Abstract
Interleukin-1 (IL-1) mediates diverse neurophysiological and neuropathological effects in the CNS through type I IL-1 receptor (IL-1R1). However, identification of IL-1R1-expressing cell types and cell-type-specific functions of IL-1R1 remains challenging. In this study, we created a novel genetic mouse model in which IL-1R1 gene expression is disrupted by an intronic insertion of a loxP flanked disruptive sequence that can be deleted by Cre recombinase, resulting in restored IL-1R1 gene expression under its endogenous promoters. A second mutation was introduced at stop codon of the IL-1R1 gene to allow tracking of the restored IL-1R1 protein by a 3HA tag and IL-1R1 mRNA by tdTomato fluorescence. These animals were designated as IL-1R1r/r and exhibited an IL-1R1 knock-out phenotype. We used IL-1R1 globally restored mice (IL-1R1GR/GR) as an IL-1R1 reporter and observed concordant labeling of IL-1R1 mRNA and protein in brain endothelial cells. Two cell-type-specific IL-1R1 restore lines were generated: Tie2Cre-IL-1R1r/r and LysMCre-IL-1R1r/r. Brain endothelial COX-2 expression, CNS leukocyte infiltration, and global microglia activation induced by intracerebroventricular injection of IL-1β were not observed in IL-1R1r/r or LysMCre-IL-1R1r/r mice, but were restored in Tie2Cre-IL-1R1r/r mice. These results reveal IL-1R1 expression in endothelial cells alone is sufficient to mediate these central IL-1-induced responses. In addition, ex vivo IL-1β stimulation increased IL-1β expression in bone marrow cells in wild-type, Tie2Cre-IL-1R1r/r, and LysMCre-IL-1R1r/r, but not IL-1R1r/r mice. These results demonstrate this IL-1R1 restore model is a valuable tool for studying cell-type-specific functions of IL-1R1.
Keywords: blood–brain barrier, endothelial cells, interleukin 1, knock-in, neuroinflammation, restore
Introduction
Interleukin 1 (IL-1) is involved in many neuroimmune responses such as fever, HPA-axis activation, sickness behavior, and prolonged slow-wave sleep (Sheng et al., 1994; Dunn, 2000; Rothwell, 2003; Dubé et al., 2005). In addition, IL-1 signaling contributes to CNS wound-healing processes (Griffin et al., 1994; Mason et al., 2001), stress-induced learning and memory impairments (Schneider et al., 1998; Rachal Pugh et al., 2001), and neurodegenerative processes (Shaftel et al., 2008). Most of these actions are mediated through the type I IL-1 receptor (IL-1R1). One explanation for the diverse range of IL-1R- mediated activities is that IL-1R1 produces distinct responses in different cell types. For example, in cultured human microglia, numerous proinflammatory cytokines such as IL-1β, IL-6, and TNF-α are produced after IL-1 stimulation (Lee et al., 1993). These proinflammatory cytokines can cause exacerbation of neuroinflammation. Conversely, in cultured rat astrocytes, a study showed that IL-1 stimulates astrocytes to release nerve growth factor which can mediate neuroprotective effects (Gadient et al., 1990). In addition, administration of IL-1 in the cerebral ventricle induced cyclooxygenase 2 (COX-2) exclusively in endothelial cells comprising brain blood vessels to mediate fever (Cao et al., 2001). Therefore, depending on the cell type, IL-1 induces different neuromodulatory molecules and functions. The precise mechanisms of how IL-1 activates specific cell types in vivo remains to be determined.
One difficulty in the analysis of IL-1R1-mediated functions is visualization of IL-1R1-expressing cells. In situ hybridization histochemistry (ISHH) has found IL-1R1 mRNA in rat brain endothelial cells (Konsman et al., 2004) and neurons in select brain regions, including basolateral nucleus of the amygdala, arcuate nucleus of the hypothalamus, trigeminal and hypoglossal motor nuclei, and area postrema (Cunningham and De Souza, 1993; Ericsson et al., 1995). However, studies using immunohistochemistry (IHC) to detect IL-1R1 protein have generated discrepant results. IL-1R1 immunoreactivity (IL-1R1-ir) has been found in endothelial cells in both rats and mice (Konsman et al., 2004; Matsuwaki et al., 2014) and in astrocytes in rats (Ravizza and Vezzani, 2006). On the contrary, there is a report showing that IL-1R1-ir was exclusively found in neurons but not endothelial cells (French et al., 1999). These discrepancies could result from the current limitations of IHC for detecting low levels of IL-1R1. However, it is known that <20 IL-1R1 molecules per cell are sufficient to mediate IL-1 signaling (Sims et al., 1993). Therefore, increasing the sensitivity for the detection of IL-1R1 protein could significantly facilitate the visualization of these molecules.
In this study, we created a novel genetic mouse model (i.e., IL-1R1 restore) that allows the selective expression of IL-1R1 on a defined cell type using its endogenous promoters. IL-1R1 mRNA and protein expression can be tracked simultaneously in this model by genetic insertion of tdTomato and 3HA tag, respectively. After characterization of this newly generated mouse model, we have identified endothelial cells as the main producer of IL-1R1 in the brain and IL-1R1 expression in endothelial cells alone is sufficient to mediate several IL-1-induced responses in the CNS.
Materials and Methods
Generation of IL-1R1 restore mice.
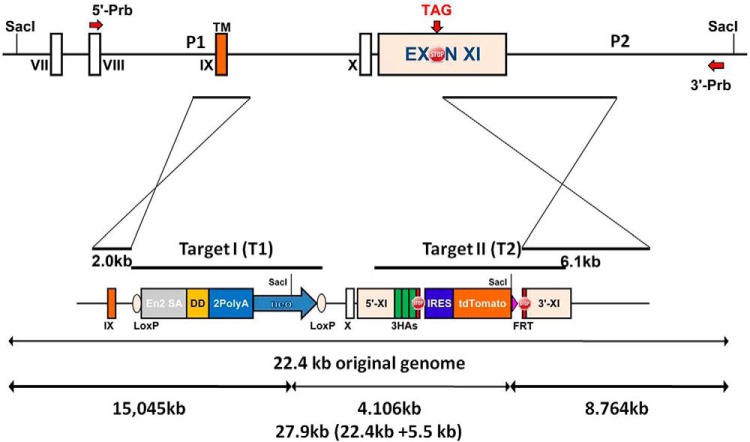
The bacterial artificial chromosome clone bMQ-81D08 containing the full mouse genomic IL-1R1 region was purchased from Source BioScience. A 22.4 kb DNA fragment containing IL-1R1 exon VII to exon XI was retrieved into the vector PL253 (ATCC PTA-4998) by homologous recombineering. Two intended mutation DNA segments were designated as knock-in target I and target II (see Fig. 1). The target I segment contained a mouse Engrailed-2 splice acceptor sequence (En2), a destabilization domain (DD), and 2PolyA signal sequence followed by a neocassette. The En2 sequence has been used to disrupt gene expression when inserted into an intron (Tsakiridis et al., 2007). The DD sequence encodes a peptide tag that would destabilize the IL-1R1 molecule (Iwamoto et al., 2010) even if the En2 insertion fails to interrupt IL-1R1 gene expression; the 2PolyA sequence causes a transcriptional stop to prevent further synthesis of downstream IL-1R1 preRNA (Proudfoot, 2011). The neocassette was added for later positive selection of the inserted sequence. These interfering insertional elements were flanked by two loxP sites. The target I sequence was generated by PCR amplification of the above described sequences and sequential subcloning into the pBluescript II SK(+) vector (Agilent Technologies). Using the same strategy, the knock-in target II was constructed. In knock-in target II, the original IL-1R1 stop codon was replaced by 3HA-STOP-IRES-tdTomato-STOP. The 3HA tag has been used to facilitate the detection of proteins with low expression level (Lobbestael et al., 2010). When added after a stop codon, the IRES-tdTomato can track the mRNA of targeted gene independent of its translation (Hellen and Sarnow, 2001; Shaner et al., 2004). Target I and target II were incorporated into PL253 vector by two rounds of homologous recombineering. Gene allele containing both target I and target II sequences was designated as IL-1R1 restore (IL-1R1r) allele. By this design, mice containing the knock-in target I in the IL-1R1 intron IX will not be able to produce functional IL-1R1 protein. However, once this mouse is mated with a mouse containing cell-type-specific Cre, the target I will be excised, ultimately restoring IL-1R1 only in the Cre-targeted cell type. The restored IL-1R1 gene will have its mRNA tracked by tdTomato fluorescence and its protein tracked by 3HA epitopes (Fig. 1).
Figure 1.
Schematic illustrations of IL-R1 wild-type (top) and restore knock-in (bottom) alleles. Top, Relevant sequence of IL-1R1 gene containing exons IX, X, and XI. Short arm for homologous recombination is designed from intron VIII to intron IX. Long arm for homologous recombination is designed from exon XI to the 3′ noncoding region. In the restore allele, knock-in Target I and knock-in Target II are inserted into the IL-1R1 locus. Target I DNA segment contains an En2 sequence, a destabilization domain (DD), and 2 polyA signal sequence followed by a neocassette. Target I segment is flanked by two loxP sites. Knock-in Target II is placed at the stop codon of the IL-1R1 sequence. The original stop codon is replaced by 3HA-STOP-IRES-tdTomato-STOP. Positions of the probes for Southern blot analysis are shown with the red arrows. The 5′ probe and 3′ probe detects a 15 kb fragment and an 8.7 kb fragment respectively in the restore allele after SacI digestion, but a 22.4 kb fragment in the wild-type. TM, Transmembrane; 5′-Prb, 5′ probe; 3′-Prb, 3′ probe; En2 SA, Engrailed-2 splice acceptor sequence; 2PolyA, 2PolyA sequence; neo, neocassette; 3HAs, 3HA tag sequence; IRES, IRES sequence; tdTomato, tdTomato sequence; SacI, restriction endonuclease SacI cleavage site.
The entire construct was verified by sequencing, then transfected into mouse embryonic stem (ES) cells. ES cell colonies were screened by SacI digestion followed by Southern blot analysis using a 5′ probe and a 3′ probe. The 5′ probe detects a 15 kb DNA fragment and the 3′ probe detects an 8.7 kb DNA fragment in the target allele according to the design in Figure 1. Both probes detect a 22 kb fragment in the wild-type allele. ES cells with successful homologous recombination were injected into blastocysts to generate a mouse carrying an IL-1R1r allele. Mice in which the IL-1R1r allele was transmitted to the germ line was obtained and confirmed by Southern blot.
Animals.
Once a C57BL/6 founder mouse with IL-1R1r germline transmission was obtained, it was bred to generate IL-1R1r/+ and IL-1R1r/r mice. Cell-type-specific Tie2Cre-IL-1R1r/r mice and LysMCre-IL-1R1r/r mice were generated by crossbreeding Tie2Cre-IL-1R1+/+ transgenic mice (stock #004128; Jackson Laboratories) or LysMCre-IL-1R1+/+ mice (stock #018956; Jackson Laboratories) with IL-1R1r/r mice. In a previous study, Koni et al. (2001) had characterized the Tie2Cre transgenic line used in the present study. They found the male Tie2Cre transgene carrier deletes loxP flanked sequence in hematopoietic cells and endothelial cells, whereas female Tie2Cre carrier deletes the loxP-flanked sequence in the germline of the offspring, which resulted in a global deletion of the loxP-flanked sequence in the next generation without further need of Cre-mediated deletion. Therefore, to maintain our Tie2Cre-IL-1R1r/r line, only male Tie2Cre-IL-1R1r/r mice were used as the Tie2Cre carrier. A female Tie2Cre-IL-1R1r/r animal was also used to mate with a male IL-1R1r/r animal to generate an IL-1R1 globally restored heterozygote (IL-1R1GR/r) animals. The designation GR indicates that the IL-1R1 restored sequence is in all cell types. Animals with IL-1R1GR will have the IL-1R1 restored without the presence of Cre. IL-1R1 globally restored homozygote (IL-1R1GR/GR) animals were obtained by breeding the IL-1R1GR/r animals with each other. All procedures were approved by The Ohio State University Animal Care and Use Committee.
Genotyping.
Genomic DNA was purified from mouse tail tissue using a protocol described previously (An et al., 2014). Briefly, a 5 mm mouse tail biopsy was incubated in 500 μl of lysis buffer for 2 h at 56°C with repeated agitation. The lysis buffer contained 10 mm Tris-HCl, pH 8.0, 100 mm EDTA, 0.5% SDS, 0.2 mg/ml ribonuclease A (Invitrogen) and 1 mg/ml proteinase K (Invitrogen). Samples were centrifuged at 13,000 rpm for 10 min to remove tissue residue. Supernatants were removed and gently mixed with 500 μl of isopropanol. Genomic DNA was then pelleted and washed with ice-cold 70% ethanol. DNA pellets were air dried and dissolved in 50 μl of DNase ad libitum water.
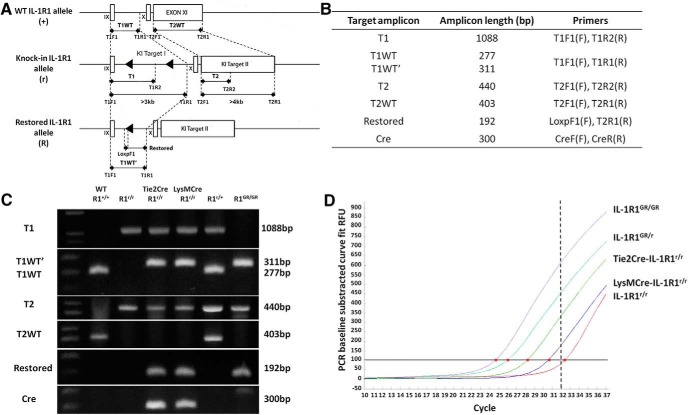
To detect the knock-in IL-1R1 allele (IL-1R1r), the inserted DNA segment target I and target II were separately detected using the following primers: for target 1 (sequence T1, see Fig. 3A), T1F1 5′-ATAAGAATGCGGCCGCTAAAGTATGTTTTTGAAATAGAAAAGATTG-3′ and T1R2 5′-ACCGGAATTCCGCGTTCTTCTTCTTTGGTTTTCGGG-3′; for target 2 (sequence T2, see Fig. 3A), T2F1 5′-GCAGAAACACGGAGTCATTTGCTGGTCAGG-3′ and T2R2 5′-CTCGTCAAGAAGACAGGGCCAG-3′. Two additional sets of primers were designed to detect wild-type IL-1R1 sequence to distinguish homozygote (IL-1R1r/r) from heterozygote (IL-1R1r/+) animals: for wild-type sequence corresponding to the target 1 locus (sequence T1WT, see Fig. 3A), T1F1 and T1R1 5′-ACCGCTCGAGATCAGAGATGAAATGACTACAAGCTGTC-3′; for wild-type sequence corresponding to the target 2 locus (sequence T2WT, see Fig. 3A), T2F1 and T2R1 5′-GAGCAGCATCCCTGAGCAACCATGGCTCTCTG-3′ (Note the forward primers T1F1 and T2F1 were the same as those used for T1 and T2, respectively). After Cre-mediated excision of target 1, the restored IL-1R1 allele (IL-1R1R) was detected by the following primers: LoxPF1 5′-ACAGTCTCTGCAGATAACTTCGTATA-3′ and T1R1 (described above). To detect the presence of Cre recombinase, the following primers were used: CreF 5′-CGATGCAACGAGTGATGAGG-3′ and CreR 5′-CGCATAACCAGTGAAACAGC-3′.
Figure 3.
Primer design for detection of knock-in allele and genotyping identification. A, Top, Genomic organization of exon IX, X, and XI in the wild-type IL-1R1 allele. Middle, Knock-in IL-1R1 allele after homologous recombination. Bottom, Restored IL-1R1 allele after Cre-recombinase-mediated excision of the disrupting sequence. Black triangles indicate loxP sequence and primer design is shown by black arrows. Sequence length represented by bars in the schematic has been adjusted for better illustration. Dashed lines indicate primer locations in the allele. B, Table of designations of target amplicons, their corresponding sizes, and primers. C, Genotyping results of animals carrying different IL-1R1 alleles. D, SYBER green quantitative RT-PCR with primers LoxPF1 and T1R1 on 1 μg of mouse tail DNA from animals with different genotypes. Different amount of the Restore amplicon were detected in the same amount of total genomic DNA from either an IL-1R1GR/GR animal or a cell-type-specific IL-1R1 restored animals. Red dots indicate the coordinate (amplification cycle number) of the amplification curve that crossed the threshold of SYBER green detection level. The threshold detection level of specific amplification of our real-time PCR machine is 32 cycles as shown by the dashed line.
Quantitative RT-PCR was performed to compare IL-1R1 globally restored genotype (IL-1R1GR) with cell-type-specific IL-1R1 restored genotype. The restored amplicon (primers LoxPF1 and T1R1) was amplified using SYBER green quantitative PCR from 1 μg of tail genomic DNA from IL-1R1GR/GR, IL-1R1GR/r, Tie2Cre-IL-1R1r/r, LysMCre-IL-1R1r/r, and IL-1R1r/r mice. Amplification curves were recorded by an ABI PRISM 7500 sequence detection system (Applied Biosystems).
Intracerebroventricular injection of IL-1β.
Intracerebroventricular injections were conducted as described previously (Ching et al., 2007). Briefly, 8- to 10-week-old animals weighing 20–30 g were anesthetized by intraperitoneal injection of 100 mg/kg Nebutal (Abbott Laboratories). Animals were then fastened onto an animal stereotaxic (David Kopf Instruments). A 28 gauge guide cannula was inserted into the right lateral ventricle [anteroposterior (AP), −1 mm; mediolateral (ML), −0.7 mm; dorsoventral (DV), −2.7 mm]. Using a microinjection system, 20 ng of IL-1β (R&D Systems) dissolved in 10 μl of sterile PBS was infused into the ventricle at rate of 0.8 μl/min (KDS Scientific). Wounds were closed with surgical staples. After animals recovered from the surgery, they were killed at 4, 12, or 24 h after surgery.
Immunohistochemistry.
For 3HA, Iba-1, GFAP, and Ly6C labeling, animals were perfusion fixed with 4% formaldehyde. Brains were removed, postfixed in 4% formaldehyde for 24 h and incubated in 20% sucrose at 4°C for 24 h. Brains were then quickly frozen on dry ice and 30-μm-thick coronal sections were generated with a cryostat. Sections were directly examined for tdTomato fluorescence by fluorescent microscopy. For double labeling of other cell markers, sections were incubated with 1% sodium borohydride and 0.5% hydrogen peroxide to reduce background. They were incubated with anti-3HA (1:500; rabbit anti-mouse, catalog #3724; Cell Signaling Technology), anti-Iba1 (1:1000; rabbit anti-mouse, catalog #019-19741; Wako Chemicals), anti-GFAP (1:1000; rabbit anti-mouse, catalog #z0334; Dako), or anti-Ly6C antibody (1:500; rat anti-mouse, catalog #ab54223; Abcam), followed by biotinylated secondary antibody and visualized by Cy2-streptavidin (Jackson Immunoresearch). For 3HA staining, tyramide signal amplification procedure was performed (PerkinElmer).
For COX-2 and CD45 labeling, mice brains were collected at 4 h (COX-2) and 12 h (CD45) after intracerebroventricular injections and frozen coronal sections were fixed in acetone/alcohol mixture, followed by incubation in a glucose oxidase and sodium azide solution. After incubation with anti-COX-2 (1:200; rabbit anti-mouse, catalog #160106; Cayman Chemicals) or anti-CD45 (1:500; rat anti-mouse, catalog #550539; BD PharMingen) primary antibody and anti-rabbit/anti-rat secondary antibody, labeling was amplified using an ABC solution (Vector Laboratories) and then visualized with a diaminobenzidine (DAB) peroxidase substrate kit (Vector Laboratories).
Immunofluorescence imaging and processing.
IHC results were examined using different microscopes. For sections visualized by DAB peroxidase substrate kit, results were obtained using a Leica DM5000B microscope connected to a Leica DFC300 FX camera and imaging software. For all of the immunofluorescence imaging, confocal images were captured using a Zeiss LSM510. Images were optimized for color, brightness, and contrast for best clarity. Multiple-channel images were overlaid and 3D reconstructed using ImageJ. For IL-1R1 mRNA distribution mapping, confocal images with overlapping anatomical marks were taken from an IL-1R1GR/GR animal. Then images were combined and spliced to produce a collage of the brain sections using Adobe Photoshop 6.0.
Quantification and proportional area analysis of microglia.
For proportional area analysis of microglia, a protocol from previous literature (Donnelly et al., 2009) was performed. In brief, representative images were obtained at 20× magnification. A threshold for positive staining was determined for each image and processed with densitometric scanning using ImageJ. Average percentage area in the positive threshold was calculated for all the pictures.
Numbers of microglia cell bodies were counted from Iba-1-stained sections. Pictures were adjusted to similar background level with ImageJ before counting. Three randomly selected regions containing >30 microglia in the area of interest were examined.
Brain endothelial cell enrichment and flow cytometry.
Brain endothelial cells were enriched using a modified protocol described by Motoike et al. (Motoike et al., 2000). In brief, IL-1R1r/r, IL-1R1GR/GR, or IL-1R1GR/r animals were first intracardially perfused with PBS. Brains were then dissected and minced into 1 mm fragments with a razor blade. Tissue was then incubated on a shaker plate at 37°C in prewarmed PBS with Collagenase I (5 mg/ml) and DNase I (120 U/ml) (Worthington) for ∼70 min until all the tissue was digested. Cells were pipetted every 10 min during the incubation. At the end of digestion, DMEM containing 10% FBS was added to the cell suspension to neutralize the collagenase activity. Cells were gently pelleted (900 × g for 6 min) and resuspended with 15 ml of 25% BSA/PBS. They were then filtered through a 70 μm mesh cell strainer and centrifuged at 2000 × g for 20 min at room temperature. The dense white myelin layer on the surface was carefully removed by pipetting. Cells were resuspended in PBS containing 0.5 mm EDTA, 60 U/ml DNase I, and 3% FBS and kept on ice. All solutions and tissue were kept in dark or covered with a black box during digestion and processing to minimize bleaching of tdTomato fluorescence.
For flow cytometric analysis, Fc receptors were blocked with anti-CD16/CD32 antibody (eBioscience) for 10 min. Cells were then labeled with fluorescent monocolonal anti-CD31 antibody (eBioscience) or isotype control for 45 min at 4°C. Cells were then washed and resuspended in FACS buffer containing 2% FBS in HBSS and 1 μg/ml sodium azide. Cell suspensions were filtered again before cytometric analysis. Approximately 1 × 104 events were analyzed using a FACSCaliber (BD Biosciences). Data were subsequently analyzed using FlowJo (TreeStar).
Bone marrow cell isolation and ex vivo IL-1β challenge.
Femurs from experimental animals were excised and epiphyses were removed. Bone marrow cells were carefully flushed out using a 23 gauge needle with ice-cold HBSS and filtered through a 70 μm nylon cell strainer. Single cell suspensions were then centrifuged and resuspended in 1 ml of red blood cell lysing buffer (0.16 m NH4Cl, 10 mm KHCO3, and 0.13 mm EDTA) for 5 min, followed by the addition of 5 ml of FBS and centrifuged. Cell pellets were resuspended with DMEM containing 10% FBS. Cell concentrations were determined using a Beckman Z2 coulter counter.
Cells (1 × 107) were preincubated with signal transduction inhibitors or culture media. SB203580 (p38 MAPK inhibitor, 20 μm; Cell Signaling Technology) or BAY11-7082 (NF-κB pathway inhibitor 5 μm; Cayman Chemicals) was first dissolved in dimethyl sulfoxide and then added to the culture medium 1 h before IL-1β stimulation. Cells were then incubated with 100 ng/ml IL-1β or culture media for control samples. After IL-1β stimulation, cells were washed and pelleted. Total RNA was extracted with TRIzol according to the manufacturer's protocol (Invitrogen). RNA was reverse transcribed to cDNA using a reverse transcription kit (Promega). Quantitative PCR was performed using the Applied Biosystems Assay-on-Demand Gene Expression protocol. Briefly, experimental cDNA was amplified with a PRISM 7500 sequence detection system (Applied Biosystems) by real-time PCR and normalized based on reference cDNA (GAPDH). Data were analyzed using the comparative threshold cycle (Ct) method and results are expressed as the fold difference from GAPDH normalized control samples.
RNA isolation and examination of restored IL-1R1 mRNA.
To examine IL-1R1 mRNA expression in immune cells, spleen and bone marrow cells were isolated for RNA extraction. Spleens were dissected from wild-type, IL-1R1r/r, and IL-1R1GR/GR animals. Bone marrow cells from the same genotypes were collected using the protocol in the “Brain endothelial cell enrichment and flow cytometry” section. Total RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. Total RNA concentration and purity was determined by Nanodrop spectrophotometry (Denville) and reversed transcribed to cDNA using a reverse transcription kit (Promega). To detect the restored IL-1R1 mRNA, PCR was performed using the following two pairs of primers: for amplicon EXT1 (an RNA sequence between IL-1R1 Exon IX and Exon X), EXT1F 5′-GGGGCTTTATCATCCTCACG-3′ and EXT1R 5′-TCTCCCAGGGTCTTGGGATA-3′, for amplicon EXT2 (another RNA sequence between IL-1R1 Exon IX and Exon X), EXT2F 5′-CCTCACGGCTACAATTGTATGC-3′ and EXT2R 5′-CAAACTGTCCCTCCAAGACC-3′.
Statistical analysis.
Data were analyzed by one-way ANOVA followed by post hoc Student's t tests. When appropriate, significant main and interaction effects were subjected to Fisher's PLSD post hoc analyses for further comparison. An α level of p ≤ 0.05 was used as the criterion for the rejection of the null hypothesis. All data were analyzed using StatView statistical software (SAS Institute). Results are reported as treatment means ± SEM.
Results
Genomic DNA from putative candidate founders were digested with SacI and labeled by the 3′ probe outside of the homologous recombination sequence. Blots were stripped and rehybridized with the 5′ external probe. Animals with the IL-1R1r allele showed corresponding fragments consistent with the design. Detailed 3′ probe and 5′ probe positions are illustrated in Figure 1. The correct founder animal (i.e., no. 2 in Fig. 2) was used to breed for all subsequent lines.
Figure 2.
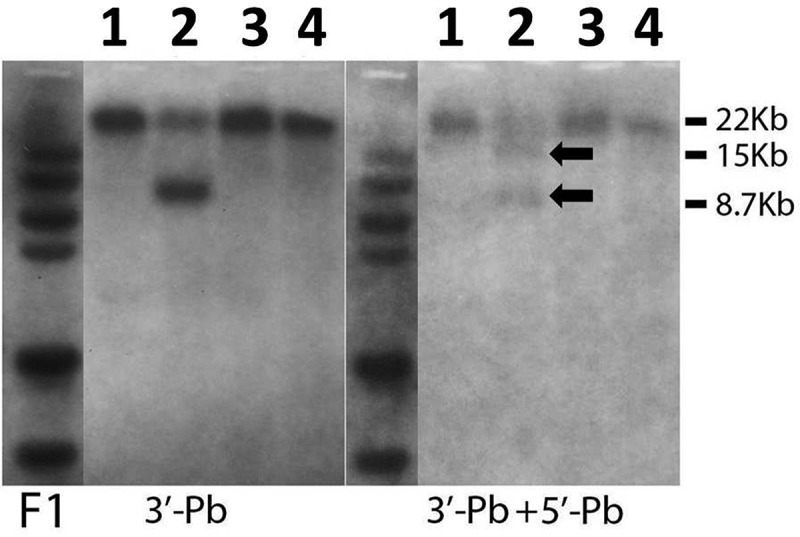
Southern blot results of homologous recombination in the putative founders. Genomic DNA from putative founder animals (1, 2, 3, and 4 are animal numbers) was probed with 3′ probe first (left). Blots were then stripped and reprobed with 5′ probe (right). Candidate animal number 2 carried the targeted knock-in construct evidenced by a 15 kb fragment and an 8.7 kb fragment after SacI digestion. Black arrows indicate bands at corresponding sizes of DNA fragments.
The IL-1R1r allele was detected also by PCR using two sets of primers (I, T1F1 and T1R2; II, T2F1 and T2R2). IL-1R1r was indicated by the presence of T1 amplicon (1088 bp) and T2 (440 bp) amplicon (Fig. 3A–C). Using the same forward primers (T1F1, T2F1) in combination with different reverse primers (T1R1, T2R1), the wild-type sequences that correspond to the T1 and T2 locations in the IL-1R1r allele were PCR amplified and designated as T1WT (277 bp) and T2WT (403 bp), respectively (Fig. 3A,B). Wild-type animals were detected by the presence of T1WT and T2WT but the absence of T1 and T2. Conversely, homozygote IL-1R1r/r animals were detected by the presence of T1 and T2 but not T1WT or T2WT. Heterozygote IL-1R1r/+ animals were determined by the presence of both sets of amplicons. The IL-1R1 r/r mice were crossed with a Tie2Cre or a LysMCre transgenic mouse to generate Tie2Cre-IL-1R1r/r and LysMCre-IL-1R1r/r mice. In Tie2Cre-IL-1R1r/r mice, the Tie2 promoter restricts Cre recombinase expression in endothelial cells and hematopoietic cells during embryogenesis and adulthood (Constien et al., 2001), whereas in the LysMCre-IL-1R1r/r mice, Cre recombinase expression is restricted in the myeloid-lineage cells (Narasimha et al., 2010). Therefore, the interfering segment target 1 was deleted in vascular and hematopoietic cells (Tie2Cre) or peripheral myeloid cells (LysMCre), respectively. In these animals, a restored IL-1R1 allele (IL-1R1R) was generated in the corresponding cell type. Cre-mediated deletion leaves one loxP sequence in the IL-1R1 intron IX in the restore allele. Therefore, PCR amplification of the restored allele by T1F1 and T1R1 at the T1WT location generates an amplicon (T1WT′) with a different size. The IL-1R1R was also specifically detected by the primers LoxPF1 and T2R1, which would generate an amplicon only from the restored allele (Restored amplicon). The presence of the Cre was detected using primers Cre300F and Cre300R. IL-1R1 globally restored animals (IL-1R1GR/GR) contained homozygote IL-1R1R in the absence of Cre. Figure 3D shows SYBR green quantitative RT-PCR of the Restored amplicon with 1 μg of genomic DNA isolated from mouse tail tissue. The cycle number at which the amplification curve crossed the threshold detection level reflected the amount of the restored allele in the total DNA. DNA from IL-1R1GR/GR animals had the earliest amplification, indicating that IL-1R1R in this animal was most abundant. Amplification of the Restore amplicon from an IL-1R1GR/r animal was ∼1 cycle behind an IL-1R1GR/GR animal. Therefore, the amount of IL-1R1R in an IL-1R1GR/r animal was approximately half of that in an IL-1R1GR/GR animal, as expected. More IL-1R1R was found in the Tie2Cre-IL-1R1r/r animal than in the LysMCre-IL-1R1r/r. This was also expected because Tie2Cre should generate IL-1R1R in all hematopoietic cells, whereas LysMCre should generate IL-1R1R only in a subpopulation of hematopoietic cells. Because there was no Cre-mediated generation of IL-1R1R in the IL-1R1r/r samples, its amplification curve was considered background noise.
The IL-1R1GR/GR animal can be used as an IL-1R1 mRNA reporter animal. Figure 4 is a collage of confocal images of coronal sections at 0.50 mm anterior to bregma taken from an IL-1R1GR/GR animal. The tdTomato fluorescence shows distinct endothelial cell morphology and distribution patterns. Interestingly, the indicated IL-1R1 mRNA was mostly produced by endothelial cells from medium size blood vessels and very rarely from capillaries. They tended to be distributed throughout the brain rather than in a region-specific manner. The same distribution of tdTomato fluorescence was detected in the brain tissue of Tie2Cre-IL-1R1r/r animals, but no tdTomato fluorescence was detected in wild-type, IL-1R1r/r, or LysMCre-IL-1R1r/r mice (data not shown).
Figure 4.
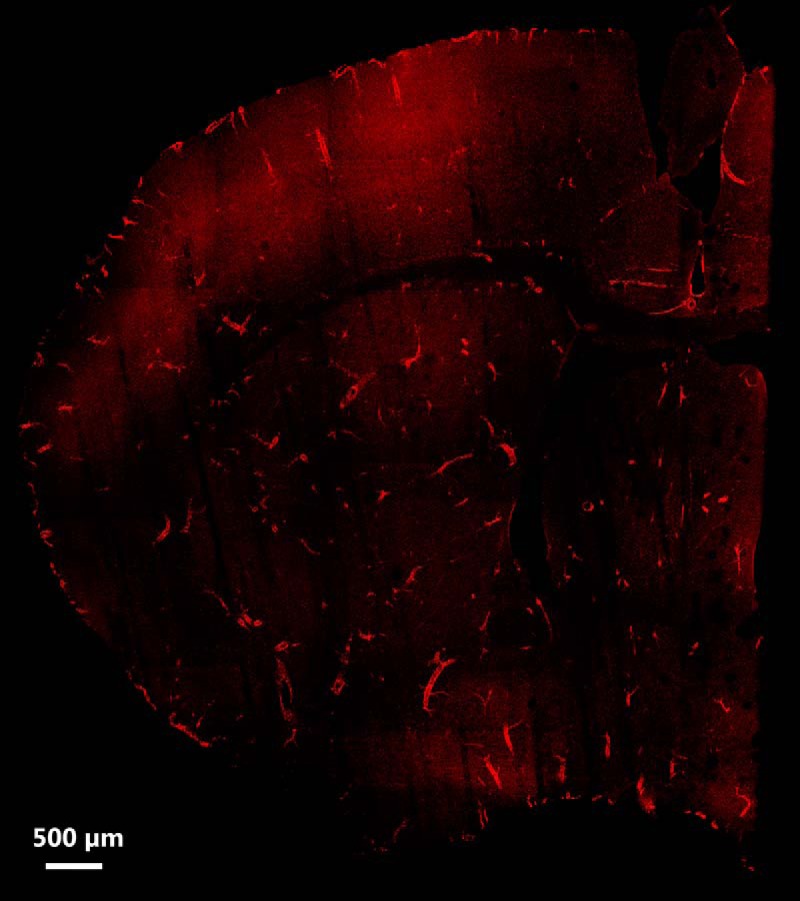
Representative distribution map of IL-1R1 mRNA-expressing cells. IL-1R1 mRNA distribution was mapped by a collage of a representative brain sections obtained from an IL-1R1GR/GR animal. IL-1R1 mRNA expression was tracked by tdTomato fluorescence. Scale bar, 500 μm.
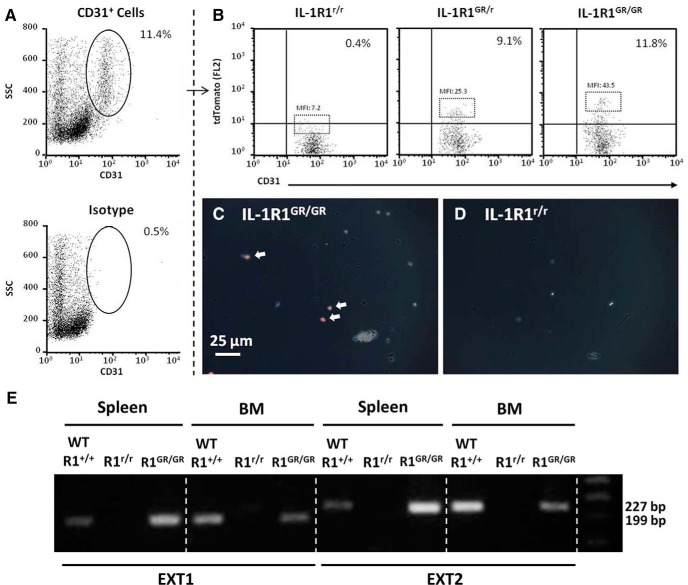
To further characterize the cell type expressing tdTomato, brain endothelial cells were extracted from IL-1R1GR/GR, IL-1R1GR/r, and IL-1R1r/r mice and examined with FACS analysis. Figure 5A shows that ∼11% of the enriched brain cells were CD31+ endothelial cells and that only 0.5% of isotype labeled cells fell within the CD31+ gate (Fig. 5A, bottom). It was noted that the CD31+ population was comprised of cells with different side-scattered (SSC) distribution. These cells also displayed different forward-scattered (FSC) distribution proportional to SSC (data not shown). It is possible some of the isolated endothelial cells adhered to each other, causing increased SSC and FSC signals. Looking within the CD31+ population, Figure 5B shows that there was a unique CD31+/tdTomato+ cell population only present in the IL-1R1GR/GR and IL-1R1GR/r mice. Subjective observation reveals that tdTomato expression was brighter in cells from IL-1R1GR/GR mice compared with IL-1R1GR/r mice. However, quantifying this by mean fluorescence intensity (MFI) was complicated by the absence of a distinct threshold between fluorescence background and actual tdTomato expression. To address this, the MFIs of the top 5% brightest cells were determined. MFI of IL-1R1r/r mice (MFI: 7.2) was considered to be fluorescent background. After subtracting the background from each value, the resulting values revealed that tdTomato expression was approximately twice as bright in IL-1R1GR/GR (MFI minus background: 36.3) relative to IL-1R1GR/r (MFI minus background: 18.1). These data are consistent with the genotyping results and also indicate that the IL-1R1 gene and tdTomato transgene exhibit a 1:1 expression ratio between copy number and gene product. Moreover, these cells also show robust tdTomato fluorescence under the microscope (Fig. 5C,D). Immune cell populations were also examined for IL-1R1 mRNA expression in the IL-1R1GR/GR animals by RT-PCR. Spleen and bone marrow cells from IL-1R1GR/GR animals had undetectable tdTomato fluorescence (data not shown). To detect IL-1R1 mRNA in these cells, two different target sequences, EXT1 and EXT2, were amplified. IL-1R1 mRNA was detected in spleen and bone marrow cells from wild-type and IL-1R1GR/GR animals but not IL-1R1r/r animals (Fig. 5E), confirming restoration of IL-1R1 in immune cells from IL-1R1GR/GR animals.
Figure 5.
Analysis of IL-1R1 mRNA producing cells in the CNS and periphery. A, Percentage of CD31 positive brain endothelial cells. B, Representative flow bivariate dot plots of CD31/tdTomato labeling of enriched brain endothelial cells from IL-1R1GR/GR, IL-1R1GR/r, or IL-1R1r/r animals. MFI was calculated for the top 5% brightest cells labeled with tdTomato. C, D, Enriched brain endothelial cells taken from IL-1R1GR/GR and IL-1R1r/r animals. White arrows indicate the tdTomato fluorescence in endothelial cells from an IL-1R1GR/GR animal. Scale bar, 25 μm. E, PCR results of IL-1R1 mRNA amplification in spleen and bone marrow cells from wild-type, IL-1R1r/r, or IL-1R1GR/GR animals. BM, Bone marrow; EXT1, an IL-1R1 mRNA amplicon from Exon IX to Exon X, 199 bp; EXT2, another IL-1R1 mRNA amplicon from Exon IX to Exon X, 227 bp.
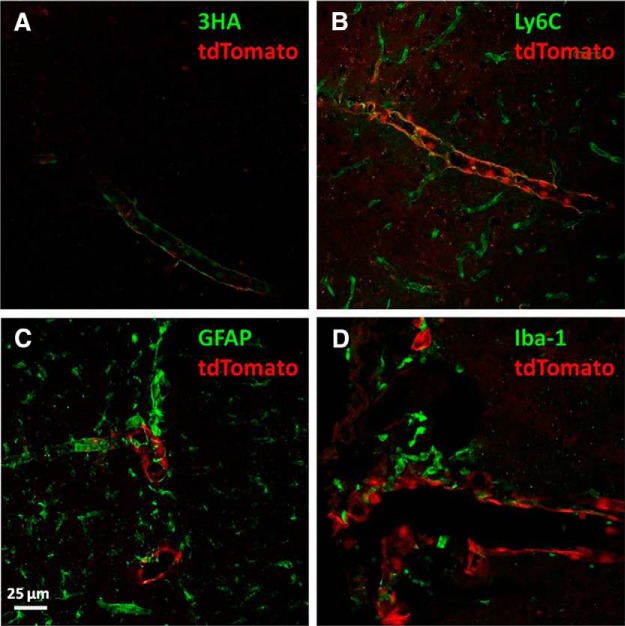
IHC was conducted in IL-1R1GR/GR animals to identify the cell type that expressed IL-1R1. Results are shown in Figure 6. Multiple-channel confocal images were 3D reconstructed using ImageJ. In Figure 6A, 3HA tag staining (green) revealed that IL-1R1 mRNA (red, tdTomato) and protein were expressed in the same cells but different locations at the subcellular level, consistent with the fact that IL-1R1 mRNA is present in the cytoplasm whereas IL-1R1 protein is cell membrane anchored. Double labeling of tdTomato with the endothelial cell marker Ly6C shows that tdTomato-expressing cells were exclusively endothelial cells (Fig. 6B). Nevertheless, endothelial cells comprising capillaries did not express tdTomato. Double labeling of tdTomato with astrocyte cell marker GFAP and the microglia cell marker Iba-1 showed no colocalization, indicating that astrocytes or microglia do not have detectable IL-1R1 expression under homeostatic conditions (Fig. 6C,D).
Figure 6.
Identification of brain IL-1R1 mRNA-expressing cell types. Brain sections from an IL-1R1GR/GR animal were labeled with 3HA (A) Ly6C (B), GFAP (C), and Iba-1(D). Pictures were taken using a confocal microscope and were 3D reconstucted with image stacks. Scale bar, 25 μm.
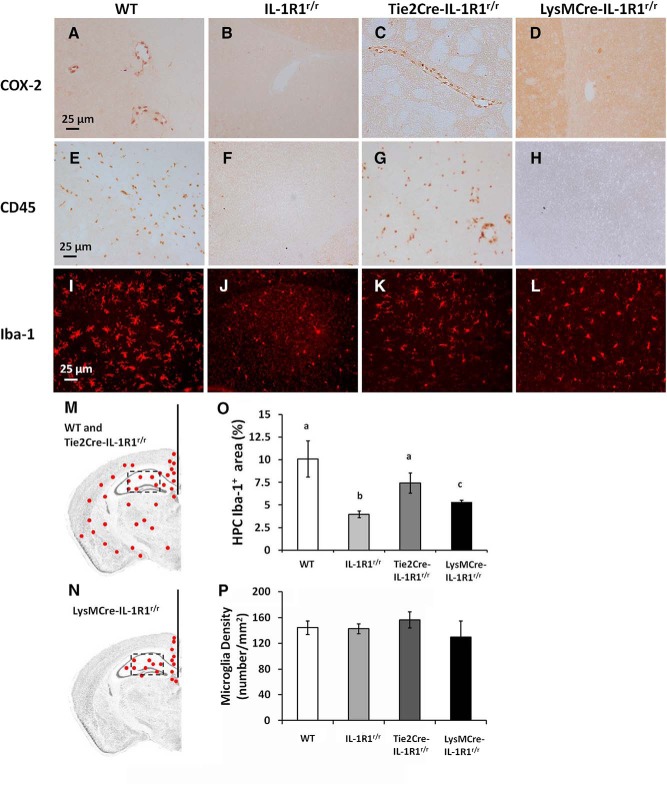
To explore cell-type-specific IL-1R1 functions, several responses to intracerebroventricular IL-1β injections were examined in wild-type, IL-1R1r/r, Tie2Cre-IL-1R1r/r, and LysMCre-IL-1R1r/r mice. Figure 7, A–D, shows IL-1β induced COX-2 expression in endothelial cells 4 h after an intracerebroventricular IL-1β injection in wild-type and Tie2Cre-IL-1R1r/r mice but not IL-1R1r/r or LysMCre-IL-1R1r/r mice. Similar to COX-2 induction, Figure 7, E–H, shows leukocyte infiltration characterized by CD45+ cells trafficking to the brain parenchyma were also detected in wild-type and Tie2Cre-IL-1R1r/r mice but not IL-1R1r/r or LysMCre-IL-1R1r/r mice 12 h after an intracerebroventricular IL-1β injection. Iba-1 staining of microglia 24 h after the intracerebroventricular IL-1β injection is shown in Figure 7, I–L. Microglia in the wild-type, Tie2Cre-IL-1R1r/r, and LysMCre-IL-1R1r/r mice exhibited deramified morphology, indicating an activated phenotype. In the IL-1R1r/r mice, microglia exhibited ramified morphology, indicating that they were in an unstimulated resting stage. Results from IL-1-stimulated IL-1R1r/r mice were not different from those obtained in PBS-injected wild-type mice (data not shown). Figure 7, M and N, shows that in wild-type and Tie2Cre-IL-1R1r/r mice, activated microglia were detected throughout the brain parenchyma, whereas in LysMCre-IL-1R1r/r mice, activated microglia were confined only in the hippocampus region. Figure 7O shows the increased proportional areas of Iba-1 after IL-1 stimulation were highest in the wild-type animals, intermediate in Tie2Cre-IL-1R1r/r animals, and lowest in LysMCre-IL-1R1r/r animals (F(3,8) = 5.209; p ≤ 0.05). To exclude the possibility that the increased proportional area of Iba-1 was not due to reduction of microglia number in the IL-1R1r/r animals, quantification of microglia number in the hippocampus was conducted. No significant difference was found in microglia number among the four genotypes (Fig. 7P).
Figure 7.
IHC labeling of COX-2, CD45, and Iba-1 following an intracerebroventricular injection of IL-1β. Brain sections from a wild-type animal, IL-1R1r/r animal, Tie2Cre-IL-1R1r/r animal, and LysMCre-IL-1R1r/r animal after an intracerebroventricular injection of IL-1β were labeled with COX-2 (A–D), CD45 (E–H), and Iba-1 (I–L). Representative distribution pattern of Iba-1 labeled cells in a wild-type or Tie2Cre-IL-1R1r/r animal (M) and a LysMCre-IL-1R1r/r animal (N). Red indicates Iba-1-labeled cells. Coronal sections of mouse brains were taken at 1.70 mm posterior to bregma. Scale bar, 25 μm. O, Quantification of fluorescence intensity in the hippocampal area (square box). Bars represent group means ± SEM. Differing letters (i.e., a, b, or c) represent significant difference between groups (p ≤ 0.05). HPC, Hippocampus. P, Quantification of microglia numbers in the hippocampal area. Bars represent group means ± SEM.
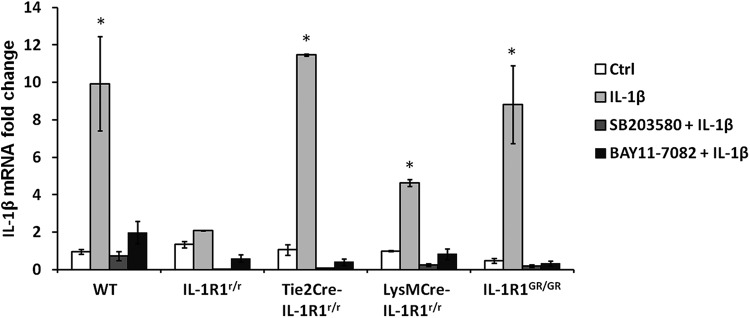
It has been reported previously that IL-1β gene expression is upregulated by IL-1β stimulation (Dinarello, 1997). This self-amplification is mediated via IL-1R1 through NF-κB and p38 MAPK pathways. We examined IL-1β mRNA expression level in bone marrow cells from wild-type, IL-1R1r/r, Tie2Cre-IL-1R1r/r, LysMCre-IL-1R1r/r, and IL-1R1GR/GR mice after ex vivo IL-1β challenge (100 ng/ml). Figure 8 shows that significant elevation of IL-1β mRNA was detected in the bone marrow cells from wild-type, Tie2Cre-IL-1R1r/r, LysMCre-IL-1R1r/r, and IL-1R1GR/GR mice but not IL-1R1r/r mice 24 h after IL-1β stimulation (p ≤ 0.05 for wild-type, Tie2Cre-IL-1R1r/r, LysMCre-IL-1R1r/r, and IL-1R1GR/GR mice). The induction of IL-1β mRNA was blocked by NF-κB pathway inhibitor BAY11–7082 and p38 MAPK pathway inhibitor SB203580, indicating that these signal transduction pathways were also restored in specific cell types.
Figure 8.
RT-PCR analysis of IL-1β mRNA levels in bone marrow cells after ex vivo IL-1β stimulation. Bone marrow cells isolated from wild-type, IL-1R1r/r Tie2Cre-IL-1R1r/r, LysMCre-IL-1R1r/r, and IL-1R1GR/GR animals were incubated with SB203580 (p38 MAPK inhibitor), BAY11–7082 (NF-κB inhibitor) or culture media for 1 h and challenged with 100 ng/ml IL-1β. IL-1β mRNA was analyzed 24 h after IL-1β treatment. Bars represent group means ± SEM. *Significant difference between control and experimental group (p ≤ 0.05).
Discussion
In the present study, we produced an IL-1R1r/r animal model using a knock-in strategy. In the IL-1R1r/r mouse, IL-1-induced CNS effects were abolished, consistent with our design that the IL-1R1r/r animals should display an IL-1R1-deficient phenotype. In addition, IL-1R1 was restored in the endothelial cells and hematopoietic cells in the Tie2Cre-IL-1R1r/r and myeloid cells in the LysMCre-IL-1R1r/r mice, as expected. An IL-1R1GR/GR mouse was also obtained and worked as an IL-1R1 mRNA and protein reporter animal. Therefore, this genetic model fulfilled all of the goals of our design.
The IL-1R1r/r mouse could serve as a more complete IL-1R1 gene knock-out. Previously, two IL-1R1 knock-out mouse lines were generated (Glaccum et al., 1997; Labow et al., 1997). However, a recent study showed that, in these models, a shortened IL-1R1 mRNA can still be expressed due to the existence of an internal promoter in the IL-1R1 gene. This shortened IL-1R1 mRNA can produce IL-1R3 in neural tissues, which is involved in IL-1-induced electrophysiological effects (Qian et al., 2012). In our IL-1R1 restore construct, an interfering DNA segment comprised of three disrupting elements was inserted into the intron IX of IL-1R1 gene, ensuring the disruption of IL-1R1/3 transcription and translation. By this design, the IL-1R1r/r animal should not have any residual IL-1R1 gene product and therefore is a better IL-1R1 knock-out.
Using the IL-1R1GR/GR animal as an IL-1R1 mRNA reporter animal, the IL-1R1 mRNA distribution in the brain was mapped by tdTomato fluorescence with high-resolution confocal microscopy. Results reveal IL-1R1 mRNA-producing cells in the brain have endothelial cell morphology and are present in medium-size blood vessels. Cells with tdTomato expression were not found in other cell types. This observation is consistent with previous results that IL-1R1 mRNA is found in endothelial cells by ISHH, but inconsistent with the detection of IL-1R1 mRNA in neurons in select regions (Ericsson et al., 1995). This discrepancy could be explained by the following facts. First, the previous ISHH studies were done on rat tissues, whereas our study was conducted in mice. The species differences may contribute to the variation of IL-1R1 mRNA level in neurons. Second, the identification of IL-1R1 mRNA in neurons by ISHH was based on morphology. It is possible that ISHH identification of neuronal IL-1R1 mRNA was not definitive. Another possibility is that the riboprobe used by Ericsson et al. (1995) recognized both IL-1R1 on endothelial cells and IL-1R3 on neurons. In contrast, in the present study, expression of extremely low levels of IL-1R3 may be below the detection limit of tdTomato, leaving neuronal IL-1R3 undetected. Nevertheless, the high IL-1R1 mRNA-expressing cells in the CNS appear to be endothelial cells. In addition, IL-1R1 mRNA expression levels were shown to be upregulated or downregulated under different conditions (Proescholdt et al., 2002; Nishida et al., 2004). With the tdTomato tracer for IL-1R1 mRNA, we can now perform visual quantitative analysis of IL-1R1 mRNA regulation. Colocalization of the tdTomato-expressing cells with endothelial cell marker Ly6C confirmed that cells expressing IL-1R1 mRNA are definitively brain endothelial cells. In the periphery, it is well known that IL-1R1 is expressed in the macrophages, T cells, and other immune cells (Sims et al., 1988). In our model, the restoration of IL-1R1 expression in the immune cells was confirmed by the presence of IL-1R1 mRNA in spleen and bone marrow cells from the IL-1R1GR/GR animals. In addition, IL-1R1-mediated function was restored in these cells from LysMCre-IL-1R1r/r mice, which restores IL-1R1 in nonendothelial immune cells. We did not observe any tdTomato fluorescence in these cells, indicating that IL-1R1 expression is maintained at very low levels under physiological conditions in these cells. In terms of immunohistochemical detection of IL-1R1 protein, most previous studies failed to detect IL-1R1-ir in the normal brain. After upregulation of IL-1R1 protein expression following electrical stimulation in the hippocampus or immune challenge by intraperitoneal LPS injection, IL-1R1-ir became detectable (Ravizza and Vezzani, 2006; Matsuwaki et al., 2014). In the present study, the restored IL-1R1 was tagged by the 3HA epitope and readily detected by IHC in the normal brain. Notably, the labeling patterns of 3HA in the normal brain resemble the pattern of IL-1R1-ir observed by Matsuwaki et al. (2014) in LPS-treated animals. Therefore, the detection of IL-1R1 protein is significantly enhanced by the use of 3HA epitope tracer. In regard to the identification of IL-1R1-expressing cell types, previous IHC studies have generated discrepant results. IL-1R1-ir was found only in endothelial cells in some studies (Konsman et al., 2004; Ravizza and Vezzani, 2006; Ching et al., 2007), only in hippocampal neurons in others (French et al., 1999), or in hippocampal neurons and astrocytes in others (Ravizza and Vezzani, 2006). These discrepancies are still difficult to explain. Nonetheless, in our IL-1R1GR/GR animal, 3HA-tagged IL-1R1 protein is expressed in the same cells in which IL-1R1 mRNA is tracked by tdTomato fluorescence. Therefore, the concordant labeling of IL-1R1 mRNA and protein achieved in this model could help avoid potential artifacts in IL-1R1-ir detection.
Our results show the IL-1R1 mRNA-producing cells from the IL-1R1GR/GR animal can be enriched and analyzed by FACS analysis. Robust tdTomato fluorescence was detected in a small percentage of isolated brain endothelial cells. Previously, isolated CNS cell types in primary cultures were used to study IL-1R1-mediated signaling (Srinivasan et al., 2004). In these cultures, it is likely that IL-1R1-expressing cells are mixed with IL-1R1 nonexpressing cells, rendering assessment of direct IL-1R1 signaling effects difficult. Because IL-1R1-expressing cells from our IL-1R1GR/GR animal can be enriched and FACS sorted, it is now possible to specifically study IL-1R1-mediated effects from IL-1R1-expressing cells.
Interesting results emerged when we compared IL-1-stimulated effects in the Tie2Cre-IL-1R1r/r with LysMCre-IL-1R1r/r animals. After intracerebroventricular IL-1β injection, endothelial COX-2 induction, leukocyte infiltration, and global activation of brain microglia, which have been found previously in wild-type animals (Basu et al., 2002; Proescholdt et al., 2002), disappeared in the IL-1R1r/r animals but reemerged in the Tie2Cre-IL-1R1r/r animals. In contrast, intracerebroventricular IL-1β failed to induce COX-2 expression and leukocyte infiltration in the LysMCre-IL-1R1r/r animals. In peripheral bone marrow cells, IL-1β-stimulated IL-1β expression was absent in the IL-1R1r/r animals but was rescued in both Tie2Cre-IL-1R1r/r and LysMCre-IL-1R1r/r animals. These results indicate that restoring IL-1R1 in peripheral immune cells is not sufficient for central IL-1β-stimulated leukocyte infiltration and COX-2 expression, whereas restoring IL-1R1 on brain endothelial cells is both sufficient and necessary for the central IL-1β-induced COX-2 expression, leukocyte infiltration, and global microglia activation. These results corroborate the conclusions from previous studies using two endothelial specific IL-1R1 knockdown mouse models that endothelial IL-1R1 mediates central IL-1-induced COX-2 expression and leukocyte infiltration (Ching et al., 2007; Li et al., 2011). This is further supported by a recent report that peripheral IL-1β-induced COX-2 expression in brain endothelial cells mediates febrile response (Wilhelms et al., 2014). A novel observation in the present study is that endothelial IL-1R1 alone is sufficient to mediate microglia activation. It should be noted that Serrats et al. (2010) have suggested that perivascular macrophages, in addition to endothelial cells, contribute significantly to central IL-1-mediated HPA-axis activation (Saper, 2010; Serrats et al., 2010), whereas IL-1R1 expression was not detected in perivascular macrophages in the present study. The studies by Serrats et al. (2010), however, were performed in the rats. In a recently published study, Matsuwaki et al. (2014) showed that IL-1R1 on nonhematopoietic cells (most likely endothelial cells), not perivascular macrophages, are responsible for IL-1β-induced HPA-axis activation in mice. Therefore, our results are consistent with this more recent report, indicating that perivascular macrophages do not play a critical role in mediating central IL-1 effects, at least in mice. The lack of detection of tdTomato fluorescence in nonendothelial CNS cell types in our IL-1R1GR/GR animals, however, does not exclude the possibility that IL-1R1 might still be expressed at very low levels in other CNS cell types, as in peripheral immune cells. Therefore, in the future, combining IL-1R1r/r animals with other CNS cell-type-specific Cre animals, such as CX3CR1-Cre or GFAP-Cre mice, may lead to discoveries of additional IL-1R1 mediated cell-type-specific effects.
Surprisingly, in the LysMCre-IL-1R1r/r animals, intracerebroventricular IL-1β induced microglia activation that was confined to the hippocampal area. One possibility for this observation was that a subpopulation of microglia cells in the hippocampus expressed a small amount of IL-1R1 after LysMCre-mediated restore. It has been shown previously that peripheral monocytes may migrate into the brain parenchyma and differentiate into a microglia-like population under certain conditions (Varvel et al., 2012). Because LysM promoter is not active in the yolk-sac-derived microglia or other cell types in the brain (Fantin et al., 2010), it is possible that hippocampus harbors a unique subpopulation of hematopoietically derived microglia during development. These microglia may have their IL-1R1 restored by LysMCre and become activated after intracerebroventricular IL-1β injection. This interesting possibility remains to be verified.
Footnotes
This work was supported by the National Institutes of Health (Grants R21-MH099482 to N.Q.) and National Institute of Dental and Craniofacial Research (Training Grant T32-DE014320 to J.F.S.).
The authors declare no competing financial interests.
References
- An Y, Belevych N, Wang Y, Zhang H, Nasse JS, Herschman H, Chen Q, Tarr A, Liu X, Quan N. Prostacyclin mediates endothelial COX-2-dependent neuroprotective effects during excitotoxic brain injury. J Inflamm Res. 2014;7:57–67. doi: 10.2147/JIR.S63205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu A, Krady JK, O'Malley M, Styren SD, DeKosky ST, Levison SW. The type 1 interleukin-1 receptor is essential for the efficient activation of microglia and the induction of multiple proinflammatory mediators in response to brain injury. J Neurosci. 2002;22:6071–6082. doi: 10.1523/JNEUROSCI.22-14-06071.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao C, Matsumura K, Shirakawa N, Maeda M, Jikihara I, Kobayashi S, Watanabe Y. Pyrogenic cytokines injected into the rat cerebral ventricle induce cyclooxygenase-2 in brain endothelial cells and also upregulate their receptors. Eur J Neurosci. 2001;13:1781–1790. doi: 10.1046/j.0953-816x.2001.01551.x. [DOI] [PubMed] [Google Scholar]
- Ching S, Zhang H, Belevych N, He L, Lai W, Pu XA, Jaeger LB, Chen Q, Quan N. Endothelial-specific knockdown of interleukin-1 (IL-1) type 1 receptor differentially alters CNS responses to IL-1 depending on its route of administration. J Neurosci. 2007;27:10476–10486. doi: 10.1523/JNEUROSCI.3357-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constien R, Forde A, Liliensiek B, Gröne HJ, Nawroth P, Hämmerling G, Arnold B. Characterization of a novel EGFP reporter mouse to monitor Cre recombination as demonstrated by a Tie2 Cre mouse line. Genesis. 2001;30:36–44. doi: 10.1002/gene.1030. [DOI] [PubMed] [Google Scholar]
- Cunningham ET, Jr, De Souza EB. Interleukin 1 receptors in the brain and endocrine tissues. Immunol Today. 1993;14:171–176. doi: 10.1016/0167-5699(93)90282-P. [DOI] [PubMed] [Google Scholar]
- Dinarello CA. Interleukin-1. Cytokine and growth factor reviews. 1997;8:253–265. doi: 10.1016/S1359-6101(97)00023-3. [DOI] [PubMed] [Google Scholar]
- Donnelly DJ, Gensel JC, Ankeny DP, van Rooijen N, Popovich PG. An efficient and reproducible method for quantifying macrophages in different experimental models of central nervous system pathology. J Neurosci Methods. 2009;181:36–44. doi: 10.1016/j.jneumeth.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubé C, Vezzani A, Behrens M, Bartfai T, Baram TZ. Interleukin-1beta contributes to the generation of experimental febrile seizures. Ann Neurol. 2005;57:152–155. doi: 10.1002/ana.20358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn AJ. Cytokine activation of the HPA axis. Ann N Y Acad Sci. 2000;917:608–617. doi: 10.1111/j.1749-6632.2000.tb05426.x. [DOI] [PubMed] [Google Scholar]
- Ericsson A, Liu C, Hart RP, Sawchenko PE. Type 1 interleukin-1 receptor in the rat brain: distribution, regulation, and relationship to sites of IL-1-induced cellular activation. J Comp Neurol. 1995;361:681–698. doi: 10.1002/cne.903610410. [DOI] [PubMed] [Google Scholar]
- Fantin A, Vieira JM, Gestri G, Denti L, Schwarz Q, Prykhozhij S, Peri F, Wilson SW, Ruhrberg C. Tissue macrophages act as cellular chaperones for vascular anastomosis downstream of VEGF-mediated endothelial tip cell induction. Blood. 2010;116:829–840. doi: 10.1182/blood-2009-12-257832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French RA, VanHoy RW, Chizzonite R, Zachary JF, Dantzer R, Parnet P, Bluthé RM, Kelley KW. Expression and localization of p80 and p68 interleukin-1 receptor proteins in the brain of adult mice. J Neuroimmunol. 1999;93:194–202. doi: 10.1016/S0165-5728(98)00224-0. [DOI] [PubMed] [Google Scholar]
- Gadient RA, Cron KC, Otten U. Interleukin-1 beta and tumor necrosis factor-alpha synergistically stimulate nerve growth factor (NGF) release from cultured rat astrocytes. Neurosci Lett. 1990;117:335–340. doi: 10.1016/0304-3940(90)90687-5. [DOI] [PubMed] [Google Scholar]
- Glaccum MB, Stocking KL, Charrier K, Smith JL, Willis CR, Maliszewski C, Livingston DJ, Peschon JJ, Morrissey PJ. Phenotypic and functional characterization of mice that lack the type I receptor for IL-1. J Immunol. 1997;159:3364–3371. [PubMed] [Google Scholar]
- Griffin WS, Sheng JG, Gentleman SM, Graham DI, Mrak RE, Roberts GW. Microglial interleukin-1 alpha expression in human head injury: correlations with neuronal and neuritic beta-amyloid precursor protein expression. Neurosci Lett. 1994;176:133–136. doi: 10.1016/0304-3940(94)90066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellen CU, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- Iwamoto M, Björklund T, Lundberg C, Kirik D, Wandless TJ. A general chemical method to regulate protein stability in the mammalian central nervous system. Chem Biol. 2010;17:981–988. doi: 10.1016/j.chembiol.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koni PA, Joshi SK, Temann UA, Olson D, Burkly L, Flavell RA. Conditional vascular cell adhesion molecule 1 deletion in mice: impaired lymphocyte migration to bone marrow. J Exp Med. 2001;193:741–754. doi: 10.1084/jem.193.6.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konsman JP, Vigues S, Mackerlova L, Bristow A, Blomqvist A. Rat brain vascular distribution of interleukin-1 type-1 receptor immunoreactivity: relationship to patterns of inducible cyclooxygenase expression by peripheral inflammatory stimuli. J Comp Neurol. 2004;472:113–129. doi: 10.1002/cne.20052. [DOI] [PubMed] [Google Scholar]
- Labow M, Shuster D, Zetterstrom M, Nunes P, Terry R, Cullinan EB, Bartfai T, Solorzano C, Moldawer LL, Chizzonite R, McIntyre KW. Absence of IL-1 signaling and reduced inflammatory response in IL-1 type I receptor-deficient mice. J Immunol. 1997;159:2452–2461. [PubMed] [Google Scholar]
- Lee SC, Liu W, Dickson DW, Brosnan CF, Berman JW. Cytokine production by human fetal microglia and astrocytes. Differential induction by lipopolysaccharide and IL-1 beta. J Immunol. 1993;150:2659–2667. [PubMed] [Google Scholar]
- Li Q, Powell N, Zhang H, Belevych N, Ching S, Chen Q, Sheridan J, Whitacre C, Quan N. Endothelial IL-1R1 is a critical mediator of EAE pathogenesis. Brain Behav Immun. 2011;25:160–167. doi: 10.1016/j.bbi.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobbestael E, Reumers V, Ibrahimi A, Paesen K, Thiry I, Gijsbers R, Van den Haute C, Debyser Z, Baekelandt V, Taymans JM. Immunohistochemical detection of transgene expression in the brain using small epitope tags. BMC Biotechnol. 2010;10:16. doi: 10.1186/1472-6750-10-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason JL, Suzuki K, Chaplin DD, Matsushima GK. Interleukin-1beta promotes repair of the CNS. J Neurosci. 2001;21:7046–7052. doi: 10.1523/JNEUROSCI.21-18-07046.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuwaki T, Eskilsson A, Kugelberg U, Jönsson JI, Blomqvist A. Interleukin-1beta induced activation of the hypothalamus-pituitary-adrenal axis is dependent on interleukin-1 receptors on non-hematopoietic cells. Brain Behav Immun. 2014;40:166–173. doi: 10.1016/j.bbi.2014.03.015. [DOI] [PubMed] [Google Scholar]
- Motoike T, Loughna S, Perens E, Roman BL, Liao W, Chau TC, Richardson CD, Kawate T, Kuno J, Weinstein BM, Stainier DY, Sato TN. Universal GFP reporter for the study of vascular development. Genesis. 2000;28:75–81. doi: 10.1002/1526-968X(200010)28:2<75::AID-GENE50>3.0.CO%3B2-S. [DOI] [PubMed] [Google Scholar]
- Narasimha AJ, Watanabe J, Ishikawa TO, Priceman SJ, Wu L, Herschman HR, Reddy ST. Absence of myeloid COX-2 attenuates acute inflammation but does not influence development of atherosclerosis in apolipoprotein E null mice. Arterioscler Thromb Vasc Biol. 2010;30:260–268. doi: 10.1161/ATVBAHA.109.198762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida M, Nasu K, Fukuda J, Kawano Y, Narahara H, Miyakawa I. Down-regulation of interleukin-1 receptor type 1 expression causes the dysregulated expression of CXC chemokines in endometriotic stromal cells: a possible mechanism for the altered immunological functions in endometriosis. J Clin Endocrinol Metab. 2004;89:5094–5100. doi: 10.1210/jc.2004-0354. [DOI] [PubMed] [Google Scholar]
- Proescholdt MG, Chakravarty S, Foster JA, Foti SB, Briley EM, Herkenham M. Intracerebroventricular but not intravenous interleukin-1beta induces widespread vascular-mediated leukocyte infiltration and immune signal mRNA expression followed by brain-wide glial activation. Neuroscience. 2002;112:731–749. doi: 10.1016/S0306-4522(02)00048-9. [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ. Ending the message: poly(A) signals then and now. Genes Dev. 2011;25:1770–1782. doi: 10.1101/gad.17268411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian J, Zhu L, Li Q, Belevych N, Chen Q, Zhao F, Herness S, Quan N. Interleukin-1R3 mediates interleukin-1-induced potassium current increase through fast activation of Akt kinase. Proc Natl Acad Sci U S A. 2012;109:12189–12194. doi: 10.1073/pnas.1205207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachal Pugh C, Fleshner M, Watkins LR, Maier SF, Rudy JW. The immune system and memory consolidation: a role for the cytokine IL-1beta. Neurosci Biobehav Rev. 2001;25:29–41. doi: 10.1016/S0149-7634(00)00048-8. [DOI] [PubMed] [Google Scholar]
- Ravizza T, Vezzani A. Status epilepticus induces time-dependent neuronal and astrocytic expression of interleukin-1 receptor type I in the rat limbic system. Neuroscience. 2006;137:301–308. doi: 10.1016/j.neuroscience.2005.07.063. [DOI] [PubMed] [Google Scholar]
- Rothwell N. Interleukin-1 and neuronal injury: mechanisms, modification, and therapeutic potential. Brain Behav Immun. 2003;17:152–157. doi: 10.1016/S0889-1591(02)00098-3. [DOI] [PubMed] [Google Scholar]
- Saper CB. The dance of the perivascular and endothelial cells: mechanisms of brain response to immune signaling. Neuron. 2010;65:4–6. doi: 10.1016/j.neuron.2009.12.029. [DOI] [PubMed] [Google Scholar]
- Schneider H, Pitossi F, Balschun D, Wagner A, del Rey A, Besedovsky HO. A neuromodulatory role of interleukin-1beta in the hippocampus. Proc Natl Acad Sci U S A. 1998;95:7778–7783. doi: 10.1073/pnas.95.13.7778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrats J, Schiltz JC, García-Bueno B, van Rooijen N, Reyes TM, Sawchenko PE. Dual roles for perivascular macrophages in immune-to-brain signaling. Neuron. 2010;65:94–106. doi: 10.1016/j.neuron.2009.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaftel SS, Griffin WS, O'Banion MK. The role of interleukin-1 in neuroinflammation and Alzheimer disease: an evolving perspective. J Neuroinflammation. 2008;5:7. doi: 10.1186/1742-2094-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Sheng JG, Boop FA, Mrak RE, Griffin WS. Increased neuronal beta-amyloid precursor protein expression in human temporal lobe epilepsy: association with interleukin-1 alpha immunoreactivity. J Neurochem. 1994;63:1872–1879. doi: 10.1046/j.1471-4159.1994.63051872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims JE, March CJ, Cosman D, Widmer MB, MacDonald HR, McMahan CJ, Grubin CE, Wignall JM, Jackson JL, Call SM, et al. cDNA expression cloning of the IL-1 receptor, a member of the immunoglobulin superfamily. Science. 1988;241:585–589. doi: 10.1126/science.2969618. [DOI] [PubMed] [Google Scholar]
- Sims JE, Gayle MA, Slack JL, Alderson MR, Bird TA, Giri JG, Colotta F, Re F, Mantovani A, Shanebeck K, et al. Interleukin 1 signaling occurs exclusively via the type I receptor. Proc Natl Acad Sci U S A. 1993;90:6155–6159. doi: 10.1073/pnas.90.13.6155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan D, Yen JH, Joseph DJ, Friedman W. Cell type-specific interleukin-1beta signaling in the CNS. J Neurosci. 2004;24:6482–6488. doi: 10.1523/JNEUROSCI.5712-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsakiridis A, Tzouanacou E, Larralde O, Watts TM, Wilson V, Forrester L, Brickman JM. A novel triple fusion reporter system for use in gene trap mutagenesis. Genesis. 2007;45:353–360. doi: 10.1002/dvg.20301. [DOI] [PubMed] [Google Scholar]
- Varvel NH, Grathwohl SA, Baumann F, Liebig C, Bosch A, Brawek B, Thal DR, Charo IF, Heppner FL, Aguzzi A, Garaschuk O, Ransohoff RM, Jucker M. Microglial repopulation model reveals a robust homeostatic process for replacing CNS myeloid cells. Proc Natl Acad Sci U S A. 2012;109:18150–18155. doi: 10.1073/pnas.1210150109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelms DB, Kirilov M, Mirrasekhian E, Eskilsson A, Kugelberg UÖ, Klar C, Ridder DA, Herschman HR, Schwaninger M, Blomqvist A, Engblom D. Deletion of prostaglandin E2 synthesizing enzymes in brain endothelial cells attenuates inflammatory fever. J Neurosci. 2014;34:11684–11690. doi: 10.1523/JNEUROSCI.1838-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]