Abstract
Myasthenia gravis (MG) is a neuromuscular autoimmune disorder resulting from autoantibodies attacking components of the neuromuscular junction. Recent studies have implicated the aberrant expression of microRNAs (miRNAs) in the pathogenesis of MG; however, the underlying mechanisms remain largely unknown. This study aimed to identify key genes regulated by miRNAs in MG. Six dysregulated pathways were identified through differentially expressed miRNAs and mRNAs in MG, and significant crosstalk was detected between five of these. Notably, crosstalk between the “synaptic long-term potentiation” pathway and four others was mediated by five genes involved in the MAPK signaling pathway. Furthermore, 14 key genes regulated by miRNAs were detected, of which six—MAPK1, RAF1, PGF, PDGFRA, EP300, and PPP1CC—mediated interactions between the dysregulated pathways. MAPK1 and RAF1 were responsible for most of this crosstalk (80%), likely reflecting their central roles in MG pathogenesis. In addition, most key genes were enriched in immune-related local areas that were strongly disordered in MG. These results provide new insight into the pathogenesis of MG and offer new potential targets for therapeutic intervention.
1. Introduction
Myasthenia gravis (MG) is a neuromuscular autoimmune disorder caused by antibodies that attack the acetylcholine receptor (AChR), leading to muscle fatigue and weakness. The pathogenesis of MG is not fully understood but involves the interaction of genetic, environmental, and immunological factors, thymic abnormalities, and age [1]. Despite advances in treatments, there are none that specifically target the autoimmune deficiency in MG. Several recent studies have shown that various microRNAs (miRNAs) are aberrantly expressed in MG, presenting new possibilities for understanding disease pathogenesis as well as for diagnosis and treatment.
miRNAs are small (~22 nucleotide) noncoding RNA molecules that regulate the expression of their target mRNAs at the posttranscriptional level. miRNAs regulate a wide range of biological processes, including development, cell differentiation and proliferation, metabolism, and apoptosis [2, 3], and contribute to the pathogenesis of a variety of autoimmune disorders including MG [4]. For example, miR-320a and let-7c are downregulated in MG patients relative to healthy control subjects [5, 6], and miRNA-146a is upregulated in MG patients and was found to act on B cells expressing AChR, thereby contributing to the development of MG [7]. A recent study showed that serum levels of a set of miRNAs were reduced in MG patients, and some were differentially expressed in early and late onset MG [8]. However, it remains unclear how the dysregulation of miRNAs leads to MG.
Most association studies aiming to identify candidate genes in MG have focused on a specific gene or several independent genes [9–12]. However, there is increasing evidence to suggest that MG arises from the interaction of multiple genes. For instance, several members of the nuclear factor- (NF-) κB signaling pathway work cooperatively to trigger an immune response in MG [13], and a recent study found that miR-155 was upregulated in MG patients, while suppressing miR-155 impaired NF-κB signaling [14]. Since functionally connected pathways crosstalk through common sets of genes [15], identifying miRNAs and their target genes that are dysregulated in MG is critical for developing effective treatment strategies.
In this study, pathways that are dysregulated in MG were identified by pathway enrichment analysis. Some MG-related genes were found to mediate crosstalk between these pathways. Key genes regulated by miRNAs were identified by combining computational predictions with mRNA expression profiles, and these were found to mediate crosstalk and were enriched in disordered local areas of pathways (LAPs) related to MG. The findings reveal a set of genes that play key roles in the pathogenesis of MG and can potentially serve as novel therapeutic targets.
2. Materials and Methods
2.1. Data Sources
2.1.1. Expression Profiles of mRNA and miRNA
The mRNA expression profiles for MG were obtained from ArrayExpress database (accession number E-MEXP-518, Raw data in Optional Supplementary Material, available online at http://dx.doi.org/10.1155/2014/724715) and comprised 30 samples from 20 MG patients and 10 control subjects, for a total of 12,814 genes [16]. The miRNA expression profiles have been previously published and were from three MG patients and three matched healthy controls [5], for a total of 866 mature human miRNAs.
2.1.2. miRNA Target Genes
miRNA target genes were determined using the following miRNA target prediction tools: miRanda (August 2010 release) [17], TargetScan (release 6.2) [18], DIANA-microT (version 5.0) [19], mirSVR [20], PicTar (five-way) [21], RNA22 [22], RNAhybrid [23], PITA (version 6) [24], MirTarget2 (version 4.0) [25], and TargetMiner [26]. To minimize the false positives, only genes predicted by at least four tools were selected as putative miRNA targets for pathway analysis.
2.2. Methods
2.2.1. Identification of Differentially Expressed mRNAs and miRNAs
Raw data from mRNA expression profiles were quantified using the global locally weighted scatterplot smoothing (Lowess) transformation and were transformed in log2-scale; differentially expressed mRNAs were identified by significance analysis of microarray data. A false discovery rate (FDR) < 0.05 and fold change (FC) > 2 were considered statistically significant. Background subtraction and normalization were performed for miRNA expression profiles and FC > 2 were selected as differentially expressed miRNAs.
2.2.2. Pathway Enrichment and Crosstalk Analysis
The Database for Annotation, Visualization, and Integrated Discovery was used for the KEGG pathway enrichment analysis [27]. P values were calculated with Fisher's exact test. A cutoff value of P < 0.05 was used to define significantly enriched pathways.
The crosstalk between pathways was calculated based on a cumulative hypergeometric distribution using the following formula:
| (1) |
where N is the total number of genes in the human genome, n is the number of genes in one pathway, m is the number of genes in a different pathway, and x is the number of genes that are common to both pathways. P < 0.05 was defined as the cutoff value for significant crosstalk between pathways.
2.2.3. Gene Ontology Enrichment Analysis
The cumulative hypergeometric distribution was also used for the gene ontology (GO) functional enrichment analysis. Gene sets were mapped to GO terms according to biological process (BP), cellular component (CC), and molecular function (MF). Using the above formula, N is the total number of genes in the human genome, n is the number of gene sets in one pathway, m is the number of genes annotated with a particular GO term, and x is the number of genes that show overlap between the pathway and the GO term. The P value was adjusted using the Benjamini and Hochberg false discovery rate (FDR) to determine statistical significance and GO terms were selected based on an FDR < 0.05.
2.2.4. Cluster Analysis of Pathways
Significant crosstalk was determined for each pair of pathways based on a hypergeometric test, and corresponding −log 10 P values formed the crosstalk matrix. Hierarchical clustering was implemented to group pathways into clusters based on the crosstalk matrix. A heatmap was generated to visualize the results using the gplots2 package of R software.
2.2.5. Identification of LAPs
The k-clique method in the SubpathwayMiner software was used to mine LAPs based on targets of differentially expressed miRNA as well as mRNA expression data [28, 29]. This method identified LAPs based on the closeness of genes in pathways as measured by the distance parameter k such that the distance between any genes in an LAP was <k. The default value k = 4 was used to identify significantly enriched LAPs.
3. Results
3.1. miRNAs Affecting Pathways Dysregulated in MG
A total of 1135 differentially expressed mRNAs were obtained from the screen, including 551 upregulated mRNAs and 584 downregulated mRNAs that were enriched in 13 (P1) and 8 (P2) pathways, respectively. A total of 46 differentially expressed miRNAs were identified, including 21 upregulated miRNAs and 25 downregulated miRNAs, and their predicted targets were enriched in 64 (P3) and 68 (P4) pathways, respectively (Tables S1, S2, and S3).
Since miRNAs are negative regulators of mRNAs [30], pathways enriched for differentially expressed mRNAs were used to filter predicted targets based on inverse miRNA-mRNA regulation. The intersections of P1 + P4 and P2 + P3 were defined as up- and downregulated pathways, respectively. Two upregulated and four downregulated pathways potentially targeted by two downregulated and 12 upregulated miRNAs were identified (Figure 1). Of the 14 dysregulated miRNAs found in this study, all but miR-634 have been validated in other autoimmune diseases based on the Human MicroRNA Disease Database (HMDD) and miR2Disease databases (Table S4) [31, 32]. For example, miR-21 is upregulated in several autoimmune diseases such as multiple sclerosis [33], psoriasis [34], and systemic lupus erythematosus [35], which shares some etiological features with MG, implying that the miRNAs identified for MG are common features of autoimmune diseases.
Figure 1.
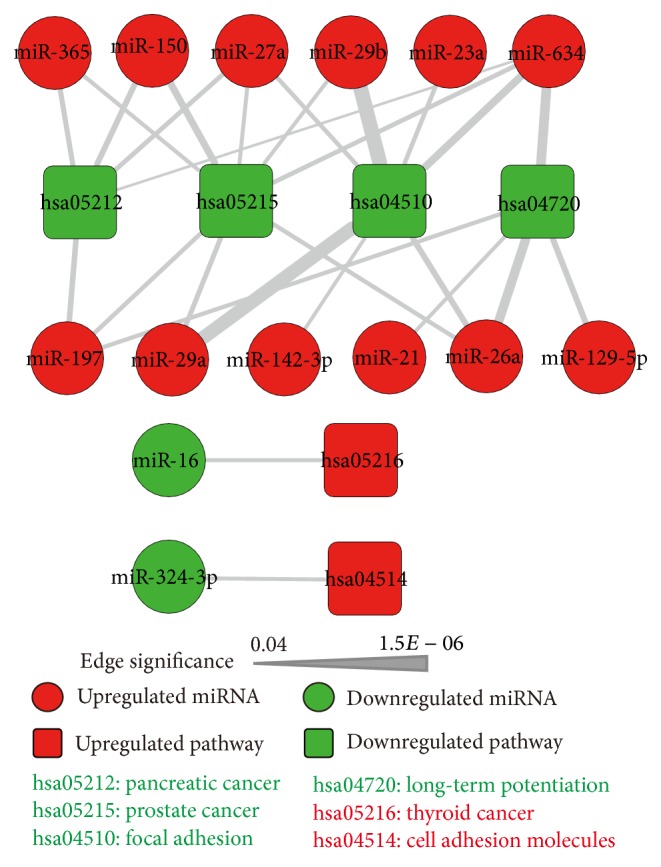
miRNA-regulated pathways in MG. Red and green rectangles represent up- and downregulated pathways, respectively; red and green circles represent up- and downregulated miRNAs, respectively. Lines represent regulatory interactions between miRNAs and their target pathways, with wider lines representing a more significant interaction.
A total of 14 dysregulated miRNAs were mapped to chromosomes using miRBase (release 20) [36]. Consistent with the findings of a previous study [37], miRNA genes affecting the same pathway were clustered together. A cluster was defined as a maximum distance of 10 kb between two miRNA genes. We found hsa-miR-29a/29b (mapped to chromosome 7q32.3) regulated the pathway hsa05215 and along with hsa-miR-27a/23a (mapped to chromosome 19p13.13) coregulated hsa04510, suggesting a synergistic interaction.
3.2. MG-Related Genes Mediate Pathway Crosstalk
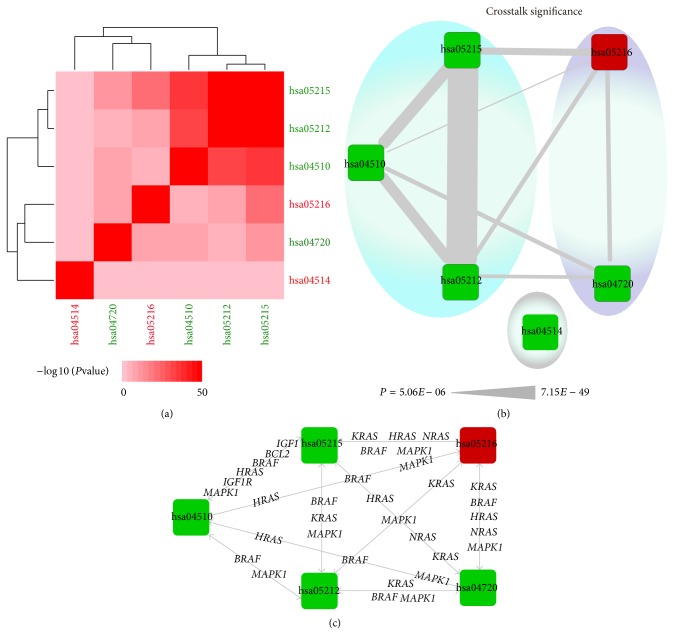
Crosstalk was observed between all dysregulated pathways except hsa04514 (Table S5). Two clusters were identified: one consisting of pathways hsa05215, hsa05212, and hsa04510 and the other of hsa05216 and hsa04720 (Figures 2(a) and 2(b)). Importantly, genes such as HRAS, MAPK1, and BRAF were engaged in crosstalk (Figure 2(c)) and have been previously found to be associated with MG [38, 39]. These results suggest that crosstalk between multiple dysregulated pathways is responsible for the heterogeneity of MG.
Figure 2.
Pathway crosstalk analysis and functional clustering. (a) Hierarchical clustering of dysregulated pathways based on −log 10 P values of their interactions; stronger interactions are represented by a more intense color. (b) Crosstalk between dysregulated pathways. Rectangular nodes represent pathways and edges denote crosstalk between two pathways, with wider edges representing greater interaction between two pathways. Two clusters (blue and purple) were identified; one pathway did not cluster with the others (grey). (c) Crosstalk between pathways mediated by MG-related genes.
It is worth noting that pathway hsa04720 (synaptic long-term potentiation (LTP))—which is important for long-lasting increases in synaptic efficacy that serve as the basis for learning and memory—interacted with each of the other four pathways via five immune-related genes, that is, BRAF, MAP2K1 and MAP2K2, and MAPK1 and MAPK3. The signaling cascade involving these genes, that is, BRAF-MEK1/2 (MAP2K1/2)-ERK1/2 (MAPK1/3), is part of the MAPK signaling pathway, which is involved in cell proliferation, differentiation, and migration [40] and was recently found to be dysregulated in MG [41].
3.3. Detecting Key Genes Regulated by miRNAs in Crosstalk Pathways
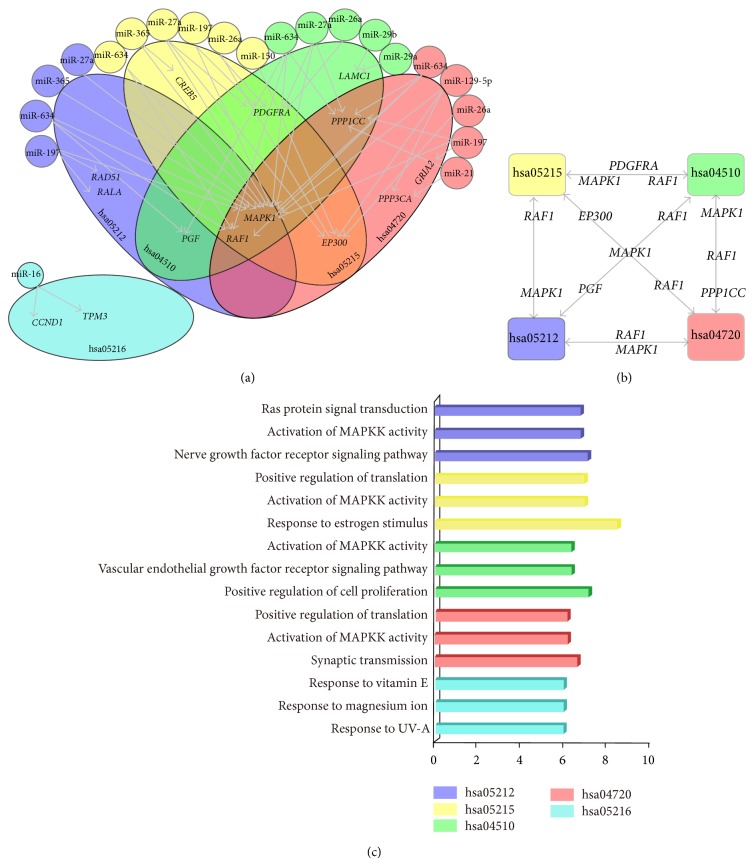
miRNAs and their target genes that were enriched in the identified pathways and whose differential expression was inversely related were selected, for a total of 14 key genes in five pathways targeted by 11 miRNAs (Figure 3(a)). Six miRNA-gene pairs were already experimentally verified, including miR-16 and CCND1 [42], miR-16 and TPM3 [43], miR-29a/b and LAMC1 [44], miR-197 and RAD51 [45], and miR-150 and EP300 [46]. Moreover, six genes (MAPK1, RAF1, PGF, PDGFRA, EP300, and PPP1CC) were common to more than one MG-related pathway (Figure 3(b)). In particular, four downregulated pathways interacted through MAPK1 and RAF1, reflecting their potential involvement in MG pathogenesis.
Figure 3.
Key genes regulated by miRNAs involved in dysregulated pathways. (a) Key genes regulated by miRNAs. Circles represent miRNAs and large ellipses represent pathways with key genes. miRNAs and their target pathways are represented by the same color; arrows denote key genes regulated by miRNAs. (b) Crosstalk between downregulated pathways through key genes. (c) Top three GO functions of key genes in each pathway; the histogram represents −log 2 (FDR).
Key genes were organized into hierarchical categories by functional enrichment analysis. A total of 133 GO terms, including 84 BP, 35 MF, and 14 CC terms, were obtained (Table S6). Key genes were primarily involved in BP such as biological regulation, response to stimulus, metabolic, single-organism, cell, and developmental processes, establishment of localization, signaling, and cellular component organization; CC such as membrane, cell part, and macromolecular complex; and MF such as catalytic activity and binding.
3.4. Key Genes Enriched in LAPs
Recent studies have suggested that pathways are not globally disrupted, but rather only in a local area [47, 48]; thus, key genes enriched in 36 LAPs were considered as being important for MG (Table S7). Though, of the MG-related crosstalk pathways, three were cancer related, some key genes were enriched in immune-related LAPs within these pathways, which could explain the association between MG and cancer-related pathways (Figures S1, S2, and S4). Key genes and their regulatory miRNAs were mapped to specific pathways and LAPs, and their contributions to the molecular mechanism of MG are discussed below.
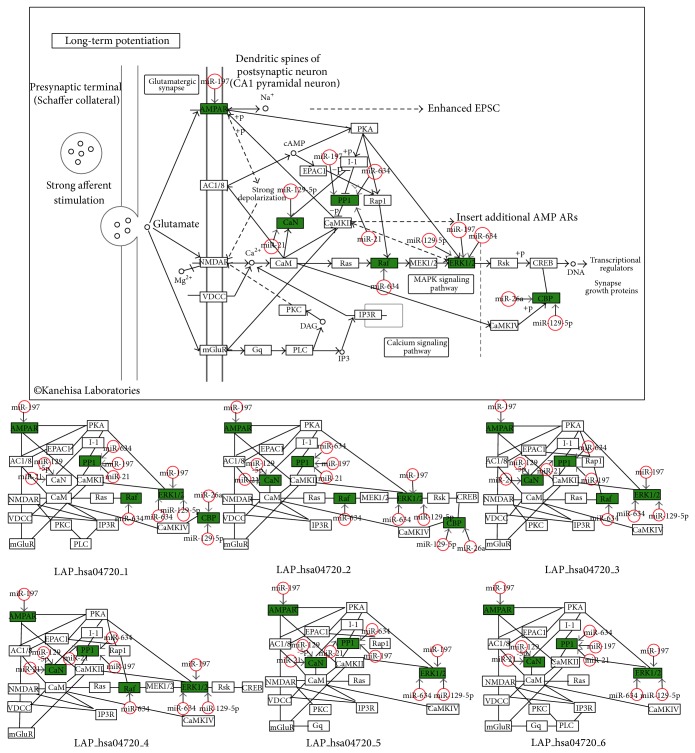
Synaptic LTP (hsa04720). This pathway is implicated in various neurological disorders, including Parkinson's disease [49], stroke [50], multiple sclerosis [51], and Alzheimer's disease [52]. Six downregulated hsa04720-related genes (RAF1, EP300, PPP1CC, MAPK1, GRIA2, and PPP3CA) were predicted to be targeted by five upregulated miRNAs (hsa-miR-634, hsa-miR-26a, hsa-miR-129-5p, hsa-miR-197, and hsa-miR-21). The structures of the six LAPs were similar, indicating that genes in the whole pathway and LAPs were tightly linked (Figure 4). All six genes have been directly or indirectly implicated in the regulation of LTP; thus, their dysregulation by miRNAs would be expected to result in neurological diseases.
Figure 4.
Representative illustration of key gene distribution (hsa04720 and LAP_hsa04720s). Downregulated key genes are shown in green; red open circles represent their regulatory miRNAs. LAP_hsa04720s and associated key genes are shown below the hsa04720 pathway.
Prostate Cancer (hsa05215). Five downregulated genes (EP300, RAF1, PDGFRA, CREB5, and MAPK1) were predicted to be targeted by six miRNAs (hsa-miR-150, hsa-miR-634, hsa-miR-197, hsa-miR-26a, hsa-miR-365, and hsa-miR-27a). The regulation of EP300 by miR-150 has been validated [46], and miR-150 is overexpressed in human lymphocyte cells and lymphocyte-derived exosomes [53]. MiR-150 is involved in immune regulation, including B and T cell differentiation and NK cell development [54, 55]; therefore, miR-150 may activate T cell immune responses by targeting EP300. CREB5 has been implicated in Alzheimer's and Parkinson's diseases [56]. Here it was predicted that the downregulation of CREB5 was due to regulation by miR-365; this was supported by a recent study showing reduced CREB activity in an experimental MG model [57]. Key genes were also enriched in immune-related LAPs that are potentially related to MG; for example, PDGFRA, MAPK1, and RAF1 were enriched in LAP_05215_7 and LAP_05215_1, both of which are involved in cytokine-cytokine receptor interactions and MAPK signaling, which play important roles in MG pathogenesis (Figure S1) [41].
Pancreatic Cancer (hsa05212). Five downregulated genes (RALA, MAPK1, RAF1, RAD51, and PGF) were predicted to be targeted by four miRNAs (hsa-miR-634, hsa-miR-197, hsa-miR-365, and hsa-miR-27a). LAP_05212_10 comprised RAD51 and BRCA2. The regulation of RAD51 by miR-197 has been experimentally verified [45]. Four of the genes were enriched in immune-related LAPs. For example, MAPK1, RAF1, and RALA were enriched in LAP_05212_3, LAP_05212_4, and LAP_05212_5, which is linked to the PI3K-Akt and MAPK signaling pathways. PGF was enriched in LAP_05212_1, which is involved in ErbB and Jak-STAT signaling pathways (Figure S2).
Focal Adhesion (hsa04510). This pathway has been implicated in nervous system disorders such as depression in genomewide molecular pathway analyses [58, 59]. Six downregulated genes (LAMC1, RAF1, PDGFRA, PPP1CC, PGF, and MAPK1) were predicted to be targeted by five miRNAs (hsa-miR-29b, hsa-miR-29a, hsa-miR-634, hsa-miR-26a, and hsa-miR-27a). The regulation of LAMC1 by miR-29a/b has been validated [44]. These genes were enriched in immune-related LAPs that are linked to cytokine-cytokine receptor interaction pathway, MAPK signaling pathway, and PI3K-Akt signaling pathway (Figure S3).
Thyroid Cancer (hsa05216). This pathway was upregulated in MG and is potentially regulated by miR-16 and its predicted targets CCND1 and TPM3. MiR-16 is downregulated in peripheral blood mononuclear cells and sera of MG patients [6, 8]. The regulatory interaction between miR-16 and CCND1 and TPM3 has been experimentally verified in humans [42, 43]. CCND1—a downstream gene in LAP_hsa05216_4—is a cell cycle-related gene that controls the G1/S transition and is a target of miR-16 [42]. TPM3—an upstream gene in LAP_hsa05216_2—is an actin-binding protein that mediates myosin-actin interaction and stabilizes microfilaments in muscle [60]. Thus, the upregulation of these two genes by miR-16 dysregulation is likely to be involved in MG (Figure S4).
4. Discussion
MG is an autoimmune disease resulting from the failure of neuromuscular transmission due to binding of autoantibodies to proteins involved in signaling at the neuromuscular junction (NMJ). Available treatments include acetylcholinesterase drugs, immunomodulatory agents, plasma exchange, intravenous immunoglobulin administration, and thymectomy. However, some MG patients are unresponsive to conventional therapies or suffer adverse reactions from long-term use of immunomodulatory drugs such as steroids or immunosuppressants [61]. Accordingly, there is a need for treatments that target specific factors involved in MG.
The present study identified six dysregulated pathways potentially targeted by 14 miRNAs. The LTP pathway is associated with multiple neurological disorders [49–52] and was found here to be involved in MG. The NMJ connects neurons to target muscles, and this communication is similar to that between two neurons in LTP. With the exception of miR-634, all miRNAs associated with pathways dysregulated in MG were autoimmune disease-related ones. For instance, miR-23a expression is upregulated in Crohn's disease [62] and lupus nephritis [63] in a pattern similar to that observed in MG. miRNAs dysregulated in MG have known functions as activators of the T cell phenotype and promote T cell-driven B cell differentiation into immunoglobulin-secreting plasma cells [35]; given that MG is a T cell-regulated disorder [64], it can be speculated that miRNA dysfunction in MG can activate T cell-induced immune responses and stimulate the synthesis of autoantibodies by B cells.
Essential genes are often implicated in multiple pathways, which can interact to give rise to a disease state [65]. For example, in MG, NF-κB signaling regulates immune responses [13], and the activation of this pathway depends on PI3K-Akt signaling [66]. In the present study, significant crosstalk was detected between pathways dysregulated in MG, which likely accounts for the heterogeneity of the disease. Notably, members of the Ras gene family were implicated in this crosstalk; it has been previously reported that some Ras gene family members are linked to the acceleration of clinical features of MG [38].
A set of key genes regulated by miRNAs associated with pathways dysregulated in MG was identified; six of these miRNA-gene pairs were already experimentally validated. For example, LAMC1 in the “focal adhesion” pathway has been implicated in processes such as cell adhesion, differentiation, migration, signaling, neurite outgrowth, and metastasis; miR-29a/b regulates this pathway and inhibits cancer cell migration and invasion by targeting LAMC1 [44]. The key genes that were identified mediated crosstalk between the dysregulated pathways and included MAPK1 and RAF1, two factors in MAPK signaling that play important roles in the immune response [39], neurotransmission [67], and neurological diseases such as Alzheimer's disease and Parkinson's disease as well as amyotrophic lateral sclerosis [68]. Thus, the pathogenesis of MG likely involves these key genes and their dysregulation by aberrantly expressed miRNAs, which affects multiple signaling pathways.
LAPs enriched in key genes were also examined to obtain more detailed information on their specific roles in each pathway. The genes were enriched in immune-related LAPs. For instance, previous studies have indicated that cytokines mediate immune responses in MG [69]; it was found here that PDGFRA—an upstream key gene in LAP_04510_13 and LAP_05215_7 whose product binds another key gene product, PGF—triggers cytokine-cytokine receptor interactions.
5. Conclusions
In this study, pathway enrichment, pathway crosstalk, functional enrichment, and LAP analyses were combined to elucidate miRNA-gene interactions in the context of pathways relevant to MG. Crosstalk between pathways dysregulated in MG was detected, and key genes in these pathways were identified that are likely involved in MG pathogenesis, providing potential new targets for therapeutic interventions.
Supplementary Material
Table S1: MiRNAs and mRNAs differentially expressed in MG Table S1 lists the miRNAs and mRNAs differentially expressed in MG. A total of 46 differentially expressed miRNAs were identified, including 21 upregulated miRNAs and 25 downregulated miRNAs. A total of 1135 differentially expressed mRNAs were obtained, including 551 upregulated mRNAs and 584 downregulated mRNAs.
Table S2: The predicted targets of differentially expressed miRNAs
Table S2 lists the predicted targets of differentially expressed miRNAs. A total of 46 differentially expressed miRNAs, 9567 predicted targets, 34963 miRNA-target regulations were obtained.
Table S3: Pathways enriched in differentially expressed mRNAs and predicted target genes of differentially expressed miRNAs
Table S3 presents significant pathways enriched by differentially expressed mRNAs and predicted genes of differentially expressed miRNAs. The up- and down-regulated mRNAs were significantly enriched in 13 pathways (define as P1) and 8 pathways (define as P2), respectively. The predicted targets of up- and down-regulated miRNAs were significantly enriched in 64 pathways (define as P3) and 68 pathways (define as P4), respectively.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant nos. 81171122, 81371324, and 31401090), the National Science Foundation of Heilongjiang Province (Grant no. ZD201208), and the Specialized Research Fund for the Doctoral Program of Higher Education of China (Grant no. 20132307110008).
Conflict of Interests
The authors declare no conflict of interests.
Authors' Contribution
Yuze Cao and Jianjian Wang contributed equally to this work.
References
- 1.Meriggioli M. N., Sanders D. B. Autoimmune myasthenia gravis: emerging clinical and biological heterogeneity. The Lancet Neurology. 2009;8(5):475–490. doi: 10.1016/s1474-4422(09)70063-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo H., Ingolia N. T., Weissman J. S., Bartel D. P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466(7308):835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kloosterman W. P., Plasterk R. H. A. The diverse functions of microRNAs in animal development and disease. Developmental Cell. 2006;11(4):441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 4.Le Panse R., Berrih-Aknin S. Autoimmune myasthenia gravis: autoantibody mechanisms and new developments on immune regulation. Current Opinion in Neurology. 2013;26(5):569–576. doi: 10.1097/wco.0b013e328364d6cd. [DOI] [PubMed] [Google Scholar]
- 5.Cheng Z., Qiu S., Jiang L., et al. MiR-320a is downregulated in patients with Myasthenia gravis and modulates inflammatory cytokines production by targeting mitogen-activated protein kinase 1. Journal of Clinical Immunology. 2013;33(3):567–576. doi: 10.1007/s10875-012-9834-5. [DOI] [PubMed] [Google Scholar]
- 6.Jiang L., Cheng Z., Qiu S., et al. Altered let-7 expression in Myasthenia gravis and let-7c mediated regulation of IL-10 by directly targeting IL-10 in Jurkat cells. International Immunopharmacology. 2012;14(2):217–223. doi: 10.1016/j.intimp.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 7.Lu J., Yan M., Wang Y., et al. Altered expression of miR-146a in myasthenia gravis. Neuroscience Letters. 2013;555:85–90. doi: 10.1016/j.neulet.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 8.Nogales-Gadea G., Ramos-Fransi A., Suarez-Calvet X., et al. Analysis of serum miRNA profiles of myasthenia gravis patients. PLoS ONE. 2014;9(3) doi: 10.1371/journal.pone.0091927.e91927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Auret J., Abrahams A., Prince S., Heckmann J. M. The effects of prednisone and steroid-sparing agents on decay accelerating factor (CD55) expression: implications in myasthenia gravis. Neuromuscular Disorders. 2014;24(6):499–508. doi: 10.1016/j.nmd.2014.02.010. [DOI] [PubMed] [Google Scholar]
- 10.Kellermayer B., Polgar N., Pal J., et al. Association of myasthenia gravis with polymorphisms in the gene of histamine N-methyltransferase. Human Immunology. 2013;74(12):1701–1704. doi: 10.1016/j.humimm.2013.07.016. [DOI] [PubMed] [Google Scholar]
- 11.Wang X. B., Pirskanen R., Giscombe R., Lefvert A. K. Two SNPs in the promoter region of the CTLA-4 gene affect binding of transcription factors and are associated with human myasthenia gravis. Journal of Internal Medicine. 2008;263(1):61–69. doi: 10.1111/j.1365-2796.2007.01879.x. [DOI] [PubMed] [Google Scholar]
- 12.Lefvert A. K., Zhao Y., Ramanujam R., Yu S., Pirskanen R., Hammarström L. PTPN22 R620W promotes production of anti-AChR autoantibodies and IL-2 in myasthenia gravis. Journal of Neuroimmunology. 2008;197(2):110–113. doi: 10.1016/j.jneuroim.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 13.Avidan N., Le Panse R., Berrih-Aknin S., Miller A. Genetic basis of myasthenia gravis—a comprehensive review. Journal of Autoimmunity. 2014;52:146–153. doi: 10.1016/j.jaut.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y.-Z., Tian F.-F., Yan M., et al. Delivery of an miR155 inhibitor by anti-CD20 single-chain antibody into B cells reduces the acetylcholine receptor-specific autoantibodies and ameliorates experimental autoimmune myasthenia gravis. Clinical and Experimental Immunology. 2014;176(2):207–221. doi: 10.1111/cei.12265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen L., Wang H., Zhang L., et al. Uncovering packaging features of co-regulated modules based on human protein interaction and transcriptional regulatory networks. BMC Bioinformatics. 2010;11, article 392 doi: 10.1186/1471-2105-11-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Panse R., Cizeron-Clairac G., Bismuth J., Berrih-Aknin S. Microarrays reveal distinct gene signatures in the thymus of seropositive and seronegative myasthenia gravis patients and the role of CC chemokine ligand 21 in thymic hyperplasia. The Journal of Immunology. 2006;177(11):7868–7879. doi: 10.4049/jimmunol.177.11.7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.John B., Enright A. J., Aravin A., Tuschl T., Sander C., Marks D. S. Human microRNA targets. PLoS Biology. 2004;2(11, article e363) doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia D. M., Baek D., Shin C., Bell G. W., Grimson A., Bartel D. P. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nature Structural and Molecular Biology. 2010;18(10):1139–1146. doi: 10.1038/nsmb.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paraskevopoulou M. D., Georgakilas G., Kostoulas N., et al. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Research. 2013;41(1):W169–W173. doi: 10.1093/nar/gkt393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Betel D., Koppal A., Agius P., Sander C., Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biology. 2010;11(8, article R90) doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krek A., Grün D., Poy M. N., et al. Combinatorial microRNA target predictions. Nature Genetics. 2005;37(5):495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 22.Miranda K. C., Huynh T., Tay Y., et al. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126(6):1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 23.Krüger J., Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Research. 2006;34:W451–W454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kertesz M., Iovino N., Unnerstall U., Gaul U., Segal E. The role of site accessibility in microRNA target recognition. Nature Genetics. 2007;39(10):1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 25.Wang X. miRDB: a microRNA target prediction and functional annotation database with a wiki interface. RNA. 2008;14(6):1012–1017. doi: 10.1261/rna.965408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bandyopadhyay S., Mitra R. TargetMiner: microRNA target prediction with systematic identification of tissue-specific negative examples. Bioinformatics. 2009;25(20):2625–2631. doi: 10.1093/bioinformatics/btp503. [DOI] [PubMed] [Google Scholar]
- 27.Dennis G., Jr., Sherman B. T., Hosack D. A., et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biology. 2003;4(5, article P3) doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 28.Li C., Li X., Miao Y., et al. SubpathwayMiner: a software package for flexible identification of pathways. Nucleic Acids Research. 2009;37(19, article e131) doi: 10.1093/nar/gkp667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu B., Li C., Zhang P., et al. Dissection of miRNA-miRNA interaction in esophageal squamous cell carcinoma. PLoS ONE. 2013;8(9) doi: 10.1371/journal.pone.0073191.e73191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farh K. K.-H., Grimson A., Jan C., et al. Biochemistry: The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310(5755):1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 31.Li Y., Qiu C., Tu J., et al. HMDD v2.0: a database for experimentally supported human microRNA and disease associations. Nucleic Acids Research. 2014;42(1):D1070–D1074. doi: 10.1093/nar/gkt1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang Q., Wang Y., Hao Y., et al. miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Research. 2009;37(1):D98–D104. doi: 10.1093/nar/gkn714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fenoglio C., Cantoni C., de Riz M., et al. Expression and genetic analysis of miRNAs involved in CD4+ cell activation in patients with multiple sclerosis. Neuroscience Letters. 2011;504(1):9–12. doi: 10.1016/j.neulet.2011.08.021. [DOI] [PubMed] [Google Scholar]
- 34.Meisgen F., Xu N., Wei T., et al. miR-21 is up-regulated in psoriasis and suppresses T cell apoptosis. Experimental Dermatology. 2012;21(4):312–314. doi: 10.1111/j.1600-0625.2012.01462.x. [DOI] [PubMed] [Google Scholar]
- 35.Stagakis E., Bertsias G., Verginis P., et al. Identification of novel microRNA signatures linked to human lupus disease activity and pathogenesis: MiR-21 regulates aberrant T cell responses through regulation of PDCD4 expression. Annals of the Rheumatic Diseases. 2011;70(8):1496–1506. doi: 10.1136/ard.2010.139857. [DOI] [PubMed] [Google Scholar]
- 36.Kozomara A., Griffiths-Jones S. MiRBase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Research. 2011;39(1):D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altuvia Y., Landgraf P., Lithwick G., et al. Clustering and conservation patterns of human microRNAs. Nucleic Acids Research. 2005;33(8):2697–2706. doi: 10.1093/nar/gki567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arimori S., Song Z. J. Expression of c-myc, c-Ki-ras and c-Ha-ras oncogene products in peripheral blood mononuclear cells from patients with myasthenia gravis. Internal Medicine. 1993;32(7):519–522. doi: 10.2169/internalmedicine.32.519. [DOI] [PubMed] [Google Scholar]
- 39.Roskoski R., Jr. ERK1/2 MAP kinases: structure, function, and regulation. Pharmacological Research. 2012;66(2):105–143. doi: 10.1016/j.phrs.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 40.Yang S.-H., Sharrocks A. D., Whitmarsh A. J. Transcriptional regulation by the MAP kinase signaling cascades. Gene. 2003;320(1-2):3–21. doi: 10.1016/s0378-1119(03)00816-3. [DOI] [PubMed] [Google Scholar]
- 41.Colombara M., Antonini V., Riviera A. P., et al. Constitutive activation of p38 and ERK1/2 MAPKs in epithelial cells of myasthenic thymus leads to IL-6 and RANTES overexpression: effects on survival and migration of peripheral T and B cells. The Journal of Immunology. 2005;175(10):7021–7028. doi: 10.4049/jimmunol.175.10.7021. [DOI] [PubMed] [Google Scholar]
- 42.Liu Q., Fu H., Sun F., et al. miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Research. 2008;36(16):5391–5404. doi: 10.1093/nar/gkn522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Selbach M., Schwanhäusser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455(7209):58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 44.Nishikawa R., Goto Y., Kojima S., et al. Tumor-suppressive microRNA-29s inhibit cancer cell migration and invasion via targeting LAMC1 in prostate cancer. International Journal of Oncology. 2014;45(1):401–410. doi: 10.3892/ijo.2014.2437. [DOI] [PubMed] [Google Scholar]
- 45.Weber F., Teresi R. E., Broelsch C. E., Frilling A., Eng C. A limited set of human MicroRNA is deregulated in follicular thyroid carcinoma. The Journal of Clinical Endocrinology and Metabolism. 2006;91(9):3584–3591. doi: 10.1210/jc.2006-0693. [DOI] [PubMed] [Google Scholar]
- 46.Duan Y., Zhou B., Su H., Liu Y., Du C. MiR-150 regulates high glucose-induced cardiomyocyte hypertrophy by targeting the transcriptional co-activator p300. Experimental Cell Research. 2013;319(3):173–184. doi: 10.1016/j.yexcr.2012.11.015. [DOI] [PubMed] [Google Scholar]
- 47.Freilich R. W., Woodbury M. E., Ikezu T. Integrated expression profiles of mRNA and miRNA in polarized primary murine microglia. PLoS ONE. 2013;8(11) doi: 10.1371/journal.pone.0079416.e79416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li X., Jiang W., Li W., et al. Dissection of human miRNA regulatory influence to subpathway. Briefings in Bioinformatics. 2012;13(2):175–186. doi: 10.1093/bib/bbr043.bbr043 [DOI] [PubMed] [Google Scholar]
- 49.Kojovic M., Bologna M., Kassavetis P., et al. Functional reorganization of sensorimotor cortex in early Parkinson disease. Neurology. 2012;78(18):1441–1448. doi: 10.1212/WNL.0b013e318253d5dd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takeuchi N., Chuma T., Matsuo Y., Watanabe I., Ikoma K. Repetitive transcranial magnetic stimulation of contralesional primary motor cortex improves hand function after stroke. Stroke. 2005;36(12):2681–2686. doi: 10.1161/01.str.0000189658.51972.34. [DOI] [PubMed] [Google Scholar]
- 51.Weiss S., Mori F., Rossi S., Centonze D. Disability in multiple sclerosis: when synaptic long-term potentiation fails. Neuroscience and Biobehavioral Reviews. 2014;43:88–99. doi: 10.1016/j.neubiorev.2014.03.023. [DOI] [PubMed] [Google Scholar]
- 52.Chen Q.-S., Kagan B. L., Hirakura Y., Xie C.-W. Impairment of hippocampal long-term potentiation by Alzheimer amyloid β-peptides. Journal of Neuroscience Research. 2000;60(1):65–72. doi: 10.1002/(sici)1097-4547(20000401)60:1x003C;65::aid-jnr7x0003e;3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 53.de Candia P., Torri A., Pagani M., Abrignani S. Serum microRNAs as biomarkers of human lymphocyte activation in health and disease. Frontiers in Immunology. 2014;5, article 43 doi: 10.3389/fimmu.2014.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiao C., Calado D. P., Galler G., et al. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131(1):146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 55.Fallah P., Arefian E., Naderi M., et al. MiR-146a and miR-150 promote the differentiation of CD133+ cells into T-lymphoid lineage. Molecular Biology Reports. 2013;40(8):4713–4719. doi: 10.1007/s11033-013-2567-6. [DOI] [PubMed] [Google Scholar]
- 56.Sakamoto K., Karelina K., Obrietan K. CREB: a multifaceted regulator of neuronal plasticity and protection. Journal of Neurochemistry. 2011;116(1):1–9. doi: 10.1111/j.1471-4159.2010.07080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen W., Zeng X., Luo F., Lv T., Zhou X., Bai J. The decreased expression of thioredoxin-1 in brain of mice with experimental autoimmune myasthenia gravis. Neuromuscular Disorders. 2014;24(8):726–735. doi: 10.1016/j.nmd.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 58.Kao C.-F., Jia P., Zhao Z., Kuo P.-H. Enriched pathways for major depressive disorder identified from a genome-wide association study. International Journal of Neuropsychopharmacology. 2012;15(10):1401–1411. doi: 10.1017/s1461145711001891. [DOI] [PubMed] [Google Scholar]
- 59.Drago A., Crisafulli C., Serretti A. The genetics of antipsychotic induced tremors: a genome-wide pathway analysis on the STEP-BD SCP sample. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2011;156(8):975–986. doi: 10.1002/ajmg.b.31245. [DOI] [PubMed] [Google Scholar]
- 60.Pieples K., Arteaga G., John Solaro R., et al. Tropomyosin 3 expression leads to hypercontractility and attenuates myofilament length-dependent Ca2+ activation. The American Journal of Physiology—Heart and Circulatory Physiology. 2002;283(4):H1344–H1353. doi: 10.1152/ajpheart.00351.2002. [DOI] [PubMed] [Google Scholar]
- 61.Dalakas M. C. Novel future therapeutic options in Myasthenia Gravis. Autoimmunity Reviews. 2013;12(9):936–941. doi: 10.1016/j.autrev.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 62.Paraskevi A., Theodoropoulos G., Papaconstantinou I., Mantzaris G., Nikiteas N., Gazouli M. Circulating MicroRNA in inflammatory bowel disease. Journal of Crohn's and Colitis. 2012;6(9):900–904. doi: 10.1016/j.crohns.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 63.Dai Y., Sui W., Lan H., Yan Q., Huang H., Huang Y. Comprehensive analysis of microRNA expression patterns in renal biopsies of lupus nephritis patients. Rheumatology International. 2009;29(7):749–754. doi: 10.1007/s00296-008-0758-6. [DOI] [PubMed] [Google Scholar]
- 64.Conti-Fine B. M., Milani M., Kaminski H. J. Myasthenia gravis: past, present, and future. The Journal of Clinical Investigation. 2006;116(11):2843–2854. doi: 10.1172/jci29894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li Y., Agarwal P., Rajagopalan D. A global pathway crosstalk network. Bioinformatics. 2008;24(12):1442–1447. doi: 10.1093/bioinformatics/btn200. [DOI] [PubMed] [Google Scholar]
- 66.Hoesel B., Schmid J. A. The complexity of NF-κB signaling in inflammation and cancer. Molecular Cancer. 2013;12(1, article 86) doi: 10.1186/1476-4598-12-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brys Z., Pluhar A., Tibor Kis J., Buda B., Szabo A. Network analysis of neurotransmitter related human kinase genes. Possible role of SRC, RAF1, PTK2B? Neuropsychopharmacologia Hungarica. 2013;15(3):165–171. [PubMed] [Google Scholar]
- 68.Kim E. K., Choi E. J. Pathological roles of MAPK signaling pathways in human diseases. Biochimica et Biophysica Acta. 2010;1802(4):396–405. doi: 10.1016/j.bbadis.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 69.Uzawa A., Kawaguchi N., Himuro K., Kanai T., Kuwabara S. Serum cytokine and chemokine profiles in patients with myasthenia gravis. Clinical & Experimental Immunology. 2014;176(2):232–237. doi: 10.1111/cei.12272. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: MiRNAs and mRNAs differentially expressed in MG Table S1 lists the miRNAs and mRNAs differentially expressed in MG. A total of 46 differentially expressed miRNAs were identified, including 21 upregulated miRNAs and 25 downregulated miRNAs. A total of 1135 differentially expressed mRNAs were obtained, including 551 upregulated mRNAs and 584 downregulated mRNAs.
Table S2: The predicted targets of differentially expressed miRNAs
Table S2 lists the predicted targets of differentially expressed miRNAs. A total of 46 differentially expressed miRNAs, 9567 predicted targets, 34963 miRNA-target regulations were obtained.
Table S3: Pathways enriched in differentially expressed mRNAs and predicted target genes of differentially expressed miRNAs
Table S3 presents significant pathways enriched by differentially expressed mRNAs and predicted genes of differentially expressed miRNAs. The up- and down-regulated mRNAs were significantly enriched in 13 pathways (define as P1) and 8 pathways (define as P2), respectively. The predicted targets of up- and down-regulated miRNAs were significantly enriched in 64 pathways (define as P3) and 68 pathways (define as P4), respectively.