquine (CQ) to sulphadoxine-pyrimethamine (SP) in 1998 and to artemether-lumefantrine (AL) in 2004. ITN use was scaled-up in the Asembo, Gem and Karemo areas of western Kenya in 1997, 1999 and 2006, respectively.
Methods
Smear-positive samples (N = 253) collected from a 2007 cross-sectional survey among children in Asembo, Gem and Karemo were genotyped for mutations in pfcrt and pfmdr1 (CQ), dhfr and dhps (SP), and at pfmdr-N86 and the gene copy number in pfmdr1 (lumefantrine). Results were compared among the three geographic areas in 2007 and to retrospective molecular data from children in Asembo in 2001.
Results
In 2007, 69 and 85% of samples harboured the pfmdr1-86Y mutation and dhfr/dhps quintuple mutant, respectively, with no significant differences by study area. However, the prevalence of the pfcrt-76T mutation differed significantly among areas (p <0.02), between 76 and 94%, with the highest prevalence in Asembo. Several 2007 samples carried mutations at dhfr-164L, dhps-436A, or dhps-613T. From 2001 to 2007, there were significant increases in the pfcrt-76T mutation from 82 to 94% (p <0.03), dhfr/dhps quintuple mutant from 62 to 82% (p <0.03), and an increase in the septuple CQ and SP combined mutant haplotype, K76Y86I51R59N108G437E540, from 28 to 39%. The prevalence of the pfmdr1-86Y mutation remained unchanged. All samples were single copy for pfmdr1.
Conclusions
Molecular markers associated with lumefantrine resistance were not detected in 2007. More recent samples will be needed to detect any selective effects by AL. The prevalence of CQ and SP resistance markers increased from 2001 to 2007 in the absence of changes in transmission intensity. In 2007, only the prevalence of pfcrt-76T mutation differed among study areas of varying transmission intensity. Resistant parasites were most likely selected by sustained drug pressure from the continued use of CQ, SP, and mechanistically similar drugs, such as amodiaquine and cotrimoxazole. There was no clear evidence that differences in transmission intensity, as a result of ITN scale-up, influenced the prevalence of drug resistance molecular markers.
Electronic supplementary material
The online version of this article (doi:10.1186/s12936-015-0588-4) contains supplementary material, which is available to authorized users.
Keywords: Anti-malarial drug resistance, ITNs, Chloroquine resistance, Sulphadoxine-pyrimethamine resistance, ACT resistance, Vector control
Background
Case management of malaria currently relies heavily on the use of a limited set of effective anti-malarial drugs which may become compromised by the development and spread of drug resistance [1]. Parasite resistance to anti-malarial drugs is driven mainly by drug pressure but it has been hypothesized that changes in malaria transmission intensity due to the scale up of vector control interventions, such as insecticide-treated bed nets (ITNs), could also affect anti-malarial drug resistance [2]. In areas where drug policy has changed and transmission-reducing malaria control interventions have achieved high coverage, monitoring the prevalence of molecular markers for drug resistance to current treatment drugs and to previously used drugs could provide a better understanding of how changes in drug pressure and the intensity of malaria transmission influence the profile of molecular markers for anti-malarial drug resistance.
The role of drug pressure on the emergence of parasite resistance to anti-malarials has been well described [3]. Due to widespread parasite resistance to chloroquine (CQ) and, subsequently, sulphadoxine-pyrimethamine (SP), all malaria-endemic countries in sub-Saharan Africa have adopted artemisinin-based combination therapy (ACT) as the first-line policy for treatment of uncomplicated Plasmodium falciparum infection [4]. However, the emergence of reduced sensitivity to artemisinins in focal areas of Southeast Asia has prompted global concern [5-10]. Although artemisinin resistance has not yet been documented on the African continent [11], monitoring parasite resistance to artemisinins or to ACT partner drugs is vital for malaria control. In addition, long-term monitoring of parasite sensitivity to previously withdrawn anti-malarial drugs, such as CQ, can provide useful surveillance information if these drugs target similar resistance markers to current or candidate ACT partner drugs [12].
In addition to drug pressure, many other factors can contribute to the development and spread of drug resistance, notably the pharmacodynamic properties of anti-malarials, substandard drugs, inadequate dosing, incomplete compliance, as well as human host factors such as age/immunity, vector and ecological characteristics [3]. Among these factors, the relationship between reductions in malaria transmission intensity by vector control tools and drug resistance is complex and remains unclear due to the limited empirical evidence available from field studies [13]. In Tanzania, the short-term use of ITNs was linked to decreased prevalence of a molecular marker associated with resistance to SP [14] and, in Zimbabwe, the implementation of indoor residual spraying (IRS) was associated with a lower risk of CQ treatment failure and reduced prevalence of CQ-resistant parasites [15]. However, evaluations of insecticide-treated curtains in Burkina Faso [16] and ITNs in western Kenya five years after their introduction indicated no clear effect of transmission reduction on the prevalence of resistance markers for SP and CQ [17]. Despite these conflicting findings, these studies suggest that changes in malaria transmission by vector control interventions, which are key elements of malaria control, might influence anti-malarial drug resistance.
Molecular markers are useful for monitoring parasite resistance to anti-malarial drugs. The combination of the dihydrofolate reductase (dhfr) triple mutant (S108T/N51I/C59R) with the dihydropteroate synthase (dhps) double mutant (A437G/K540E) has been associated with in vivo SP treatment failure [18]. Several single nucleotide polymorphisms (SNPs) in P. falciparum CQ transporter (pfcrt) gene (C72S, M74I, N75E, N75K and K76T) and P. falciparum multidrug-resistance 1 (pfmdr1) gene (N86Y, S1034C, N1042D, and D1246Y) have been linked to CQ resistance [19]. Among these SNPs, K76T and N86Y remain the most important [19,20]. Pfcrt-76T has also been associated with in vivo resistance to amodiaquine (AQ), a 4-aminoquinolone anti-malarial drug that is both structurally related to and shares a similar mechanism of action to CQ [21-23]. In addition, increases in the pfmdr1 copy number have been associated with in vivo and/or in vitro decreased sensitivity to mefloquine and lumefantrine, the ACT partner drug in artesunate-mefloquine (AM) and the artemether-lumefantrine (AL), respectively [24-26]. Several in vitro and in vivo studies, conducted mainly in Africa, have shown that the selection of pfmdr1-N86, N1042, D1246 and pfcrt-K76 susceptible alleles and mutant type of pfmdr1-184F was associated with the development of parasite resistance to lumefantrine [27-32]. Collectively, these findings suggest that the detection of pfmdr1 copy number and/or pfmdr1 SNPs and pfcrt-K76 wild type could be useful molecular markers to monitor resistance to mefloquine and lumefantrine.
As part of a 2007 annual malaria survey conducted in three areas of western Kenya, this study assessed the prevalence of resistance markers to currently and previously used first-line anti-malarials within the context of drug policy change and scale-up of ITNs. The specific objectives were to: 1) determine the prevalence of drug-resistant molecular markers for CQ, SP and lumefantrine after the introduction of AL; 2) assess the relationship between the observed prevalence of drug resistance markers and differences in transmission intensity, as a result of ITN scale-up, among three geographically contiguous study areas in 2007; and, 3) compare the prevalence of the molecular markers from a subset of 2007 samples with molecular data from a 2001 survey from the same study area [17].
Methods
Study sites, population and design
This study was integrated into a 2007 annual malaria parasitaemia and anaemia cross-sectional survey conducted by the Kenya Medical Research Institute (KEMRI) and the US Centers for Disease Control and Prevention (CDC) in three contiguous areas in western Kenya: the Asembo area (formerly Rarieda Division, Bondo District), Gem (formerly Yala and Wagai Divisions, Siaya District) and Karemo (formerly Karemo Division, Siaya District) [33]. One of the primary aims of the survey was to evaluate the effect of malaria control interventions on the community-level prevalence of malaria parasitaemia and anaemia among children. History of anti-malarial drugs and bed net use and dried blood spots (DBS) were obtained from 1,271 (Asembo N = 327, Gem N = 451, Karemo N = 492) participants during the survey. For the study described here, 253 P. falciparum smear-positive samples, 61 in Asembo, 97 in Gem and 95 in Karemo, collected from participants <15 years old were available for genotyping of anti-malarial drug resistance molecular markers. In order to provide a temporal description in drug resistance markers, the data from the 2007 survey in Asembo were compared with molecular data collected from samples obtained in a 2001 community cross-sectional survey conducted in Asembo area in children < five years old as part of a randomized controlled trial of ITNs between 1996 and 2001 [17,34,35].
ITN use has been high in the Asembo and Gem areas since 1997 and 1999, respectively, when nets were first distributed to every household as part of an ITN efficacy trial. ITNs were introduced in Karemo in 2004 and scaled up in 2006 as part of a mass campaign to distribute free bed nets to children under the age of five. At the time of the 2007 survey, the parasitaemia prevalence in participants <15 years old was different among the three study areas: 35.8% (95% confidence interval (CI) 29.0-43.0%) in Asembo, 45.4% (95% CI 39.3-51.6%) in Gem and 50.3% (95% CI 43.3-57.2%) in Karemo (KEMRI/CDC, unpublished data). In the 2001 survey, the prevalence of parasitaemia in children < five years old was 34% in the Asembo area [34]. The entomologic inoculation rate (EIR) was estimated at four infective bites per person per year in Asembo and Gem and 20 infective bites per person per year in Karemo in the 2007 survey (KEMRI/CDC, unpublished data) and at 1.3 infective bites per person per year in Asembo area in 2001 [36].
In 1998, SP replaced CQ as first-line treatment policy for uncomplicated malaria in Kenya. National policy change from SP to AL was officially announced in 2004, although AL was not used in the study areas until mid-2006. AQ, the second-line drug during 1998–2004, temporarily became the first-line drug policy for treatment of uncomplicated malaria prior to the implementation of AL in 2006 [37,38]. SP remains the recommended drug for intermittent preventive treatment for malaria in pregnancy (IPTp) in the country. In addition, cotrimoxazole (CTX, trimethoprim-sulphamethoxazole) was commonly used for first line treatment of acute respiratory illnesses and also for prevention of opportunistic infection as a component of HIV care in western Kenya. CTX, a common antifolate antimicrobial, has similar anti-malarial properties to SP, by inhibiting dhps enzyme in the folic acid biosynthetic pathway.
This study was approved by the Ethical Review Committee of KEMRI, Nairobi, Kenya, the Institutional Review Board of Michigan State University, and the Institutional Review Board of CDC, Atlanta, Georgia. Written informed consent was obtained from parents or legal guardians of children and written assent was also obtained from the participants between 13 and 18 years of age.
Laboratory methods
DNA extraction
Genomic DNA was purified from DBS using the commercial DNA purification QIAamp® blood mini-kit (Qiagen Inc, Valencia, CA, USA).
Genotyping of mutations in drug resistance genes
SNPs at dhfr-50, 51, 59, 108, 164, and dhps-436, 437, 540, 581, 613 were detected using published pyrosequencing procedures [39]. Pfcrt codons 72, 73, 74, 75, 76, pfmdr1 codons 86, 184, 1034, 1042, 1246 and ambiguous results at dhfr and dhps genes were examined using direct sequencing [40].
Pyrosequencing
Primary and secondary (nested) polymerase chain reaction (PCR) primers and pyrosequencing primers were synthesized at the CDC Biotechnology Core Facility, Atlanta, USA. Wild type and mutant positive controls and no DNA template negative controls were included in all experiments. Samples were run in pyrosequencing 96-well plates on PyroMark ID (Biotage AB, Uppsala, Sweden) and SNPs were called using the PyroMark ID 1.0 SNP software (Biotage AB, Uppsala, Sweden) [39].
Direct sequencing
Nested PCR sequence procedures were used [40] and product sequences were read using Big Dye Terminator v3.1 cycle sequencing kit on iCycler thermal cycler (Bio-Rad, Hercules, CA, USA). Dye terminators were cleaned by precipitating reactions in 70% ethanol and rehydrating in 10 μL HiDi formamide. Reactions were sequenced on the 3130 ABI Genetic Analyzer (ABI Prism, Foster City, CA, USA). The predominant sequence in each sample was called based on the highest peak for each SNP.
Pfmdr1 gene copy number
Copy number amplification was performed on the samples that were single infection as determined by the highly polymorphic, neutral microsatellite (MS) marker Poly-α [41]. Amplification of pfmdr1 was detected using real-time PCR (Stratagene MX3005P; Agilent Technologies, Santa Clara, CA) using published reaction conditions, primer sequences and probes labelled with 3′ black hole quencher (BHQ) and 5′6-carboxyfluorescein (FAM), for pfmdr1 or 5′ hexachlorofluorescein (HEX), for β-tubulin [25]. Each experiment included a standard curve with five-fold serial dilutions of 3D7, 3D7 calibrator (one copy), the positive controls Dd2 (three to four copies) and W2Mef (two copies), no DNA template control, and field samples run in triplicate. Copy number was calculated using the comparative ΔΔCt method and rounded to the nearest integer. Samples were repeated if the threshold cycle (Ct) value was above 32 or if the two-tailed 95% CI (determined from the individual replicate ΔΔCt calculations around the copy number estimation) exceeded 0.4 [25,42].
Genetic definitions
All SNPs genotyped were classified as wild type or mutant and analysed individually first. Genotypes related to SP resistance markers (dhfr, dhps and combined dhfr/dhps) were defined based on the criteria outlined by Kublin and colleagues [18]. The dhfr genotype reflected mutations in dhfr-51, 59 and 108, classified as wild type, single, double and triple mutants. The dhps genotype described mutations in dhps-437 and 540, categorized as wild type, single and double mutants. The combined dhfr/dhps genotype (mutations in dhfr-51, 59, 108, and dhps-437, 540) was classified as wild type, single, double, triple, quadruple, and quintuple mutants.
Haplotypes were also constructed for dhfr codons 50, 51, 59, 108, and 164 and dhps codons 436, 437, 540, 581, and 613 in the samples that were single infection based on the neutral MS marker, poly-α. The dhfr/dhps haplotypes offer information on mutant variants in parasite populations that cannot be assessed from the genotypes described above. Pfcrt haplotypes based on codons 72, 73, 74, 75, and 76 and pfmdr1 haplotypes, including codons 86, 184, 1034, 1042, and 1246, were reported based on direct sequencing data. In addition, CQ and SP combined haplotypes were assembled based on pfcrt-76, pfmdr1-86, dhfr-51, 59,108, and dhps-437, 540 [43,44] in the samples that were single infection by poly-α. The CQ and SP combined haplotypes provide information on the genetic linkage of mutations in parasites carrying drug resistance to multiple drugs and also an insight into potential synergistic effects by the different drug resistance genes on the recovery of sensitive parasites to individual drug [44,45].
Data and statistical analysis
Differences in participant characteristics among the three study areas in the 2007 survey were analysed using Chi-square or Fisher’s exact test (when expected cell counts were less than five) for categorical variables and analysis of variance (ANOVA) for continuous, normally distributed variables. Parasite density was log transformed. Anaemia was defined as haemoglobin level < 11 g/dL. Statistical comparisons in the prevalence of all SNP mutations in dhfr, dhps, pfcrt and pfmdr1 genes and SP resistance genotypes in samples among the three areas and between the 2001 and 2007 surveys were made using Chi-square tests. All statistical tests were considered as independent for each molecular marker. Statistical significance was defined by a two-sided p-value <0.05. Data and statistical analyses were performed using SAS software version 9.2 (SAS Institute Inc., Cary, NC, USA).
Results
Characteristics of smear-positive participants at the 2007 survey by study area
Smear-positive participants included in this study from Asembo, Gem and Karemo were similar in sex and age distribution, proportion anaemic, presence of gametocytes, fever within two weeks prior to survey, and anti-malarial drugs taken for fever. Geometric mean parasite density was low and also similar among the three areas. However, bed net and ITN use differed significantly between the study areas (p <0.0001). Reported bed net (treated or untreated) use in Karemo, Gem and Asembo, respectively, was 20, 44 and 51%, while ITN use was 7, 31 and 49% (Table 1). In addition, reported fever in last 24 hours differed significantly among the three areas (p <0.01), 15% for Asembo, 21% for Gem and 36% for Karemo (Table 1). Notably, self-reported AL use was low in all study areas, ranging from 0 to 3% (Table 1).
Table 1.
Characteristics of Plasmodium falciparum smear-positive participants at the 2007 survey in western Kenya, by study area
| Characteristic | Asembo (n = 61) | Gem (n = 97) | Karemo (n = 95) | p-value a |
|---|---|---|---|---|
| Male sex | 32/61 (53) | 47/96 (49) | 43/91 (46) | 0.75 |
| Age, years | ||||
| ≤5 | 24/61 (39) | 40/97 (41) | 40/95 (42) | 0.94 |
| 6-15 | 37/61 (61) | 57/97(59) | 55/95 (58) | |
| Anaemic, Hb < 11 g/dL | 24 (39) | 33 (34) | 46 (48) | 0.12 |
| Parasite density, geometric mean, per uL (95% CI)b | 548 (351–935) | 513 (374–750) | 704 (488–1,091) | 0.54 |
| Gametocytes present | 9/61 (15) | 19/97 (20) | 12/95 (13) | 0.40 |
| Fever in last two weeks | 31/61 (51) | 62/95 (64) | 47/95 (50) | 0.088 |
| Fever in last 24 hours | 9/59 (15) | 20/95 (21) | 34/95 (36) | 0.0083* |
| Drug taken for feverc | ||||
| Coartem | 1/31 (3) | 1/61 (2) | 0/47 (0) | -- |
| Sulphadoxine-pyrimethamine | 4/31 (13) | 2/61 (3) | 2/47 (4) | 0.20 |
| Antifolated | 7/31(23) | 4/61 (7) | 3/47 (6) | 0.055 |
| Chloroquine | 2/31 (7) | 3/61 (5) | 0/47 (0) | -- |
| Amodiaquine | 4/31 (13) | 5/61 (8) | 2/47 (4) | 0.37 |
| Other (non-anti-malarial) | 24/31(77) | 38/61 (62) | 28/47 (60) | 0.24 |
| Bed net usage | ||||
| Anye (treated or untreated) | 31/61 (51) | 42/95 (44) | 19/95 (20) | <0.0001* |
| ITNf | 30/61 (49) | 29/95 (31) | 7/95 (7) | <0.0001* |
Note. Data are proportion (%) of P. falciparum smear-positive participants with molecular data, unless otherwise indicated.
Hb, haemoglobin; CI, confidence interval; ITN, insecticide-treated bed net.
aDerived from overall Chi-square or Fisher’s exact (where expected cell counts were below 5) test for categorical variables or analysis of variance (ANOVA) for continuous, normally distributed variables.
bAsexual parasite density and the presence of gametocytes were detected by microscopy. Parasite density was log transformed prior to statistical analysis.
cSelf-reported drug use was asked for participants who reported being febrile in the two weeks prior to the survey; participant may have taken multiple drugs to treat fever.
dSulphadoxine-pyrimethamine or Septrin (cotrimoxazole). eReported sleeping under a bed net the night prior to survey.
fITN use among participants who reported sleeping under a bed net the night prior to survey.
*P <0.05, statistically significant.
Prevalence of drug resistance molecular markers
At the 2007 survey
Since there were differences in the age distributions between the 2001 and 2007 surveys, the molecular profiles in children < five and participants between five and fifteen years of age were compared in 2007. No significant differences in molecular profiles were observed between the different age groups (Additional file 1) and therefore, subsequent comparisons between 2001 and 2007 were not adjusted for age.
CQ resistance mutations were highly prevalent; at least 73% of samples in all study areas harboured mutations at pfcrt-74I, 75K/E, or 76T, and mutations at pfmdr1-86Y, 184F, and 1246Y were detected at 69, 23 and 40%, overall, respectively (Figure 1). No samples carried mutations at pfmdr1-1034C, 1042D or pfcrt-72S. The prevalence of pfcrt-76T mutation, but not pfmdr1-86Y, differed significantly among study areas (p <0.02): 94% in Asembo, 81% in Gem and 76% in Karemo (Figure 1).
Figure 1.
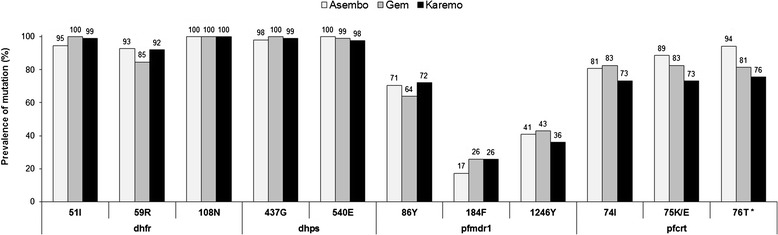
Comparison of prevalence of point mutations among study areas. Point mutations in dhfr-51, 59, 108, dhps-437, 540, pfmdr1-86, 184, 1246 and pfcrt-74, 75, 76 by study area. As few mutations were detected at dhfr-50, 164, dhps-436, 581, 613, pfcrt-72, 73 and pfmdr1-1034, 1042, the data are presented in the Results section, but not shown in this Figure. Statistical analysis performed using Chi-square tests. *p <0.05 difference in prevalence mutations between study areas.
The overall prevalence of key SP resistance mutations at dhfr-51I, 59R, 108N and dhps-437G, 540E exceeded 85% and there was no significant difference in these five SNPs by study area (Figure 1). All samples were wild type at dhfr-C50 and dhps-A581. Several mutations that are usually uncommon in Africa were observed in two samples at dhfr-164L (one sample in Gem, one in Karemo), three at dhps-436A (two samples in Gem, one in Karemo), and one at dhps-613T (Karemo). Combining all areas, the prevalence of the dhfr triple mutant (dhfr 51I/59R/108N) was 88%, dhps double mutant (dhps 437G/540E) was 96% and combined dhfr/dhps quintuple mutant (dhfr 51I/59R/108N + dhps 437G/540E) was 85%. These genotypes did not differ significantly by study area (Figure 2).
Figure 2.
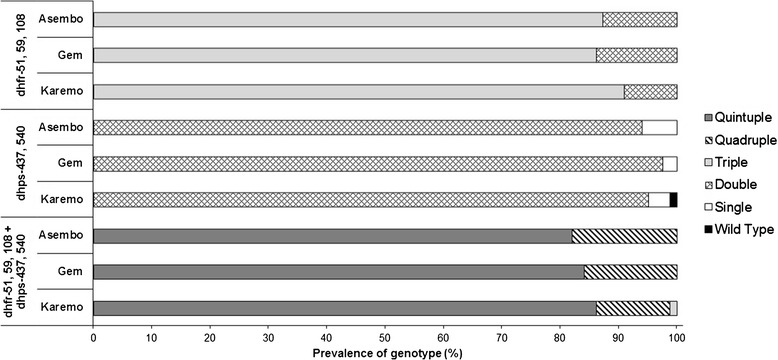
Comparison of prevalence SP drug resistance genotypes among study areas. Dhfr, dhps, and combined dhfr/dhps genotypes by study area. Statistical analysis performed using Chi-square tests. *p <0.05 difference in prevalence mutations between study areas.
Comparison of 2001 and 2007 Asembo surveys
Molecular marker data collected from a 2001 cross-sectional survey in Asembo area [17] were compared with the 2007 molecular data in Asembo only. The prevalence of mutations at pfmdr1-86Y,dhfr-51I,108N and dhps-437G did not change between time points. A significant increase in mutations at pfcrt-76T from 82 to 94% (p <0.03), dhfr-59R from 82 to 93% (p <0.05), and dhps-540E from 83 to 100% (p <0.002) was observed from 2001 to 2007, respectively (Figure 3). Overall, the dhps and combined dhfr/dhps genotypes differed significantly between 2001 and 2007 (p <0.03). The dhps double mutant (dhps-437G/540E) increased considerably from 79% (2001) to 94% (2007) and the combined dhfr/dhps quintuple mutant (dhfr 51I/59R/108N + dhps 437G/540E) was 20 percentage points higher in 2007 compared to the 2001 prevalence of 62% (Figure 4). Pfmdr1 gene copy number was determined in the samples that were single infection determined by the MS marker Poly-α at years 2001 and 2007. All samples were single copy for pfmdr1.
Figure 3.
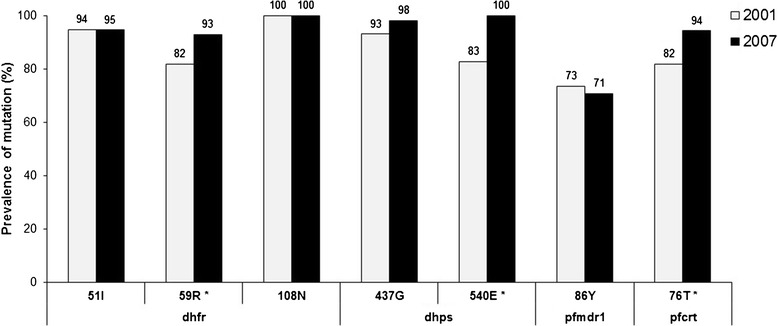
Comparison of prevalence of point mutations in Asembo area between 2001 and 2007 surveys. Point mutations in dhfr 51, 59, 108, dhps 437, 540, pfmdr1-86 and pfcrt-76 by study year. Statistical analysis performed using Chi-square tests. *p <0.05 difference in prevalence of mutations between 2001 and 2007.
Figure 4.
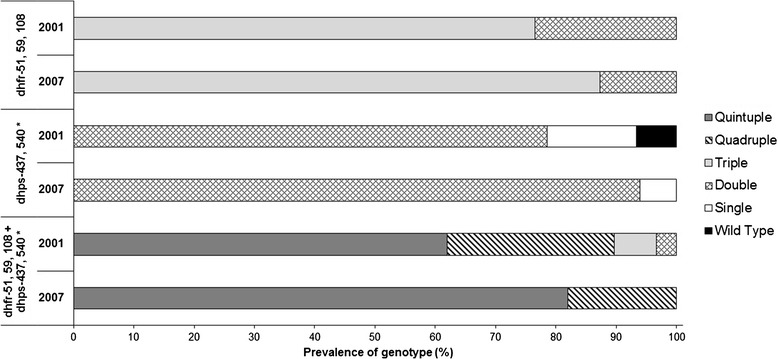
Comparison of prevalence SP drug resistance genotypes in Asembo area between 2001 and 2007 surveys. Dhfr, dhps and combined dhfr/dhps genotypes by study year. Statistical analysis performed using Chi-square tests. *p <0.05 difference in prevalence of mutations between 2001 and 2007.
Haplotypes of CQ, SP, and CQ and SP markers combined
Haplotypes of CQ resistance markers in the 2007 survey
The results of pfcrt and pfmdr1 haplotypes are presented by study area (Tables 2 and 3). For pfcrt, 11 different haplotypes were observed: eight in Asembo, six in Gem and seven in Karemo. The majority of samples in all study areas were triple mutant type for pfcrt-74, 75, and 76, haplotype C72V73I74E75T76 (75% in Asembo, 80% in Gem, 69% in Karemo). The haplotype harbouring an alternative mutant codon K at 75, C72V73I74K75T76, was only detected in one sample from Asembo. Importantly, in Gem and Karemo, 15 and 21% of samples, respectively, were wild type for all five pfcrt codons (C72V73M74N75K76) while substantially fewer samples from Asembo were the wild type haplotype (4%). The remaining eight haplotype variants carrying either single mutation or double mutations at different codons were present from 1 to 8% in three study areas.
Table 2.
Pfcrt haplotypes at the 2007 survey in western Kenya, by study area
| pfcrt haplotype a | 72 | 73 | 74 | 75 | 76 | No. mutations | Asembo, N (%) | Gem, N (%) | Karemo, N (%) |
|---|---|---|---|---|---|---|---|---|---|
| (n = 52) | (n = 86) | (n = 89) | |||||||
| Wild type | C tgt | V gta | M atg | N aat | K aaa | ||||
| Mutant | S agt | I att | E gaa | T aca | |||||
| S tct | K aaa | ||||||||
| C | V | M | N | K | 0 | 2 (4) | 13 (15) | 19 (21) | |
| C | V | I | N | K | 1 | 0 (0) | 1 (1) | 2 (2) | |
| C | V | M | E | K | 1 | 0 (0) | 1 (1) | 0 (0) | |
| C | V | M | N | T | 1 | 3 (6) | 1 (1) | 1 (1) | |
| C | V | M | K | T | 2 | 4 (8) | 0 (0) | 0 (0) | |
| C | V | M | E | T | 2 | 1 (2) | 0 (0) | 3 (3) | |
| C | V | I | N | T | 2 | 1 (2) | 0 (0) | 2 (2) | |
| C | V | I | K | K | 2 | 0 (0) | 1 (1) | 0 (0) | |
| C | V | I | E | K | 2 | 1 (2) | 0 (0) | 1 (1) | |
| C | V | I | K | T | 3 | 1 (2) | 0 (0) | 0 (0) | |
| C | V | I | E | T | 3 | 39 (75) | 69 (80) | 61 (69) |
Note. No. = number. Wild type amino acids are shown in normal font, while mutated amino acids are depicted in bold font. aThe construction of the pfcrt haplotype included codons 72, 73, 74, 75, and 76. All isolates coded for wild type C72 and V73.
Table 3.
Pfmdr1 haplotypes at the 2007 survey in western Kenya, by study area
| pfmdr1 haplotype a | 86 | 184 | 1034 | 1042 | 1246 | No. mutations | Asembo, N (%) | Gem, N (%) | Karemo, N (%) |
|---|---|---|---|---|---|---|---|---|---|
| (n = 47) | (n = 75) | (n = 70) | |||||||
| Wild type | N aat | Y tat | S agt | N aat | D gat | ||||
| Mutant | Y tat | F ttt | C tgt | D gat | Y tat | ||||
| N | Y | S | N | D | 0 | 5 (11) | 10 (13) | 9 (13) | |
| N | Y | S | N | Y | 1 | 2 (4) | 8 (11) | 2 (3) | |
| N | F | S | N | D | 1 | 5 (11) | 6 (8) | 7 (10) | |
| Y | Y | S | N | D | 1 | 16 (34) | 20 (27) | 22 (31) | |
| N | F | S | N | Y | 2 | 1 (2) | 3 (4) | 1 (1) | |
| Y | F | S | N | D | 2 | 2 (4) | 6 (8) | 7 (10) | |
| Y | Y | S | N | Y | 2 | 16 (34) | 17 (23) | 19 (27) | |
| Y | F | S | N | Y | 3 | 0 (0) | 5 (7) | 3 (4) |
Note. No. = number. Wild type amino acids are shown in normal font, while mutated amino acids are depicted in bold font.
a The construction of the pfmdr1 haplotype included codons 86, 184, 1034, 1042, and 1246. All isolates coded for wild type S1034 and N1042.
A total of eight haplotypes were found for pfmdr1, with all eight of these haplotypes present in Gem and Karemo but seven present in Asembo. The wild type haplotype N86Y184S1034N1042D1246 was detected in 11% of samples from Asembo and 13% of samples in Gem and Karemo. The predominant haplotypes in all three areas contained a single mutation at codon 86, Y86Y184S1034N1042D1246 (34% in Asembo, 27% in Gem, 31% in Karemo), or double mutations at codons 86 and 1246, Y86Y184S1034N1042Y1246 (34% in Asembo, 23% in Gem, and 27% in Karemo). The triple mutant at pfmdr1-86, 184 and 1246, haplotype Y86F184S1034N1042Y1246, was found in 7% of samples from Gem, 4% of samples from Karemo and, of note, no samples from Asembo. The remaining four haplotype variants with single mutation (codon 184 or 1246) or double mutations (codons 184 and 1246 or 86 and 184) were present from 1 to 11% in three areas.
Haplotypes of SP resistance markers in the 2007 survey
In total, eight different haplotypes were detected at the 2007 survey across all study areas. Four haplotypes were observed in Asembo, five in Gem and three in Karemo. Overall, approximately 72% of isolates in all three study areas were C50I51R59N108I164S436G437E540A581A613, carrying quintuple mutations at the key dhfr and dhps codons. Another major haplotype observed was the quadruple mutant C50I51C59N108I164S436G437E540A581A613 (19%, overall). The remaining quadruple haplotypes with different combination at five codons (I51R59N108A437E540) were present in four samples in three areas. In Gem, one sample contained a mutation at dhps-A436 in quintuple mutant and another sample had the dhfr-L164 mutation in quadruple haplotype. The presence of the dhfr triple mutant without any additional mutations, C50I51R59N108I164S436A437K540A581A613, was only observed in one sample from Karemo (Table 4).
Table 4.
Haplotypes for dhfr and dhps resistance markers combined at the 2007 survey in western Kenya, by study area
| SP haplotype a | dhfr 51 | dhfr 59 | dhfr 108 | dhfr 164 | dhps 436 | dhps 437 | dhps 540 | No. mutations | Asembo, N (%) (n = 17) | Gem, N (%) (n = 32) | Karemo, N (%) (n = 25) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Wild type | N aat | C tgt | S agc | I ata | S tct | A gct | K aaa | ||||
| Mutant | I att | R cgt | N aac | L tta | A gct | G ggt | E gaa | ||||
| I | R | N | I | S | A | K | 3 | 0 (0) | 0 (0) | 1 (4) | |
| N | R | N | I | S | G | E | 4 | 2 (12) | 0 (0) | 0 (0) | |
| I | C | N | I | S | G | E | 4 | 3 (18) | 6 (19) | 5 (20) | |
| I | R | N | I | S | G | K | 4 | 0 (0) | 1 (3) | 0 (0) | |
| I | R | N | I | S | A | E | 4 | 1 (6) | 0 (0) | 0 (0) | |
| I | R | N | I | S | G | E | 5 | 11 (65) | 23 (72) | 19 (76) | |
| I | C | N | L | S | G | E | 5 | 0 (0) | 1 (3) | 0 (0) | |
| I | R | N | I | A | G | E | 6 | 0 (0) | 1 (3) | 0 (0) |
Note. SP = sulphadoxine-pyrimethamine; No. = number. Wild type amino acids are shown in normal font, while mutated amino acids are depicted in bold font. All samples were single infection for the microsatellite marker poly-α.
aThe construction of the SP haplotype included dhfr-50, 51, 59, 108, 164 and dhps-436, 437, 540, 581, 613. All samples coded for wild type C at dhfr-50, A at dhps-581, and A at dhps-613.
Comparison of haplotypes of CQ and SP resistance markers combined between 2001 and 2007
CQ and SP combined haplotypes, based on pfcrt-76, pfmdr1-86, dhfr-51, 59, 108, and dhps-437, 540, were constructed to determine if there is linkage between CQ resistance genes and the accumulation of SP resistance genes. The combined haplotypes were compared among study areas in 2007 and between 2001 and 2007. Overall, fewer haplotypes were observed at 2007 (seven haplotypes) compared to 2001 (23 haplotypes). In the 2007 survey, two novel haplotypes were observed: T76Y86I51R59N108A437E540 (six mutations) in Asembo and K76N86I51R59N108G437K540 (four mutations) in Gem. Most notably, the prevalence of the septuple mutant haplotype (T76Y86I51R59N108G437E540) increased from 28% in 2001 to 39% in 2007 in the Asembo area. It was also noted that 48 and 41% of parasite isolates from Gem and Karemo, respectively, carried the septuple mutant haplotype in 2007 (Table 5).
Table 5.
Haplotypes for CQ and SP resistance markers combined at the 2001 (Asembo) and 2007 (Asembo, Gem, and Karemo) western Kenya surveys
| CQ and SP haplotype a | crt 76 | mdr1 86 | dhfr 51 | dhfr 59 | dhfr 108 | dhps 437 | dhps 540 | No. mutations | Asembo 2001* N (%) (n = 111) | Asembo 2007, N (%) (n = 18) | Gem 2007, N (%) (n = 31) | Karemo 2007, N (%) (n = 22) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wild type | K aaa | N aat | N aat | C tgt | S agc | A gct | K aaa | |||||
| Mutant | T aca | Y tat | I att | R cgt | N aac | G ggt | E gaa | |||||
| K | N | I | C | N | A | K | 2 | 1 (1) | 0 (0) | 0 (0) | 0 (0) | |
| T | N | I | C | N | A | K | 3 | 1 (1) | 0 (0) | 0 (0) | 0 (0) | |
| T | Y | I | C | N | A | K | 4 | 3 (3) | 0 (0) | 0 (0) | 0 (0) | |
| T | Y | N | R | N | A | K | 4 | 1 (1) | 0 (0) | 0 (0) | 0 (0) | |
| K | Y | I | R | N | A | K | 4 | 1 (1) | 0 (0) | 0 (0) | 0 (0) | |
| T | N | I | R | N | A | K | 4 | 2 (2) | 0 (0) | 0 (0) | 0 (0) | |
| T | N | I | C | N | G | K | 4 | 1 (1) | 0 (0) | 0 (0) | 0 (0) | |
| T | N | N | R | N | G | K | 4 | 2 (2) | 0 (0) | 0 (0) | 0 (0) | |
| K | N | I | R | N | G | K | 4 | 0 (0) | 0 (0) | 1 (3) | 0 (0) | |
| K | N | I | C | N | G | E | 4 | 1 (1) | 1 (6) | 1 (3) | 0 (0) | |
| T | Y | I | R | N | G | K | 5 | 2 (2) | 0 (0) | 0 (0) | 0 (0) | |
| T | Y | I | C | N | G | K | 5 | 2 (2) | 0 (0) | 0 (0) | 0 (0) | |
| K | Y | I | R | N | G | K | 5 | 1 (1) | 0 (0) | 0 (0) | 0 (0) | |
| T | N | I | R | N | G | K | 5 | 1 (1) | 0 (0) | 0 (0) | 0 (0) | |
| K | Y | I | C | N | G | E | 5 | 3 (3) | 0 (0) | 2 (7) | 1 (5) | |
| T | N | I | C | N | G | E | 5 | 7 (6) | 0 (0) | 3 (10) | 1 (5) | |
| T | N | N | R | N | G | E | 5 | 4 (4) | 1 (6) | 0 (0) | 0 (0) | |
| K | N | I | R | N | G | E | 5 | 3 (3) | 0 (0) | 0 (0) | 1 (5) | |
| T | Y | I | R | N | G | K | 6 | 4 (4) | 0 (0) | 0 (0) | 0 (0) | |
| T | Y | I | R | N | A | E | 6 | 0 (0) | 1 (6) | 0 (0) | 0 (0) | |
| T | Y | N | R | N | G | E | 6 | 5 (5) | 1 (6) | 0 (0) | 0 (0) | |
| T | Y | I | C | N | G | E | 6 | 11 (10) | 2 (11) | 1 (3) | 3 (14) | |
| K | Y | I | R | N | G | E | 6 | 1 (1) | 0 (0) | 0 (0) | 3 (14) | |
| T | N | I | R | N | G | E | 6 | 23 (21) | 5 (28) | 8 (26) | 4 (18) | |
| T | Y | I | R | N | G | E | 7 | 31 (28) | 7 (39) | 15 (48) | 9 (41) |
Note. CQ = chloroquine; SP = sulphadoxine-pyrimethamine; No. = number. Wild type amino acids are shown in normal font, while mutated amino acids are depicted in bold font. All samples were single infection for the microsatellite marker poly-α.
aThe construction of the CQ-SP haplotype included pfcrt-76, pfmdr1 -86, dhfr-51, 59, 108, and dhps-437, 540.
*Data are derived from a previous study [17].
Discussion
The objective of this study was to assess the effect of changes in drug treatment policy and ITN scale-up on resistance markers for CQ, SP and lumefantrine. At the 2007 survey, only the prevalence of pfcrt-76T mutation differed among study areas. The prevalence of CQ and SP resistance markers increased from 2001 to 2007 despite drug policy change. No molecular marker evidence for lumefantrine resistance was detected.
In order to address the study objective, qualitative rankings of transmission intensity were determined based on the proxy measures of ITN use, EIR, years since ITN scale-up and malaria prevalence. Collectively, these measures indicate that in 2007 a higher level of transmission in Karemo (ITN scale-up in 2006, ITN usage 7%, EIR 20 and parasite prevalence 50%) than in Asembo and Gem (ITN scale-up in 1997 and 1999, ITN usage 31-49%, EIR 4 and parasite prevalence 36-45%). In Asembo, there was no significant change in parasite prevalence and EIR between 2001 and 2007, suggesting that the intensity of malaria transmission was similar between the two time points. In 1998, SP was introduced as first-line treatment for uncomplicated malaria, followed by AL in 2006. While these changes occurred in an uncontrolled manner, it is useful to describe how these broad programmatically induced shifts in policies may have affected molecular profiles of anti-malarial drug resistance in this study.
A previous study conducted in Malawi observed a return of wild type pfcrt-K76 molecular marker and CQ efficacy 12 years after CQ was withdrawn [46]. In Kenya, CQ was replaced by SP as the national first-line treatment policy in 1998. A study conducted in coastal Kenya from 1993 to 2006 reported that following the policy change to SP, the prevalence of wild type pfcrt-K76 also increased, although at a slower rate than in Malawi [47]. However, this study showed a significant increase in the prevalence of pfcrt-76T mutation from 76% in 2001 to 94% in 2007. Data from a study conducted between 1996 and 2001 in the same area of western Kenya [17] and this study (Table 1) indicate that CQ was still used in this study area despite policy change. The continued use of CQ after policy change, although limited, could account for the observed significant increase in prevalence of pfcrt-76T mutation. Between 2004 and 2006, AQ was used temporarily as the first-line drug for treatment of uncomplicated malaria prior to the implementation of AL in 2006 and self-reported use of AQ was also observed in the current study (Table 1). Previous reports showed that the pfcrt haplotype, S72V73M74N75T76, is associated with AQ treatment failure [22,23]. It is possible that the continued increase in the prevalence of pfcrt-76T mutation over time could be partially driven by the drug pressure derived from the use of AQ even though all samples were wild type at pfcrt -C72 (Table 2). In contrast, the prevalence of pfmdr1-86Y mutation remained unchanged over time. The significant increase in prevalence of parasites harbouring pfcrt-76T mutation but not pfmdr1-86Y mutation over time further support that CQ and/or AQ use selects for mutations in pfcrt [19,21,22,48,49].
Similarly, significant increases in the prevalence of the dhps double mutant and the dhfr/dhps quintuple mutant genotypes were observed (Figure 4) from 2001 to 2007, as well as the emergence of mutations at dhfr-164L, dhps-436A and dhps-613T in several 2007 samples. In the haplotype analysis of 2007 samples, 72% overall contained quintuple mutant haplotype parasites, C50I51R59N108I164S436G437E540A581A613 (Table 4). The increased selection of SP drug resistance parasites over time could be attributed to several factors. First, although at government health facilities, AL replaced SP as the first-line treatment for uncomplicated malaria in children in mid-2006, SP was still being used in the study area through 2007 (Table 1). This may have resulted in sustained drug pressure on these markers. Second, as SP remains the recommended drug for IPTp in the country, it is possible that pregnant women could serve as infectious reservoir for sustaining and circulating SP resistance parasites [50]. Third, the high use of CTX as an antibiotic treatment for respiratory infections and for the prevention of opportunistic infections in HIV-infected individuals might have also selected for SP drug resistance markers. Therefore, it is necessary to monitor SP molecular markers for an extended period in both the general population and pregnant women after a drug policy change leading to reduced SP drug pressure on the whole population.
The selection of wild type pfmdr1-N86, N1042, D1246, pfcrt-K76 and mutant type of pfmdr1-184F, and increased pfmdr1 gene copy number have been associated with decreased lumefantrine sensitivity [27-30,32]. From 2001 to 2007, the prevalence of the pfmdr1-N86 wild type remained statistically unchanged from 26.6 to 29.4% (Table 1), while a significant increase in the pfcrt-76T mutation was found. Multiple copies of pfmdr1 were not detected in samples at either time point. All 2007 samples were wild type pfmdr1-N1042 while 23.3% carried the mutation pfmdr1-184F and 40% had pfmdr1-1246Y, although 2001 samples were not genotyped for these three codons. The unchanged wild type pfmdr1-N86 from 2001 to 2007 along with low usage of AL (Table 1) detected in this study and too soon after the policy switch to AL is not sufficient to conclude that drug pressure by lumefantrine might select for N86 sensitive parasites. The studies conducted in Mozambique and Uganda reported an increase in parasite population with both pfcrt-K76 and pfmdr1-N86 wild types five years after policy change to AL [27,51]. In addition, Henriques et, al. observed the selection of pfcrt-K76 and pfmdr1-N86 wild types on day 3 in their ACT efficacy trial (including AL arm) conducted in Mbita, southwestern border of Kenya [28]. The difference in the pfcrt-76 results between this study and the report from Mozambique, Uganda and Mbita of Kenya could be mainly due to continued use of CQ and AQ as observed by the self-reported anti-malarial drug use in this study population (Table 1). Continuous monitoring of both pfmdr1-86 and pfcrt-76 in addition to pfmdr1-184, 1042, 1246 and pfmdr1 gene copy number will be essential in detecting the emergence of lumefantrine resistance.
In this study, the drug resistance profiles were also assessed within the context of scale up of ITNs. Theoretical models suggest that changes in malaria transmission intensity, such as by the use of vector control interventions, can indirectly influence the spread of drug resistance through three main clinical/epidemiological ‘mediators’. As proposed by Hastings and Watkins, the three mediators could regulate several effector variables: 1) the number of parasite clones per host, which, in turn, determines the intrahost dynamics and rate of sexual recombination; 2) the perceived threat of infection that affects the level of community/population drug use; or, 3) immunity against malaria, which can change levels of therapeutic drug use and the biomass (number of parasites) per infection [2]. Additionally, the relationship between transmission intensity and spread of drug resistance depends on whether resistance to a drug is encoded by single gene or by multiple genes [2]. The genetic basis of SP is considered monogenetic as SP are encoded by dhps and dhfr genes, respectively. Mathematical models predict that the monogenic-based SP resistance spreads faster in high-transmission areas compared to medium- and low-transmission areas [2]. In contrast, CQ resistance, encoded by two genes, pfcrt and pfmdr1 [19,49], is multigenic and follows a concave curve such that the prevalence of resistance markers is highest in low- and high-transmission areas and lowest in areas of intermediate transmission [2,52].
This study observed the prevalence of the pfcrt-76T mutation differed among areas of varying transmission intensity in 2007 with the highest prevalence in the lowest transmission area (Asembo) and lowest prevalence in highest transmission area (Karemo) (Figure 1). In addition, haplotype analysis of pfcrt showed that fewer samples from Asembo were the wild type (C72V73M74N75K76) haplotype compared to Gem and Karemo (Table 2). However, there were no significant differences in the prevalence of pfmdr1-86Y mutation and SP dhfr/dhps quintuple mutant genotype among the three areas (Figures 1 and 2). There are several reasons why the prevalence data for both CQ and SP markers from this study do not fit the theoretical models described above. First, the similarity in prevalence of pfmdr1, dhfr and dhps markers among the three areas could be explained by the essentially medium to high transmission and small relative differences in transmission intensity between areas. As noted, the difference between an EIR of 4 (Asembo and Gem) and an EIR of 20 (Karemo) would have minimal impact on parasite population genetic structure, resulting in reduced ability to determine the relationship between transmission intensity and the prevalence of molecular markers for the three genes. It is also possible that the gene flow due to migration of human and mosquito populations between the areas or from other parts of Kenya, unmeasured in this study, might contribute to the lack of difference in prevalence of the pfmdr1, dhfr and dhps gene markers among the three areas. Second, the observed higher prevalence of pfcrt-76T mutation and loss of more wild type (C72V73M74N75K76) haplotype in Asembo compared to the other two areas are weak although notable. The lack of information of CQ and AQ use at the population level for these three study areas and small sample sizes in the reported CQ and AQ use for fever in this study, limited the ability to explore drug pressure, influenced by transmission intensity, on the observed higher prevalence of pfcrt-76T mutation and loss of pfcrt wild haplotype in Asmebo. However, MS data from another study using the same set of samples showed that the overall proportion of multiple clone infections remained high in 2007 but relatively lower in Asembo (88%) compared to Karemo (97%). The relatively lower proportion of multiple clone infections in Asembo might be associated with the occurrence of intrahost dynamics, in which resistant parasite clones bearing pfcrt molecular markers benefit from removal of co-infecting drug-sensitive clones on treatment of human host [2], resulting in high prevalence of pfcrt-76T mutation and loss of pfcrt wild haplotype.
In this study, haplotypes based on key SNPs in pfcrt, pfmdr1, dhfr, and dhps [44,45] were also further compared between 2001 and 2007 surveys and by areas in 2007. In 2001, 23 haplotypes were found which reduced to seven in 2007, with fewer unique haplotypes observed in 2007. The results suggest selection of haplotype variants of multiple drug resistance genes possibly by genetic recombination during the sexual reproduction stage in mosquitoes [53] and the selection pressure could reduce the parasite population size resulting in a more likely loss of rare haplotypes. More importantly, an increase in the septuple mutant (K76Y86I51R59N108G437E540) was detected from 28% in 2001 to 39% in 2007 (Table 5) in Asembo. In addition, in 2007, other two neighbouring areas, Gem and Karemo, showed the prevalence of the septuple haplotype mutant similar to Asembo. These results suggest a strong linkage among mutations of pfcrt, pfmdr1, dhfr, and dhps in the parasite population and a possible effect by the strong linkage among the resistance genes on preventing the recovery of sensitive parasites to individual drug. The increase in the septuple mutant haplotype is alarming as sustained selection pressure could promote an increase in the prevalence of septuple mutant parasites to the point where both CQ and SP resistance mutations eventually reach fixation in the parasite population.
This study had a few limitations. The qualitative ranking of transmission intensity based on proxies, such as parasite prevalence, is subject to wide variation over time. The geographic proximity of the three study areas and evaluation of the effect of transmission intensity on drug resistance at a single point may have also limited the ability to detect differences in the molecular profiles of drug resistance among the areas. The small number of samples analysed in this study might have also reduced the statistical power to detect differences in the prevalence of mutations in all the molecular markers among the areas. In addition, the data presented here were from samples collected in 2001 and 2007 and may not reflect the most recent prevalence of drug resistance markers in the study areas. Additional surveys are currently underway to genotype samples collected after 2007 from the same study areas in order to provide more recent estimates and analyse trends in molecular marker prevalence after a longer period of time after AL implementation. Finally, molecular data on the prevalence of artemisinin resistance markers at baseline and in more recent parasite samples from this area will be critically important.
Conclusion
Molecular markers associated with lumefantrine resistance were not detected in 2007. More recent samples will be needed to detect any selective effects by AL. The prevalence of CQ and SP resistance markers increased from 2001 to 2007 in the absence of changes in transmission intensity. Among these mutations, in 2007, only the prevalence of pfcrt-76T mutation differed among study areas of varying transmission intensity, with the highest prevalence in Asembo. Resistant parasites were most likely selected by sustained drug pressure from the continued use of CQ, SP, despite drug policy change to AL, and mechanistically similar drugs such as AM and CTX. There was no clear evidence that difference in transmission intensity, as a result of ITN scale-up, influenced prevalence of drug resistance molecular markers.
Acknowledgements
We express gratitude to all the study participants for their involvement in the current study. We would like to thank Kimberly Lindblade for providing data from the 2001 survey. Our appreciation extends to the KEMRI/CDC field workers and malaria laboratory staff in Kisumu who assisted with this study and phase II of ITN trial. We thank the Director, Kenya Medical Research Institute for permission to publish this paper. This study is financially supported by Malaria Branch, Division of Parasitic Diseases and Malaria, Center for Global Health, Centers for Disease Control and Prevention and by US National Science Foundation, Ecology of Infectious Diseases grant # EF-0723770.
Disclaimer
The findings and conclusions in this manuscript are those of the authors and do not necessarily represent the opinions of the Centers for Disease Control and Prevention of the US Government.
Additional file
Comparison of prevalence of point mutations, by age.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MS, YOO, AL, and WG carried out genotyping work for the current study. CO, KO, SM, JG, EW, LS, and MH implemented the 2007 cross-sectional survey and JG, FOK, WH, and BN carried out the 2001 survey of ITN trial. MS conducted the statistical analysis of the current study. YPS conceived and designed the current study. MS and YPS wrote the paper. All authors contributed to the interpretation of results and critical discussion of the conclusion and approved the final manuscript.
Contributor Information
Monica Shah, Email: MShah2@cdc.gov.
Yusuf Omosun, Email: gup4@cdc.gov.
Ashima Lal, Email: Ashima_Lal11@gmail.com.
Christopher Odero, Email: COdero@kemricdc.org.
Wangeci Gatei, Email: wgg3@cdc.gov.
Kephas Otieno, Email: KOtieno@kemricdc.org.
John E Gimnig, Email: hzg1@cdc.gov.
Feiko ter Kuile, Email: Feiko.TerKuile@lstmed.ac.uk.
William A Hawley, Email: byh0@cdc.gov.
Bernard Nahlen, Email: bnahlen@usaid.gov.
Simon Kariuki, Email: SKariuki@kemricdc.org.
Edward Walker, Email: walker@msu.edu.
Laurence Slutsker, Email: lms5@cdc.gov.
Mary Hamel, Email: mlh8@cdc.gov.
Ya Ping Shi, Email: yps0@cdc.gov.
References
- 1.White NJ. Antimalarial drug resistance. J Clin Invest. 2004;113:1084–92. doi: 10.1172/JCI21682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hastings IM, Watkins WM. Intensity of malaria transmission and the evolution of drug resistance. Acta Trop. 2005;94:218–29. doi: 10.1016/j.actatropica.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 3.Wongsrichanalai C, Pickard AL, Wernsdorfer WH, Meshnick SR. Epidemiology of drug-resistant malaria. Lancet Infect Dis. 2002;2:209–18. doi: 10.1016/S1473-3099(02)00239-6. [DOI] [PubMed] [Google Scholar]
- 4.WHO . Guidelines for the treatment of malaria. 2. Geneva: World Health Organization; 2010. [PubMed] [Google Scholar]
- 5.Carrara VI, Zwang J, Ashley EA, Price RN, Stepniewska K, Barends M, et al. Changes in the treatment responses to artesunate-mefloquine on the northwestern border of Thailand during 13 years of continuous deployment. PLoS One. 2009;4:e4551. doi: 10.1371/journal.pone.0004551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, et al. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2009;361:455–67. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Noedl H, Se Y, Schaecher K, Smith BL, Socheat D, Fukuda MM, et al. Evidence of artemisinin-resistant malaria in western Cambodia. N Engl J Med. 2008;359:2619–20. doi: 10.1056/NEJMc0805011. [DOI] [PubMed] [Google Scholar]
- 8.Noedl H, Se Y, Sriwichai S, Schaecher K, Teja-Isavadharm P, Smith B, et al. Artemisinin resistance in Cambodia: a clinical trial designed to address an emerging problem in Southeast Asia. Clin Infect Dis. 2010;51:e82–9. doi: 10.1086/657120. [DOI] [PubMed] [Google Scholar]
- 9.Rogers WO, Sem R, Tero T, Chim P, Lim P, Muth S, et al. Failure of artesunate-mefloquine combination therapy for uncomplicated Plasmodium falciparum malaria in southern Cambodia. Malar J. 2009;8:10. doi: 10.1186/1475-2875-8-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Phyo AP, Nkhoma S, Stepniewska K, Ashley EA, Nair S, McGready R, et al. Emergence of artemisinin-resistant malaria on the western border of Thailand: a longitudinal study. Lancet. 2012;379:1960–6. doi: 10.1016/S0140-6736(12)60484-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor SM, Parobek CM, DeConti DK, Kayentao K, Coulibaly SO, Greenwood BM, et al. Absence of putative artemisinin resistance mutations among Plasmodium falciparum in sub-Saharan Africa: a molecular epidemiologic study. J Infect Dis. 2014; jiu467: 1-9. [DOI] [PMC free article] [PubMed]
- 12.Plowe CV, Roper C, Barnwell JW, Happi CT, Joshi HH, Mbacham W, et al. World Antimalarial Resistance Network (WARN) III: molecular markers for drug resistant malaria. Malar J. 2007;6:121. doi: 10.1186/1475-2875-6-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Talisuna AO, Okello PE, Erhart A, Coosemans M, D’Alessandro U. Intensity of malaria transmission and the spread of Plasmodium falciparum resistant malaria: a review of epidemiologic field evidence. Am J Trop Med Hyg. 2007;77:170–80. [PubMed] [Google Scholar]
- 14.Alifrangis M, Lemnge MM, Ronn AM, Segeja MD, Magesa SM, Khalil IF, et al. Increasing prevalence of wildtypes in the dihydrofolate reductase gene of Plasmodium falciparum in an area with high levels of sulfadoxine/pyrimethamine resistance after introduction of treated bed nets. Am J Trop Med Hyg. 2003;69:238–43. [PubMed] [Google Scholar]
- 15.Mharakurwa S, Mutambu SL, Mudyiradima R, Chimbadzwa T, Chandiwana SK, Day KP. Association of house spraying with suppressed levels of drug resistance in Zimbabwe. Malar J. 2004;3:35. doi: 10.1186/1475-2875-3-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diallo DA, Sutherland C, Nebie I, Konate AT, Ord R, Pota H, et al. Sustained use of insecticide-treated curtains is not associated with greater circulation of drug-resistant malaria parasites, or with higher risk of treatment failure among children with uncomplicated malaria in Burkina Faso. Am J Trop Med Hyg. 2007;76:237–44. [PubMed] [Google Scholar]
- 17.Shah M, Kariuki S, Vanden Eng J, Blackstock AJ, Garner K, Gatei W, et al. Effect of transmission reduction by insecticide-treated bednets (ITNs) on antimalarial drug resistance in western Kenya. PLoS One. 2011;6:e26746. doi: 10.1371/journal.pone.0026746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kublin JG, Dzinjalamala FK, Kamwendo DD, Malkin EM, Cortese JF, Martino LM, et al. Molecular markers for failure of sulfadoxine-pyrimethamine and chlorproguanil-dapsone treatment of Plasmodium falciparum malaria. J Infect Dis. 2002;185:380–8. doi: 10.1086/338566. [DOI] [PubMed] [Google Scholar]
- 19.Wellems TE, Plowe CV. Chloroquine-resistant malaria. J Infect Dis. 2001;184:770–6. doi: 10.1086/322858. [DOI] [PubMed] [Google Scholar]
- 20.Dorsey G, Kamya MR, Singh A, Rosenthal PJ. Polymorphisms in the Plasmodium falciparum pfcrt and pfmdr-1 genes and clinical response to chloroquine in Kampala, Uganda. J Infect Dis. 2001;183:1417–20. doi: 10.1086/319865. [DOI] [PubMed] [Google Scholar]
- 21.Olliaro P, Nevill C, LeBras J, Ringwald P, Mussano P, Garner P, et al. Systematic review of amodiaquine treatment in uncomplicated malaria. Lancet. 1996;348:1196–201. doi: 10.1016/S0140-6736(96)06217-4. [DOI] [PubMed] [Google Scholar]
- 22.Sa JM, Twu O. Protecting the malaria drug arsenal: halting the rise and spread of amodiaquine resistance by monitoring the PfCRT SVMNT type. Malar J. 2010;9:374. doi: 10.1186/1475-2875-9-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sa JM, Twu O, Hayton K, Reyes S, Fay MP, Ringwald P, et al. Geographic patterns of Plasmodium falciparum drug resistance distinguished by differential responses to amodiaquine and chloroquine. Proc Natl Acad Sci U S A. 2009;106:18883–9. doi: 10.1073/pnas.0911317106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lim P, Alker AP, Khim N, Shah NK, Incardona S, Doung S, et al. Pfmdr1 copy number and arteminisin derivatives combination therapy failure in falciparum malaria in Cambodia. Malar J. 2009;8:11. doi: 10.1186/1475-2875-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Price RN, Uhlemann AC, Brockman A, McGready R, Ashley E, Phaipun L, et al. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet. 2004;364:438–47. doi: 10.1016/S0140-6736(04)16767-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sidhu AB, Uhlemann AC, Valderramos SG, Valderramos JC, Krishna S, Fidock DA. Decreasing pfmdr1 copy number in Plasmodium falciparum malaria heightens susceptibility to mefloquine, lumefantrine, halofantrine, quinine, and artemisinin. J Infect Dis. 2006;194:528–35. doi: 10.1086/507115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conrad MD, LeClair N, Arinaitwe E, Wanzira H, Kakuru A, Bigira V, et al. Comparative impacts over 5 years of artemisinin-based combination therapies on Plasmodium falciparum polymorphisms that modulate drug sensitivity in Ugandan children. J Infect Dis. 2014;210:344–53. doi: 10.1093/infdis/jiu141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henriques G, Hallett RL, Beshir KB, Gadalla NB, Johnson RE, Burrow R, et al. Directional selection at the pfmdr1, pfcrt, pfubp1, and pfap2mu loci of Plasmodium falciparum in Kenyan children treated with ACT. J Infect Dis. 2014;210:2001–8. doi: 10.1093/infdis/jiu358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mungthin M, Khositnithikul R, Sitthichot N, Suwandittakul N, Wattanaveeradej V, Ward SA, et al. Association between the pfmdr1 gene and in vitro artemether and lumefantrine sensitivity in Thai isolates of Plasmodium falciparum. Am J Trop Med Hyg. 2010;83:1005–9. doi: 10.4269/ajtmh.2010.10-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mwai L, Kiara SM, Abdirahman A, Pole L, Rippert A, Diriye A, et al. In vitro activities of piperaquine, lumefantrine, and dihydroartemisinin in Kenyan Plasmodium falciparum isolates and polymorphisms in pfcrt and pfmdr1. Antimicrob Agents Chemother. 2009;53:5069–73. doi: 10.1128/AAC.00638-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nkhoma S, Nair S, Mukaka M, Molyneux ME, Ward SA, Anderson TJ. Parasites bearing a single copy of the multi-drug resistance gene (pfmdr-1) with wild-type SNPs predominate amongst Plasmodium falciparum isolates from Malawi. Acta Trop. 2009;111:78–81. doi: 10.1016/j.actatropica.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sisowath C, Ferreira PE, Bustamante LY, Dahlstrom S, Martensson A, Bjorkman A, et al. The role of pfmdr1 in Plasmodium falciparum tolerance to artemether-lumefantrine in Africa. Trop Med Int Health. 2007;12:736–42. doi: 10.1111/j.1365-3156.2007.01843.x. [DOI] [PubMed] [Google Scholar]
- 33.Hamel MJ, Adazu K, Obor D, Sewe M, Vulule J, Williamson JM, et al. A reversal in reductions of child mortality in western Kenya, 2003–2009. Am J Trop Med Hyg. 2011;85:597–605. doi: 10.4269/ajtmh.2011.10-0678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindblade KA, Eisele TP, Gimnig JE, Alaii JA, Odhiambo F, ter Kuile FO, et al. Sustainability of reductions in malaria transmission and infant mortality in western Kenya with use of insecticide-treated bednets: 4 to 6 years of follow-up. JAMA. 2004;291:2571–80. doi: 10.1001/jama.291.21.2571. [DOI] [PubMed] [Google Scholar]
- 35.Phillips-Howard PA, ter Kuile FO, Nahlen BL, Alaii JA, Gimnig JE, Kolczak MS, et al. The efficacy of permethrin-treated bed nets on child mortality and morbidity in western Kenya II. Study design and methods. Am J Trop Med Hyg. 2003;68:10–5. [PubMed] [Google Scholar]
- 36.Lindblade KA, Gimnig JE, Kamau L, Hawley WA, Odhiambo F, Olang G, et al. Impact of sustained use of insecticide-treated bednets on malaria vector species distribution and culicine mosquitoes. J Med Entomol. 2006;43:428–32. doi: 10.1093/jmedent/43.2.428. [DOI] [PubMed] [Google Scholar]
- 37.Amin AA, Zurovac D, Kangwana BB, Greenfield J, Otieno DN, Akhwale WS, et al. The challenges of changing national malaria drug policy to artemisinin-based combinations in Kenya. Malar J. 2007;6:72. doi: 10.1186/1475-2875-6-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sasi P, Abdulrahaman A, Mwai L, Muriithi S, Straimer J, Schieck E, et al. In vivo and in vitro efficacy of amodiaquine against Plasmodium falciparum in an area of continued use of 4-aminoquinolines in East Africa. J Infect Dis. 2009;199:1575–82. doi: 10.1086/598862. [DOI] [PubMed] [Google Scholar]
- 39.Zhou Z, Poe AC, Limor J, Grady KK, Goldman I, McCollum AM, et al. Pyrosequencing, a high-throughput method for detecting single nucleotide polymorphisms in the dihydrofolate reductase and dihydropteroate synthetase genes of Plasmodium falciparum. J Clin Microbiol. 2006;44:3900–10. doi: 10.1128/JCM.01209-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alam MT, de Souza DK, Vinayak S, Griffing SM, Poe AC, Duah NO, et al. Selective sweeps and genetic lineages of Plasmodium falciparum drug -resistant alleles in Ghana. J Infect Dis. 2011;203:220–7. doi: 10.1093/infdis/jiq038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gatei W, Kariuki S, Hawley W, ter Kuile F, Terlouw D, Phillips-Howard P, et al. Effects of transmission reduction by insecticide-treated bed nets (ITNs) on parasite genetics population structure: I. The genetic diversity of Plasmodium falciparum parasites by microsatellite markers in western Kenya. Malar J. 2010;9:353. doi: 10.1186/1475-2875-9-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yuan JS, Reed A, Chen F, Stewart CN., Jr Statistical analysis of real-time PCR data. BMC Bioinformatics. 2006;7:85. doi: 10.1186/1471-2105-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Menegon M, Talha AA, Severini C, Elbushra SM, Mohamedani AA, Malik EM, et al. Frequency distribution of antimalarial drug resistance alleles among Plasmodium falciparum isolates from Gezira State, central Sudan, and Gedarif State, eastern Sudan. Am J Trop Med Hyg. 2010;83:250–7. doi: 10.4269/ajtmh.2010.09-0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Osman ME, Mockenhaupt FP, Bienzle U, Elbashir MI, Giha HA. Field-based evidence for linkage of mutations associated with chloroquine (pfcrt/pfmdr1) and sulfadoxine-pyrimethamine (pfdhfr/pfdhps) resistance and for the fitness cost of multiple mutations in P. falciparum. Infect Genet Evol. 2007;7:52–9. doi: 10.1016/j.meegid.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 45.Mockenhaupt FP, Bousema JT, Eggelte TA, Ehrhardt S, Otchwemah RN, Sauerwein RW, et al. Concurrence of Plasmodium falciparum dhfr and crt mutations in northern Ghana. Malar J. 2005;4:42. doi: 10.1186/1475-2875-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laufer MK, Thesing PC, Eddington ND, Masonga R, Dzinjalamala FK, Takala SL, et al. Return of chloroquine antimalarial efficacy in Malawi. N Engl J Med. 2006;355:1959–66. doi: 10.1056/NEJMoa062032. [DOI] [PubMed] [Google Scholar]
- 47.Mwai L, Ochong E, Abdirahman A, Kiara SM, Ward S, Kokwaro G, et al. Chloroquine resistance before and after its withdrawal in Kenya. Malar J. 2009;8:106. doi: 10.1186/1475-2875-8-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Djimde A, Doumbo OK, Cortese JF, Kayentao K, Doumbo S, Diourte Y, et al. A molecular marker for chloroquine-resistant falciparum malaria. N Engl J Med. 2001;344:257–63. doi: 10.1056/NEJM200101253440403. [DOI] [PubMed] [Google Scholar]
- 49.Sidhu AB, Verdier-Pinard D, Fidock DA. Chloroquine resistance in Plasmodium falciparum malaria parasites conferred by pfcrt mutations. Science. 2002;298:210–3. doi: 10.1126/science.1074045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iriemenam NC, Shah M, Gatei W, van Eijk AM, Ayisi J, Kariuki S, et al. Temporal trends of sulphadoxine-pyrimethamine (SP) drug-resistance molecular markers in Plasmodium falciparum parasites from pregnant women in western Kenya. Malar J. 2012;11:134. doi: 10.1186/1475-2875-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raman J, Mauff K, Muianga P, Mussa A, Maharaj R, Barnes KI. Five years of antimalarial resistance marker surveillance in gaza province, Mozambique, following artemisinin-based combination therapy roll out. PLoS One. 2011;6:e25992. doi: 10.1371/journal.pone.0025992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Talisuna AO, Langi P, Mutabingwa TK, Van Marck E, Speybroeck N, Egwang TG, et al. Intensity of transmission and spread of gene mutations linked to chloroquine and sulphadoxine-pyrimethamine resistance in falciparum malaria. Int J Parasitol. 2003;33:1051–8. doi: 10.1016/S0020-7519(03)00156-5. [DOI] [PubMed] [Google Scholar]
- 53.Read AF, Anwar M, Shutler D, Nee S. Sex allocation and population structure in malaria and related parasitic protozoa. Proc Biol Sci. 1995;260:359–63. doi: 10.1098/rspb.1995.0105. [DOI] [PubMed] [Google Scholar]


