Abstract
Oxygen-depleted bodies of water are becoming increasingly common in marine ecosystems. Solutions to reverse this trend are needed and under development, for example, by the Baltic deep-water OXygenation (BOX) project. In the framework of this project, the Swedish Byfjord was chosen for a pilot study, investigating the effects of an engineered oxygenation on long-term anoxic bottom waters. The strong stratification of the water column of the Byfjord was broken up by pumping surface water into the deeper layers, triggering several inflows of oxygen-rich water and increasing oxygen levels in the lower water column and the benthic zone up to 110 μmol l−1.We used molecular ecologic methods to study changes in bacterial community structure in response to the oxygenation in the Byfjord. Water column samples from before, during and after the oxygenation as well as from two nearby control fjords were analyzed. Our results showed a strong shift in bacterial community composition when the bottom water in the Byfjord became oxic. Initially dominant indicator species for oxygen minimum zones such as members of the SUP05 clade declined in abundance during the oxygenation event and nearly vanished after the oxygenation was accomplished. In contrast, aerobic species like SAR11 that initially were restricted to surface waters could later be detected deep into the water column. Overall, the bacterial community in the formerly anoxic bottom waters changed to a community structure similar to those found in oxic waters, showing that an engineered oxygenation of a large body of anoxic marine water is possible and emulates that of a natural oxygenation event.
Introduction
Oxygen-limited or -depleted bodies of marine water have become increasingly common in recent years (Stramma et al., 2008). These hypoxic zones (O2<2 ml l−1 or <90 μmol l−1), also called oxygen minimum zones (OMZs), have been observed all around the globe (Stramma et al., 2008) and have been extensively studied off the coasts of Chile (for example, Morales et al., 1999; Stevens and Ulloa, 2008; Canfield et al., 2010), Benguela (for example, Kuypers et al., 2005; Woebken et al., 2007), Oregon (for example, Chan et al., 2008) and the Arabian Sea (for example, Morrison et al., 1999; Fuchs et al., 2005). In OMZs, the biogeochemical cycling of elements changes significantly, which, in the case of the water turning completely anoxic, also has potential effects on the climate. For example, it has been shown that anoxia (Naqvi et al., 2000) and sulfidic conditions (Dalsgaard et al., 2013) can increase the production of the greenhouse gas N2O. Further it has been estimated that up to 50% of the fixed nitrogen in oceans can be lost in OMZs, and therefore expanding OMZs potentially affects future primary production and carbon cycling (Codispoti et al., 2001; Lam et al., 2009; Ward et al., 2009). Climate models predict an overall decline in dissolved oxygen in the oceans due to an increase in water temperature in the future (Matear and Hirst, 2003), which will further intensify the development of OMZs.
Hypoxia is not only a problem on upwelling-prone shelves, in coastal areas and the open ocean, but also large masses of bottom waters in marine basins are affected. Examples are found in Saanich Inlet (Anderson and Devol 1973), the Cariaco Basin (Scranton et al., 2001), the Black Sea (Jorgensen 1982) and the Baltic Sea (Conley et al., 2009), where naturally occurring hypoxia is intensified by anthropogenic eutrophication (Diaz and Rosenberg, 2008; Conley et al., 2011).
Prokaryotic community structure in OMZs
Microbial community structures are studied in many OMZs (for example, Stevens and Ulloa, 2008; Ward et al., 2008; Woebken et al., 2008; Jayakumar et al., 2009; Zaikova et al., 2010). Wright et al. (2012) summarized the current knowledge about the bacterial diversity in OMZs and found that there are conserved patterns between all different OMZs studied. The authors also identified the most abundant bacterial phyla in OMZs to be Proteobacteria, Bacteroidetes, Actinobacteria, Planctomycetes and the candidate phylum SAR406 (Marine Group A) (Wright et al., 2012). Most of the phylogenetic groups of bacteria in OMZs are found in other environments as well; however, some microbial clades seem to be rather specific to OMZs, for example, the SUP05 clade (Walsh et al., 2009).
Microbes in the Baltic Sea have been studied for many years, but only recently have analyses of the total bacterial community structure been undertaken. They revealed unique communities adapted to the brackish water prevalent in the Baltic Sea (Riemann et al., 2008; Koskinen et al., 2011). The knowledge of bacterial diversity in the Baltic Sea has further been improved with two studies generating high-resolution bacterial community profiles from the seasonal samples at one station (Andersson et al., 2010) and a transect along the salinity gradient of the Baltic (Herlemann et al., 2011).
The Baltic Sea OMZ
The special geological features of the Baltic Sea, with sills at its entrance and its characteristic deep basins, make it prone to hypoxia, which has already occurred during previous climatic warm periods (Zillen et al., 2008). In the past ∼100 years, hypoxia has been increasingly observed in the Baltic Sea, which is probably caused by nutrient-driven eutrophication (Conley et al., 2009).
During the 1990s, a natural event decreased the phosphorus content in the Baltic proper substantially. It coincided with a decrease in the anoxic deepwater volume, manifested in a lowering of the top of the halocline from 60 to 100 m, which increased the oxygen contents between 80 and 120 m depth. After a few years, the halocline rose, and the oxygen content below 80 m depth decreased. The Baltic proper went back to a state with high phosphorus content and strong summertime blooms of cyanobacteria. This event demonstrated that it should be possible to counteract hypoxia and restore the Baltic proper into a less eutrophic state by using artificial oxygenation of the deepwater by enforced mixing (Stigebrandt and Gustafsson, 2007). These authors estimated that the deepwater of the Baltic proper may be kept oxygenated by pumping about 10 000 m3 s−1 of cold, oxygen saturated water from ∼50 m depth down to ∼120 m depth in the deepwater. It was suggested that the pumping should be performed by ∼100 floating windmills equipped with pumps.
Because the dynamics of phosphorus is not sufficiently understood (for example, Gustafsson and Stigebrandt, 2007), it is not known if it would be possible to keep the Baltic proper in a less eutrophic state by a continuous supply of oxygen to the deepwater. This was one of the main topics to be investigated within the frame of the Baltic deep-water OXygenation (BOX) project (http://www.marsys.se/lang/gb/about-us/research/baltic-deepwater-oxygenation-box/).
The study site
Part of the BOX project is a pilot oxygenation study in the Swedish Byfjord in order to determine the effects of oxygenation on long-term anoxic bottom waters. The Byfjord, located on the Swedish west coast with the city Uddevalla at its end, is in a similar trophic condition as the Baltic Sea. It is about 4 km long, 1.5 km wide, and its greatest depth is 51 m. The entrance of the fjord has a sill at a depth of 13.5 m, connecting it to the Havstensfjord (Supplementary Figure S1). The fjord has a strong permanent halocline just below the sill depth due to the vertical stratification in the Havstensfjord and the coastal water outside of the fjord, which is dominated by an ∼15-m-thick layer of relatively fresh surface water from the Kattegat flowing northwards along the coast. Further strengthening of the stratification comes from the river Bäveån, which brings ∼80% of all freshwater into the fjord. Owing to the weak vertical mixing, stagnant conditions prevail in the Byfjord and the degradation of organic matter leads to oxygen deficits in the unventilated deep water. Over the past 40 years, anoxic (<1 μmol l−1 O2) conditions prevailed at 30 m interrupted by short periods every 2–5 years where oxygen levels were increased to hypoxic (<90 μmol l−1 O2) or higher (180 μmol l−1, Supplementary Figure S2) due to inflows of dense water from Havstensfjord. Anoxic periods were accompanied by the accumulation of hydrogen sulfide (Supplementary Figure S3). The Byfjord is the only Swedish fjord that has long-term anoxia in deepwater like the Baltic proper. This and the fact that it is relatively small, which made oxygenation by pumping manageable, were the main reasons for choosing it for the pilot oxygenation study.
The goal was to permanently increase the oxygen concentration of the water column to reduce the leakage of nutrients (mainly phosphorus) from the sediments. Computations described in Stigebrandt and Liljebladh (2011) showed that it should be sufficient to pump about 2 m3 s−1 of surface water to the deeper layers (35 m) to keep these oxic. Depending on season, the pumping can supply 30–100% of the oxygen consumed. It, however, should weaken the stratification, resulting in an increased frequency and intensity of natural occurring inflows from the oxic Havstensfjord. When implemented, this proved to be correct and a stronger mixing of the water column was observed during times of pumping as predicted in Stigebrandt and Liljebladh (2011). There have been four complete and a few partial water exchanges of the basin water (below sill level) between the first pumping in October 2010 and spring 2012 (Stigebrandt et al., 2014).
Here we present our analysis of microbial community dynamics during the oxygenation process. In contrast to all the currently available studies from OMZs of permanently anoxic and seasonally hypoxic sites, the study presented here documents changes in the microbial community that occurred when a long-term anoxic basin became oxic as a result of a geo-engineering experiment.
Materials and methods
Sample collection
Samples from the Byfjord (BF: 58°20.109 N, 11°52.799 O), Havstensfjord (HF: 58°18.72 N, 11°46.25 O) and Koljöfjord (KF: 58°13.733 N, 11°34.559 O) were collected with Niskin bottles aboard the R/V Skagerak during eight time points between March 2010 and April 2012, covering times before, during and after the oxygenation (see Figure 1 and Supplementary Table S1 for details). Between 5 and 20 l of water per depth from multiple depths (spanning oxic, oxycline and anoxic waters; see Supplementary Table S1 for details) were filtered through 0.22 μm Supor PES Membrane Disc Filters (PALL, Dreieich, Germany) to collect the biomass. Each membrane was stored in 5 ml sucrose lysis buffer (0.75 M sucrose, 0.4 M NaCl, 50 mM Tris pH 9.0 and 200 mM EDTA pH 8.0) at −20 °C until DNA extraction.
Figure 1.
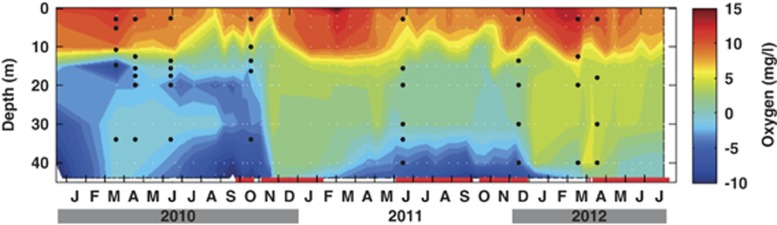
Changes in oxygen concentrations in the water column of the Byfjord between 2010 and 2012. Pumping water from the halocline to 35 m (pump run times are indicated by red lines at the bottom of the plot) triggered several inflows. In the beginning of 2010 also a small natural inflow occurred. Oxygen is plotted in mg/l, with negative values indicating the oxygen necessary to oxidize the hydrogen sulfide present. Black dots indicate time and depths of sampling for molecular work.
Nutrient collection and analysis
The Orust–Tjörn fjord system, to which the studied fjords belong to, has been monitored monthly since 1990 by the Bohuskustens Vattenvårdsförbund (www.bvvf.se) in collaboration with subcontractors (mainly the Swedish Meteorological and Hydrological Institute, SMHI). Sampling frequency has been doubled during the BOX project. All samples were taken using a Seabird rosette sampler (Sea-Bird Electronics, Bellevue, WA, USA) with a SB19+ CTD (Conductivity, Temperature and Depth probe). Initial accuracy of conductivity is 0.005 mS m−1 and of temperature 0.005°C. The hydrographic parameters used in this study and their accuracy are oxygen (iodometric titration, 0.6−63±10%, 63−469±5% μmol l−1), hydrogen sulfide (1–300±9% μmol l−1), phosphate (0.02−0.2±12%, 0.2−10±3% μmol l−1), nitrate (0.1–1.5±7%, 1.5−50±4% μmol l−1), nitrite (0.02−0.2±11%, 0.2−10±3%) and ammonium (0.2−100±14% μmol l−1) (Grasshof et al., 1999). See Supplementary Table S2 for details.
DNA extraction
DNA was extracted as described previously (Giovannoni et al., 1996; Treusch et al., 2009) with minor modifications. Membranes were allowed to thaw at RT, 500 μl 10% (w/v) sodium dodecyl sulfate (SDS) and 500 μl Proteinase K (10 mg ml−1) were added, followed by incubation steps at 37 °C for 30 min and 55 °C for 30 min under continuous mixing. Nucleic acids were purified by using phenol:chloroform:isoamylalcohol (IAA) (25:24:1) and chloroform:IAA (24:1) extractions, each followed by a centrifugation at 2500 r.c.f. for 10 and 5 min, respectively. Genomic DNA was precipitated with 0.1 volume of 2 M sodium acetate and 2 volumes of 96% ethanol, incubation at −20 °C for 8 h and centrifugation at 10 000 r.c.f. for 45 min. DNA pellets were washed with 70% ice-cold ethanol and centrifuged at 10 000 r.c.f. for 30 min. The DNA was air dried at 28 °C and resuspended in 500 μl TE buffer (pH 7.5). Impurities were removed with the PowerClean Clean-Up Kit (MoBio, Carlsbad, CA, USA) according to the manufacturer's instructions. DNA concentrations, quality and purity were quantified spectrophotometrically (NanoDrop, Wilmington, DE, USA) and by agarose gel electrophoresis.
Terminal restriction fragment length polymorphism (T-RFLP)
To profile the bacterial community we used terminal restriction fragment length polymorphism (T-RFLP) (Liu et al., 1997) targeting the first ∼500 bp of the 16S rRNA gene with the fluorescence-labeled forward primer B27F FAM (5′-AGRGTTYGATYMTGGCTCAG-3′) and reverse primer 519R (5′-GWATTACCGCGGCKGCTG-3′). Polymerase chain reaction (PCR) conditions were as follows: 1 min at 94 °C followed by 29 cycles of 94 °C for 15 s, 54 °C for 45 s, 72 °C for 45 s and a final extension at 72 °C for 5 min. Each 50 μl PCR reaction contained 5–10 ng template DNA, 20 pmol of each primer, 1.25 U Taq polymerase (Thermo Scientific, Slangerup, Denmark), 40 nmol deoxynucleotides and 125 nmol MgCl2 in 1 × Taq reaction buffer (Thermo Scientific). Ten microliters of each PCR product were hydrolyzed with 5 U of BsuRI (Thermo Scientific) at 37 °C for 8 h, followed by purification with the GeneJET PCR Purification Kit (Thermo Scientific). One hundred nanograms DNA of each sample were sent to the Uppsala Genome Center (Uppsala, Sweden) for capillary electrophoresis.
Pretreatment of T-RFLP data
The Peak Scanner software (Applied Biosystems, Foster City, CA, USA) was used to determine T-RF sizes. Noise filtration (factor: 1.0) and binning (clustering threshold factor: 0.8) were performed with T-RFLP analysis Expedited (http://trex.biohpc.org/; Culman et al., 2009). Operational taxonomic units (OTUs) that were present in only one sample and had a relative abundance of less than 1% were removed manually.
Statistical analysis
The T-RFLP data were analyzed with the Primer-E software package (PRIMER-E Ltd, Ivybridge, UK; Clarke and Gorley, 2006). To down-weigh highly abundant species, the matrix was log transformed. Similarities between samples were calculated using the Bray–Curtis algorithm. The resulting resemblance matrix was finally illustrated through non-metric multi-dimensional scaling (MDS) (Clarke and Ainsworth, 1993) in a two-dimensional plot. Analysis of similarities (ANOSIM) and analyses for species contributions (SIMPER) for a priori defined groups of samples were performed on the Bray–Curtis resemblance matrix.
Clone libraries
To identify OTUs observed in the T-RFLP analysis, clone libraries of bacterial 16S rRNA genes were constructed from two samples (BF April 2010 18 m, BF June 2011 3 m), which covered the observed diversity best. Five to ten nanograms of template DNA were amplified in 50-μl PCR reactions that contained 20 pmol (each) of the primers B27F (5′-AGRGTTYGATYMTGGCTCAG-3′) and U1492R (5′-GGYTACCTTGTTACGACTT-3′), 5 U Taq polymerase (Sigma-Aldrich, St Louis, MO, USA) and 25 nmol deoxynucleotides in 1 × Taq reaction buffer including MgCl2 (Sigma-Aldrich) with the following conditions: 2 min at 94 °C followed by 30 cycles of 94 °C for 1 min, 54 °C for 1 min, 72 °C for 2 min and a final extension at 72 °C for 10 min.
Four microliters of each PCR product were cloned into a pCR4-TOPO vector using the TOPO TA Cloning Kit for Sequencing (Invitrogen, Carlsbad, CA, USA) and chemically transformed into TOP10 cells according to the manufacturer's instructions.
A total of 200 clones were screened by colony PCR with the primers M13F (5′-GTAAAACGACGGCCAG-3′) and M13R (5′-CAGGAAACAGCTATGAC-3′) under the following PCR conditions: 10 min at 94 °C followed by 30 cycles at 94 °C for 30 s, 50 °C for 30 s, 72 °C for 2 min and a final extension at 72 °C for 10 min. Each 50 μl reaction contained 20 pmol of each primer, 1 U Taq polymerase (Thermo Scientific), 40 nmol deoxynucleotides and 125 nmol MgCl2 in 1 × Taq reaction buffer (Thermo Scientific). Obtained PCR products were digested with 5 U BsuRI (Thermo Scientific) for 8 h and analyzed by 2% agarose gel electrophoresis in 1 × TAE along with 5 μg BsuRI hydrolyzed ΦX174 DNA (Thermo Scientific) as a size standard. Clones with similar RFLPs were grouped together and representative clones from each group were sequenced by Macrogen Europe (Amsterdam, Netherlands) using the primers T3 (5′-ATTAACCCTCACTAAAGGGA-3′) and T7 (5′-TAATACGACTCACTATAGGG-3′). In total, 96 clones were sequenced.
Phylogenetic analysis
Forward and reverse reads were assembled, screened for chimeras using Bellerophon (Huber et al., 2004) and imported into the SSU Reference 111 SILVA database (Pruesse et al., 2007). Phylogenetic reconstructions were performed using the ARB software package (Ludwig et al., 2004). TRF-CUT (Ricke et al., 2005) was used for in silico T-RF predictions. All sequences have been submitted to GenBank under the accession numbers KC545702–KC545786.
Results
Oxygenation of the Byfjord
Water from the surface layer of the Byfjord was mixed in the anoxic bottom water by several pump runs starting in October 2010. This triggered inflows of oxygen-rich water from the Havstensfjord into the Byfjord (Figure 1). The whole water column turned oxic, but during 2011 the oxycline started to rise and the bottom water turned anoxic again. A second inflow in April 2011 was observed, but with minor impact on the oxygen concentrations. The oxycline, however, remained deeper in the water column than in the previous years and a third inflow in the beginning of 2012 lowered the oxycline again. After a fourth inflow in March 2012, the whole water column was oxic with oxygen levels up to 180 μmol−l−1 and metal oxides were traceable in the upper 2 cm of the sediment (P Hall, unpublished results). As a result of these inflows from the Havstensfjord, ∼50% of the water in the Byfjord below 15 m has been successively replaced by the water originating from the Havstensfjord. For detailed information about the physical background of the oxygenation, please refer to Stigebrandt et al. (2014).
Bacterial community fingerprinting
To analyze the response of the bacterial community to the oxygenation process, we generated community fingerprints from water column biomass samples using 16S rRNA gene-based T-RFLP. In addition to the 40 Byfjord samples, eight samples from the Havstensfjord and seven samples from the Koljöfjord were analyzed. Havstensfjord served as control station because of its role as source of the oxic water inflows into the Byfjord and for the fact that no artificial mixing was performed here. Koljöfjord was the second control station without artificial mixing.
We identified a total of 188 OTUs in the T-RFLP dataset, of which 152 were detected in all three fjords and contributed to 90% (BF), 97% (KF) and 98% (HF) of the total bacterial abundances observed. The number of OTUs varied from 42 to 84 per sample without any observed correlation between number of OTUs and depths or oxygen concentrations.
To compare the different community fingerprints, we used the PRIMER-E software package to visualize the similarities and differences between the samples as distances between them in a two-dimensional MDS plot (Figure 2). Samples with similar microbial composition were thereby clustered together, whereas samples with different microbial compositions ordinated farther apart. Environmental data were overlain as vectors onto the resulting MDS plot to display which samples or clusters are associated with which environmental parameter.
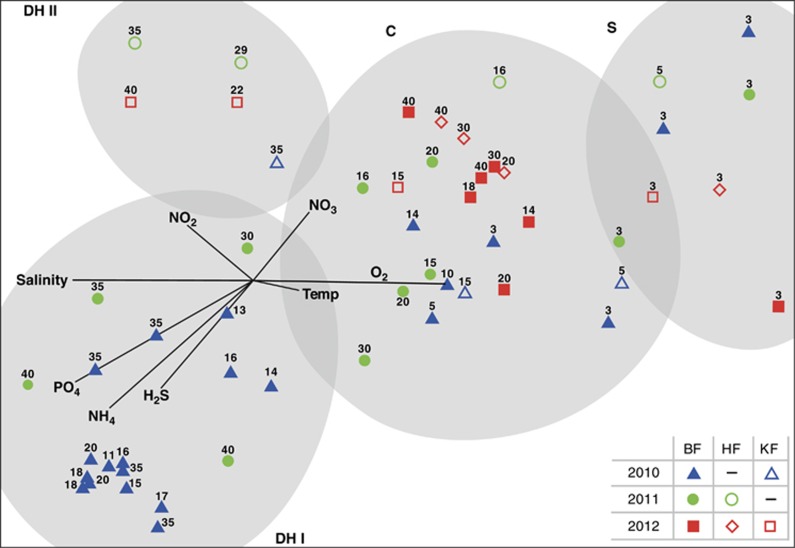
Figure 2.
Multivariate statistical analysis of microbial community structure based on 16S rRNA gene T-RFLP data. The similarities between the 40 Byfjord (BF), 8 Havstensfjord (HF) and 7 Koljöfjord (KF) samples were analyzed using Bray–Curtis similarity index and plotted by MDS. The microbial communities in the Byfjord have been separated into three different clusters: surface (S), chemocline (C) and deep hypoxic cluster one (DH I). Samples from HF and KF were present in S, C and formed a second deep hypoxic cluster (DH II). Clusters are indicated by gray shading and samples in each cluster showed >45% similarity with each other. The plot was overlain with vectors for environmental parameters using Pearson correlation. 2D stress=0.11.
Four clusters could be distinguished, defined by samples with >45% similarity in OTU composition. The surface cluster (S) included exclusively samples from 3 and 5 m, whereas samples below these depths were split up between the other three clusters independent of their actual depth. Samples from 2010 taken below 5 m in the Byfjord were mainly affiliated with the deep hypoxic cluster I (DH I), while samples from the same depths taken in 2011 were evenly distributed between the chemocline (C) and the DH I cluster. Samples from 2012 were all affiliated with the C cluster except the surface samples, which ordinated in the surface cluster. Samples from the control fjords were found in the surface and the C cluster but not in the DH I cluster, instead forming a separate deep hypoxic cluster (DH II). Although in 2011 the hypoxic communities were different in BF and HF, we observed that the 30- and 40-m samples from 2012 clustered together after the water column in both fjords turned oxic. This is notable considering the different microbial and geochemical histories of both fjords.
Vectors for environmental parameters showed that salinity and oxygen concentrations were mainly responsible for separating the surface and the chemocline cluster from the deep hypoxic clusters. Ammonium, hydrogen sulfide and phosphate were part of this differentiation as well and could additionally explain the differences between DH I and DH II. Higher concentrations of nitrate and nitrite were mainly affiliated with the DH II and C clusters.
In order to confirm the separation of samples into different clusters, we used an ANOSIM. All samples were therefore a priori divided into three groups according to oxygen and hydrogen sulfide concentrations: oxic (> 90 μmol l−1 O2), hypoxic (< 90 μmol l−1 O2) and sulfidic (H2S detected). These groups were comparable but not identical to the groups found in the cluster analysis (Supplementary Figure S4), which could not be used in the ANOSIM analysis in order to avoid circular argumentation. The ANOSIM revealed a global R-value of 0.569, supporting that the samples in the three defined groups have different microbial communities with some overlap. Pairwise comparison of the sulfidic group with the hypoxic resp. oxic group revealed R-values of 0.593 and 0.798, indicating differences between the sulfidic and hypoxic groups, with some overlap and clear distinction between the sulfidic and oxic groups. The oxic and hypoxic groups shared the highest overlap, as indicated by an R-value of 0.238.
Environmental data
To obtain a detailed picture of how the environmental data match the microbial community, we displayed the biogeochemical data as a heat map separately for each sample (Supplementary Figure S5).
The surface cluster was mainly associated with the highest oxygen levels, but lowest salinity and nutrient concentrations. Moderate to high oxygen and low nutrients but the highest nitrate concentrations defined the chemocline cluster. Salinity and phosphate levels were also sporadically elevated in this cluster. Finally, low oxygen and nitrate concentrations in combination with high ammonium, sulfide, phosphate and salinity levels characterize the deep hypoxic cluster I. The deep hypoxic cluster II seemed to be lower in hydrogen sulfide, moderate in ammonium, oxygen and nitrate concentrations, but higher than average in nitrite. However, only five samples were present in this cluster, and considering the variation in some of the environmental data between those samples it remains unclear how representative they are.
Spatial and temporal abundance of individual OTUs
To reveal the most abundant bacterial species before, during and after the oxygenation, we analyzed the relative abundance of individual OTUs within the T-RFLP data set. As we were aware that oxygen is most likely the main shaping factor, we first divided samples from each water column profile into oxic (>90 μmol l−1 O2) and hypoxic (<90 μmol l−1 O2) zones to assess these very different layers separately. Relative abundances of each OTU were then integrated among samples from the same profile and zone and finally sorted after the most abundant OTUs (Supplementary Table S4). The five most abundant OTUs from each integrated area were assembled among all samples. This led to a total of 38 different OTUs, which we considered to have the highest contribution to the microbial community in the Byfjord and the two control fjords. Within individual samples, the summarized relative abundance of these 38 OTUs reached 74% on average, with at least 57% and at most 83%, supporting our assumption. Repeating this analysis with the 10 most abundant OTUs in each sample added only 8 OTUs to the previously mentioned 38 OTUs. Furthermore, the additional 8 OTUs increased the average relative abundance only by ∼5% and were generally close to the detection limit of our method (∼1% of the total fluorescence) (Nocker et al., 2007) in individual samples. Hence we focus on the 38 most important OTUs. Additional analyses for species contributions (SIMPER) confirmed that the chosen 38 OTUs included the most abundant OTUs in the Byfjord (Supplementary Table S3), represent a major part of the bacterial community and are mainly responsible for the differentiation of oxic, hypoxic and sulfidic communities displayed in the MDS plot (Figure 2).
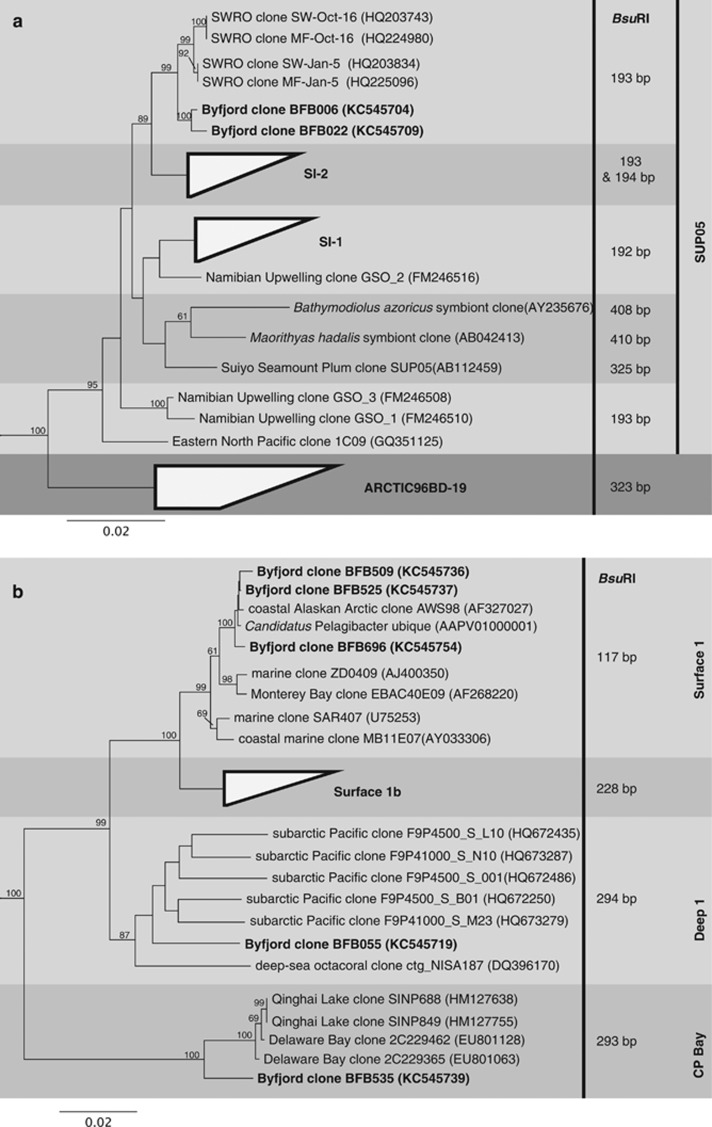
We furthermore predicted a phylogenetic placement for the observed OTUs with 16S rDNA sequences obtained from the clone libraries. Sequences were therefore placed into phylogenetic trees and in silico digested with BsuRI (Supplementary Table S5). This showed that the two clones BFB006 and BFB022 were affiliated with the SUP05 cluster within the gamma proteobacteria (Figure 3a). The SUP05 clones from the Byfjord clustered neither with SUP05 subclades observed in the Saanich Inlet (SI-1 and SI-2) nor within those from the Baltic Sea (BBAL-1 also SI-2), but were closest related to sequences from a seawater reverse osmosis pre-treatment system. We furthermore identified several SAR11 clones that, interestingly, originated from three different subclades (Surface 1, Deep 1 and Chesapeake Bay) but could be separated in the T-RFLP dataset based on the differences in their terminal restriction fragment sizes (Figure 3b).
Figure 3.
Neighbor-joining 16S rRNA phylogenetic reconstructions of (a) members of the SUP05 and the ARTIC96BD-19 clades, based on 1255 positions of the 16S rRNA sequence alignment and Congregibacter litoralis KT71(AAOA01000004) as outgroup. (b) Members of the SAR11 clade based on 1466 positions with Rickettsia bellii (U11014) as outgroup. Bootstrap values over 60% are shown and indicate the percentage of 1000 repetitions. Predicted terminal restriction fragment sizes are shown for each clone and subclade. CP Bay=Chesapeake Bay.
Combined results from the phylogenetic trees and the T-RFLP analyses showed that during the timeframe of this study members of the SAR11 Surface 1 clade (OTU 113 bp) were the most abundant bacterial species in the oxic zone independent of season and fjord, whereas members of the SUP05 cluster (OTU 193 bp) were dominating the hypoxic parts. Owing to the oxygenation in 2011 and 2012, the hypoxic parts declined or disappeared, which was also reflected in the depression of SUP05 and proliferation of SAR11 Surface 1 and Chesapeake Bay subclade populations.
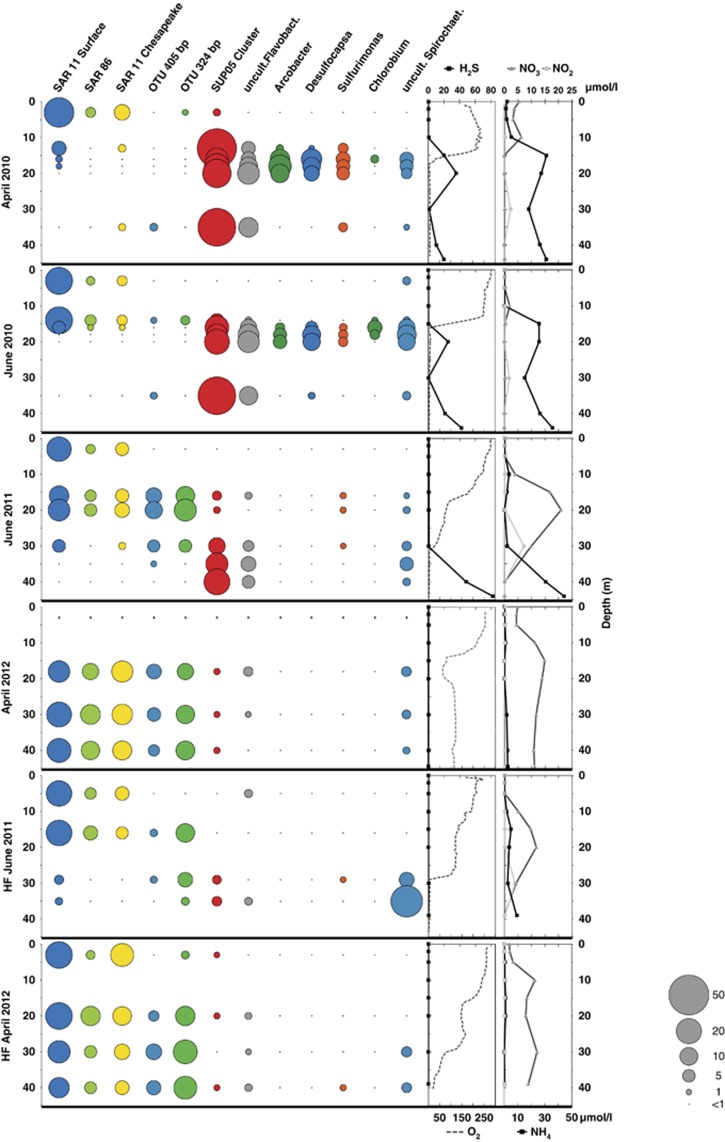
SAR11 subclades, SUP05 and the remaining OTUs that were revealed to be highly abundant were furthermore analyzed in detail for spatial and temporal abundance and directly related to environmental data. Representative OTUs are shown in Figure 4, while all 38 OTUs are shown in Supplementary Figure S6. This showed that all three SAR11 subclades and SAR86 (Supplementary Table S5) were mainly present in regions where oxygen was detectable in significant amounts (>25 μmol/l). These OTUs were initially restricted to layers above 15 m but were later, following oxygenation, also highly abundant below this depth. In contrast, SUP05, uncultured members of the Flavobacteriaceae and Spirochaetes, Arcobacter, Sulfurimonas, Chlorobium and Desulfocapsa were mainly highly abundant in oxygen-free areas and became lower in abundance when oxygen concentrations increased. However, the high abundance of SUP05 above the oxycline in April 2010 seems to be inconsistent with this observation.
Figure 4.
Changes in relative abundances of the most important OTUs in response to the oxygenation. Ten identified and two unidentified OTUs relative abundances in the Byfjord (four time points) and the Havstensfjord (two time points) are shown in comparison with environmental parameters (H2S, NH4, NO3, NO2, oxygen; all in μmol l−1). Relative abundance is displayed as bubble size. The correlation between bubble size and abundance is linear from 50 down to 1%. Not detectable abundances and such below 1% are binned together (see the legend).
Arcobacter, Desulfocapsa, Flavobacteriaceae, Chlorobium, Sulfurimonas and SUP05 were furthermore absent or in low abundance in the hypoxic region of the Havstensfjord. Additionally, this region was different from the hypoxic zone in the Byfjord due to the absence of sulfide in Havstensfjord, while the remaining geobiochemical data were comparable. Spriochaetes were found to be more abundant in the Havstensfjord as long as ammonium was detectable and sulfide absent.
Unidentified OTUs 405 and 324 bp were close to the detection limit in 2010 but became highly abundant in 2011/2012 and were restricted to areas where nitrate and/or nitrite were present. This is strongly supported by their increased relative abundance in June 2011 and April 2012, when nitrite levels in the water column were elevated. The same observation was made in Havstensfjord.
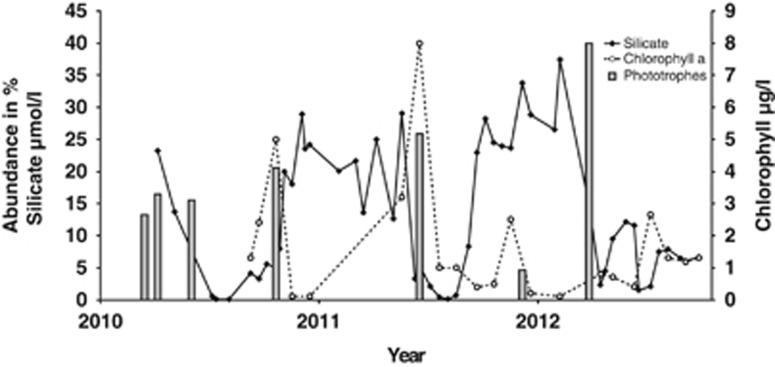
Four OTUs were identified as chloroplasts and exclusively observed in samples from 3 and 5 m where light conditions promote phototrophic growth. The OTUs 263, 278, 393, 485 and 519 bp were also exclusively observed in the surface but could not be identified. Individual OTUs showed strong seasonal changes, whereas the summarized abundance of all OTUs in this group of potential phototrophic organisms behaved more consistent and was well inversely and directly correlated with silica and chlorophyll a, respectively (Figure 5). In December 2011 the abundance of this group strongly decreased, whereas in March 2012 it increased to 41%. March 2012 was also the first time during the oxygenation experiment at which the sediment surface turned oxic. We could not show the further development of this group owing to the fact that the surface sample from April 2012 was an outlier.
Figure 5.
Combined relative abundance of OTUs of potential phototrophs (gray bars) over time in comparison to silicate (black line) and chlorophyll a (dotted line) concentrations in the surface layer of the Byfjord.
Discussion
Changes in the microbial community structure
Oxygen is known to be a major shaping factor for microbial communities in marine ecosystems. Although studies on OMZs often only capture snapshots of the community structure, these communities constantly change in response to changing oxygen levels. The fate of anoxic microbial communities in response to enhanced oxygen levels, however, is rarely described in marine systems. The transition of the anaerobic community to a common aerobic community is always assumed but has yet not been observed. To study these transitions, a controlled experimental setup with continuous sampling is required and the Byfjord oxygenation pilot provided this opportunity. Because of the pumping and subsequently triggered inflows of oxygen-rich water, the oxygen levels in the Byfjord rose to an extent that the anoxic bottom water first regressed and eventually entirely disappeared. The MDS analysis (Figure 2) and the ANOSIM results showed that the amount of oxygen available shaped the bacterial communities, regardless of the depths they were collected from or the season they were collected in. This observation is similar to earlier studies in which the composition of OMZ, surface and deep oxycline communities were compared (for example, Stevens and Ulloa, 2008). In the Byfjord, a bacterial community similar to the communities previously observed in the oxycline replaced the declining hypoxic community.
Considering that water from 20 m depths of the Havstensfjord is the main source of water for the inflows into the Byfjord, it is reasonable that the communities in both fjords showed greater similarity during the inflow periods. The fact that the community structure in the Byfjord was still similar, but not identical, to the Havstensfjord in March 2012, several weeks after the inflow happened, shows that this community could establish itself. That environmental parameters, and not the inoculum, shape the microbial community was also recently shown by Sjostedt et al. (2012).
Interestingly, hypoxic samples from the Byfjord and from the control fjords form distinct clusters in the MDS analysis (DH I and DH II, Figure 2). This separation can be explained by higher sulfide and nutrient concentrations in the Byfjord. Sulfide, ammonium and phosphate were four, six and two times, respectively, higher in the hypoxic samples from the Byfjord compared with the control stations (Supplementary Table S2). Other environmental parameters (salinity, temperature and oxygen) could not explain the differences in microbial communities observed.
In our analysis, it became obvious that the microbial communities in the surface cluster are shaped by high oxygen, low salinity and low nutrient levels. Although there are no data for light, OTUs associated with chloroplasts and their correlations with the chlorophyll data (Figure 5) are good indications that eukaryotic phototrophs play a role in separating the surface cluster from the other clusters.
Identification of observed OTUs
T-RFLP is a well-established method that was recently shown to be as reliable as 454 pyrotaq sequencing (Pilloni et al., 2012), although the detection limit is higher and the resolution lower. For example, OTUs that are lower abundant than 1% cannot be detected reliably (Nocker et al., 2007) and not in all cases clear separations between subclades can be observed. Further, the construction of clone libraries is necessary in order to determine the organisms represented by the OTUs. Clones can be analyzed either by in silico predictions of T-RF lengths or by T-RF determination using T-RFLP or a combination of both. This leads to reliable identifications of the observed OTUs. Pure in silico predictions based on public 16S rRNA databases are feasible but often have ambiguous results.
We could identify most of the abundant OTUs with our clone libraries. The comparison of the environmental T-RFLP data set with predicted T-RF lengths from the clones led in most cases to a direct match. However, for some OTUs we observed a phenomenon known as T-RF drift (Kaplan and Kitts, 2003), where the predicted T-RF size from the clone is different from the observed OTU in the environmental data. Hence, we allowed for 1 base pair difference between predicted and actual fragment size. The only exception was the SAR11 Surface 1 clade OTU, where we observed a 4-bp difference. This phenomenon is documented in the literature (Morris et al., 2005) and was also confirmed by T-RF analysis of one of our SAR11 Surface 1 clones (data not shown).
Spatial and temporal distribution of species before the oxygenation
Our survey shows that the bacterial community in the Byfjord before the oxygenation was comparable with communities observed in other OMZs, which normally comprise SAR11, SAR86, SUP05 and other sulfur oxidizers, sulfur reducers and Flavobacteriaceae (Stevens and Ulloa, 2008; Canfield et al., 2010; Stewart et al., 2012).
Members of the SAR11 Surface 1 and SAR86 clades were dominant in the oxygen-rich layers of the Byfjord, showing abundances of up to 38% and 12%, respectively. These results are in line with previous reports that describe both clades as important members of aerobic marine microbial communities (for example, Giovannoni and Rappé, 2000; Stevens and Ulloa, 2008).
The hypoxic layers were dominated by members of the SUP05 clade, similar to reports from other OMZs (Wright et al., 2012). In April 2010, however, they were also surprisingly abundant (47%) above the oxycline. It has been reported that members of the SUP05 clade have genes for a sulfur-oxidizing (SOX) pathway, which potentially enables S2O3 reduction after abiotic oxidation of hydrogen sulfide with oxygen (Walsh et al., 2009). In the Byfjord, we observed high sulfide concentrations just below the oxycline, which could have led to significant S2O3 concentrations above the oxycline where enhanced SUP05 growth was observed. In addition, the simultaneous decline in nitrate concentrations supports this assumption since nitrate serves as an electron acceptor for SUP05 members (Stevens and Ulloa, 2008; Walsh et al., 2009). It is also possible that members of the SUP05 clade present here can use oxygen as electron acceptor, similar to SUP05 bacteria observed in hydrothermal vent plumes in the Guaymas Basin (Anantharaman et al., 2013), or as indicated by the presence of genes for a complete oxidative phosphorylation complex in a SUP05/ARCTIC96BD-19 metagenome in the eastern South Pacific (Murillo et al., 2014). However, the two types of cytochrome c oxidase genes observed in the SUP05 clam symbionts and the Guaymas Basin were not found in the SUP05 metagenome from the hypoxic waters of the Saanich Inlet (Walsh et al., 2009; Anantharaman et al., 2013), making the use of oxygen by OMZ SUP05 highly speculative. It might be that the high SUP05 population above the oxycline in the Byfjord represents an ecotype of the SUP05 cluster that we were not able to differentiate from the other Byfjord SUP05 members by using T-RFLP; hence further studies will be needed to resolve this observation in detail.
An additional observation from April 2010 shows that SUP05 abundance decreased to 15% directly below the oxycline and rose back to 43% at 35 m. This was most likely due to a small natural inflow of oxygen-rich water 1 month earlier that created conditions at 35 m that were comparable to the region above the oxycline. Nitrification at this depth was enhanced as well, indicated by a small but significant nitrite peak and the decline of ammonium. Missing accumulation of nitrate indicated a faster reduction of nitrate than oxidation of nitrite. It is, however, surprising that we could observe significant SUP05 abundance in layers with high hydrogen sulfide concentrations (up to 82 μmol/l), while recent findings from the Baltic Sea indicate that the abundance of SUP05 members is negatively correlated with hydrogen sulfide (Glaubitz et al., 2013). That some Byfjord SUP05 members seem to behave differently may indicate a not described ecotype, which is furthermore supported by our phylogenetic analysis where Byfjord SUP05 sequences were not affiliated with the subcluster BBAL-1 (also known as SI-2) described in the Baltic Sea (Glaubitz et al., 2013). As mentioned earlier, further experiments are necessary to resolve potential SUP05 ecotypes and their ecological function in the Byfjord.
We could further detect sulfide oxidizers like Arcobacter (Wirsen et al., 2002), Sulfurimonas (Inagaki et al., 2003) and Chlorobium (Musat et al., 2008) in sulfidic areas but also, with significant lower abundances, above the oxycline. Sievert et al. (2007) showed in culture experiments that Arcobacter grows as a microaerophile in the vicinity of oxic–anoxic interfaces, supporting our findings. Arcobacter has been described previously in hypoxic and anoxic estuary regions like the Saanich Inlet (Wright et al., 2012) and the Baltic Sea (Koskinen et al., 2011).
Sulfurimonas has been previously detected in the oxycline of the Baltic Sea (Labrenz et al., 2005) and one strain, Sulfurimonas gotlandica GD1T, has been isolated and characterized (Labrenz et al., 2013). The strain showed chemolithoautotrophical growth, performing dark CO2 fixation while coupling denitrification with the oxidation of reduced sulfur compounds (Grote et al., 2012). The closest relatives of the phototrophic green sulfur bacterium detected in the Byfjord are capable of oxidizing S2O3, but like all green sulfur bacteria are obligate anaerobic (Musat et al., 2008). Deltaproteobacterial Desulfocapsa species (Finster et al., 1998) were also abundant (max. 13%) in the anoxic bottom water, indicating an active sulfur cycle in the water column of the Byfjord.
We also found members of the Flavobacteriaceae and Spirochaete to be abundant in the Byfjord (max. 14% resp. 11%). Their roles within OMZs remain unclear. However, we observed increased Spirochaete abundance in the sulfide-free Havstensfjord as long as ammonium was present, indicating possible involvement in nitrogen cycling.
Oxygenation
The relative abundances of species associated with the OMZ and the chemocline region decreased substantially after oxygen levels rose. Arcobacter, Desulfocapsa, Sulfurimonas and Chlorobium were more sensitive to the changing conditions than SUP05, the uncultured Spirochaetes and the uncultured Flavobacteriaceae, which is not surprising considering the physiological characteristics of these species (Finster et al., 1998; Wirsen et al., 2002; Grote et al., 2008; Musat et al., 2008).
Benefitting from the rise in oxygen were the aerobic species like members of the SAR11 Surface 1, SAR11 Cheasapeak Bay and SAR86 clades, which increased in abundance in these formerly anoxic layers. The unidentified OTUs with 405 and 324 bp also became more abundant. This was accompanied by rising nitrate and nitrite concentrations, indicating possible involvement in nitrogen cycling. Unfortunately, these OTUs were not present in the samples the clone libraries were constructed from and therefore were not represented in them. In an in silico analysis, a significant number of sequences affiliated with the SAR324 (Marine Group B) clade were predicted to have terminal fragments of 405/406 bp besides a group of Flavobacteriaceae with 405 bp. Although some uncertainty remains, these are strong indications that the OTU 405 bp is representing SAR324 clade organisms. Similarly, predictions for OTU 324 bp revealed sequences belonging to the Rikenellaceae (predicted 324 bp), a group of fecal bacteria (Worthen et al., 2006), and ARCTIC96BD-19 clade (323 bp) as possible identifications. ARCTIC96BD-19 clade organisms are often observed in OMZs (Stevens and Ulloa, 2008; Wright et al., 2012) and in the bottom boundary layer of hypoxia-prone costal shelves (Bertagnolli et al., 2011). It was suggested that they require specific ecological conditions, namely gradients in oxygen and nitrite (Stevens and Ulloa, 2008). Interestingly, we observed an increase in abundance for OTU 324 bp after the oxygenation in the Byfjord, indicating an aerobic or microaerophilic lifestyle. Contrary to Stevens and Ulloa (2008), however, this OTU was correlated with the presence of nitrate and not nitrite.
Conclusions
The large-scale engineered oxygenation is one possibility to restore hypoxic or anoxic bodies of marine waters. However, little experience is available if these ecosystems can return to a natural oxic state. With our study in the Swedish Byfjord, we provide the first evidence that it is possible to change a long-term anaerobic marine bacterial community by engineered oxygenation. With the inflow of oxygen-rich water, a community structure similar to those in seasonal oxic fjords established itself, with no unexpected blooms of members of the rare biosphere. Because the bacterial community structure in the Byfjord is comparable to those in well-studied OMZs like the Saanich Inlet and the Baltic Sea, we can expect that a larger-scale artificial oxygenation, for example, in the Baltic would result in a similar shift in bacterial community structure as we observed here. How stable this established community is in the long run has still to be determined.
Acknowledgments
We thank the scientific and the ship crew of University of Gothenburg R/V Skagerak for all their help on the cruises. This study was partially financed by the Swedish Environmental Protection Agency through the Baltic deep-water OXygenation (BOX) project and by the Danish National Research Foundation (grant number: DNRF53). We further thank Donald Canfield, Sean Crowe and Loreto De Brabandere for fruitful scientific discussions.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Anantharaman K, Breier JA, Sheik CS, Dick GJ. Evidence for hydrogen oxidation and metabolic plasticity in widespread deep-sea sulfur-oxidizing bacteria. Proc Natl Acad Sci USA. 2013;110:330–335. doi: 10.1073/pnas.1215340110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JJ, Devol AH. Deep water renewal in Saanich Inlet, an intermittently anoxic basin. Estuar Coast Mar Sci. 1973;1:10. [Google Scholar]
- Andersson AF, Riemann L, Bertilsson S. Pyrosequencing reveals contrasting seasonal dynamics of taxa within Baltic Sea bacterioplankton communities. ISME J. 2010;4:171–181. doi: 10.1038/ismej.2009.108. [DOI] [PubMed] [Google Scholar]
- Bertagnolli AD, Treusch AH, Mason OU, Stingl U, Vergin KL, Chan F, et al. Bacterial diversity in the bottom boundary layer of the inner continental shelf of Oregon, USA. Aquatic Microb Ecol. 2011;64:15–25. [Google Scholar]
- Canfield DE, Stewart FJ, Thamdrup B, De Brabandere L, Dalsgaard T, Delong EF, et al. A cryptic sulfur cycle in oxygen-minimum-zone waters off the Chilean coast. Science. 2010;330:1375–1378. doi: 10.1126/science.1196889. [DOI] [PubMed] [Google Scholar]
- Chan F, Barth JA, Lubchenco J, Kirincich A, Weeks H, Peterson WT, et al. Emergence of anoxia in the California current large marine ecosystem. Science. 2008;319:920–920. doi: 10.1126/science.1149016. [DOI] [PubMed] [Google Scholar]
- Clarke KR, Ainsworth M. A method of linking multivariate community structure to environmental variables. Mar Ecol Prog Ser. 1993;92:205–219. [Google Scholar]
- Clarke KR, Gorley RN. PRIMER v6: User Manual/Tutorial. Plymouth: PRIMER-E Ltd; 2006. [Google Scholar]
- Codispoti LA, Brandes JA, Christensen JP, Devol AH, Naqvi SWA, Paerl HW, et al. The oceanic fixed nitrogen and nitrous oxide budgets: moving targets as we enter the anthropocene. Sci Mar. 2001;65:85–105. [Google Scholar]
- Conley DJ, Bonsdorff E, Carstensen J, Destouni G, Gustafsson BG, Hansson LA, et al. Tackling hypoxia in the Baltic Sea: is engineering a solution. Environ Sci Technol. 2009;43:3407–3411. doi: 10.1021/es8027633. [DOI] [PubMed] [Google Scholar]
- Conley DJ, Carstensen J, Aigars J, Axe P, Bonsdorff E, Eremina T, et al. Hypoxia is increasing in the coastal zone of the Baltic Sea. Environ Sci Technol. 2011;45:6777–6783. doi: 10.1021/es201212r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culman SW, Bukowski R, Gauch HG, Cadillo-Quiroz H, Buckley DH. T-REX: software for the processing and analysis of T-RFLP data. BMC Bioinformatics. 2009;10:171. doi: 10.1186/1471-2105-10-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalsgaard T, De Brabandere L, Hall POJ. Denitrification in the water column of the central Baltic Sea. Geochim Cosmochim Acta. 2013;106:247–260. [Google Scholar]
- Diaz RJ, Rosenberg R. Spreading dead zones and consequences for marine ecosystems. Science. 2008;321:926–929. doi: 10.1126/science.1156401. [DOI] [PubMed] [Google Scholar]
- Finster K, Liesack W, Thamdrup B. Elemental sulfur and thiosulfate disproportionation by Desulfocapsa sulfoexigens sp nov, a new anaerobic bacterium isolated from marine surface sediment. Appl Environ Microbiol. 1998;64:119–125. doi: 10.1128/aem.64.1.119-125.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs BM, Woebken D, Zubkov MV, Burkill P, Amann R. Molecular identification of picoplankton populations in contrasting waters of the Arabian Sea. Aquatic Microbial Ecol. 2005;39:145–157. [Google Scholar]
- Giovannoni SJ, Rappe MS, Vergin KL, Adair NL. 16S rRNA genes reveal stratified open ocean bacterioplankton populations related to the Green non-sulfur bacteria. Proc Natl Acad Sci USA. 1996;93:7979–7984. doi: 10.1073/pnas.93.15.7979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni SJ, Rappé MS.2000Evolution, diversity and molecular ecology of marine prokaryotesIn: Kirchman DL, (ed)Microb Ecol Oceans Wiley-Liss: New York; 47–85. [Google Scholar]
- Glaubitz S, Kiesslich K, Meeske C, Labrenz M, Jurgens K. SUP05 dominates the gammaproteobacterial sulfur oxidizer assemblages in pelagic redoxclines of the Central Baltic and Black Seas. Appl Environ Microbiol. 2013;79:2767–2776. doi: 10.1128/AEM.03777-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasshof K, Kremling K, Ehrhardt M.1999Methods of Seawater Analysis3rd edn.Wiley VCH [Google Scholar]
- Grote J, Jost G, Labrenz M, Herndl GJ, Juergens K. Epsilonproteobacteria represent the major portion of chemoautotrophic bacteria in sulfidic waters of pelagic redoxclines of the Baltic and Black Seas. Appl Environ Microbiol. 2008;74:7546–7551. doi: 10.1128/AEM.01186-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote J, Schott T, Bruckner CG, Glockner FO, Jost G, Teeling H, et al. Genome and physiology of a model Epsilonproteobacterium responsible for sulfide detoxification in marine oxygen depletion zones. Proc Natl Acad Sci USA. 2012;109:506–510. doi: 10.1073/pnas.1111262109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson BG, Stigebrandt A. Dynamics of nutrients and oxygen/hydrogen sulfide in the Baltic Sea deep water. J Geophys Res-Biogeo. 2007;112 [Google Scholar]
- Herlemann DPR, Labrenz M, Jurgens K, Bertilsson S, Waniek JJ, Andersson AF. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 2011;5:1571–1579. doi: 10.1038/ismej.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber T, Faulkner G, Hugenholtz P. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics. 2004;20:2317–2319. doi: 10.1093/bioinformatics/bth226. [DOI] [PubMed] [Google Scholar]
- Inagaki F, Takai K, Hideki KI, Nealson KH, Horikishi K. Sulfurimonas autotrophica gen. nov., sp nov., a novel sulfur-oxidizing Epsilon-proteobacterium isolated from hydrothermal sediments in the mid-Okinawa Trough. Int J Syst Evol Microbiol. 2003;53:1801–1805. doi: 10.1099/ijs.0.02682-0. [DOI] [PubMed] [Google Scholar]
- Jayakumar A, O'Mullan GD, Naqvi SWA, Ward BB. Denitrifying bacterial community composition changes associated with stages of denitrification in oxygen minimum zones. Microb Ecol. 2009;58:350–362. doi: 10.1007/s00248-009-9487-y. [DOI] [PubMed] [Google Scholar]
- Jorgensen BB. Ecology of the bacteria of the sulphur cycle with special reference to anoxic-oxic interface environments. Philos Trans R Soc Lond B. 1982;298:543–561. doi: 10.1098/rstb.1982.0096. [DOI] [PubMed] [Google Scholar]
- Kaplan CW, Kitts CL. Variation between observed and true Terminal restriction fragment length is dependent on true TRF length and purine content. J Microbiol Meth. 2003;54:121–125. doi: 10.1016/s0167-7012(03)00003-4. [DOI] [PubMed] [Google Scholar]
- Koskinen K, Hultman J, Paulin L, Auvinen P, Kankaanpaa H. Spatially differing bacterial communities in water columns of the northern Baltic Sea. FEMS Microbiol Ecol. 2011;75:99–110. doi: 10.1111/j.1574-6941.2010.00987.x. [DOI] [PubMed] [Google Scholar]
- Kuypers MMM, Lavik G, Woebken D, Schmid M, Fuchs BM, Amann R, et al. Massive nitrogen loss from the Benguela upwelling system through anaerobic ammonium oxidation. Proc Natl Acad Sci USA. 2005;102:6478–6483. doi: 10.1073/pnas.0502088102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrenz M, Jost G, Pohl C, Beckmann S, Martens-Habbena W, Jurgens K. Impact of different in vitro electron donor/acceptor conditions on potential chemolithoautotrophic communities from marine pelagic redoxclines. Appl Environ Microbiol. 2005;71:6664–6672. doi: 10.1128/AEM.71.11.6664-6672.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrenz M, Grote J, Mammitzsch K, Boschker HTS, Laue M, Jost G, et al. Sulfurimonas gotlandica sp nov., a chemoautotrophic and psychrotolerant Epsilonproteobacterium isolated from a pelagic redoxcline, and an emended description of the genus Sulfurimonas. Int J Syst Evol Micr. 2013;63:4141–4148. doi: 10.1099/ijs.0.048827-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam P, Lavik G, Jensen MM, van de Vossenberg J, Schmid M, Woebken D, et al. Revising the nitrogen cycle in the Peruvian oxygen minimum zone. Proc Natl Acad Sci USA. 2009;106:4752–4757. doi: 10.1073/pnas.0812444106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WT, Marsh TL, Cheng H, Forney LJ. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol. 1997;63:4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liungman O, Rydberg L, Goransson CG. Modeling and observations of deep water renewal and entrainment in a Swedish sill fjord. J Phys Oceanogr. 2001;31:3401–3420. [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhu K, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matear RJ, Hirst AC. Long-term changes in dissolved oxygen concentrations in the ocean caused by protracted global warming. Global Biogeochem Cycles. 2003;17:4. [Google Scholar]
- Morales CE, Hormazabal SE, Blanco JL. Interannual variability in the mesoscale distribution of the depth of the upper boundary of the oxygen minimum layer off northern Chile (18-24S): implications for the pelagic system and biogeochemical cycling. J Mar Res. 1999;57:909–932. [Google Scholar]
- Morris RM, Vergin KL, Cho JC, Rappe MS, Carlson CA, Giovannoni SJ. Temporal and spatial response of bacterioplankton lineages to annual convective overturn at the Bermuda Atlantic Time-series Study site. Limnol Oceanogr. 2005;50:1687–1696. [Google Scholar]
- Morrison JM, Codispoti LA, Smith SL, Wishner K, Flagg C, Gardner WD, et al. The oxygen minimum zone in the Arabian Sea during 1995. Deep-Sea Res Part II-Top Studies Oceanogr. 1999;46:1903–1931. [Google Scholar]
- Murillo AA, Ramírez-Flandes S, DeLong EF, Ulloa O.2014Enhanced metabolic versatility of planktonic sulfur-oxidizing γ-proteobacteria in an oxygen-deficient coastal ecosystem Front Mar Scie-pub ahead of print 7 July 2014doi: 10.3389/fmars.2014.00018 [DOI]
- Musat N, Halm H, Winterholler B, Hoppe P, Peduzzi S, Hillion F, et al. A single-cell view on the ecophysiology of anaerobic phototrophic bacteria. Proc Natl Acad Sci USA. 2008;105:17861–17866. doi: 10.1073/pnas.0809329105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqvi SWA, Jayakumar DA, Narvekar PV, Naik H, VVSS Sarma, D'Souza W, et al. Increased marine production of N2O due to intensifying anoxia on the Indian continental shelf. Nature. 2000;408:346–349. doi: 10.1038/35042551. [DOI] [PubMed] [Google Scholar]
- Nocker A, Burr M, Camper AK. Genotypic microbial community profiling: a critical technical review. Microb Ecol. 2007;54:276–289. doi: 10.1007/s00248-006-9199-5. [DOI] [PubMed] [Google Scholar]
- Pilloni G, Granitsiotis MS, Engel M, Lueders T. Testing the limits of 454 pyrotag sequencing: reproducibility, quantitative assessment and comparison to T-RFLP fingerprinting of aquifer microbes. PLoS One. 2012;7:e40467. doi: 10.1371/journal.pone.0040467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig WG, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricke P, Kolb S, Braker G. Application of a newly developed ARB software-integrated tool for in silico terminal restriction fragment length polymorphism analysis reveals the dominance of a novel pmoA cluster in a forest soil. Appl Environ Microbiol. 2005;71:1671–1673. doi: 10.1128/AEM.71.3.1671-1673.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemann L, Leitet C, Pommier T, Simu K, Holmfeldt K, Larsson U, et al. The native bacterioplankton community in the central Baltic sea is influenced by freshwater bacterial species. Appl Environ Microbiol. 2008;74:503–515. doi: 10.1128/AEM.01983-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scranton MI, Astor Y, Bohrer R, Ho TY, Muller-Karger F. Controls on temporal variability of the geochemistry of the deep Cariaco Basin. Deep-Sea Res Part I-Oceanogr Res Pap. 2001;48:1605–1625. [Google Scholar]
- Sievert SM, Wieringa EBA, Wirsen CO, Taylor CD. Growth and mechanism of filamentous-sulfur formation by Candidatus Arcobacter sulfidicus in opposing oxygen-sulfide gradients. Environ Microbiol. 2007;9:271–276. doi: 10.1111/j.1462-2920.2006.01156.x. [DOI] [PubMed] [Google Scholar]
- Sjostedt J, Koch-Schmidt P, Pontarp M, Canback B, Tunlid A, Lundberg P, et al. Recruitment of members from the rare biosphere of marine bacterioplankton communities after an environmental disturbance. Appl Environ Microbiol. 2012;78:1361–1369. doi: 10.1128/AEM.05542-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens H, Ulloa O. Bacterial diversity in the oxygen minimum zone of the eastern tropical South Pacific. Environ Microbiol. 2008;10:1244–1259. doi: 10.1111/j.1462-2920.2007.01539.x. [DOI] [PubMed] [Google Scholar]
- Stewart FJ, Ulloa O, DeLong EF. Microbial metatranscriptomics in a permanent marine oxygen minimum zone. Environ Microbiol. 2012;14:23–40. doi: 10.1111/j.1462-2920.2010.02400.x. [DOI] [PubMed] [Google Scholar]
- Stigebrandt A, Gustafsson BG. Improvement of Baltic proper water quality using large-scale ecological engineering. AMBIO. 2007;36:280–286. doi: 10.1579/0044-7447(2007)36[280:iobpwq]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Stigebrandt A, Liljebladh B.2011Oxygenation of large volumes of natural waters by geoengineering: with particular reference to a pilot experiment in ByfjordenIn: Badescu V, Cathcart RB, (eds)Macroengineering Seawater in Unique Environments Springer: Berlin, Heidelberg [Google Scholar]
- Stigebrandt A, Liljebladh B, de Brabandere L, Forth M, Granmo A, Hall P, et al. 2014An experiment with forced oxygenation of the deepwater of the anoxic by fjord Western Sweden; Ambio. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stramma L, Johnson GC, Sprintall J, Mohrholz V. Expanding oxygen-minimum zones in the tropical oceans. Science. 2008;320:655–658. doi: 10.1126/science.1153847. [DOI] [PubMed] [Google Scholar]
- Treusch AH, Vergin KL, Finlay LA, Donatz MG, Burton RM, Carlson CA, et al. Seasonality and vertical structure of microbial communities in an ocean gyre. ISME J. 2009;3:1148–1163. doi: 10.1038/ismej.2009.60. [DOI] [PubMed] [Google Scholar]
- Walsh DA, Zaikova E, Howes CG, Song YC, Wright JJ, Tringe SG, et al. Metagenome of a versatile chemolithoautotroph from expanding oceanic dead zones. Science. 2009;326:578–582. doi: 10.1126/science.1175309. [DOI] [PubMed] [Google Scholar]
- Ward BB, Tuit CB, Jayakumar A, Rich JJ, Moffett J, Naqvi SWA. Organic carbon, and not copper, controls denitrification in oxygen minimum zones of the ocean. Deep-Sea Res Part I-Oceanogr Res Pap. 2008;55:1672–1683. [Google Scholar]
- Ward BB, Devol AH, Rich JJ, Chang BX, Bulow SE, Naik H, et al. Denitrification as the dominant nitrogen loss process in the Arabian Sea. Nature. 2009;461:78–U77. doi: 10.1038/nature08276. [DOI] [PubMed] [Google Scholar]
- Wirsen CO, Sievert SM, Cavanaugh CM, Molyneaux SJ, Ahmad A, Taylor LT, et al. Characterization of an autotrophic sulfide-oxidizing marine Arcobacter sp that produces filamentous sulfur. Appl Environ Microbiol. 2002;68:316–325. doi: 10.1128/AEM.68.1.316-325.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woebken D, Teeling H, Wecker P, Dumitriu A, Kostadinov I, DeLong EF, et al. Fosmids of novel marine planctomycetes from the Namibian and Oregon coast upwelling systems and their cross-comparison with planctomycete genomes. Int Soc Microbial Ecol J. 2007;1:419–435. doi: 10.1038/ismej.2007.63. [DOI] [PubMed] [Google Scholar]
- Woebken D, Lam P, Kuypers MMM, Naqvi SWA, Kartal B, Strous M, et al. A microdiversity study of anammox bacteria reveals a novel Candidatus Scalindua phylotype in marine oxygen minimum zones. Environ Microbiol. 2008;10:3106–3119. doi: 10.1111/j.1462-2920.2008.01640.x. [DOI] [PubMed] [Google Scholar]
- Worthen PL, Gode CJ, Graf J. Culture-independent characterization of the digestive-tract microbiota of the medicinal leech reveals a tripartite symbiosis. Appl Environ Microbiol. 2006;72:4775–4781. doi: 10.1128/AEM.00356-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JJ, Konwar KM, Hallam SJ. Microbial ecology of expanding oxygen minimum zones. Nat Rev Microbiol. 2012;10:381–394. doi: 10.1038/nrmicro2778. [DOI] [PubMed] [Google Scholar]
- Zaikova E, Walsh DA, Stilwell CP, Mohn WW, Tortell PD, Hallam SJ. Microbial community dynamics in a seasonally anoxic fjord: Saanich Inlet, British Columbia. Environ Microbiol. 2010;12:172–191. doi: 10.1111/j.1462-2920.2009.02058.x. [DOI] [PubMed] [Google Scholar]
- Zillen L, Conley DJ, Andren T, Andren E, Bjorck S. Past occurrences of hypoxia in the Baltic Sea and the role of climate variability, environmental change and human impact. Earth-Sci Rev. 2008;91:77–92. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.