Abstract
Microbiota provide their hosts with a range of beneficial services, including defense from external pathogens. However, host-associated microbial communities themselves can act as a source of opportunistic pathogens depending on the environment. Marine poikilotherms and their microbiota are strongly influenced by temperature, but experimental studies exploring how temperature affects the interactions between both parties are rare. To assess the effects of temperature, temperature stress and infection on diversity, composition and dynamics of the hemolymph microbiota of Pacific oysters (Crassostrea gigas), we conducted an experiment in a fully-crossed, three-factorial design, in which the temperature acclimated oysters (8 or 22 °C) were exposed to temperature stress and to experimental challenge with a virulent Vibrio sp. strain. We monitored oyster survival and repeatedly collected hemolymph of dead and alive animals to determine the microbiome composition by 16s rRNA gene amplicon pyrosequencing. We found that the microbial dynamics and composition of communities in healthy animals (including infection survivors) were significantly affected by temperature and temperature stress, but not by infection. The response was mediated by changes in the incidence and abundance of operational taxonomic units (OTUs) and accompanied by little change at higher taxonomic levels, indicating dynamic stability of the hemolymph microbiome. Dead and moribund oysters, on the contrary, displayed signs of community structure disruption, characterized by very low diversity and proliferation of few OTUs. We can therefore link short-term responses of host-associated microbial communities to abiotic and biotic factors and assess the potential feedback between microbiota dynamics and host survival during disease.
Introduction
Over the last couple of decades, it has become clear that microbiota are of vital importance for survival, homeostasis and development of animals (McFall-Ngai et al., 2013). Tight relationships between hosts and their symbionts even inspired a hologenome theory of evolution (Rosenberg et al., 2007), proposing a holobiont—a host together with the associated microorganisms—as the unit of selection. One prominent service that microbiota provide for their hosts is protection from pathogens (Kamada et al., 2013). However, in compromised hosts or under (un)favorable environmental conditions, the symbionts themselves can act as opportunistic pathogens (Garnier et al., 2007; Cerf-Bensussan and Gaboriau-Routhiau, 2010; Olson et al., 2014). As disease has a large impact on population dynamics and evolution of affected organisms (Altizer et al., 2003), it is important to understand how the environmental factors and stress affect the composition and function of microbiota and the outcome of host–microbe interactions.
Tissues of healthy marine invertebrates usually harbor species-rich microbial communities (Prieur et al., 1990; Gomez-Gil et al., 1998; Romero et al., 2002; Gomez-Gil et al., 2010; King et al., 2012; Trabal et al., 2012; Wegner et al., 2013). This also applies to the hemolymph (Olafsen et al., 1993; Garnier et al., 2007, Wendling et al., 2014)—the functional analog of blood in vertebrates (Bachere et al., 2004). The presence of viable bacteria in the hemolymph of healthy oysters can influence the outcome of pathogen infections either by stimulating immunity or by competitive exclusion (Schmitt et al., 2012); isolation of antimicrobial compounds of bacterial origin from oyster hemolymph has provided support for the latter hypothesis (Defer et al., 2013). Yet oyster hemolymph microbiota have rarely been studied and so far only by means of culture-dependent methods (Olafsen et al., 1993; Garnier et al., 2007; Wendling et al., 2014). However, neither cultivation nor molecular fingerprinting methods provide realistic estimates of community diversity and composition (Pedros-Alio, 2006; Bent and Forney, 2008). Next-generation sequencing, although by no means free of biases (Fierer and Lennon, 2011; Sergeant et al., 2012; Cai et al., 2013), enables detailed characterization of microbial community composition and dynamics, including rare phylotypes (Huse et al., 2008) that can act as a seed bank and mediate community response to environmental change (Caporaso et al., 2012; Pedros-Alio, 2012; Sjostedt et al., 2012). Recently, amplicon sequencing has been used to characterize oyster gut (King et al., 2012; Trabal Fernandez et al., 2013) and gill microbiomes (Wegner et al., 2013), resulting in higher estimates of α-diversity and challenging the long-held view that the oyster microbiota are dominated mainly by pseudomonads and vibrios (Prieur et al., 1990; Olafsen et al., 1993; Garnier et al., 2007).
Temperature is an important factor in shaping microbial communities in marine abiotic and biotic habitats (for example, Fuhrman et al., 2008). Shifts from mutualist- to pathogen-dominated communities due to global warming have already been reported (Ritchie, 2006), as well as an increase in occurrence of infectious diseases (Altizer et al., 2013). At lower latitudes, warming usually implies crossing the upper limits of thermal tolerance and is almost certain to have adverse effects on the affected organisms (Lafferty et al., 2004; Fan et al., 2013). The outcome in temperate regions can be much less predictable, depending on thermal optima and ranges of hosts and pathogens (Thomas and Blanford, 2003). For example, temperatures over 20 °C are necessary for oyster summer mortalities to occur (Samain et al., 2007; Watermann et al., 2008), but it is low temperatures (<14 °C) that promote development of brown ring disease in clams (Paillard et al., 2004).
Many temperature-dependent disease outbreaks have been linked to various Vibrio species (Lacoste et al., 2001; Lee et al., 2001; Garnier et al., 2007; Cervino et al., 2008; Elston et al., 2008) that are commonly isolated from healthy marine animals (Prieur et al., 1990; Olafsen et al., 1993; Gomez-Gil et al., 1998; Iida et al., 2000; Vega Thurber et al., 2009; Trabal et al., 2012; Zhao et al., 2012). The effects of temperature on Vibrio spp. virulence have also been demonstrated experimentally (Kushmaro et al., 1998; Wendling and Wegner, 2013), making this group a suitable candidate for experimentally exploring temperature-dependent host–pathogen interactions in the marine environment.
So far, most of the research on microbial dynamics in marine poikilotherms has been observational and focused on seasonal changes (Beleneva and Zhukova, 2009; Preheim et al., 2011; Zurel et al., 2011; Erwin et al., 2012; Carlos et al., 2013; Mahalaxmi et al., 2013). However, seasonality does not equal temperature (Farcy et al., 2007; Gilbert et al., 2012) and experimental studies addressing the temperature-dependent short-term microbial dynamics, which may be highly relevant to development of disease, are mostly confined to corals and sponges (Vega Thurber et al., 2009; Simister et al., 2012; Fan et al., 2013).
Although higher temperatures usually promote microbial growth including pathogens, acclimated eurythermic hosts may be well prepared to deal with them. In vitro experiments on oyster hemocytes revealed little change in immunocompetence over wide range of environmentally relevant temperatures (Ashton-Alcox and Ford, 1998; Gagnaire et al., 2006). Temperature stress, on the other hand, severely compromised host defenses (Malham et al., 2009), illustrating the need to examine temperature effects in broader context of animal condition and history. Mortalities observed in wild populations of marine poikilotherms are often due to complex interplay of multiple stressors, such as in the well-documented case of oyster summer mortalities, (for example, Samain et al., 2007; Wendling and Wegner, 2013) or Vibrio harveyi infection in abalones (Travers et al., 2008). Controlled experiments are therefore the only way to assess the importance and relative contributions of individual factors.
To examine how infection, temperature and temperature stress affect oyster survival and the composition and dynamics of hemolymph microbiota, we experimentally challenged Pacific oysters with a virulent Vibrio spp. strain and exposed them to different temperature treatments. We thus present experimental data describing the short-term microbial dynamics in response to abiotic and biotic stress in Pacific oysters. With the combination of the above experimental factors and a high temporal resolution of microbial community dynamics, we can now try to link changes in microbial communities to different stressors and host survival.
Materials and methods
Biological material
All oysters were collected in a Wadden Sea tidal flat in Königshafen, Germany (55° 1′ 44′′ N, 8° 26′ 3′′ E) and subsequently kept in flow-through aquaria in climate chambers set to either 22 °C (warm-acclimated) or 8 °C (cold-acclimated). To avoid temperature shock during the transfer to the laboratory, the warm-acclimated oysters were collected in late august 2010, while the cold-acclimated oysters of matching size were collected 10 days before the experiment (beginning of November 2010). Warm-acclimated oysters were fed three times a week using shellfish diet instant algae mix (Varicon Aqua Solutions Ltd., Malvern, UK).
For infection, we used the isolate Vibrio sp. D29w affiliated to Vibrio orientalis/tubiashii clade based on multilocus sequence typing sequencing (Thieltges et al., 2013), which was obtained from hemolymph of the local oyster population. This isolate was shown to induce intermediate levels of mortality upon injection in adults at ambient temperatures (Thieltges et al., 2013), but was highly virulent for larvae at elevated temperatures (Wendling et al., 2014). We cultured bacteria overnight in 8 ml of soya-peptone medium at 25 °C and shaking at 240 r.p.m. Bacterial cells were then collected by centrifugation for 10 min at 5000 r.p.m., resuspended in fresh soya-peptone medium and adjusted to the concentration of 2 × 108 cells per ml.
Experimental design and hemolymph sampling
The experiment was designed to examine the effects of temperature, temperature stress and infection in a fully crossed three-factorial design for duration of 7 days. A total of 48 oysters (24 animals from each acclimation group) were kept singly in aerated 2.5 l glass jars with no flow-through, after being randomly assigned to experimental treatments. The oysters were either left at their acclimation temperature or stressed by non-abrupt transfer (together with seawater, allowing for gradual temp. equalization) to the opposite temperature, resulting in four temperature treatments: cold-acclimated (CC), warm-acclimated (WW), cold-stressed (WC) and warm-stressed (CW). Half of the oysters in each of the four groups were injected either with Vibrio sp. D29w strain or with the pure soya-peptone medium. Injections of 500 μl (∼108 bacteria) were applied into the adductor muscle with 23 × 1/4 gauge (dm 0.60 × 30 mm) needle introduced via a notch drilled on a ventral shell side. During the experiment, the oysters were kept individually in fully-aerated 2.5 jars filled with the filtered seawater (Supplementary File 1).
To examine the composition and dynamics of hemolymph microbiota, 200 μl of hemolymph was drawn from the adductor muscle just before the experiment and on the third, fifth and seventh day of the experiment. Samples were immediately stored on ice and transferred to −80 °C as soon as possible. The oysters were checked for survival during hemolymph collection. To have a proxy for the abundance of vibrios, 4 μl of hemolymph was streaked on vibrio-selective TCBS (thiosulfate-citrate-bile-salts-sucrose) agar immediately after sampling, the plates were grown at 25 °C, and colony forming units were counted after 24 h cultivation.
DNA extraction and amplicon sequencing
DNA was extracted from 174 whole hemolymph samples (Table 1; Supplementary File 1) with the Illustra TriplePrep kit (GE Healthcare Life Sciences, Hamburg, Germany) according to the manufacturer's protocol. DNA concentration and purity were checked with a Nanodrop ND-1000 spectrometer (peqlab, Erlangen, Germany) and all samples were adjusted to equal DNA concentration (5 ng μl−1).
Table 1. Number of oysters and hemolymph samples (analyzed/collected) per treatment and sampling point.
| Treatmenta |
Sampling point (day) |
Oysters per treatment | Samples per treatment | |||
|---|---|---|---|---|---|---|
| 0 | 3 | 5 | 7 | |||
| CC_0 | 6/6 | 6/6 | 6/6 | 6/6 | 6 | 24/24 |
| CC_I | 6/6 | 6/6b | 6/6b,c | 4/4c | 6 | 22/22b,c |
| CW_0 | 6/6 | 6/6 | 6/6 | 6/6 | 6 | 24/24 |
| CW_I | 6/6 | 6/6c | 1/1 | 1/1 | 6 | 14/14c |
| WC_0 | 6/6 | 5/6 | 5/6 | 6/6 | 6 | 22/24 |
| WC_I | 6/6 | 6/6 | 5/6b | 6/6c | 6 | 23/24b,c |
| WW_0 | 6/6 | 6/6c | 3/5 | 4/5 | 6 | 19/22c |
| WW_I | 6/6 | 6/6 | 4/4b | 4/4c | 6 | 20/20b,c |
| Total | 48/48 | 47/48b,c | 36/40b,c | 37/38c | 48 | 168/174b,c |
Abbreviations: CC, cold acclimated; CW, warm stressed; WC, cold-stressed; WW, warm-acclimated.
First letter denotes acclimation temperature, Second experimental temperature, followed by infection status.
Moribund oysters included. Total moribund samples: 5/6.
Dead oysters included. Total dead samples: 14/14.
Ribosomal 16S rDNA V1-V2 region was amplified with a barcoded universal bacterial 27F (CTATGCGCCTTGCCAGCCCGCTCAG-MID-TCAGAGTTTGATCMTGGCTCAG)-338R (CGTATCGCCTCCCTCGCGCCATCAG-MID-TGCTGCCTCCCGTAGGAGT) primer pair. Forward primers were marked with barcodes MID02 and MID03 and the reverse primers with MID01-MID98 (excluding MID09 and MID12). Each individual PCR including negative controls was coded by a unique combination of forward and reverse MIDs (Binladen et al., 2007; Wegner, 2009). In all, 20-μl PCRs contained 1.5 mM MgCl2, 0.2 mM of each nucleotide, 0.2 μm of each primer, 0.5 unit of Taq polymerase (GoTaq Flexi DNAp; Promega, Mannheim, Germany) and 20 ng of DNA template. DNA was amplified using the following protocol: 1 min initial denaturation at 94 °C, 30 cycles of 40 s at 94 °C, 30 s at 55 °C, 30 s at 72 °C and final extension 2 min at 72 °C.
Quality of PCR products was checked on the QIAxcel system using a QIAxcel DNA Screening kit (Qiagen, Hilden, Germany). Equal amounts (10 μl) of each product were then pooled together and cleaned by ethanol precipitation. Pooled samples were finally adjusted to 150 ng μl−1 and sequenced at Roche GS-FLX 454 platform at the Institute of clinical molecular biology at the Christian-Albrechts-University Kiel, Germany.
Data analysis
Processing of raw sequence data
Raw reads were demultiplexed using modified Python scripts from the cogent package (Knight et al., 2007). In short, original binary file (sff) was split into multiple sff files corresponding to individual samples allowing for only perfect matches to both barcodes and primers and translated into sfftxt-files using Mothur (Schloss et al., 2009). Quality control included denoising and chimera removal and was performed using the AmpliconNoise V1.23 pipeline (Quince et al., 2011). Only flowgrams with a minimum length of 200 bp before the first noise signal were kept for further analysis. Initial cutoff value for removing sequencing noise was 0.01 and cluster size 60. No ambiguities were allowed and maximum homopolymer length was set to 7. PCR noise removal cutoff value was 0.08 with the cluster size of 30. Chimeras were identified with Perseus and sequences with probability higher than 50% of being chimeras were excluded from further analysis. A custom perl script was then used to trim primer sequences and create an input fasta file for further analysis.
Raw demultiplexed sequence data are available at European Nucleotide Archive under the study accession number PRJEB5702 (sample accession numbers ERR457899–ERR458074).
We used the QIIME pipeline (Caporaso et al., 2010) to create operational taxonomic unit (OTU) tables and perform rarefaction, taxonomical composition assessment and phylogenetic diversity analyses. OTUs were picked with uclust (Edgar, 2010) at a 97% similarity threshold. Taxonomy was assigned with RDP classifier (Wang et al., 2007) to the genus level, with 60% confidence using the 110 Greengenes taxonomy 12_10 (McDonald et al., 2012) as a reference database. The sequences assigned to the genus Vibrio were then compared with 16S rDNA sequences obtained from cultured strains isolated from hemolymph of local oysters (courtesy of C Wendling) to identify OTU corresponding to the injected strain. We defined contaminants as OTUs with more than five reads in negative controls and removed them from further analysis. This threshold was chosen because negative control samples represented pools of several individual reactions and most sequences were represented with a single read. Sequences were aligned with mafft (Katoh et al., 2002) and a phylogenetic tree was built using fasttree (Price et al., 2010). We calculated rarefaction curves for Shannon, evenness, species richness and phylogenetic diversity (Faith, 1992) to assess the effects of sample sizes on these α-diversity metrics.
Statistical analyses
All other statistical analyses were performed in R (R Core Team, 2013). Host survival analysis was conducted using the survival package (Therneau, 2013). For bacterial communities, we analyzed relative species abundance, α- and β-diversity with the Vegan package (Oksanen et al., 2013). Differences in α-diversity patterns between treatments were analyzed using non-parametric tests and linear mixed models (nlme package; Pinheiro et al., 2013). To assess β-diversity, we calculated Bray-Curtis and weighted UniFrac distances (Hamady et al., 2010) between the samples and analyzed them by non-metric multidimensional scaling (NMDS) and the Adonis implementation of Permanova (non-parametric permutational multivariate analysis of variance; Anderson, 2001). Results are reported for Bray-Curtis distances if not explicitly stated otherwise. We also determined how abundant or dominant (⩾1% of sample) community members contributed to explaining the variation between the treatments. To statistically examine taxonomical composition and identify phylotypes associated with individual treatments and conditions, we applied indicator species approach implemented in indicspecies package (Cáceres and Legendre, 2009).
Results
Oyster survival upon infection
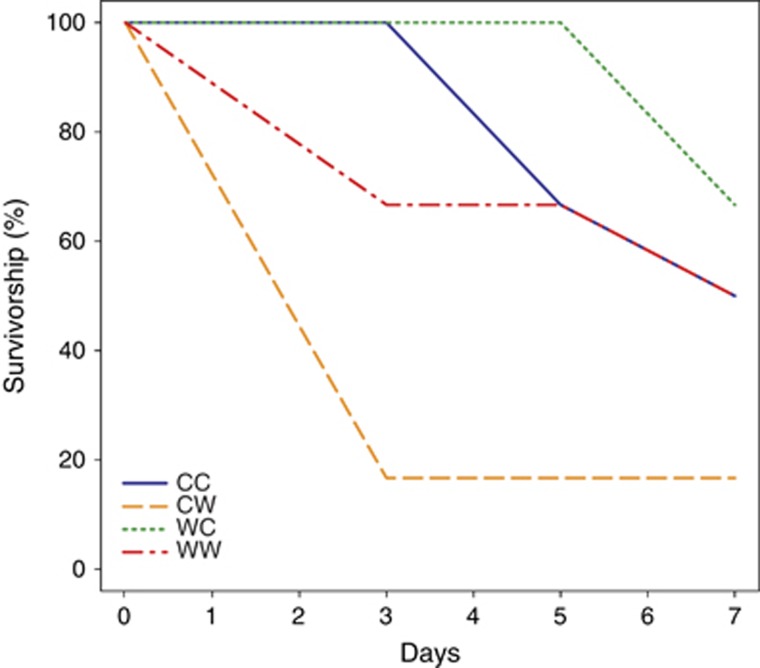
Experimental challenge with Vibrio sp. D29w resulted in 54% mortality of infected oysters, as opposed to a single death event in the control treatment (χ2(1)=14.52, P<0.001, odds=25.33 (3.10, 1195.27)). Survival analysis of infected oysters revealed significant differences between warm-stressed animals and all the other groups (Peto & Peto modification of the Gehan-Wilcoxon test, χ2(3)=9, P=0.029). Not only did the warm-stressed oysters experience the highest mortality, but they also died earlier, within the first 3 days of the experiment (Figure 1).
Figure 1.
Survival curves for infected oysters. CC, cold acclimated; CW, warm stressed; WC, cold-stressed; WW, warm-acclimated.
Hemolymph microbiome: general characteristics
Results of sequence data processing are shown in Supplementary Table 1. Singletons (OTUs represented with a single read in the data set) and the samples with fewer than 100 reads were excluded from further analyses. It is noteworthy that four out of six low-coverage libraries came from two uninfected oysters (Table 1; Supplementary File 1), suggesting that low number of reads in these cases may reflect the true absence of bacteria.
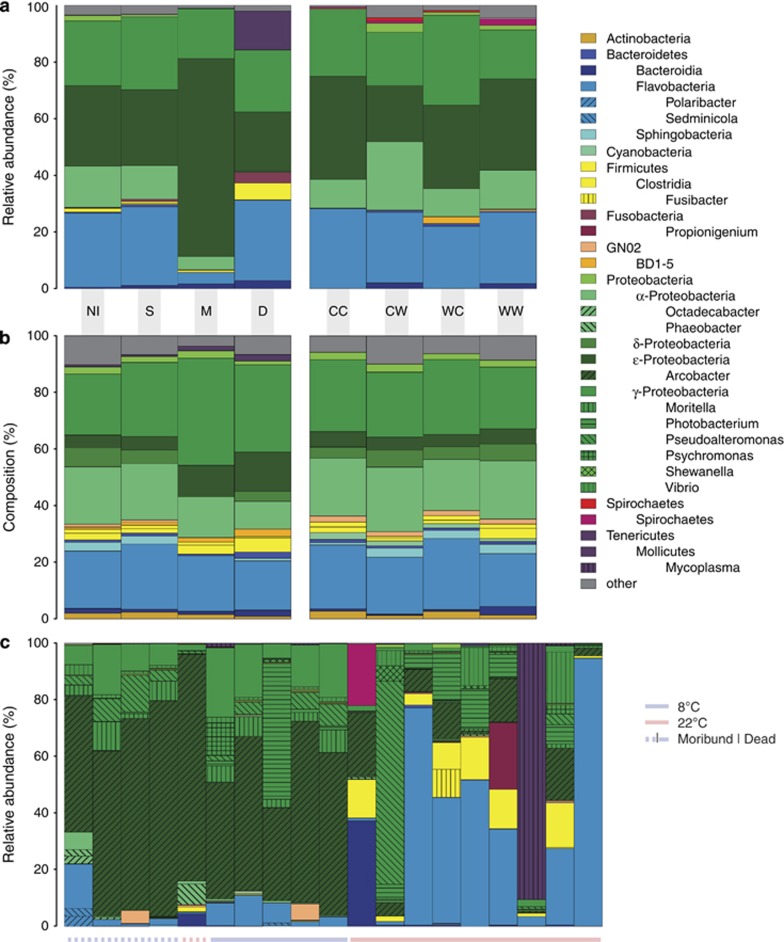
Phyla Proteobacteria and Bacteroidetes encompassed the bulk of OTU diversity and abundance (Figures 2a and b; Supplementary Figure 1). Relative OTU abundances fitted well to Fisher's log-series distribution, both in the data set as a whole and in the individual samples (Kolmogorov–Smirnoff test, D=0.000–0.196, P>0.881, details not shown), indicating that few OTUs accounted for the majority of reads. The amount of variability in community composition explained by treatments was slightly higher when only abundant OTUs (⩾1%), and not the complete data set were considered (Supplementary Figure 2).
Figure 2.
(a) Relative abundance and (b) taxonomical composition (number of OTUs per taxon) at the class level of hemolymph microbiomes (days 3–7) grouped by oyster condition (left, all oysters) and temperature treatments (right, alive oysters). (c) Relative abundance of genera in individual moribund and dead oysters. CC, cold acclimated; CW, warm stressed; WC, cold-stressed; WW, warm-acclimated; NI, non-infected; S, survivors; M, moribund; D, dead.
Rarefaction curves of α-diversity metrics show that Shannon's H and evenness—the indices we based our conclusions upon—were relatively stable even at the sampling depths of <100 reads. Furthermore, the relative differences between the treatments were fairly constant over a large range of sampling depths (Supplementary Figure 3). To assure negligible effects of sampling effort on α- and β-diversity analyses, we generated 10 random subsets of 100 reads per sample and calculated correlations between the α-diversity metrics (Spearman) and NMDS ordinations (Procrustes) based on subsets and the original data set. High correlation values indicated that our results were not influenced by differences in sampling depths of individual libraries (Supplementary Tables 2 and 3). The main analysis was thus based on the complete data set (excluding singletons) to keep the estimates of α-diversity realistic and to avoid information loss.
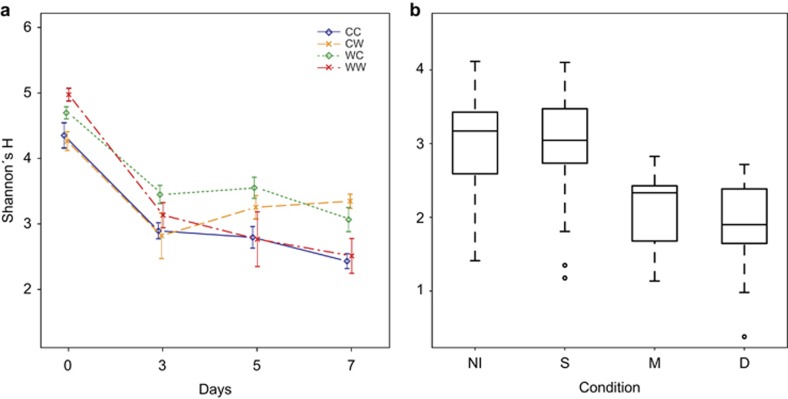
The transfer from the flow-through system to the experimental conditions was followed by significant drop in OTU diversity of alive oysters (39% OTUs were found only on day 0; Figure 3a) contributing to a clear distinction between pre-experimental and experimental communities (Supplementary Figure 4a). Loss of rare OTUs (mean relative abundance of OTUs present only on day 0 was 0.29%, quartiles: 0.14%, 0.36%) and increase in dominance were reflected in substantially lower Shannon's H (median: day 0=4.545, days 3–7=3.059, W=4931, P<0.001, r=−0.718) and evenness (median: day 0=0.899, days 3–7=0.675, W=4836, P<0.001, r=−0.720), regardless of treatment (Figure 3a; Supplementary Figure 5). Since pre-experimental communities could not be meaningfully assigned to the tested groups, they were omitted from the analyses concerning the effects of experimental treatments and oyster condition.
Figure 3.
Alpha-diversity, expressed as Shannon's H, in response to (a) temperature treatments over time and (b) to oyster condition. Infection is not shown for clarity. Only alive oysters are shown in (a); day 0 samples are omitted from (b). Error bars represent standard error of mean. CC, cold acclimated; CW, warm stressed; WC, cold-stressed; WW, warm-acclimated; NI, non-infected; S, survivors; M, moribund; D, dead.
Hemolymph microbiome: effects of infection
To describe effects of infection on hemolymph microbiota, we grouped the infected oysters into three categories: surviving (S, alive at the next sampling point), moribund (M, dead at the following sampling point) and dead (D). We refer to control and surviving oysters together as healthy (H).
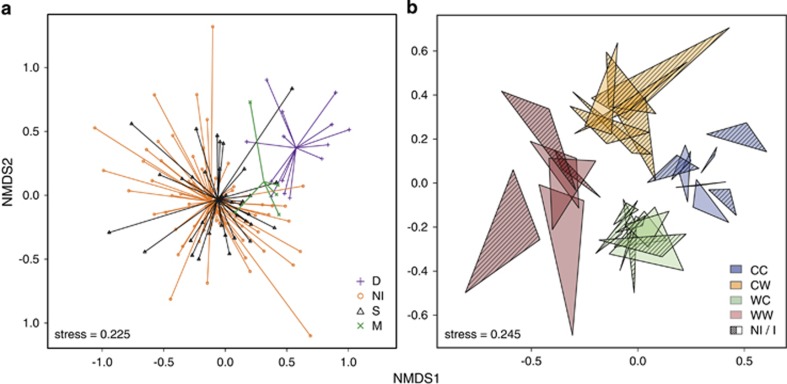
Experimental challenge barely affected α- (Table 2) and β-diversity (Table 3). Microbiomes from healthy oysters (H) formed largely overlapping clusters (Figure 4b), reflecting a comparatively small but significant effect of infection on β-diversity (Table 3). In contrast, moribund (M) and dead (D) oysters formed separate groups from healthy oysters in the NMDS plot (Figure 4a). M and D communities were characterized by proliferation of one or very few OTUs and therefore displayed very low α-diversity (Figure 3b). At 8 °C, the microbiomes of dead oysters (D) closely resembled those of moribund animals (M, Figure 2c), whereas we observed more variation in community composition and an increase in anaerobic bacteria soon after death at 22 °C.
Table 2. Linear mixed model for treatment and condition effects on Shannon's H, day 0 samples excluded (AIC=275.599, BIC=320.717, logLik (d.f.=17)=−120.800).
| Factors | d.f. | Fa | Significant contrasts | Estimate | s.e. | 2.5% CI | 97.5% CI | |
|---|---|---|---|---|---|---|---|---|
| Main effects | Acclimation temp. | 1, 43 | 1.776 | |||||
| Experimental temp. | 1, 43 | 2.547 | ||||||
| Time | 2, 62 | 0.735 | ||||||
| Health status | 2, 62 | 13.569*** | Moribund and dead vs healthy | 0.298 | 0.06 | 0.177 | 0.418 | |
| Infection | 1, 43 | 0.735 | ||||||
| Interaction terms | A. temp. × E. temp. | 1, 43 | 14.280*** | Stressed vs acclimated | −0.421 | 0.11 | −0.646 | −0.196 |
| A. temp. × Time | 2, 62 | 1.407 | ||||||
| E. temp. × Time | 2, 62 | 4.954* | E. temp. on (day 3 vs days 5–7) | 0.148 | 0.05 | 0.042 | 0.253 | |
| A. temp. × E. temp. × Time | 2, 62 | 3.361* | (Stress. vs acc.) on day 3 vs on days 5–7 | −0.178 | 0.07 | −0.321 | −0.035 | |
| Random intercept | Oyster | 0.113 | 0.005 | 2.671 |
Abbreviations: AIC, akaike information criterion; BIC, bayesian information criterion; CI, confidence interval; d.f., degrees of freedom.
Significance levels: *⩽0.05, ***<0.001.
Table 3. Adonis (Permanova) results for experimental communities (day 0 excluded) based on Bray-Curtis distances.
| d.f. | SS | MS | pseudoF | R2a,b | |
|---|---|---|---|---|---|
| Main effects | |||||
| Acclimation temp. | 1 | 3.308 | 3.308 | 12.152 | 0.076*** |
| Experimental temp. | 1 | 4.062 | 4.062 | 14.923 | 0.093*** |
| Time | 2 | 1.435 | 0.717 | 2.636 | 0.033*** |
| Infection | 1 | 0.853 | 0.853 | 3.133 | 0.020*** |
| Health status | 2 | 1.769 | 0.885 | 3.25 | 0.041*** |
| Interaction terms | |||||
| A. temp. × E. temp. | 1 | 1.154 | 1.154 | 4.239 | 0.026*** |
| A. temp. × Time | 2 | 0.729 | 0.365 | 1.34 | 0.017** |
| E. temp. × Time | 2 | 1.034 | 0.517 | 1.899 | 0.024*** |
| A. temp. × E. temp. × Time | 2 | 0.672 | 0.336 | 1.235 | 0.015** |
| Residuals | 105 | 28.579 | 0.272 | 0.656 | |
| Total | 119 | 43.594 | 1 | ||
Abbreviations: MS, mean sum of squares; SS, sum of squares.
Significance values based on 999 permutations.
Significance levels: **<0.01, ***<0.001.
Figure 4.
NMDS plots of Bray-Curtis distances between the microbial communities (day 0 excluded): (a) grouped by oyster condition and (b) showing temporal stability and effects of treatments (alive oysters only). Each triangle represents an oyster with vertices representing days 3, 5 and 7 after the onset of experiment. CC, cold acclimated; CW, warm stressed; WC, cold-stressed; WW, warm-acclimated; NI, non-infected; S, survivors; M, moribund; D, dead.
In contrast to our expectation, the microbiomes of moribund oysters were dominated by the genus Arcobacter and not, as expected, by Vibrio spp. (Figure 2c; Supplementary File 2). Overall, the infected oysters harbored more strains from other genera with described pathogenic species, such as Photobacterium and Shewanella (Supplementary File 2).
The indicator species analysis revealed that higher Vibrio sp. D29w abundances were associated with infection (IndVal=0.769, P=0.017), but not directly with mortality or disease (Supplementary File 2). Overall, the OTUs assigned to Vibrionaceae resp. genus Vibrio (including the injected strain) were common (present in 98.85%, resp. 92.26% of the samples), but not very abundant (5.1%, resp. 1.6% mean relative abundance). The presence of vibrios (other Vibrionales occasionally grow on TCBS agar, hence the taxonomically non-specific term) was lower (87.36%) when calculated from colony forming unit counts (Supplementary File 1). The discrepancy was most likely due to the low abundance coupled with the low volume of hemolymph plated. Unlike relative read abundances, higher colony forming unit counts were related to disease and death (generalized linear mixed model: Z=−2.61, P=0.009 (moribund and dead) vs healthy parameter estimate: −0.249±0.095, confidence interval (CI)=(−0.436, −0.062); Supplementary Table 4).
Hemolymph microbiome: effects of temperature and temperature stress
Before the experiment, Shannon's H (median: 8 °C=4.228, 22 °C=4.711, W=117, P<0.001, r=−0.525) and phylogenetic diversity indices (median: 8 °C=19.697, 22 °C=28.216, W=81, P<0.001, r=−0.652) were higher in warm-acclimated communities, with no difference in evenness between the groups (median: 8 °C=0.653, 22 °C=0.717, W=214, P<0.128). During the experiment, only the phylogenetic component (phylogenetic diversity) of α-diversity remained positively correlated with higher ambient temperature (Supplementary Figure 5; Supplementary Tables 5 and 6), indicating the presence of more rare, divergent phylotypes in warmer environment.
Both directions of temperature stress prevented a steady decrease in diversity and evenness that we observed in the acclimated communities (Figure 3a; Table 2; Supplementary Figure 5). The effect became more pronounced toward the end of the experiment, indicating its persistence and reflecting the gradual and lagged microbiome response to stress. We expected that the OTUs present or dominating the acclimated communities were already established in hemolymph before the experiment, thus getting a head start; and that, in contrast, the temperature stress would facilitate colonization by new species and promote growth of rare or dormant OTUs. However, we found little support for this hypothesis, as the proportion of the OTUs that were dominant in experimental communities and already present at the beginning did not significantly differ between the warm-acclimated and stressed oysters, although it was higher for cold-acclimated microbiomes (Supplementary Table 7).
Acclimation and experimental temperature, as well as their interaction, considerably affected the composition of hemolymph microbiota throughout the experiment (Table 3). Ordination by NMDS clearly separated the communities by temperature treatments (Figure 4b). Ordination and Permanova based on weighted UniFrac showed that little of phylogenetic β-diversity was attributable to the treatments (Supplementary Figures 2 and 4), reflecting similarity of the microbiome composition at above-OTU taxonomic levels. However, we found some indication for increase in potentially pathogenic genera in stressed oysters and at 22 °C in general (Supplementary File 2).
To estimate how experimental treatments affected microbial dynamics, we used the area of polygons connecting all samples from a single oyster in the NMDS plot as a proxy for temporal stability: the smaller the area, the more stable the community. Analysis of variance confirmed the visual impression (Figure 4b) that microbiomes in the cold environment were more stable compared with microbiomes from the warm environment (F1,26=17.86, P<0.001). Stress and infection, on the other hand, had no significant effect on temporal microbiome dynamics (F1,26=0.157, P=0.695, F1,26=0.89, P=0.367).
Discussion
Host–pathogen interactions have an important role in population dynamics and evolution of organisms. Although microbes inherently differ in their pathogenic potential, a disease usually arises from a complex interplay of multiple factors. Here, we show that temperature represents a notable determinant of microbial dynamics in oyster hemolymph, and propose that the lower temporal stability of microbiomes at higher temperatures may have contributed to the higher mortality of the heat-stressed hosts upon infection. We also show that a decrease in diversity and proliferation of opportunistic pathogens precede death, thus representing a good indicator of declining health.
Infection, microbiota and oyster health
Higher mortality at 22 °C can be partially attributed to faster growth of the injected strain (Supplementary Figure 6) and/or to temperature-dependent increase in expression of virulence factors (Kimes et al., 2012). The effect of absolute temperature, however, cannot account for the difference in mortality between stressed and acclimated oysters. Wang et al. (2012) reported synergic effects of heat stress and infection on scallop survival and attributed it to energetic stress. A similar mechanism may have played a role for the oyster mortality, because only heat stress, but not cold stress has been shown to increase energy consumption in Pacific oysters (Bougrier et al., 1995; Malham et al., 2009).
Injection of the virulent Vibrio sp. strain clearly caused mortality, but was also associated with an increased number of cultivable vibrios in the oyster hemolymph. In contrast, we could not directly link incidence or abundance of the injected strain in the sequenced libraries to disease (Supplementary File 2). For once, we cannot discriminate between sequences from active and inactive cells that would not cause disease (Williams et al., 2009). Moreover, Vibrio spp. can have significant influence on the host health, despite the low relative abundance (Vega Thurber et al., 2009). Still, low abundance of the injected strain suggests that other vibrios could have contributed to the mortalities, as the exogenous bacteria can be cleared quickly from the hemolymph (Parisi et al., 2008), while stimulating growth of inactive residents (Froelich and Oliver, 2013).
The abundance of Arcobacter spp. in moribund oysters also suggests a pronounced role of the indirect effects for the mortalities (Hauton et al., 1997). Arcobacter spp. is often found in association with marine organisms—ranging from bottle-nosed dolphins (Lima et al., 2012), marine seaweeds (Hollants et al., 2011), crabs (Givens et al., 2013), mussels (Collado et al., 2009), abalones (Tanaka et al., 2004) and oysters (Romero et al., 2002). The ɛ-Proteobacteria are usually rare in coastal seawater and sediments (Campbell et al., 2011; Gobet et al., 2012), and they were rare in recent studies of oyster stomach (King et al., 2012), gut (Trabal Fernandez et al., 2013) and gill microbiota (Wegner et al., 2013). Thus, the high abundance of Arcobacter spp. here could mean that these strains represent hemolymph-specific symbionts, which were not investigated in previous studies of oyster microbiota. Arcobacter spp. are often microaerophilic (Vandamme and Deley, 1991) and growth in the oysters could be facilitated by periodical valve closing (Sow et al., 2011) or high variation in respiratory time activity (Bougrier et al., 1998)—both of which may promote microaerophilic conditions. Dominance of Arcobacter spp. in moribund oysters, on the other hand, might have been a consequence of increased hypoxia due to disease-induced reduction of filtration activity (McHenery and Birkbeck, 1986). Nevertheless, the high abundance of Arcobacter spp. strains in moribund oysters, starved abalones (Tanaka et al., 2004) and necrotic sponges (Fan et al., 2013) may suggest their potential as opportunistic pathogens when occurring in high enough densities (Olson et al., 2014). Dominance of Arcobacter spp. in unhealthy animals resulted in low microbial diversity, in contrast to the diverse microbiomes of infection survivors, which were barely distinguishable from controls (Figures 3b and 4b). This might reflect the crossing of a resilience threshold that a healthy community has against disturbance (Lozupone et al., 2012). We cannot directly relate microbiome stability to disease resistance, but low diversity has repeatedly been found to coincide with impaired health in various animals (Garnier et al., 2007; Chang et al., 2008; Green and Barnes, 2010).
Microbial community dynamics in relation to infection and temperature stress
Warm temperatures can lead to higher heterogeneity in microbiome composition (Erwin et al., 2012; Boutin et al., 2013) and stress can favor shift toward more pathogen-dominated communities (Boutin et al., 2013). Thus, the lower community stability at 22 °C (Figure 4b) and temperature- or stress-related increase in potentially pathogenic bacteria (Supplementary File 2) could have contributed to the observed mortality pattern. However, microbiomes often remain stable and pathogen-free even in stressful conditions (Erwin et al., 2012; Simister et al., 2012; Pita et al., 2013; Wegner et al., 2013), indicating their potential role in host acclimation (Rosenberg et al., 2007). Phylogenetic similarity of the microbiomes suggests a common origin from the local seawater microbiota (Lozupone et al., 2007). The drop in diversity following the transfer to the non-flow-through conditions was associated with a loss of rare and probably transient OTUs, underlining the importance of immigration for the assembly of the hemolymph microbiome. However, the persistence of relatively rare, presumably resident bacteria—such as Vibrio spp. (Pruzzo et al., 2005)—contrasts the loss of transient OTUs (Romero et al., 2002) and indicates that the hemolymph microbiome is not a simple result of filter-feeding lifestyle. While the individual stability indicates the importance of host genotype for the community assembly (Wegner et al., 2013), the fine-scale differences between the treatments also illustrate acclimation potential of the hemolmyph microbiome. Multiple competing ecotypes usually coexist in bacterial populations (Cohan and Koeppel, 2008) and even the isolates with virtually identical 16s rRNA sequences are sometimes adapted to very different conditions (Cohan and Koeppel, 2008; Hall et al., 2010), We cannot determine bacterial activity (Campbell et al., 2011) nor function (Salles et al., 2012) based on 16s rDNA sequences; moreover, our coverage is insufficient to capture very rare bacteria and thus estimate the full potential for community acclimation (Sjostedt et al., 2012). Nevertheless, relatively high persistence of bacterial residents and the adjustments in fine-scale community composition following the environmental change could represent the microbiome's way to buffer the impact of environmental stress. Shifts in composition of the endogenous OTUs can thus prevent growth of external pathogens (Sjostedt et al., 2012; Froelich and Oliver, 2013) and contribute to the maintenance of homeostasis.
The microbiome of warm-acclimated oysters might have been influenced by their extended time to acclimate to laboratory conditions. This bias would mainly influence day 0 samples, where we cannot discriminate between effects of temperature or acclimation. Since we can assume that cold acclimated oysters recovered from handling stress at the start of the experiment (Thompson et al., 2012), we mainly focus on those results where oysters could uneqivocally be assigned to experimental treatments (days 3–7) for our interpretation to avoid any such bias. During those stages of the experiment, the quick shift of communities in response to experimental conditions, the strong effect fo experimental temperature (Supplementary Figures 2 and 3) and the similarity of microbiomes from surviving oysters in response to infection indicate that oyster history did not largely affect our results throughout the experiment.
Our detailed insight into short-term microbial dynamics of Pacific oyster hemolymph microbiota in response to environmental conditions and infection shows that temperature is indeed a master switch determining community structure and dynamics of oyster-associated microbiota. Microbial communities responded quickly to environmental change, but remained relatively stable within individuals for the duration of the experiment. Community disturbance by heat stress, coupled with host stress and faster bacterial growth at 22 °C may have acted in concert to cause high mortality rates. Stress can also amplify the direct immediate effects by giving rise to secondary opportunistic pathogens (for example, Arcobacter). Heat stress alone was not sufficient to cause mortality, showing that direct or indirect effects of pathogenic bacteria are necessary to induce mortality. To disentangle direct from indirect effects mediated by resident microbial communities further studies are needed for increasing the temporal resolution during the early phases of an infection to cover more bacterial communities associated with moribund oysters. The robustness of microbial communities against infection and plasticity in response to temperature suggest that the hemolymph microbiome can indeed have a vital role for host defense in changing environments.
Acknowledgments
We thank Carolin C Wendling for Vibrio sp. sequences and the sequencing group of the Institute of Clinical Molecular Biology (IKMB) at the Christian-Albrechts-University Kiel for performing 454 sequencing. The group received infrastructural funding from the DFG Clusters of Excellence Future Ocean and Inflammation at Interfaces. This study was supported by the DFG (Deutsche Forschungsgemeinschaft) Emmy Noether Programme (We4641/1-3) and International Max Planck Research School for Evolutionary Biology.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Altizer S, Harvell D, Friedle E. Rapid evolutionary dynamics and disease threats to biodiversity. Trends Ecol Evol. 2003;18:589–596. [Google Scholar]
- Altizer S, Ostfeld RS, Johnson PT, Kutz S, Harvell CD. Climate change and infectious diseases: from evidence to a predictive framework. Science. 2013;341:514–519. doi: 10.1126/science.1239401. [DOI] [PubMed] [Google Scholar]
- Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26:32–46. [Google Scholar]
- Ashton-Alcox KA, Ford SE. Variability in molluscan hemocytes: a flow cytometric study. Tissue Cell. 1998;30:195–204. doi: 10.1016/s0040-8166(98)80068-2. [DOI] [PubMed] [Google Scholar]
- Bachere E, Gueguen Y, Gonzalez M, De Lorgeril J, Garnier J, Romestand B. Insights into the anti-microbial defense of marine invertebrates: the penaeid shrimps and the oyster Crassostrea gigas. Immunol Rev. 2004;198:149–168. doi: 10.1111/j.0105-2896.2004.00115.x. [DOI] [PubMed] [Google Scholar]
- Beleneva I, Zhukova N. Seasonal dynamics of cell numbers and biodiversity of marine heterotrophic bacteria inhabiting invertebrates and water ecosystems of the Peter the Great Bay, Sea of Japan. Microbiology. 2009;78:369–375. [PubMed] [Google Scholar]
- Bent SJ, Forney LJ. The tragedy of the uncommon: understanding limitations in the analysis of microbial diversity. ISME J. 2008;2:689–695. doi: 10.1038/ismej.2008.44. [DOI] [PubMed] [Google Scholar]
- Binladen J, Gilbert MTP, Bollback JP, Panitz F, Bendixen C, Nielsen R, et al. The use of coded PCR primers enables high-throughput sequencing of multiple homolog amplification products by 454 parallel sequencing. PLoS One. 2007;2:e197. doi: 10.1371/journal.pone.0000197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bougrier S, Geairon P, Deslous-Paoli J, Bacher C, Jonquières G. Allometric relationships and effects of temperature on clearance and oxygen consumption rates of Crassostrea gigas (Thunberg) Aquaculture. 1995;134:143–154. [Google Scholar]
- Bougrier S, Collet B, Geairon P, Geffard O, Heral M, Deslous-Paoli JM. Respiratory time activity of the Japanese oyster Crassostrea gigas (Thunberg) J Exp Mar Biol Ecol. 1998;219:205–216. [Google Scholar]
- Boutin S, Bernatchez L, Audet C, Derôme N. Network analysis highlights complex interactions between pathogen, host and commensal microbiota. PLoS One. 2013;8:e84772. doi: 10.1371/journal.pone.0084772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cáceres M, Legendre P. Associations between species and groups of sites: indices and statistical inference. Ecology. 2009;90:3566–3574. doi: 10.1890/08-1823.1. [DOI] [PubMed] [Google Scholar]
- Cai L, Ye L, Tong A, Lok S, Zhang T. Biased diversity metrics revealed by bacterial 16S pyrotags derived from different primer sets. PLoS One. 2013;8:e53649. doi: 10.1371/journal.pone.0053649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell BJ, Yu L, Heidelberg JF, Kirchman DL. Activity of abundant and rare bacteria in a coastal ocean. Proc Natl Acad Sci USA. 2011;108:12776–12781. doi: 10.1073/pnas.1101405108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Paszkiewicz K, Field D, Knight R, Gilbert JA. The Western English Channel contains a persistent microbial seed bank. ISME J. 2012;6:1089–1093. doi: 10.1038/ismej.2011.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlos C, Torres TT, Ottoboni LM. Bacterial communities and species-specific associations with the mucus of Brazilian coral species. Sci Rep. 2013;3:1624. doi: 10.1038/srep01624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerf-Bensussan N, Gaboriau-Routhiau V. The immune system and the gut microbiota: friends or foes. Nat Rev Immunol. 2010;10:735–744. doi: 10.1038/nri2850. [DOI] [PubMed] [Google Scholar]
- Cervino JM, Thompson FL, Gomez-Gil B, Lorence EA, Goreau TJ, Hayes RL, et al. The Vibrio core group induces yellow band disease in Caribbean and Indo-Pacific reef-building corals. J Appl Microbiol. 2008;105:1658–1671. doi: 10.1111/j.1365-2672.2008.03871.x. [DOI] [PubMed] [Google Scholar]
- Chang JY, Antonopoulos DA, Kalra A, Tonelli A, Khalife WT, Schmidt TM, et al. Decreased diversity of the fecal Microbiome in recurrent Clostridium difficile-associated diarrhea. J Infect Dis. 2008;197:435–438. doi: 10.1086/525047. [DOI] [PubMed] [Google Scholar]
- Cohan FM, Koeppel AF. The origins of ecological diversity in prokaryotes. Curr Biol. 2008;18:R1024–R1034. doi: 10.1016/j.cub.2008.09.014. [DOI] [PubMed] [Google Scholar]
- Collado L, Cleenwerck I, Van Trappen S, De Vos P, Figueras MJ. Arcobacter mytili sp. nov., an indoxyl acetate-hydrolysis-negative bacterium isolated from mussels. Int J Syst Evol Microbiol. 2009;59:1391–1396. doi: 10.1099/ijs.0.003749-0. [DOI] [PubMed] [Google Scholar]
- Defer D, Desriac F, Henry J, Bourgougnon N, Baudy-Floc'h M, Brillet B, et al. Antimicrobial peptides in oyster hemolymph: the bacterial connection. Fish Shellfish Immunol. 2013;34:1439–1447. doi: 10.1016/j.fsi.2013.03.357. [DOI] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Elston RA, Hasegawa H, Humphrey KL, Polyak IK, Hase CC. Re-emergence of Vibrio tubiashii in bivalve shellfish aquaculture: severity, environmental drivers, geographic extent and management. Dis Aquat Organ. 2008;82:119–134. doi: 10.3354/dao01982. [DOI] [PubMed] [Google Scholar]
- Erwin PM, Pita L, Lopez-Legentil S, Turon X. Stability of sponge-associated bacteria over large seasonal shifts in temperature and irradiance. Appl Environ Microbiol. 2012;78:7358–7368. doi: 10.1128/AEM.02035-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith DP. Systematics and conservation - on predicting the feature diversity of subsets of taxa. Cladistics. 1992;8:361–373. doi: 10.1111/j.1096-0031.1992.tb00078.x. [DOI] [PubMed] [Google Scholar]
- Fan L, Liu M, Simister R, Webster NS, Thomas T. Marine microbial symbiosis heats up: the phylogenetic and functional response of a sponge holobiont to thermal stress. ISME J. 2013;7:991–1002. doi: 10.1038/ismej.2012.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farcy E, Voiseux C, Lebel JM, Fievet B. Seasonal changes in mRNA encoding for cell stress markers in the oyster Crassostrea gigas exposed to radioactive discharges in their natural environment. Sci Total Environ. 2007;374:328–341. doi: 10.1016/j.scitotenv.2006.11.014. [DOI] [PubMed] [Google Scholar]
- Fierer N, Lennon JT. The generation and maintenance of diversity in microbial communities. Am J Bot. 2011;98:439–448. doi: 10.3732/ajb.1000498. [DOI] [PubMed] [Google Scholar]
- Froelich B, Oliver J. Increases in the amounts of Vibrio spp. in oysters upon addition of exogenous bacteria. Appl Environ Microbiol. 2013;79:5208–5213. doi: 10.1128/AEM.01110-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman JA, Steele JA, Hewson I, Schwalbach MS, Brown MV, Green JL, et al. A latitudinal diversity gradient in planktonic marine bacteria. Proc Natl Acad Sci USA. 2008;105:7774–7778. doi: 10.1073/pnas.0803070105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnaire B, Frouin H, Moreau K, Thomas-Guyon H, Renault T. Effects of temperature and salinity on haemocyte activities of the Pacific oyster, Crassostrea gigas (Thunberg) Fish Shellfish Immunol. 2006;20:536–547. doi: 10.1016/j.fsi.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Garnier M, Labreuche Y, Garcia C, Robert M, Nicolas JL. Evidence for the involvement of pathogenic bacteria in summer mortalities of the Pacific oyster Crassostrea gigas. Microb Ecol. 2007;53:187–196. doi: 10.1007/s00248-006-9061-9. [DOI] [PubMed] [Google Scholar]
- Gilbert JA, Steele JA, Caporaso JG, Steinbruck L, Reeder J, Temperton B, et al. Defining seasonal marine microbial community dynamics. ISME J. 2012;6:298–308. doi: 10.1038/ismej.2011.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givens CE, Burnett KG, Burnett LE, Hollibaugh JT. Microbial communities of the carapace, gut, and hemolymph of the Atlantic blue crab, Callinectes sapidus. Mar Biol. 2013;160:2841–2851. [Google Scholar]
- Gobet A, Boer SI, Huse SM, van Beusekom JE, Quince C, Sogin ML, et al. Diversity and dynamics of rare and of resident bacterial populations in coastal sands. ISME J. 2012;6:542–553. doi: 10.1038/ismej.2011.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Gil B, Tron-Mayen L, Roque A, Turnbull J, Inglis V, Guerra-Flores A. Species of Vibrio isolated from hepatopancreas, haemolymph and digestive tract of a population of healthy juvenile Penaeus vannamei. Aquaculture. 1998;163:1–9. [Google Scholar]
- Gomez-Gil B, Roque A, Lacuesta B, Rotllant G. Diversity of vibrios in the haemolymph of the spider crab Maja brachydactyla. J Appl Microbiol. 2010;109:918–926. doi: 10.1111/j.1365-2672.2010.04718.x. [DOI] [PubMed] [Google Scholar]
- Green TJ, Barnes AC. Bacterial diversity of the digestive gland of Sydney rock oysters, Saccostrea glomerata infected with the paramyxean parasite, Marteilia sydneyi. J Appl Microbiol. 2010;109:613–622. doi: 10.1111/j.1365-2672.2010.04687.x. [DOI] [PubMed] [Google Scholar]
- Hall EK, Singer GA, Kainz MJ, Lennon JT. Evidence for a temperature acclimation mechanism in bacteria: an empirical test of a membrane-mediated trade-off. Funct Ecol. 2010;24:898–908. [Google Scholar]
- Hamady M, Lozupone C, Knight R. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 2010;4:17–27. doi: 10.1038/ismej.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauton C, Williams J, Hawkins L. The effects of a live in vivo pathogenic infection on aspects of the immunocompetence of the common shore crab, Carcinus maenas (L) J Exp Mar Biol Ecol. 1997;211:115–128. [Google Scholar]
- Hollants J, Leroux O, Leliaert F, Decleyre H, De Clerck O, Willems A. Who is in there? Exploration of endophytic bacteria within the siphonous green seaweed Bryopsis (Bryopsidales, Chlorophyta) PLoS One. 2011;6:e26458. doi: 10.1371/journal.pone.0026458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse SM, Dethlefsen L, Huber JA, Mark Welch D, Relman DA, Sogin ML. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 2008;4:e1000255. doi: 10.1371/journal.pgen.1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida Y, Honda R, Nishihara M, Muroga K. Bacterial flora in the digestive tract of cultured Pacific oyster. Fish Pathol. 2000;35:173–177. [Google Scholar]
- Kamada N, Chen GY, Inohara N, Nunez G. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol. 2013;14:685–690. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimes NE, Grim CJ, Johnson WR, Hasan NA, Tall BD, Kothary MH, et al. Temperature regulation of virulence factors in the pathogen Vibrio coralliilyticus. ISME J. 2012;6:835–846. doi: 10.1038/ismej.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King GM, Judd C, Kuske CR, Smith C. Analysis of stomach and gut microbiomes of the eastern oyster (Crassostrea virginica) from coastal Louisiana, USA. PLoS One. 2012;7:e51475. doi: 10.1371/journal.pone.0051475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight R, Maxwell P, Birmingham A, Carnes J, Caporaso JG, Easton BC, et al. PyCogent: a toolkit for making sense from sequence. Genome Biol. 2007;8:R171. doi: 10.1186/gb-2007-8-8-r171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushmaro A, Rosenberg E, Fine M, Ben Haim Y, Loya Y. Effect of temperature on bleaching of the coral Oculina patagonica by Vibrio AK-1. Mar Ecol Prog Ser. 1998;171:131–137. [Google Scholar]
- Lacoste A, Jalabert F, Malham S, Cueff A, Gelebart F, Cordevant C, et al. A Vibrio splendidus strain is associated with summer mortality of juvenile oysters Crassostrea gigas in the Bay of Morlaix (North Brittany, France) Dis Aquat Organ. 2001;46:139–145. doi: 10.3354/dao046139. [DOI] [PubMed] [Google Scholar]
- Lafferty KD, Porter JW, Ford SE. Are diseases increasing in the ocean?*. Annu Rev Ecol Evol Syst. 2004;35:31–54. [Google Scholar]
- Lee KK, Liu PC, Chen YC, Huang CY. The implication of ambient temperature with the outbreak of vibriosis in cultured small abalone Haliotis diversicolor supertexta Lischke. J Therml Biol. 2001;26:585–587. [Google Scholar]
- Lima N, Rogers T, Acevedo-Whitehouse K, Brown MV. Temporal stability and species specificity in bacteria associated with the bottlenose dolphins respiratory system. Environ Microbiol Rep. 2012;4:89–96. doi: 10.1111/j.1758-2229.2011.00306.x. [DOI] [PubMed] [Google Scholar]
- Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol. 2007;73:1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahalaxmi B, Revathy K, Raghunathan C, Anjalai K, Suabshini A. Distribution of microbial population associated with crabs from Ennore seacoast Bay of Bengal north east coast of India. Int J Curr Microbiol Appl Sci. 2013;2:290–305. [Google Scholar]
- Malham S, Cotter E, O'Keeffe S, Lynch S, Culloty S, King J, et al. Summer mortality of the Pacific oyster, Crassostrea gigas, in the Irish Sea: the influence of temperature and nutrients on health and survival. Aquaculture. 2009;287:128–138. [Google Scholar]
- McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai M, Hadfield MG, Bosch TC, Carey HV, Domazet-Loso T, Douglas AE, et al. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci USA. 2013;110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHenery JG, Birkbeck TH. Inhibition of filtration in Mytilus edulis L. by marine vibrios. J Fish Dis. 1986;9:257–261. [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara RB, et al. 2013. vegan: Community Ecology Package, R package version 2.0-7 edn.
- Olafsen JA, Mikkelsen HV, Glaever HM, Hansen GH. Indigenous bacteria in hemolymph and tissues of marine bivalves at low-temperatures. Appl Environ Microbiol. 1993;59:1848–1854. doi: 10.1128/aem.59.6.1848-1854.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson JB, Thacker RW, Gochfeld DJ. Molecular community profiling reveals impacts of time, space, and disease status on the bacterial community associated with the Caribbean sponge Aplysina cauliformis. FEMS Microbiol Ecol. 2014;87:268–279. doi: 10.1111/1574-6941.12222. [DOI] [PubMed] [Google Scholar]
- Paillard C, Allam B, Oubella R. Effect of temperature on defense parameters in manila clam Ruditapes philippinarum challenged with Vibrio tapetis. Dis Aquat Organ. 2004;59:249–262. doi: 10.3354/dao059249. [DOI] [PubMed] [Google Scholar]
- Parisi MG, Li H, Jouvet LB, Dyrynda EA, Parrinello N, Cammarata M, et al. Differential involvement of mussel hemocyte sub-populations in the clearance of bacteria. Fish Shellfish Immunol. 2008;25:834–840. doi: 10.1016/j.fsi.2008.09.005. [DOI] [PubMed] [Google Scholar]
- Pedros-Alio C. Marine microbial diversity: can it be determined. Trends Microbiol. 2006;14:257–263. doi: 10.1016/j.tim.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Pedros-Alio C. The rare bacterial biosphere. Ann Rev Mar Sci. 2012;4:449–466. doi: 10.1146/annurev-marine-120710-100948. [DOI] [PubMed] [Google Scholar]
- Pinheiro J, Bates D, DebRoy S, Sarkar D, R Development Core Team 2013. nlme: Linear and Nonlinear Mixed Effects Models, R package version 3.1-110 edn.
- Pita L, Erwin PM, Turon X, Lopez-Legentil S. Till Death Do Us Part: stable sponge-bacteria associations under thermal and food shortage stresses. PLoS One. 2013;8:e80307. doi: 10.1371/journal.pone.0080307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preheim SP, Boucher Y, Wildschutte H, David LA, Veneziano D, Alm EJ, et al. Metapopulation structure of Vibrionaceae among coastal marine invertebrates. Environ Microbiol. 2011;13:265–275. doi: 10.1111/j.1462-2920.2010.02328.x. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieur D, Nicolas JL, Plusquellec A, Vigneulle M. Interactions between bivalve mollusks and bacteria in the marine-environment. Oceanogr Mar Biol. 1990;28:277–352. [Google Scholar]
- Pruzzo C, Gallo G, Canesi L. Persistence of vibrios in marine bivalves: the role of interactions with haemolymph components. Environ Microbiol. 2005;7:761–772. doi: 10.1111/j.1462-2920.2005.00792.x. [DOI] [PubMed] [Google Scholar]
- Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ. Removing noise from pyrosequenced amplicons. BMC Bioinformatics. 2011;12:38. doi: 10.1186/1471-2105-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria; 2013. [Google Scholar]
- Ritchie KB. Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar Ecol Prog Ser. 2006;322:1–14. [Google Scholar]
- Romero J, Garcia-Varela M, Laclette JP, Espejo RT. Bacterial 16S rRNA gene analysis revealed that bacteria related to Arcobacter spp. constitute an abundant and common component of the oyster microbiota (Tiostrea chilensis) Microb Ecol. 2002;44:365–371. doi: 10.1007/s00248-002-1063-7. [DOI] [PubMed] [Google Scholar]
- Rosenberg E, Koren O, Reshef L, Efrony R, Zilber-Rosenberg I. The role of microorganisms in coral health, disease and evolution. Nat Rev Microbiol. 2007;5:355–362. doi: 10.1038/nrmicro1635. [DOI] [PubMed] [Google Scholar]
- Salles JF, Le Roux X, Poly F. Relating phylogenetic and functional diversity among denitrifiers and quantifying their capacity to predict community functioning. Front Microbiol. 2012;3:209. doi: 10.3389/fmicb.2012.00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samain JF, Degremont L, Soletchnik P, Haure J, Bedier E, Ropert M, et al. Genetically based resistance to summer mortality in the Pacific oyster (Crassostrea gigas) and its relationship with physiological, immunological characteristics and infection processes. Aquaculture. 2007;268:227–243. [Google Scholar]
- Sow M, Durrieu G, Briollais L, Ciret P, Massabuau JC. Water quality assessment by means of HFNI valvometry and high-frequency data modeling. Environ Monit Assess. 2011;182:155–170. doi: 10.1007/s10661-010-1866-9. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt P, Rosa RD, Duperthuy M, de Lorgeril J, Bachere E, Destoumieux-Garzon D. The Antimicrobial Defense of the Pacific Oyster, Crassostrea gigas. How diversity may compensate for scarcity in the regulation of resident/pathogenic microflora. Front Microbiol. 2012;3:160. doi: 10.3389/fmicb.2012.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergeant MJ, Constantinidou C, Cogan T, Penn CW, Pallen MJ. High-throughput sequencing of 16S rRNA gene amplicons: effects of extraction procedure, primer length and annealing temperature. PLoS One. 2012;7:e38094. doi: 10.1371/journal.pone.0038094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simister R, Taylor MW, Tsai P, Fan L, Bruxner TJ, Crowe ML, et al. Thermal stress responses in the bacterial biosphere of the Great Barrier Reef sponge, Rhopaloeides odorabile. Environ Microbiol. 2012;14:3232–3246. doi: 10.1111/1462-2920.12010. [DOI] [PubMed] [Google Scholar]
- Sjostedt J, Koch-Schmidt P, Pontarp M, Canback B, Tunlid A, Lundberg P, et al. Recruitment of members from the rare biosphere of marine bacterioplankton communities after an environmental disturbance. Appl Environ Microbiol. 2012;78:1361–1369. doi: 10.1128/AEM.05542-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka R, Ootsubo M, Sawabe T, Ezura Y, Tajima K. Biodiversity and in situ abundance of gut microflora of abalone (Haliotis discus hannai) determined by culture-independent techniques. Aquaculture. 2004;241:453–463. [Google Scholar]
- Therneau TM.2013. A Package for Survival Analysis in S, R package version 2.37-4 edn.
- Thieltges DW, Engelsma MY, Wendling CC, Wegner KM. Parasites in the Wadden Sea food web. J Sea Res. 2013;82:122–133. [Google Scholar]
- Thomas MB, Blanford S. Thermal biology in insect-parasite interactions. Trends Ecol Evol. 2003;18:344–350. [Google Scholar]
- Thompson EL, Taylor DA, Nair SV, Birch G, Coleman R, Raftos DA. Optimal acclimation periods for oysters in laboratory-based experiments. J Mollus Stud. 2012;78:304–307. [Google Scholar]
- Trabal N, Mazon-Suastegui JM, Vazquez-Juarez R, Asencio-Valle F, Morales-Bojorquez E, Romero J. Molecular analysis of bacterial microbiota associated with oysters (Crassostrea gigas and Crassostrea corteziensis) in different growth phases at two cultivation sites. Microb Ecol. 2012;64:555–569. doi: 10.1007/s00248-012-0039-5. [DOI] [PubMed] [Google Scholar]
- Trabal Fernandez N, Mazon-Suastegui JM, Vazquez-Juarez R, Ascencio-Valle F, Romero J. Changes in the composition and diversity of the bacterial microbiota associated with oysters (Crassostrea corteziensis, Crassostrea gigas and Crassostrea sikamea) during commercial production. FEMS Microbiol Ecol. 2013;88:69–83. doi: 10.1111/1574-6941.12270. [DOI] [PubMed] [Google Scholar]
- Travers MA, Le Goic N, Huchette S, Koken M, Paillard C. Summer immune depression associated with increased susceptibility of the European abalone, Haliotis tuberculata to Vibrio harveyi infection. Fish Shellfish Immunol. 2008;25:800–808. doi: 10.1016/j.fsi.2008.08.003. [DOI] [PubMed] [Google Scholar]
- Vandamme P, Deley J. Proposal for a New Family, Campylobacteraceae. Int J Syst Bacteriol. 1991;41:451–455. [Google Scholar]
- Vega Thurber R, Willner-Hall D, Rodriguez-Mueller B, Desnues C, Edwards RA, Angly F, et al. Metagenomic analysis of stressed coral holobionts. Environ Microbiol. 2009;11:2148–2163. doi: 10.1111/j.1462-2920.2009.01935.x. [DOI] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang L, Zhang H, Ji Q, Song L, Qiu L, et al. Immune response and energy metabolism of Chlamys farreri under Vibrio anguillarum challenge and high temperature exposure. Fish Shellfish Immunol. 2012;33:1016–1026. doi: 10.1016/j.fsi.2012.08.026. [DOI] [PubMed] [Google Scholar]
- Watermann BT, Herlyn M, Daehne B, Bergmann S, Meemken M, Kolodzey H. Pathology and mass mortality of Pacific oysters, Crassostrea gigas (Thunberg), in 2005 at the East Frisian coast, Germany. J Fish Dis. 2008;31:621–630. doi: 10.1111/j.1365-2761.2008.00953.x. [DOI] [PubMed] [Google Scholar]
- Wegner KM. Massive parallel MHC genotyping: titanium that shines. Mol Ecol. 2009;18:1818–1820. doi: 10.1111/j.1365-294X.2009.04173.x. [DOI] [PubMed] [Google Scholar]
- Wegner KM, Volkenborn N, Peter H, Eiler A. Disturbance induced decoupling between host genetics and composition of the associated microbiome. BMC Microbiol. 2013;13:252. doi: 10.1186/1471-2180-13-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendling CC, Batista FM, Wegner MK. Persistence, seasonal dynamics and pathogenic potential of Vibrio communities from Pacific oyster hemolymph. PLoS One. 2014;9:e94256. doi: 10.1371/journal.pone.0094256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendling CC, Wegner KM. Relative contribution of reproductive investment, thermal stress and Vibrio infection to summer mortality phenomena in Pacific oysters. Aquaculture. 2013;412:88–96. [Google Scholar]
- Williams HR, Macey BM, Burnett LE, Burnett KG. Differential localization and bacteriostasis of Vibrio campbellii among tissues of the Eastern oyster, Crassostrea virginica. Dev Comp Immunol. 2009;33:592–600. doi: 10.1016/j.dci.2008.10.008. [DOI] [PubMed] [Google Scholar]
- Zhao J, Shi B, Jiang QR, Ke CH. Changes in gut-associated flora and bacterial digestive enzymes during the development stages of abalone (Haliotis diversicolor) Aquaculture. 2012;338:147–153. [Google Scholar]
- Zurel D, Benayahu Y, Or A, Kovacs A, Gophna U. Composition and dynamics of the gill microbiota of an invasive Indo-Pacific oyster in the eastern Mediterranean Sea. Environ Microbiol. 2011;13:1467–1476. doi: 10.1111/j.1462-2920.2011.02448.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.