Abstract
Thiomicrospira species are ubiquitously found in various marine environments and appear particularly common in hydrothermal vent systems. Members of this lineage are commonly classified as sulfur-oxidizing chemolithoautotrophs. Although sequencing of Thiomicrospira crunogena's genome has revealed genes that encode enzymes for hydrogen uptake activity and for hydrogenase maturation and assembly, hydrogen uptake ability has so far not been reported for any Thiomicrospira species. We isolated a Thiomicrospira species (SP-41) from a deep sea hydrothermal vent and demonstrated that it can oxidize hydrogen. We show in vivo hydrogen consumption, hydrogen uptake activity in partially purified protein extracts and transcript abundance of hydrogenases during different growth stages. The ability of this strain to oxidize hydrogen opens up new perspectives with respect to the physiology of Thiomicrospira species that have been detected in hydrothermal vents and that have so far been exclusively associated with sulfur oxidation.
Introduction
Thiomicrospira species are widespread in marine environments and have been isolated from hydrothermal vents, intertidal mud flats, marine Arctic sediments and continental shelf sediments (Kuenen and Veldkamp, 1972; Ruby and Jannasch, 1982; Jannasch et al., 1985; Brinkhoff and Muyzer, 1997; Brinkhoff et al., 1999a; 1999b; Knittel et al., 2005; Takai et al., 2004). All so far isolated Thiomicrospira species are capable of using reduced sulfur compounds as electron donors and are described as chemolithoautotrophs. However, according to Takai et al. (2004) T. thermophila and two T. crunogena strains, namely L-12 and TH-55, should rather be described as chemolithomixotrophs. Although Thiomicrospira's close relative Hydrogenovibrio can use hydrogen as an electron donor for growth (Nishihara et al., 1998, 2001), attempts to cultivate Thiomicrospira with hydrogen as sole electron donor have remained unsuccessful (Nishihara et al., 1991). Sequencing of T. crunogena XCL-2's genome has revealed that genes encoding the large and small subunit of the NiFe hydrogenase as well as those of enzymes necessary for the assembly and maturation of the structural hydrogenase are available suggesting that under certain conditions XCL-2 may indeed be able to use hydrogen (Scott et al., 2006).
Thiomicrospira species have been repeatedly recognized in hydrothermal environments (for example, Brazelton and Baross, 2010, Perner et al., 2011) where reduced hydrothermal fluids transport energy-rich inorganic electron donors like hydrogen or reduced sulfur compounds to the surface (Jannasch and Mottl, 1985). Phylogenetically diverse microorganisms can oxidize these reduced substrates whereby they gain energy that some can use for autotrophic CO2 fixation (Campbell et al., 2006, Jannasch and Mottl, 1985, Miroshnichenko and Bonch-Osmolovskaya, 2006). If Thiomicrospira indeed cannot utilize hydrogen in habitats like Lost City, where sulfide is particularly scarce, but hydrogen is highly abundant, the widespread predominance of Thiomicrospira is somewhat puzzling (Brazelton et al., 2006, 2010; Brazelton and Baross, 2010).
We collected low-temperature hydrothermal fluids (⩽16 °C) emanating near the Sisters Peak chimney on the Mid-Atlantic Ridge where mixed fluids had a pH of 6.6, 1.2 μM hydrogen and 82 μM sulfide (Perner et al., 2013). From these emissions we isolated a Thiomicrospira species (SP-41) that can oxidize hydrogen. In the following we present in vivo hydrogen consumption rates, hydrogen uptake activity of partially purified protein extracts and hydrogenase transcript abundance at different growth stages of SP-41.
Materials and Methods
Sample collection
Low-temperature hydrothermal fluids emanating near the Sisters Peak chimney structure were collected (Perner et al., 2013). The Sisters Peak area is hosted in basalt and is located at 4°48′S Mid-Atlantic Ridge at a water depth of roughly 3000 m (Koschinsky et al., 2008). Hydrothermal fluids were retrieved during dives with the remotely operated vehicle Kiel ROV 6000 (GEOMAR, Kiel, Germany) during the cruise MAR-SUED V (April/May 2009) with the RV Meteor. Hydrothermal fluids were collected using the pumped flow-through system KIPS (Kiel Pumping System) (Garbe-Schönberg et al., 2006). To ensure that the retrieved fluids were indeed hydrothermally influenced liquids, we monitored the in situ temperature of the fluids online during sampling—temperature is a tracer for mixing of hydrothermal fluids with ambient seawater. Prior to fluid sampling, the fluid collection system was flushed for several minutes with the vent fluids, which exhibited a stable above ambient seawater temperature (⩽16 °C), before the valves were closed to collect the actual sample. Fluid chemistry conducted on these fluid samples evidenced that these collected fluids were hydrothermal fluids and not ambient seawater (cf. Perner et al., 2013). For further details on sampling with the KIPS, see Perner et al. (2009).
Enrichment and isolation of Thiomicrospira sp. SP-41
Initially an enrichment culture containing SP-41 was grown by inoculating hydrogen-spiked artificial seawater medium (MJ medium, 10 ml) with 1 ml of low-temperature hydrothermal fluids. MJ medium was prepared as described before (Sako et al., 1996), without yeast extract and without trypticase peptone, but 10 ml of vitamin solution was added (Balch et al., 1979). A volume of 50 ml of medium was filled into 120 ml serum bottles closed with rubber stoppers and purged with a H2:CO2:O2 (79:20:1) gas mixture (Westfalen AG, Münster, Germany). These flasks were inoculated with 2 ml of the 10 ml pre-culture. The reduction state of the medium was monitored with resazurin (0.5 mg l−1). According to the color change in the medium (from blue to colorless), oxygen was being rapidly used up within 24 h. After 24 h incubation at 28 °C, the culture was transferred to fresh MJ medium as described above. In the home lab Thiomicrospira sp. SP-41 was isolated by plating the culture on T-ASW plates (containing 40 mM thiosulfate) (Dobrinski et al., 2005) and then streaked on fresh T-ASW plates two more times to obtain isolated colonies. The purity of SP-41 was verified with fluorescence in situ hybridization and cloning and sequencing of 16S rRNA gene fragments (details see below). SP-41 was then routinely cultivated on MJ medium as described above, but with the addition of 0.6 mM thiosulfate (MJ-T medium).
Cultivation of other Thiomicrospira species
TH-55 and T. thermophila were obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ, Braunschweig, Germany). When the experiments were performed T. crunogena XCL-2 was not available from public culture collections (for example, DSMZ and ATCC) and was therefore not included in our growth experiments. TH-55 was grown on T-ASW medium at 28 °C (Dobrinski et al., 2005) and T. thermophila on DSMZ medium 1011 at 37 °C, using a H2:CO2 (80:20) gas mix (Westfalen AG) instead of N2:CO2.
In vivo hydrogen consumption experiments
For hydrogen consumption experiments all bacteria were grown in serum bottles on the respective media and a gas mix of H2:CO2:O2:He (2:20:1:77) (Westfalen AG). TH-55 was grown on T-ASW medium and T. thermophila was grown on DSMZ 1011 medium as described above. For T. thermophila grown in DSMZ medium 1011 subsequently 20% of the headspace was replaced by sterile air. For comparison with SP-41 both strains were additionally grown in MJ-T medium (MJ medium with 0.6 mM thiosulfate) as described above. SP-41 cells were initially grown in MJ-T medium to gain sufficient inoculation material and incubated for 1–2 days at 28 °C. Cultures were then transferred 1:100 into MJ medium: (i) with 0.6 mM thiosulfate (MJ-T medium), (ii) with 0.5 mM cysteine but no thiosulfate (MJ-C medium) and (iii) without thiosulfate or cysteine (MJ medium). To monitor pH changes in the medium, 2 ml of filter-sterilized 0.5% phenol red solution was added. Hydrogen consumption was measured in vivo using a Trace GC Ultra gas chromatograph (Thermo Fisher Scientific Inc., Waltham, MA, USA) equipped with a ShinCarbon ST 100/120 column (Restek Corporation, Bellefonte, PA, USA) and a Pulse Discharge Detector (Vici Valco Instruments, Houston, TX, USA) with helium 6.0 as carrier gas (Linde Group, München, Germany). For every measurement 1 ml of the headspace was taken, diluted 1:87 and from this dilution 2 ml injected into the GC. Hydrogen consumption rates were calculated as described before (Perner et al., 2010). Cell numbers were determined by direct cell counting using a counting chamber (Hawksley, Sussex, UK). The cultures were checked for purity at the end of the experiments with fluorescence in situ hybridization and/or PCR and subsequent sequencing of 16S rRNA or hydrogenase genes. All experiments were performed at least in triplicate, controls were performed in duplicate. Mean values and standard deviations are from three independent measurements.
Hydrogen uptake activity for Thiomicrospira protein extracts
For enzyme assays SP-41 and TH-55 were grown in MJ-T medium (0.6 mM thiosulfate) as described above and regassed with H2:CO2:O2 (79:20:1) (Westfalen AG). Filter-sterilized 0.5% phenol red solution (2 ml) was added to monitor pH changes in the medium. The purity of the strains was controlled before performing the enzyme assays. All further steps were performed in an anaerobic chamber (Coy Laboratory Products, Grass Lake, MI, USA) to prevent direct contact of hydrogenases with oxygen. After ∼2.5 days of incubation, cells were harvested by centrifugation at 17 000 g (Sorvall RC 6 Plus Centrifuge, Thermo Fisher Scientific Inc.) and washed once in anoxic 50 mM Tris buffer pH 8. Hydrogenases were partially purified as described before (Adams and Hall, 1979) with a few variations: throughout the assay anoxic 50 mM Tris buffer pH 8 with 5 mM 1,4-dithiothreitol (Tris-DTT) were used in all steps. The cells were broken by sonication (UP50H, Hielscher Ultrasonics GmbH, Teltow, Germany) and cell debris and membranes harvested by ultracentrifugation at 40 000 g (LM-8 Ultracentrifuge, Beckman Coulter Inc., Brea, CA, USA) for 1 h. Membrane proteins were solubilized by using 3% sodium deoxycholate. Ammonium sulfate precipitation was performed by saturating the buffer consecutively at different concentrations (15%, 30%, 50% and 70%) and pelleting of proteins by ultracentrifugation with subsequent resuspension in Tris-DTT buffer.
Protein concentrations of the fractions were determined using the method from Bradford (1976). Hydrogen uptake activity was measured following previous descriptions (Muth et al., 1987; Payne et al., 1993) with oxidized methyl viologen as artificial electron acceptor. The change in absorption was monitored spectrophotometrically (V-630 UV-Vis Spectrophotometer equipped with an EHCS-760 Peltier Cell Holder, Jasco, Gross-Umstadt, Germany) in reaction buffer (50 mM Tris pH 8, 5 mM DTT and 10 mM methyl viologen dichloride) at 602 nm and 25 °C. The extinction coefficient was 12.04 mM-1 cm−1 at assay conditions. The anoxic reaction buffer was filled into cuvettes (Hellma GmbH & Co. KG, Müllheim, Germany) sealed with rubber stoppers during gassing with N2 5.0 (⩾99.999%) (Westfalen AG) and was reduced by adding 5 mM sodium dithionite solution. Protein fractions were added from an anoxic solution. Controls consisted of reaction buffer with protein solution where the enzymatic reaction was monitored under a N2 atmosphere. For determining the hydrogen uptake activity of the protein extracts, subsequently nitrogen was replaced by H2 5.0 (⩾99.999%) (Westfalen AG). Hydrogen uptake activity calculations were performed from three independent measurements.
Hydrogenase transcripts at different growth stages
To quantify the hydrogenase large subunit transcripts of SP-41 during growth, cells were grown on MJ-T medium with H2:CO2:O2:He (2:20:1:77) (Westfalen AG) in the headspace for up to 172 h. Cells were harvested at different time points (8, 22.5, 46.5 and 172 h) of growth. For reverse transcription quantitative PCR (RT-qPCR) the following kits were used according to the manufacturer's protocols: Presto Mini RNA Bacteria Kit (Genaid Biotech Ltd., New Taipei City, Taiwan) for total RNA isolation, RTS DNase Kit (Mo Bio Laboratories Inc., Carlsbad, CA, USA), RNA Clean & Concentrator-5 (Zymo Research Corporation, Irvine, CA, USA), SuperScript VILO Master Mix (Life Technologies, Carlsbad, CA, USA) for synthesizing cDNA and SYBR Select Master Mix for CFX (Life Technologies). The primers 40 F (5′-CCA GTC ACC CGA ATT GAA GG-3′) and 189 R (5′-AGC AAT CAA ACC GGT ATC CC-3′) targeting the SP-41 hydrogenase large subunit gene were used (initial denaturation at 95 °C for 2 min, 40 cycles of denaturation at 95 °C for 15 s, annealing at 52 °C for 20 s, elongation at 72 °C for 15 s). Controls of the template lacking reverse transcriptase were performed by PCR. RT-qPCR results were evaluated with the Bio-Rad CFX Manager software (Bio-Rad Laboratories, Hercules, CA, USA). Transcription experiments were performed in triplicate.
Fluorescence in situ hybridization
Approximately 2 × 106 cells of each culture were fixed with formaldehyde (4% v/v) for 4 h, concentrated on polycarbonate filters (type: GTTP, pore size 0.2 μm, Millipore, Eschborn, Germany), washed twice with 1 × PBS (phosphate-buffered saline) and stored at 4 °C until further treatment. In addition, 250 ml of low-temperature hydrothermal fluids were treated as described above and fixed on a polycarbonate filter. Fluorescence in situ hybridization analyses with cy-3 labeled oligonucleotide probes (final concentration 50 ng μl−1) and 4′,6-diamidino-2-phenylindole (5 μg ml−1) coloring were performed as stated in Glöckner et al. (1999). A new probe, Tcr69 (5′-CTT GTC GTT TCC GTC CG-3′) was designed based on the 16S rRNA gene sequences of several T. crunogena species, namely SP-41, TH-55, XCL-2, L-12 and JB-B2 as well as T. halophila and required a concentration of 35% formamide for successfully targeting T. crunogena species. The probe has one mismatch to T. thermophila for which it does not give any signals. In addition, the nonsense probe NON338 (Wallner et al., 1993) was applied to all filters, which did not give any signals. Filter sections were inspected using an Axio Imager.M2 microscope (Carl Zeiss AG, Oberkochen, Germany).
DNA extraction and processing of 16S rRNA and hynL genes
DNA from SP-41 and TH-55 was extracted with the ultra clean microbial DNA isolation kit (Mo Bio Laboratories Inc.) according to the manufacturer's instructions. 16S rRNA genes were PCR amplified using the primers 27 F and 1492 R (Lane, 1991) and the conditions described previously (Perner et al., 2009). Details on the cloning for genes have been described before (Perner et al., 2009). For sequencing of both DNA strands of the 16S rRNA genes from SP-41 and TH-55, the primers 27 F, 1492 R, 562 (5′-TAA TCT TGC GAC CGT ACT CC-3′), 719 (5′-CTG ACG CTG AGG TGC GAA AG-3′) and 1149 (5′-CCC AGT GTG GCT GAT CAT CC-3′) were used. The hynL (large subunit of the structural membrane-bound uptake hydrogenase) gene from TH-55 was partially amplified using the primers hynl110F and hynl410R (Takai et al., 2005). PCR conditions were: initial denaturation 95 °C for 5 min followed by 32 cycles of denaturation at 95 °C for 45 s, annealing at 45 °C for 45 s and elongation for 70 s. The PCR product was cloned as described above. For double-stranded sequencing, the primers hynl110F, hynl410R, 208 (5′-TTA ACG CAG GGC GTT TAG GG-3′), 338 (5′-GCT CAC TTC CCG CTG AAT CC-3′), 614 (5′-TGC CAG CGG CAT TAT TTG GG-3′) and 714 (5′-CTT CGT CGG CAT ACC AGG AG-3′) were used. To sequence SP-41's full hynL gene, a fosmid clone containing the whole gene was used as template. The fosmid library was constructed with the CopyControl Fosmid Library Production Kit (Epicentre, Madison, WI, USA) according to the manufacturer's instructions. For double-stranded sequencing, the primers hynl110F, hynl410R, 90 (5′-TCC TGC ACG GCT GCC AAA TC-3′), 502 (5′-TGA ACC GTT GTT TGC CAG TG-3′), 571 (5′-AGT GTC GTC AAA CGG TTC TG-3′) and 656 (5′-ACC GAT ATG AAT GCG GAT GG-3′) were used. Sequencing was performed with a 3730xl DNA Analyzer (Applied Biosystems, Life Technologies). All sequences were edited with Lasergene Software SeqMan (DNAStar, Madison, WI, USA). 16S rRNA and hynL gene sequences were edited and analyzed as previously described (Perner et al., 2009, 2010). Phylogenetic trees were calculated using PhyML (Guindon and Gascuel, 2003) and settings as described before (Perner et al., 2009, 2010).
Data deposition
The 16S rRNA and hynL gene sequences from SP-41 and TH-55 were deposited in the National Center for Biotechnology Information under the accession numbers KJ573628, KJ573629, KJ573630 and KJ573631, respectively. The new strain SP-41 has been submitted to the DSMZ and is designated DSM 28671.
Results and Discussion
Phylogeny of Thiomicrospira sp. SP-41
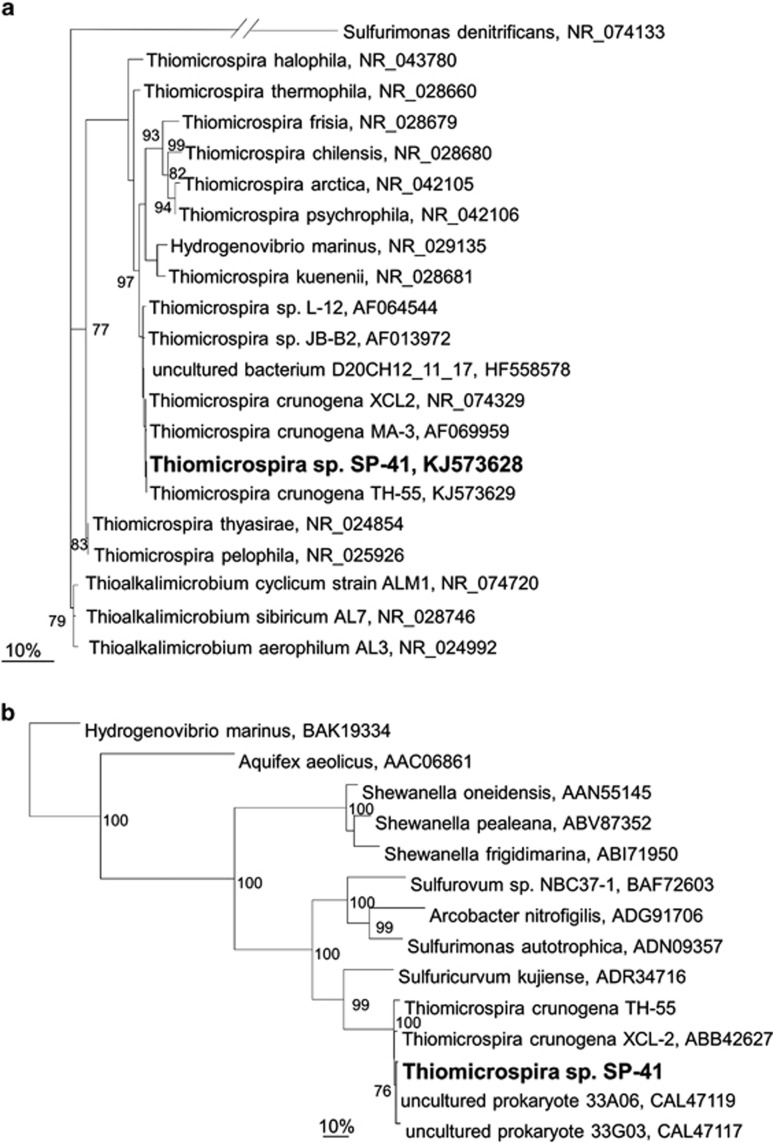
Our isolate SP-41 was phylogenetically classified as a Thiomicrospira species (Figure 1a). On the basis of 16S rRNA genes, SP-41's closest relatives were three T. crunogena strains, namely, TH-55, MA-3 and XCL-2 (99% DNA identity), with 1, 2 and 3 nt differences, respectively. The large subunit of SP-41's NiFe uptake hydrogenase (hynL) resembled hynL of XCL-2 (90% DNA and 98% AA identity) and the partial hynL sequence of TH-55 (89% DNA identity and 97% AA identity) (Figure 1b). Interestingly, the cluster containing the hynL genes of SP-41, XCL-2 and TH-55 exhibited the highest relatedness to hynL genes from Epsilonproteobacteria (⩽71% AA identity). The best hit to non-Epsilonproteobacteria was to Shewanella (Gammaproteobacteria, 70% DNA and 49% AA identity). Moreover, Hydrogenovibrio, which belongs to the same family as Thiomicrospira, showed only 40% AA similarity (Figure 1b). As SP-41, TH-55 and XCL-2 were all isolated from hydrothermal vent environments (Jannasch et al., 1985; Wirsen et al., 1998) and Epsilonproteobacteria are one of the most dominant groups found in these habitats (Campbell et al., 2006), these Thiomicrospira hynL genes may have been acquired by a common ancestor through lateral gene transfer from Epsilonproteobacteria. Likewise, other genes in XCL-2 have also been posited to have been obtained through lateral gene transfer from different organisms (Scott et al., 2006). This may indicate rapid evolution in this type of environments, but more information need to be gathered to investigate this hypothesis in detail.
Figure 1.
Phylogenetic relationships of Thiomicrospira sp. SP-41 (a) 16S rRNA genes and (b) the large subunits of the uptake hydrogenase (hynL) based on maximum likelihood analysis. The percentage of bootstrap resamplings is indicated at the nodes and only bootstrap values >75 are shown. The scale bar represents the expected number of changes per nucleotide (a) or amino-acid (b) position.
Growth, H2 consumption, hydrogenase activity and hynL expression of SP-41 with thiosulfate
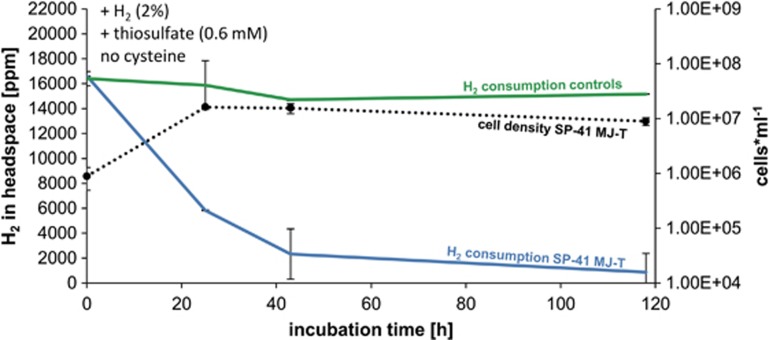
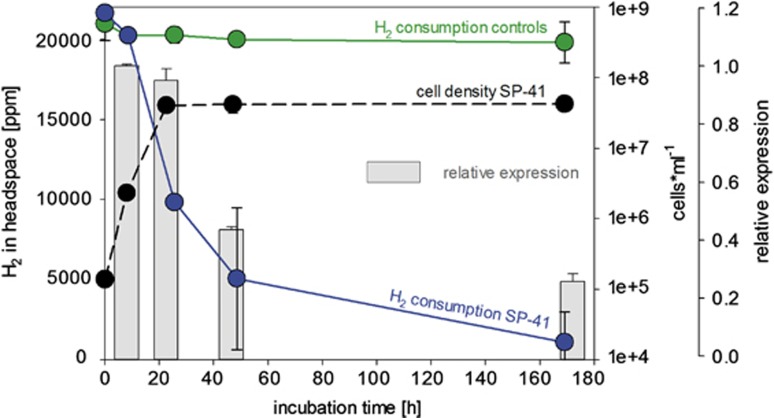
SP-41 grown on artificial seawater medium supplemented with 0.6 mM thiosulfate (MJ-T medium) and hydrogen, consumed most of the hydrogen (∼86%) within the first 40 h of incubation (Figure 2). After ∼24 h of incubation 65% of the hydrogen was used up and cell density reached a maximum (2 × 107 cells ml−1) (Figure 2). Despite no further increase in cell density, hydrogen was still being consumed until approximately only 5% of the originally present hydrogen was available. The hydrogen uptake activity of SP-41's partially purified membrane fraction was 1.26±0.13 μmol H2 per min per mg protein (Supplementary Figure S1). This is in the range of what has been measured for other hydrogenases from deep sea hydrothermal microorganisms (Takai et al., 2005) and is ∼10-fold higher than the activity of T. crunogena's close relative H. marinus (Nishihara et al., 2001). Hydrogen uptake activity of SP-41's soluble proteins was negligible (Supplementary Figure S1). We also tested the transcription level of the hynL gene at different time points during growth on MJ-T medium with hydrogen, which demonstrated that the relative expression of the hynL gene varied in SP-41 with incubation time (Figure 3). The relative expression of the hynL gene transcript decreased to ∼45% after 46.5 h and ∼25% after 172 h of incubation. The reduction in hynL transcript products correlated with decreasing hydrogen concentration (23% and 5% of hydrogen were left after 46.5 and 172 h of incubation, respectively) and the cells entering the stationary phase (Figure 3). In summary, SP-41 expresses an active hydrogen-uptake hydrogenase enzyme and can consume hydrogen when thiosulfate is present.
Figure 2.
In vivo hydrogen consumption of SP-41 and cell numbers in MJ-T medium (0.6 mM thiosulfate) and H2:CO2:O2:He (2:20:1:77) in the headspace. SP-41's hydrogen consumption is shown in blue, respective cell numbers as broken black line and controls (only MJ-T medium) in green.
Figure 3.
Transcription experiments showing hydrogen consumption, cell numbers and hynL gene expression level for SP-41 grown in MJ-T medium (0.6 mM thiosulfate). SP-41's hydrogen consumption is shown in blue, respective cell numbers as broken black line and controls (only MJ-T medium) in green. Relative expressions of the hynL gene from SP-41 at different time points during growth are indicated as gray bars.
Growth and H2 consumption of SP-41 without thiosulfate
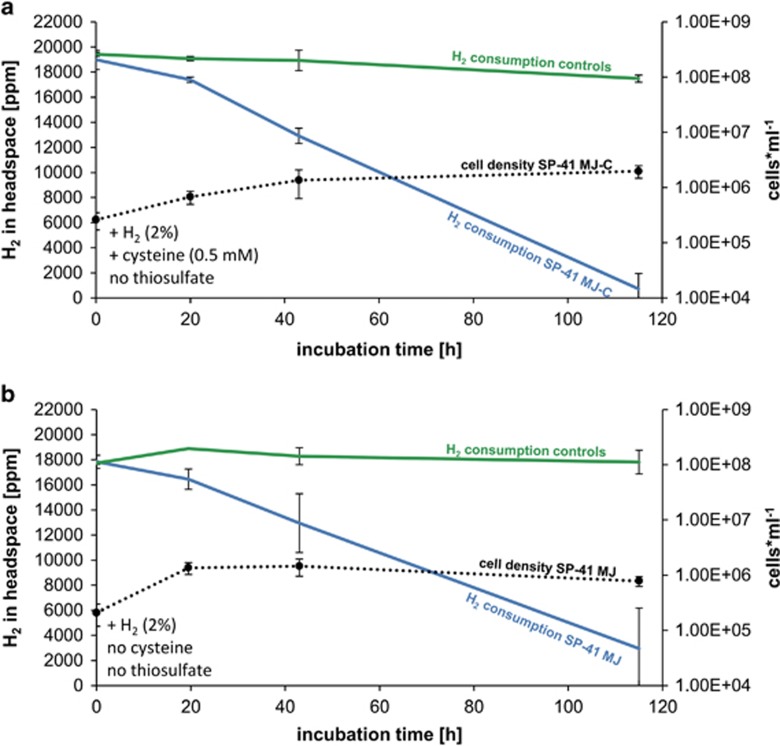
As the genome of XCL-2 lacks genes encoding enzymes of assimilatory sulfate reduction (APS reductase and ATP sulfurylase), which are essential for cysteine biosynthesis in the absence of thiosulfate or sulfide, it has been posited that XCL-2 cannot grow without adding respective sulfur sources (Scott et al., 2006). To test how dependent SP-41 is for its growth with hydrogen on thiosulfate and cysteine, hydrogen growth experiments were also performed in MJ medium (no thiosulfate) with and without supplemented cysteine (0.5 mM). In both experimental setups most of the hydrogen was consumed within 118 h of incubations (Figure 4). While offering cysteine (0.5 mM), SP-41 consumed 95% of the available hydrogen (Figure 4a), which compares with what we observed for SP-41 when thiosulfate (0.6 mM) was added (Figure 2). In contrast, when neither cysteine nor thiosulfate were supplemented, only 84% of the total hydrogen was used up within 118 h of cultivation (Figure 4b). The results indicate that SP-41 can consume hydrogen even in the absence of thiosulfate and cysteine.
Figure 4.
In vivo hydrogen consumption of SP-41 grown under different conditions: (a) SP-41 grown in MJ-C medium (without thiosulfate but with 0.5 mM cysteine), and in (b) MJ medium (without thiosulfate and without cysteine). Hydrogen consumption of SP-41 is shown in blue, respective cell numbers as broken black lines and controls (only MJ-C medium or MJ medium) in green.
Comparing different growth conditions of SP-41
When hydrogen is oxidized aerobically to water (H2+1/2O2≫H2O) the reaction yields 237 kJ mol-1 (Fuchs et al., 2007). Theoretically, a relation should exist between the oxidation of electron donors (catabolism) and autotrophic CO2 fixation (anabolism), governed by an empirical Gibbs energy dissipation coefficient (Heijnen and Van Dijken, 1992). For hydrogenotrophy ∼1060 kJ catabolic energy is required to fix 1 mol of carbon in biomass (Heijnen and Van Dijken, 1992). Given that we have 237 kJ mol−1 hydrogen oxidized aerobically to water, we would expect a molar ratio of hydrogen consumption to CO2 fixation on the order of five. In our experiments, during the exponential growth phase on MJ-T medium 1194 nmol H2 per h or 29 860 nmol H2 were oxidized—generating an estimated amount of 0.007 kJ energy—and 7.7 × 108 new cells were formed (Table 1). Assuming that all cells in the incubation consumed hydrogen, SP-41's hydrogen consumption rate in the exponential growth phase would be 1.47±0.03 fmol H2 per h per cell (Table 1), which is in the range of the consumptions of other bacterial strains (Häring and Conrad, 1991; Klüber and Conrad, 1993; Olson and Maier, 2002). If all these cells also fixed CO2 autotrophically then an estimated amount of 0.29 fmol CO2 per h per cell or 103 fg carbon per cell could be fixed in these incubations. This corresponds well with carbon contents measured for bacterial cells in the exponential growth phase in batch cultures (depending on the culture conditions between 39±3 fg carbon per cell and 149±8 fg of carbon per cell) (Vrede et al., 2002). Conclusively, the amount of hydrogen consumed in the MJ-T medium is theoretically sufficient to form the number of cells that we counted.
Table 1. Hydrogen consumption and cell numbers in different media during exponential growth of SP-41 and TH-55.
|
SP-41 |
TH-55 |
|||
|---|---|---|---|---|
|
Type of medium |
Medium |
|||
| MJ-T | MJ-C | MJ | MJ-T | |
| H2 consumption | ||||
| Total (nmol) | 29 860 | 3655 | 7421 | 21 102 |
| Per hour (nmol h−1) | 1194 | 183 | 381 | 898 |
| Per cell and time (fmol H2 per h per cell) | 1.47 | 6.10 | 5.90 | 0.73 |
| Energy (kJ) | 0.0071 | 0.0009 | 0.0018 | 0.005 |
| Total cell numbers | 8.2 × 108 | 3.4 × 107 | 6.8 × 107 | 1.2 × 109 |
In cultures without thiosulfate but with cysteine less hydrogen was consumed, that is, 3655 nmol hydrogen (or 183 nmol h-1), less energy generated (estimated 0.001 kJ) and fewer new cells (2.1 × 107) formed during exponential growth (Table 1). During exponential growth in cultures without thiosulfate and without cysteine only 7421 nmol H2 was used (that is, 381 nmol h-1), theoretically generating 0.002 kJ energy and producing 5.8 × 107 new cells (Table 1). Owing to lower cell densities in these cultures (Table 1) the estimated hydrogen consumption per cell was fourfold higher than in the incubations where thiosulfate was present: hydrogen consumption in the incubations without thiosulfate and with and without cysteine was 6.1 and 5.9 fmol H2 per h per cell, respectively (Table 1). Assuming that all these cells fix CO2 autotrophically and assuming a molar ratio of hydrogen consumption to primary production on the order of 5, theoretically 1.22 and 1.18 fmol CO2 per h per cell, respectively, could be fixed in these incubations by SP-41.
In summary, conclusions drawn from these three types of growth experiments, that is, artificial seawater and hydrogen (i) with thiosulfate (MJ-T medium) (Figure 2), (ii) with cysteine but no thiosulfate (MJ-C medium) (Figure 4a) and (iii) without cysteine or thiosulfate (MJ medium) (Figure 4b), are manifold: SP-41 can grow on hydrogen without supplementing thiosulfate and cysteine and hydrogen consumption per cell is comparable for both types of experiments (Table 1). However, cell growth was significantly higher when thiosulfate was added to the medium (P-value <0.001). As cell growth in MJ-T medium was higher than in the MJ-C and MJ media, but hydrogen consumption per cell and per hour was ∼fourfold lower in the MJ-T medium than in the other experiments, some of the newly formed biomass in the MJ-T medium is likely related to thiosulfate oxidation.
In vivo hydrogen consumption of reference strains and uptake hydrogenase activity of TH-55
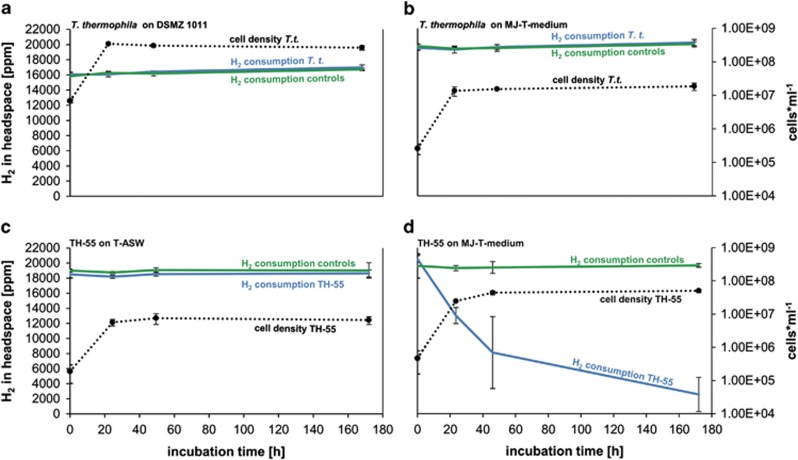
To investigate whether the ability to consume hydrogen is a unique feature of our new strain SP-41, we tested experimentally if T. thermophila and TH-55 can consume hydrogen in the medium they were originally grown in and in the medium (MJ-T medium) we used to grow SP-41. T. thermophila grew on both media tested, that is, DSMZ medium 1011 with hydrogen and MJ-T medium with hydrogen (Figures 5a and b). It grew significantly denser in the DSMZ medium 1011 than in the MJ-T medium (P-value <0.001), which is likely a consequence of higher cell density in the inoculum (Figures 5a and b). However, in neither experiments T. thermophila consumed hydrogen under the provided conditions (Figures 5a and b). This coincides with previous studies where it was shown that T. thermophila could not grow on hydrogen but required thiosulfate, elemental sulfur or sulfide as sole energy source (Takai et al., 2004). In line with this observation were our unsuccessful efforts to amplify hynL genes from T. thermophila with degenerate primers.
Figure 5.
In vivo hydrogen consumption of TH-55 and T. thermophila. (a) T. thermophila in DSMZ medium 1011, (b) T. thermophila in MJ-T medium (0.6 mM thiosulfate), (c) TH-55 in T-ASW medium and (d) TH-55 in MJ-T medium (0.6 mM thiosulfate). Hydrogen consumptions are shown as blue lines, respective cell numbers as broken black lines and controls (only DSMZ medium 1011, T-ASW medium or MJ-T medium) as green lines.
TH-55 was also not capable of consuming hydrogen when grown on the medium it was originally isolated on, that is, T-ASW medium (Figure 5c), which compares with previous attempts to cultivate T. crunogena including TH-55 on hydrogen (cf. Nishihara et al., 1991; Takai et al., 2004). In contrast, while cultivating TH-55 on MJ-T medium, hydrogen consumption and growth were observed (Figure 5d). The maximum consumption of TH-55 on MJ-T medium in the exponential growth phase was 0.73±0.13 fmol H2 per h per cell, which was half of SP-41's hydrogen consumption rate. As in the thiosulfate-rich medium T-ASW (40 mM thiosulfate) no hydrogen was consumed but in the MJ-T medium with nearly 67-fold lower thiosulfate concentration (0.6 mM) hydrogen was used, TH-55 may only revert to hydrogen oxidation under thiosulfate limitation. In both types of experiments 0.9 mM hydrogen was available at the start of the experiment. Besides the large discrepancies between thiosulfate concentrations in the two tested media, other major differences between T-ASW and MJ-T medium include the pH (T-ASW pH 8 and MJ-T pH 6.9) and NaHCO3 concentrations (T-ASW 2.4 mM and MJ-T 12 mM), which may have profound impact on TH-55's hydrogen oxidation ability. In other bacteria the hydrogen uptake ability has been shown to depend on a number of factors including growth and reaction conditions, such as availability of thiosulfate, hydrogen or CO2 (Rákhely et al., 2007). For Thiomicrospira species this would need to be tested in further experiments.
Interestingly, cell growth of TH-55 was significantly higher while offering hydrogen in MJ-T medium (with 0.6 mM thiosulfate) than while growing cells in T-ASW medium (with 40 mM thiosulfate) (P-value 0.001) (Figures 5c and d). We estimated whether the cultures would be oxygen-limited in the T-ASW medium faster than in the MJ-T medium. Cells in the T-ASW medium, where no hydrogen is consumed, reach lower cell densities than cells in the MJ-T medium, where hydrogen is consumed, because of the increased oxygen demand of thiosulfate oxidation; aerobic thiosulfate oxidation requires two oxygen molecules but aerobic hydrogen oxidation theoretically only needs half an oxygen molecule per reaction (Fuchs et al., 2007). Our MJ-T and T-ASW incubations hold 1% oxygen in the headspace, which equals ∼30 000 nmol oxygen per incubation. Given that one oxygen molecule is theoretically required to oxidize two hydrogen molecules (Fuchs et al., 2007) and a maximum of 60 000 nmol of hydrogen was provided in the MJ-T medium then 30 000 nmol of oxygen would be needed to aerobically fully oxidize all available hydrogen in these incubations, if hydrogen is the sole electron donor. Hence, the cells growing with hydrogen are theoretically not limited by oxygen during incubation. For T-ASW incubations supplemented with thiosulfate (2000 μmol in the incubations), oxygen would become a limiting factor with thiosulfate oxidation as the prevailing metabolism because aerobic thiosulfate oxidation requires two molecules of oxygen (Fuchs et al., 2007) and thus 4000 μmol of oxygen would be needed to oxidize the available thiosulfate fully. On the basis of the stoichiometry, the oxygen availability (30 000 nmol) in these incubations would support oxidation of at most 15 000 nmol thiosulfate. In addition to growth stagnation related to oxygen limitation in the T-ASW medium, the higher cell densities in the MJ-T medium may be related to considerably higher carbonate concentrations in the MJ-T medium, which have been shown to stimulate autotrophic CO2 fixation in Thiomicrospira (Dobrinski et al., 2010). TH-55 also exhibited uptake hydrogenase activity (Supplementary Figure S1). The activity of partially purified membrane-associated proteins was 0.51±0.05 μmol H2 per min per mg protein, which is roughly half of the activity determined for SP-41 (Supplementary Figure S1). This is in line with the findings that TH-55's hydrogen consumption in the exponential phase is roughly half of that from SP-41. Soluble proteins from TH-55 did not show uptake hydrogenase activity and coincided with the findings for soluble proteins from SP-41.
Environmental implications
Our findings can explain the previously puzzling high abundance of Thiomicrospira species in sulfide-poor but hydrogen-rich hydrothermal habitats (Lost City) (Brazelton et al., 2006, Brazelton and Baross, 2010; Brazelton et al., 2010). They can also explain the hydrogen consumption measured in incubations with Lilliput hydrothermal fluids, which were enriched with T. crunogena (28%) at the end of the experiments (Perner et al., 2011). In fact, in these incubations hydrogen consumption to CO2 fixation ratios suggested that hydrogen would be fueling biomass production, but due to the absence of experiments demonstrating that Thiomicrospira can indeed oxidize hydrogen and due to relatively high sulfide concentrations in the hydrothermal fluids used for inoculating the incubations, oxidation of residual sulfide was postulated to be responsible for primary production (Perner et al., 2011).
In the environmental hydrothermal fluid samples emanating at Sisters Peak, Thiomicrospira accounted for 0.7% of DAPI-stained cells and counts of cell numbers suggested 1.4 × 105 cells per ml hydrothermal fluids. This amounts to ∼990 Thiomicrospira cells in 1 ml Sisters Peak fluid sample. If all these cells can consume hydrogen (depending on the strain and conditions between 0.73 and 6.1 fmol H2 per h per cell) then between 0.72 and 6.02 nmol H2 per h could be consumed in 1 l of Sisters Peak low-temperature hydrothermal fluids. Given the molar ratio between hydrogen consumption to CO2 fixation in autotrophic hydrogen oxidizers to be on the order of five, 0.14–1.2 nmol carbon could be fixed autotrophically if the gained energy would be used for biomass synthesis. It is very difficult to estimate fluxes from venting environments as diffused fluid emanations are highly variable. For the Sisters Peak hydrothermal fluid emission site no fluid flow rates are available, but flow rates for low-temperature venting sites have been estimated at 116–17 580 l h−1 (Sarrazin et al., 2009; Wankel et al., 2011). Assuming that the Sisters Peak low-temperature fluids emanate at a rate as has been estimated for other venting sites and assuming that all the local Thiomicrospira species are active and consume hydrogen at the above mentioned rates then between 84 nmol (0.00002 kJ) and 105 μmol hydrogen (0.025 kJ) could be consumed and subsequently 17 nmol and 21 μmol CO2 could be fixed at one venting site in 1 h by Thiomicrospira. For comparison, Petersen et al. (2011) calculated that 435 μmol hydrogen could be consumed per hour by endosymbionts inhabiting a single vent mussel. The Sisters Peak endmember fluids—that is, 100% hydrothermal fluids without any mixed ambient seawater—holds an estimated 1.6 mM hydrogen (Perner et al., 2014). The Sisters Peak low-temperature fluids, containing 6.9% hydrothermal endmember, have 1167 nM hydrogen (Perner et al., 2013). However, if converting the hydrogen concentrations of the endmember fluids to mixed fluids with a 6.9% endmember portion, considerably more hydrogen should be available in the low-temperature emissions, namely 110 μM, suggesting that hydrogen is being consumed biologically. Likewise, in other low-temperature diffuse fluids hydrogen concentrations are 50–80% lower than predicted and the loss of hydrogen in such fluids has been attributed to biological activity (Wankel et al., 2011). Conclusively, sufficient hydrogen is available in the Sisters Peak hydrothermal system to support our hydrogen consumption rates. Thiomicrospira species could account for considerable hydrogen consumption and respective primary production in this vent environment and possibly others as well.
Conclusion
This is the first report of a Thiomicrospira strain, namely SP-41, that expresses active hydrogenases and can consume hydrogen. In fact, hydrogen uptake ability does not appear to be a unique feature of our strain SP-41, but is also found for TH-55 and may even be valid for other Thiomicrospira species not tested here. Experimental data further suggest that under conditions of low thiosulfate concentrations hydrogen may become a promising substrate for some Thiomicrospira species. Future physiological experiments will have to address a set of key questions to better understand the role that environmental parameters have on Thiomicrospira's presence and its hydrogen metabolism. This includes elucidating how widespread hydrogen consumption is among the culturable species of the ubiquitous Thiomicrospira lineage. Further, it will be vital to understand the role that the availability of thiosulfate and other reduced sulfur sources have for enhancing or reducing Thiomicrospira's hydrogen oxidation ability.
Acknowledgments
We thank the captain and crew members of the RV Meteor as well as the ROV Kiel6000 (GEOMAR, Kiel) for helping us to obtain deep-sea vent samples. We thank Wenke Bahnsen and Dagmar Svensson for technical assistance in the laboratory. The work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG) priority program 1144 ‘From Mantle to Ocean: Energy-, Material- and Life-cycles at Spreading Axes'.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Adams MW, Hall DO. Purification of the membrane-bound hydrogenase of Escherichia coli. Biochem J. 1979;183:11–22. doi: 10.1042/bj1830011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch WE, Fox GE, Magrum LJ, Woese CR, Wolfe RS. Methanogens: reevaluation of a unique biological group. Microbiol Rev. 1979;43:260–296. doi: 10.1128/mr.43.2.260-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brazelton WJ, Schrenk MO, Kelley DS, Baross JA. Methane- and sulfur-metabolizing microbial communities dominate the Lost City hydrothermal field ecosystem. Appl Environ Microbiol. 2006;72:6257–6270. doi: 10.1128/AEM.00574-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazelton WJ, Baross JA. Metagenomic comparison of two Thiomicrospira lineages inhabiting contrasting deep-sea hydrothermal environments. PLoS One. 2010;5:e13530. doi: 10.1371/journal.pone.0013530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazelton WJ, Ludwig KA, Sogin ML, Andreishcheva EN, Kelley DS, Shen CC, et al. Archaea and bacteria with surprising microdiversity show shifts in dominance over 1,000-year time scales in hydrothermal chimneys. Proc Natl Acad Sci USA. 2010;107:1612–1617. doi: 10.1073/pnas.0905369107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkhoff T, Muyzer G. Increased species diversity and extended habitat range of sulfur-oxidizing Thiomicrospira spp. Appl Environ Microbiol. 1997;63:3789–3796. doi: 10.1128/aem.63.10.3789-3796.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkhoff T, Muyzer G, Wirsen CO, Kuever J. Thiomicrospira kuenenii sp. nov. and Thiomicrospira frisia sp. nov., two mesophilic obligately chemolithoautotrophic sulfur-oxidizing bacteria isolated from an intertidal mud flat. Int J Syst Bacteriol. 1999;49 (Pt 2:385–392. doi: 10.1099/00207713-49-2-385. [DOI] [PubMed] [Google Scholar]
- Brinkhoff T, Muyzer G, Wirsen CO, Kuever J. Thiomicrospira chilensis sp. nov., a mesophilic obligately chemolithoautotrophic sulfuroxidizing bacterium isolated from a Thioploca mat. Int J Syst Bacteriol. 1999;49:875–879. doi: 10.1099/00207713-49-2-875. [DOI] [PubMed] [Google Scholar]
- Campbell BJ, Engel AS, Porter ML, Takai K. The versatile ɛ-proteobacteria: key players in sulphidic habitats. Nat Rev Microbiol. 2006;4:458–468. doi: 10.1038/nrmicro1414. [DOI] [PubMed] [Google Scholar]
- Dobrinski KP, Longo DL, Scott KM. The carbon-concentrating mechanism of the hydrothermal vent chemolithoautotroph Thiomicrospira crunogena. J Bacteriol. 2005;187:5761–5766. doi: 10.1128/JB.187.16.5761-5766.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrinski KP, Boller AJ, Scott KM. Expression and function of four carbonic anhydrase homologs in the deep-sea chemolithoautotroph Thiomicrospira crunogena. Appl Environ Microbiol. 2010;76:3561–3567. doi: 10.1128/AEM.00064-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs G, Eitinger T, Heider J, Kemper B, Kothe E, Schink B, et al. Allgemeine Mikrobiologie. Georg Thieme Verlag: Stuttgart, Germany; 2007. [Google Scholar]
- Garbe-Schönberg D, Koschinsky A, Ratmeyer V, Jähmlich H, Westernströer U. KIPS - A new multiport valve-based all-teflon fluid sampling system for ROVs. Geophys Res Abstr. 2006;8:07032. [Google Scholar]
- Glöckner FO, Fuchs BM, Amann R. Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl Environ Microbiol. 1999;65:3721–3726. doi: 10.1128/aem.65.8.3721-3726.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Häring V, Conrad R. Kinetics of H2 oxidation in respiring and denitrifying Paracoccus denitrificans. FEMS Microbiol Lett. 1991;78:259–264. [Google Scholar]
- Heijnen JJ, Van Dijken JP. In search of a thermodynamic description of biomass yields for the chemotrophic growth of microorganisms. Biotechnol Bioeng. 1992;39:833–858. doi: 10.1002/bit.260390806. [DOI] [PubMed] [Google Scholar]
- Jannasch HW, Mottl MJ. Geomicrobiology of deep-sea hydrothermal vents. Science. 1985;229:717–725. doi: 10.1126/science.229.4715.717. [DOI] [PubMed] [Google Scholar]
- Jannasch HW, Wirsen CO, Nelson DC, Robertson LA. Thiomicrospira crunogena sp. nov., a colorless, sulfur-oxidizing bacterium from a deep-sea hydrothermal vent. Int J Syst Bacteriol. 1985;35:422–424. [Google Scholar]
- Klüber HD, Conrad R. Ferric iron-reducing Shewanella putrefaciens and N2-fixing Bradyrhizobium japonicum with uptake hydrogenase are unable to oxidize atmospheric H2. FEMS Microbiol Lett. 1993;111:337–341. [Google Scholar]
- Knittel K, Kuever J, Meyerdierks A, Meinke R, Amann R, Brinkhoff T. Thiomicrospira arctica sp. nov. and Thiomicrospira psychrophila sp. nov., psychrophilic, obligately chemolithoautotrophic, sulfur-oxidizing bacteria isolated from marine Arctic sediments. Int J Syst Evol Microbiol. 2005;55:781–786. doi: 10.1099/ijs.0.63362-0. [DOI] [PubMed] [Google Scholar]
- Koschinsky A, Garbe-Schönberg D, Sander S, Schmidt K, Gennerich H-H, Strauss H. Hydrothermal venting at pressure-temperature conditions above the critical point of seawater, 5°S on the Mid-Atlantic Ridge. Geology. 2008;36:615–618. [Google Scholar]
- Kuenen JG, Veldkamp H. Thiomicrospira pelophila, gen. n., sp. n., a new obligately chemolithotrophic colourless sulfur bacterium. Antonie Van Leeuwenhoek. 1972;38:241–256. doi: 10.1007/BF02328096. [DOI] [PubMed] [Google Scholar]
- Lane DJ.199116S/23S rRNA sequencingIn: Stackebrandt E, Goodfellow M, (eds)Nucleic Acid Techniques in Bacterial Systematics John Wiley: Chichester, UK; 115–175. [Google Scholar]
- Miroshnichenko ML, Bonch-Osmolovskaya EA. Recent developments in the thermophilic microbiology of deep-sea hydrothermal vents. Extremophiles. 2006;10:85–96. doi: 10.1007/s00792-005-0489-5. [DOI] [PubMed] [Google Scholar]
- Muth E, Morschel E, Klein A. Purification and characterization of an 8-hydroxy-5-deazaflavin-reducing hydrogenase from the archaebacterium Methanococcus voltae. Eur J Biochem. 1987;169:571–577. doi: 10.1111/j.1432-1033.1987.tb13647.x. [DOI] [PubMed] [Google Scholar]
- Nishihara H, Igarashi Y, Kodama T. Hydrogenovibrio marinus gen. nov., sp. nov., a Marine Obligately Chemolithoautotrophic Hydrogen-Oxidizing Bacterium. Int J Syst Bacteriol. 1991;41:130–133. [Google Scholar]
- Nishihara H, Yaguchi T, Chung SY, Suzuki K, Yanagi M, Yamasato K, et al. Phylogenetic position of an obligately chemoautotrophic, marine hydrogen-oxidizing bacterium, Hydrogenovibrio marinus, on the basis of 16 S rRNA gene sequences and two form I RuBisCO gene sequences. Arch Microbiol. 1998;169:364–368. doi: 10.1007/s002030050584. [DOI] [PubMed] [Google Scholar]
- Nishihara H, Miyata Y, Miyashita Y, Bernhard M, Pohlmann A, Friedrich B, et al. Analysis of the molecular species of hydrogenase in the cells of an obligately chemolithoautotrophic, marine hydrogen-oxidizing bacterium Hydrogenovibrio marinus. Biosci Biotechnol Biochem. 2001;65:2780–2784. doi: 10.1271/bbb.65.2780. [DOI] [PubMed] [Google Scholar]
- Olson JW, Maier RJ. Molecular hydrogen as an energy source for Helicobacter pylori. Science. 2002;298:1788–1790. doi: 10.1126/science.1077123. [DOI] [PubMed] [Google Scholar]
- Payne MJ, Chapman A, Cammack R. Evidence for an [Fe]-type hydrogenase in the parasitic protozoan Trichomonas vaginalis. FEBS Lett. 1993;317:101–104. doi: 10.1016/0014-5793(93)81500-y. [DOI] [PubMed] [Google Scholar]
- Perner M, Bach W, Hentscher M, Koschinsky A, Garbe-Schönberg D, Streit WR, et al. Short-term microbial and physico-chemical variability in low-temperature hydrothermal fluids near 5°S on the Mid-Atlantic Ridge. Environ Microbiol. 2009;11:2526–2541. doi: 10.1111/j.1462-2920.2009.01978.x. [DOI] [PubMed] [Google Scholar]
- Perner M, Petersen JM, Zielinski F, Gennerich HH, Seifert R. Geochemical constraints on the diversity and activity of H2-oxidizing microorganisms in diffuse hydrothermal fluids from a basalt- and an ultramafic-hosted vent. FEMS Microbiol Ecol. 2010;74:55–71. doi: 10.1111/j.1574-6941.2010.00940.x. [DOI] [PubMed] [Google Scholar]
- Perner M, Hentscher M, Rychlik N, Seifert R, Strauss H, Bach W. Driving forces behind the biotope structures in two low-temperature hydrothermal venting sites on the southern Mid-Atlantic Ridge. Environ Microbiol Rep. 2011;3:727–737. doi: 10.1111/j.1758-2229.2011.00291.x. [DOI] [PubMed] [Google Scholar]
- Perner M, Hansen M, Seifert R, Strauss H, Koschinsky A, Petersen S. Linking geology, fluid chemistry, and microbial activity of basalt- and ultramafic-hosted deep-sea hydrothermal vent environments. Geobiology. 2013;11:340–355. doi: 10.1111/gbi.12039. [DOI] [PubMed] [Google Scholar]
- Perner M, Gonnella G, Kurtz S, LaRoche J. Handling temperature bursts reaching 464 degrees C: different microbial strategies in the Sisters Peak hydrothermal chimney. Appl Environ Microbiol. 2014;80:4585–4598. doi: 10.1128/AEM.01460-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen JM, Zielinski FU, Pape T, Seifert R, Moraru C, Amann R, et al. Hydrogen is an energy source for hydrothermal vent symbioses. Nature. 2011;476:176–180. doi: 10.1038/nature10325. [DOI] [PubMed] [Google Scholar]
- Rákhely G, Laurinavichene TV, Tsygankov AA, Kovács KL. The role of Hox hydrogenase in the H2 metabolism of Thiocapsa roseopersicina. Biochim Biophys Acta. 2007;1767:671–676. doi: 10.1016/j.bbabio.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Ruby EG, Jannasch HW. Physiological characteristics of Thiomicrospira sp. Strain L-12 isolated from deep-sea hydrothermal vents. J Bacteriol. 1982;149:161–165. doi: 10.1128/jb.149.1.161-165.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sako Y, Takai K, Ishida Y, Uchida A, Katayama Y. Rhodothermus obamensis sp. nov., a modern lineage of extremely thermophilic marine bacteria. Int J Syst Bacteriol. 1996;46:1099–1104. doi: 10.1099/00207713-46-4-1099. [DOI] [PubMed] [Google Scholar]
- Sarrazin J, Rodier P, Tivey MK, Singh H, Schultz A, Sarradin PM. A dual sensor device to estimate fluid flow velocity at diffuse hydrothermal vents. Deep Sea Res I: Oceanogr Res Papers. 2009;56:2065–2074. [Google Scholar]
- Scott KM, Sievert SM, Abril FN, Ball LA, Barrett CJ, Blake RA, et al. The genome of deep-sea vent chemolithoautotroph Thiomicrospira crunogena XCL-2. PLoS Biol. 2006;4:e383. doi: 10.1371/journal.pbio.0040383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai K, Hirayama H, Nakagawa T, Suzuki Y, Nealson KH, Horikoshi K. Thiomicrospira thermophila sp. nov., a novel microaerobic, thermotolerant, sulfur-oxidizing chemolithomixotroph isolated from a deep-sea hydrothermal fumarole in the TOTO caldera, Mariana Arc, Western Pacific. Int J Syst Evol Microbiol. 2004;54:2325–2333. doi: 10.1099/ijs.0.63284-0. [DOI] [PubMed] [Google Scholar]
- Takai K, Campbell BJ, Cary SC, Suzuki M, Oida H, Nunoura T, et al. Enzymatic and genetic characterization of carbon and energy metabolisms by deep-sea hydrothermal chemolithoautotrophic isolates of Epsilonproteobacteria. Appl Environ Microbiol. 2005;71:7310–7320. doi: 10.1128/AEM.71.11.7310-7320.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrede K, Heldal M, Norland S, Bratbak G. Elemental composition (C, N, P) and cell volume of exponentially growing and nutrient-limited bacterioplankton. Appl Environ Microbiol. 2002;68:2965–2971. doi: 10.1128/AEM.68.6.2965-2971.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallner G, Amann R, Beisker W. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry. 1993;14:136–143. doi: 10.1002/cyto.990140205. [DOI] [PubMed] [Google Scholar]
- Wankel SD, Germanovich LN, Lilley MD, Genc G, DiPerna CJ, Bradley AS, et al. Influence of subsurface biosphere on geochemical fluxes from diffuse hydrothermal fluids. Nature Geosci. 2011;4:461–468. [Google Scholar]
- Wirsen CO, Brinkhoff T, Kuever J, Muyzer G, Molyneaux S, Jannasch HW. Comparison of a new Thiomicrospira strain from the mid-atlantic ridge with known hydrothermal vent isolates. Appl Environ Microbiol. 1998;64:4057–4059. doi: 10.1128/aem.64.10.4057-4059.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.