Abstract
Although uncultured microorganisms have important roles in ecosystems, their ecophysiology in situ remains elusive owing to the difficulty of obtaining live cells from their natural habitats. In this study, we employed a novel magnetic nanoparticle-mediated isolation (MMI) method to recover metabolically active cells of a group of previously uncultured phenol degraders, Burkholderiales spp., from coking plant wastewater biosludge; five other culturable phenol degraders—Rhodococcus sp., Chryseobacterium sp. and three different Pseudomonas spp.—were also isolated from the same biosludge using traditional methods. The kinetics of phenol degradation by MMI-recovered cells (MRCs) was similar to that of the original sludge. Stable isotope probing (SIP) and pyrosequencing of the 16S rRNA from the ‘heavy' DNA (13C-DNA) fractions indicated that Burkholderiales spp. were the key phenol degraders in situ in the biosludge, consistent with the results of MRCs. Single-cell Raman micro-spectroscopy was applied to probe individual bacteria in the MRCs obtained from the SIP experiment and showed that 79% of them were fully 13C-labelled. Biolog assays on the MRCs revealed the impact of various carbon and nitrogen substrates on the efficiency of phenol degradation in the wastewater treatment plant biosludge. Specifically, hydroxylamine, a metabolite of ammonia oxidisation, but not nitrite, nitrate or ammonia, inhibited phenol degradation in the biosludge. Our results provided a novel insight into the occasional abrupt failure events that occur in the wastewater treatment plant. This study demonstrated that MMI is a powerful tool to recover live and functional cells in situ from a complex microbial community to enable further characterisation of their physiology.
Introduction
Around half of the total carbon in global biomass is present in microbes that have crucial roles not only in mediating global carbon and nitrogen cycles but also in regulating our climate (Schleifer, 2004). However, the majority of microorganisms present in the environment remain uncultivated (Whitman et al., 1998; Rappe and Giovannoni, 2003), making it difficult to study their physiology. In addition, it is equally, if not more, important to study their functionalities and ecological roles in the biological context within their native microbial community.
It is a great challenge to understand the microbial physiology and in situ ecological roles of as yet uncultured bacteria. Several methods have been developed to study uncultured bacteria. Meta-approaches (for example, metagenomics, metatranscriptomics, metaproteomics and metabolomics) (Handelsman, 2004) circumvent the cultivation issue by extracting the total nucleic acids, proteins or chemicals from an environmental sample, and by directly analysing them as a whole. These meta-approaches have given an unprecedented view of the diversity and complexity of microbial communities. Stable isotope probing (SIP) links uncultured microbial cells with the metabolism of specific stable isotope (13C or 15N)-labelled substrates (Radajewski et al., 2000; Manefield et al., 2002). SIP combined with metagenomics is able to establish a connection between bacterial identity and ecological function (Chen and Murrell, 2010; Wang et al., 2012). SIP requires that stable isotopes such as 13C and 15N be incorporated into biomass (DNA, RNA or protein) and therefore has limited success in processes that have no incorporation of stable isotopes into biomass, such as nitrification, denitrification, sulphate reduction, iron reduction, methanogenesis, co-metabolism in consortia or mixed organic carbon anabolism (Bombach et al., 2010; Nelson and Carlson, 2012). Fluorescence in situ hybridisation and immunomagnetic cell capture have been used to isolate uncultured anaerobic methane-oxidising Archaea (Pernthaler et al., 2008). More recently, single-cell approaches have been developed to sort individual uncultured bacterial cells based on Raman or fluorescence signals, which can be subsequently coupled to single-cell genomic analyses (Huang et al., 2009; Rinke et al., 2013; Wang et al., 2013).
Although powerful, all these technologies are unable to recover live, functional cells of uncultured bacteria for further physiological study. A true understanding of uncultured microorganisms requires the study of live cells in their natural environment. Given the complexity of the natural microbial community, it is difficult to target individual members of populations and separate them from the rest of the community. Various techniques have been developed to isolate and cultivate uncultured microorganisms, including the dilution and modification of nutrient media, encapsulation of cells into beads or stimulation of cell growth (Kaeberlein et al., 2002; Zengler et al., 2002; Vartoukian et al., 2010). Although these techniques have some success, one of the limitations of these techniques is the inability to study uncultured bacteria in situ. It is thus desirable to identify and isolate functionally active, but as yet uncultured bacteria directly from their natural environment.
To address these challenges, in this study, a magnetic nanoparticle (MNP)-mediated isolation (MMI) method was developed and employed to reveal the active microbial cells that perform in situ phenol degradation—Burkholderiales spp.—from coking plant wastewater. MMI-recovered live cells (MRCs), which were dominated by Burkholderiales spp., were able to degrade phenol, showing similar degradation kinetics to the original biosludge. The results of DNA SIP and pyrosequencing of the ‘heavy' DNA fractions confirmed that Burkholderiales spp. were key degraders, whose sequences were >99% identical to the dominant species in MRCs. Single-cell Raman micro-spectroscopy was used to examine individual cells in MRCs obtained from the SIP experiment, which indicated that the majority (79%) of the individual cells in the MRCs were fully 13C-labelled.
Furthermore, Biolog assays were applied to MRCs using various carbon and nitrogen substrates and they revealed that a metabolite of the ammonia oxidisation pathway, hydroxylamine (NH2OH), was a key inhibitor that caused failure of phenol degradation in the coking wastewater treatment plant.
Materials and methods
Site description and sample collection
The coke oven biological wastewater treatment plant is operated by Tata Steel at Scunthorpe, UK. Main contaminants in the plant's influent are phenolic compounds, thiocyanate, polycyclic aromatic hydrocarbons and ammonia (50–70 mg l−1), which are listed in Supplementary Table S1. The average concentration of the major contaminant, phenol, was 250 mg l−1. The operational temperature of the biological treatment unit was 25 °C. Settled biosludge, of normal and poor performance, was sampled from the activated sludge treatment tank and processed to set up microcosms on the same day. There was no detectable phenol in the supernatant of the settled sludge (Supplementary Table S2). Biosludges of good and poor performance were designated as G-BS and P-BS, respectively. The G-BS was sampled during periods of regular operation at the plant, whereas the P-BS was sampled just before a failure of water treatment that was associated with a sudden increase in nitrite concentration in the aeration tank (Supplementary Figure S1).
Isolation of phenol degraders from the biosludges by enrichment and cultivation
The raw biosludge was directly spread onto mineral medium (MM) agar (MMA) plates with phenol (250 mg l−1) as the sole carbon source. One litre of MM contained 2.5 g Na2HPO4, 2.5 g KH2PO4, 1.0 g NH4Cl, 0.1 g MgSO4•7H2O, 10 μl saturated CaCl2 solution, 10 μl saturated FeSO4 solution and 1 ml Bauchop & Elsden solution (Bauchop and Elsden, 1960). Noble agar (1% w/v) was used to prepare the MM agar (MMA) plates. The plates were incubated in the dark for 48–72 h, and single colonies were identified and re-spread onto a MMA–phenol plate for further purification. The isolated strains were cultivated in MM-–phenol liquid medium for nucleic acid extraction.
MNP synthesis and functionalisation of biosludges
MNP synthesis was carried out as previously described (Zhang et al., 2011) with the following modifications. Briefly, 1 ml of FeCl2 (1.0 M dissolved in 2.0 M HCl) and 2 ml of FeCl3 (2.0 M dissolved in 2.0 M HCl) were mixed, to which 25 ml of NaOH (2.0 M) was slowly added drop by drop, with constant vortex mixing, for 30 min. The synthesised MNPs were harvested by a permanent magnet and the supernatant was replaced with deionised water of the same volume. This washing step was repeated six times until the pH was neutral. Five millilitres of MNPs were then mixed with 45 ml of polyallylamine hydrochloride solution (10 mg ml−1), which was then stabilised for 60 min in an ultrasound bath with 40 kHz and an output energy of 75 W (Langford Electronics Ltd, Coventry, UK). After centrifugation at 10 000 g for 10 min, the pellet was re-suspended in 50 ml deionised water and dispersed by vortex mixing. The final solution was passed through a 0.2-μm filter (Millipore, Billerica, MA, USA) and was then ready for bacterial functionalisation. Polyallylamine hydrochloride is a cationic polyelectrolyte, contributing positive charge to the MNPs and maintaining their dispersion in the water.
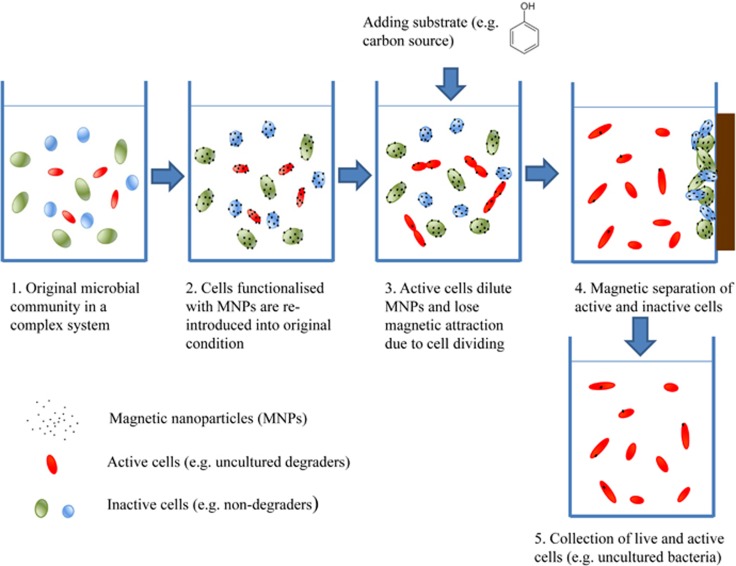
A schematic for cultivating bacteria from biosludge through MMI is shown in Figure 1. First, all cells from the biosludge were functionalised by mixing biosludge with biocompatible MNPs. Ten millilitres of biosludge (G-BS and P-BS) was centrifuged at 3000 r.p.m. for 10 min and the bacterial pellet was harvested and resuspended in the same volume of deionised water. The cell suspension was mixed with 10 ml of polyallylamine hydrochloride -stabilised MNP solution. The bacteria–MNP mixture was incubated at room temperature for 20 min with shaking (150 r.p.m.). The MNP-functionalised bacteria were then separated from the aqueous phase by a permanent magnet, followed by resuspension in deionised water. The washing step was repeated three times to remove those bacteria that were not functionalised by MNPs. Subsequently, the MNP-functionalised bacteria were resuspended in 10 ml filter-sterilised wastewater. To prepare filtered-sterilised wastewater, G-BS was centrifuged at 4000 r.p.m. for 10 min and the supernatant was passed through 0.2 μm filters twice to remove cells.
Figure 1.
Schematic process of recovering live bacteria from their natural environment through MNP functionalisation and separation.
The MNP cells were then re-introduced into filter-sterilised wastewater in which 250 mg l−1 phenol (13C6 or 12C phenol) was added as the carbon source. In the presence of phenol, active phenol degraders divided, causing the MNPs coated onto the cells to gradually get diluted and their magnetic attraction eventually lost. After phenol degradation was completed, a permanent magnet was re-applied. The active phenol degraders were freely suspended in the water phase, whereas the rest of cells (non-dividing or inactive phenol degraders) were attracted and immobilised by the magnet (Figure 1). In this way live bacterial cells responsible for phenol degradation in situ can be recovered by this MMI method.
Phenol degradation in microcosms
For in situ phenol degradation experiments, a series of treatments were carried out including: phenol blank control (no biosludge), original biosludge control (no phenol), original biosludge supplemented with water (negative control), 13C- or 12C-phenol (final concentration 250 mg l−1), and filter-sterilised biosludge mixed with MNP cells supplemented with water (negative control), 13C- or 12C-phenol (final concentration 250 mg l−1). Three replicates were carried out for each treatment. Samples were taken every 30 min from the incubations using G-BS and every hour from those using P-BS to determine the residual phenol concentrations. A subset of samples (0.5 ml) was taken from 13C and 12C-phenol amended G-BS microcosms at t=0, 2.5, 5 and 7 h, which were used for later DNA-SIP analyses of active microbial populations involved in phenol degradation. Phenol concentration was measured by a spectrophotometric method described by the American Public Health Association (Greenberg et al., 2005). Briefly, 100 μl of cell-free sample was diluted in 900 μl deionised water, dosed in the following order with 400 μl of 2.0 M NH4OH, 200 μl of 2% (w/w) aminoantipyrine and 400 μl of 2% (w/w) K3Fe(CN)6. The absorbance of the mixture (200 μl) was then measured at 500 nm wavelength using a microplate reader (Synergy II multimode, BioTek Instruments, Inc., Winooski, VT, USA).
MNP-mediated cell isolation and counting
After completion of phenol degradation, MRCs in the suspension were stained by 4′,6-diamidino-5-phenyl-indole (DAPI) (Kubista et al., 1987) and counted under a Zeiss Axioplan 2 epifluorescence microscope. MRCs in the suspension were centrifuged at 3000 r.p.m. for 10 min and resuspended in the same volume of phosphate-buffered saline (54.44 mg KH2PO4 and 106.8 mg Na2HPO4·12 H2O in 10 ml deionised water). To enumerate the population of MRCs, 20 μl of the cell suspension was diluted to 1 ml with autoclaved ultra-high quality water, buffered with phosphate-buffered saline. This dilution was then incubated in the dark with DAPI stain, corresponding to a working concentration of 12.5 μg ml−1, filtered onto a 0.2-mm black polycarbonate filter paper and mounted onto a glass slide with Fluoroshield mounting medium (Sigma-Aldrich, Co., Dorset, UK). The resultant slides were analysed and imaged under fluorescent light using the microscope with a DAPI filter cube. The cells were detected using 358-nm UV light for excitation and 461 nm for emission. In order to determine the cell density of the supernatant, cell counts were performed for 15 randomly chosen fields of view. For means of comparison, a 10−4 dilution of the original biosludge was enumerated using the same approach.
Physiological testing of MRCs using the Biolog high-throughput phenotypic assay
Biolog plate (Biolog, Hayward, CA, USA) analyses were undertaken to examine the carbon and nitrogen metabolism of MRCs. Pseudomonas putida XY5 isolated from the same biosludge was used as a control. Biolog PM1 plate was used for carbon metabolism and PM3 for nitrogen metabolism in accordance with the manufacturer's instructions. The coking wastewater naturally contains a high concentration of ammonia and operator experience found that a failure of treatment was always associated with the presence of nitrite in the aeration tank, which suggests that the nitrogen source could affect phenol biodegradation. Hence, for the PM3 nitrogen assay, 250 mg l−1 phenol was used as the carbon source to reveal the effect of nitrogen sources on phenol degradation.
The effect of nitrogen source on phenol biodegradation
Ammonium chloride, sodium nitrite, sodium nitrate and hydroxylamine were added into filter-sterilised wastewater (from G-BS) as nitrogen sources. The background concentrations of phenol and nitrogen sources are shown in Supplementary Table S2. All samples were set up in triplicate. Phenol was added into all treatments with a final concentration of 250 mg l−1. The final concentrations of NH3-N, NO2− and NO3− were 232 mg l−1 (100 mg l−1 amendment with background 132 mg l−1 shown in Supplementary Table S2), 50 mg l−1 and 100.23 mg l−1 (100 mg l−1 amendment with background 0.23 mg l−1 shown in Supplementary Table S2), respectively. The final concentrations of NH2OH were 0.1, 0.2, 0.5, 1.0, 2.0, 5.0 and 10 mg l−1. Samples were added into a 96-well microplate: each well contained 160 μl of filter-sterilised wastewater, 20 μl of appropriate phenol with nitrogen compounds (or water), and 20 μl of cell suspension of MRCS (or water). The microplate was incubated at 25 °C in the microplate reader and OD600 was recorded every 15 min. At the end of incubation, the pH values in each sample were measured using indicator strips. The residual phenol concentration was measured according to the method described above, and the percentage of phenol degradation was calculated from the final concentration relative to the three cell-free controls on the same plate.
Detection of single cells in MRCs by Raman micro-spectroscopy
Raman micro-spectroscopy was employed to quantify 13C-incorporation of MRCs at the single cell level (Huang et al., 2004, 2007, 2009). MRCs were harvested from the above treatments. MRCs were centrifuged at 3500 r.p.m. for 10 min, and washed with water three times. Each cell suspension (2∼5 μl) was spread onto a calcium fluoride (CaF2) slide and allowed to air dry before Raman analysis. Single-cell Raman spectra (SCRS) were acquired using a confocal Raman microscope (LabRAM HR, HORIBA Scientific, London, UK). A × 100 magnifying dry objective (NA=0.90, Olympus, UK) was used to observe and acquire Raman signals from single cells. The Raman scattering was excited with a 532-nm Nd:YAG laser (Torus Laser, Laser Quantum, UK). The laser power on a single cell was about 15 mW. Each Raman spectrum was acquired between the range of 1989 and 336 cm−1, with 1021 data points and a resolution of ∼1 cm−1. LabSpec software (HORIBA Scientific) was used to operate the Raman system and acquire Raman spectra. Acquisition time was 20 s for measurement of each single cell.
Nucleic acid extraction
The biosludge samples were collected at the beginning and the end of phenol degradation. Total nucleic acids were extracted from 1.0 ml of each biosludge sample or culture of the isolated phenol degraders using a PowerSoil DNA Isolation Kit (MO BIO, Carlsbad, CA, USA) according to the manufacturer's instructions.
DNA-SIP
13C-labelled ‘heavy' DNA (13C-DNA) was separated from the unlabelled ‘light' DNA (12C-DNA) by equilibrium density gradient centrifugation using a protocol described by Neufeld et al. (2007). Briefly, 1 μg of DNA was mixed with the CsCl gradient buffer to a volume of 1.2 ml, to which 4.6 ml of 7.163 M CsCl solution was added to obtain a final density of 1.725 g ml−1. The mixture was inverted gently and transferred into a 5.8-ml ultracentrifuge tube (Beckman, High Wycombe, UK). After balancing and sealing, the tubes were spun in an ultracentrifuge (Optima L-80 XP, Beckman Coulter) at 44 100 r.p.m. (∼177 000 g, VTi65.2 rotor, Beckman) for 40 h. The DNA of different density was retrieved by gradient collection into 12 fractions of 400 μl volume from the bottom of the ultracentrifuge tube. The injection of deionised water was manipulated to push down fractions from the top of the ultracentrifuge tube by a low-flow peristaltic pump (Watson Marlow Ltd., Falmouth, UK). The DNA fractions were then purified by glycogen and PEG 6000 solution for 2 h, washed with 70% ethanol, air dried and dissolved in 50 μl DNase-free water. The DNA from the 12C phenol control was manipulated with the same centrifugation, fractionation and purification procedures.
PCR amplification of 16S rRNA genes and phenol hydroxylase genes
PCR amplification was carried out in a C1000 thermal cycler (Bio-Rad Laboratories Ltd, Hertfordshire, UK). Each reaction (50 μl) contained 0.3 μl Dream Taq DNA polymerase (Fermentas, Fisher Scientific Ltd, Loughborough, UK), 2 μl deoxynucleotide triphosphates at a concentration of 5 mM, 2 μl of each primer (5 μM), 1 μl DNA template and 37.7 μl molecular water. For MRCs, 1 μl cell suspension was directly used as the DNA template for the amplification of 16S rRNA genes. DNA fragments of 16S rRNA genes were amplified by the primer pair of 63f and 1387r, as listed in Table 1. After 94 °C for 10 min, 35 cycles were undertaken with 94 °C for 1 min, 56 °C for 1 min, and 72 °C for 1 min, followed by a final extension at 72 °C for 10 min. The largest subunit of the multi-component phenol hydroxylases (LmPH) was amplified using different primer sets targeting three different types of phenol hydroxylases (Futamata et al., 2001). For the primer pair of phe1f/phe3r (Table 1), the PCR program used was as follows: 10 min at 94 °C; 35 cycles of 94 °C for 1 min, 56 °C for 1 min, and 72 °C for 1.5 min; final extension at 72 °C for 10 min. The PCR program for the primer pheUf/pheMHr and pheUf/pheHr was as follows: 94 °C for 10 min; 5 cycles of 94 °C for 1 min, 58 °C for 1 min and 72 °C for 1 min; 5 cycles of 94 °C for 1 min, 57 °C for 1 min and 72 °C for 1 min; 25 cycles of 94 °C for 1 min, 56 °C for 1 min and 72 °C for 1 min; 72 °C for 10 min as final extension. For the primer set pheUf/pheLr, the program was: 94 °C for 10 min; 5 cycles of 94 °C for 1 min, 55 °C for 1 min and 72 °C for 1 min; 5 cycles of 94 °C for 1 min, 54 °C for 1 min and 72 °C for 1 min; 25 cycles of 94 °C for 1 min, 53 °C for 1 min and 72 °C for 1 min; 72 °C for 10 min as final extension. The PCR products were checked by electrophoresis on a 1.5% (w/v) agarose gel (Sigma-Aldrich, Co.) using TBE buffer.
Table 1. Primers used for the PCR of 16S rRNA and phenol hydroxylase gene.
| Primer | Sequence (5′–3′) | Reference |
|---|---|---|
| 63f | 5′-CAGGCCTAACACATGCAAGTC-3′ | Marchesi et al., 1998 |
| 1387r | 5′-GGGCGGWGTGTACAAGGC-3′ | Marchesi et al., 1998 |
| phe1f | 5′-GA(G/A)GGCATCAA(A/G)AT(C/T)-3′ | Futamata et al., 2001 |
| phe3r | 5′-CAG(C/G)CG(A/G)T(A/T)ACC(G/T)CGCCAGAACC-3′ | Futamata et al., 2001 |
| pheUf | 5′-CCAGG(C/G)(C/G/T)GA(G/A)AA(A/G)GAGA(A/G)GAA(G/A)CT-3′ | Futamata et al., 2001 |
| pheLr | 5′-GG(A/G/C)A(G/T/C)(A/G)TTG(C/T)CCGGGTC-3′ | Futamata et al., 2001 |
| pheMHr | 5′-GAT(T/C/G)GGCAC(A/G)TTGTCTTC-3′ | Futamata et al., 2001 |
| pheHr | 5′-GTGGCCATGTCGCCATTGA-3′ | Futamata et al., 2001 |
To determine the diversity of microbes in MRCs, the 16S rRNA PCR products from MRCs were cloned into pGEM-T vector (Fermentas, Fisher Scientific Ltd), and transferred into Escherichia coli JM109 competent cells by heat shock. Thirty clones were randomly selected for plasmid extraction and the 16S rRNA inserts were sequenced.
Nucleotide sequencing and computational analysis
PCR amplicon libraries of the hypervariable V1-V3 region of the 16S rRNA genes (corresponding to E. coli positions 5–534) were generated for ‘heavy' and ‘light' DNA fractions of the 13C6-phenol incubated DNA-SIP microcosms (t=0, 2.5, 5 and 7 h, respectively). PCR was performed using the forward primer (5′-NNNNNNN-TGGAGAGTTTGATCCTGGCTCAG-3′) and reverse primer (5′-NNNNNNN-TACCGCGGCTGCTGGCAC-3′). Unique heptad-nucleotide sequences (seven bases) were synthesised at the 5′ end of each pair of primers as barcodes, which were used to assign sequences to the different samples. Pyrosequencing was carried out using a Genome Sequencer FLX Titanium platform (454 Life Sciences Roche, Branford, CT, USA) where reads of on average 400 bp in length were produced.
In quality filtering, reads were discarded if they were shorter than 150 bp, or longer than 1000 bp, had an average quality score of <35 in each 50-bp window rolling along the whole read, or contained primer mismatches, uncorrectable barcodes, ambiguous bases or homopolymer runs in excess of eight bases. Sequences that passed the quality filters were then analysed using the MOTHUR software package (Schloss et al., 2009). Sequences were assigned to operational taxonomic units (OTUs) with a 97% pairwise identity as the threshold, and then classified taxonomically using the Greengenes 16S rRNA reference database (McDonald et al., 2012) with a confidence threshold of >80%. The Greengenes taxonomies were used to generate summaries of the taxonomic distributions at different phylogenetic levels. To standardise sequence counts across samples with uneven sampling, we randomly selected 2008 sequences per sample (rarefaction) and used this as a basis to compare the abundances of OTUs across samples. The Bray–Curtis metric was used to generate distance matrices from samples, which were visualised as a PCoA (principal coordinates analysis) plot.
To assess the abundance of the 13C-DNA fraction in the samples, pyrosequencing reads were aligned with BLASTN to the reference sequence (an uncultivated Burkholderiales spp. Genbank accession no. KM067154) of the dominant OTU in MRCs. The sequence similarities of these alignments were visualised as histograms, in which the Burkholderiales spp. reference sequence was compared to 12C-DNA and 13C-DNA fractions at the two time points at 0 and 7 h of phenol degradation.
The 454 reads that were taxonomically assigned to the order Burkholderiales using Greengenes database were clustered into OTUs with a 97% pairwise identity as the threshold. The representative sequences of all 12 OTUs in the 13C-DNA fraction were then aligned to the Burkholderiales spp. reference sequence (Genbank accession no. KM067154), which was identified in MRCs using MUSCLE. For phylogenetic analysis, a maximum likelihood (ML) tree was built with 1000 bootstrappings in MEGA6.
DNA sequences of 16S-rRNA gene and phenol hydroxylase from this study are available in Genbank (accession number KM067152 to KM067154 and KJ174591 to KJ174607).
Results
Isolation of culturable phenol-degrading microorganisms
Rhodococcus sp. XY2, Chryseobacterium sp. XY3 and three different Pseudomonas strains (XY4, XY5 and XY6) were isolated on agar plates from the biosludge samples (Supplementary Table S3) using a minimal medium with phenol as the sole carbon source. The 16S rRNA sequences of Pseudomonas sp. XY4 and XY5 are identical (100%) to that of P. pseudoalcaligenes and P. putida KT2440, respectively, whereas the 16S rRNA of Pseudomonas sp. XY6 is 99.6% identical to that of P. plecoglossicida. However, subsequent culture-independent analyses suggest that these isolates were not responsible for phenol biodegradation in situ.
MNP functionalisation of biosludges does not affect the rates of phenol degradation
The rates of phenol degradation of two biosludge samples of contrasting performances (G-BS and P-BS) were determined in order to assess the impact of MNP on phenol degradation. The data are shown in Figure 2a. After 7 h, the microbial community in G-BS degraded phenol completely (Figure 2a). In contrast, no phenol degradation occurred in P-BS in the initial 18 h and phenol degradation was not completed until 36 h (Figure 2a). The use of stable isotope-labelled phenol (13C-phenol) had no impact on the performance of phenol degradation using the two biosludges samples (Figure 2a). MNP-functionalised biosludges had similar rates of phenol degradation compared with those of the raw biosludges, confirming that MNP functionalisation is biocompatible (Figure 2a).
Figure 2.
(a) Kinetics of phenol biodegradation in biosludges of good and poor performance (G-BS and P-BS, respectively). Phenol degradation in G-BS was completed in 7 h (▪, ♦ and •), whereas in P-BS phenol degradation completed in 36 h (□, ◊ and o). Neither the use of isotope-labelled phenol (13C-phenol, ♦ and ◊) nor MNP functionalisation (• and o) had significant impact on bacterial phenol degradation in G-BS and P-BS samples. No phenol degradation occurred without the biosludge (▴ and ▵). A subset of samples were withdrawn from the G-BS incubations with 13C-phenol and 12C-phenol at t=0, 2.5, 5, 7 h for DNA-SIP analyses and the data are presented in Figure 4, Supplementary Figures S2 and S3). (b) The phenol degradation performances of MRCs (•), the initial biosludges, G-BS (▪) and negative controls (▴, no biosludge added).
MRCs had a similar performance as the whole biosludge for phenol degradation
The MNP-functionalised cells were introduced into filter-sterilised wastewater to allow for propagation of active phenol degraders in their natural environment. Either 13C-labelled or 12C-phenol or water as positive and negative controls were added to the MNP-functionalised cells. In these experiments, there were no free cells in treatments at time point t=0. After phenol degradation was completed (Figure 2a), MRCs were magnetically separated from the treatments. DAPI staining of MRCs obtained from the 13C- or 12C-phenol treatment showed that the cell population was 1.48±0.49 × 105 cells ml−1. In contrast, the cell density in controls where phenol was not added was two orders of magnitude lower (<103 cells ml−1), which excludes the possibility that MNPs were lost owing to some random reason. In comparison, the total cell population in the original biosludge was 1.05±0.64 × 109 cells ml−1. The DAPI counting approach may underestimate the cell population because the cell-harvesting step using centrifugation at 3000 r.p.m. for 10 min may miss small cells that are not easily pelleted.
The MRCs derived from MNP-treated biosludges were incubated with phenol (250 mg l−1) to determine whether they were functionally active. The data presented in Figure 2b demonstrate that the degradation pattern was similar to that of the original raw biosludges (G-BS) after a 2-h delay (Figure 2b). MRCs from the controls (no phenol amendment) had no phenol degradation activity (data not shown). This suggested that the active phenol degraders responsible for phenol degradation in the original raw G-BS should be recovered from MRCs.
Raman micro-spectroscopy analyses of MRCs confirmed 13C incorporation at the single cell level
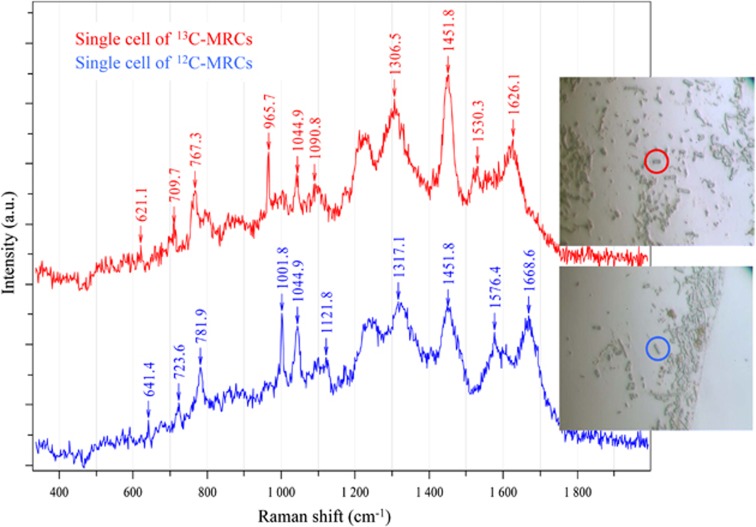
MRCs from 13C- and 12C-phenol treatments were examined by Raman micro-spectroscopy at the single-cell level. SCRS of MRCs were acquired to examine their 13C incorporation based on the fact that some Raman bands of 13C-labelled cells shift to lower wavenumbers upon the incorporation of 13C from a growth substrate (Huang et al., 2004, 2007, 2009; Li et al., 2013). SCRS of MRCs from 12C-phenol treatments were used as controls. Microscopic images indicated that cells from MRCs were more uniform than those in the original biosludge. MRCs were mostly rod shaped and of similar sizes and Raman spectral patterns (Figure 3). SCRS from the cells with 13C incorporation showed significant Raman shifts in the marker bands, namely, the phenylalanine band from 1001.8 to 965.7 cm−1 and the protein band from 1668.6 to 1626.1 cm−1 (Figure 3). Other Raman bands such as 641, 723, 781, 1121, 1317, and 1576 cm−1 also shifted in a similar way owing to the incorporation of 13C into the cells (Figure 3). Analysis of Raman spectra of 135 randomly selected single cells in the MRCs from the 13C-phenol treatment indicated that 79% of MRCs were fully labelled with 13C (Figure 3). These results indicated that most freely suspended cells from the MRCs were indeed active phenol degraders.
Figure 3.
Raman micro-spectroscopy identification of 13C-stable isotope incorporation into MRCs.
Uncultured Burkholderiales spp. were responsible for phenol degradation
A clone library of 16S rRNA genes was obtained from PCR products using MRCs as the DNA template. Thirty clones were randomly selected and the 16S rRNA genes were sequenced. The data suggested that 67% of bacteria in MRCs were uncultivated Burkholderiales spp. (number: KM067154 and KM067153). Interestingly, this is a new group of Burkholderiales spp., showing <95% identity to all prokaryotic 16S rRNAs in the NCBI database.
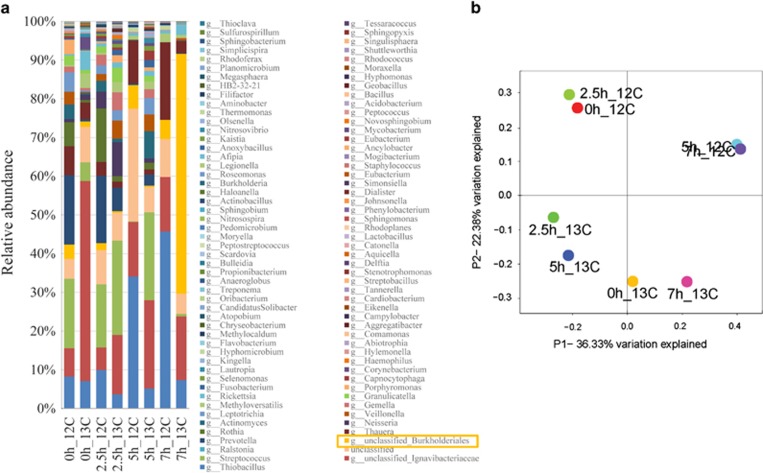
Pyrosequencing of ‘heavy' fractions of DNA-SIP samples indicated that the dominant bacteria at the family level (∼62%454 reads) in the 13C-DNA fractions at t=7 h were unclassified Burkholderiales spp. (Figure 4a). Microbial community structure changed over time and by the time of complete phenol degradation at 7 h, the microbial structure had changed significantly in that the 13C-DNA fraction was different from the rest of the microbial community structures (Figure 4b).
Figure 4.
(a) Relative abundance of taxonomic classification of microbial community at the family level in the 12C- and 13C-DNA fractions of 13C6-phenol incubated biosludges. Taxonomic assignment was carried out using the Greengenes 16S rRNA database. DNA-SIP experiments indicated that Burkholderiales spp. were dominant species in the 13C-DNA fraction after 7 h phenol degradation, suggesting they were key phenol degraders in situ. Sample code example: 2.5h_13C means the 13C-DNA fraction at time t=2.5 h degradation shown in Figure 2. (b) Bacterial communities during phenol degradation are clustered using PCoA of the Bray-Curtis matrix. The percentage of variation explained by the principal coordinates is indicated on the axes.
The 16S rRNA sequence of Burkholderiales sp. (accession number: KM067154) retrieved from MRC was used as the reference for the alignment of all 454 reads of 12C- and 13C-DNA fractions at t=0, 2.5, 5 and 7 h. The histogram presented in Supplementary Figure S2 clearly showed that >60% reads of the 13C-DNA fraction at t=7 h were identical (>99% identity) to dominant sequence (KM067154) retrieved from MRC (Supplementary Figure S2).
Further analyses of the retrieved Burkholderiales sequences from the heavy fraction at t=7 h revealed the presence of 12 OTUs, of which OTU 34 was dominant (Supplementary Figure S3A). A comparison between the sequence of KM067154 and OTU 34 showed that they are 99% identical. A phylogenetic tree was generated using the sequences of the 12 OTUs representing Burkolderiales spp. and the dominant Burkholderiales spp. (accession number: KM067154) independently retrieved from the MRCs (Supplementary Figure S3). It showed that DNA-SIP and MRC derived data are highly consistent, indicating that the uncultivated Burkholderiales spp. was active and responsible for phenol degradation in situ (Figure 4 andSupplementary Figure S3). Thus, the data indicated that the dominant species in MRCs were the same as those in the 13C-fraction. The advantage of MRCs is that live cells were obtained that can be used for further physiological study.
Diversity of functional genes for phenol degradation
A key functional gene for phenol degradation is phenol hydroxylase, which converts phenol into catechol before the TCA cycle. Phenol hydroxylase can be recovered by PCR using degenerate primers (Table 1). Supplementary Table S3 summarises the occurrence of phenol hydroxylase genes in different samples.
In the original biosludges (both the BS-G and BS-P samples), all four types of LmPH, phe1, pheMH, pheL and pheH, were found. The phenol hydroxylase genes in the MRCs and 13C-fraction belong to the Burkholderiales order; more specifically, the phe1 and pheL (accession number KJ174604 and KJ174605) genes in the MRCs are identical to those of Cupriavidus metallidurans CH34 (formerly Ralstonia metallidurans, Janssen et al., 2010) (Supplementary Table S3).
In the cultivated species isolated from the biosludges, however, the phenol hydroxylase genes were of different types. Specifically, Chryseobacterium sp. XY3 has an identical pheL (a phenol hydroxylase subunit gene) to that of Comamonas sp. J5-66 (Sun et al., 2012). The isolated P. pseudoalcaligenes XY4 and P. plecoglossicida XY6 have two domains of LmPH (phe1 and pheMH) (accession number KJ174598 and KJ174600), which were both identical to that in Pseudomonas sp. (Supplementary Table S3) (Kim et al., 2005). Interestingly, it was found that P. putida XY5 contained novel types of phenol hydroxylases, phe1 and pheMH (accession number KJ174599) that have not been reported previously.
Phenotyping MRCs
A remarkable advantage of MMI is that it enables the isolation of live bacteria for ecophysiological analysis. Biolog high-throughput phenotypic microarrays were used for the phenotypic analysis of MRCs. They served two purposes: the characterisation of phenotypes and the identification of key factors affecting the performance of phenol degradation. The MRCs and P. putida XY5 showed different phenotypic patterns for carbon metabolism (Supplementary Table S4), providing additional evidence that the bacteria isolated using the MMI technique were different from those readily cultivated phenol degraders such as P. putida. Specifically for MRCs, the carbon sources, such as D-alanine, α-D-glucose, tyramine and L-glutamine, promoted cell growth, whereas the carbon sources, such as D-galactonic acid-γ-lactone, L-galactonic acid-γ-lactone, m-tartaric acid and D-threonine, inhibited cell growth (Supplementary Table S4).
To examine the impact of nitrogen sources on biodegradation performance, phenol (250 mg l−1) was used as the carbon source in the PM3 nitrogen test plates. Data presented in Supplementary Table S5 showed that MRCs and P. putida XY5 had different response patterns to different nitrogen sources. NH2OH and D,L-α-amino-caprylic acid significantly inhibited phenol degradation in both MRCs and P. putida XY5, whereas ammonia, nitrite and nitrate did not show any repression effect on phenol degradation (Supplementary Table S5).
Hydroxylamine is an inhibitor for phenol degradation in coke oven biosludges
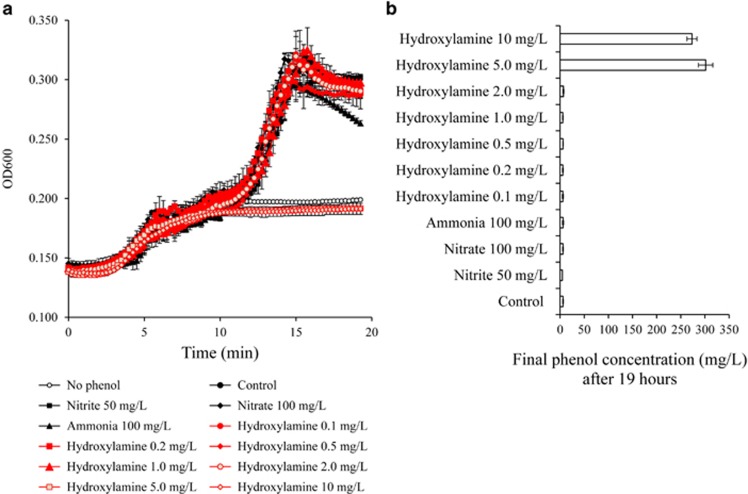
The ammonia concentration in the influent wastewater was 50–70 mg l− (Supplementary Table S1) and 132 mg l−1 in the settled sludge (Supplementary Table S2) generated from thiocyanate (SCN−) degradation. It was observed that the failure of wastewater treatment was associated with a sudden increase of nitrite (Supplementary Figure S1). Ammonia and its metabolic intermediate compounds NH2OH, NO2− and NO3− were added into the filter-sterilised wastewater along with MRCs within which Burkholderiales spp. were enriched. During 19 h of incubation, the pH in the media of all treatments remained between 6.4 and 6.8, indicating that metabolism of NH2OH, nitrite, nitrate and ammonia did not alter the pH of the media. Cell growth in the treatment with NH2OH >5 mg l−1 was inhibited, whereas cells with 232 mg l−1 NH3-N, 50 mg l−1 NO2−, 100.23 mg l−1 NO3− and <2 mg l−1 NH2OH continued to grow (Figure 5a). Phenol concentration remained unchanged after 19 h incubation when NH2OH was >5 mg l−1, whereas phenol was completely degraded in the other treatments (Figure 5b). These results indicated that a NH2OH concentration >5 mg l−1 completely inhibited phenol degradation by Burkholderiales spp., whereas 50 mg l−1 NO2−, 100.23 mg l−1 NO3− and 232 mg l−1 NH3-N did not inhibit phenol degradation (Figure 5).
Figure 5.
(a) Growth curves of MRCs in the presence of hydroxylamine, ammonia, nitrite or nitrate. (b) The remained phenol concentration after 19 h during phenol degradation by MRCs.
Discussion
It is increasingly evident that as yet uncultured bacteria can have key roles in the biodegradation and bioremediation of environmental pollutants (Huang et al., 2009; Chen and Murrell, 2010; Wang et al., 2012). In this study, we demonstrated that MMI can be used to recover live cells of key pollutant degraders from a complex microbial community, such as biosludge, and that MRCs can be used for further eco-physiological studies. MRCs were able to degrade phenol and had a similar degradation pattern to the original biosludge. Fully 13C-labelled phenol of ambient concentration was introduced into the biosludge to probe the in situ active degraders. Subsequent recovery of MRCs and Raman micro-spectroscopy analyses at the single-cell level demonstrated that the majority of MRCs were indeed labelled by 13C, indicating that they have a key role in phenol degradation. These data are consistent with the results from the DNA-SIP analyses: sequencing and phylogenetic analyses indicated that the major species in the 13C-DNA fraction of the biosludge was related to a group of so-far uncultivated Burkholderiales spp., which showed high sequence identity (>99%) to the predominant 16S rRNA gene retrieved from clone library analysis of MRCs. Collectively, our results demonstrated that the MMI method was powerful in identifying and isolating a new group of Burkholderiales spp. as the key phenol degraders in these biosludges.
This methodology builds on the fact that cell division of the active bacterial cells will dilute MNP coatings and ultimately result in a loss of magnetic attraction. Conversely, the metabolically inactive bacteria keep their MNPs and thus remain magnetically attractive. To enable effective isolation of these two groups of cells in a complex community, the following properties for MNPs are essential (Zhang et al., 2011)—they need to be: (1) biocompatible—MNPs should have minimal impact on cell physiology in terms of growth and enzymatic activities; (2) magnetically controllable—MNP-functionalised cells can be easily manipulated by a magnetic field, which requires a suitable MNP size and MNP-to-cell ratio; (3) highly efficient for functionalisation—MNP coating efficiency is >99.9%, ensuring that almost all cells in a microbial community can be magnetically functionalised; (4) dilutable—MNPs coated on cells can be diluted and eventually lost after cell divisions. Although the MMI approach has been shown to be powerful in this study, it has its own limitations. So far, the MMI approach is only effective in the recovery of actively dividing and rapidly growing bacteria that are capable of escaping the MNPs within a given time. It remains to be established whether MMI can be used to separate active, but slow-growing bacteria or those that can turn over the substrate without cell division.
The operation data of the coking plant's wastewater treatment suggested that a sudden increase of NO2− in the wastewater was often associated with a sudden drop in the removal efficiency of chemical oxygen demand and subsequent failure of water treatment (Supplementary Figure S1). It was observed that the treatment often failed when NO2− concentration was greater than a threshold of 10 mg l−1. For example, at the point of high NO2− concentration (>10 mg l−1) in Nov 2012, a failure of water treatment occurred along with the appearance of P-BS (Supplementary Figure S1). The P-BS was still able to degrade phenol but there was a long lag time (18 h) before phenol degradation occurred (Figure 2a). This implied that nitrogen metabolism by the biosludge microbial community affected wastewater treatment performance. Hence, in the Biolog PM3 nitrogen metabolism test, phenol (250 mg l−1) was used as the sole carbon source to examine the impact of nitrogen metabolism on the phenol degrading ability of uncultured but metabolically active microbial cells. Figure 5a indicated that NH2OH >5 mg l−1 completely inhibited phenol degradation, whereas 50 mg l−1 NO2− did not inhibit phenol degradation. Both ammonia-oxidising Archaea and ammonia-oxidising Bacteria oxidise NH3 into NH2OH (Arp et al., 2002; Vajrala et al., 2013), which is then further oxidised to nitrite (NO2−) and finally nitrate (NO3−). The experimental data clearly indicated that it was NH2OH, but not NO2−, NO3− or NH3, that inhibited phenol degradation in the coking wastewater. The threshold of this NH2OH inhibition effect was between 2 and 5 mg l−1 (Figure 5). It is likely, therefore, that the high concentration of NO2− within the wastewater treatment facility was a result of NH2OH accumulation and it was NH2OH that led to the failure in wastewater treatment. NH2OH is a volatile and unstable compound and in fact, when its concentration decreased below the threshold, phenol biodegradation resumed (Figure 2a). Presumably, the degradation of SCN- (120–250 mg l−1 in the influent, Supplementary Table S1) would produce ammonia via SCN−→SO42−+NH3+CO2, which could lead to an increase in ammonia during the wastewater treatment (for example, 132 mg l−1 in settled sludge shown in Supplementary Table S2) compared with 50–70 mg l−1 ammonia in the influent (Supplementary Table S1). The accumulation of NH2OH is likely due to the low activity of hydroxylamine oxidoreductase that catalyses the formation of NO2−.
To summarise, we demonstrate that live and uncultured bacteria can be recovered using this novel MMI approach. It is foreseeable that this MMI approach will greatly accelerate the pace of exploration for as yet uncultured microbes, and help our understanding of the diversity, physiology, functional potential, evolution, adaptation and ecophysiology of the microbes present in the environment. MMI-enriched uncultured cells can be subjected to single-cell isolation and genome assembly.
Acknowledgments
We thank EU ECOWATER project (RFCR-CT-2010-00010) and EPSRC Grant EP/H04986X/1 for financial support. WEH and JX acknowledge the support from the Soil Microbiota Program from the Chinese Academy of Sciences (XDB15040100) and Methodology Innovation Program from the Ministry of Science and Technology of China (MOST 2011IM030100). YC is supported by NERC (NE/H016236/1) and GBMF (GBMF3303). The authors thank Andrew Fairburn in Sheffield and Cunpei Bo and Fei Teng in CAS for technical support.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Author contributions
WEH, DaZ and EA designed the research. DaZ, WEH, JPB, DiZ, HL, BJ, GL and YC performed the experiments. DaZ and WEH analysed data. YW, SH and JX undertook computational analysis. WEH, DaZ, PAD and YC wrote the paper.
Supplementary Material
References
- Arp DJ, Sayavedra-Soto LA, Hommes NG. Molecular biology and biochemistry of ammonia oxidation by Nitrosomonas europaea. Arch Microbiol. 2002;178:250–255. doi: 10.1007/s00203-002-0452-0. [DOI] [PubMed] [Google Scholar]
- Bauchop T, Elsden SR. The growth of micro-organisms in relation to their energy supply. J Gen Microbiol. 1960;23:457–469. doi: 10.1099/00221287-23-3-457. [DOI] [PubMed] [Google Scholar]
- Bombach P, Chatzinotas A, Neu TR, Kaestner M, Lueders T, Vogt C. Enrichment and characterization of a sulfate-reducing toluene-degrading microbial consortium by combining in situ microcosms and stable isotope probing techniques. FEMS Microbiol Ecol. 2010;71:237–246. doi: 10.1111/j.1574-6941.2009.00809.x. [DOI] [PubMed] [Google Scholar]
- Chen Y, Murrell JC. When metagenomics meets stable-isotope probing: progress and perspectives. Trends Microbiol. 2010;18:157–163. doi: 10.1016/j.tim.2010.02.002. [DOI] [PubMed] [Google Scholar]
- Futamata H, Harayama S, Watanabe K. Group-specific monitoring of phenol hydroxylase genes for a functional assessment of phenol-stimulated trichloroethylene bioremediation. Appl Environ Microbiol. 2001;67:4671–4677. doi: 10.1128/AEM.67.10.4671-4677.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg AE, Clesceri LS, Eaton AD. Standard methods for the examination of water and wastewater. American Public Health Association (APHA): Washington, DC, USA; 2005. [Google Scholar]
- Handelsman J. Metagenomics: application of genomics to uncultured microorganisms. Microbiol Mol Biol Rev. 2004;68:669–685. doi: 10.1128/MMBR.68.4.669-685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang WE, Griffiths RI, Thompson IP, Bailey MJ, Whiteley AS. Raman microscopic analysis of single microbial cells. Anal Chem. 2004;76:4452–4458. doi: 10.1021/ac049753k. [DOI] [PubMed] [Google Scholar]
- Huang WE, Stoecker K, Griffiths R, Newbold L, Daims H, Whiteley AS, et al. Raman-FISH: combining stable-isotope Raman spectroscopy and fluorescence in situ hybridization for the single cell analysis of identity and function. Environ Microbiol. 2007;9:1878–1889. doi: 10.1111/j.1462-2920.2007.01352.x. [DOI] [PubMed] [Google Scholar]
- Huang WE, Ferguson A, Singer AC, Lawson K, Thompson IP, Kalin RM, et al. Resolving genetic functions within microbial populations: in situ analyses using rRNA and mRNA stable isotope probing coupled with single-cell Raman-fluorescence in situ yybridization. Appl Environ Microbiol. 2009;75:234–241. doi: 10.1128/AEM.01861-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen PJ, Van Houdt R, Moors H, Monsieurs P, Morin N, Michaux A, et al. The complete genome sequence of Cupriavidus metallidurans strain CH34, a master survivalist in harsh and anthropogenic environments. PLoS ONE. 2010;5:e10433. doi: 10.1371/journal.pone.0010433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein T, Lewis K, Epstein SS. Isolating "uncultivable" microorganisms in pure culture in a simulated natural environment. Science. 2002;296:1127–1129. doi: 10.1126/science.1070633. [DOI] [PubMed] [Google Scholar]
- Kim JY, Kim JK, Lee SO, Kim CK, Lee K. Multicomponent phenol hydroxylase-catalysed formation of hydroxyindoles and dyestuffs from indole and its derivatives. Lett Appl Microbiol. 2005;41:163–168. doi: 10.1111/j.1472-765X.2005.01734.x. [DOI] [PubMed] [Google Scholar]
- Kubista M, Akerman B, Norden B. Characterization of interaction between DNA and 4',6-diamidino-2-phenylindole by optical spectroscopy. Biochemistry (Mosc) 1987;26:4545–4553. doi: 10.1021/bi00388a057. [DOI] [PubMed] [Google Scholar]
- Li MQ, Huang WE, Gibson CM, Fowler PW, Jousset A. Stable isotope probing and Raman apectroscopy for monitoring carbon flow in a food chain and revealing metabolic pathway. Anal Chem. 2013;85:1642–1649. doi: 10.1021/ac302910x. [DOI] [PubMed] [Google Scholar]
- Manefield M, Whiteley AS, Griffiths RI, Bailey MJ. RNA stable isotope probing, a novel means of linking microbial community function to phylogeny. Appl Environ Microbiol. 2002;68:5367–5373. doi: 10.1128/AEM.68.11.5367-5373.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchesi JR, Sato T, Weightman AJ, Martin TA, Fry JC, Hiom SJ, et al. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl Environ Microbiol. 1998;64:795–799. doi: 10.1128/aem.64.2.795-799.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CE, Carlson CA. Tracking differential incorporation of dissolved organic carbon types among diverse lineages of Sargasso Sea bacterioplankton. Environ Microbiol. 2012;14:1500–1516. doi: 10.1111/j.1462-2920.2012.02738.x. [DOI] [PubMed] [Google Scholar]
- Neufeld JD, Vohra J, Dumont MG, Lueders T, Manefield M, Friedrich MW, et al. DNA stable-isotope probing. Nat Protoc. 2007;2:860–866. doi: 10.1038/nprot.2007.109. [DOI] [PubMed] [Google Scholar]
- Pernthaler A, Dekas AE, Brown CT, Goffredi SK, Embaye T, Orphan VJ. Diverse syntrophic partnerships from-deep-sea methane vents revealed by direct cell capture and metagenomics. Proc Natl Acad Sci USA. 2008;105:7052–7057. doi: 10.1073/pnas.0711303105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radajewski S, Ineson P, Parekh NR, Murrell JC. Stable-isotope probing as a tool in microbial ecology. Nature. 2000;403:646–649. doi: 10.1038/35001054. [DOI] [PubMed] [Google Scholar]
- Rappe MS, Giovannoni SJ. The uncultured microbial majority. Annu Rev Microbiol. 2003;57:369–394. doi: 10.1146/annurev.micro.57.030502.090759. [DOI] [PubMed] [Google Scholar]
- Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng JF, et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499:431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- Schleifer KH. Microbial diversity: Facts, problems and prospects. Syst Appl Microbiol. 2004;27:3–9. doi: 10.1078/0723-2020-00245. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JQ, Xu L, Tang YQ, Chen FM, Wu XL. Simultaneous degradation of phenol and n-hexadecane by Acinetobacter strains. Bioresour Technol. 2012;123:664–668. doi: 10.1016/j.biortech.2012.06.072. [DOI] [PubMed] [Google Scholar]
- Vajrala N, Martens-Habbena W, Sayavedra-Soto LA, Schauer A, Bottomley PJ, Stahl DA, et al. Hydroxylamine as an intermediate in ammonia oxidation by globally abundant marine archaea. Proc Natl Acad Sci USA. 2013;110:1006–1011. doi: 10.1073/pnas.1214272110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartoukian SR, Palmer RM, Wade WG. Strategies for culture of 'unculturable' bacteria. FEMS Microbiol Lett. 2010;309:1–7. doi: 10.1111/j.1574-6968.2010.02000.x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Chen Y, Zhou Q, Huang S, Ning K, Xu J, et al. A culture-independent approach to unravel uncultured bacteria and functional genes in a complex microbial community. PLoS ONE. 2012;7:e47530. doi: 10.1371/journal.pone.0047530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ji YT, Wharfe ES, Meadows RS, March P, Goodacre R, et al. Raman activated cell ejection for isolation of single cells. Anal Chem. 2013;85:10697–10701. doi: 10.1021/ac403107p. [DOI] [PubMed] [Google Scholar]
- Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proc Natl Acad Sci USA. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zengler K, Toledo G, Rappe M, Elkins J, Mathur EJ, Short JM, et al. Cultivating the uncultured. Proc Natl Acad Sci USA. 2002;99:15681–15686. doi: 10.1073/pnas.252630999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Fakhrullin RF, Ozmen M, Wang H, Wang J, Paunov VN, et al. Functionalization of whole-cell bacterial reporters with magnetic nanoparticles. Microbial Biotech. 2011;4:89–97. doi: 10.1111/j.1751-7915.2010.00228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.