Abstract
Understanding environmental and biological influences on the dynamics of microbial communities has received great attention in microbial ecology. Here, utilizing large time-series 16S rRNA gene data, we show that in activated sludge of an environmentally important municipal wastewater treatment plant, 5-year temporal dynamics of bacterial community shows no significant seasonal succession, but is consistent with deterministic assemblage by taxonomic relatedness. Biological interactions are dominant drivers in determining the bacterial community assembly, whereas environmental conditions (mainly sludge retention time and inorganic nitrogen) partially explain phylogenetic and quantitative variances and indirectly influence bacterial assembly. We demonstrate a correlation-based statistical method to integrate bacterial association networks with their taxonomic affiliations to predict community-wide co-occurrence and co-exclusion patterns. The results show that although taxonomically closely related bacteria tend to positively co-occur (for example, out of a cooperative relationship), negative co-excluding correlations are deterministically observed between taxonomically less related species, probably implicating roles of competition in determining bacterial assembly. Overall, disclosures of the positive and negative species–species relations will improve our understanding of ecological niches occupied by unknown species and help to predict their biological functions in ecosystems.
Introduction
As the most popular biological wastewater treatment application worldwide, activated sludge has been in use for exactly a century to treat a large variety of municipal and industrial wastewaters to protect our environment and human health (Wagner and Loy, 2002; Seviour and Nielsen, 2010). Activated sludge is a unique artificial microbial ecosystem with high diversity (over 700 genera and thousands of operational taxonomic units (OTUs)) (Zhang et al., 2012) and with high biomass concentration (generally 2–10 g l−1) (Grady et al., 2011). The highly diverse bacterial communities in this engineered ecosystem efficiently aggregate themselves in the heterogeneous structure of activated sludge flocs to guarantee stable and good performance of biological wastewater treatment (Daims et al., 2006; Nielsen et al., 2012; Zhang et al., 2012; Ju et al., 2013a).
In the past two decades, great efforts have been made to isolate, characterize or quantify functional microorganisms directly involved in removing nutrients (nitrogen and phosphorus) (Bond et al., 1995; Juretschko et al., 1998; Daims et al., 2006), hydrolyzing and fermenting bacteria (Juretschko et al., 1998; Xia et al., 2008), floc-forming bacteria (Shin et al., 1993; Schmid et al., 2003) and detrimental microorganisms that raise bulking and foaming problems (Wanner, 1994; Guo and Zhang, 2012) in activated sludge. Despite a rapidly increasing knowledge concerning the biochemical and ecological characteristics of these key microbes in wastewater treatment, full-scale activated sludge-based wastewater treatment plants (WWTPs) with nutrient removal still suffer from a series of operational problems, such as process instability (Eikelboom, 2000), sludge settling problems (Jenkins et al., 2004) and poor performance in nutrient removal (Seviour and Nielsen, 2010). Therefore, more fundamental knowledge regarding the microbial structure is essential to elucidate the biological mechanisms behind the problems.
Recently, high-throughput culture-independent sequencing tools, such as 16S rRNA-based pyrosequencing, have been widely used to survey and improve our understanding of biodiversity in various municipal or industrial WWTPs (Kwon et al., 2010; McLellan et al., 2010; Ibarbalz et al., 2013). However, these descriptive studies have left many unanswered questions regarding the underlying species–species interactions and environment–species relations driving bacterial community assembly and dynamics (Ju et al., 2013b). Further, the long-term temporal variation in microbial constituents and interactions in a WWTP over environmental gradients remains largely unknown due to either the unavailability of sufficient time-series samples and long-term physicochemical and biological monitoring data, or the lack of a powerful analytical method for mining the huge sequence data derived from high-throughput sequencing.
In this study, we applied a correlation-based network analysis, which was based on over 570 000 bacterial 16S rRNA gene sequences from 58 activated sludge samples collected monthly from a typical municipal WWTP over 5 years (2007–2012), to explore the long-term bacterial assembly and temporal species–species associations (SSAs) in activated sludge. We described a correlation-based statistical method to integrate bacterial SSA networks with their taxonomic affiliations to reveal the non-random assembly patterns among species. Creatively interpreting OTUs in an SSA network may help to transit the microbial ecology of activated sludge from a plain description of microbial components to a framework of potential microbial interactions, in which ecological rules guiding the microbial assembly and functions could be speculated for the encouragement of further validation via specific experimental designs or directly applied to guide the system toward an optimized performance. Moreover, the synchronous and time-lagged correlations between potential influential factors (for example, plant operational parameters and wastewater quality) and bacterial species, as well as the contributions of influential factors on the temporal variability of biodiversity, were also explored in the network interface and correlated with the functional stability of activated sludge, which is a good model of the artificial microbial ecosystems.
Materials and Methods
Sample collection
The sampling site is a full-scale municipal WWTP (216 000 m3 day−1) in Shatin, Hong Kong (22°23′N 114°11′E), which treats saline domestic sewage containing ∼30% seawater. The plant is designed as an anoxic/oxic (A/O) process for carbon and nitrogen removal. Activated sludge samples were collected monthly from the middle of aerobic (oxic) tank from July 2007 to July 2012. The samples were fixed on site using an equal volume of 100% (v/v) ethanol. Then, the fixed samples in 50% ethanol were immediately delivered to the laboratory and stored in a −20 °C refrigerator. Throughout the entire sampling period, the plant could effectively remove 95–98% of CBOD (carbonaceous biochemical oxygen demand) in the sewage; however, the plant usually encountered unstable ammonium removal (47–98%) from every December to the next March, during which period, severe foaming (level 4–5, 5–10 cm, highly stable foams; Seviour and Nielsen, 2010) of activated sludge was usually observed (Supplementary Figure S1). Other detailed information concerning variation of plant operational parameters and physicochemical conditions, the sampling dates and treatment performance were summarized in Supplementary Table S1 and Supplementary Information S1.
DNA extraction and 454 pyrosequencing
For each activated sludge sample, DNA was first extracted from 2.0 ml sludge using a FastDNA @ SPIN Kit for Soil (MP Biomedicals, LLC, Illkirch, France), then the V3-V4 regions (∼465 nucleotides) of the 16S rRNA genes were amplified with 338F and 802R, and purified PCR amplicons were finally send out for pyrosequencing (see Supplementary Information S2 for detailed information). All 16S rRNA sequences from pyrosequencing have been deposited into the NCBI short-reads archive database with accession number SRR1154613.
Sequence processing
The raw sequencing data from 454 pyrosequencing were processed using the QIIME pipeline v 1.7.0 (Caporaso et al., 2010). In brief, the raw sequences were first quality trimmed into different samples, denoised by Denoiser (Reeder and Knight, 2010) and chimera checked using ChimeraSlayer (Quince et al., 2011) to yield clear reads. Then, the normalization of the clear sequences was conducted by randomly extracting 10 000 clean sequences from each sample data set (except for two samples from March and April, 2011, which have clean sequences of 9524 and 6257, respectively) to fairly compare all samples at the same sequencing depth. Next, the normalized sequences from all samples were clustered into OTUs using the Uclust algorithm at identity thresholds of 0.90 and 0.97 (Edgar, 2010), which approximately corresponding to the taxonomic levels of family and species for bacteria, respectively. Both the final 0.90 and 0.97-OTU tables consisted of 575 781 clear sequences, which were distributed into 2192 family-level and 5136 species-level bacterial OTUs, respectively. Of those OTUs, 861 and 2075, respectively, were represented by at least five OTUs. Finally, the taxonomic assignment of the representative sequences was conducted using the RDP Classifier program (80% confidence level) and GreenGenes database newly released in May 2013 (McDonald et al., 2012).
Statistical and network analysis
The core diversity analyses (using QIIME v 1.7.0), BIO-ENV analysis (using PRIMER-E v6 software (PRIMER-E Ltd, Ivybridge, UK), Spearman's rank correlation method and a significance test of 99 permutations to determine the combination of environmental variables that best explain community patterns) and correlation analysis (using R; Ihaka and Gentleman, 1996) between α-diversity indices and 15 environmental variables (Supplementary Table S1) were described in Supplementary Information S3.
For the network analysis, we first used extended Local Similarity Analysis to find the time-dependent correlations between species-level OTUs and environmental variables (Ruan et al., 2006; Xia et al., 2011). The Local Similarity Analysis calculates synchronous and time-delayed correlations based on the normalized ranked data and produces correlation coefficients that are analogous to a Spearman's ranked correlation (Ruan et al., 2006). Then, we used Cytopscape v2.8.3 (Shannon et al., 2003) for network visualization and topological analysis, as described in Supplementary Information S3. Here, we developed a python script to check statistically the observed (O) and random incidences (R) of bacterial co-occurrence and co-exclusion (Supplementary Information S5). The degree of the lack of agreement between O and R (O/R ratio; Supplementary Table S7; Supplementary information S7) is used as a benchmark for checking non-random assembly patterns in complex bacterial communities.
Results
Monthly, seasonal and inter-annual variability of bacterial structure
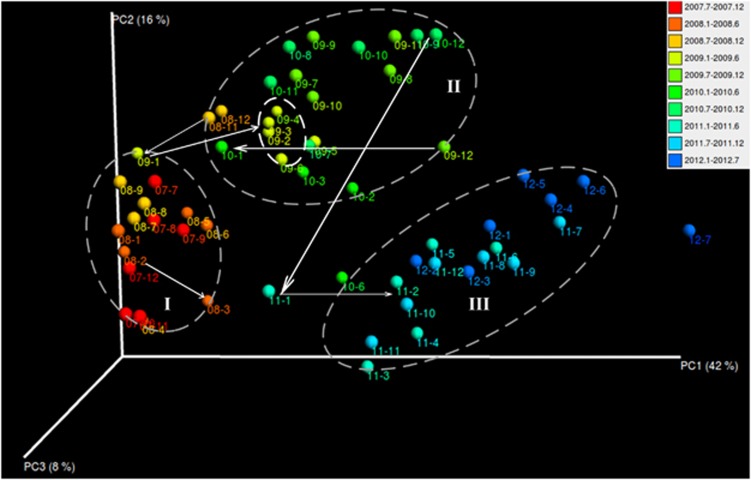
The seasonal succession of bacterial communities over various environmental gradients has been widely observed in many natural ecosystems, including soil (Lipson and Schmidt, 2004), oceans (Gilbert et al., 2011), lakes (Eiler et al., 2011; Paver et al., 2013). However, in an artificially controlled, half-close engineered biological wastewater treatment system, such as activated sludge, whether the dynamics of the bacterial community structure still follow a seasonal succession remains to be explored. Here, we compared the monthly, seasonal and inter-annual variations in bacterial diversity and abundance between time-series activated sludge samples using weighted UniFrac distances (considering both species phylogeny and abundance). Overall, temporal changes in the phylogenetic composition and abundance of family-level OTUs were quite high across the 5-year sampling period (Figure 1; see Supplementary Figure S3 for similar trends at the species level). Unlike the aforementioned natural ecosystems, there was no obvious seasonal succession of bacterial communities in the artificially controlled biotechnical ecosystem (for example, activated sludge) because samples collected from the same or adjacent months in different years were hardly clustered together. Instead, an annual shift in the communities proceeded in large leaps, that is, from ellipse I (2007–2008), via ellipse II (2009–2010) and finally into ellipse III (2011–2012), at particular time slots, which were usually found in winter months from around December to the next February (as illustrated by the white arrows within one or across different ellipses). This period was exactly the time when severe filamentous foaming was usually observed at the surface of the aeration tank (AT) of Shatin WWTP (also as indicated by the prevalence of bacterial OTUs potentially related to bulking and foaming; Supplementary Figure S6), accompanied by higher levels of NH3-N (Supplementary Figure S2a) and relatively lower η(NH3-N) (that is, removal efficiency of NH3-N; Supplementary Figure S2d; Supplementary Table S1) in the AT. Strikingly, the largest monthly variability was observed between November 2010 and February 2011, exactly when low F/M (Supplementary Figure S2b) bulking, together with filamentous foaming, occurred in the AT, indicating that these detrimental events greatly affect both the phylogenic and quantitative profiles of bacterial communities in activated sludge.
Figure 1.
Three-dimensional principal coordinate analysis (PCoA) plot showing the bacterial community difference of the 5-year activated sludge samples. The analysis was performed using the abundance matrix of family-level (0.90 similarity) OTUs in different samples, and pairwise community distances were determined using the weighted UniFrac algorithm.
Environmental influences on bacterial diversity and abundance
Correlation analysis between bacterial α-diversity matrices and physicochemical and operational variables showed that on the one hand, bacterial α-diversity in activated sludge was most closely positively correlated with sludge retention time (SRT) (coefficients of 0.60±0.03) and mixed liquor suspended solids (0.55±0.02), followed by NO3-N (0.55±0.03) and η(NH3-N) (0.54±0.10) (cluster I, Supplementary Figure S4; Supplementary Table S2); on the other hand, NO2-N-AT (NO2-N concentration in the AT, −0.61±0.05), influent CBOD (−0.57±0.03), F/M (−0.51±0.03) and NH3-N-AT (−0.51±0.08) showed significantly negative correlations with the α-diversity matrices (Supplementary Table S2 and cluster III, Supplementary Figure S4). Additionally, other physicochemical (for example, temperature, salinity and pH) and operational parameters (hydraulic retention time and dissolved oxygen) had either little or no statistically significant (P-value>0.05) correlation with the α-diversity matrices (cluster III, Supplementary Figure S4), indicating that these parameters may have little impact on activated sludge bacterial diversity.
Once we demonstrated that bacterial α-diversity in activated sludge could be influenced by environmental variables, a BIO-ENV trend correlation analysis was conducted to identify which combination of variables best explained changes in the bacterial abundance and in the diversity over time (that is, β-diversity). Supplementary Table S3 shows that operational parameters, in general, explained much better variation in the change in the bacterial structure than did physiochemical variables in the influent (for example, NH3-N and CBOD) or in the AT (for example, temperature, pH and salinity). The variables that best explained the weighted Unifrac distance between all pairs of 0.97 or 0.90-OTUs were identical and included SRT and F/M (BEST: 0.97-OTUs, rho=0.483; P-value=0.01; 0.90-OTUs, rho=0.471, P-value=0.01, respectively). Both level OTUs still correlated best with these two operational parameters when unweighted (without considering abundance) Unifrac distance matrices were used instead, except for the incorporation of some physiochemical parameters, which included influent NH3-N and CBOD and NO3-N in the AT.
Defining the bacterial community by frequency and functionality
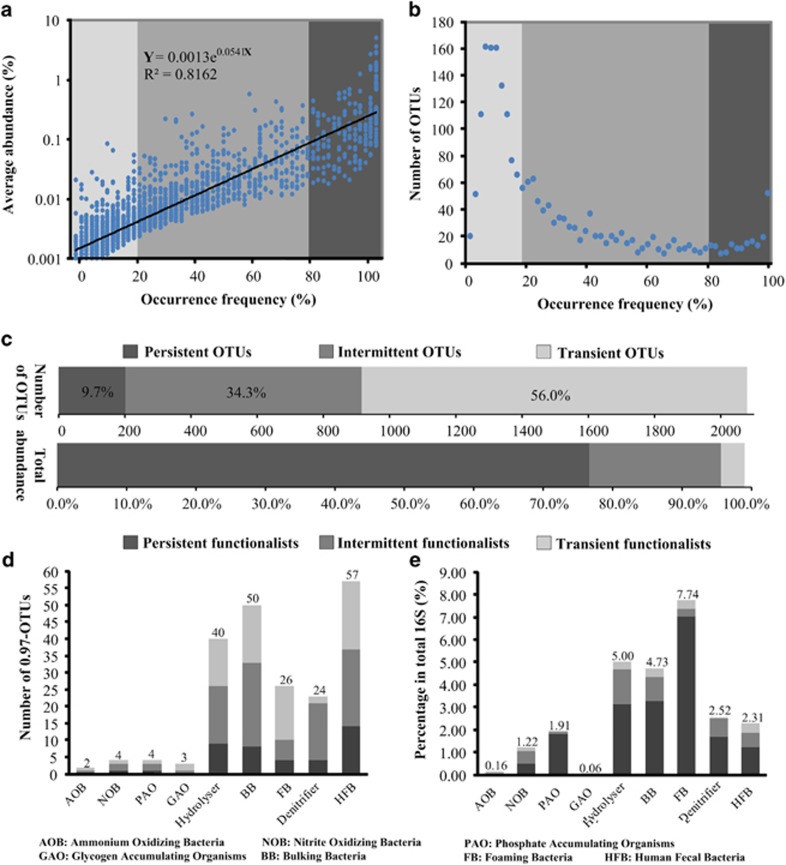
Partitioning ecological communities by their abundance and by their occurring frequency facilitate the exploration of the core and satellite species in many temporal or spatial scale data sets. In general, satellite species were typically transient and low in abundance, whereas core species were persistent in a given habitat and high in abundance (van der Gast et al., 2010). On the basis of the occurrence frequency, we divided the bacterial community of activated sludge into the following three arbitrarily defined ecological categories: persistent (⩾80% of months), intermittent (20–80% exclusive) and transient (⩽20%) OTUs (Figure 2). Overall, positive relations between the mean abundance and occurrence frequency have been observed, which were best fitted using the following exponential equation: Y=0.0013e0.0932X (R2=0.82) (Figure 2a). This revealed that persistent OTUs were generally more abundant than intermittent and transient OTUs, although the former ecological categories included a much lower number of OTUs (Figure 2b). Specifically, persistent OTUs merely occupied 9.7% of 2075 bacterial OTUs but accounted for 76.6% of all 16S rRNA gene sequences, implicating the existence of a high proportion of longstanding core species in activated sludge to sustain its long-term functional stability. By contrast, transient OTUs composed over 56.0% of all bacterial OTUs but merely occupied a minor proportion (3.4%) (Figure 2c), suggesting an extremely high diversity of minority species in activated sludge.
Figure 2.
Defining the 5-year activated sludge microbiome at the Shatin WWTP. The mean abundance of OTUs (a) and the number of OTUs (b) are shown relative to the 0.97-OTU's percentage occurrence (X axis). The occurrence frequency was calculated by dividing the number of months in which an OTU was detected by the number of total months. The abundance and the number of OTUs of different occurrence frequencies (Persistent, Intermittent and Transient) are shown in (c). Diversity (d) and mean abundance (e) of potential functional bacterial groups (in the monthly samples they were detected) of different occurrence frequencies (see Supplementary Table S4 for the list of the functional bacteria).
The taxonomic distribution of persistent, intermittent and transient OTUs was slightly different from each other (Supplementary Figure S5). Some bacterial classes, such as Alphaproteobacteria, Actinobacteria and Nitrospira, tended to be more persistent, whereas others (for example, Deltaproteobacteria, most sulfate-reducing bacteria in this class, which depend on the aerobic/anaerobic condition) were more transitory over the 5-year sampling period. Persistent OTUs were primarily affiliated with 12 classes, such as Alphaproteobacteria, Actinobacteria, Gammaproteobacteria, Acidimicrobia, Sphingobacteria and Anaerolineae, which are also key bacterial groups commonly found in activated sludge of different municipal WWTPs (Wagner and Loy, 2002; Sanapareddy et al., 2009; Xia et al., 2010; Zhang et al., 2012). By contrast, intermittent and transient OTUs included a larger proportion (30–35%, Supplementary Table S4) of populations from other bacterial classes, such as TM7-1, TM7-3, Synergistia, Verrucomicrobiae and Chlamydia (Supplementary Table S4). Noteworthy, the predominance of Alphaproteobacteria predominated over Betaproteobacteria is mainly attributed by the salinity (∼1%) of wastewater in Shatin WWTPs, as we discussed previously (Zhang et al., 2012).
Further comparison with previous studies (Jenkins et al., 2004; McLellan et al., 2010; Seviour and Nielsen, 2010; Guo et al., 2013) implicated high diversity (209 OTUs, Figure 2d) and a considerable proportion (averaged 25.7% of bacterial 16S rRNA sequences, Figure 2e) of potentially functional bacteria (see Supplementary Table S4 for a full list) in activated sludge, including nitrifying bacteria (two OTUs of ammonia-oxidizing bacteria (AOB); four OTUs of nitrite-oxidizing bacteria (NOB)), phosphate accumulating organisms (four OTUs), glycogen accumulating organisms (three OTUs), hydrolyzers (40 OTUs), bulking and foaming bacteria (BFB, 76 OTUs), denitrifiers (24 OTUs) and fermentative human-fecal bacteria (57 OTUs). Although persistent functionalists (42 OTUs, 20% of the total number of functional OTUs) were represented by much fewer OTUs than intermittent (94) and transient functionalists (74), they accounted for over 70% of the 16S sequences of all functional bacteria, implicating the longstanding co-existence of a core set of functional bacteria in activated sludge.
Figures 3e and 4f show OTUs related to bulking and foaming (averaged 12.5% of bacterial 16S rRNA sequences), which primarily consisted of filamentous Microthrixaceae (averaged 2.5%, 7 OTUs), Caldilinea in the phylum Chloroflexi (1.5%, 23 OTUs), hydrophobic Mycobacterium (3.9%, 18 OTUs) and filamentous, hydrophobic Gordonia (3.8%, 7 OTUs). Although these notorious and always filamentous BFB, if present outside the bioflocs, can cause settling (bulking) and foaming problems and deteriorate effluent quality, it is believed that BFB-related filaments are usually presented in ‘well-behaved' activated sludge and have versatile roles (for example, bioflocs formation; Kragelund et al., 2007; lipids or oleic acid degradation, Nielsen et al., 2010) other than being detrimental. Moreover, the protein hydrolyzers Saprospiraceae (phylum Bacteroidetes) were highly diverse (40 OTUs, Figure 2d) and abundant (5.0%, Figure 2e) in activated sludge. Intriguingly, the poor representativeness of AOB in activated sludge hardly hindered NH3-N oxidization (as indicated by the continuously detected nitrite in the AT; Supplementary Table S1), most likely justifying previous findings that Nitrosomonas has high transcription activity in spite of its low abundance in activated sludge (Yu and Zhang, 2012). It is also possible that there are unassigned or unidentified AOB.
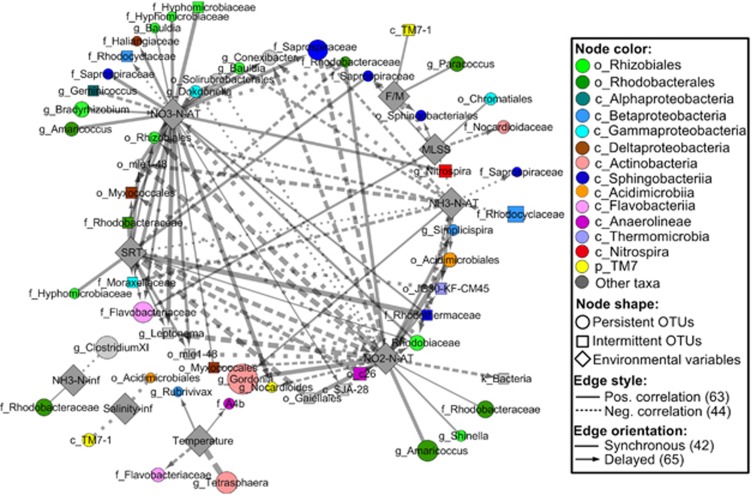
Figure 3.
Environment–species network uncovered synchronous and delayed relations between bacteria and environmental variables in activated sludge. Only local similarity that were statistically significant (P-value ⩽0.05, Q-value ⩽0.01) and strong (local similarity ⩾0.6 or ⩽−0.6) are shown, resulting in networks composed of 67 nodes and 115 edges. Node label stands for an environmental variable or the lowest classifiable taxonomic rank (p_, c_, o_, f_ and g_ representing phylum, class, order, family and genus, respectively) of 0.97-OTU, and node size of each OTU is proportional to its average abundance in the samples it was detected. The line thickness is proportional to the absolute value of local similarity, and line arrows indicate a 1-month shift/delay in the correlation.
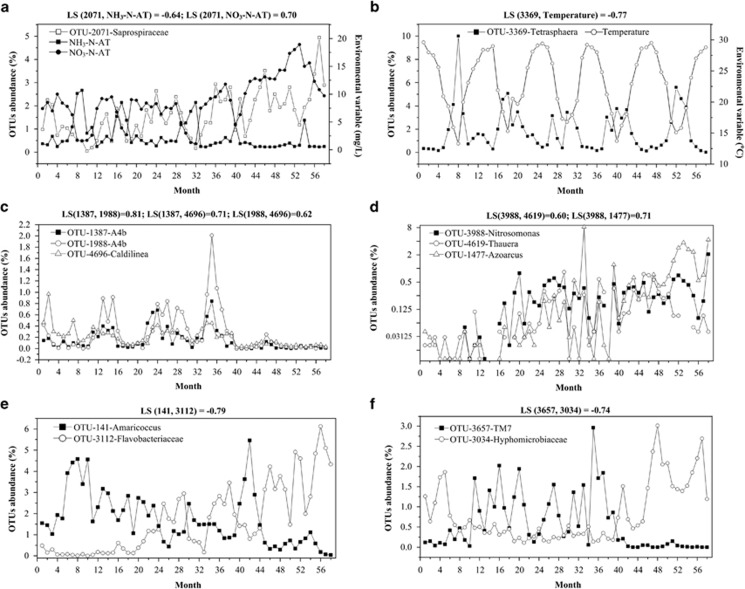
Figure 4.
Examples of strong and significant correlations between species and environment (a, b) and between species and species (c, d, e and f) in 58 activated sludge during 5-year sampling period. An local similarity is considered as strong and significant when local similarity ⩾0.6 or ⩽−0.6, and P-value⩽0.05 and Q-value⩽0.01. The OTU abundance is calculated as the number of sequences assigned to each OTU divided by the total number of 16S rRNA gene sequences in that sample. The missing points in the (d) represent OTUs abundance of 0.
Environment–species association and SSA
Tracking correlations between microorganisms and between microorganisms and their surrounding environments in a network interface provide insights into microbial interactions, as well as an awareness of the conditions that favor or disfavor particular microbes. Restricting the analysis to the environment–species association networks (Figure 3; Supplementary Figure S7), strong correlations between variables including NH3-N-AT, NO2-N-AT, NO3-N-AT and SRT and bacterial taxa were the most frequent, followed by those correlations between taxa and other variables, such as mixed liquor suspended solids, temperature and F/M. Few significant and no significant, strong correlations were observed between bacterial taxa and other variables, including hydraulic retention time, dissolved oxygen, pH and SCOD influent (Supplementary Figure S9). Strikingly, variables with more edges connected to bacterial taxa, that is, NH3-N-AT, NO2-N-AT, NO3-N-AT and SRT, tend to have much better correlation with bacterial α-diversity than those variables with fewer edges (for example, hydraulic retention time, dissolved oxygen and pH) (Supplementary Figure S4), confirming that SRT and inorganic nitrogen in the AT, compared with other environmental variables, may much more significantly affect the community structure.
The mathematical statistics of the environment–species association network (Figure 3) indicate that bacterial taxa were primarily connected to SRT and NO3-N-AT via positive correlations (both synchronous and delayed). Thus, the increase in SRT and NO3-N concentrations may promote the accumulation of many bacterial OTUs, such as seven Rhizobiales-affiliated OTUs (f_Hyphomicrobiaceae, g_Bauldia, g_Bradyrhizobium and f_Rhodobiaceae), two hydrolyzer-affiliated OTUs (f_Saprospiraceae), two Chloroflexi-affiliated OTUs (o_mle1-48) and one NOB-affiliated OTU (g_Nitrospira). In contrast, negative correlations dominated the correlations between NO2-N, NH3-N and bacterial OTUs, revealing that the buildup of NO2-N and NH3-N in the AT tends to reduce the abundances of certain bacterial groups. In addition, some bacterial OTUs, such as those bacterial OTUs affiliated with g_Conexibacter, o_Solirubrobacterales, f_Saprospiraceae (Figure 4a) and f_Moraxellaceae, were positively correlated with NO3-N-AT with delay, on the one hand, and negatively correlated with NO2-N-AT and/or NH3-N-AT on the other hand, indicating that these bacteria thrive when AT is relatively high in nitrate but low in nitrite and ammonium.
The analysis of the integrated network (Supplementary Figure S7) composed of positive-correlated nodes extracted from the environment–species association network and the SSA network shows that two groups of environmental variables, that is, (I) NH3-N-AT & NO2-N-AT and (II) SRT and NO3-N, exerted considerable impacts on the overall co-occurrence patterns of the bacterial community, but to different degrees. Topological partitioning shows that the network could be divided into two large clusters (or modules). In the upper cluster, NH3-N-AT and NO2-N-AT were connected to only a small proportion of OTUs (12 nodes) on the right corner of the cluster, reflecting their limited influence on the cluster structure. By contrast, in the lower cluster, SRT and NO3-N were correlated with more OTUs (28 nodes), many of which are hub OTUs (nodes with a high number of connections, also known as high degree nodes) in the center of the cluster (comparing Supplementary Table S6 against Supplementary Figure S7). Via hubs, the impacts of these parameters could spread rapidly to reach neighboring OTUs in other parts of the cluster. Further topological and taxonomic comparison lends interesting and novel insights into the community structure. For instance, almost all OTUs of three bacterial phyla, including TM7, Chloroflexi (classes Anaerolineae and Thermomicrobia) and Actinobacteria (classes Actinobacteria and Acidimicrobiia), only occurred in the relatively loosely (70 OTU nodes, 441 edges) connected upper cluster, whereas almost all OTUs of two classes, Sphingobacteria and Gammaproteobacteria, were only found in the densely connected (70 OTU nodes, 441 edges) lower cluster.
Preferential attachment of bacterial nodes revealed deterministic co-occurrence and co-exclusion patterns
We constructed correlation-based SSA networks and used a statistical method to test both the co-occurrence and co-exclusion patterns between bacterial communities. The resulting entire SSA network (local similarity ⩾0.6 or ⩽−0.6) consisted of 150 nodes and 913 edges (average degree of 12.17 and average shortest path length of 2.866, Table 1; see Supplementary Figure S10 for the cumulative degree distribution and exponentially decreased average shortest path length with increasing node degree), with 557 positive interactions between 145 OTUs compared with 356 negative interactions between 117 OTUs (Table 1). The higher clustering coefficients of the entire and positive SSA networks, compared with Erdös-Réyni random networks, with small characteristic shortest path lengths (Table 1), similar to random graphs, suggest that the network has ‘small-world' properties, that is, nodes that are highly interconnected (clustered) more than would be expected by chance alone. By contrast, the negative SSA, which reflects species–species exclusion patterns, tends to be unclustered (an average clustering coefficient of 0) and less modularized (modularity: 0.457, Table 1; values >0.4 suggest that the network has a modular structure; Newman 2006), compared with the highly clustered, more modularized (modularity: 0.586) positive SSA, revealing distinct characteristics of positive and negative interactions between species.
Table 1. Comparison of topological properties of species–species association (SSA) networks of activated sludge with their corresponding Erdös-Réyni random networks of identical size.
| SSAa | Avg. clustering coefficient | Avg. shortest path length | Network heterogeneity | Network centralization | Network diameter | Modularity | Graph density | Avg. degree |
|---|---|---|---|---|---|---|---|---|
| Whole | 0.466 | 2.866 | 0.785 | 0.176 | 8 | 0.424±0.002 | 0.082 | 12.170 |
| Random | 0.083±0.00 | 2.262±0.01 | 0.271±0.022 | 0.060 | 4 | 0.239±0.006 | 0.082 | 12.170 |
| Positive | 0.477 | 4.615 | 0.818 | 0.135 | 14 | 0.586±0.001 | 0.053 | 7.630 |
| Random | 0.053±0.00 | 2.667±0.00 | 0.331±0.014 | 0.052 | 5 | 0.316±0.004 | 0.053 | 7.630 |
| Negative | 0 | 3.491 | 0.732 | 0.140 | 9 | 0.457±0.001 | 0.052 | 6.085 |
| Random | 0.053±0.00 | 2.830±0.00 | 0.390±0.013 | 0.061 | 6 | 0.357±0.004 | 0.052 | 6.085 |
Whole SSA: 150 nodes, 913 edges; Positive SSA: 145 nodes, 557 edges; Negative SSA: 117 nodes, 356 edge.
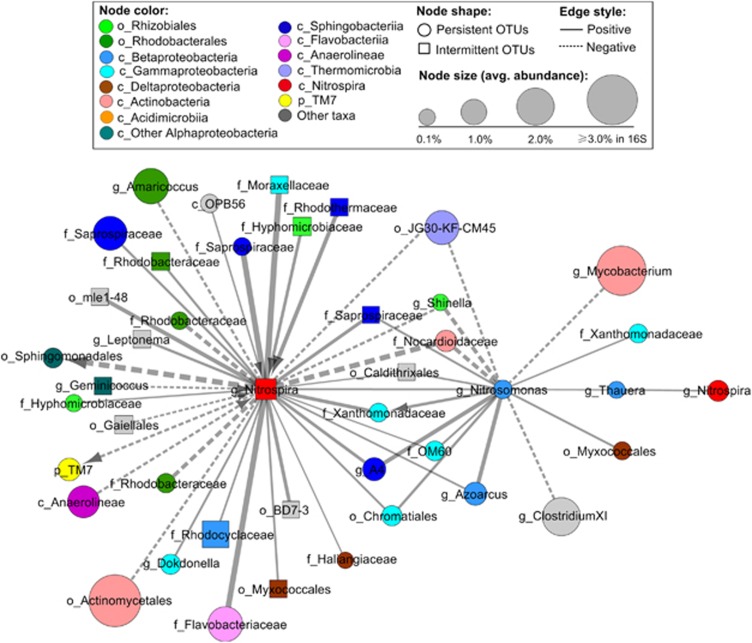
The further structural and statistical analysis showed that OTUs from the same taxa (from phylum down to the order level) tended to co-occur (positive correlations, Supplementary Figure S7) and that OTUs from different taxa tended to co-exclude (negative correlation, Supplementary Figure S8) more than would be expected by chance when considering taxa frequency and random associations, although their degrees of co-occurrence or co-exclusion (as measured by O/R ratio) differed (Supplementary Tables S7 and S8). On the one hand, statistical and structural analysis of the positive SSA network showed that OTUs within two orders, that is, Rhizobiales (O/R=4.3) and Rhodobacterales (O/R=2.0, family Rhodobacteraceae), and four classes, that is, Sphingobacteria (O/R=5.1, primarily family Saprospiraceae, Supplementary Figure S11b), Anaerolineae (O/R=3.6; Figure 4c; Supplementary Figure S11a), Gammaproteobacteria (O/R=4.3) and Betaproteobacteria (O/R=2.5), tended to co-occur more than would be expected by chance (Supplementary Table S6). On the other hand, the statistical analysis of the negative SSA network demonstrated that OTUs from different taxa, (I) Anaerolineae and Rhodobacterales (O/R=2.5, Supplementary Figure S11a), (II) Anaerolineae and Betaproteobacteria (particularly family Xanthomonadaceae) (O/R=2.7, Supplementary Figure S11a), (III) Flavobacteria and Thermomicrobia (O/R=7.1) and (IV) Rhizobiales (particularly family Hyphomicrobiaceae, for example, Figure 4f) and TM7 (O/R=1.9), tend to co-exclude more than would be expected by chance (Supplementary Table S6). Strikingly, apart from the deterministic patterns of intra-taxon co-occurrence and inter-taxa co-exclusion, higher incidences of inter-taxa co-occurrence, more than would be expected by chance, were also observed between OTUs of different taxa, including (I) Sphingobacteria (primarily Saprospiraceae) and Gammaproteobacteria (Supplementary Figure S11b), (II) Anaerolineae and TM7 (Supplementary Figure S11a), (III) Nitrospira and Sphingobacteria or Gammaproteobacteria (Figure 5) and (IV) other pairs of taxa.
Figure 5.
Preferential attachment of bacterial nodes in the species–species association network revealed deterministic bacterial co-occurrence (solid edges) and co-exclusion (dash edges) patterns in activated sludge. Only correlations that were statistically significant (P-value ⩽0.05, Q-value⩽0.01) and strong (local similarity ⩾0.6 or ⩽−0.6) are shown. Node label stands for the lowest classifiable taxonomic rank (p_, c_, o_, f_ and g_ representing phylum, class, order, family and genus, respectively) of 0.97-OTU. Nitrite-oxidizing bacteria Nitrospira tends to co-occur with OTUs of Sphingobacteria, Gammaproteobacteria and Betaproterobacteria (for example Nitrosomonas, Thauera and Azoarcus), but co-exclude with OTUs of Actinobacteria. The line thickness is proportional to the absolute value of local similarity. The arrow indicates the time-lagged correlations with arrow pointing to the lagged OTU.
Discussion
It has long been assumed that differences in species abundance in microbial communities reflect changes in environmental conditions. Although this statement emphasizes the significance of environmental impacts, it ignores the influences of interior species–species interactions on the community assembly. Currently, it remains difficult to predict which bacteria can co-exist or co-exclude steadily over temporal gradients of environmental variables, let alone the cooperative or competitive relations among these bacteria, causing the artificial and purposeful manipulation of engineered microorganisms (for example, in biological WWTPs) extremely challenging. In this study, utilizing large time-series 16S rRNA gene sequencing data, we constructed a bacterial SSA network consisting of 3899 pairwise significant SSA correlations (among which 913 correlations are strong, with coefficients ⩾0.6) connecting 170 species-level OTUs. We find that although taxonomically closely related bacteria tend to co-occur out of cooperative relations or a similar niche preference, co-excluding negative correlations are usually deterministically observed between taxonomically less related species, most likely implicating a role of competition in community assembly. Moreover, the highly clustered and modularized structure (also characterized by nodes connected by many closed triangle or polygonal loops) of the positive SSA network is completely different from the unclustered and less modularized structure of the negative SSA network. This result indicated that positive interactions (primarily cooperative relations) among bacteria are usually established by a cluster of multiple highly interacted species with similar ecological niches, whereas bacteria are likely to form relatively simple and open ‘one-to-many' or ‘one-to-one' negative interactions (most likely competition) with one another.
The non-random co-occurrence patterns between taxonomically closely related bacteria can be derived from taxa sharing similar ecological niches or cooperative relations, as noted elsewhere (Barberán et al., 2011; Ju et al., 2013b). One typical example is the intra-taxon co-occurrence observed among Betaproteobacteria AOB, Nitrosomonas and two other Betaproteobacteria-affiliated denitrifying bacteria, that is, Thauera and Azoarcus (Figures 4d and 5), which resulted from a syntrophic relation, in which nitrite released by the former is utilized by the latter. Other examples are the strong intra-taxon co-occurrences evident in the class Anaerolineae or in the order Sphingobacteria, which are likely derived from the preference for similar niches, as supported by the assemblage of all OTUs of Anaerolineae in the upper module and all OTUs of Sphingobacteria in the lower module (Supplementary Figure S7). Overall, the deterministic intra-taxon co-occurrence patterns evident between taxonomically closely related species were in agreement with the widespread ecological phenomenon of phylogenetic clustering (that is, co-occurring species being more closely related than would be expected by chance), which most likely implicate the importance of environmental filtering and niche differentiation in shaping the assembly of bacterial communities in activated sludge (Losos, 2008; Philippot et al., 2010).
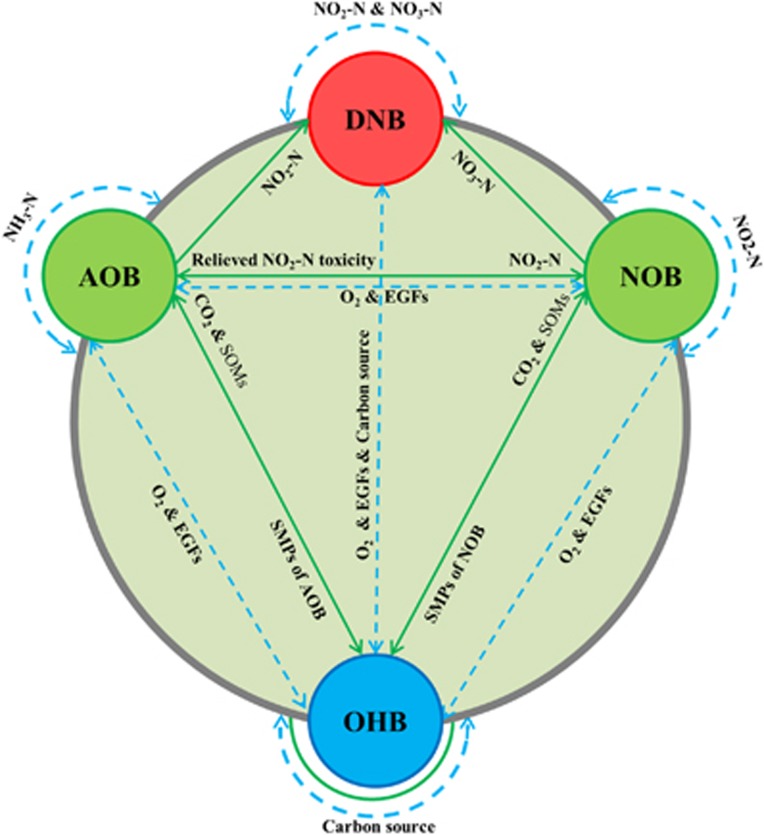
Moreover, deterministic inter-taxa co-exclusion patterns are prevalent among taxonomically less related (or distanced) species. This phenomenon, together with our observation of almost no significant negative correlations between OTUs with the same taxa, suggested that co-exclusion primarily occurred between bacteria that were taxonomically distanced. In a high-biomass, resource-limited biotechnical system, such as activated sludge, negative interactions are ubiquitous within or between functional and detrimental bacteria (for example, nitrifiers vs heterotrophs (Nitrosomonas vs Clostridium XI; Nitrospira vs TM7; Figure 5); BFB vs nutrient-removal organisms (Mycobacterium vs Nitrosomonas; Caldilinea vs Azoarcus; Figure 5) and floc-forming vs filamentous microbes), which, in general, reflect fierce competition between these bacteria for limited resources of essential growth factors, dissolved oxygen, carbon source or other substrates (Daims et al., 2006, Seviour and Nielsen, 2010) (Figure 6).
Figure 6.
Schematic diagram of potential bacterial interactions among autotrophic and heterotrophic bacteria in a nitrogen-removal activated sludge treatment plant. Positive and negative interactions are namely illustrated by green and blue lines with arrows revealing an exchange or a competition for substrates or nutrients. Mutualistic symbiosis: AOB provide nitrite (NO2-N) for NOB, which in turn remove NO2-N and thus relieve its inhibitory effects on AOB. Commensalism: biodegradation of macromolecules (for example, protein hydrolysis by Saprospiraceae) into small organic molecules, which are easily available to other heterotrophic bacteria (OHB). Other cooperative interactions: (I) AOB and NOB provide NO2-N and NO3-N for denitrifying bacteria (DNB), (II) HB release CO2 which is assimilated by autotrophic AOB and NOB, and (III) AOB and NOB release soluble microbial products (SMPs), which are utilized by HB as carbon sources. Competition: (I) AOB and NOB compete with each other for carbon sources and oxygen and with HB for oxygen and essential growth factors (EGFs); (II) different AOB, NOB or DNB compete with each other for NH3-N, NO2-N or NO2-N/NO3-N, respectively; and (III) different heterotrophic DNB or OHB compete with each other for carbon sources. A color version of this figure is available on The ISME Journal online.
Strikingly, non-random inter-taxa co-occurrence patterns between taxonomically distanced bacteria in activated sludge most likely suggest species interactions, such as mutualism and commensalism. For example, AOB Nitrosomonas co-occurs with NOB Nitrospira (Figure 5) out of a relation of mutualistic symbiosis, in which AOB provides nitrite for NOB, and in return, NOB removes nitrite to prevent its inhibition on AOB. The co-occurrence between the commensal bacteria Flavobacteria and the protein-hydrolyzing bacteria Saprospiraceae (Supplementary Figure S11b) is a typical instance of commensalism, that is, the former cross-feed on amino acids from protein hydrolyzed by the latter, as is often found in biodegradation (Faust and Raes, 2012). On the basis of these meaningful observations, we predict that the deterministic co-occurrence observed between TM7 and Chloroflexi (Anaerolineae; Supplementary Figure S11a) could be derived from a cooperative relation. These two types of bacteria have been detected by FISH to co-occur in filament epiphytic protein-hydrolyzing communities of five full-scale WWTPs (Xia et al., 2007). The recent construction and analysis of TM7 genomes indicate that microaerophilic TM7 often buries its coccus cells deeply in flocs and primarily ferments glucose and other sugars in bioreactors (Albertsen et al., 2013). On the basis of this knowledge, it is speculated that TM7 may colonize to filamentous Chloroflexi in activated sludge bioflocs because of the benefits of minimized oxygen exposure and the easier adsorption of organic molecules (usually in the form of colloids and small particles) caught by the filaments from the bulk wastewater. In return, fermentative TM7 may provide substrates to its filamentous host. Overall, our observation that co-occurring bacteria tend to be taxonomically less related essentially resembles the phylogenetic overdispersion of co-occurring species of plants or animals observed in many studies (Losos, 2008; Bennett et al., 2013), revealing that negative interactions (such as competition) have an important impact on the community assembly of large varieties of biological communities from microorganisms (for example, bacteria) to macroscopic plants and animals.
Finally, the dominance of species–species correlations over those between environment and species, as well as the lack of strong correlations between environmental variables and many persistent OTUs, may relate to the fact that the activated sludge is kept operated/cultured in the artificially controlled (thus relatively stable) Shatin WWTP, where climates show no significant seasonal variations, thus, indicating that the variations in bacterial abundance were primarily driven more by biological interactions than by temporal changes in the physico-chemical and operational parameters. It is also possible that unmeasured influential variables could exist and contribute to instances of bacterial occurrence or changes in abundance. Among all 15 measured variables, SRT and inorganic nitrogen (for example, NH3-N and NO3-N) in the AT best explain partial phylogenetic and quantitative variances and indirectly affect bacterial assembly. Generally, SRT selects microbial populations based on their growth rates, and thus can strongly select against slowly growing nitrifying bacteria (especially NOB), particularly in the case of low temperatures in winter when growth rate is lower. The positive correlations between (I) SRT, mixed liquor suspended solids, NO3-N, η(NH3-N) and (II) bacterial α-diversity indicate that appropriately extending SRT or maintaining sufficient biomass is beneficial for improving bacterial biodiversity and ammonium removal in activated sludge. This result, in turn, helps us to explain or to predict how the system performance (for example, NH3-N removal) can respond to changes in operational conditions, considering a close link between microbial diversity and process robustness. From an engineer's perspective, realizing that (I) maintaining rationally assembled microbial community structure (in terms of both diversity and abundance) is critical to sustaining long-term satisfactory and steady performance and that (II) the community structure is highly dependent on the biological species–species interactions, which can be manipulated indirectly via the control of certain key operational parameters (for example, SRT and organic loadings) and physicochemical conditions (for example, inorganic nitrogen concentrations), can change ways of thinking when we operate WWTPs. For example, in the case of an incomplete nitrification event, priority could be given to thinking of operational or chemical measures to inhibit the growth of potential competing bacteria (for example, BFB) to promote the representativeness and effectiveness of functional nitrifying bacteria. Overall, the fulfillment of these innovative attempts in microbial manipulation toward better process performance should be established by acquiring more fundamental knowledge regarding the complex interactions among microbial communities. More studies or scientific attempts toward this inspiring goal are warranted.
Acknowledgments
We would like to thank GRF of Hong Kong for financial support (7195/06E, 7197/08E, 7202/09E, 7198/10E, 7201/11E and 7190/12E) on this long-term research project. FJ would like to thank the University of Hong Kong for the postgraduate scholarship. We would like to thank Dr Feng GUO for his help on DNA extraction and PCR, plus Dr Lin YE and Miss Qingmei YAN for help on sludge sampling. We also wish to thank DSD (Drainage Services Department) for sharing of operation data.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Author contributions
TZ designed the experiments, supervised the data analysis and revised the final manuscript. FJ performed the sampling, bioinformatics, statistics and network analysis of the raw sequencing data, wrote python scripts for data analysis and wrote the manuscript draft.
Supplementary Material
References
- Albertsen M, Hugenholtz P, Skarshewski A, Nielsen KL, Tyson GW, Nielsen PH. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol. 2013;31:533–538. doi: 10.1038/nbt.2579. [DOI] [PubMed] [Google Scholar]
- Barberán A, Bates ST, Casamayor EO, Fierer N. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 2011;6:343–351. doi: 10.1038/ismej.2011.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett JA, Lamb EG, Hall JC, Cardinal-McTeague WM, Cahill JF. Increased competition does not lead to increased phylogenetic overdispersion in a native grassland. Ecol Lett. 2013;16:1168–1176. doi: 10.1111/ele.12153. [DOI] [PubMed] [Google Scholar]
- Bond PL, Hugenholtz P, Keller J, Blackall LL. Bacterial community structures of phosphate-removing and non-phosphate-removing activated sludges from sequencing batch reactors. Appl Environ Microbiol. 1995;61:1910–1916. doi: 10.1128/aem.61.5.1910-1916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daims H, Taylor MW, Wagner M. Wastewater treatment: a model system for microbial ecology. Trends Biotechnol. 2006;24:483–489. doi: 10.1016/j.tibtech.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Eikelboom DH. Process Control of Activated Sludge Plants by Microscopic Investigation. IWA Publishing: London, UK; 2000. [Google Scholar]
- Eiler A, Heinrich F, Bertilsson S. Coherent dynamics and association networks among lake bacterioplankton taxa. ISME J. 2011;6:330–342. doi: 10.1038/ismej.2011.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust K, Raes J. Microbial interactions: from networks to models. Nat Rev Microbiol. 2012;10:538–550. doi: 10.1038/nrmicro2832. [DOI] [PubMed] [Google Scholar]
- Gilbert JA, Steele JA, Caporaso JG, Steinbrück L, Reeder J, Temperton B, et al. Defining seasonal marine microbial community dynamics. ISME J. 2011;6:298–308. doi: 10.1038/ismej.2011.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grady Jr, CL, Daigger GT, Love NG, Filipe CD, Leslie Grady C. Biological Wastewater Treatment. IWA Publishing: London, UK; 2011. [Google Scholar]
- Guo F, Zhang T. Profiling bulking and foaming bacteria in activated sludge by high throughput sequencing. Water Res. 2012;46:2772–2782. doi: 10.1016/j.watres.2012.02.039. [DOI] [PubMed] [Google Scholar]
- Guo F, Ju F, Cai L, Zhang T. Taxonomic precision of different hypervariable regions of 16S rRNA gene and annotation methods for functional bacterial groups in biological wastewater treatment. PLoS One. 2013;8:e76185. doi: 10.1371/journal.pone.0076185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarbalz FM, Figuerola EL, Erijman L. Industrial activated sludge exhibit unique bacterial community composition at high taxonomic ranks. Water Res. 2013;47:3854–3864. doi: 10.1016/j.watres.2013.04.010. [DOI] [PubMed] [Google Scholar]
- Ihaka R, Gentleman R. R: a language for data analysis and graphics. J Comput Graph Stat. 1996;5:299–314. [Google Scholar]
- Jenkins D, Richard MG, Daigger GT. Manual on the Causes and Control of Activated Sludge Bulking, Foaming, and Other Solids Separation Problems. IWA Publishing: London, UK; 2004. [Google Scholar]
- Ju F, Guo F, Ye L, Xia Y, Zhang T. Metagenomic analysis on seasonal microbial variations of activated sludge from a full-scale wastewater treatment plant over 4 years. Environ Microbiol Rep. 2013;6:80–89. doi: 10.1111/1758-2229.12110. [DOI] [PubMed] [Google Scholar]
- Ju F, Xia Y, Guo F, Wang Z, Zhang T. Taxonomic relatedness shapes bacterial assembly in activated sludge of globally distributed wastewater treatment plants. Environ Microbiol. 2013;16:2421–2432. doi: 10.1111/1462-2920.12355. [DOI] [PubMed] [Google Scholar]
- Juretschko S, Timmermann G, Schmid M, Schleifer K-H, Pommerening-Röser A, Koops H-P, et al. Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl Environ Microbiol. 1998;64:3042–3051. doi: 10.1128/aem.64.8.3042-3051.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kragelund C, Remesova Z, Nielsen JL, Thomsen TR, Eales K, Seviour R, et al. Ecophysiology of mycolic acid-containing Actinobacteria (Mycolata) in activated sludge foams. FEMS Microbiol Ecol. 2007;61:174–184. doi: 10.1111/j.1574-6941.2007.00324.x. [DOI] [PubMed] [Google Scholar]
- Kwon S, Kim T-S, Yu GH, Jung J-H, Park H-D. Bacterial community composition and diversity of a full-scale integrated fixed-film activated sludge system as investigated by pyrosequencing. J Microbiol Biotechnol. 2010;20:1717–1723. [PubMed] [Google Scholar]
- Lipson DA, Schmidt SK. Seasonal changes in an alpine soil bacterial community in the Colorado Rocky Mountains. Appl Environ Microbiol. 2004;70:2867–2879. doi: 10.1128/AEM.70.5.2867-2879.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losos JB. Phylogenetic niche conservatism, phylogenetic signal and the relationship between phylogenetic relatedness and ecological similarity among species. Ecol Lett. 2008;11:995–1003. doi: 10.1111/j.1461-0248.2008.01229.x. [DOI] [PubMed] [Google Scholar]
- McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan S, Huse S, Mueller-Spitz S, Andreishcheva E, Sogin M. Diversity and population structure of sewage-derived microorganisms in wastewater treatment plant influent. Environ Microbiol. 2010;12:378–392. doi: 10.1111/j.1462-2920.2009.02075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MEJ. Modularity and community structure in networks. Proc Natl Acad Sci. 2006;103:8577–8582. doi: 10.1073/pnas.0601602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen PH, Mielczarek AT, Kragelund C, Nielsen JL, Saunders AM, Kong Y, et al. A conceptual ecosystem model of microbial communities in enhanced biological phosphorus removal plants. Water Res. 2010;44:5070–5088. doi: 10.1016/j.watres.2010.07.036. [DOI] [PubMed] [Google Scholar]
- Nielsen PH, Saunders AM, Hansen AA, Larsen P, Nielsen JL. Microbial communities involved in enhanced biological phosphorus removal from wastewater—a model system in environmental biotechnology. Curr Opin Biotechnol. 2012;23:452–459. doi: 10.1016/j.copbio.2011.11.027. [DOI] [PubMed] [Google Scholar]
- Paver SF, Hayek KR, Gano KA, Fagen JR, Brown CT, Davis-Richardson AG, et al. Interactions between specific phytoplankton and bacteria affect lake bacterial community succession. Environ Microbiol. 2013;15:2489–2504. doi: 10.1111/1462-2920.12131. [DOI] [PubMed] [Google Scholar]
- Philippot L, Andersson SG, Battin TJ, Prosser JI, Schimel JP, Whitman WB, et al. The ecological coherence of high bacterial taxonomic ranks. Nat Rev Microbiol. 2010;8:523–529. doi: 10.1038/nrmicro2367. [DOI] [PubMed] [Google Scholar]
- Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ. Removing noise from pyrosequenced amplicons. BMC Bioinformatics. 2011;12:38. doi: 10.1186/1471-2105-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder J, Knight R. Rapid denoising of pyrosequencing amplicon data: exploiting the rank-abundance distribution. Nat Methods. 2010;7:668–689. doi: 10.1038/nmeth0910-668b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Q, Dutta D, Schwalbach MS, Steele JA, Fuhrman JA, Sun F. Local similarity analysis reveals unique associations among marine bacterioplankton species and environmental factors. Bioinformatics. 2006;22:2532–2538. doi: 10.1093/bioinformatics/btl417. [DOI] [PubMed] [Google Scholar]
- Sanapareddy N, Hamp TJ, Gonzalez LC, Hilger HA, Fodor AA, Clinton SM. Molecular diversity of a North Carolina wastewater treatment plant as revealed by pyrosequencing. Appl Environ Microbiol. 2009;75:1688–1696. doi: 10.1128/AEM.01210-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Thill A, Purkhold U, Walcher M, Bottero JY, Ginestet P, et al. Characterization of activated sludge flocs by confocal laser scanning microscopy and image analysis. Water Res. 2003;37:2043–2052. doi: 10.1016/S0043-1354(02)00616-4. [DOI] [PubMed] [Google Scholar]
- Seviour R, Nielsen PH. Microbial Ecology of Activated Sludge. IWA Publishing: London, UK; 2010. [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin YK, Hiraishi A, Sugiyama J. Molecular systematics of the genus Zoogloea and emendation of the genus. Int J Syst Bacteriol. 1993;43:826–831. doi: 10.1099/00207713-43-4-826. [DOI] [PubMed] [Google Scholar]
- van der Gast CJ, Walker AW, Stressmann FA, Rogers GB, Scott P, Daniels TW, et al. Partitioning core and satellite taxa from within cystic fibrosis lung bacterial communities. ISME J. 2010;5:780–791. doi: 10.1038/ismej.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M, Loy A. Bacterial community composition and function in sewage treatment systems. Curr Opin Biotechnol. 2002;13:218–227. doi: 10.1016/s0958-1669(02)00315-4. [DOI] [PubMed] [Google Scholar]
- Wanner J. Activated Sludge Bulking and Foaming Control. CRC Press: Florida, USA; 1994. [Google Scholar]
- Xia L, Steele J, Cram J, Cardon Z, Simmons S, Vallino J, et al. Extended local similarity analysis (eLSA) of microbial community and other time series data with replicates. BMC Syst Biol. 2011;5:S15. doi: 10.1186/1752-0509-5-S2-S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S, Duan L, Song Y, Li J, Piceno YM, Andersen GL, et al. Bacterial community structure in geographically distributed biological wastewater treatment reactors. Environ Sci Technol. 2010;44:7391–7396. doi: 10.1021/es101554m. [DOI] [PubMed] [Google Scholar]
- Xia Y, Kong Y, Nielsen PH. In situ detection of protein-hydrolysing microorganisms in activated sludge. FEMS Microbiol Ecol. 2007;60:156–165. doi: 10.1111/j.1574-6941.2007.00279.x. [DOI] [PubMed] [Google Scholar]
- Xia Y, Kong Y, Thomsen TR, Nielsen PH. Identification and ecophysiological characterization of epiphytic protein-hydrolyzing Saprospiraceae (“Candidatus Epiflobacter” spp.) in activated sludge. Appl Environ Microbiol. 2008;74:2229–2238. doi: 10.1128/AEM.02502-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Zhang T. Metagenomic and metatranscriptomic analysis of microbial community structure and gene expression of activated sludge. PLoS One. 2012;7:e38183. doi: 10.1371/journal.pone.0038183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Shao M-F, Ye L. 454 Pyrosequencing reveals bacterial diversity of activated sludge from 14 sewage treatment plants. ISME J. 2012;6:1137–1147. doi: 10.1038/ismej.2011.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.