Abstract
RIO protein kinases (RIOKs) are a relatively conserved family of enzymes implicated in cell cycle control and ribosomal RNA processing. Despite their functional importance, they remain a poorly understood group of kinases in multicellular organisms. Here, we show that the C. elegans genome contains one member of each of the three RIOK sub-families and that each of the genes coding for them has a unique tissue expression pattern. Our analysis showed that the gene encoding RIOK-1 (riok-1) was broadly and strongly expressed. Interestingly, the intestinal expression of riok-1 was dependent upon two putative binding sites for the oxidative and xenobiotic stress response transcription factor SKN-1. RNA interference (RNAi)-mediated knock down of riok-1 resulted in germline defects, including defects in germ line stem cell proliferation, oocyte maturation and the production of endomitotic oocytes. Taken together, our findings indicate new functions for RIOK-1 in post mitotic tissues and in reproduction.
Introduction
Protein kinases form a large family of diverse regulatory enzymes that are encoded by approximately two percent of the genes in most metazoan genomes [1, 2]. Through the phosphorylation of protein targets, they regulate various cellular processes, including transcription, translation and cell-cycle progression [1]. Of the 518 protein kinases encoded in the human genome, 478 form a single superfamily known as the eukaryotic protein kinase (ePK) family [1]. These enzymes are defined by their conserved, bi-lobed catalytic core which contains 12 subdomains involved in substrate binding, ATP binding and catalysis [3]. A second, smaller superfamily of 40 ‘atypical’ protein kinases (aPK) share structural homology to the ePK catalytic core, but lack overall sequence similarity [4]. The aPKs are divided into 13 small homology groups, one of which is the right open reading frame (RIO) family of kinases.
Two members of the RIO kinase (RIOK) family, designated Rio1p and Rio2p, were identified in the yeast Saccharomyces cerevisiae [5, 6]. Subsequent studies have identified these kinases in various organisms, ranging from ancient single-celled archaea to complex multicellular eukaryotes [4, 7]. A third member named RIOK-3 that has greater similarly to RIOK-1 was first identified as a homolog of Aspergillus nidulans SUDD [8]. To date, RIOK-3 is only known to exist within multicellular eukaryotes [9]. The RIOK family feature a distinctive RIO domain that contains motifs typical of ePKs, including ATP-binding, catalytic and metal-binding loops and a hinge region [9, 10], but lacks motifs involved in substrate binding and the ‘activation’ domain. The truncated RIO catalytic domain and the inability to identify in vitro substrates for RIOK-1 and RIOK-2 have led to speculation that the RIOKs do not function as kinases in vivo, and are instead ATPases with vestigial kinase-related structures [11]. While RIOKs have been demonstrated to undergo autophosphorylation, their in vivo targets are unknown [9].
RIOKs have been reported to function in multiple pathways and links to various cancers and other human diseases are emerging [11–13]. RIOK-1 and RIOK-2 are non-ribosomal factors individually required for normal ribosomal RNA biogenesis and cell cycle progression [5, 14, 15]. In yeast, depletion of either RIOK-1 or RIOK-2 results in defects in 20S pre-ribosomal RNA processing. In human cells, RIOK-2 is required for the production of 18S pre-rRNA [15] and RIOK-3 is require for 21S pre-rRNA processing [16]. RIO-2 has also been identified to be a ribosomal assembly factor that prevents premature translation initiation on the small (40S) subunit [17, 18]. Depletion of yeast RIOK-1 results in a dramatic increase in the number of binucleated and anucleated cells and a disruption to G1 to S and anaphase progression [5]. In contrast, yeast cells depleted of RIOK-2 do not feature any stage specific cell cycle arrest; however, they show accelerated mitotic exit and a correlated increase in the degradation of the cell cycle regulator cyclin B1 [6]. Recently, RIOK-3 was shown to be an adapter protein required for NF-κB signaling [19] and for antiviral immune responses via the type I interferon pathway [20].
Although RIOKs have been studied in yeast and mammalian cell lines, presently little is known about them in a developmental and organismal context. Here, we report that the C. elegans genome contains three riok genes and that each of them has a distinct tissue expression pattern and that riok-1 and-2 are essential for development. We also show that riok-1 is essential for reproduction, where it is required for oogenesis, but not spermatogenesis. Knockdown of riok-1 by RNA interference (RNAi) results in the formation of endomitotic oocytes, suggesting a new role for RIOKs in meiosis.
Materials and Methods
Strains
C. elegans strains were cultured using standard techniques [21]. The wild-type strain Bristol N2 and the following mutant strains were used: fog-2(q71)V, rrf-1(pk1417)I, VP303 rde-1(ne219); kbIs7[pnhx-2::rde-1, rol-6]. The riok-1(tm3775) and riok-2(tm3803) deletion mutants where generated by the National BioResource Project for the Nematode, and were outcrossed at least seven times to N2 and maintained as balanced strains riok-1(tm3775)/hT2 and riok-2(tm3803)/hIn1.
RNAi
RNAi clones to riok-1, dpy-13, car-1 and skn-1 were obtained from the C. elegans ORFeome library [22]. We generated RNAi feeding constructs for riok-2 and riok-3 by cloning 1000 bp and 1200 bp amplicons produced by reverse transcription PCR (REF) into the feeding vector pL4440 and transformation into the RNAi feeding bacterial strain HT115(DE3). For feeding RNAi, synchronised L1s were placed on RNAi plates until they grew to adult hermaphrodites. As a negative control, we used the plasmid pCB19, which encodes a portion of the Arabidopsis thaliana gene Lhcb4.3 that has no homology to C. elegans.
Analysis of brood size
Brood size analysis was completed at 20°C, 23°C and 25°C. Synchronised L1 worms were placed on RNAi plates and grown to L4 stage. At the L4 stage, were singled on to individual plates and transferred to a new small plate every 12 h. For the fog-2 brood size, worms were grown on RNAi plates from the L1 stage and then one L4 female was transferred to a plate along with 10 males worms and the worms moved every 12 hours to a fresh plate
Constructing transgenic worms
To create Priok-1_2SKN-1::GFP, a 2403 bp region 5’ of the ATG site and the first two exons of riok-1 was amplified and cloned into the PstI and BamHI sites in pPD95.75 (total insert size: 2591 bp). A second construct containing one predicted SKN-1 site (Priok-1_1SKN-1::GFP), was created by amplifying a 1917 bp-region 5’ of the ATG site including the first two exons of riok-1 followed by cloning into pPD95.75 (total insert size: 2142 bp). A third construct that lacked any SKN-1 site (Priok-1_ ΔSKN-1::GFP) was made following site-directed mutagenesis of Priok-1_1SKN-1::GFP using a QuikChange II site—directed mutagenesis kit (Stratagene). To achieve this, primers were designed to mutagenise the predicted SKN-1 site from attGtCAT to attCtGCA, which has been shown to inhibit SKN-1 binding [23]. To create Priok-2::GFP, a 703 bp region upstream of the ATG site and including the first two exons was amplified and cloned into the PstI and BamHI sites in pPD95.75 (total insert size: 2098 bp). To create Priok-3::GFP, 592 bp upstream to the ATG and the first four exon was amplified and cloned into the PstI and BamHI sites in pPD95.75 (total insert size: 2056 bp). All constructs were confirmed by sequencing. Wild-type worms were microinjected with the GFP reporter construct (50 ng/μl) and rol-6 marker pRF4 construct (50 ng/μl) using standard methods [24].
Imaging
For the immunostaining of gonads, worms were anesthetised in 0.001% tetramisole in M9 buffer. Gonads were dissected, snap frozen in liquid nitrogen and stained as described previously [25]. Slides were examined using an Olympus 1X81/1X2-UCB microscope. Primary antibodies used were: anti-PH3 (1/2000; rabbit) (Upstate Biotechnology), mAB414 (1/200; mouse) (Covance), MAPK (1/200; mouse) (Sigma). Secondary antibodies Alexa A488 and A555 (Invitrogen) were used at dilutions 1/1000 and 1/1200, respectively.
For live worm imaging, worms were anesthetized in a minimum of 0.001% tetramisole in M9 on a cover slip and mounted on a 1% agarose pad. Live worm slides were examined under the Olympus 1X81/1X2-UCB microscope or a Zeiss LSM 510 Meta confocal microscope. To observe sensory neurons in live RIOK::GFP transgenic worms, they were incubated with the lipophilic fluorescent dye Dil (Life Technologies) in the dark for three hours. Worms were then washed once with M9, plated and destained overnight and live worms were mounted on 1% agarose pads and examined by confocal microscopy. All images were processed using Adobe Photoshop and figures drawn using Adobe Illustrator.
Results
The C. elegans genome contains three riok genes
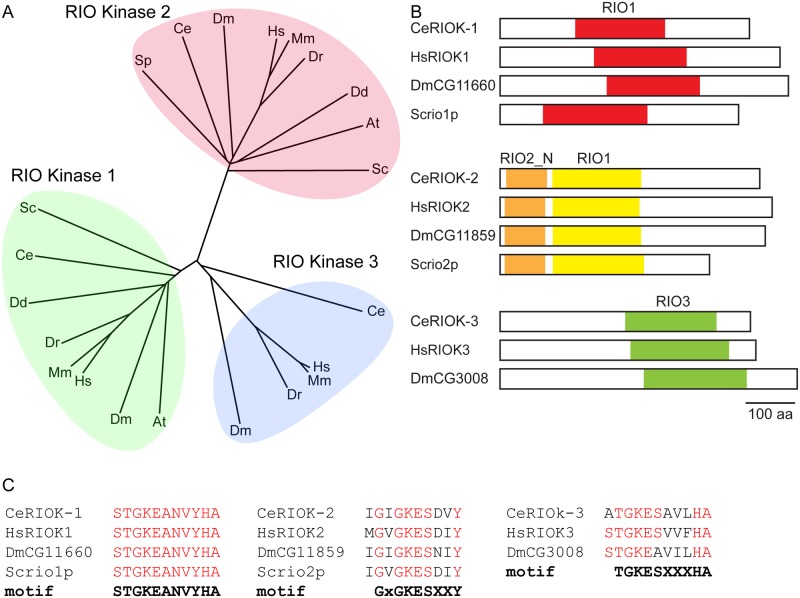
The proteins M01B12.5/RIOK-1 and Y105E8B.3/RIOK-2 were encoded in the C. elegans genome, and have 61% and 56% similarity to the yeast homologs Rio1p and Rio2p, respectively. A third RIOK, protein ZK632.3/RIOK-3, was 48% similar to Rio1p. Phylogenetic analysis places C. elegans RIOK-1 and RIOK-2 with RIOK orthologs from yeast to humans, and RIOK-3 to a metazoan-specific group of RIOKs (Fig. 1A). Each of the three RIOKs has distinct domains (Fig. 1B) and regions of conservation (data not shown). RIOK-1 has a highly conserved “RIOK-like kinase” domain [10] in the middle of the protein. RIOK-2 kinases are most similar in the amino terminal half of the protein where two domains are identified: the RIOK-2 N-terminal domain which is similar to winged helix domains and may be able to bind DNA [10] and the RIOK-1 catalytic domain. RIOK-3 kinases are less conserved in sequence, but contain the “RIOK-like kinase” domain in the carboxy-terminal half of the protein.
Fig 1. The RIO family of kinases are evolutionary conserved.
(A) Unrooted phylogenetic tree of RIO proteins derived from a Clustal Omega alignment of whole proteins sequences (European Bioinformatics Institute). Tree was drawn using Philodendron Phylogenetic tree printer. Hs, H. sapiens; Mm, M. musculus; Dm, melanogaster; Ce, C. elegans; Xl, X. Laevis; Dr, D. reri; Dd, D. discoideum; At, A. thaliana; Sc, S. cerevisiae (B) Schematic representing the domain organisation in RIO kinases. Each of the RIO kinase sub-families has a conserved domain organisation; Hs, H. sapiens; Dm, melanogaster; Ce, C. elegans; Sc, S. cerevisiae examined. (C) Alignment of the ATP binding loop in the RIO domain.
The phosphate-binding loop (P-loop), which binds nucleotides, has been used to differentiate among the three RIOK families [26]. The P-loop of RIOK-1 kinase of S. cerevisiae, Drosophila, C. elegans and humans are identical in sequence and match the canonical RIOK-1 motif [26] (Fig. 1C). The P-loop of RIOK-2 kinases are less well conserved, but adheres to the GxGKESxxY motif characteristic of RIOK-2 proteins. The RIOK-3 P-loop motif is the least well conserved, with both the C. elegans and Drosophila proteins having a serine to alanine change from the canonical motif for RIOK-3 P-loops (Fig. 1C). Taken together, these findings indicate that the C. elegans genome has a single member of each of the three RIOKs.
Localisation of riok expression
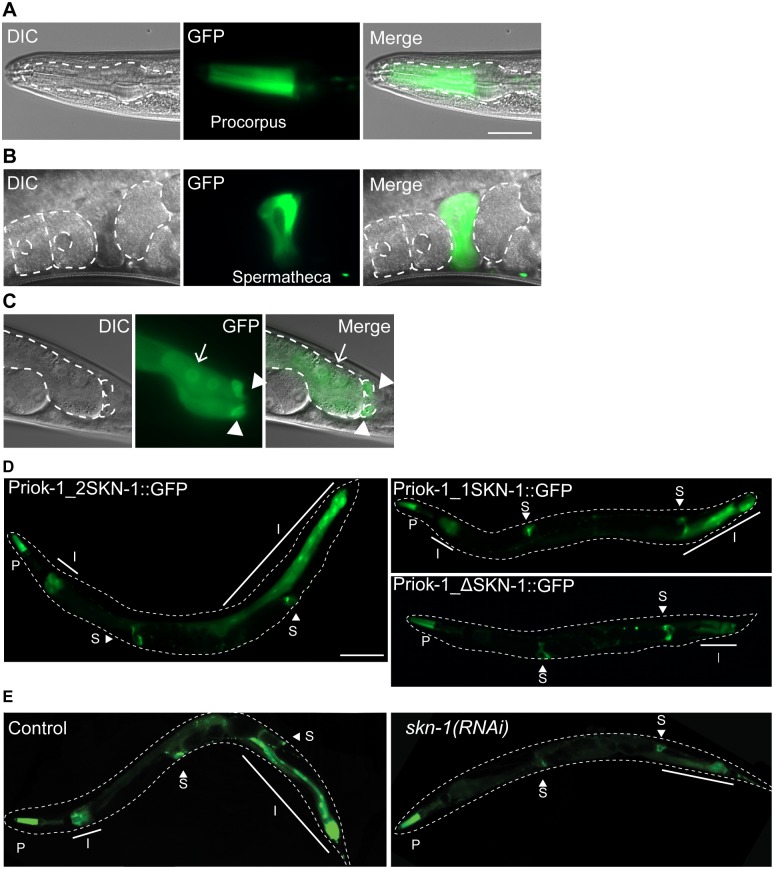
The tissue expression pattern of the three members of the RIOK family is not known for any metazoans. Therefore, we generated transcriptional fusion reporters that express GFP under the control of each of the predicted riok promoter sequences. We defined the promoter of each gene as the intergenic region between the ATG site of the riok gene and the nearest 5’ gene. A previous study [27] showed that knockdown of riok-1 by RNAi induced expression of several target genes of the stress response transcription factor SKN-1. Given this connection we examined the putative riok-1 promoter for SKN-1 binding sites. We identified two putative SKN-1 sites 512 (TTTATCAT) and 2312 nucleotides (ATTGTCAT) from the start of the riok-1 open reading frame. The GFP expression pattern of both Priok-1_2SKN-1::GFP and Priok-1::GFP were similar in all developmental stages. Expression was detected in the pharynx (procorpus), the spermatheca, intestine, some neurons, rectal gland and the rectal valve (Fig. 2 and data not shown). In the intestine, GFP expression by the promoter containing two SKN-1 binding sites (Priok-1_2SKN-1::GFP) was slightly more extensive compared to the Priok-1_1SKN-1 promoter (Fig. 2D). We generated additional transgenic lines in which the predicted SKN-1 binding site from the Priok-1_1SKN-1::GFP construct was changed from ATTGTCAT to ATTCTGCA, as these mutations have been shown to abolish SKN-1 binding [23]. In these transgenic lines GFP expression was unchanged in non-intestinal cells, while intestinal cells showed dramatically reduced GFP expression (Fig. 2D). To further examine the requirement for SKN-1 for riok-1 gene expression, we used RNAi to knockdown SKN-1 levels. In skn-1(RNAi) worms, the intestinal expression of the Priok-1_2SKN-1::GFP reporter gene was significantly reduced in the intestine cells, while expression in the other somatic tissues was unchanged (Fig. 2E). We conclude that high levels of GFP expression in the intestine, but not other tissues, requires SKN-1 and SKN-1-binding sites in the riok-1 promoter.
Fig 2. The putative riok-1 promoter drives GFP expression in multiple tissues.
(A) procorpus region of the pharynx; pharynx is outlined (B) spermatheca; oocytes and eggs are outlined. (C) intestine (arrow) and the rectal gland (arrow heads); intestinal cells and rectal gland outlined. Scale bar = 25μm. (D) Intestinal expression is largely SKN-1 dependent. Representative images of GFP expression in Priok-1_2SKN-1, Priok-1_1SKN-1 and Priok-1_ΔSKN-1 worms. I, intestine; P, Pharynx; S, spermatheca. (E) SKN-1 is required for normal expression of riok-1 in the intestine. RNAi knockdown of skn-1, but not control (pCB19), leads to a reduction in the Priok-1_2SKN-1 intestinal expression. Worms are outlined with a dashed line and the solid line indicates the extent of GFP expression in the intestine. Scale bar = 100 μm.
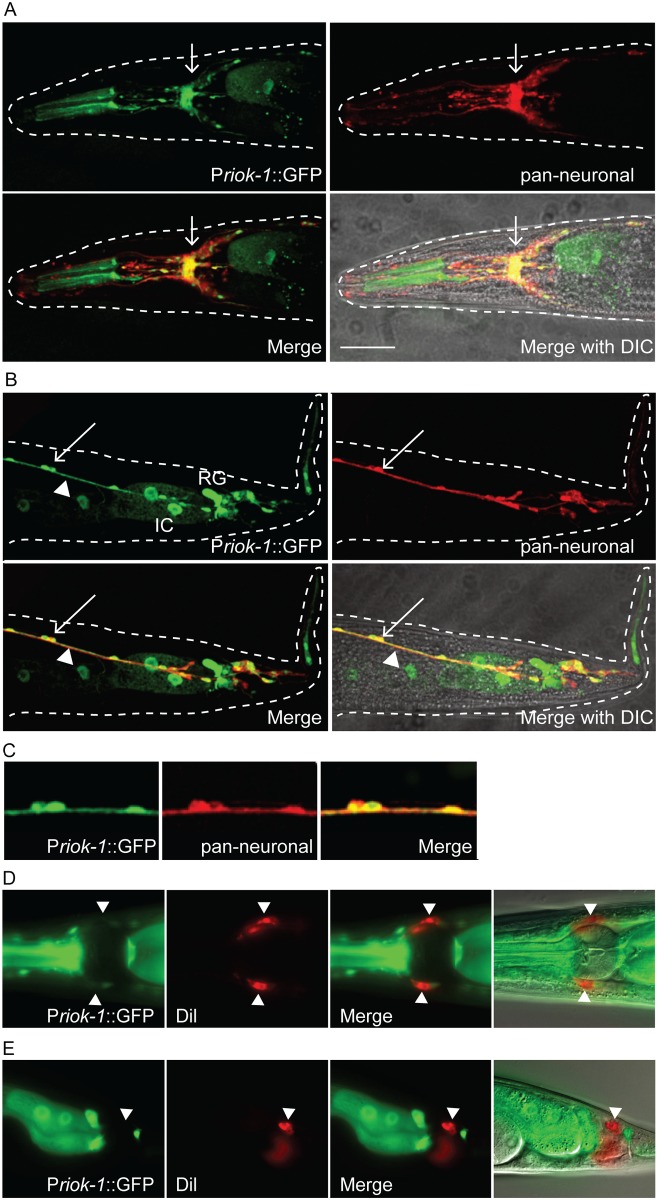
To confirm the neuronal expression pattern of Priok-1::GFP, we generated transgenic worms expressing both Priok-1::GFP and a pan-neuronal marker (Prgef- 1::DsRed2). Co-localisation of GFP with the pan-neuronal marker was observed in tail and longitudinal neurons and in all D-type neurons (Fig. 3). Priok-1_1SKN-1::GFP reporter gene expression was observed in a subset of neurons within the head and tail regions of worms. To determine whether these neurons are sensory, we conducted dye-filling assays. No co-localisation of the lipophilic dye (Dil) with GFP was detected (Fig. 3 D-E), indicating that RIOK-1 is not expressed in the head (amphid) or the tail (phasmid) sensory neurons in C. elegans.
Fig 3. Priok-1_2SKN-1 drives GFP expression in a small subset of neurons.
(A–C) Single plane confocal images from adult worms. (A) GFP was expressed in a subset of head neurons and is highly expressed in the nerve ring displaying strong co-localisation with the pan-neuronal marker (arrow). (B) Co-localisation of GFP with pan-neuronal dsRED. D-type neurons (arrow) and longitudinal nerves (arrow head). Intestinal cells (IC) and the rectal gland (RG) can also be observed. (C) Zoomed in image of the D-type neurons displaying co-localisation. Scale bar = 25μm. (D–E) Priok-1_2SKN-1 GFP expression does not co-localise with the lipophilic dye Dil, in head neurons (D) or tail neurons (E). Scale bar = 15 μm.
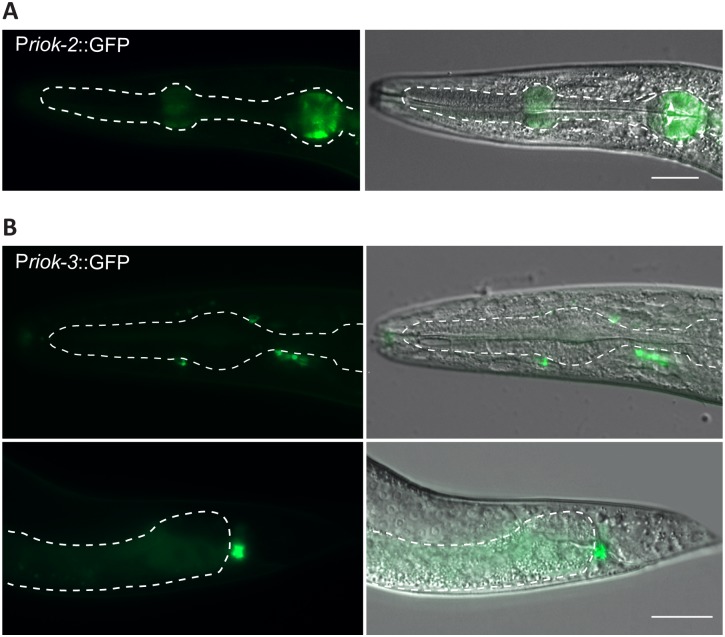
In contrast to RIOK-1, both RIOK-2 and-3 have a restricted tissue expression pattern. Worms expressing GFP under the control of the riok-2 promoter showed consistent GFP expression only in adult staged worms, where it localised to the metacorpus and posterior bulbus of the pharynx (Fig. 4A). GFP expression under control of the riok-3 promoter was first observed in embryos and continued throughout larval development. GFP intensity was highest in adult worms, where it was found in a small number of tail neurons whose positioning was consistent with PVQ, PHAL/PQR neurons (Fig. 4B).
Fig 4. The putative promoters of riok-2 and -3 drive GFP in distinct tissues.
(A) riok-2 transgenic worms displayed weak GFP expression in the metacorpus and posterior bulb of the pharynx. Scale bar = 25μm (B) riok-3 transgenic worms revealed weak GFP expression, in some head and was strongly expressed in a tail neuron which may be the PVQ, PHAL or PQR neuron. Scale bar = 25μm.
Functional analysis of RIOKs
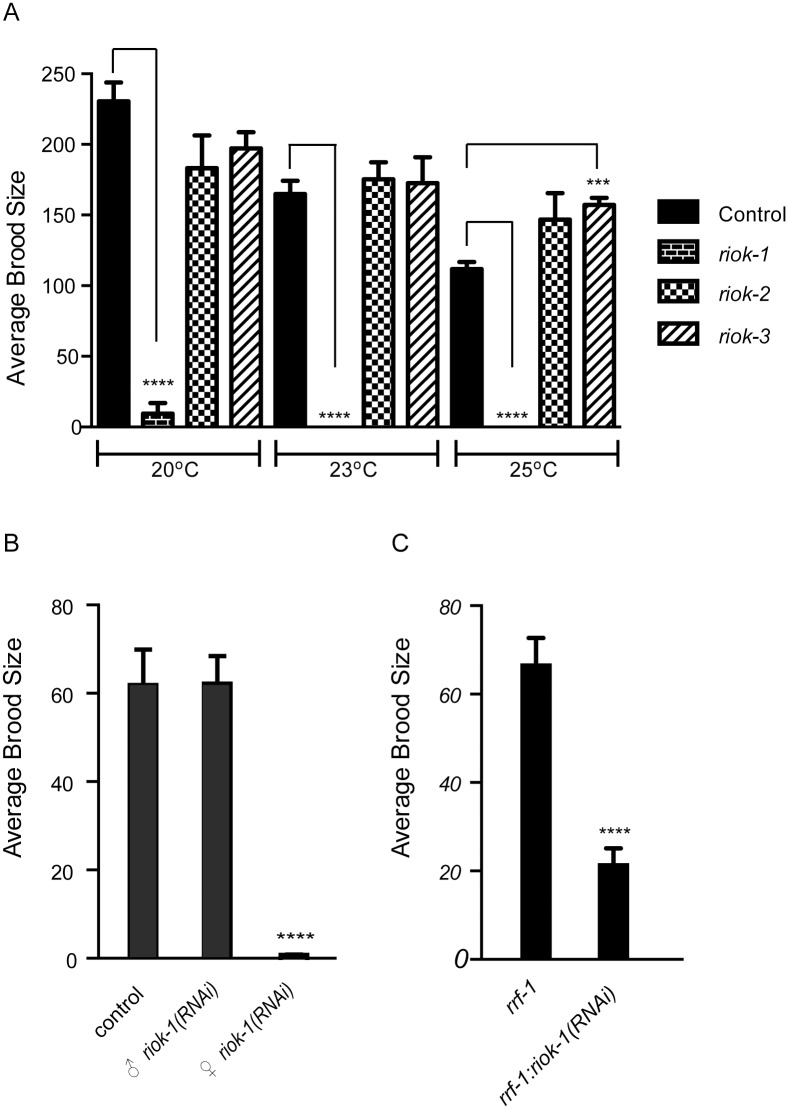
The functions of RIOKs in C. elegans are not yet understood. Predicted genetic null mutants are available for all three rioks; homozygote mutants for riok-1 and riok-2 both had 100% penetrance early larval arrest, while riok-3 worms had no obvious phenotype (data not shown). To characterise the functions of riok-1, riok-2 and riok-3, we used RNAi to knockdown each kinase. RNAi by microinjection led to early larval arrest similar to the null mutants representing riok-1 and riok-2, but no overt phenotypes for riok-3. RNAi using the feeding approach produced worms that developed to adulthood without any obvious developmental deficits. However, knockdown of riok-1 resulted in a significant reduction in the number of progeny produced at 20°C and sterility at 23°C and 25°C. Knockdown of riok-2 and riok-3 produced similar numbers of progeny compared with their controls (Fig. 5A). Given the neuronal expression of riok-1, we also conducted behavioral touch assays using neuronal RNAi sensitive strains. However, we did not observe any touch response defects (data not shown), suggesting that riok-1 is not essential for this response. The relatively broad tissue expression pattern of riok-1 in adult worms and its requirement for reproduction indicate that this kinase may have diverse functions in C. elegans.
Fig 5. riok-1(RNAi) sterility is associated with an oogenesis defect.
(A) Total average brood size for wild-type worms knocked down for riok-1, riok-2 or riok-3 at 20°C, 23°C and 25°C. riok-1(RNAi) worms have a significant reduction in brood size at 20°C and are sterile at 23°C and 25°C. Error bars represent the SEM; n ≥6. **** = P values <0.0001. *** = P values 0.0003 for riok-3 at 25°C (B) Average number of progeny produced in 17 hours from crossing fog-2(q71) male and female worms. Crosses were; control, ♀ fog-2(q71) x ♂ fog-2(q71); ♀ fog-2(q71) x ♂riok-1(RNAi); ♀ fog-2(q71)riok-1(RNAi) x ♂ fog-2(q71). Thirteen individual crosses per treatment were conducted at 25°C with 10 males mated per L4 female. **** = P <0.0001. (C) Knockdown of riok-1 in rrf-1 worms resulted in significantly reduced progeny. Total brood size for rrf-1 worms grown on riok-1 and pCB19 (negative control) RNAi at 25°C. Error bars represent SEM. n = 23, **** = P <0.0001.
To establish whether sterility linked to RIOK-1 was due to a defect in spermatogenesis or oogenesis, we knocked down riok-1 in fog-2(q70), which exists as a male and female worms. When fog-2 female worms were knocked down for riok-1 and crossed with untreated male worms, very few (10 ± 2, n = 10) progeny were generated. In contrast, riok-1(RNAi) male worms crossed with untreated females produced high levels of progeny (Fig. 5B). Based on these findings, we conclude that riok-1 is essential for normal oogenesis but dispensable for spermatogenesis. To determine whether the riok-1(RNAi) oogenesis defect was somatic gonad or germ cell-derived, we took advantage of a mutant in rrf-1 which is resistant to RNAi in most somatic tissues, but capable of RNAi in germ cells. Knockdown of riok-1 in rrf-1 worms resulted in 48% of worms being sterile and 52% producing few progeny (21.7 ± 3.3 n = 34, P<0.01) (Fig. 5C). The rrf-1 strain does have some intestinal RNAi capacity [28], therefore we knocked down riok-1 in the stain VP303, which is only capable of RNAi in intestinal cells. When riok-1 was knocked down in this background the worms had a normal brood size (data not shown). We concluded that the riok-1 sterility likely results from a defect in germ cell function during oogenesis.
RIOK-1 is required for normal germ cell proliferation
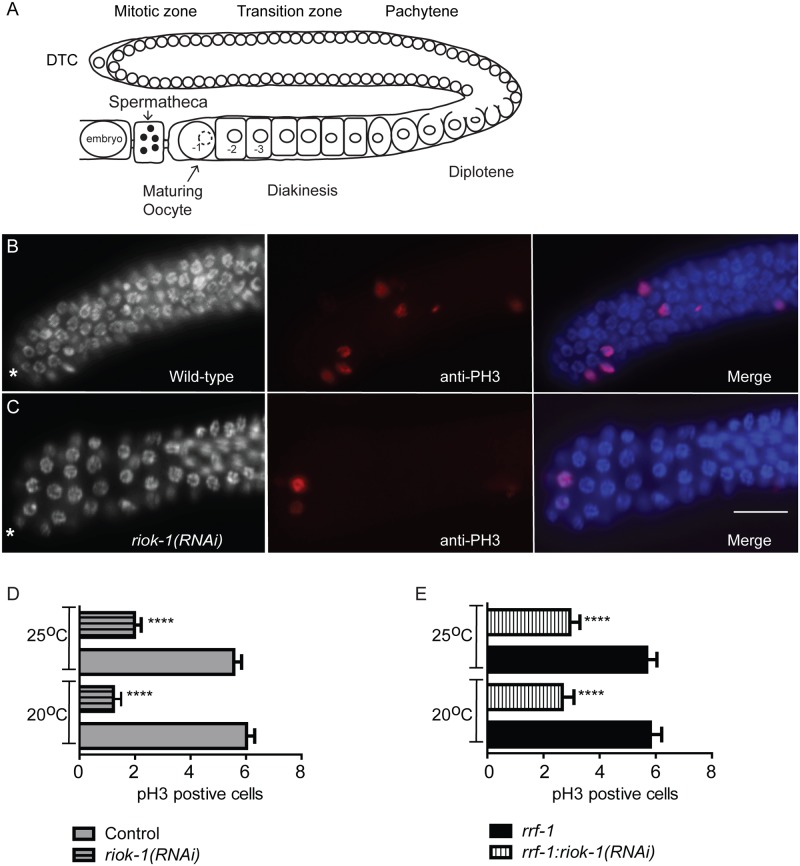
We next examined the gonads of one-day old hermaphrodites depleted of RIOK-1, -2 or-3. Gonads of riok-2(RNAi) and riok-3(RNAi) worms displayed grossly normal morphology, whereas riok-1(RNAi) worms exhibited smaller mitotic and transitions zones in the distal gonad compared with controls (Table 1). We then examined whether knockdown of riok-1 caused a reduction in cellular proliferation by immunostaining dissected gonads with the proliferation marker anti-phosphohistone H3 (PH3). Worms knocked down for riok-1 had a significantly reduced number of PH3-positive cells in the mitotic zone at both 20°C and 25°C (Fig. 6B-D), indicating decreased germ cell proliferation. To examine whether this decrease was due to riok-1 expression in somatic or germ cells, we knocked down riok-1 in a strain lacking the rrf-1 gene. Germ cell proliferation was indeed reduced in rrf-1:riok-1(RNAi) compared with rrf-1 worms (Fig. 6E), suggesting that the observed gonadal defects are likely to be due to germ rather than somatic cell abnormalities. Interestingly, no defects in chromosomal organisation were detected when gonads of riok-1(RNAi) worms were examined for defects in transition zone and pachytene stage germ cells using antibodies to the synaptonemal complex proteins SYP-1 [29](S1 Fig.), suggesting that riok-1 is not required for the pachytene stage of germ cell development. Together, these data indicate that riok-1 is required for the early stages of germ cell development.
Table 1. Worms depleted of riok-1 had a significant reduction in the size of the mitotic zone.
| Distal germ cells count (diameter) | ||
|---|---|---|
| Mitotic zone (MZ) | Transition zone (TZ) | |
| Wild-type | 19 cells ± 0.4 (n = 10) | 9 cells ± 0.3 (n = 10) |
| riok-1(RNAi) | 12 cells ± 0.7 (n = 10) | 7 cells ± 0.4 (n = 10) |
Values represent Mean and SEM (P <0.0001).
Fig 6. riok-1 is required for normal germ cell proliferation.
(A) Overview of the adult C. elegans hermaphrodite gonad. One of the two gonad U-shaped tubular gonad is shown. The single somatic distal tip cell (DTC) maintains a population of self-renewing germline stem cells. Germ cells enter into the meiotic prophase I in the transition zone and then progress to the pachytene stage. During these stages, germ cells are only partially enclosed in a membrane and share a common cytoplasmic core. As germ cells exit pachytene, they move through the “loop” region and enter diplotene and begin to fully cellularise. A signal from sperm directs the most proximal oocytes to undergo maturation and fertilisation occurs are the oocyte moves through the spermatheca into the uterus. (B-C) Dissected gonads were stained with anti-pH3 staining to detect proliferating germ cells and DAPI to visualise DNA. Asterisks indicate the distal end of the gonad. Scale bar = 15μm. (D-E) riok-1(RNAi) gonads had a reduced number of pH3 stained cells in the mitotic zone (D) Wild-type worms with riok-1 knocked down at 20°C and 25°C (n = 50) (E)) rrf-1;pCB19(RNAi) and rrf-1:riok-1(RNAi) worms at 20°C and 25°C (n = 50). Error bars represent SEM; **** = P values <0.0001.
RIOK-1 is required for normal germ cell progression in the proximal gonad
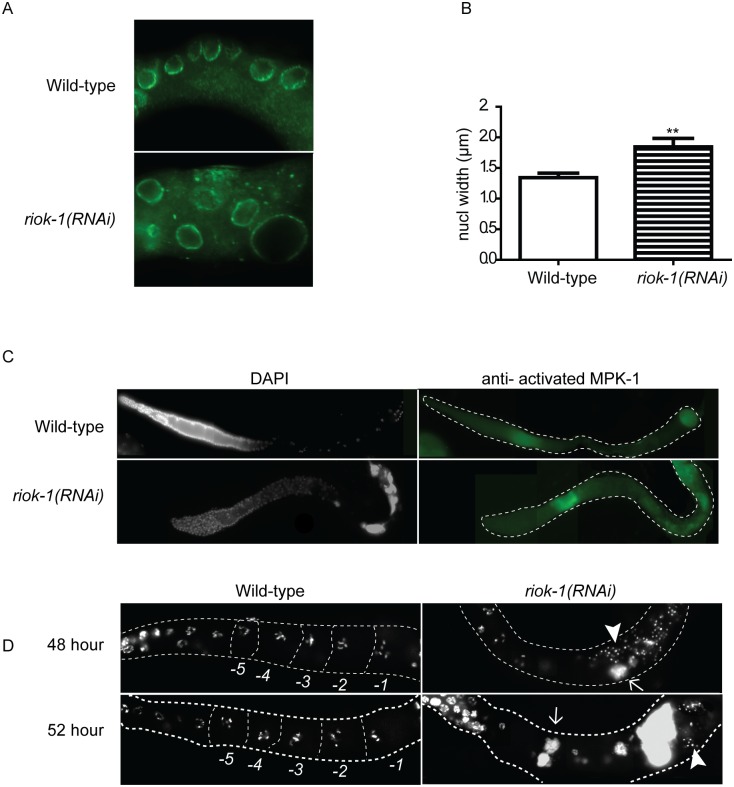
After germ cells exit pachytene, they enter the loop region of the gonad where they progress into the diplotene stage. In wild-type worms, germ cells are organised into a single row as they move through diplotene (Fig. 6A). When riok-1 was knocked down, germ cells entering diplotene were disorganised and their nuclei were abnormally enlarged. We immunostained dissected gonads with a nuclear pore complex marker and found that germ cell nuclei of riok-1(RNAi) worms were significantly larger (P<0.05) than wild-type worms (Fig. 7A-B). As germ cells exit pachytene, MPK-1 mitogen activated protein kinase (MAPK) is inactivated and only reactivated in the most proximal oocytes. To examine whether this down regulation of MAPK activity occurs in the absence of riok-1, we immunostained gonads from one-day old riok-1(RNAi) worms with a monoclonal antibody that specifically binds to the activated, diphosphorylated form of MAPK (dpMPK-1). In wild-type worms, MAPK staining was predominantly present in the pachytene germ cells and the most proximal oocytes (Fig. 7C). Interestingly, in 50% (n = 120) of riok-1(RNAi) gonads, dpMPK-1 staining did not decrease as germ cell exit pachytene, instead it was evident throughout diplotene and diakinesis (Fig. 7C). These findings indicate that RIOK-1 is directly or indirectly required to inhibit the MAPK pathway as germ cells exit pachytene or limit the activation in late stage oocytes.
Fig 7. Proximal gonad organization requires riok-1.
(A) Representative images of wild-type and riok-1(RNAi) dissected gonads immunostained stained for nuclear pores. (B) The nuclear diameter of germ cells in the proximal loop region of riok-1(RNAi) worms are significantly increased in size (n = 12). Error bars represent SEM; ** = P values 0.0045. (C) Wild-type and riok-1(RNAi) dissected gonads immunostained stained for activated MAPK. Increased MAPK staining is seen in pachytene and throughout diplotene. (D) Progression of the riok-1(RNAi) EMO phenotype was examined at time points 48 and 52 hours post L1. Gonads from dissected wild-type and riok-1(RNAi) worms were stained with DAPI to visualise DNA. The gonad and oocytes are outlined and proximal wild-type oocytes have been outlined and labelled (-1 to-5). At all time points wild-type worms displayed the typical six bivalent chromosomes in their maturing oocytes within the proximal gonad (n = 30). riok-1(RNAi) worms (n = 30) showed DNA aggregates (arrow) in the proximal region of the gonad and abnormal localised sperm (arrow head). Scale bar = 25 μm; worms grown at 25°C.
Oocytes in riok-1(RNAi) worms appeared abnormal and did not display the typical rectangular shape seen in wild-type worms. Large DNA aggregates were detected in the proximal gonad of riok-1 knockdown worms (Fig. 7D). These DNA clumps were consistent with an endomitotic oocyte (EMO) phenotype [30]. This finding suggests that riok-1 might be involved in regulating normal oocyte maturation or possibly ovulation. To examine the onset and progression of the endomitotic oocytes, gonads were dissected from wild-type and riok-1(RNAi) adult worms (48, 52, 56 and 60 h following the L1 stage). Wild-type worms displayed the typical six bivalent chromosomes within their oocytes at all time points (Fig. 7D). At 48 h, riok-1(RNAi) worms displayed small DNA aggregates (Fig. 7D, arrow) and they continued to get larger over time. At all time-points, riok-1(RNAi) worms had abnormally located sperm near the DNA clumps in the proximal gonad (Fig. 7D, data not shown). This result suggested that the spermatheca did not function normally when riok-1 was perturbed. Temporal analysis showed that the riok-1(RNAi) DNA aggregates enlarged over time, indicating the EMO phenotype is aggravated.
To examine whether the EMO phenotype was sperm dependent, riok-1 was knocked down in the male and female fog-2(q70) strain. In the hermaphrodite gonad spermatogenesis occurs prior to oogenesis, and oocyte maturation and ovulation occurs constitutively in adults until sperm stores are depleted [31]. In unmated female worms oocytes arrest in meiotic prophase I. No endomitotic oocytes were observed in fog-2 (mated or unmated) control worms (Table 2). Within unmated fog-2: riok-1(RNAi) females, most worms did not display an EMO phenotype (Table 2). Surprisingly, mated fog-2 females also did not display an EMO phenotype but were sterile (Table 2). Together these data suggest that the EMO phenotype is dependent upon hermaphrodite derived sperm, but not male derived sperm.
Table 2. Ovulation is required for endomitotic oocytes in riok-1 worms.
| Genotype | % EMO | n-value |
|---|---|---|
| Wild-type | 0% | 25 |
| riok-1(RNAi) | 100% | 25 |
| fog-2(q71) female unmated | 0% | 30 |
| fog-2(q71):riok-1(RNAi) female unmated | 0% | 25 |
| fog-2(q71) female mated | 0% | 27 |
| fog-2(q71):riok-1(RNAi) female mated | 0.1% | 28 |
Discussion
In this study, we showed that the C. elegans genome encodes three members of the RIOK family, and that RIOK-1 and-2 are essential for larval development. RIOKs from yeast and mammalian cells have been shown to be required for fundamental steps in rRNA maturation and cell cycle regulation [26]. Therefore, we were surprised that our transgenic analysis of riok gene expression revealed detectable expression in relatively few cells. It is possible that riok gene(s) are expressed in the germline, however our transgenic analysis would have not supported germline expression due to the silencing of the extrachromosomal transgene arrays in the germline [32]. Both riok-1 and -3 were expressed throughout development and adulthood, while riok-2 was only detected in adult-stage worms. It is possible that the tissue expression pattern of the RIOKs is wider than our analysis showed when the expression specific levels are very low. Indeed, both Rio1P and Rio2P are reported to be expressed at very low levels in yeast [6, 33]. Therefore, it is possible that riok-1 and-2 are expressed widely, but at very low levels in C. elegans and may function in rRNA processing, as is the case in other species. Null mutants in other genes that affect rRNA processing in C. elegans also show a similar larval arrest as riok-1 and-2 mutants [34].
The riok-1 promoter drove strong GFP expression in several neurons, the spermatheca and the intestine. The intestine is the primary detoxification organ in C. elegans. Expression of the riok-1 reporter was high in the first anterior and the last few posterior cells of the intestine, a pattern similar to several genes that function in the oxidative stress pathway [27]. The riok-1 intestinal expression was dependent on the predicted SKN-1 binding sites in its promoter. SKN-1 is a key transcription factor that is involved in oxidative and xenobiotic stress responses in C. elegans [35, 36]. Knockdown of riok-1 has been reported to result in the induction of several genes in the SKN-1 stress response pathway, suggesting that riok-1 may function in regulating cellular metabolism, although riok-1(RNAi) worms showed only a modest decrease in stress resistance [27]. The genomic locus for riok-1 encodes two isoforms of RIOK-1 [37]. The small isoform lacks the complete RIO domain, including the putative catalytic site. It will be interesting to develop tools to examine which of the two isoforms of RIOK-1 are expressed in a SKN-1-dependent manner, as the short isoform could act as an endogenous dominant-negative as has been reported for other protein kinases [38]. Together, these data suggest a new role for RIOKs in stress pathways.
An unexpected discovery was that riok-1 is required for fertility. Worms in which riok-1 was knocked down displayed an oogenesis-specific sterility that was associated with several defects in gonad organisation. When riok-1 was knocked down, the number of proliferating mitotic cells was significantly reduced, and the nuclei of germ cells entering the diplotene stage of meiosis prophase I had abnormally organised and large nuclei. In the proximal gonad, oocyte maturation appeared abnormal, and oocytes underwent multiple rounds of DNA replication, resulting in the generation of endomitotic oocytes. Interestingly, pachytene stage germ cells exhibited no obvious abnormalities, suggesting riok-1 is required for several specific steps in germ cell development. As endomitotic oocytes are often associated with defects in oocyte ovulation and cell cycle regulation, some of the riok-1 associated defects could be due to a direct role in cell cycle regulation as has been reported in mitotic cells of other organisms [5, 14, 15].
Normally, oocytes and sperm are kept physically separated, and they meet only as the oocytes enter the spermatheca during ovulation. The presence of sperm in the proximal gonad of riok-1(RNAi) worms might have contributed to the endomitotic phenotype through an inappropriate activation of the MAPK signaling, which was abundant throughout the proximal gonad (Fig. 7B). The riok-1 reporter was highly expressed in the spermatheca but not in other tissues of the somatic gonad. RIOK-1 may play an important role in regulating the ability of the spermatheca to retain sperm, and thereby indirectly restrict MAPK signaling. The analysis of riok-1 knockdown in the somatic RNAi deficient strain rrf-1 led to 48% of worms being sterile, with the remainder producing very few progeny. Given that riok-1 knockdown in wild-type worms consistently produced 100% sterility, it is possible that RIOK-1 does function in both somatic and germ cells, with the latter having a more dominant contribution to the sterility. Determining the gonad expression pattern of RIOK-1 might help resolve the relative contribution of these tissues. Taken together, our findings suggest that RIOKs in C. elegans have functional roles in both somatic and germ cells.
Supporting Information
High magnification images of wild-type and riok-1(RNAi) germline nuclei at the indicated stages stained with DAPI and SYP-1. Bars, 5 μm.
(TIF)
Acknowledgments
The pan-neuronal DsRed2 marker (rgef-1) was obtained from Massimo Hilliard, University of Queensland.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by a National Health and Medical Research Council grant, APP1044022 (RBG, PRB & AH). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S (2002) The protein kinase complement of the human genome. Science 298: 1912–1934. [DOI] [PubMed] [Google Scholar]
- 2. Manning G (2005) Genomic overview of Protein Kinases (December 13, 2005), WormBook, ed. The C. elegans Research Community, WormBook, Available: http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hanks SK, Hunter T (1995) Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J 9: 576–596. [PubMed] [Google Scholar]
- 4. Scheeff ED, Bourne PE (2005) Structural evolution of the protein kinase-like superfamily. PLOS Comput Biol 1: e49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vanrobays E, Gleizes PE, Bousquet-Antonelli C, Noaillac-Depeyre J, Caizergues-Ferrer M, et al. (2001) Processing of 20S pre-rRNA to 18S ribosomal RNA in yeast requires Rrp10p, an essential non-ribosomal cytoplasmic protein. EMBO J 20: 4204–4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Geerlings TH, Faber AW, Bister MD, Vos JC, Raue HA (2003) Rio2p, an evolutionarily conserved, low abundant protein kinase essential for processing of 20 S Pre-rRNA in Saccharomyces cerevisiae . J Biol Chem 278: 22537–22545. [DOI] [PubMed] [Google Scholar]
- 7. Krupa A, Srinivasan N (2002) Lipopolysaccharide phosphorylating enzymes encoded in the genomes of Gram-negative bacteria are related to the eukaryotic protein kinases. Protein Sci 11: 1580–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Anaya P, Evans SC, Dai C, Lozano G, May GS (1998) Isolation of the Aspergillus nidulans sudD gene and its human homologue. Gene 211: 323–329. [DOI] [PubMed] [Google Scholar]
- 9. Laronde-Leblanc N, Guszczynski T, Copeland T, Wlodawer A (2005) Structure and activity of the atypical serine kinase Rio1. Febs J 272: 3698–3713. [DOI] [PubMed] [Google Scholar]
- 10. LaRonde-LeBlanc N, Wlodawer A (2004) Crystal structure of A. fulgidus Rio2 defines a new family of serine protein kinases. Structure 12: 1585–1594. [DOI] [PubMed] [Google Scholar]
- 11. Kimmelman AC, Hezel AF, Aguirre AJ, Zheng H, Paik JH, et al. (2008) Genomic alterations link Rho family of GTPases to the highly invasive phenotype of pancreas cancer. Proc Natl Acad Sci U S A 105: 19372–19377. 10.1073/pnas.0809966105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Read RD, Fenton TR, Gomez GG, Wykosky J, Vandenberg SR, et al. (2013) A kinome-wide RNAi screen in Drosophila Glia reveals that the RIO kinases mediate cell proliferation and survival through TORC2-Akt signaling in glioblastoma. PLoS Genet 9: e1003253 10.1371/journal.pgen.1003253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tariki M, Wieczorek SA, Schneider P, Banfer S, Veitinger S, et al. (2013) RIO kinase 3 acts as a SUFU-dependent positive regulator of Hedgehog signaling. Cell Signal 25: 2668–2675. 10.1016/j.cellsig.2013.08.037 [DOI] [PubMed] [Google Scholar]
- 14. Angermayr M, Bandlow W (2002) RIO1, an extraordinary novel protein kinase. FEBS letters 524: 31–36. [DOI] [PubMed] [Google Scholar]
- 15. Vanrobays E, Gelugne JP, Gleizes PE, Caizergues-Ferrer M (2003) Late cytoplasmic maturation of the small ribosomal subunit requires RIO proteins in Saccharomyces cerevisiae . Mol Cell Biol 23: 2083–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Baumas K, Soudet J, Caizergues-Ferrer M, Faubladier M, Henry Y, et al. (2012) Human RioK3 is a novel component of cytoplasmic pre-40S pre-ribosomal particles. RNA Biol 9: 162–174. 10.4161/rna.18810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Strunk BS, Novak MN, Young CL, Karbstein K (2012) A translation-like cycle is a quality control checkpoint for maturing 40S ribosome subunits. Cell 150: 111–121. 10.1016/j.cell.2012.04.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ferreira-Cerca S, Sagar V, Schafer T, Diop M, Wesseling AM, et al. (2012) ATPase-dependent role of the atypical kinase Rio2 on the evolving pre-40S ribosomal subunit. Nat Struct Mol Biol 19: 1316–1323. 10.1038/nsmb.2403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shan J, Wang P, Zhou J, Wu D, Shi H, et al. (2009) RIOK3 interacts with caspase-10 and negatively regulates the NF-kappaB signaling pathway. Mol Cell Biochem 332: 113–120. 10.1007/s11010-009-0180-8 [DOI] [PubMed] [Google Scholar]
- 20. Feng J, De Jesus PD, Su V, Han S, Gong D, et al. (2014) RIOK3 Is an Adaptor Protein Required for IRF3-Mediated Antiviral Type I Interferon Production. J Virol 88: 7987–7997. 10.1128/JVI.00643-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stiernagle T (2006) Maintenance of C. elegans (February 11, 2006), WormBook, ed. The C. elegans Research Community, WormBook, Available: http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vaglio P, Lamesch P, Reboul J, Rual JF, Martinez M, et al. (2003) WorfDB: the Caenorhabditis elegans ORFeome Database. Nucleic Acids Res 31: 237–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. An JH, Blackwell TK (2003) SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev 17: 1882–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mello C, Fire A (1995) DNA transformation. Methods Cell Biol 48: 451–482. [PubMed] [Google Scholar]
- 25. Boag PR, Atalay A, Robida S, Reinke V, Blackwell TK (2008) Protection of specific maternal messenger RNAs by the P body protein CGH-1 (Dhh1/RCK) during Caenorhabditis elegans oogenesis. J Cell Biol 182: 543–557. 10.1083/jcb.200801183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. LaRonde-LeBlanc N, Wlodawer A (2005) A family portrait of the RIO kinases. J Biol Chem 280: 37297–37300. [DOI] [PubMed] [Google Scholar]
- 27. Wang J, Robida-Stubbs S, Tullet JM, Rual JF, Vidal M, et al. (2010) RNAi screening implicates a SKN-1-dependent transcriptional response in stress resistance and longevity deriving from translation inhibition. PLoS Genet 6 10.1371/journal.pgen.1001260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kumsta C, Hansen M (2012) C. elegans rrf-1 mutations maintain RNAi efficiency in the soma in addition to the germline. PLoS One 7: e35428 10.1371/journal.pone.0035428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. MacQueen AJ, Colaiacovo MP, McDonald K, Villeneuve AM (2002) Synapsis-dependent and-independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans . Genes Dev 16: 2428–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Clandinin TR, DeModena JA, Sternberg PW (1998) Inositol trisphosphate mediates a RAS-independent response to LET-23 receptor tyrosine kinase activation in C. elegans . Cell 92: 523–533. [DOI] [PubMed] [Google Scholar]
- 31. McCarter J, Bartlett B, Dang T, Schedl T (1999) On the control of oocyte meiotic maturation and ovulation in Caenorhabditis elegans . Dev Biol 205: 111–128. [DOI] [PubMed] [Google Scholar]
- 32. Kelly WG, Xu S, Montgomery MK, Fire A (1997) Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics 146: 227–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Angermayr M, Schwerdffeger K, Bandlow W (2003) A nucleosome-free dG-dC-rich sequence element promotes constitutive transcription of the essential yeast RIO1 gene. Biol Chem 384: 1287–1292. [DOI] [PubMed] [Google Scholar]
- 34. Kudron MM, Reinke V (2008) C. elegans nucleostemin is required for larval growth and germline stem cell division. PLoS Genet 4: e1000181 10.1371/journal.pgen.1000181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Papp D, Csermely P, Soti C (2012) A role for SKN-1/Nrf in pathogen resistance and immunosenescence in Caenorhabditis elegans . PLoS Pathog 8: e1002673 10.1371/journal.ppat.1002673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tullet JM, Hertweck M, An JH, Baker J, Hwang JY, et al. (2008) Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans . Cell 132: 1025–1038. 10.1016/j.cell.2008.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Breugelmans B, Jex AR, Korhonen PK, Mangiola S, Young ND, et al. (2014) Bioinformatic exploration of RIO protein kinases of parasitic and free-living nematodes. Int J Parasitol 44:827–836. 10.1016/j.ijpara.2014.06.005 [DOI] [PubMed] [Google Scholar]
- 38. McCormick JA, Ellison DH (2011) The WNKs: atypical protein kinases with pleiotropic actions. Physiol Rev 91: 177–219. 10.1152/physrev.00017.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
High magnification images of wild-type and riok-1(RNAi) germline nuclei at the indicated stages stained with DAPI and SYP-1. Bars, 5 μm.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.