Abstract
Previous studies have shown that chronic cocaine administration induces SIRT1, a Class III histone deacetylase, in the nucleus accumbens (NAc), a key brain reward region, and that such induction influences the gene regulation and place conditioning effects of cocaine. To determine the mechanisms by which SIRT1 mediates cocaine-induced plasticity in NAc, we used chromatin immunoprecipitation followed by massively parallel sequencing (ChIP-seq), 1 d after 7 daily cocaine (20 mg/kg) or saline injections, to map SIRT1 binding genome-wide in mouse NAc. Our unbiased results revealed two modes of SIRT1 action. First, despite its induction in NAc, chronic cocaine causes depletion of SIRT1 from most affected gene promoters in concert with enrichment of H4K16ac (itself a deacetylation target of SIRT1), which is associated with increased expression of these genes. Second, we deduced the forkhead transcription factor (FOXO) family to be a downstream mechanism through which SIRT1 regulates cocaine action. We proceeded to demonstrate that SIRT1 induction causes the deacetylation and activation of FOXO3a in NAc, which leads to the induction of several known FOXO3a gene targets in other systems. Finally, we directly establish a role for FOXO3a in promoting cocaine-elicited behavioral responses by use of viral-mediated gene transfer: we show that overexpressing FOXO3a in NAc enhances cocaine place conditioning. The discovery of these two actions of SIRT1 in NAc in the context of behavioral adaptations to cocaine represents an important step forward in advancing our understanding of the molecular adaptations underlying cocaine action.
Keywords: addiction, behavior, ChIP-Seq, cocaine, genomics, SIRT1
Introduction
Long-term exposure to drugs of abuse induces persistent changes in gene expression, neuronal morphology, and synaptic function in the nucleus accumbens (NAc), a primary region in the brain's reward circuitry (Robison and Nestler, 2011). Epigenetic mechanisms may underlie the dysregulation of gene expression observed in addiction, and cocaine-induced post-translational modifications of histone proteins, including acetylation, have been shown to contribute to drug action (Maze and Nestler, 2011). Acetylation, a well-studied modification in the context of drugs of abuse, is catalyzed by histone acetyltransferases and reversed by histone deacetylases, both of which regulate gene expression in postmitotic cells. For example, acetylation of several lysine residues on the N-terminal tails of histones, specifically histone H4 Lys16 acetylation (H4K16ac), H3K9ac, and H3K27ac, is associated with increased gene expression, whereas their deacetylation results in inhibition of gene expression.
Sirtuins (SIRTs) are categorized as Class III histone deacetylases, which rely on NAD+ to catalyze the deacetylase reaction. There are seven sirtuins, each with distinct subcellular localizations and enzymatic activities. Sirtuins regulate a wide range of biological processes and affect the structural organization of the brain through axon elongation (Li et al., 2013), dendritic plasticity (Codocedo et al., 2012), and neuronal precursor cell fate determination (Rafalski et al., 2013). Sirtuins have also being implicated in hypothalamic function, circadian rhythmicity (Asher et al., 2008; Chang and Guarente, 2013), endocrine regulation (Cohen et al., 2009), feeding behaviors (Ramadori et al., 2011), and the synaptic plasticity underlying learning and memory (Gao et al., 2010; Michán et al., 2010). Our laboratory has provided evidence for the involvement of sirtuins in the actions of drugs of abuse. Chronic exposure to cocaine induces SIRT1 and SIRT2 expression in NAc, with chronic morphine also inducing SIRT1 (Ferguson et al., 2013). No other SIRT family member is regulated in this brain region by either drug. Drug induction of SIRT1 in NAc is mediated by the Fos family transcription factor, ΔFosB (Ferguson et al., 2013). Furthermore, SIRT1 induction in NAc promotes behavioral responses to cocaine and to morphine: viral-mediated overexpression of the protein in NAc increases cocaine- and morphine-elicited conditioned place preference, whereas its local knockdown has the opposite effect (Ferguson et al., 2013).
The goal of the present study is to understand the downstream mechanisms by which SIRT1 mediates cocaine-induced plasticity in NAc. We used a genome-wide approach, namely, chromatin immunoprecipitation (ChIP) followed by massively parallel sequencing (ChIP-seq), to map SIRT1 binding in NAc from chronic cocaine- or chronic saline-treated mice. Our results identified depletion of SIRT1 coupled with increased H4K16ac, as well as activation of the forkhead transcription factor FOXO3a, as being two key mechanisms of SIRT1-mediated gene activation in this brain region. Together, these studies identify several novel substrates of cocaine-induced neural and behavioral plasticity.
Materials and Methods
Animals.
C57BL/6J male mice (The Jackson Laboratory), 7–9 weeks old, weighing 25–30 g were group housed five per cage and habituated to the animal facility 1 week before use. They were maintained at 23°C to 25°C on a 12 h light-dark cycle (lights on from 7:00 A.M. to 7:00 P.M.) with ad libitum access to food and water. All animal protocols were approved by the Institutional Animal Care and Use Committee at the Icahn School of Medicine at Mount Sinai.
Drugs.
Cocaine-HCl was obtained from Sigma-Aldrich or National Institute on Drug Abuse (Bethesda, MD). Drugs were given as daily intraperitoneal injections for 7 d at 20 mg/kg in their home cage. Control groups received 7 d of saline. Animals were analyzed 24 h after the last injection.
RNA isolation and PCR.
Bilateral 14-gauge NAc punch dissections were obtained from 1-mm-thick coronal brain sections and frozen immediately on dry ice. Dissections were thawed and processed in TriZol (Invitrogen) according to the manufacturer's guidelines. RNA was isolated and purified using RNeasy Micro columns (QIAGEN). We confirmed purity of our samples by spectroscopy at 260/280 and 260/230 > 1.8. RNA was reverse transcribed to cDNA using iScript Kit (Bio-Rad). cDNA was quantified by qPCR using SYBR green. qPCR was performed using an Applied Biosystems 7500 system. Reactions were run in triplicate and analyzed using the ΔΔCt method (Livak and Schmittgen, 2001) with GAPDH as a normalization control.
ChIP.
ChIP was performed as described previously (Kumar et al., 2005) with minor modifications. Briefly, for each ChIP, anterior and posterior bilateral 14-gauge NAc punches were pooled (5 animals, 20 NAc punches per sample). Punches were fixed for 10 min with 1% formaldehyde and then quenched with 2 m glycine for 5 min. Samples were homogenized using a desktop sonicator at low settings (amplitude 40%) 2× for 7 s on ice. Next, samples were sheered using a diogenode bioruptor XL at 4°C at high sonication intensity for 30 s on/30 s off for 25 min, followed by 25 min rest and an additional 25 min of sonication. Fragment size range of 250–1000 bp was verified with an Agilient Bioanalyzer. Before sonication of samples, magnetic sheep anti-rabbit or anti-mouse beads (Invitrogen) were prepared with the respective antibody of interest at 4°C overnight on a rotator. Following washing of the magnetic bead/antibody complex, 7.5 mg of the magnetic bead/antibody complex was added to 400 μl of sheared chromatin for histone ChIPs, or 1000 μl for SIRT1 ChIP, for 16 h at 4°C; 80 μl of each sample of sheared chromatin was used as input controls. Samples were washed with LiCl and Tris-EDTA buffers. Reverse cross-linking was performed at 65°C overnight, and proteins were removed with proteinase K (Invitrogen). DNA was purified using a DNA purification kit (QIAGEN). Additionally, a normal IgG control was performed to test for nonspecific binding.
Nuclear-cytoplasmic fractionation.
NAc punches were homogenized with a glass Dounce tissue grinder and loose pestle in Buffer A (1 m Tris-HCl, 1 m sucrose, 1 m DTT, with protease and phosphatase inhibitors); 10% of the lysate was reserved to assay total protein levels, and the rest was centrifuged at 3700 rpm for 10 min. The supernatants were then removed, centrifuged at 7500 rpm for 7 min, and the resulting supernatants were stocked as the cytoplasmic fraction. Buffer B (1 m Tris-HCl pH 7.5, 0.1 m EDTA, 0.1 m EGTA, 1 m sucrose, 1 m DTT, 10% NP-40, protease and phosphatase inhibitors) was added to the pellets from the first centrifugation, and samples were kept on ice for 10 min before centrifuging again at 3700 rpm for 10 min. The supernatants were discarded, and Buffer C (1 m Tris-HCl, 37.5% glycerol, 5 m NaCl, 0.1 m EGTA, 1 m DTT, 10% NP-40, with protease and phosphatase inhibitors) was used to resuspend the nuclear fraction. The fractions were then processed for Western blotting as below or further separated into chromatin and nonchromatin nuclear fractions. Tubulin and total H3 were used as loading controls and to verify appropriate cytoplasmic and nuclear enrichment.
ChIP-seq.
Animals were treated chronically with cocaine (20 mg/kg) daily for 7 d; and 24 h later, NAc tissue pooled from ∼5 mice was processed for ChIP as described previously (Renthal and Nestler, 2009; Maze et al., 2010). Antibodies were all ChIP grade from Abcam or Cell Signaling Technology. Approximately 10 ng of input DNA or pull-down DNA was used for the preparation of sequencing libraries following the instructions of Illumina's ChIP-seq sample prep kit (catalog #IP-102-1001). In brief, DNA fragment overhangs were converted into phosphorylated blunt ends using T4 DNA polymerase, Klenow polymerase, and T4 polynucleotide kinase. An “A” base was then added to the DNA fragments to enable ligation to the adapters, which have a single “T” overhang. The libraries were analyzed on a 2% agarose gel and size selected between 175 and 300 bp. Gel-extracted DNA was further enriched by PCR and run on a bioanalyzer to validate size distribution and concentration. All libraries were sequenced on Illumina Hi-seq 2000 machines at Mount Sinai and analyzed according to published methods (Maze et al., 2011; Feng et al., 2014).
ChIP-seq data processing.
Raw ChIP-seq data were processed through initial image analysis, base calling, quality filtering, and short read alignment using Illumina's CASAVA pipeline. Only the uniquely aligned short reads were kept for further analysis. Global visualization of a ChIP-seq sample is often useful to determine the enrichment of the protein of interest and to assess data quality. We therefore used a new program called ngs.plot (Shen et al., 2014). Briefly, the alignment files are used to derive genomic coverage, which is normalized to “aligned Reads Per Million mapped reads” at single base resolution. A database is built into ngs.plot, which contains genomic coordinates of interesting biological features, such as TSS, transcription end sites, and gene body for a few model organisms. Additionally, ngs.plot allows input of customized genomic regions. Differential analysis was performed by diffReps (Shen et al., 2013) with window size of 200 bp and moving size of 20 bp. FDR < 10% was used as significance cutoff. Basal level peak calling was performed using MACS (Zhang et al., 2008) with the three saline replicates pooled and DNA input samples used as background.
Immunoblotting.
Immunoblotting was performed using standard procedures on NAc punch dissections from individual animals (Maze et al., 2010). Briefly, frozen NAc tissue was homogenized in 30 μl of homogenization buffer containing 320 mm sucrose, 5 mm HEPES buffer, 1% SDS, phosphatase inhibitor cocktails I and II (Sigma), and protease inhibitors (Roche) with an ultrasonic processor (Cole Parmer). 10–30 μg of protein was loaded onto 4%–15% gradient Tris-HCl polyacrylamide gels for electrophoresis fractionation (Bio-Rad). Proteins were transferred to nitrocellulose membranes, blocked with 5% (w/v) BSA, and incubated overnight at 4°C with primary antibodies in 5% BSA. After thorough washing with TBS plus 0.1% Tween 20, membranes were incubated with secondary antibodies (1:40,000–1:60,000) dissolved in 5% BSA blocking buffer for 1 h at room temperature. Final blots were developed by chemiluminescence analysis using supersignal dura ECL (Pierce Biotechnology; Thermo Fisher Scientific). Images were quantified using densitometry with ImageJ (National Institutes of Health, Bethesda, MD), and samples were normalized using GAPDH or β-tubulin, which were not affected by cocaine or SIRT1 overexpression.
Immunoprecipitations.
A coimmunoprecipitation kit (Roche) was used as follows. Bilateral 14-gauge NAc punches were lysed by douncing in 300 μl of provided lysis buffer and centrifuged, and the supernatant was transferred to a clean tube. Preclearance was accomplished by incubation with protein G-agarose for 3 h on a rotator at 4°C. The beads were centrifuged, and the supernatant was transferred to fresh tubes to be incubated with 5 μl of anti-acetylysine antibody (Abcam) for 1 h before 50 μl of a homogeneous protein G-agarose suspension was added and incubated overnight at 4°C on a rotator. The complexes were centrifuged, and the supernatant was removed, the beads were washed two times with lysis buffer 1, two times with lysis buffer 2, and once with lysis buffer 3. Protein sample buffer was added, and the samples were boiled for 3 min. Complexes were then analyzed by Western blotting.
Viral-mediated gene transfer.
Mice were anesthetized with a ketamine/xylazine mixture (ketamine 100 mg/kg and xylazine 10 mg/kg) and prepared for sterotaxic surgery. Thirty-three gauge syringe needles (Hamilton) were used to bilaterally infuse 0.5 μl of virus into NAc at a 10° angle at a rate of 0.1 μl/min at 1.6 mm anteroposterior, 1.5 mm lateral, and 4.4 mm dorsoventral from bregma. We used bicistronic p1005 HSV (Herpes simplex virus) vectors expressing GFP alone or GFP plus FOXO3a, FOXO3a-TM, or FOXO3a-DM. In this system, GFP expression is driven by a cytomegalovirus promoter, whereas the gene of interest is driven by the IE4/5 promoter (Maze et al., 2010). Animals receiving HSV injections were allowed to recover for at least 3 d after surgery. Viral injection sites were verified by confirming the GFP signal in NAc slices using a fluorescence dissecting microscope.
Conditioned place preference (CPP).
CPP was performed as described previously (Ferguson et al., 2013). Several visual and nonvisual (tactile) cues enabled the animals to distinguish two separate chambers of the apparatus. All conditioning and test sessions were performed under dim illumination. Briefly, animals were first prescreened in a photo-beam monitored box to detect any baseline bias to the two chambers of the CPP apparatus. Mice that showed statistically significant preference to one of the two chambers were removed from further analysis (<10% of all animals). Mice were then divided into control and experimental groups with equivalent pretest scores. During the training phase, animals were injected with saline in the morning and placed in one chamber for 30 min, then with cocaine (5 mg/kg i.p.) and placed in the other chamber in the afternoon. This training regimen was conducted for 2 d followed by a test day in which mice were placed back into the apparatus without treatment for 20 min and evaluated for chamber preference. For all groups, baseline locomotion in response to saline was assessed. CPP scores were calculated by subtracting time spent in the saline-paired chamber from time spent in the cocaine-paired chamber.
Statistical analysis.
One- and two-way ANOVAs were performed to determine significance for conditions in which there were more than two groups or two factors. Unpaired Student's t test with a two-tailed p value were used for other comparisons, including qPCR, Western blotting, comparing HSV-GFP to HSV-FOXO3a vectors and ChIP experiments. All values included in the figure legends represent mean ± SEM.
Results
ChIP-seq reveals transcriptional targets of SIRT1 in NAc
To explore the mechanisms by which SIRT1 mediates cocaine-induced plasticity in NAc, we performed ChIP-seq for SIRT1 on NAc extracts from chronic cocaine- or saline-treated mice. We used a treatment regimen (7 daily injections, with animals analyzed 24 h later), which has been shown to induce robust changes in gene expression in the NAc (Renthal et al., 2009; Maze et al., 2010; Feng et al., 2014). After preprocessing, normalization, and identification of SIRT1 binding sites using MACs peak calling software (Zhang et al., 2008), statistical analysis of genomic regions that displayed cocaine-induced increases or decreases in SIRT1 binding (i.e., differential sites) was performed using diffReps (Shen et al., 2013), which uses a sliding window strategy to generate a high-resolution differential binding profile of SIRT1. These analyses provided a genome-wide map of downstream SIRT1 targets under control and drug-treated conditions.
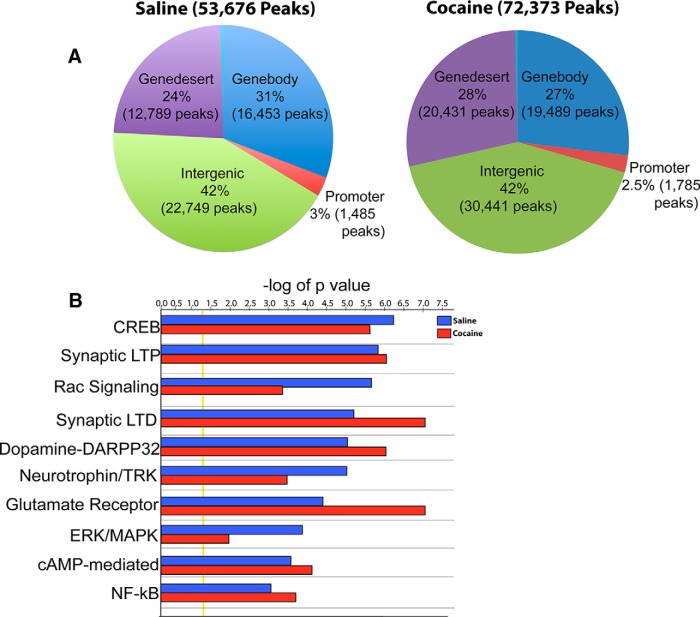
Results from the average of three independent replicate SIRT1 ChIP-seq experiments identified 53,676 peaks in NAc of saline-treated mice and 72,373 peaks in cocaine-treated mice (Fig. 1A). The larger number of significant SIRT1 peaks under cocaine conditions is consistent with the global increase in SIRT1 expression seen in NAc under these conditions (Ferguson et al., 2013). The genomic binding profile for saline and cocaine conditions was very similar, with ∼70% of binding occurring in gene desert and intergenic regions and the other ∼30% binding in promoter and gene body regions (Fig. 1A). Gene annotation mapping revealed a relatively even distribution of peaks across various genomic regions (i.e., gene deserts, gene promoters, gene bodies, and intergenic regions) in both saline and cocaine conditions (Fig. 1A). Molecular pathway analysis using Ingenuity software on genes that display significant SIRT1 binding (Fig. 1B) revealed that many SIRT1 target genes are significantly (using Fisher's exact test) associated with signaling pathways previously linked to cocaine action (Robison and Nestler, 2011), for example, pathways related to CREB signaling in neurons (−log p value saline = 6.1 cocaine = 5.541), synaptic long-term potentiation (−log p value saline = 5.7 cocaine = 5.9), synaptic long-term depression (−log p value saline = 5.1 cocaine = 7.34), glutamatergic signaling (−log p value saline = 4.3 cocaine = 7.1), and ERK/MAPK signaling (−log p value saline = 4.2 cocaine = 2.0).
Figure 1.
ChIP-seq reveals genomic target regions for SIRT1 binding in NAc under saline and cocaine conditions. ChIP-seq reveals relatively even distribution of SIRT1 binding across genomic regions in NAc. A, Genome-wide occupancy of SIRT1 in saline (53,676 peaks) and chronic cocaine (72,373 peaks) (20 mg/kg) conditions. Regional analysis of genomic regions in NAc from both saline- and cocaine-treated mice reveals that ∼70% and 30% of SIRT1 peaks reside in intergenic/gene desert and promoter/gene body regions of the genome, respectively. B, SIRT1 regulates canonical signaling pathways implicated in synaptic and behavioral plasticity. The analyses shown were performed on averages of 3 saline and 3 cocaine replicates. Pearson's correlation analysis revealed strong consistency (>75%) between each replicate in the saline and cocaine groups.
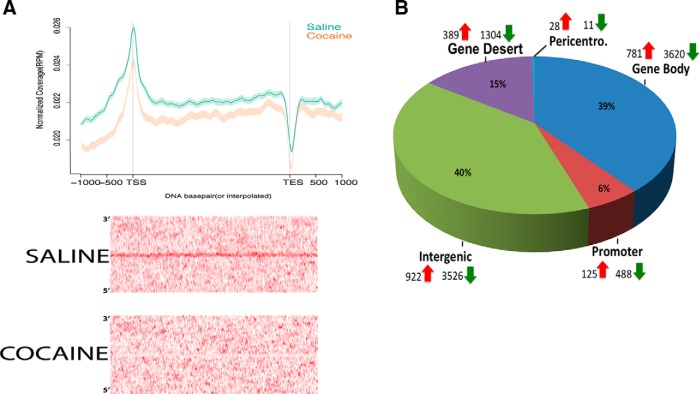
We observed a decrease in SIRT1 binding in NAc after chronic cocaine across gene regions genome-wide and particularly at the TSSs of genes. This reduction was surprising in light of the increased global levels of SIRT1 in NAc after cocaine treatment (Ferguson et al., 2013). First, MACs-based peak distribution analysis indicated that chronic cocaine induced a small decrease in the fraction of peaks that occur at or near gene-coding regions, from 34% in saline mice to 29.5% in cocaine mice (Fig. 1A). Additionally, plotting SIRT1 binding across all gene regions genome-wide, by use of ngs.plot (Shen et al., 2014), indicated that chronic cocaine causes a depletion of SIRT1 across entire gene regions compared with saline conditions (Fig. 2A, top). A differential site heatmap of SIRT1 binding after cocaine further supports the observed SIRT1 depletion and illustrates that this depletion is particularly dramatic around TSS regions after cocaine treatment (Fig. 2A, bottom). The differential site analysis of all genes that displayed altered SIRT1 binding after exposure to cocaine revealed that ∼84% of these genes showed a decrease in SIRT1 binding: 3620 showed decreased SIRT1 binding versus 781 that showed increased binding (Fig. 2B), supporting the relative depletion of SIRT1 from genic regions (Fig. 2A). A complete list of genes that show cocaine-induced changes in SIRT1 binding is available online (http://neuroscience.mssm.edu/nestler/nidappg/nacchroniccocaine.html). Genome-wide, diffReps analysis identified a total of 11,196 differential SIRT1 sites in NAc between saline- and cocaine-treated animals; 8949 were downregulated, whereas only 2245 were upregulated (Fig. 2B). Approximately 55% of these sites were present in intergenic and gene desert regions and the other 45% were present in promoter and gene body regions of the genome (Fig. 2B).
Figure 2.
Cocaine-induced changes in SIRT1 binding in NAc. A, Top, SIRT1 binding in NAc across gene regions genome-wide as visualized by ngs.plot. Binding at upstream promoters, TSSs, gene bodies, and transcription end sites (TESs) in normalized reads per million (RPM). Cocaine causes SIRT1 depletion across gene regions, but particularly at promoters and TSSs relative to saline. Bottom, Differential peak heatmap illustrates a dramatic depletion of SIRT1 binding in NAc after exposure to cocaine. B, Cocaine induces significantly more sites of reduced SIRT1 binding across genomic regions, for example, with 82% of genic sites showing a decrease in SIRT1 binding and 18% showing an increase in SIRT1 binding.
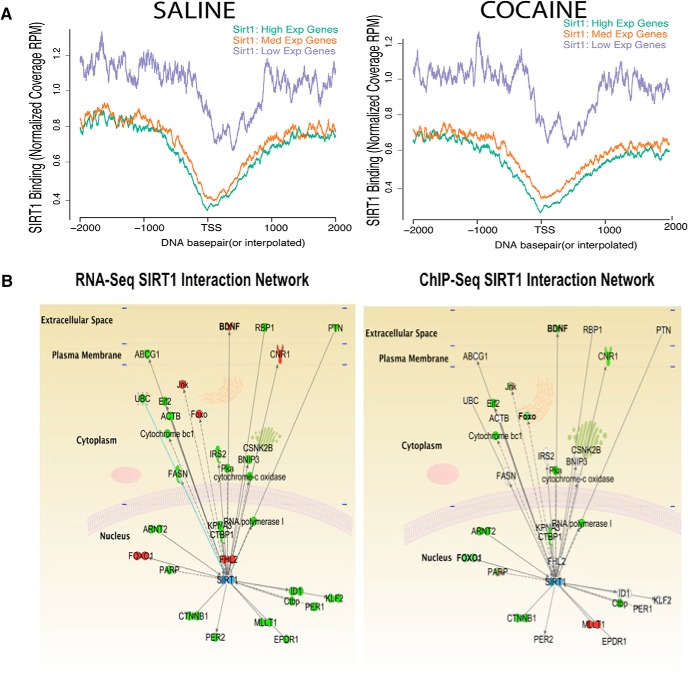
To test whether SIRT1 acts primarily as a transcriptional repressor in NAc in vivo, as observed in peripheral tissues and cultured cells (Luo et al., 2001), we mapped SIRT1 binding to genes based on their levels of mRNA expression using RNA sequencing (RNA-seq) data (Feng et al., 2014) derived from NAc, with genes grouped by their levels of mRNA expression: low, medium, and high. We observed dramatically higher levels of SIRT1 binding to genes that are expressed at low levels (Fig. 3A), a finding seen in NAc of both saline- and cocaine-treated mice. Among genes that show cocaine-induced depletion of SIRT1 binding in concert with increased mRNA levels is Bdnf. Indeed, independent quantitative qPCR and qChIP analysis of Bdnf in NAc of saline- and chronic cocaine-treated mice validated the RNA-seq and ChIP-seq results (Bdnf mRNA levels: 1.25 ± 0.10 relative to control; SIRT1 binding at Bdnf: 0.75 ± 0.13 relative to control; p < 0.05). Such findings are of particular note because of the well-known role of BDNF in NAc in regulating cocaine action (Lobo et al., 2010), and a recent study showing that SIRT1 indirectly regulates BDNF levels in hippocampus (Gao et al., 2010). Molecular pathway analysis of genes that display altered SIRT1 binding after cocaine, overlaid with those that show altered mRNA expression, reveals SIRT1 at the center of a major hub of biological regulation (Fig. 3B).
Figure 3.
Correlations of SIRT1 enrichment with gene expression in NAc. A, Correlation of SIRT1 binding by ChIP-seq (displayed by ngs.plot) with gene expression levels by RNA-seq shows that, under saline and cocaine conditions, genes expressed at lowest levels display dramatically higher enrichment of SIRT1. B, Ingenuity Pathway Analysis of the overlaid SIRT1 ChIP-seq and RNA-seq data identifies a network of genes regulated by cocaine in NAc. Green represents a decrease in gene expression (left) or in SIRT1 binding (right) in response to cocaine; red represents an increase. SIRT1 network interaction maps illustrate numerous SIRT1 targets. For example, cocaine increases the expression levels of Bdnf in NAc, an effect associated with decreased SIRT1 binding to the Bdnf promoter.
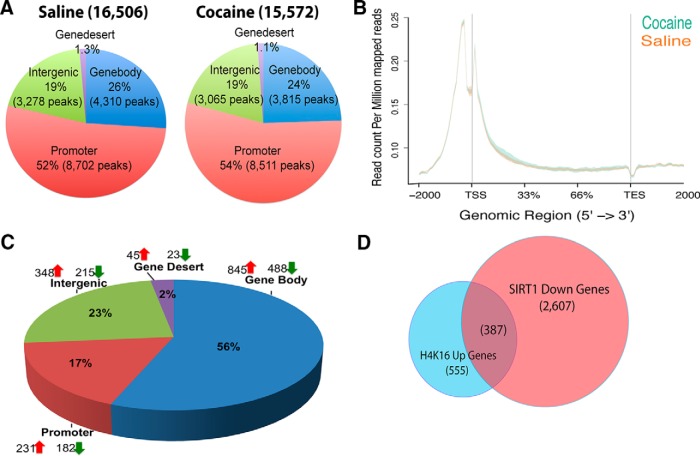
Overall, our results suggest that exposure to chronic cocaine induces a more permissive environment for transcription via the displacement of SIRT1 from the promoter/gene body of numerous downstream gene targets. H4K16ac is a well-known, direct target of SIRT1 in other systems, whereby the deacetylation of H4K16 by SIRT1 contributes to SIRT1's repressive effects on gene expression (Vaquero et al., 2004, 2007; O'Hagan et al., 2008; Hajji et al., 2010; Peng et al., 2012; Neumayer and Nguyen, 2014; Noguchi et al., 2014). Consistent with this literature, we showed recently that overexpressing SIRT1 in NAc decreases total cellular levels of H4K16ac (Ferguson et al., 2013). We therefore examined how the genome-wide enrichment of H4K16ac in NAc correlates with SIRT1 enrichment. ChIP-seq revealed strong enrichment of H4K16ac in gene promoter regions in both saline and cocaine conditions, with ∼50% of peaks residing at promoters (Fig. 4A,B). Importantly, cocaine induced an increase in H4K16ac enrichment at gene body and promoter regions (TSSs), which complements the observed decrease in SIRT1 binding at these regions (Fig. 4C). To further explore the networks associated with genes that show both increased H4K16ac and decreased SIRT1 binding, we subjected this dataset (387 genes) to Ingenuity Pathway Analysis. Such evaluation of these 387 genes revealed significant enrichment in signaling pathways associated with gene expression (p = 1.74E-05), nervous system development and function (p = 4.28E-05), and behavior (p = 1.30E-04).
Figure 4.
H4K16ac, a downstream target of SIRT1, is regulated in NAc by cocaine. A, ChIP-seq reveals prominent binding of H4K16ac to promoter/gene body regions (∼80%) in NAc, with much less binding to nongenic regions. Pie charts represent genome-wide occupancy of H4K16ac in saline (16,506 peaks) and chronic cocaine (15,562 peaks) conditions. B, H4K16ac binding to gene regions genome-wide, as visualized by ngs.plot, in NAc under saline and chronic cocaine conditions. C, Analysis of cocaine-induced changes in H4K16ac binding genome-wide, showing most regulation in promoter and gene body regions. D, Venn diagram showing genes that display both a decrease in SIRT1 binding and an increase in H4K16ac binding in NAc after chronic cocaine.
Further analysis of the ChIP-seq list of genes that show a corresponding increase in acH4K16 binding and decreased SIRT1 binding reveals several genes that have been previously shown to be regulated in the NAc by cocaine. Examples include the following: Mef2c (myocyte enhancer factor 2C) (Pulipparacharuvil et al., 2008; Host et al., 2011; Dietrich et al., 2012), Nlgn1 (neuroligin 1) (Tiruchinapalli et al., 2008), Kalrn (kalirin) (Ma et al., 2012), Ntrk2 (neurotrophic tyrosine kinase, receptor, type 2) (Graham et al., 2009; Lobo et al., 2010), and Grip1 (glutamate receptor interacting protein 1) (Briand et al., 2014), among others. As well, we observed cocaine regulation of several additional neural genes that control synaptic function in other systems and might contribute to cocaine-induced behavioral and synaptic plasticity, such as Nlgn2 (neuroligin 2) (van der Kooij et al., 2014), Synpo (synaptopodin) (Korkotian et al., 2014), Neurl1 (neuralized ligase) (Timmusk et al., 2002), Nedd4l (neural precursor cell expressed, developmentally downregulated 4-like, E3 ubiquitin protein ligase) (Ekberg et al., 2014), Striatin (a calmodulin binding protein (Chen et al., 2012), Ncam2 (neural cell adhesion molecule 2) (Bajor et al., 2012), Kif13b (kinesin family member 13B) (Yoshimura et al., 2010), Nf1 (neurofibromin 1) (Oliveira and Yasuda, 2014), Shank2 (SH3 and multiple ankyrin repeat domains 2) (Schneider et al., 2014), Wasl (Wiskott-Aldrich syndrome-like) (Wegner et al., 2008), Kif1a (kinesin family member 1A) (Lee et al., 2015), Kif5c (kinesin family member 5C) (Willemsen et al., 2014), Contactin 6 (a neural adhesion molecule) (Mercati et al., 2013), and Negr1 (neuronal growth regulator 1) (Pischedda et al., 2014). Also represented on the list of H4K16ac/SIRT1-affected genes were several whose products regulate chromatin, including Mll2 (an H3K4 methyltransferase), Kdm5b (an H3K4 demethylase), Prmt3 (a protein arginine methyltransferase), Atf7 (activating transcription factor 7), and Med27 (mediator complex subunit 27).
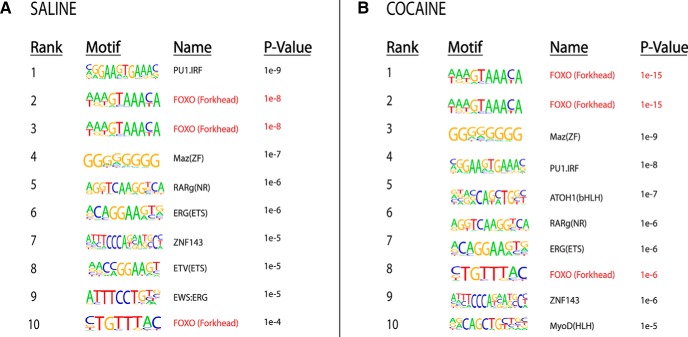
Given the fact that SIRT1 does not possess any intrinsic sequence-specific DNA-binding domains and is thought to be recruited to genomic regions through interactions with DNA-binding proteins, we speculated that SIRT1 might interact with transcription factors or related proteins to regulate gene expression in NAc. Therefore, we performed a de novo motif analysis of SIRT1-bound sequences within 3 kb of the TSSs from saline and cocaine conditions and compared these results to known transcription factor motifs. Of the 10 most significant SIRT1-associated motifs in NAc of either saline- or cocaine-treated mice, 30% corresponded to consensus binding sites for the FOXO family of transcriptions factors (motif: AAGTAAACA) (Fig. 5), which play important roles in numerous cellular functions (Brunet et al., 2004) but have not to date been studied in addiction models.
Figure 5.
Significant enrichment of FOXO binding motifs at genomic sites that shows significant SIRT1 binding by ChIP-seq in NAc under saline (A) and cocaine (B) conditions. Motif analysis of SIRT1-bound sequences from saline and cocaine conditions. Of the 10 most significant SIRT1-associated motifs in NAc of saline- and of cocaine-treated mice, 30% correspond to consensus binding sites for the FOXO family of transcription factors.
SIRT1 regulates the FOXO3a signaling pathway in NAc
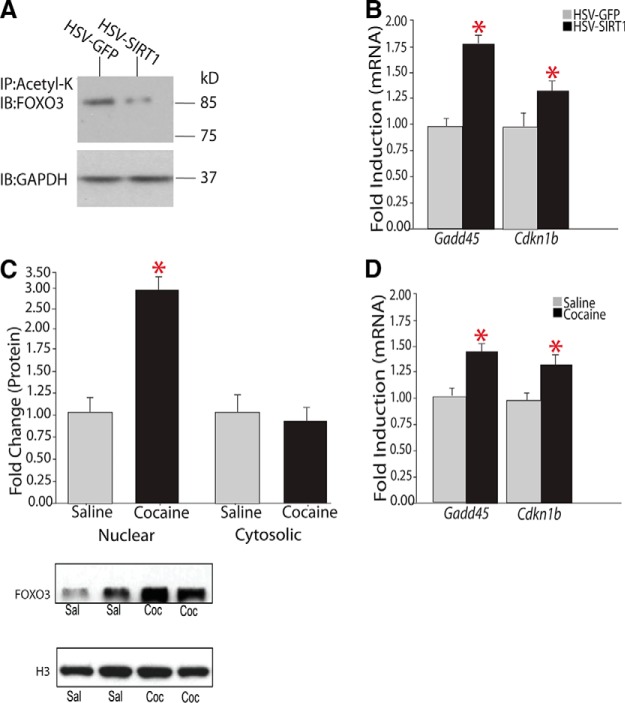
Based on this motif analysis, we explored interactions between SIRT1 and FOXO proteins in NAc, focusing on FOXO3a, because it is the predominant family member expressed throughout the brain (Hoekman et al., 2006) and because several prior studies have demonstrated SIRT1-FOXO3a interactions in other systems (Tissenbaum and Guarente, 2001; Patterson, 2003; Brunet et al., 2004; Motta et al., 2004). First, we found that FOXO3a is a downstream target of SIRT1 in NAc: immunoprecipitation of total acetylated proteins followed by immunoblotting for FOXO3a revealed a significant decrease in acetyl-FOXO3a in NAc overexpressing SIRT1 (Fig. 6A). Because such deacetylation of FOXO3a would increase its transcriptional activity (Nemoto et al., 2004), we performed qPCR on two established targets of FOXO3a in other systems, Gadd45α (growth arrest and DNA-damage-inducible protein-45a) and Cdkn1b (cyclin-dependent kinase inhibitor 1B; also known as p27Kip1) (Nemoto et al., 2004), and observed a significant increase in expression of both targets in NAc overexpressing SIRT1 (Fig. 6B). Second, although chronic cocaine administration had no effect on FOXO3a mRNA levels in NAc (data not shown), we observed increased levels of FOXO3a protein in nuclear fractions of NAc after chronic cocaine, with no change observed in cytosolic fractions (Fig. 6C). Third, we demonstrated that chronic cocaine increased expression levels of both FOXO3a targets, Gadd45α and Cdkn1b, in NAc (Fig. 6E). These results support a scheme wherein cocaine activates the FOXO3a signaling pathway in NAc via the induction of SIRT1 and consequent deacetylation of FOXO3a, leading to FOXO3a's activation and transcriptional activation of downstream target genes. Note that, via this mechanism, induction of SIRT1 leads to the induction, not repression, of a set of genes through indirect activation of FOXO3a.
Figure 6.
Regulation of the FOXO3a signaling pathway in NAc by SIRT1 and cocaine. A, Representative Western blot of immunoprecipitated pan-acetyl-K from NAc of mice injected with HSV-GFP or HSV-SIRT1 and probed with an anti-FOXO3a or -GAPDH antibody. HSV-SIRT1 deacetylates FOXO3a. Each lane represents pooled tissue from bilateral NAc punches from 5 mice. Results are representative of 5 replications. B, HSV-SIRT1 in NAc increases the induction of two known FOXO3a target genes, Gadd45α (HSV-GFP, 1.00 ± 0.10, n = 10; vs HSV-SIRT1, 1.81 ± 0.10, n = 9) and Cdkn1b (HSV GFP, 1.00 ± 0.05, n = 9; vs HSV-SIRT1, 1.68 ± 0.06, n = 9). C, Western blotting of nuclear or cytosolic fractions of NAc of mice treated chronically with saline or cocaine reveals a significant increase in FOXO3a protein levels in nuclear fractions (HSV GFP, 1.00 ± 0.29, n = 7; vs HSV-SIRT1, 2.93 ± 0.38, n = 7), with no differences in cytosolic fractions. D, Chronic cocaine administration induces downstream FOXO3a targets in NAc: Gadd45α (saline, 1.00 ± 0.02, n = 8; vs cocaine, 1.426 ± 0.06, n = 9) and Cdkn1b (saline, 1.00 ± 0.032, n = 8; vs cocaine, 1.301 ± 0.06, n = 9). *p < 0.05.
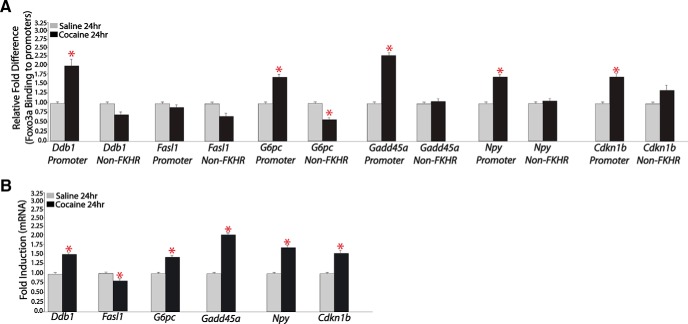
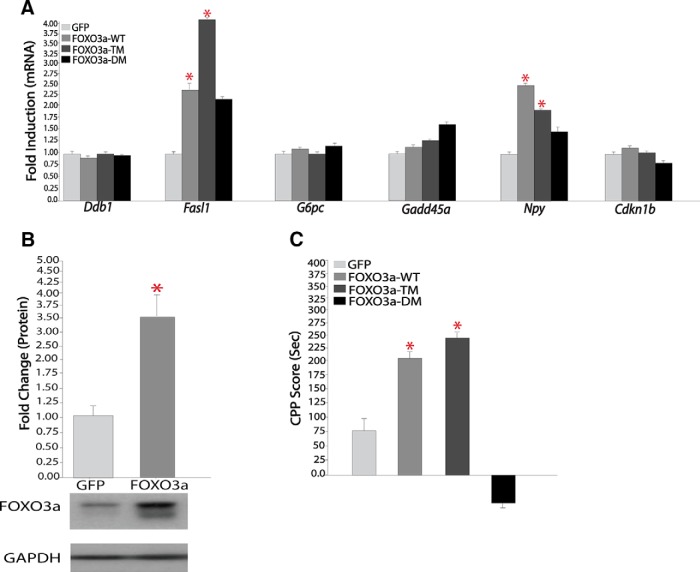
To provide further evidence for this scheme, we assessed whether chronic cocaine alters FOXO3a binding to the promoter regions of the Gadd45α and Cdkn1b genes as well as to several other genes inferred as FOXO3a targets from our motif analysis. As shown in Figure 7, Gadd45α and Cdkn1b, as well as most of the other investigated genes, showed cocaine-induced increases both in FOXO3a binding to their promoter regions (Figure 7A) and in mRNA expression levels (Figure 7B) in NAc. The other genes that displayed such regulation include Ddb1 (damage-specific DNA binding protein 1), Npy (neuropeptide Y), and G6pc (glucose 6-phosphatase). Interestingly, another inferred target, Fasl (Fas ligand), a known target of FOXO3a in other systems, did not show cocaine induction of FOXO3a to its gene promoter; rather, its mRNA expression was suppressed in NAc in response to chronic cocaine administration. We showed further that HSV-mediated overexpression of FOXO3a in NAc increased the expression of Npy and Fasl without influencing the other genes analyzed, compared with HSV-GFP injected control animals (Fig. 8A,B). Overexpressing a constitutively active mutant of FOXO3a (FOXO3a-TM) exerted equivalent effects, whereas expressing a dominant negative mutant of FOXO3a (FOXO3a-DM) had no effect. The fact that FOXO3a overexpression per se is not sufficient to mimic cocaine induction of Ddbl1, G6pc, Gadd45a, and Cdkn1b, but could induce Fasl, which is not induced by cocaine, indicates that other cocaine-induced factors are required, in concert with FOXO3a activation, to control the expression of these various genes in the NAc.
Figure 7.
Chronic cocaine increases FOXO3a binding to, and mRNA expression levels of, several of its downstream targets in NAc. A, qChIP for FOXO3a in NAc of mice treated chronically with cocaine or saline on several known downstream targets of FOXO3a (n = 5 or 6). B, qPCR analysis of NAc of mice treated chronically with cocaine or saline for FOXO3a downstream targets (n = 7 or 8). *p < 0.05.
Figure 8.
Effect of FOXO3a overexpression in NAc on gene expression and cocaine-induced place conditioning. A, HSV-FOXO3a vectors regulate downstream targets in the NAc, Fasl (HSV-GFP, 1.00 ± 0.12, n = 8; vs HSV-FOXO3a WT, 2.28 ± 0.59, n = 7; vs HSV-FOXO3aTM, 3.98 ± 0.83, n = 7); and Npy (HSV-GFP, 1.02 ± 0.11, n = 11; vs HSV-FOXO3aWT, 2.38 ± 0.64, n = 10; vs HSV-FOXO3aTM, 1.79 ± 0.28, n = 12), with no effect on other targets studied. B, Western blotting of NAc injected with HSV-GFP or HSV-FOXO3a reveals significant induction of FOXO3a protein with the latter vector (HSV-GFP, 1.00 ± 0.09, n = 6; vs HSV-FOXO3a, 3.35 ± 0.60). C, Mice were trained to cocaine (5 mg/kg)- or saline-paired sides of a chamber for 3 d after viral-mediated gene transfer with HSV-GFP, HSV-FOXO3a, HSV-FOXO3a-TM, or HSV-FOXO3a-DM. CPP scores are determined as the difference in time spent between cocaine- and saline-paired chambers. Data are presented as mean ± SEM (n = 8–11). *p < 0.05.
In any event, our data implicate FOXO3a in the transcriptional effects that cocaine exerts in NAc. To directly test the relevance of FOXO3a and its transcriptional actions in cocaine-related behaviors, we used HSV vectors to overexpress wild-type FOXO3a, FOXO3a-TM, or FOXO3a-DM in the mouse NAc and assessed cocaine CPP (Fig. 8C). We observed a dramatic increase in CPP scores to a subthreshold dose of cocaine in mice overexpressing FOXO3a or FOXO3a-TM relative to a GFP control vector (F(3,31) = 6.94, p = 0.001). In contrast, overexpression of FOXO3a-DM trended to decrease the small preference seen under control conditions. These data demonstrate that FOXO3a activity in NAc promotes rewarding responses to cocaine.
Discussion
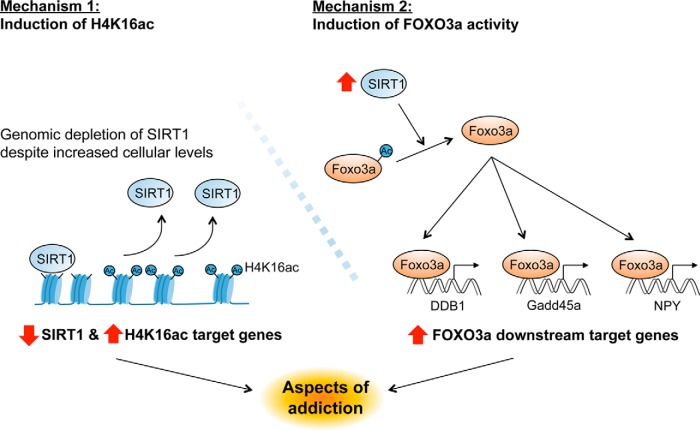
We recently showed that SIRT1 is induced in NAc by chronic cocaine administration and that SIRT1 overexpression in this region increases behavioral responses to cocaine, whereas its local knockdown has the opposite effect (Ferguson et al., 2013). The objective of the present study was to determine the downstream effects through which SIRT1 promotes cocaine action. To accomplish this goal, we used an unbiased approach (ChIP-seq) to create a genome-wide binding profile of SIRT1 and one of its primary downstream deacetylation targets, H4K16ac, in NAc of mice treated chronically with saline or cocaine. The results indicated that the induction of SIRT1, although associated with increased total numbers of SIRT1 binding sites, is surprisingly depleted from genic regions, in particular, gene promoters. This depletion occurs in concert with increased levels of H4K16ac and gene activation. We also used these unbiased data to identify FOXO transcription factors as the most highly regulated transcriptional mechanism involved in SIRT1 action and subsequently confirmed that indeed FOXO3a, the predominant FOXO protein in brain, is regulated in NAc by chronic cocaine administration, where it contributes to cocaine regulation of several downstream target genes and to cocaine's behavioral effects. Together, these data identify two main modes of action of SIRT1 in NAc (Fig. 9) and highlight the importance of using unbiased experimental approaches as a way to identify new transcriptional and molecular mechanisms underlying the chronic effects of cocaine on the brain.
Figure 9.
SIRT1 regulates gene expression in NAc via an H4K16ac- and a FOXO3a-dependent pathway. Cocaine regulation of SIRT1 induces gene expression in the NAc via two distinct actions. Mechanism 1, Despite the global increase in SIRT1 levels in NAc after cocaine, there is a significant depletion, through unknown mechanisms, of SIRTI binding to the promoter of certain genes, which leads to increased H4K16ac enrichment at those genes. Mechanism 2, SIRT1 deacetylates FOXO3a resulting in activation of FOXO3a's transcriptional activity and increased expression of its downstream targets. Such cocaine regulation of SIRT1, and SIRT1's regulation of gene expression through these distinct mechanisms, contributes to aspects of cocaine-induced neural and behavioral plasticity.
SIRT1 is the mammalian homolog of Sir2 in Saccharomyces cerevisiae, which was discovered as an inhibitor of the silent mating type loci, HML and HMR (Klar et al., 1979) and found to be localized to highly repressed regions of DNA, such as pericentromeres, heterochromatin, and major satellite repeats (Kanellopoulou et al., 2005). In mammalian systems, where SIRT1 has likewise been shown to exert a generally repressive effect on transcription, SIRT1 has also been demonstrated to bind significantly to euchromatic regions of the genome, including promoter and gene body regions in non-nervous tissues (Oberdoerffer et al., 2008; Bolasco et al., 2012). For example, an earlier study using ChIP-chip revealed SIRT1 binding to the promoter regions of genes in embryonic stem cells under basal conditions and reported a dramatic redistribution of SIRT1 away from gene promoters to other genomic regions in the presence of oxidative stress (e.g., H2O2) (Oberdoerffer et al., 2008). We observed a similar pattern of SIRT1 regulation in NAc: SIRT1 is broadly distributed across the genome in NAc at rest; and after chronic administration of cocaine, there is a dramatic displacement of SIRT1 from promoter regions of genes. These findings demonstrate that cocaine acts as a potent stimulus to reshape the SIRT1 epigenomic landscape.
One interesting question raised by these findings is as follows: Why is the increased amount of SIRT1 protein expressed in NAc after chronic cocaine not reflected by overall increased binding at genic regions, with manyfold more genes showing depletion of SIRT1 than showing recruitment of SIRT1? As well, what is the mechanism that depletes SIRT1 from promoter and gene body regions despite the elevated global levels of SIRT1 in the cells? The answers to these questions remain unknown, but it is interesting to speculate that such induction and redistribution of SIRT1, with an increase seen in the total number of SIRT1 sites, might help to compensate for the loss in heterochromatin that we have shown occurs in NAc in response to chronic cocaine administration (Maze et al., 2011). This loss of heterochromatin is mediated by reduced binding of repressive histone methylation, H3K9me3 and H3K9me2, to nongenic regions (Maze et al., 2010, 2011; Feng et al., 2014). Future work is needed to study this and many alternative hypotheses.
The general depletion of SIRT1 binding to genes after cocaine would be expected to contribute to an increase in those genes' expression. In the present study, we confirmed this hypothesis by demonstrating that Bdnf, a gene well characterized for its role in NAc in mediating cocaine-induced neural and behavioral plasticity (Lobo et al., 2010), is disinhibited by the loss of SIRT1 binding. We show that chronic cocaine administration decreases SIRT1 binding to the Bdnf gene, as found first by ChIP-seq and confirmed by qChIP, in concert with Bdnf induction under these conditions. These findings are consistent with a recent study, which demonstrated that knockout of SIRT1 leads to elevated levels of BDNF in hippocampus (Gao et al., 2010). By overlaying SIRT1 ChIP-seq data with an RNA-seq dataset, we identified a network of genes that show similar patterns of SIRT1-mediated regulation in NAc after chronic cocaine exposure (e.g., Fig. 3B).
One mechanism by which loss of SIRT1 would be expected to increase gene expression is through increased levels of H4K16ac, given that this mark is one of the primary substrates for SIRT1 in diverse tissues (Vaquero et al., 2007), including NAc (Ferguson et al., 2013). Consistent with these expectations, ChIP-seq analysis for H4K16ac in NAc showed prominent binding to promoter regions, as previously described in cultured cells (Wang et al., 2009; Füllgrabe et al., 2013), along with significant overlap of gene promoters that displayed increased H4K16ac binding plus decreased SIRT1 binding in NAc after chronic cocaine. Molecular pathway analysis of this set of genes revealed several interesting networks that now warrant further investigation for their role in cocaine action (see Results).
Aside from H4K16ac, additional substrates for SIRT1 in other experimental systems are FOXO transcription factors, which are deacetylated and activated by SIRT1 (Brunet et al., 2004; Daitoku et al., 2004; Giannakou and Partridge, 2004; Nemoto et al., 2004). Indeed, SIRT1 has been shown to thereby regulate expression levels of several FOXO gene targets (Daitoku et al., 2004; Giannakou and Partridge, 2004; Motta et al., 2004). We identified FOXO transcription factors as an important mediator of SIRT1 in NAc through a completely unbiased motif analysis by studying regions of the genome that show altered SIRT1 binding after cocaine exposure. This analysis was based on the knowledge that SIRT1 does not bind to DNA in a sequence-specific manner, indicating the involvement of DNA-binding proteins in mediating the downstream effects of SIRT1 induction by cocaine. We found that the FOXO DNA-binding motif was the most highly represented in our SIRT1 ChIP-seq dataset. Interestingly, the other most highly represented motifs correspond to binding sites for the PU.1-IRF (interferon-regulatory factor)-ETS (E26 transformation-specific) and the Maz(ZP) (Myc-associated zinc finger protein) families of transcription factors, which have been implicated in several peripheral cancers but have not to date been studied in brain. It would be interesting to evaluate the functioning of these factors in cocaine models.
FOXO proteins influence a range of functions, including cell cycle, differentiation, metabolism, apoptosis, stress resistance, DNA repair, and aging (Brunet et al., 1999; Birkenkamp and Coffer, 2003). However, the involvement of FOXO in molecular and behavioral adaptations to cocaine has not been previously examined. Here, based on our unbiased motif analysis, we explored a role for FOXO3a, the FOXO isoform most highly expressed in brain, in cocaine action on NAc. We demonstrate that SIRT1 overexpression deacetylates FOXO3a in NAc and that chronic cocaine administration induces the nuclear translocation of FOXO3a in this brain region. We demonstrate further that FOXO3a induction in NAc enhances rewarding responses to cocaine. We also show that cocaine exposure induces several known gene targets of FOXO3a in concert with increased binding of FOXO3a to those genes' promoter regions. Among these cocaine-induced, FOXO3a downstream targets are Gadd45α and Cdkn1b. Cdkn1b has been shown previously to be a target of FOXO3a in other tissues: one group showed inhibition of Cdkn1b by FOXO3a in HeLa cells, whereas other groups reported activation, similar to the findings in this study, in mouse embryonic fibroblasts (Brunet et al., 2004; Motta et al., 2004; van der Horst et al., 2004). These observations underscore the highly cell type-specific actions of most transcription factors. The induction of Gadd45α in NAc by chronic cocaine is particularly interesting based on a recent study showing that GADD45 regulates memory and synaptic plasticity (Sultan et al., 2012). Our finding that chronic cocaine, through a SIRT1-FOXO3a network, induces Gadd45α and Ddb1, another DNA repair protein, in NAc implicates for the first time proteins that are involved in DNA repair in other systems in the long-term actions of cocaine on the brain. Further work is needed to determine whether actual DNA injury is involved or whether these effects reflect distinct functions of the proteins within fully differentiated adult brain tissue.
In conclusion, the present study demonstrates the power of unbiased epigenomic assays to reveal fundamentally new mechanisms by which chronic cocaine induces molecular and behavioral plasticity. We show two mechanisms by which chronic cocaine regulation of SIRT1 induces gene transcription in NAc. First, the loss of SIRT1 from certain gene promoters increases H4K16ac at those promoters to activate transcription. Second, the induction of global levels of SIRT1 deacetylates and activates FOXO3a, which then induces many of its downstream gene targets. The discovery of these novel mechanisms in the context of cocaine exposure advances our understanding of the molecular basis of cocaine addiction.
Footnotes
This work was supported by National Institute on Drug Abuse, National Alliance for Research on Schizophrenia and Depression, and UNCF/MERCK.
The authors declare no competing financial interests.
References
- Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- Bajor M, Michaluk P, Gulyassy P, Kekesi AK, Juhasz G, Kaczmarek L. Synaptic cell adhesion molecule-2 and collapsin response mediator protein-2 are novel members of the matrix metalloproteinase-9 degradome. J Neurochem. 2012;122:775–788. doi: 10.1111/j.1471-4159.2012.07829.x. [DOI] [PubMed] [Google Scholar]
- Birkenkamp KU, Coffer PJ. Regulation of cell survival and proliferation by the FOXO (Forkhead box, class O) subfamily of Forkhead transcription factors. Biochem Soc Trans. 2003;31:292–297. doi: 10.1042/bst0310292. [DOI] [PubMed] [Google Scholar]
- Bolasco G, Calogero R, Carrara M, Banchaabouchi MA, Bilbao D, Mazzoccoli G, Vinciguerra M. Cardioprotective mIGF-1/SIRT1 signaling induces hypertension, leukocytosis and fear response in mice. Aging (Albany NY) 2012;4:402–416. doi: 10.18632/aging.100464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briand LA, Kimmey BA, Ortinski PI, Huganir RL, Pierce RC. Disruption of glutamate receptor-interacting protein in nucleus accumbens enhances vulnerability to cocaine relapse. Neuropsychopharmacology. 2014;39:759–769. doi: 10.1038/npp.2013.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/S0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- Chang HC, Guarente L. SIRT1 mediates central circadian control in the SCN by a mechanism that decays with aging. Cell. 2013;153:1448–1460. doi: 10.1016/j.cell.2013.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YK, Chen CY, Hu HT, Hsueh YP. CTTNBP2, but not CTTNBP2NL, regulates dendritic spinogenesis and synaptic distribution of the striatin-PP2A complex. Mol Biol Cell. 2012;23:4383–4392. doi: 10.1091/mbc.E12-05-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codocedo JF, Allard C, Godoy JA, Varela-Nallar L, Inestrosa NC. SIRT1 regulates dendritic development in hippocampal neurons. PLoS One. 2012;7:e47073. doi: 10.1371/journal.pone.0047073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen DE, Supinski AM, Bonkowski MS, Donmez G, Guarente LP. Neuronal SIRT1 regulates endocrine and behavioral responses to calorie restriction. Genes Dev. 2009;23:2812–2817. doi: 10.1101/gad.1839209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daitoku H, Hatta M, Matsuzaki H, Aratani S, Ohshima T, Miyagishi M, Nakajima T, Fukamizu A. Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc Natl Acad Sci U S A. 2004;101:10042–10047. doi: 10.1073/pnas.0400593101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich JB, Takemori H, Grosch-Dirrig S, Bertorello A, Zwiller J. Cocaine induces the expression of MEF2C transcription factor in rat striatum through activation of SIK1 and phosphorylation of the histone deacetylase HDAC5. Synapse. 2012;66:61–70. doi: 10.1002/syn.20988. [DOI] [PubMed] [Google Scholar]
- Ekberg JA, Boase NA, Rychkov G, Manning J, Poronnik P, Kumar S. Nedd4–2 (NEDD4L) controls intracellular Na(+)-mediated activity of voltage-gated sodium channels in primary cortical neurons. Biochem J. 2014;457:27–31. doi: 10.1042/BJ20131275. [DOI] [PubMed] [Google Scholar]
- Feng J, Wilkinson M, Liu X, Purushothaman I, Ferguson D, Vialou V, Maze I, Shao N, Kennedy P, Koo J, Dias C, Laitman B, Stockman V, LaPlant Q, Cahill ME, Nestler EJ, Shen L. Chronic cocaine-regulated epigenomic changes in mouse nucleus accumbens. Genome Biol. 2014;15:R65. doi: 10.1186/gb-2014-15-4-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson D, Koo JW, Feng J, Heller E, Rabkin J, Heshmati M, Renthal W, Neve R, Liu X, Shao N, Sartorelli V, Shen L, Nestler EJ. Essential role of SIRT1 signaling in the nucleus accumbens in cocaine and morphine action. J Neurosci. 2013;33:16088–16098. doi: 10.1523/JNEUROSCI.1284-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Füllgrabe J, Lynch-Day MA, Heldring N, Li W, Struijk RB, Ma Q, Hermanson O, Rosenfeld MG, Klionsky DJ, Joseph B. The histone H4 lysine 16 acetyltransferase hMOF regulates the outcome of autophagy. Nature. 2013;500:468–471. doi: 10.1038/nature12313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Wang WY, Mao YW, Gräff J, Guan JS, Pan L, Mak G, Kim D, Su SC, Tsai LH. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature. 2010;466:1105–1109. doi: 10.1038/nature09271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannakou ME, Partridge L. The interaction between FOXO and SIRT1: tipping the balance towards survival. Trends Cell Biol. 2004;14:408–412. doi: 10.1016/j.tcb.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Graham DL, Krishnan V, Larson EB, Graham A, Edwards S, Bachtell RK, Simmons D, Gent LM, Berton O, Bolanos CA, DiLeone RJ, Parada LF, Nestler EJ, Self DW. Tropomyosin-related kinase B in the mesolimbic dopamine system: region-specific effects on cocaine reward. Biol Psychiatry. 2009;65:696–701. doi: 10.1016/j.biopsych.2008.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajji N, Wallenborg K, Vlachos P, Füllgrabe J, Hermanson O, Joseph B. Opposing effects of hMOF and SIRT1 on H4K16 acetylation and the sensitivity to the topoisomerase II inhibitor etoposide. Oncogene. 2010;29:2192–2204. doi: 10.1038/onc.2009.505. [DOI] [PubMed] [Google Scholar]
- Hoekman MF, Jacobs FM, Smidt MP, Burbach JP. Spatial and temporal expression of FoxO transcription factors in the developing and adult murine brain. Gene Expr Patterns. 2006;6:134–140. doi: 10.1016/j.modgep.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Host L, Dietrich JB, Carouge D, Aunis D, Zwiller J. Cocaine self-administration alters the expression of chromatin-remodelling proteins: modulation by histone deacetylase inhibition. J Psychopharmacol. 2011;25:222–229. doi: 10.1177/0269881109348173. [DOI] [PubMed] [Google Scholar]
- Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar AJ, Fogel S, Lusnak K. Gene conversion of the mating-type locus in Saccharomyces cerevisiae. Genetics. 1979;92:777–782. doi: 10.1093/genetics/92.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkotian E, Frotscher M, Segal M, Zhang XL, Poschel B, Faul C, Upreti C, Stanton PK, Mundel P. Synaptopodin regulates spine plasticity: mediation by calcium stores. J Neurosci. 2014;34:11641–11651. doi: 10.1523/JNEUROSCI.0381-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Choi KH, Renthal W, Tsankova NM, Theobald DE, Truong HT, Russo SJ, Laplant Q, Sasaki TS, Whistler KN, Neve RL, Self DW, Nestler EJ. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron. 2005;48:303–314. doi: 10.1016/j.neuron.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Lee JR, Srour M, Kim D, Hamdan FF, Lim SH, Brunel-Guitton C, Décarie JC, Rossignol E, Mitchell GA, Schreiber A, Moran R, Van Haren K, Richardson R, Nicolai J, Oberndorff KM, Wagner JD, Boycott KM, Rahikkala E, Junna N, Tyynismaa H, et al. De novo mutations in the motor domain of KIF1A cause cognitive impairment, spastic paraparesis, axonal neuropathy and cerebellar atrophy. Hum Mutat. 2015;36:69–78. doi: 10.1002/humu.22709. [DOI] [PubMed] [Google Scholar]
- Li XH, Chen C, Tu Y, Sun HT, Zhao ML, Cheng SX, Qu Y, Zhang S. Sirt1 promotes axonogenesis by deacetylation of Akt and inactivation of GSK3. Mol Neurobiol. 2013;48:490–499. doi: 10.1007/s12035-013-8437-3. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lobo MK, Covington HE, 3rd, Chaudhury D, Friedman AK, Sun H, Damez-Werno D, Dietz DM, Zaman S, Koo JW, Kennedy PJ, Mouzon E, Mogri M, Neve RL, Deisseroth K, Han MH, Nestler EJ. Cell type-specific loss of BDNF signaling mimics optogenetic control of cocaine reward. Science. 2010;330:385–390. doi: 10.1126/science.1188472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/S0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- Ma XM, Huang JP, Xin X, Yan Y, Mains RE, Eipper BA. A role for kalirin in the response of rat medium spiny neurons to cocaine. Mol Pharmacol. 2012;82:738–745. doi: 10.1124/mol.112.080044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maze I, Nestler EJ. The epigenetic landscape of addiction. Ann N Y Acad Sci. 2011;1216:99–113. doi: 10.1111/j.1749-6632.2010.05893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maze I, Covington HE, 3rd, Dietz DM, LaPlant Q, Renthal W, Russo SJ, Mechanic M, Mouzon E, Neve RL, Haggarty SJ, Ren Y, Sampath SC, Hurd YL, Greengard P, Tarakhovsky A, Schaefer A, Nestler EJ. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science (New York) 2010;327:213–216. doi: 10.1126/science.1179438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maze I, Feng J, Wilkinson MB, Sun H, Shen L, Nestler EJ. Cocaine dynamically regulates heterochromatin and repetitive element unsilencing in nucleus accumbens. Proc Natl Acad Sci U S A. 2011;108:3035–3040. doi: 10.1073/pnas.1015483108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercati O, Danckaert A, André-Leroux G, Bellinzoni M, Gouder L, Watanabe K, Shimoda Y, Grailhe R, De Chaumont F, Bourgeron T, Cloëz-Tayarani I. Contactin 4, -5 and -6 differentially regulate neuritogenesis while they display identical PTPRG binding sites. Biol Open. 2013;2:324–334. doi: 10.1242/bio.20133343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michán S, Li Y, Chou MM, Parrella E, Ge H, Long JM, Allard JS, Lewis K, Miller M, Xu W, Mervis RF, Chen J, Guerin KI, Smith LE, McBurney MW, Sinclair DA, Baudry M, de Cabo R, Longo VD. SIRT1 is essential for normal cognitive function and synaptic plasticity. J Neurosci. 2010;30:9695–9707. doi: 10.1523/JNEUROSCI.0027-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, Gu W, Bultsma Y, McBurney M, Guarente L. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–563. doi: 10.1016/S0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- Nemoto S, Fergusson MM, Finkel T. Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science. 2004;306:2105–2108. doi: 10.1126/science.1101731. [DOI] [PubMed] [Google Scholar]
- Neumayer G, Nguyen MD. TPX2 impacts acetylation of histone H4 at lysine 16: implications for DNA damage response. PLoS One. 2014;9:e110994. doi: 10.1371/journal.pone.0110994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi A, Kikuchi K, Zheng H, Takahashi H, Miyagi Y, Aoki I, Takano Y. SIRT1 expression is associated with a poor prognosis, whereas DBC1 is associated with favorable outcomes in gastric cancer. Cancer Med. 2014;3:1553–1561. doi: 10.1002/cam4.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberdoerffer P, Michán S, McVay M, Mostoslavsky R, Vann J, Park SK, Hartlerode A, Stegmuller J, Hafner A, Loerch P, Wright SM, Mills KD, Bonni A, Yankner BA, Scully R, Prolla TA, Alt FW, Sinclair DA. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135:907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hagan HM, Mohammad HP, Baylin SB. Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island. PLoS Genet. 2008;4:e1000155. doi: 10.1371/journal.pgen.1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira AF, Yasuda R. Neurofibromin is the major ras inactivator in dendritic spines. J Neurosci. 2014;34:776–783. doi: 10.1523/JNEUROSCI.3096-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson GI. Aging: new targets, new functions. Curr Biol. 2003;13:R279–R281. doi: 10.1016/S0960-9822(03)00201-X. [DOI] [PubMed] [Google Scholar]
- Peng L, Ling H, Yuan Z, Fang B, Bloom G, Fukasawa K, Koomen J, Chen J, Lane WS, Seto E. SIRT1 negatively regulates the activities, functions, and protein levels of hMOF and TIP60. Mol Cell Biol. 2012;32:2823–2836. doi: 10.1128/MCB.00496-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pischedda F, Szczurkowska J, Cirnaru MD, Giesert F, Vezzoli E, Ueffing M, Sala C, Francolini M, Hauck SM, Cancedda L, Piccoli G. A cell surface biotinylation assay to reveal membrane-associated neuronal cues: Negr1 regulates dendritic arborization. Mol Cell Proteomics. 2014;13:733–748. doi: 10.1074/mcp.M113.031716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulipparacharuvil S, Renthal W, Hale CF, Taniguchi M, Xiao G, Kumar A, Russo SJ, Sikder D, Dewey CM, Davis MM, Greengard P, Nairn AC, Nestler EJ, Cowan CW. Cocaine regulates MEF2 to control synaptic and behavioral plasticity. Neuron. 2008;59:621–633. doi: 10.1016/j.neuron.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafalski VA, Ho PP, Brett JO, Ucar D, Dugas JC, Pollina EA, Chow LM, Ibrahim A, Baker SJ, Barres BA, Steinman L, Brunet A. Expansion of oligodendrocyte progenitor cells following SIRT1 inactivation in the adult brain. Nat Cell Biol. 2013;15:614–624. doi: 10.1038/ncb2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadori G, Fujikawa T, Anderson J, Berglund ED, Frazao R, Michán S, Vianna CR, Sinclair DA, Elias CF, Coppari R. SIRT1 deacetylase in SF1 neurons protects against metabolic imbalance. Cell Metab. 2011;14:301–312. doi: 10.1016/j.cmet.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renthal W, Nestler EJ. Chromatin regulation in drug addiction and depression. Dialogues Clin Neurosci. 2009;11:257–268. doi: 10.31887/DCNS.2009.11.3/wrenthal. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renthal W, Kumar A, Xiao G, Wilkinson M, Covington HE, 3rd, Maze I, Sikder D, Robison AJ, LaPlant Q, Dietz DM, Russo SJ, Vialou V, Chakravarty S, Kodadek TJ, Stack A, Kabbaj M, Nestler EJ. Genome-wide analysis of chromatin regulation by cocaine reveals a role for sirtuins. Neuron. 2009;62:335–348. doi: 10.1016/j.neuron.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robison AJ, Nestler EJ. Transcriptional and epigenetic mechanisms of addiction. Nat Rev Neurosci. 2011;12:623–637. doi: 10.1038/nrn3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider K, Seemann E, Liebmann L, Ahuja R, Koch D, Westermann M, Hübner CA, Kessels MM, Qualmann B. ProSAP1 and membrane nanodomain-associated syndapin I promote postsynapse formation and function. J Cell Biol. 2014;205:197–215. doi: 10.1083/jcb.201307088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Shao NY, Liu X, Maze I, Feng J, Nestler EJ. diffReps: detecting differential chromatin modification sites from ChIP-seq data with biological replicates. PLoS One. 2013;8:e65598. doi: 10.1371/journal.pone.0065598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Shao N, Liu X, Nestler E. ngs.plot: quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics. 2014;15:284. doi: 10.1186/1471-2164-15-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan FA, Wang J, Tront J, Liebermann DA, Sweatt JD. Genetic deletion of Gadd45b, a regulator of active DNA demethylation, enhances long-term memory and synaptic plasticity. J Neurosci. 2012;32:17059–17066. doi: 10.1523/JNEUROSCI.1747-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmusk T, Palm K, Belluardo N, Mudò G, Neuman T. Dendritic localization of mammalian neuralized mRNA encoding a protein with transcription repression activities. Mol Cell Neurosci. 2002;20:649–668. doi: 10.1006/mcne.2002.1148. [DOI] [PubMed] [Google Scholar]
- Tiruchinapalli DM, Caron MG, Keene JD. Activity-dependent expression of ELAV/Hu RBPs and neuronal mRNAs in seizure and cocaine brain. J Neurochem. 2008;107:1529–1543. doi: 10.1111/j.1471-4159.2008.05718.x. [DOI] [PubMed] [Google Scholar]
- Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- van der Horst A, Tertoolen LG, de Vries-Smits LM, Frye RA, Medema RH, Burgering BM. FOXO4 is acetylated upon peroxide stress and deacetylated by the longevity protein hSir2(SIRT1) J Biol Chem. 2004;279:28873–28879. doi: 10.1074/jbc.M401138200. [DOI] [PubMed] [Google Scholar]
- van der Kooij MA, Fantin M, Kraev I, Korshunova I, Grosse J, Zanoletti O, Guirado R, Garcia-Mompó C, Nacher J, Stewart MG, Berezin V, Sandi C. Impaired hippocampal neuroligin-2 function by chronic stress or synthetic peptide treatment is linked to social deficits and increased aggression. Neuropsychopharmacology. 2014;39:1148–1158. doi: 10.1038/npp.2013.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- Vaquero A, Sternglanz R, Reinberg D. NAD+-dependent deacetylation of H4 lysine 16 by class III HDACs. Oncogene. 2007;26:5505–5520. doi: 10.1038/sj.onc.1210617. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegner AM, Nebhan CA, Hu L, Majumdar D, Meier KM, Weaver AM, Webb DJ. N-wasp and the arp2/3 complex are critical regulators of actin in the development of dendritic spines and synapses. J Biol Chem. 2008;283:15912–15920. doi: 10.1074/jbc.M801555200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen MH, Ba W, Wissink-Lindhout WM, de Brouwer AP, Haas SA, Bienek M, Hu H, Vissers LE, van Bokhoven H, Kalscheuer V, Nadif Kasri N, Kleefstra T. Involvement of the kinesin family members KIF4A and KIF5C in intellectual disability and synaptic function. J Med Genet. 2014;51:487–494. doi: 10.1136/jmedgenet-2013-102182. [DOI] [PubMed] [Google Scholar]
- Yoshimura Y, Terabayashi T, Miki H. Par1b/MARK2 phosphorylates kinesin-like motor protein GAKIN/KIF13B to regulate axon formation. Mol Cell Biol. 2010;30:2206–2219. doi: 10.1128/MCB.01181-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]