Abstract
Effects of extremely high carbon dioxide (CO2) concentrations on soil microbial communities and associated processes are largely unknown. We studied a wetland area affected by spots of subcrustal CO2 degassing (mofettes) with focus on anaerobic autotrophic methanogenesis and acetogenesis because the pore gas phase was largely hypoxic. Compared with a reference soil, the mofette was more acidic (ΔpH ∼0.8), strongly enriched in organic carbon (up to 10 times), and exhibited lower prokaryotic diversity. It was dominated by methanogens and subdivision 1 Acidobacteria, which likely thrived under stable hypoxia and acidic pH. Anoxic incubations revealed enhanced formation of acetate and methane (CH4) from hydrogen (H2) and CO2 consistent with elevated CH4 and acetate levels in the mofette soil. 13CO2 mofette soil incubations showed high label incorporations with ∼512 ng 13C g (dry weight (dw)) soil−1 d−1 into the bulk soil and up to 10.7 ng 13C g (dw) soil−1 d−1 into almost all analyzed bacterial lipids. Incorporation of CO2-derived carbon into archaeal lipids was much lower and restricted to the first 10 cm of the soil. DNA-SIP analysis revealed that acidophilic methanogens affiliated with Methanoregulaceae and hitherto unknown acetogens appeared to be involved in the chemolithoautotrophic utilization of 13CO2. Subdivision 1 Acidobacteriaceae assimilated 13CO2 likely via anaplerotic reactions because Acidobacteriaceae are not known to harbor enzymatic pathways for autotrophic CO2 assimilation. We conclude that CO2-induced geochemical changes promoted anaerobic and acidophilic organisms and altered carbon turnover in affected soils.
Introduction
Mofettes are cold exhalations of volcanic carbon dioxide (CO2), which migrates through the lower crust or upper mantle to the surface via tectonic faults (Kämpf et al., 2007). Mofette soils received increasing attention as natural analogues for the potential leakage of CO2 during carbon capture and storage and associated long-term effects on ecosystems. Although studies evaluated potential consequences of elevated CO2 (<10%) on soil organisms and processes (e.g., He et al. 2010), these findings had little relevance to environments where extreme CO2 partial pressures (>90%) alter soil chemistry and formation. Low pH, anoxic conditions and increased soil carbon contents are frequently observed in mofette soils (Ross et al., 2000; Blume and Felix-Henningsen 2009; Rennert et al. 2011).
Biological responses to high CO2 venting were mainly studied with a focus on plants, with changes in vegetation, for example, reduced growth and increased plant C/N ratios, likely due to hypoxia and/or lower nutrient availability (Pfanz et al., 2004, Vodnik et al., 2007). However, little is known about the effect of CO2 seepage on active microbial communities and associated processes. Differences in the abundance of lipid biomarkers and group-specific 16S rRNA genes were observed between mofette and reference soils at Latera Caldera, Italy (Oppermann et al., 2010) and Laacher See, Germany (Frerichs et al., 2012). Interestingly, 13C-enriched stable carbon isotopic compositions of total organic carbon (TOC) and individual lipid biomarkers in Latera Caldera mofette soil pointed to a considerable incorporation of volcanic CO2 (Oppermann et al., 2010) raising the question as to which organisms are involved in the utilization of volcanic CO2.
In the current study, we investigated a wetland area in northwestern Bohemia, Czech Republic, where spots of subcrustal degassings lead to soil gas phase concentrations of up to 100% CO2. We expected the reduction of CO2 by methanogens and/or acetogens to become the thermodynamically dominant electron accepting process in the wetlands after depletion of the usually small pools of alternative electron acceptors. Because hydrogenotrophic methanogens and metabolically versatile acetogens can tolerate slightly acidic conditions (Küsel et al., 2000; Horn et al., 2003), these anaerobes might be favored by the low pH, anoxic conditions, as well as the high availability of CO2 in mofette soil (Drake et al., 2009, 2013), thus providing an important link in the carbon flow from volcanic CO2 to soil carbon.
To resolve whether specific anaerobic microbial groups are major consumers of volcanic CO2, we analyzed differences in (i) acetogenic and methanogenic potentials, (ii) diversity of active prokaryotes and (iii) pore-water biogeochemistry in different depths of mofette and reference soil. Furthermore, we studied the potential contribution of CO2-utilizing microbial communities to mofette carbon flow by using microcosm incubations with 13C-labeled CO2. Here, we followed the incorporation of 13C-CO2 into biomass by lipid biomarkers analysis over time and identified labeled organisms by DNA stable isotope probing (SIP).
Materials and methods
Site description and sampling design
The wetland area containing several mofettes (50°08‘48‘‘ N, 12°27‘03‘‘ E) is located close to the river Plesná, in northwestern Bohemia, Czech Republic, (Flechsig et al., 2008), in a region considered to be the seismic center of the Cheb Basin (Bankwitz et al., 2003). Mofette soil gas consisted of predominantly CO2 (99.83±0.03 vol.%) with a δ13C value of -1.95±0.06‰ vs Vienna Peedee Belemnite Standard (VPDB) (Bräuer et al., 2004; Kämpf et al., 2013), a minor fraction of N2 (0.127±0.021 vol.%) and traces of Ar, O2, CH4 and He (ppmv range). Mofette soils were identified in the field by visible upcoming gas streams within a semi-circular region lacking vegetation. A reference soil lacking CO2 emissions was identified with a portable landfill gas analyser (GA2000, Ansyco, Karlsruhe, Germany) in close proximity (<5 m distance). All samplings in this study were conducted on the same mofette and the respective reference soil.
Sampling was conducted according to the following exploratory scheme: in April 2011, we studied acetogenic and methanogenic potentials in different depths of mofette and reference soils. After observing pronounced differences, in June 2011, we investigated pore-water geochemistry as well as active microbial communities in both soils. Following these observations, in November/December 2011 we evaluated the role of CO2-utilizing microbial communities by using 13CO2 labeling incubations and parallel stable carbon isotopic analysis of pore-water acetate, CH4 and CO2. Additional comparative pore-water sampling of mofette and reference soils was performed in August 2012 to confirm previous observations.
Pore-water and soil sampling
All samplings were conducted on the same mofette and respective reference soils. Pore-water samples were taken in 1-cm intervals by using a dialysis sampler with 5-ml replicate cells down to 60 cm depth after 2 weeks of equilibration (Hädrich et al., 2012). Soil samples were obtained in form of three biological replicates from 0–10 cm, 10–25 cm and 25–40 cm depth intervals with an auger (1.7 cm diameter), transported at 4 °C under an Ar-headspace for anoxic microcosms or immediately frozen in the field with liquid N2 and stored at −80 °C until RNA extraction.
Geochemical analysis
Pore-water redox potential, pH, and CH4 and acetate concentrations were measured as described previously (Reiche et al., 2008). TOC and its stable carbon isotopic composition (δ13C) was analyzed by continuous-flow elemental analyzer–isotope ratio mass spectrometer (EA-IRMS) as described previously (Heuer et al., 2010, Hädrich et al., 2012). Pore-water acetate δ13C was analyzed by isotope-ratio-monitoring liquid chromatography/mass spectrometry as described by Heuer et al. (2006, 2009). Isotope-ratio-monitoring gas chromatography/mass spectrometry was used to determine the δ13C of pore-water CH4 as described previously (Ertefai et al., 2010; Hädrich et al., 2012). All isotope ratios are given in δ13C notation (per mil, ‰) relative to the VPDB, with δ13C=[(Rsample – RVPDB) / RVPDB] × 103, with R=13C/12C and RVPDB=0.0112372±2.9 × 10–6.
Acetogenic and methanogenic potentials
Anoxic microcosms (April 2011) were constructed by adding 10 g (wet weight (ww)) replicate soil material (under Ar) to sterile 150-ml incubation flasks (Mueller and Krempel, St-Prex, Switzerland). After sealing with butyl rubber stoppers 50 ml anoxic and sterile basal mineral medium (Küsel and Drake, 1994), devoid of any additional electron acceptors, was added. The gas phase was flushed with sterile CO2 (mofette soil) or N2 (reference soil). H2 was added to half of the bottles for a final headspace concentration of 80:20 (v/v) H2:CO2 and incubated statically in the dark at 15 °C.
Nucleic acid extraction, 16S rRNA pyrosequencing and quantitative PCR
RNA was extracted from 0.5 to 3 g (ww) soil with the RNA PowerSoil Total RNA Isolation kit in combination with the DNA Elution Accessory kit (MO BIO Laboratories, Carlsbad, CA, USA) according to the manufacturer's instructions. Reverse transcription was performed as described by Herrmann et al. (2012). Aliquots of resulting cDNA were shipped on ice to Research and Testing (Lubbock, TX, USA) for tag-encoded FLX Titanium amplicon pyrosequencing with 16S rRNA primers B28F and B519R (V1-V3 region; Loy et al., 2002; Callaway et al., 2009) for Bacteria, and A341-F and A958-R for Archaea (V3-V5 region; DeLong 1992; Watanabe et al., 2002). Pyrosequencing data were denoised and analyzed in MOTHUR v1.25.1 (Schloss et al., 2009). Denoising included removal of sequences with primer mismatches or <250 bp, and screening for quality scores<30 (with MOTHUR commands qwindowaverage=30 and qwindowsize=100). According to the Schloss SOP (http://www.mothur.org/wiki/Schloss_SOP), sequences were trimmed to a length of 250 bp, screened for chimeras (Uchime) and grouped into OTUs. The alignment was checked with MOTHUR and ARB (Ludwig et al., 2004) to remove any plastid and non-16S rRNA bacterial or archaeal reads. To ensure a better comparison of samples differing in the number of sequence reads, read numbers of each sample were normalized to the size of the smallest data set (cf. Tables 1 and 2; Supplementary Tables S3 and S4) by using the subsampling function implemented in MOTHUR. Statistical analyses and taxonomic classification against the SILVA database were also performed in MOTHUR. The pyrosequencing data generated in this study were deposited in the EMBL-EBI Sequence Read Archive [PRJEB6160].
Table 1. Bacterial 16S rRNA pyrosequencing parameters, statistical estimators and gene abundance in different depths of the mofette and reference soils in June 2011.
| Sample | No. of sequencesa | No. of OTUsb | Coverage (%) | Inverse Simpson's (1/D)c | Chao 1 | Shannon-Wienerc | Bacterial 16S rRNA gene copies (g (dw)−1)d |
|---|---|---|---|---|---|---|---|
| Mofette soil | |||||||
| 0–10 cm | 1098 (3747) | 82 | 97 | 2.7 (2.5; 3.0) | 126 (105; 173) | 2.1 (2.0; 2.2) | 7.4±0.9 × 109 |
| 10–25 cm | 1098 (2200) | 124 | 95 | 5.2 (4.7; 6.0) | 154 (140; 182) | 3.0 (2.8; 3.1) | 4.9±0.1 × 109 |
| 25–40 cm | 1098 (4392) | 73 | 97 | 2.5 (2.3; 2.7) | 149 (113; 226) | 2.0 (1.8; 2.1) | 3.8±0.6 × 109 |
| Reference soil | |||||||
| 0–10 cm | 1098 (1098) | 177 | 95 | 19.8 (17.4; 23.0) | 200 (188; 223) | 4.0 (3.9; 4.1) | 1.6±0.2 × 1010 |
| 10–25 cm | 1098 (1577) | 184 | 93 | 16.2 (14.3; 18.8) | 229 (206; 269) | 3.9 (3.8; 4.0) | 4.9±0.4 × 109 |
| 25–40 cm | 1098 (2481) | 213 | 92 | 25.2 (22.0; 29.6) | 288 (258; 340) | 4.3 (4.2; 4.4) | 5.4±0.3 × 108 |
Number of denoised sequences before normalization is given in brackets.
97% sequence similarity cutoff.
Higher numbers indicate more diversity; numbers in parentheses are 95% confidence intervals.
Mean±s.d.
Table 2. Archaeal 16S rRNA pyrosequencing parameters, statistical estimators and gene abundance in different depths of the mofette and reference soils in June 2011.
| Sample | No. of sequencesa | No. of OTUsb | Coverage (%) | Inverse Simpson's (1/D)c | Chao 1 | Shannon-Wienerc | Archaeal 16S rRNA gene copies (g (dw)−1)d |
|---|---|---|---|---|---|---|---|
| Mofette soil | |||||||
| 0–10 cm | 545 (545) | 38 | 98 | 4.3 (3.9; 4.8) | 45 (40; 62) | 2.0 (1.9; 2.1) | 7.7±0.5 × 108 |
| 10–25 cm | 545 (3624) | 22 | 98 | 1.9 (1.8; 2.1) | 29 (24; 54) | 1.2 (1.1; 1.3) | 5.2±0.5 × 108 |
| 25–40 cm | 545 (4353) | 23 | 98 | 2.7 (2.4; 3.0) | 32 (25; 60) | 1.4 (1.3; 1.5) | 4.3±0.3 × 108 |
| Reference soil | |||||||
| 0–10 cm | 545 (2089) | 66 | 93 | 4.2 (3.7; 4.9) | 121 (91; 187) | 2.2 (2.1; 2.4) | 3.1±0.2 × 108 |
| 10–25 cm | 545 (676) | 70 | 94 | 8.4 (7.5; 9.6) | 93 (80; 126) | 2.8 (2.7; 2.9) | 2.7±0.1 × 108 |
| 25–40 cm | 545 (1434) | 67 | 94 | 10.4 (9.3; 11.9) | 98 (80; 141) | 2.9 (2.8; 3.0) | 7.6±0.4 × 106 |
Number of denoised sequences before normalization is given in brackets.
97% sequence similarity cutoff.
Higher numbers indicate more diversity; numbers in parentheses are 95% confidence intervals.
Mean±s.d.
Copy numbers of archaeal and bacterial 16S rRNA genes in DNA extracts co-eluted during the RNA extraction were determined by quantitative PCR. Quantitative PCR was performed on a Mx3000P instrument (Agilent, Santa Clara, CA, USA) by using the primer combinations A-806F and A-958R (V5 region; DeLong 1992; Takai and Horikoshi, 2000) and B-28F and B-338R (V1-V2 region; Daims et al., 1999; Loy et al., 2002), as described previously (Herrmann et al. 2012).
13C-CO2 labeling experiment
Microcosm incubations
13C-CO2 incubations (November 2011) were constructed by adding 8 g (ww) mofette soil from 0 to 10 cm or 25 to 40 cm depth to 120 ml serum bottles. The headspace was flushed three times with sterile N2 (100%) to avoid dilution of 13CO2. The headspace was then set to either 100% 13C-CO2 (13CO2 treatment; >99 atomic percent 13C; Sigma-Aldrich, St Louis, MO, USA) or 100% natural abundance CO2 (unlabeled control treatment; Linde Gas, Pullach, Germany). This procedure was repeated once a week. An extensive labeling was chosen, as CO2 is not considered to be a microbial energy source and low overall incorporation was expected, similar to previous reports of dark CO2 fixation in soils (e.g., Miltner et al., 2004, 2005; Yuan et al., 2012). After 0, 5, 14 and 28 days, replicate microcosms of both treatments were sampled destructively for stable carbon isotopic analysis of δ13C-TOC, lipid biomarkers, as well as for DNA-SIP.
Lipid and polar lipid derived fatty acids (PLFAs) analysis
Bacterial and archaeal lipid biomarkers were extracted from 1 to 2 g (dry weight (dw)) soil from the unlabeled and 13CO2 incubations as described in the Supplementary Information. Fatty acid methyl esters and ether-cleaved hydrocarbons were analyzed by GC–MS for identification, GC–FID for quantification and isotope-ratio-monitoring gas chromatography/mass spectrometry for stable carbon isotopic composition (Elvert et al., 2003; Lin et al., 2010). Fatty acids are presented as the total number of carbon atoms, followed by a colon and the number of double bonds. The prefixes ai and i denote methylation in anteiso- and iso-position, respectively. 10Me signifies a methyl group in C-10 position.
DNA-SIP analysis
DNA was extracted from samples of the 13CO2 incubations and unlabeled control treatment to perform SIP. We chose to analyze 0–10 cm because our incubation experiments and pore-water data suggested parallel methanogenic and acetogenic activity mainly in shallow depths, as described in Results and Discussion. DNA-SIP was chosen over RNA-SIP because DNA-labeling occurs during cell doubling and extraction yields and consistency from the organic-rich mofette soils were generally higher than those of RNA. DNA extracts from samples collected on 0, 5, 14 and 28 days were separated by using CsCl density gradient centrifugation as described previously (Date et al., 2010) with a NVT 90 rotor in a XL-70 ultracentrifuge (both Beckman Coulter, Krefeld, Germany). After centrifugation, 12 fractions of 400 μl were collected. Fraction density was determined by repeated weighing and ranged from 1.649±0.002 g ml−1 to 1.764±0.002 g ml−1 for the unlabeled control treatment and 1.669±0.004 g ml−1 to 1.770±0.002 g ml−1 for the 13CO2 treatment. After confirmation of successful gradient formation, DNA was purified by PEG precipitation as described by Neufeld et al. 2007 and resuspended in 40 μl of TE buffer prior to storage at −20 °C. Total nucleic acid concentrations were quantified by using a Nanodrop 1000 spectrophotometer (Thermo Scientific, Waltham, MA, USA). Only samples after 14 and 28 days incubation exhibited separation of labeled and unlabeled DNA. The shift towards heavier fractions for day 14 samples are presented in Supplementary Figure S1. Fractions 1–4, 5–8 and 9–12 were combined to new subfractions ‘light', ‘medium' and ‘heavy', respectively. The ‘light', ‘medium' and ‘heavy' subfractions were analyzed by FLX 454 pyrosequencing of bacterial 16S rRNA genes and qPCR targeting archaeal and bacterial 16S rRNA genes, as described above.
Genes encoding the formyltetrahydrofolate synthetase (fhs, FTHFS, EC 6.3.4.3) and the methyl coenzyme M reductase (mcrA, EC 2.8.4.1), diagnostic indicators of acetogenesis and methanogenesis, respectively, were amplified from all 12 SIP fractions of the 13CO2 and unlabeled control treatments. FTHFS is also found in nonacetogens, such as methanogens and sulfur-reducing organisms. Therefore, acetogen similarity scores (HSc) were calculated to assess potential acetogen affiliations (Henderson et al., 2010). fhs was amplified by using primers FTHFS-F and FTHFS-R (Lovell and Leaphart, 2005), whereas, mcrA was amplified with primers mcrA-F and mcrA-R (Springer et al., 1995). PCR products from each fraction were separated by agarose gel electrophoresis to verify a shift towards heavier fractions (Supplementary Figures S2 and S3). fhs and mcrA gene fragments of the expected size (1.1 kb or 0.5 kb, respectively) from fractions 5 to 10 (control) from the unlabeled control treatment, as well as fractions 5 to 6 (labeled) from the 13CO2 treatment, were purified, cloned, sequenced and analyzed as described previously (Hädrich et al., 2012). A total of 31 fhs and mcrA sequences for each combined fraction were analyzed and used for phylogenetic tree construction. Phylogenetic trees were generated on the basis of neighbor-joining and parsimony methods with 1000 bootstraps, covering amino acid positions 198–423 for fhs (Lovell et al., 1990) and 75–227 of Methanocella paludicola for mcrA. Sequences were assigned to clusters based on their position in the phylogenetic tree. All clone sequences obtained in this study were deposited in the Genbank database (KF748793-KF748915).
Thermodynamic calculations
In situ standard free energies (ΔG0in situ) for acetoclastic and hydrogenotrophic methanogenesis, as well as acetogenesis were calculated as described in the Supplementary Information.
Results
Soil and pore-water geochemistry
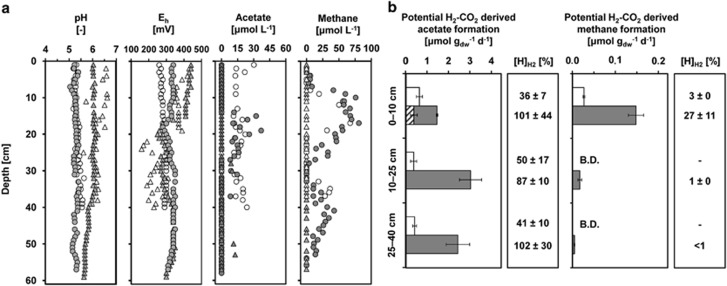
Soil and pore-water geochemistry were distinctly different between the mofette and reference site (Figure 1a; Supplementary Table S1). The mofette was highly enriched in TOC in all depths of the mofette soil with up to 32±7 w.-% C and a C/N of 24±2 compared with the reference with up to 6.3±1.2 w.-% C and a C/N of 11.6±0.5. Pore-water pH was more acidic (5.31±0.10) in the mofette compared with the reference soil (6.02±0.24). Mofette soil redox potential remained fairly stable over depth (0.32±0.02 V), suggesting microoxic to anoxic conditions throughout. In contrast, the reference soil had a distinct redox transition from oxic (0.41±0.02 V) to hypoxic conditions (0.29±0.04 V) at the 19 cm and 16 cm depths in June 2011 and August 2012, respectively. Parallel to the stable low redox potential, dissolved Fe(II) was high throughout the mofette soil (up to 604 μM; Supplementary Figure S4), whereas in the reference soil, dissolved Fe(II) was only detectable below the redox transition. Nitrate was low (<80 μM) at both sites and depleted below 5 cm depth. Sulfate decreased with depth and reached concentrations of up to 340 μM in mofette pore waters and up to 230 μM in reference pore waters. Pore-water acetate was heterogeneously distributed over the soil depth of the mofette at both sampling dates with concentrations up to 37 μM (Figure 1a). In the reference soil, acetate was not detected in pore waters except for four discrete depth levels in August. CH4 was only detected in the mofette soil pore water with two peaks in the 10 and 40 cm depth intervals. Thus, CH4 concentrations were low, but they might be flushed out by the strong CO2 upstream, which is in the range of hundreds to several thousand liters per hour (Kämpf et al., 2013).
Figure 1.
(a) Pore-water profiles of pH, Eh, acetate and CH4 concentrations in mofette (circles) and reference (triangles) soils in June 2011 (open symbols) and August 2012 (filled symbols). (b) Potential H2-CO2-derived acetate and CH4 formation in the mofette soil (filled bars) and the reference soil (empty bars) determined in April 2011 (mean±s.d., N=3). Also shown are estimated recoveries of reducing equivalents from consumed H2 ([H]H2) by H2-CO2-dependent acetogenesis (H2:acetate=4:1) and methanogenesis (H2:CH4=8:1), as well as the formations of acetate and CH4 without addition of H2 (striped bars). B.D.=below detection.
Acetogenic and methanogenic potentials
Anoxic microcosm incubations showed up to eight times higher rates of potential acetate formation from supplemental H2-CO2 in mofette compared with reference soils (Figure 1b) and it was highest in 10–25 cm depth with 3.0±0.5 μmol g (dw) soil−1 d−1. Acetate production without additional H2 was only observed in 0–10 cm of the mofette soil. The estimated recovery of reducing equivalents from H2 (H2-CO2 treatments) suggested that acetogenesis (H2:acetate=4:1) was responsible for up to 100% of H2 consumption in the mofette soil. Similarly, H2-CO2-dependent CH4 formation was generally higher in the mofette soil but only in the first 10 cm methanogenesis (H2:CH4=8:1) was a substantial part of observed H2 consumption (27%) with 0.15±0.02 μmol g (dw) soil−1 d−1 (Figure 1b). In contrast, less than 50% of reducing equivalents from H2 (H2-CO2 treatments) were recovered by methanogenesis and acetogenesis in the reference soil suggesting that other major anaerobic, H2 consuming microbial processes took place.
Structure of the active microbial community
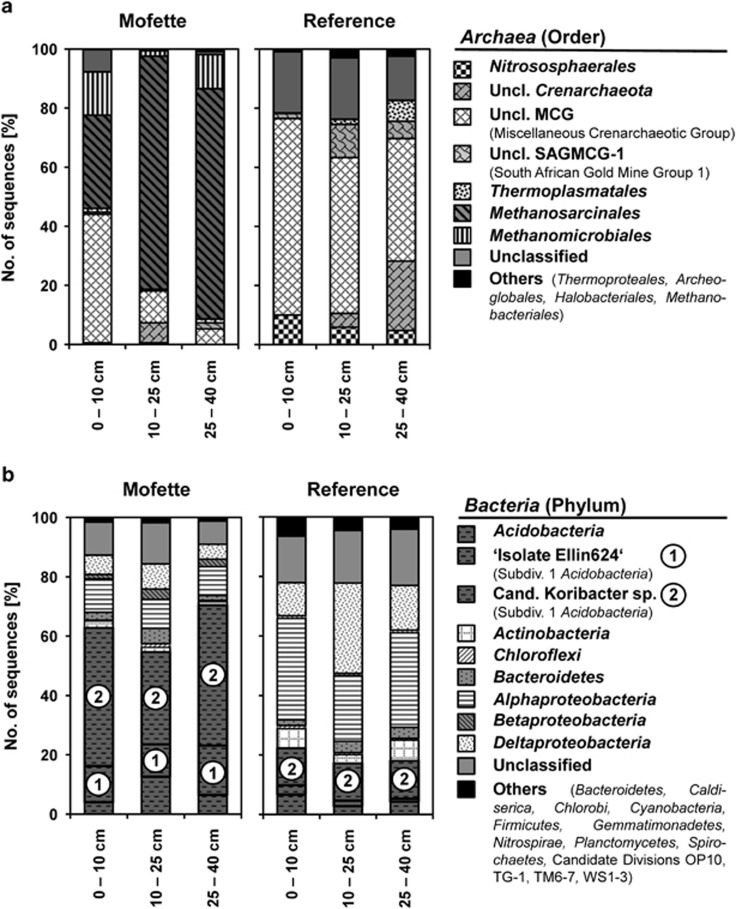
Major differences in the active archaeal community composition between the mofette and reference soils were revealed with 16S rRNA-targeted pyrosequencing (Figures 2a and b). Methanosarcinales and Methanomicrobiales were the dominant archaeal groups in all mofette depths (50–90% of total archaeal sequence reads), whereas in reference soil, methanogenic taxons represented <1% of total sequence reads (Figure 2a). Nitrososphaerales of the Thaumarchaeota were exclusively found in the reference soil where their contribution to total Archaea decreased (10–5%) with depth. Only Crenarchaeota, such as unclassified members of the Miscellaneous Crenarchaeotic Group (Inagaki et al. 2003), made up high fractions in both soils: 41–66% in the reference and 5–43% in the mofette.
Figure 2.
Structure of active (a) Archaea and (b) Bacteria communities in different depths of the mofette and reference soils sampled in June 2011. Relative abundances are based on 16S rRNA-based pyrosequencing. Patterned bars in (B) highlight the two dominant Bacteria phylotypes: Isolate strain ‘Ellin624' (1) and Cand. Koribacter sp. (2).
In contrast to the marked differences in archaeal community composition, more overlapping bacterial taxa were active in both soils. Members of the Acidobacteria (55–70%) dominated the mofette soil bacterial community (Figure 2b), with members of the Proteobacteria (17–22%) and Chloroflexi (2–5%) being also abundant. Proteobacteria (47–53%) and Acidobacteria (16–22%) were most abundant in the reference soil (Figure 2b). Surprisingly, almost all acidobacterial sequences in mofette and reference soils were related (>96% sequence identity) to Candidatus Koribacter sp. (Ward et al., 2009) or (>98% sequence identity) to isolate strain 'Ellin624' (Sait et al. 2006) of Acidobacteria Subdivision 1.
Diversity estimators revealed low bacterial and archaeal diversity in both soils (Tables 1 and 2). However, diversity estimates also suggested significantly higher prokaryotic diversity in the reference soil compared with the mofette soil. Archaeal and bacterial 16S rRNA gene copy numbers decreased with depth and were similar for both soils with 109–1010 bacterial and 108–109 archaeal 16S rRNA genes g (dw) soil −1 (Tables 1 and 2).
13CO2 incorporation into bacterial and archaeal lipids
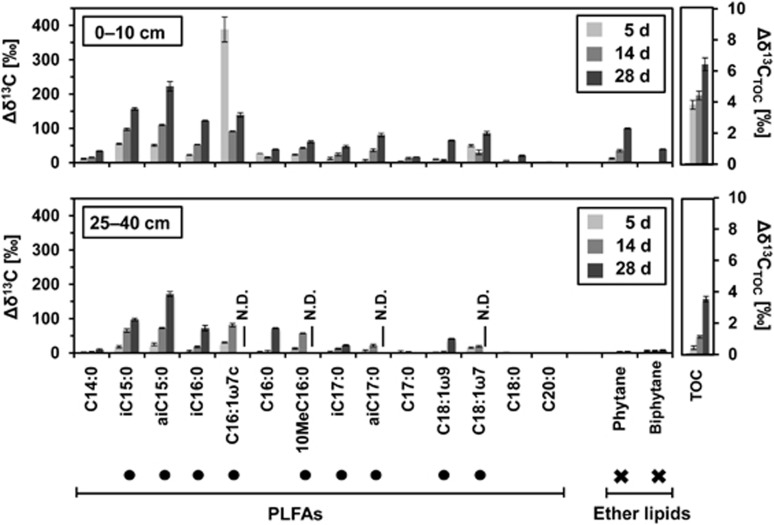
δ13C values were obtained for 14 PLFAs and 3 ether lipid-derived hydrocarbons from the 13CO2 incubations of 0, 5, 14 and 28 days. More compounds were detectable but concentrations were too low for precise δ13C analysis. Initial δ13C values of individual PLFAs averaged -28±2‰ for both depths (Supplementary Table S2), which is close to C3 plants (Clark and Fritz, 1997). The overall distribution of PLFAs was similar at both depths; concentrations were high with 453.6±40.6 μg C g (dw) soil−1 for 0–10 cm and 239.8±35.8 μg C g (dw) soil−1 in 25–40 cm.
Over 28 days of incubation with 13CO2, we observed a continuous 13C enrichment of primarily monounsaturated [C16:1ω7c, C18:1ω9, C18:1ω7] and branched [iC15:0, aiC15:0, iC16:0, 10MeC16:0, iC17:0, aiC17:0] PLFAs (Figure 3), which are considered to be bacterial in origin (Allison et al., 2007; Jin and Evans, 2010). 13C-labeling of ether lipid-derived hydrocarbons was found for archaeal lipids in 0–10 cm incubations for phytane (derived from archaeol) and biphytane (precursor caldarchaeol; Liu et al., 2011). On the basis of changes in δ13C of TOC over the incubation, total incorporations into the bulk soil were estimated to be 494±14 ng C g (dw) soil−1 d−1 in 0–10 cm and 512±10 ng C g (dw) soil−1 d−1 in 25–40 cm, respectively (Supplementary Table S2). PLFAs, which are <5% of the microbial biomass, accounted for a significant proportion of CO2 assimilation with 10.7±0.4 ng C g (dw) soil −1 d−1 in 0–10 cm and 3.9±0.3 ng C g (dw) soil−1 d−1 in 25–40 cm, respectively. Estimated incorporation of 13C into ether lipids in 0–10 cm incubations was in the same range as for single PLFAs with 0.57±0.01 ng C g (dw) soil−1 d−1 but it was negligible in 25–40 cm.
Figure 3.
Incorporation of 13CO2 into PLFAs, ether lipid-derived hydrocarbons and TOC of mofette soil microbial communities in 0–10 cm and 25–40 cm depths during a 28-day incubation. Incorporation is shown as the difference between δ13C at sampling days 5 (light grey bars), 14 (medium grey bars), 28 (dark grey bars) in the 13CO2 treatment compared with the corresponding unlabeled control treatment. Bacterial biomarkers (dots) and archaeal biomarkers (crosses) are highlighted. N.D.=not determined.
13C incorporation into the 16S rRNA, fhs and mcrA genes
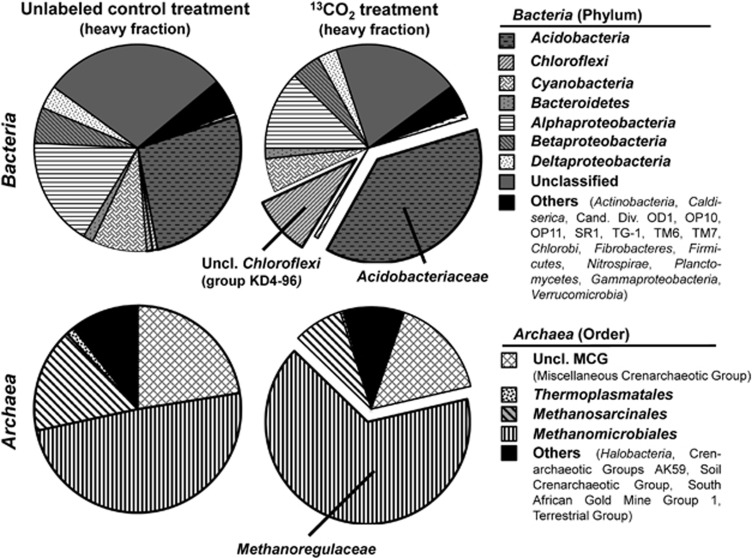
DNA-SIP on samples after 14 days of incubation with 13CO2 indicated a shift towards heavier fractions (13C-enriched) (Supplementary Figure S1). Similar to the RNA-based observations in June 2011, dominant bacterial and archaeal taxa in the SIP incubations (Figure 4; Supplementary Tables S3 and S4) were Acidobacteriaceae (Acidobacteria) and Methanoregulaceae (Methanomicrobiales), respectively. Compared with the unlabeled control treatment, we found a moderate increase in sequence abundance of 10% for Acidobacteriaceae and 9% for unclassified Chloroflexi group KD4-96 (Pruesse et al., 2007) in heavy fractions of the 13C labeled treatment (Supplementary Table S1). However, because of the dominance of Acidobacteriaceae-related 16S rRNA gene sequences, it is likely that we were not able to identify lower abundance CO2 utilizing bacteria. Methanoregulaceae-related 16S rRNA sequences showed a notable increase in medium and heavy fractions of ∼13% and ∼20%, respectively (Supplementary Table S2).
Figure 4.
Relative sequence contributions of Archaea orders and Bacteria phyla in heavy SIP fractions from the unlabeled control treatment and the 13CO2 treatment of 0–10 cm depth soil after 14 days incubation. Expanded sections represent deduced labeled taxa.
Functional genes encoding fhs and mcrA, diagnostic indicators of acetogenesis and methanogenesis, respectively, clearly shifted to heavier fractions 5–6 in the 13CO2 treatment (Supplementary Figures S2 and S3). fhs sequences were distributed among six clusters (>72% sequence similarity) and all contained sequences from the labeled fractions (Supplementary Figure S5a). The fhs clusters 1–3 branched with acetogens of 'Cluster A' (Lovel and Leaphart, 2005) and showed high HSc of 91–97%. Only sequences from cluster 3 appeared to be related to a cultured acetogen representative, Sporomusa ovata (>93% sequence similarity). The low affiliation of sequences from fhs cluster 1 and 2 with cultured acetogen representatives suggests that they originated from novel acetogens. Other labeled fhs sequences showed a maximum HSc of 79% and branched with environmental fhs homologues or with sequences from non-acetogens within the Chloroflexi and Firmicutes. Although their putative functional affiliation to acetogenesis remains unclear, the occurrence in the heavy fractions suggests direct or indirect 13C-incorporation.
mcrA sequences from 13C-labeled fractions (Supplementary Figure S5b) branched exclusively within mcrA clusters 1, 2, 4 and 5 (>90% sequence similarity) and were related to Methanoregula boonei (>92% sequence similarity) or Methanolinea mesophila (>84% sequence similarity). Sequences of mcrA cluster 2, related to Methanoregula formicica (>93% sequence similarity), and two more sequences within the Methanocellaceae and Methanosarcinaceae were only found in the density fractions containing unlabeled DNA. The higher contribution of sequences of mcrA cluster 1 in the 13C-labeled fractions (74%) compared with the unlabeled fractions (58%) was fairly consistent to that of Methanoregulaceae 16S rRNA genes and indicated a stronger labeling of associated organisms.
Stable carbon isotope geochemistry of organic metabolites in pore waters of the mofette soil
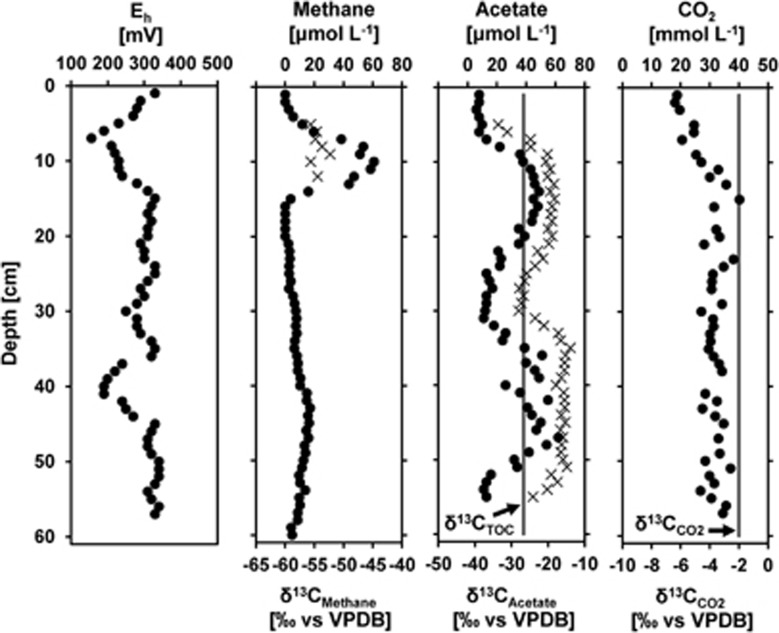
Parallel to the SIP incubations, we investigated the pore-water geochemistry in November 2011 (Figure 5; Supplementary Figure S6). Redox potentials averaged 0.28±0.05 V and had two minima at the 7 and 40 cm depth. Pore-water CH4 showed a distinct peak in ca. 5–15 cm depth with concentrations >61 μM and a fairly uniform δ13CCH4 of –54.4±1.3‰. Below 15 cm CH4 concentrations were constant, reaching up to 17 μM, but were too low for δ13C analysis. Concentrations of pore-water acetate increased with depth and were in general above the detection limit for δ13C analysis (10 μM) below 6 cm depth (Figure 5). The depth profile showed two distinct peaks of up to 50 μM at 7–25 cm and 32–52 cm, respectively. The δ13C values of acetate ranged from -33.6‰ to -13.9‰ and varied with acetate concentration over depth. Only in the shallowest samples (7–8 cm), acetate was depleted in 13C relative to TOC. Where acetate concentrations were low, δ13C values of acetate and TOC were similar whereas high acetate concentrations coincided with distinct 13C-enrichments of up to 12.8‰ relative to TOC. With –26.7±0.3‰, δ13C-TOC was uniform over depth (Figure 5). CO2 concentrations were high throughout the pore-water profile and increased from 18 mM (at 2 cm depth) to a fairly stable concentration of 32.1±3 mM (>10 cm depth), that is, close to saturation.
Figure 5.
Concentration (filled circles) and stable carbon isotopic composition (crosses; ‰ vs VPDB) of pore-water CO2, CH4 and acetate, as well as Eh from November 2011 parallel to the 13C-CO2 labeling experiment. Measured δ13CTOC and reported δ13CCO2 values (Bräuer et al., 2004; Kämpf et al., 2013) are highlighted throughout the profile as grey lines.
Discussion
Effect of upstreaming CO2 on active microbial community structure
Emanating CO2 appeared to cause alteration of two key pore-water geochemical parameters: the mofette soil was (i) more acidic than the reference soil (ΔpH=–0.73±0.24) and (ii) characterized by a low, constant redox potential over depth, as upstreaming CO2 impedes the introduction of atmospheric oxygen (Figure 1a). The high TOC contents and C/N ratios in mofette bulk soil were similar to previous observations of carbon accumulations in mofette soils and suggested restricted degradation (Blume and Felix-Henningsen, 2009; Rennert et al., 2011).
The active archaeal community contained groups that were unique to either the mofette soil or the reference soil. For example, euryarchaeotal sequences were almost exclusively found in the mofette soil where they were mainly represented by the strictly anaerobic methanogenic taxa Methanosarcinales (33–79% of the sequence reads) and Methanomicrobiales (3–15%). Only Crenarchaeota were active in high fractions in both soils and were mainly represented by unclassified members of the Miscellaneous Crenarchaeotic Group, which are thought to be involved in the anaerobic degradation of complex organic carbon compounds (Teske and Sørensen, 2008).
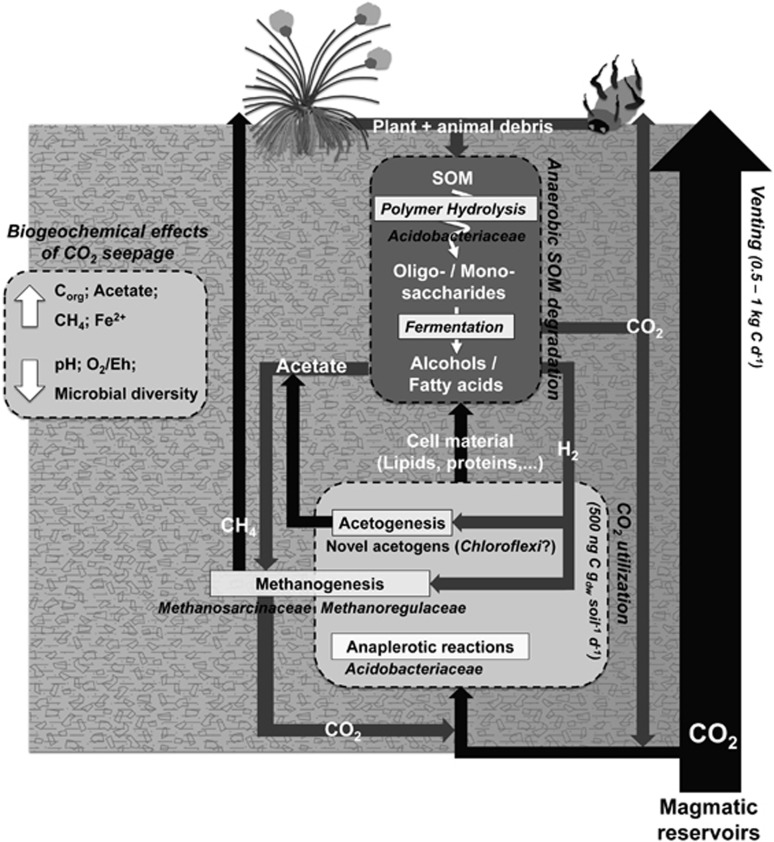
In contrast, the active bacterial community composition was similar in both soils, suggesting that these consortia are well adapted to changes in redox and pH. However, organisms related to Acidobacteria subdivision 1, isolate strain ‘Ellin624' (>98% sequence identity; Sait et al. 2006), and to Candidatus Koribacter sp. (>96% sequence identity, Ward et al., 2009), quantitatively dominated active Bacteria in the mofette soil. Members of this group are primarily considered to be heterotrophs and their abundance in soils is strongly linked to low pH (Sait et al., 2006). Incubation studies and genome sequences of subdivision 1 and 3 Acidobacteria (e.g., Candidatus Koribacter versatilis; Ward et al., 2009) suggested an important role of these organisms in the degradation of complex organic compounds such as cellulose, hemicellulose, starch and chitin. These compounds are likely a major fraction of the high carbon content in the mofette soil. Because the activity of primary degrading Eukaryotes (e.g., most fungi) is thought to be restricted under anoxic conditions (Leschine, 1995; Reith et al., 2002), the abundance, activity and metabolic capabilities of Acidobacteria might determine and consequently limit the degradation of organic matter (Figure 6).
Figure 6.
Conceptual model of carbon flow from volcanic CO2 to soil carbon in the mofette summarizing the observed effects of extreme CO2 degassing on a wetland soil. Promoted CO2 utilization and key microbial taxa are presented. Black arrows indicate pathways that are directly affected by the utilization of volcanic CO2. SOM: soil organic matter.
Bacterial groups assimilating volcanic CO2
After the 28-day incubation, all analyzed fatty acids were enriched in 13C and indicated a label distribution between taxonomically diverse groups. Given the prompt label distribution among all analyzed bacterial fatty acids after 5 days and continuous 13C enrichment over the time series of our study, this suggests that 13CO2 uptake could be attributed predominantly to primary consumers. 13CO2 assimilation rates were at least two times higher than in previous dark soil incubations (Miltner et al., 2004, 2005; Šantrůčková et al., 2005; Yuan et al., 2012). Only soil incubations under light, which promote additional phototrophic CO2 assimilation, were in the same range or higher (Yuan et al., 2012). 16S rRNA DNA-SIP analysis suggested the labeling of Acidobacteriaceae and unclassified Chloroflexi of group KD4-96 (Pruesse et al., 2007). C16:1ω7c and iC15:0, which represent the major fatty acids in subdivision 1 Acidobacteria (Foesel et al., 2013), also showed the strongest labeling in our incubations. However, the genome of Candidatus Koribacter versatilis (Acidobacteriaceae) contains no genes of enzymes to be predicted to be involved in autotrophy (Ward et al., 2009). The importance of non-obligately autotrophic, that is, anaplerotic or facultative autotrophic, CO2 fixation for bacteria in resource-limited environments, such as the ocean, was previously identified (Matin, 1978; Eiler, 2006; Hügler and Sievert, 2011). However, in soils where organic carbon is not limiting, the role of obligate autotrophy vs mixotrophic and/or heterotrophic CO2 assimilation is still unclear. Previous studies suggested a significant contribution of non-obligate autotrophic CO2 assimilation to the overall soil carbon budget (Miltner et al., 2004; Šantrůčková et al., 2005; Selesi et al., 2005, 2007), which might be as high as 3–5% of the net respiration (Miltner et al., 2005). Furthermore, addition of organic substrates to dark soil incubations stimulated CO2 fixation and correlated with respiration (Miltner et al., 2005, Šantrůčková et al., 2005). Together with our results, this suggests that, although a wide spectrum of anaerobic and aerobic soil microorganisms can assimilate CO2, intermediary metabolisms of soil heterotrophs, for example, via anaplerotic reactions of the TCA cycle strongly determine CO2 utilization in the mofette (Figure 6).
Acetogens and methanogens assimilating volcanic CO2
fhs and mcrA DNA-SIP of the 13CO2 incubations suggested that novel acetogens and Methanoregulaceae were primarily responsible for chemolithoautotrophic processes (Figure 6). DNA-SIP for 16S rRNA genes confirmed the labeling of Methanoregulaceae and suggested the labeling of unclassified Chloroflexi in group KD4-96 (Pruesse et al., 2007). To date, members of the Chloroflexi are not reported to grow acetogenically. However, genome sequencing (e.g., Wasmund et al., 2014) and metagenomic approaches (e.g., Chan et al., 2013; Hug et al., 2013) suggest that the catalytic potential to utilize CO2 via the acetyl-CoA pathway might be widespread among members of the Chloroflexi. We speculate that labeled unclassified Chloroflexi are involved in acetogenesis in the mofette soil and belong to one of the identified fhs cluster. SIP analysis of deeper depth incubations with higher acetogenic potential or comparative incubations with additional H2 might reveal the identity of key acetogens.
Consistent with the 13C incorporation into the mcrA gene pool, phytane and biphytane presumably derived from archaeol and caldarchaeol, that is, the major core lipids in methanogens (Koga, 2011), exhibited moderate 13C-enrichment in the shallow sample. Lower 13C incorporation into archaeal lipids compared with bacterial fatty acids likely reflects the combination of lower growth rates of methanogens relative to bacteria and the dilution of 13C incorporation by the large pool of fossil recalcitrant ether lipids that was not removed by the lipid separation protocol used. The hydrogenotrophic Methanoregula boonei, to which most mcrA gene sequences were related, is one of the only three acidophilic methanogens isolated to date (Bräuer et al., 2006, 2011). Methanogens and acetogens commonly have neutral pH optima as they rely on a membrane proton gradient for energy conservation (Whitman et al.,, 2001; Drake et al., 2013) and therefore must be well adapted, that is, buffered intracellularly, to survive in these environments. Their ecological importance for the flow of carbon and reductants was shown for other acidic environments (Horn et al., 2003; Cadillo-Quiroz et al., 2006; Drake et al., 2009; Hunger et al., 2011), and their adaption seems to be a competitive trait over common substrate competitors, such as H2-utilizing sulfate reducers.
We estimated the relative contribution of hydrogenotrophic and acetoclastic methanogenesis according to Conrad (2005) by using the δ13C values of CH4, CO2 and acetate, and isotopic fractionations of –71.8±8.8‰ (ɛCH4/CO2; for hydrogenotrophic methanogenesis in Methanoregulaceae dominated sediment (Liu et al., 2013) and –21‰ (ɛCH4/Acetate; for acetoclastic methanogenesis by a pure culture of Methanosarcinaceae (Gelwicks et al., 1994). The contribution of hydrogenotrophic methanogenesis increased in the mofette soil from 5% in 5 cm to 44% in 10 cm depth. Moreover, thermodynamic calculations suggested that H2 concentrations of only 0.1–0.2 nM were required for hydrogenotrophic methanogenesis with a free energy yield of at least –10 kJ mol−1. By contrast, acetoclastic methanogenesis is a thermodynamically feasible reaction throughout the sampled depth interval with a free energy yield of at least –44.9±1.9 kJ mol−1.
Assuming an average isotopic fractionation (i.e., ɛacetate/CO2) of –57‰ (Gelwicks et al., 1989; Heuer et al., 2010; Blaser et al., 2013), we would expect acetogenesis to produce acetate with δ13C-values around –59‰ in the mofette soil, where δ13CCO2 is on average –1.95±0.06‰ (Bräuer et al., 2004). However, the observed δ13C of pore-water acetate ranged from –33.6‰ to –13.9‰. A slight 13C-depletion of acetate relative to TOC was only observed at 7–8 cm depth, though thermodynamic calculations suggested that acetogenesis would yield sufficient free energy (–10 kJ mol−1) in the presence of a low threshold H2 concentration of only 7–10 nM. Previous gas measurements in this region detected H2 concentrations in the emanating gas stream of ∼1 to 30 ppm (Weinlich et al., 1998, 2003). Because of the constant flushing by the ebullition of CO2 gas and the associated stripping of any minor components H2 accumulation outside of microenvironments is unlikely. Therefore, we assume that H2 for acetogens is mainly supplied via syntrophic relationships from organisms fermenting the highly enriched organic matter in the mofette (Figure 6). Considering, that CO2 readily diffuses across the cell membrane (Lodish et al., 1995), elevated CO2 concentrations might increase the energy yield of hydrogenotrophic acetogenesis and methanogenesis (Supplementary Figure S7) and therefore significantly lower the threshold H2 concentration. The coincidence of high acetate concentrations and high δ13Cacetate is surprising, because no process is known that corresponds to 13C-enrichment during acetate formation. Instead, to date 13C-enrichment of the pore-water acetate pool is only known to result from preferential consumption of isotopically light acetate during acetoclastic methanogenesis. If the 13C-enrichment of the pore-water mofette acetate pool results from acetoclastic methanogenesis, the acetate concentrations were likely decreasing over time particularly in the depths with the highest acetate concentrations (14–21 cm and 38–54 cm). This finding and the ample free energy yield of acetoclastic methanogenesis of at least –44.9±1.9 kJ mol−1 points to a decoupling of acetate production and consumption in distinct intervals of the mofette soil via excess acetate production. This decoupling would contribute to the inhibition of anaerobic matter decomposition.
Conclusion
Extreme CO2 degassing significantly affected mofette soil chemistry (lower pH and redox potential, as well as increased carbon content) and resulted in lowered microbial diversity by favoring acidotolerant microorganisms, such as subdivision 1 Acidobacteria, and anaerobic methanogens and acetogens (Figure 6). The promotion of these organisms might be linked to substantial changes in carbon cycling that led to the accumulation of carbon in mofette soils. We observed high CO2 fixation rates, relative to previous studies, but these rates were negligible relative to the strong CO2 emanations. Our data suggest that the diversity of microbial communities at other sites with high CO2 seepages may be affected, thereby altering carbon cycling through inhibition of organic carbon decomposition and promotion of CO2-fixation.
Acknowledgments
We thank Xavier Prieto Mollar for assistance during lipid and stable isotope analysis; Jenny Wendt, Heike Geilmann and Martin Nowak for assistance with TOC and δ13C-TOC analysis; Alexander Schulze, Carsten Simon and Carolin Neubert for help during sampling; and Steffen Kolb and Anke Hädrich for helpful discussions and comments on the manuscript. This work was funded by the Deutsche Forschungsgemeinschaft through grant KU1367/10-1 and through the Gottfried Wilhelm Leibniz Prize to Kai-Uwe Hinrichs.
The research presented in this manuscript was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. Any use of trade, product, or firm names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Allison VJ, Condron LM, Peltzer DA, Richardson SJ, Turner BL. Changes in enzyme activities and soil microbial community composition along carbon and nutrient gradients at the Franz Josef chronosequence, New Zealand. Soil Biol and Biochem. 2007;39:1770–1781. [Google Scholar]
- Bankwitz P, Schneider G, Kämpf H, Bankwitz E. Structural characteristics of epicentral areas in Central Europe: study case Cheb Basin (Czech Republic) J Geodyn. 2003;35:5–32. [Google Scholar]
- Blaser MB, Dreisbach LK, Conrad R. Carbon isotope fractionation of 11 acetogenic strains grown on H2 and CO2. Appl Environ Microbiol. 2013;79:1787–1794. doi: 10.1128/AEM.03203-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blume HP, Felix-Henningsen P. Reductosols: Natural soils and Technosols under reducing conditions without an aquic moisture regime. J Plant Nutr Soil Sci. 2009;172:808–820. [Google Scholar]
- Bräuer K, Kämpf H, Niedermann S, Strauch G, Weise SM. Evidence for a nitrogen flux directly derived from the European subcontinental mantle in the Western Eger Rift, Central Europe. Geochim Cosmochim Acta. 2004;68:4935–4947. [Google Scholar]
- Bräuer SL, Cadillo-Quiroz H, Yashiro E, Yavitt JB, Zinder SH. Isolation of a novel acidiphilic methanogen from an acidic peat bog. Nature. 2006;442:192–194. doi: 10.1038/nature04810. [DOI] [PubMed] [Google Scholar]
- Bräuer SL, Cadillo-Quiroz H, Ward RJ, Yavitt JB, Zinder SH. Methanoregula boonei gen. nov., sp. nov. an acidiphilic methanogen isolated from an acidic peat bog. Int J Syst Evol Microbiol. 2011;61:45–52. doi: 10.1099/ijs.0.021782-0. [DOI] [PubMed] [Google Scholar]
- Cadillo-Quiroz H, Bräuer S, Yashiro E, Sun C, Yavitt J, Zinder S. Vertical profiles of methanogenesis and methanogens in two contrasting acidic peatlands in central New York State, USA. Environ Microbiol. 2006;8:1428–1440. doi: 10.1111/j.1462-2920.2006.01036.x. [DOI] [PubMed] [Google Scholar]
- Callaway TR, Dowd SE, Wolcott RD, Sun Y, McReynolds JL, Edrington TS, et al. Evaluation of the bacterial diversity in cecal contents of laying hens fed various molting diets by using bacterial tag-encoded FLX amplicon pyrosequencing. Poult Sci. 2009;88:298–302. doi: 10.3382/ps.2008-00222. [DOI] [PubMed] [Google Scholar]
- Chan Y, Nostrand JDV, Zhou J, Pointing SB, Farrell RL. Functional ecology of an Antarctic Dry Valley. Proc Natl Acad Sci USA. 2013;110:8990–8995. doi: 10.1073/pnas.1300643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark ID, Fritz P. Environmental isotopes in hydrogeology. CRC Press/Lewis Publishers: Boca Raton, USA; 1997. [Google Scholar]
- Conrad R. Quantification of methanogenic pathways using stable carbon isotopic signatures: a review and a proposal. Org Geochem. 2005;36:739–752. [Google Scholar]
- Daims H, Brühl A, Amann R, Schleifer KH, Wagner M. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst Appl Microbiol. 1999;22:434–444. doi: 10.1016/S0723-2020(99)80053-8. [DOI] [PubMed] [Google Scholar]
- Date Y, Nakanishi Y, Fukuda S, Kato T, Tsuneda S, Ohno H, et al. New monitoring approach for metabolic dynamics in microbial ecosystems using stable-isotope-labeling technologies. J Biosci Bioeng. 2010;110:87–93. doi: 10.1016/j.jbiosc.2010.01.004. [DOI] [PubMed] [Google Scholar]
- DeLong EF. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake HL, Horn MA, Wüst PK. Intermediary ecosystem metabolism as a main driver of methanogenesis in acidic wetland soil. Environ Microbiol Reports. 2009;1:307–318. doi: 10.1111/j.1758-2229.2009.00050.x. [DOI] [PubMed] [Google Scholar]
- Drake L, Küsel K, Matthies C.2013Acetogenic prokaryotesIn: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, (eds)The Prokaryotes: Prokaryotic Physiology and Biochemistry4th edn.Springer: Berlin Heidelberg; 3–60. [Google Scholar]
- Eiler A. Evidence for the ubiquity of mixotrophic bacteria in the upper ocean: Implications and consequences. Appl Environ Microbiol. 2006;72:7431–7437. doi: 10.1128/AEM.01559-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvert M, Boetius A, Knittel K, Jørgensen BB. Characterization of specific membrane fatty acids as chemotaxonomic markers for sulfate-reducing bacteria involved in anaerobic oxidation of methane. Geomicrobiol J. 2003;20:403–419. [Google Scholar]
- Ertefai TF, Heuer VB, Prieto-Mollar X, Vogt C, Sylva SP, Seewald J, et al. The biogeochemistry of sorbed methane in marine sediments. Geochim Cosmochim Acta. 2010;74:6033–6048. [Google Scholar]
- Flechsig C, Bussert R, Rechner J, Schütze C, Kämpf H. The Hartousov mofette soil field in the Cheb Basin, Western Eger Rift (Czech Republic): a comperative geoelectric, sedimentologic and soil gas study of a magmatic diffuse CO2-degassing structure. Z Geol Wiss. 2008;36:177–193. [Google Scholar]
- Foesel BU, Rohde M, Overmann J. Blastocatella fastidiosa gen. nov., sp. nov., isolated from semiarid savanna soil – The first described species of Acidobacteria subdivision 4. Syst Appl Microbiol. 2013;36:82–89. doi: 10.1016/j.syapm.2012.11.002. [DOI] [PubMed] [Google Scholar]
- Frerichs J, Oppermann BI, Gwosdz S, Möller I, Herrmann M, Krüger M. Microbial community changes at a terrestrial volcanic CO2 vent induced by soil acidification and anaerobic microhabitats within the soil column. FEMS Microbiol Ecol. 2012;84:60–74. doi: 10.1111/1574-6941.12040. [DOI] [PubMed] [Google Scholar]
- Gelwicks JT, Risatti JB, Hayes JM. Carbon isotope effects associated with autotrophic acetogenesis. Org Geochem. 1989;14:441–446. doi: 10.1016/0146-6380(89)90009-0. [DOI] [PubMed] [Google Scholar]
- Gelwicks JT, Risatti JB, Hayes JM. Carbon isotope effects associated with aceticlastic methanogenesis. Appl Environ Microbiol. 1994;60:467–472. doi: 10.1128/aem.60.2.467-472.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hädrich A, Heuer VB, Herrmann M, Hinrichs K-U, Küsel K. Origin and fate of acetate in an acidic fen. FEMS Microbiol Ecol. 2012;81:339–354. doi: 10.1111/j.1574-6941.2012.01352.x. [DOI] [PubMed] [Google Scholar]
- He Z, Xu M, Deng Y, Kang S, Kellogg L, Wu L, et al. Metagenomic analysis reveals a marked divergence in the structure of belowground microbial communities at elevated CO2. Ecol Lett. 2010;13:564–575. doi: 10.1111/j.1461-0248.2010.01453.x. [DOI] [PubMed] [Google Scholar]
- Henderson G, Naylor GE, Leahy SC, Janssen PH. Presence of novel, potentially homoacetogenic bacteria in the rumen as determined by analysis of Formyltetrahydrofolate Synthetase sequences from ruminants. Appl Environ Microbiol. 2010;76:2058–2066. doi: 10.1128/AEM.02580-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann M, Hädrich A, Küsel K. Predominance of thaumarchaeal ammonia oxidizer abundance and transcriptional activity in an acidic fen. Environ Microbiol. 2012;14:3013–3025. doi: 10.1111/j.1462-2920.2012.02882.x. [DOI] [PubMed] [Google Scholar]
- Heuer V, Elvert M, Tille S, Krummen M, Mollar XP, Hmelo LR, et al. Online δ13C analysis of volatile fatty acids in sediment/porewater systems by liquid chromatography-isotope ratio-mass spectrometry. Limnol Oceanogr: Methods. 2006;4:346–357. [Google Scholar]
- Heuer VB, Pohlman JW, Torres ME, Elvert M, Hinrichs KU. The stable carbon isotope biogeochemistry of acetate and other dissolved carbon species in deep subseafloor sediments at the northern Cascadia Margin. Geochim Cosmochim Acta. 2009;73:3323–3336. [Google Scholar]
- Heuer VB, Krüger M, Elvert M, Hinrichs KU. Experimental studies on the stable carbon isotope biogeochemistry of acetate in lake sediments. Org Geochem. 2010;41:22–30. [Google Scholar]
- Horn MA, Matthies C, Küsel K, Schramm A, Drake HL. Hydrogenotrophic methanogenesis by moderately acid-tolerant methanogens of a methane-emitting acidic peat. Appl Environ Microbiol. 2003;69:74–83. doi: 10.1128/AEM.69.1.74-83.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug LA, Castelle CJ, Wrighton KC, Thomas BC, Sharon I, Frischkorn KR, et al. Community genomic analyses constrain the distribution of metabolic traits across the Chloroflexi phylum and indicate roles in sediment carbon cycling. Microbiome. 2013;1:22. doi: 10.1186/2049-2618-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hügler M, Sievert SM. Beyond the Calvin Cycle: Autotrophic carbon fixation in the ocean. Annu Rev Mar Sci. 2011;3:261–289. doi: 10.1146/annurev-marine-120709-142712. [DOI] [PubMed] [Google Scholar]
- Hunger S, Schmidt O, Hilgarth M, Horn MA, Kolb S, Conrad R, et al. Competing formate- and carbon dioxide-utilizing prokaryotes in an anoxic methane-emitting fen soil. Appl Environ Microbiol. 2011;77:3773–3785. doi: 10.1128/AEM.00282-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki F, Suzuki M, Takai K, Oida H, Sakamoto T, Aoki K, et al. Microbial communities associated with geological horizons in coastal subseafloor sediments from the sea of Okhotsk. Appl Environ Microbiol. 2003;69:7224–7235. doi: 10.1128/AEM.69.12.7224-7235.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin VL, Evans RD. Microbial 13C utilization patterns via stable isotope probing of phospholipid biomarkers in Mojave Desert soils exposed to ambient and elevated atmospheric CO2. Global Change Biol. 2010;16:2334–2344. [Google Scholar]
- Kämpf H, Geissler WH, Bräuer K.2007Combined gas-geochemical and receiver function studies of the Vogtland/NW Bohemia intraplate mantle degassing field, Central EuropeIn: Ritter JRR, Christensen UR, (eds)Mantle Plumes Springer: Berlin Heidelberg; 127–158. [Google Scholar]
- Kämpf H, Bräuer K, Schumann J, Hahne K, Strauch G. CO2 discharge in an active, non-volcanic continental rift area (Czech Republic): Characterisation (δ13C, 3He/4He) and quantification of diffuse and vent CO2 emissions. Chem Geol. 2013;339:71–83. [Google Scholar]
- Koga Y. Early evolution of membrane lipids: how did the lipid divide occur. J Mol Evol. 2011;72:274–282. doi: 10.1007/s00239-011-9428-5. [DOI] [PubMed] [Google Scholar]
- Küsel K, Drake HL. Acetate synthesis in soil from a Bavarian beech forest. Appl Environ Microbiol. 1994;60:1370–1373. doi: 10.1128/aem.60.4.1370-1373.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küsel K, Dorsch T, Acker G, Stackebrandt E, Drake HL. Clostridium scatologenes strain SL1 isolated as an acetogenic bacterium from acidic sediments. Int J Syst Evol Microbiol. 2000;50:537–546. doi: 10.1099/00207713-50-2-537. [DOI] [PubMed] [Google Scholar]
- Leschine SB. Cellulose degradation in anaerobic environments. Annu Rev Microbiol. 1995;49:399–426. doi: 10.1146/annurev.mi.49.100195.002151. [DOI] [PubMed] [Google Scholar]
- Lin YS, Lipp JS, Yoshinaga M, Lin S-H, Elvert M, Hinrichs KU. Intramolecular stable carbon isotopic analysis of archaeal glycosyl tetraether lipids. Rapid Commun Mass Spectrom. 2010;24:2817–2826. doi: 10.1002/rcm.4707. [DOI] [PubMed] [Google Scholar]
- Liu XL, Lipp JS, Hinrichs KU. Distribution of intact and core GDGTs in marine sediments. Org Geochem. 2011;42:368–375. [Google Scholar]
- Liu Y, Yao T, Gleixner G, Claus P, Conrad R. Methanogenic pathways, 13C isotope fractionation, and archaeal community composition in lake sediments and wetland soils on the Tibetan Plateau. J Geophys Res Biogeosci. 2013;118:650–664. [Google Scholar]
- Lodish H, Baltimore D, Berk A, Zipursky SL, Matsudira P, Darnell J. Molecular cell biology. Palgrave Macmillan: Houndmills; 1995. Transport across cell membranes; pp. 633–668. [Google Scholar]
- Lovell CR, Przybyla A, Ljungdahl LG. Primary structure of the thermostable Formyltetrahydrofolate Synthetase from Clostridium thermoaceticum. Biochem. 1990;29:5687–5694. doi: 10.1021/bi00476a007. [DOI] [PubMed] [Google Scholar]
- Lovell CR, Leaphart AB. Community-level analysis: key genes of CO2-reductive acetogenesis. Meth Enzymol. 2005;397:454–469. doi: 10.1016/S0076-6879(05)97028-6. [DOI] [PubMed] [Google Scholar]
- Loy A, Lehner A, Lee N, Adamczyk J, Meier H, Ernst J, et al. Oligonucleotide microarray for 16S rRNA gene-based detection of all Recognized lineages of sulfate-reducing prokaryotes in the environment. Appl Environ Microbiol. 2002;68:5064–5081. doi: 10.1128/AEM.68.10.5064-5081.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matin A. Organic nutrition of chemolithotrophic bacteria. Annu Rev Microbiol. 1978;32:433–468. doi: 10.1146/annurev.mi.32.100178.002245. [DOI] [PubMed] [Google Scholar]
- Miltner A, Richnow H-H, Kopinke F-D, Kästner M. Assimilation of CO2 by soil microorganisms and transformation into soil organic matter. Org Geochem. 2004;35:1015–1024. [Google Scholar]
- Miltner A, Kopinke FD, Kindler R, Selesi D, Hartmann A, Kästner M. Non-phototrophic CO2 fixation by soil microorganisms. Plant and Soil. 2005;269:193–203. [Google Scholar]
- Neufeld JD, Vohra J, Dumont MG, Lueders T, Manefield M, Friedrich MW, et al. DNA stable-isotope probing. Nat Protocols. 2007;2:860–866. doi: 10.1038/nprot.2007.109. [DOI] [PubMed] [Google Scholar]
- Oppermann BI, Michaelis W, Blumenberg M, Frerichs J, Schulz HM, Schippers A, et al. Soil microbial community changes as a result of long-term exposure to a natural CO2 vent. Geochim Cosmochim Acta. 2010;74:2697–2716. [Google Scholar]
- Pfanz H, Vodnik D, Wittmann C, Aschan G, Raschi A.2004Plants and geothermal CO2 exhalations - Survival in and adaptation to a high CO2 environmentIn: Esser K, Lüttge U, Beyschlag W, Murata J, (eds)Progress in Botany Vol 65Springer: Berlin Heidelberg; 499–538. [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucl Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiche M, Torburg G, Küsel K. Competition of Fe(III) reduction and methanogenesis in an acidic fen. FEMS Microbiol Ecol. 2008;65:88–101. doi: 10.1111/j.1574-6941.2008.00523.x. [DOI] [PubMed] [Google Scholar]
- Reith F, Drake HL, Küsel K. Anaerobic activities of bacteria and fungi in moderately acidic conifer and deciduous leaf litter. FEMS Microbiol Ecol. 2002;41:27–35. doi: 10.1111/j.1574-6941.2002.tb00963.x. [DOI] [PubMed] [Google Scholar]
- Rennert T, Eusterhues K, Pfanz H, Totsche KU. Influence of geogenic CO2 on mineral and organic soil constituents on a mofette soil site in the NW Czech Republic. Eur J Soil Sci. 2011;62:572–580. [Google Scholar]
- Ross DJ, Tate KR, Newton PCD, Wilde RH, Clark H. Carbon and nitrogen pools and mineralization in a grassland gley soil under elevated carbon dioxide at a natural CO2 spring. Global Change Biol. 2000;6:779–790. [Google Scholar]
- Sait M, Davis KER, Janssen PH. Effect of pH on isolation and distribution of members of subdivision 1 of the phylum Acidobacteria occurring in soil. Appl Environ Microbiol. 2006;72:1852–1857. doi: 10.1128/AEM.72.3.1852-1857.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šantrůčková H, Bird MI, Elhottová D, Novák J, Picek T, Šimek M, et al. Heterotrophic fixation of CO2 in Soil. Microb Ecol. 2005;49:218–225. doi: 10.1007/s00248-004-0164-x. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selesi D, Schmid M, Hartmann A. Diversity of green-like and red-like ribulose-1,5-bisphosphate carboxylase/oxygenase large-subunit genes (cbbL) in differently managed agricultural soils. Appl Environ Microbiol. 2005;71:175–184. doi: 10.1128/AEM.71.1.175-184.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selesi D, Pattis I, Schmid M, Kandeler E, Hartmann A. Quantification of bacterial RubisCO genes in soils by cbbL targeted real-time PCR. J Microbiol Methods. 2007;69:497–503. doi: 10.1016/j.mimet.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Springer E, Sachs MS, Woese CR, Boone DR. Partial gene sequences for the A subunit of Methyl-Coenzyme M Reductase (mcrI) as a phylogenetic tool for the family Methanosarcinaceae. Int J Syst Bacteriol. 1995;45:554–559. doi: 10.1099/00207713-45-3-554. [DOI] [PubMed] [Google Scholar]
- Takai K, Horikoshi K. Rapid detection and quantification of members of the archaeal community by quantitative PCR using fluorogenic probes. Appl Environ Microbiol. 2000;66:5066–5072. doi: 10.1128/aem.66.11.5066-5072.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teske A, Sørensen KB. Uncultured archaea in deep marine subsurface sediments: have we caught them all. ISME J. 2008;2:3–18. doi: 10.1038/ismej.2007.90. [DOI] [PubMed] [Google Scholar]
- Vodnik D, Maček I, Videmšek U, Hladnik J. The life of plants under extreme CO2. Biol Acta Slov. 2007;50:31–39. [Google Scholar]
- Ward NL, Challacombe JF, Janssen PH, Henrissat B, Coutinho PM, Wu M, et al. Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl Environ Microbiol. 2009;75:2046–2056. doi: 10.1128/AEM.02294-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmund K, Schreiber L, Lloyd KG, Petersen DG, Schramm A, Stepanauskas R, et al. Genome sequencing of a single cell of the widely distributed marine subsurface Dehalococcoidia, phylum Chloroflexi. ISME J. 2014;8:383–397. doi: 10.1038/ismej.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Kodama Y, Hamamura N, Kaku N. Diversity, abundance, and activity of archaeal populations in oil-contaminated groundwater accumulated at the bottom of an underground crude oil storage cavity. Appl Environ Microbiol. 2002;68:3899–3907. doi: 10.1128/AEM.68.8.3899-3907.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinlich FH, Tesar J, Weise SM, Bräuer K, Kämpf H. Gas flux distribution in mineral springs and tectonic structure in the western Eger Rift. J Geosci. 1998;43:91–110. [Google Scholar]
- Weinlich FH, Bräuer K, Kämpf H, Strauch G, Tesař J, Weise SM. Gas flux and tectonic structure in the western Eger Rift, Karlovy Vary–Oberpfalz and Oberfranken, Bavaria. Geolines. 2003;15:181–187. [Google Scholar]
- Whitman W, Boone D, Koga Y, Keswani J. Bergey's Mannual of Systematic Bacteriology. Springer: Heidelberg; 2001. Taxonomy of methanogenic archaea; pp. 211–294. [Google Scholar]
- Yuan H, Ge T, Chen C, O'Donnell AG, Wu J. Significant role for microbial autotrophy in the sequestration of soil carbon. Appl Environ Microbiol. 2012;78:2328–2336. doi: 10.1128/AEM.06881-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.