Abstract
The charged multivesicular body proteins (Chmp1–7) are an evolutionarily conserved family of cytosolic proteins that transiently assembles into helical polymers that change the curvature of cellular membrane domains. Mutations in human CHMP2B cause frontotemporal dementia, suggesting that this protein may normally control some neuron-specific process. Here, we examined the function, localization, and interactions of neuronal Chmp2b. The protein was highly expressed in mouse brain and could be readily detected in neuronal dendrites and spines. Depletion of endogenous Chmp2b reduced dendritic branching of cultured hippocampal neurons, decreased excitatory synapse density in vitro and in vivo, and abolished activity-induced spine enlargement and synaptic potentiation. To understand the synaptic effects of Chmp2b, we determined its ultrastructural distribution by quantitative immuno-electron microscopy and its biochemical interactions by coimmunoprecipitation and mass spectrometry. In the hippocampus in situ, a subset of neuronal Chmp2b was shown to concentrate beneath the perisynaptic membrane of dendritic spines. In synaptoneurosome lysates, Chmp2b was stably bound to a large complex containing other members of the Chmp family, as well as postsynaptic scaffolds. The supramolecular Chmp assembly detected here corresponds to a stable form of the endosomal sorting complex required for transport-III (ESCRT-III), a ubiquitous cytoplasmic protein complex known to play a central role in remodeling of lipid membranes. We conclude that Chmp2b-containing ESCRT-III complexes are also present at dendritic spines, where they regulate synaptic plasticity. We propose that synaptic ESCRT-III filaments may function as a novel element of the submembrane cytoskeleton of spines.
Keywords: ESCRT filaments, frontotemporal dementia, postsynaptic scaffold, spinoskeleton, structural plasticity
Introduction
The endosomal sorting complex required for transport-III (ESCRT-III) is a ubiquitous protein complex that is crucial to cellular membrane remodeling during inward vesiculation of multivesicular endosomes, mitotic cell abscission, and viral budding (for review, see Henne et al., 2013; McCullough et al., 2013). The core subunits of ESCRT-III are a family of evolutionarily conserved proteins, called charged multivesicular body proteins (Chmp1-7). Dominant mutations in subunit CHMP2B cause frontotemporal dementia (FTD; Skibinski et al., 2005; Isaacs et al., 2011), raising the question whether some neuron-specific process is controlled by this protein. Chmp2b and other ESCRT-III subunits exist in the cytosol in a monomeric, autoinhibited state. After release of intramolecular inhibition, individual subunits polymerize into circular or helical filaments (20–200 nm wide, subject to subunit species) that adhere to lipid membranes and change their curvature (Shim et al., 2007; Hanson et al., 2008; Lata et al., 2008; Bajorek et al., 2009; Henne et al., 2012; Buchkovich et al., 2013).
We have previously shown that Chmp2b preferentially polymerizes under the plasma membrane and, when overexpressed, organizes into helices that deform patches of the cell surface into long protrusions (Bodon et al., 2011). Holomeric ESCRT-III consists of heteropolymeric Chmp filaments assembled at sites of endosomal or plasma membrane deformation, after recruitment by the upstream-acting complexes ESCRT-I and -II and/or by the Chmp4b-binding protein Alix. ESCRT-III assemblies at membranes are thought to be transient, being dissociated after interaction with the depolymerizing ATPase Vps4. This depolymerization step is coupled to scission of membrane necks modeled by ESCRT-III filaments (Henne et al., 2013).
FTD-associated CHMP2B mutants are deregulated and form aberrant polymeric structures in transfected cells (Skibinski et al., 2005). Strong expression of such a CHMP2B mutant in cultured cortical neurons is rapidly lethal (Lee et al., 2007), and transgenic mice expressing the same mutant throughout their brain display progressive neurodegenerative changes (Ghazi-Noori et al., 2012). These effects have been linked to defects in autophagic and endosomal degradation pathways (Filimonenko et al., 2007; Lee et al., 2007; Urwin et al., 2010). However, these phenotypes arise from a toxic gain rather than loss of function in the mutant allele. Indeed, degeneration was not seen in knock-out mice severely (though not completely) deficient for Chmp2b (Ghazi-Noori et al., 2012). Furthermore, Chmp2b depletion did not affect survival of cultured neurons (Lee et al., 2007; Belly et al., 2010) but significantly impaired the morphological maturation of dendritic spines (Belly et al., 2010). This, together with the fact that synaptogenesis correlates with Chmp2b gene transcription in some neuronal populations (Xu et al., 2012), led us to hypothesize that normal Chmp2b and ESCRT-III may function at synapses.
Here, we show that Chmp2b is required for maintaining the complexity of dendritic trees as well as the stability, efficacy, and plasticity of excitatory synapses. We also show that the protein preferentially localizes beneath the plasma membrane of dendritic spines and is bound to stable ESCRT-III complexes associated with the postsynaptic cytoskeleton, suggesting that ESCRT-III polymers cooperate with postsynaptic scaffolds and actin filaments to shape dendritic spines.
Materials and Methods
Plasmids.
See Table 1 for details of all constructs. The pSuper-mCherry vector was described by Belly et al. (2010) and expresses the inserted shRNA together with mCherry. The Chmp2b-specific shRNA (Lee et al., 2007; Belly et al., 2010) and the GFP-specific shRNA [previously shown by Alvarez et al. (2006) to have no effect on neuronal morphology or electrical properties] have been described and were inserted into pSuper-mCherry. cDNAs encoding wild-type (WT) or mutant versions of the RNAi-resistant Chmp2b (Belly et al., 2010) were cloned in the pBI-CMV3 vector (Clontech), yielding pBI-CMV3-Chmp2bWT, pBI-CMV3-Chmp2bR19/22/26A, and pBI-CMV3-Chmp2bL4D/F5D. In these vectors, Chmp2b is coexpressed with fluorescent protein ZsGreen from a bidirectional promoter. cDNA encoding wild-type, RNAi-resistant Chmp2b (carrying a single C-terminal Flag tag; Bodon et al., 2011) was inserted upstream of the IRES sequence of the pIRES2-EGFP vector (Clontech), yielding pIRES2-EGFP-Chmp2b (used in Fig. 6B). In this vector, Chmp2b is coexpressed with EGFP from a bicistronic transcript. All constructs were verified by sequencing (Beckman Coulter).
Table 1.
Plasmids used in this study
| Plasmid name | Cloning technique | Cloning sites | Feature | Tags | Source |
|---|---|---|---|---|---|
| pIRES2-EGFP | Starting vector | Clontech | |||
| pIRES2-EGFP-CHMP2B WT | In-Fusion (Clontech) | XhoI, XhoI | RNAi res. | C-terminal FLAG | This study |
| pBI-CMV3 | Starting vector | Clontech | |||
| pBI-CMV3-CHMP2B WT | In-Fusion (Clontech) | BamHI, HindIII | RNAi res. | This study | |
| pBI-CMV3-CHMP2B R19/22/26A | In-Fusion (Clontech) | BamHI, HindIII | RNAi res. | This study | |
| pBI-CMV3-CHMP2B L4D/F5D | In-Fusion (Clontech) | BamHI, HindIII | RNAi res. | This study | |
| pBI-CMV3-CHMP2B L207D/L210D | In-Fusion (Clontech) | BamHI, HindIII | RNAi res. | This study | |
| pSuper-mCherry | Starting vector | Belly et al., 2010 | |||
| pSuper-mCherry shRNA vs CHMP2B | Standard ligation | HindIII, BglII | Belly et al., 2010 | ||
| pSuper-mCherry shRNA vs GFP | Standard ligation | HindIII, BglII | This study | ||
| pCDNA3.1-CHMP2B WT | Topo (Invitrogen) | C-terminal HA | This study | ||
| pCDNA3.1-CHMP2B R19/22/26A | Topo (Invitrogen) | C-terminal HA | This study | ||
| pCDNA3.1-CHMP4B | Topo (Invitrogen) | C-terminal FLAG | Bodon et al., 2011 |
The cloning technique used to construct each plasmid and, if applicable, the associated kit supplier are indicated. RNAi res. Indicates the encoded mRNA is resistant to RNAi by the Chmp2b shRNA.
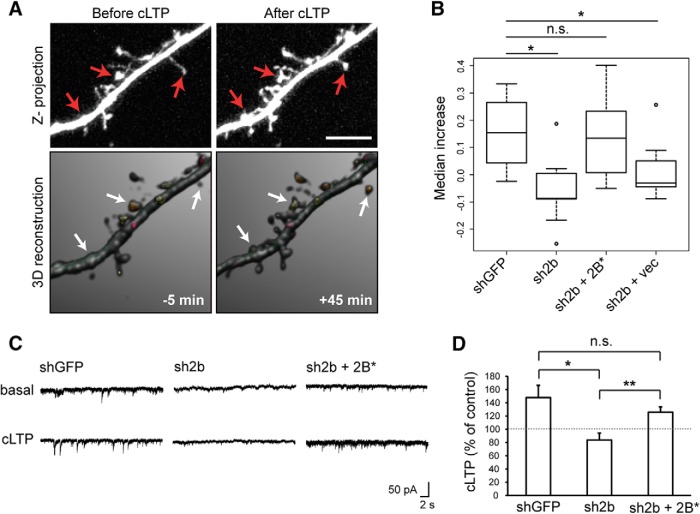
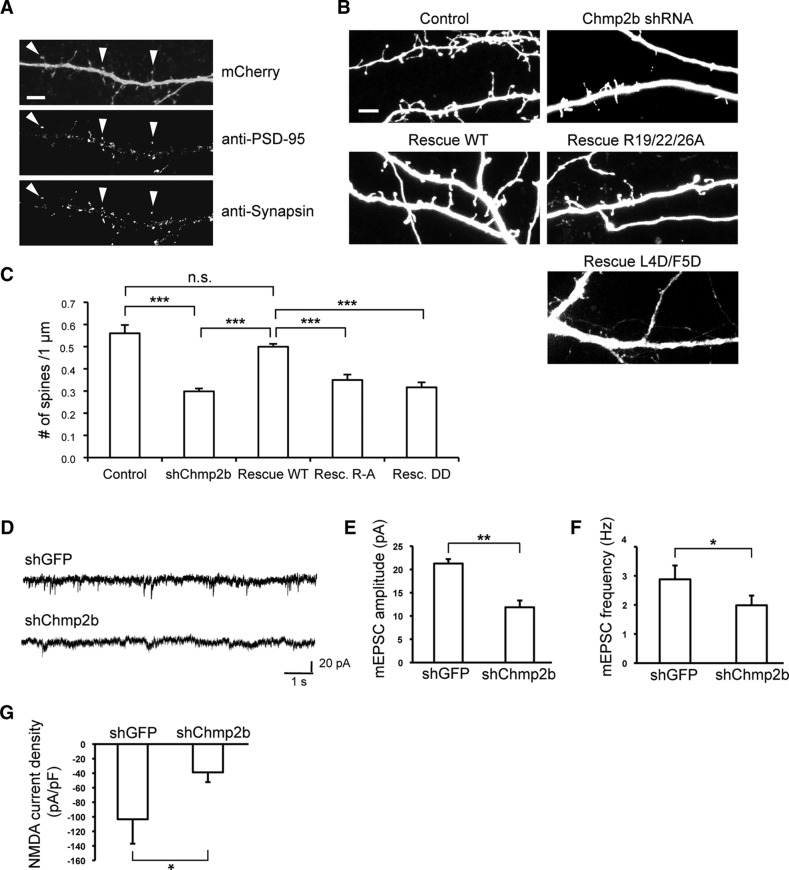
Figure 6.
Chmp2b is required for stable increase in spine volume and in synaptic efficacy during chemical LTP. A, B, Hippocampal neurons were transfected with pSuper-mCherry and rescue plasmids. Confocal image stacks of dendritic segments (visualized by mCherry fluorescence) were acquired before and after induction of cLTP. 3D models of dendritic morphology were built from the stacks with NeuronStudio and used to measure changes in spine volume. A, Maximal intensity projection of confocal images (top) and 3D models (bottom) of a representative dendritic segment before and after cLTP in a neuron transfected with control plasmid. Arrows indicate spines that have expanded. B, The percentage increase in spine volume was measured for each stable spine of a segment 5 min before and 45 min after cLTP induction, and median increases were calculated per transfected neuron. The box plot shows the distribution of median increases in neurons transfected as indicated (n = 8–9 neurons per condition, 50–100 spines per neuron). vec, Vector expressing GFP alone. The spines of control neurons were significantly expanded after cLTP induction (p = 0.0075, Wilcoxon one-sample test). The increase was abolished by Chmp2b knockdown (*p = 0.011 compared with control, Wilcoxon test with Holm correction) and reinstated by expression of Chmp2b* (p = 0.336 compared with control), but not by GFP alone (p = 0.041 vs control). C, D, Hippocampal neurons were transfected at 18 DIV with the indicated plasmids and cLTP was induced as in A. mEPSCs were recorded before and after cLTP induction by patch-clamp recording in the whole-cell configuration. C, Sample traces obtained before and after cLTP in neurons transfected as indicated. D, The histogram shows the relative potentiation of mEPSCs (mean percentage increase per neuron) compared with the mean amplitude before cLTP. The dotted line indicates 100% of basal amplitude (mean mEPSC amplitude before LTP). LTP was abolished in Chmp2b-deprived neurons (*p = 0.033 compared with control, Holm-corrected t test). Expression of Chmp2b* rescued LTP (p = 0.046 compared with basal amplitude in the same cells, paired t test; **p = 0.0062 vs nonrescued cells; p = 0.34 vs control cells, two-sided t tests). Control, n = 5 cells; Chmp2b shRNA plus vector, n = 11 cells; Chmp2b shRNA plus Chmp2b*WT, n = 9 cells.
Antibodies.
See Table 2 for details of antibodies and dilutions.
Table 2.
Antibodies used in this study
| Antibody | Species | Company (reference) | Dilutions |
||
|---|---|---|---|---|---|
| IF | WB | IHC/EM | |||
| Anti-Chmp2b | Rabbit polyclonal | Abcam (ab33174) | 500 | 10,000 | 1000 |
| Anti-Chmp2b | Mouse monoclonal | Covalab | 100 | 2000 | |
| Anti-Chmp4b | Rabbit polyclonal | Abcam (ab105767) | 500 | 2000 | |
| Anti-Chmp4b | Mouse monoclonal | Covalab | 100 | 1000 | |
| Anti-Alix | Rabbit polyclonal | Covalab (ab0204) | 6000 | ||
| Anti-PSD95 | Mouse monoclonal | NeuroMab (73-028) | 200 | 4000 | |
| Anti-GluN1 | Mouse monoclonal | Synaptic Systems (114011) | 200 | 4000 | |
| Anti-Synaptophysin | Mouse monoclonal | Merck Millipore (MAB5258) | 10,000 | ||
| Anti-HA | Mouse monoclonal | Cell Signaling (2367) | 200 | 5000 | |
| Anti-HA | Rabbit polyclonal | Sigma-Aldrich (H6908) | 5000 | ||
| Anti-Flag | Mouse monoclonal | Sigma-Aldrich (F3165) | 1000 | 10,000 | |
| Anti-Flag | Rabbit polyclonal | Sigma-Aldrich (F7425) | 1000 | 5000 | |
| Anti-Actin | Mouse monoclonal | Millipore (MAB1501R) | 20,000 | ||
| Anti-Tubulin | Mouse monoclonal | Gift from A. Andrieux's laboratory | 20,000 | ||
| Anti-Pan MAGUK | Mouse monoclonal | NeuroMab (75-029) | 4000 | ||
| Anti-GluN2B | Mouse monoclonal | NeuroMab (75-097) | 4000 | ||
| Anti-Glu2 | Mouse monoclonal | Millipore (MABN71) | 5000 | ||
| Anti-ShankIII | Rabbit polyclonal | Santa Cruz Biotechnology (30193) | 1000 | ||
| Anti-Homer | Mouse monoclonal | Santa Cruz Biotechnology (17842) | 2000 | ||
| Anti-Mouse HRP | Goat polyclonal | Jackson ImmunoResearch (115-035-166) | 20,000 | ||
| Anti-Rabbit HRP | Goat polyclonal | Jackson ImmunoResearch (115-035-044) | 20,000 | ||
| Anti-Mouse Alexa Fluor 488 | Goat polyclonal | Invitrogen (A-11029) | 1500 | ||
| Anti-Mouse Alexa Fluor 594 | Goat polyclonal | Invitrogen (A-11032) | 1500 | ||
| Anti-Mouse Cy5 | Goat polyclonal | Jackson ImmunoResearch (115-175-146) | 1000 | ||
| Anti-Rabbit Alexa Fluor 488 | Goat polyclonal | Invitrogen (A-11034) | 1500 | ||
| Anti-Rabbit Alexa Fluor 594 | Goat polyclonal | Invitrogen (A-11037) | 1500 | ||
| Anti-Rabbit Cy5 | Goat polyclonal | Jackson ImmunoResearch (111-175-144) | 1000 | ||
The antibody dilutions we used are indicated for each application. IF, Immunofluorescence; WB, Western immunoblotting; IHC, immunohistochemistry.
Animals.
Animals were handled and killed in conformity with European law and internal regulations of INSERM. Pregnant Oncins France souche A (OFA; a substrain of Sprague Dawley) rats (Charles River) were used for primary neuron cultures, and 2-month-old OFA rats of both sexes were used for biochemical analysis of brain tissue. The rats were deeply anesthetized by isoflurane inhalation before decapitation, quickly followed by cesarean section or brain dissection. Three-month-old male C57BL/6 mice and 1.3-month-old female Chmp2b KO mice (here called KD mice since knock-out is incomplete; Ghazi-Noori et al., 2012) or female WT littermates (on C57BL/6 background) were used for immunohistochemistry and electron microscopy (EM) studies. Mice were anesthetized by intraperitoneal injection of 0.1 ml of sodium pentobarbital (5.6% w/v; CEVA Santé Animale) and intracardially perfused with phosphate-buffered 0.9% NaCl (PBS), followed by 0.1 m phosphate-buffered 4% formaldehyde, pH 7.4, supplemented with 0.05% glutaraldehyde (Sigma). The brains were carefully removed and postfixed for 4 h in the same fixative, and 60-μm-thick coronal brain sections were cut with a Vibratome (VT 1000 S; Leica).
Cell culture and transfection.
Primary cultures of hippocampal neurons were prepared as described previously (Belly et al., 2010). Briefly, hippocampi were dissected from E18 rat embryos, treated with trypsin, and disrupted by seven to eight cycles of aspiration and ejection through a micropipette tip. Dissociated hippocampal cells were seeded at a density of 1.5 × 104/cm2 in 35 mm dishes on coverslips precoated for 4 h with 50 μg/ml poly-d-lysine (Sigma), in neuronal culture medium (Neurobasal medium containing B27 supplement, penicillin/streptomycin, and 0.5 mm l-glutamine; Invitrogen) supplemented with 10% heat-inactivated horse serum (Invitrogen). Fourteen-mililimeter-wide round coverslips were used for immunostaining, and 30-mm-wide coverslips were used for live imaging. Neurons were maintained in water-saturated 95% air/5% CO2 at 37°C. The seeding medium was replaced after 20 h with serum-free neuronal culture medium.
Neurons were transfected using a calcium phosphate method optimized for highly mature neuronal cultures (Goetze et al., 2004; Jiang and Chen, 2006). For cotransfection experiments, for each dish, 1 μg of pSuper-mCherry-based plasmid was mixed with 2 μg of pBI-CMV3-based or pIRES2-EGFP-based plasmid. Five microliters of 250 mm CaCl2 were added to 45 μl of water containing 3 μg of DNA, mixed, and immediately added dropwise to 50 μl of 2× N, N-Bis (2-hydroxyethyl)-2-aminoethanesulfonic acid (BES)-buffered saline solution, pH 7.1. The mixture was gently stirred by tapping the tube and bubbling air once, left to incubate for 10 min, and added to coverslips in 1 ml of transfection medium [Eagle's MEM (Invitrogen) including 2.2% NaHCO3 and supplemented with B27, 0.05 mm glutamine, and 20 mm glucose]. After 1 h in the incubator, the coverslips were transferred to dishes containing 2 ml of transfection medium (without B27) preincubated for 30 min in 10% CO2. The dishes were quickly returned to 5% CO2 and left for 5–10 min before returning the coverslips to their original (conditioned) neuronal culture medium. For immunofluorescence of exogenously expressed proteins, neurons were fixed 16–20 h after transfection. For RNAi experiments, fixation or live imaging took place 72 h after transfection.
HEK-293T and HeLa cells were grown in DMEM (Invitrogen) supplemented with 10% FBS and 2 mm l-glutamine (Sigma). Cells were transfected after 24 h of culture using jetPEI (Polyplus) for HeLa cells and calcium phosphate for HEK cells. Cells were fixed or lysed 36 h after transfection.
Immunohistochemistry.
Brain sections were incubated in 10% goat preimmune serum (GPi) diluted in 50 mm Tris buffer, pH 7.4, containing 0.9% NaCl [Tris-buffered saline (TBS)], with 0.2% Triton X-100, for 1 h. Sections were then incubated overnight at 4°C in a primary antibody at a final protein concentration of 1–2 μg/ml, which had been diluted in TBS containing 1% GPi. After several washes in TBS, the sections were further incubated for 2 h in biotinylated goat anti-rabbit IgG (Sigma Immuno Chemicals) diluted 1:100 in TBS containing 1% GPi. They were then transferred into avidin–biotin–peroxidase complex (ABC kit; Vector Laboratories) diluted 1:100 for 2 h at room temperature. Bound peroxidase enzyme activity was revealed using DAB (0.05% in TB, pH 7.4) as the chromogen and 0.01% H2O2 as the substrate. Finally, sections were air-dried and coverslipped.
Immunofluorescent staining.
Cultured neurons were fixed for 20 min at room temperature in phosphate-buffered 4% paraformaldehyde supplemented with 4% sucrose. After three washes in PBS containing 0.1% glycine, cells were permeabilized in PBS containing 0.25% Triton X-100 for 10 min at 4°C and rinsed with PBS. Coverslips were blocked for 1 h in PBS containing 5% GPi and incubated for 1–2 h at room temperature in a humidified chamber with primary antibodies diluted in a blocking solution. After washing in PBS, cells were incubated for 1 h with secondary antibodies conjugated to Alexa Fluor 488, Alexa Fluor 594, or Cyanine 5 (Cy5), diluted in the blocking solution. Coverslips were rinsed and mounted in Mowiol.
Golgi staining.
The dendritic spines of hippocampal neurons in situ were visualized by the Golgi impregnation technique. For this, we used the FD Rapid GolgiStain kit (FDNeuroTechnologies). Hippocampal tissue blocks (2.5–3 mm) were immersed in equal volumes of solutions A and B for 7 d and impregnated with solution C for 48 h at 4°C. Then, the brains were cut with a vibratome into 150-μm-thick sections embedded in agarose, washed twice in double-distilled water, and incubated for 10 min in a mixture of one part of solution D, one part of solution E, and two parts of double-distilled water. Sections were washed twice, dehydrated with increasing ethanol, and mounted with epoxy resin (Fluka) between two coverslips. The slides were analyzed by investigators without knowledge of genotype. In each of three animals per genotype, nine secondary or tertiary pyramidal neuron dendrites were randomly selected in the CA1 stratum radiatum. Selection criteria were (1) dendrites displayed complete Golgi impregnation, (2) the cell type was clearly identifiable, and (3) the minimum length of the dendrites was 50 μm from the soma. Stacks of bright-field images with 0.3 μm spacing were acquired with a Zeiss Axioskop 50 microscope with 63× oil objective (NA 1.4; Plan-Aprochromat) coupled to a CCD camera (CoolSnap ES; Roper Scientific) operated by Metaview software (Molecular Devices). Images were analyzed with ImageJ. The number of spines of each basic type per unit dendritic length was measured.
Fluorescence microscopy.
Light microscopy images were acquired with a Zeiss LSM 710 inverted confocal microscope, using a 63× Plan-Apochromat oil-immersion objective (NA 1.4; Zeiss) or (for Sholl analysis) a 40× oil objective (NA 1.3; Zeiss). The instrument was controlled by Zen software and includes acousto-optical filters for attenuating laser power, scanning mirrors with adjustable speed, and resolution of emitted light by a diffraction grating and beam guides for setting spectral collection windows. Laser lines at 488, 561, and 633 nm were used for exciting fluorescence of EGFP, ZsGreen, or Alexa Fluor 488; mCherry or Texas Red; and Cyanine 5, respectively. For multicolor imaging, collection windows were adjusted based on emission spectra to minimize cross-channel contamination. Pinhole size was usually set to generate 0.9-μm-thick optical slices (≅1 Airy unit) at all wavelengths. For fixed samples, laser power was adjusted to 0.5–1.25 mW in all channels, and the signal was averaged over four scans and digitized on an 8 bit grayscale. In the experiments shown in Figure 2, B and C, Z-stacks were acquired with 0.09 × 0.09 × 0.2 μm3 voxel size and processed by a blind deconvolution algorithm (AutoQuant; Media Cybernetics) to improve resolution and remove shot noise. The processed images were slightly contrasted with ImageJ for clarity.
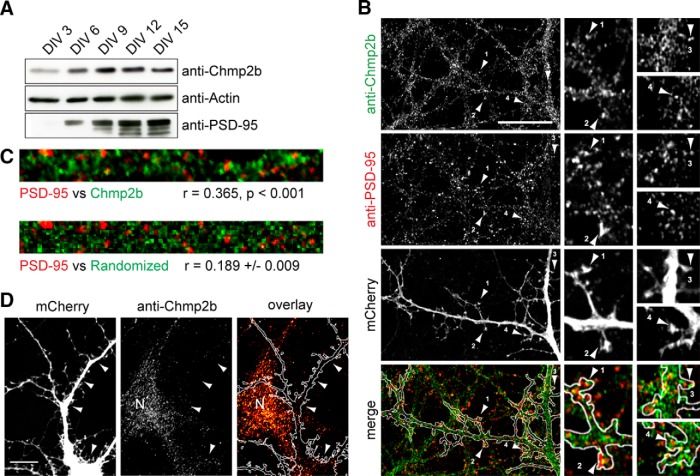
Figure 2.
Chmp2b forms clusters in dendrites. A, Cultured mouse cortical neurons were lysed at various stages of maturation, and equal amounts of lysate protein were analyzed by immunoblotting with the indicated antibodies. PSD-95 was used as a marker for synaptogenesis. B, Hippocampal neurons were transfected with empty pSuper-mCherry vector, fixed at 21 DIV, and stained by dual immunofluorescence with anti-Chmp2b and anti-PSD-95 antibodies. Left, Punctate staining of a dendritic segment in a transfected neuron (single confocal plane). Staining of nontransfected cells is also visible. Merge, Overlap (yellow) between Chmp2b and PSD-95 puncta, in dendritic shafts and spines, visualized with mCherry (white contour). The numbered arrowheads indicate spines with colocalized puncta. Right, Enlarged views of the same spines. Scale bar, 20 μm. C, Top, Dendritic segment double stained by anti-Chmp2b and anti-PSD-95. Bottom, Same after randomization of Chmp2b staining pattern. The correlation coefficient (r) between actual Chmp2b and PSD-95 staining intensities is significantly higher than the mean r value generated by randomized Chmp2b patterns (p < 0.0001, z test). D, Specificity of Chmp2b immunofluorescence. Hippocampal neurons were transfected with pSuper-mCherry expressing Chmp2b-specific shRNA, stained by immunofluorescence with anti-Chmp2b, and imaged by confocal microscopy. Left, Representative transfected neuron, visualized by mCherry. Middle, Chmp2b immunostaining of the same field. Arrowheads indicate the position of the transfected cell dendrites and soma. N, Neighboring, nontransfected neuron. Right, Immunostaining (red hot pseudocolor) is mostly absent from the transfected neuron (outlined in white), though clearly detectable in the nontransfected cell (N). Scale bar, 10 μm.
To evaluate the probability of random overlap between Chmp2b and postsynaptic densitity-95 (PSD-95) clusters, a dendritic region was selected in a way to exclude as much as possible empty pixels. Pearson's correlation coefficient (r) between Chmp2b and PSD-95 intensity values was calculated for the actual image, and the probability of obtaining the observed coefficient was calculated based on 1000 randomized images similar to the one shown using the ImageJ plug-in JACOP (Bolte and Cordelières, 2006).
For Sholl analysis, images (1024 × 1024 pixels; 350 × 350 μm; four optical sections in z) were taken from neurons coexpressing ZsGreen and mCherry, within the same fluorescence range for all neurons (40 neurons per transfection condition, three independent cultures). Sholl analysis was performed on maximum intensity projections of mCherry fluorescence using the Sholl Analysis plugin of ImageJ.
For postsynaptic spine counting in cultured neurons, transfected neurons were stained by immunofluorescence with anti-PSD-95 antibody and Cy5-conjugated secondary antibody. Triply labeled neurons (mCherry-ZsGreen-Cy5) were imaged in all three channels as above. For each neuron, 10–20 optical sections were acquired (voxel size, 0.1 × 0.1 × 0.2 μm3). Images were analyzed with ImageJ. Postsynaptic spines were manually identified in successive sections as spine-like dendritic protrusions (visualized by mCherry fluorescence) carrying PSD-95 clusters. Spines were counted along ∼540 ± 220 μm (mean ± SD) dendritic length per neuron. Similar results were obtained with two independent cultures, allowing us to pool the data. Dual immunostaining of mCherry-transfected neurons with anti-PSD-95 and anti-synapsin antibodies verified that in neurons at this stage, virtually all PSD-95 clusters located on spines were colocalized with the presynaptic marker synapsin. Images of neurons used to study RNAi effects were acquired and analyzed by investigators blind to the transfected constructs.
Electron microscopy.
Free-floating brain sections were incubated in 10% GPi diluted in TBS for 1 h at room temperature to block nonspecific staining. Sections were then incubated overnight at 4°C in TBS containing 1% GPi and a primary antibody at a final protein concentration of 1–2 μg/ml. The sections were then labeled either by immunogold or by immunoperoxidase. For immunoperoxidase labeling, we followed the immunohistochemistry procedure, as described above (starting from the secondary antibody step). For immunogold labeling, sections were incubated for 3 h with goat anti-rabbit IgG coupled to 1.4 nm gold beads (Nanoprobes) diluted 1:300 in TBS containing 1% GPi. After several washes in PBS, the sections were postfixed in 1% glutaraldehyde diluted in the same buffer for 10 min. They were washed in double-distilled water, followed by silver enhancement of the gold particles with a HQ Silver kit (Nanoprobes).
The peroxidase-reacted sections and the gold–silver-labeled sections were processed for EM. This included treatment with osmium tetroxide (1% in 0.1 m PB), block staining with uranyl acetate, dehydration through a graded series of ethanol solutions, and flat embedding on glass slides in Durcupan (Fluka) resin. Regions of interest were cut at 70 nm on an ultramicrotome (Reichert Ultracut E; Leica) and collected on one-slot copper grids. Staining was performed on drops of 1% aqueous uranyl acetate, followed by Reynolds's lead citrate. EM images were acquired in a JEOL-1200 electron microscope with a digital camera (Veleta, SIS; Olympus) and analyzed with ImageJ. Measurements of gold particle counts along the radial and tangential coordinates of the spine cytoplasm were acquired and normalized as described by Rácz and Weinberg (2008). Briefly, normalized radial position, rN (the fraction of the distance from the center to the plasma membrane), was computed according to the following equation:
where r1 is the radial distance from the membrane to the particle and r2 is the radial distance of the particle from the geometric center of the spine. To correct the radial distribution for distance-dependent increase in bin area, particle densities along the radial coordinate were divided by normalized radial distance, thus giving increased weight to particle density close to the spine center. Three-dimensional reconstruction of spine heads from serial section digital images was done using Reconstruct software (John C. Fiala, Department of Biology, Boston University, MA). For comparisons between wild-type and Chmp2b-deficient mice, acquisition of microscopy images and morphometric quantification were performed without knowledge of genotype.
Purification of synaptoneurosomes and PSD fractionation.
Subcellular fractionation was performed according to Carlin et al. (1980) with minor modifications. Samples were kept at 4°C during all procedures. In brief, one rat forebrain was homogenized in 13 ml of ice-cold isotonic lysis buffer (0.32 m sucrose, 4 mm HEPES, pH 7.4, protease inhibitors [EDTA-free Complete; Roche Molecular)] using a Teflon-glass homogenizer (15 strokes). The homogenate was centrifuged at 1400 × g for 10 min to remove nuclei. The postnuclear supernatant (S1) was spun down at 4°C for 10 min at 13,800 × g, yielding crude synaptosomes (pellet P2) and crude cytosol (supernatant S2). The P2 fraction was homogenized in lysis buffer and layered onto discontinuous sucrose density gradients (made of 3 ml each of 0.85, 1.0, and 1.2 m sucrose layers). The gradients were centrifuged at 82,500 × g (27,000 rpm in a Beckman SW41 rotor) for 120 min, and the purified synaptoneurosome fraction (SYN) was collected at the 1.0–1.2 m interface. The synaptoneurosomes were resuspended in 5 vol of lysis buffer and spun down for 20 min at 150,000 × g. The pelleted synaptoneurosomes were resuspended in Triton buffer [50 mm HEPES, pH 7.4, 0.5% Triton X-100, protease inhibitors (EDTA-free Complete; Roche Molecular), 2 mm EDTA], stirred 30 min in the cold room, and centrifuged at 32,000 × g for 20 min to obtain the PSDI pellet. PSDI was resuspended in 50 mm HEPES, pH 7.4. One-half of the suspension was solubilized again by adding 1 vol of 1% Triton X-100 and centrifuged at 200,000 × g for 20 min, yielding pellet PSDII. The other half was solubilized by adding 1 vol of 6% Sarkosyl and centrifuged at 200,000 × g for 1 h, yielding PSDIII.
Gel filtration of synaptoneurosome extract.
Size-exclusion chromatography (gel filtration) was used for separation of Chmp2b-containing molecular assemblies that had been solubilized by mild detergent extraction from gradient-purified synaptoneurosomes. Chromatography was performed at 4°C on a 120 ml HiPrep Sephacryl S-300 HR column coupled to an Akta FPLC system (GE Heathcare). The column was equilibrated, and separation was achieved in 25 mm Tris-HCl, pH 7.4, 150 mm NaCl, 0.5% Triton X-100, 10 mm NaF, 10 mm β-glycerophosphate and protease inhibitors (Roche) at a flow rate of 0.5 ml/min. Synaptoneurosomes freshly prepared from one brain were resuspended in 3 ml of deoxycholate (DOC) buffer [10 mm Tris-HCl, pH 9.0, 1% sodium deoxycholate, 150 mm NaCl, protease inhibitors (Complete; Roche Molecular), and phosphatase inhibitors (P0044; Sigma-Aldrich)]. The suspension was stirred by rotation at 4°C for 30 min and centrifuged at 100,000 × g for 20 min. The supernatant was filtered (pore size, 0.2 μm; Millipore), and 2.5 ml was loaded on the column. Fractions (2 ml) were collected, and the elution profile was analyzed by immunoblotting and by measurement of total protein concentration with BCA protein assay (Pierce). The profile was calibrated by measuring the elution volume of protein standards in the same conditions (MWGF1000; Sigma-Aldrich).
Immunoprecipitation.
For immunoprecipitation of the brain Chmp2b complex, 2 mg (total protein) of gradient-purified synaptoneurosomes were resuspended in 1 ml of DOC buffer and incubated for 30 min at 4°C. After centrifugation at 100,000 × g for 20 min, the supernatant was collected and diluted with 3 vol of immunoprecipitation buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, and 1% Triton X-100). The synaptosomal lysate was incubated for 2.5 h at 4°C with 15 μg of antibody that had been covalently coupled to superparamagnetic protein A beads (Dynabeads, 10001D; Invitrogen) by cross-linking with dimethyl-pimelimidate (Sigma) according to Harlow and Lane (1988). After extensive washing in buffer containing 50 mm Tris-HCl, pH 7.4, 0.25% sodium deoxycholate, and 0.75% Triton X-100, together with increasing concentrations of NaCl (150–500 mm), bound proteins were eluted by boiling in Laemmli buffer, resolved by SDS-PAGE, and blotted onto PVDF membranes (Millipore). For samples first resolved by gel filtration, fractions of interest were pooled and immunoprecipitated under the same conditions.
For immunoprecipitation of tagged proteins, transfected HEK-293T were washed with PBS and homogenized in isotonic lysis buffer, and the lysate was centrifuged at 1000 × g for 10 min. The postnuclear supernatant was centrifuged at 50,000 × g for 1 h, and the pelleted membranes were resuspended in DOC buffer. Immunoprecipitation was performed with anti-tag antibody immobilized on protein A Dynabeads as above.
Immunoblotting.
Cells or tissue lysates were resuspended in Laemmli buffer and resolved by SDS-PAGE in 8–12% polyacrylamide gels. Proteins were electrotransferred onto PVDF membranes. Membranes were blocked for 30 min in TBS containing 0.1% Tween 20 and 5% dry milk and incubated for 1 h with primary antibodies diluted in the blocking solution. After six washes in TBS–Tween, the membranes were further incubated for 30 min with secondary antibodies coupled to HRP, washed six more times as before, and incubated with luminescence-generating HRP substrate, and bound antibodies were revealed by luminography on film.
Mass spectrometry analysis.
A DOC extract from gradient-purified synaptoneurosomes was fractionated by gel filtration, and the high-molecular-weight (HMW) fractions were pooled and immunoprecipitated with control or anti-Chmp2b antibody. Two other, independently prepared synaptoneurosome extracts were directly immunoprecipitated with the same antibodies. The three pairs of immunoprecipitates were digested, and the resulting peptides were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) as described further below. In each of the three experiments, peptides derived from control and anti-Chmp2b immunoprecipitates were analyzed in consecutive LC-MS/MS runs performed on the same day.
For protein digestion, proteins in Laemmli buffer were stacked (2 mm) on SDS-PAGE gels (4–12% NuPAGE gels; Invitrogen) before being stained with Coomassie blue R-250 (Bio-Rad). The gel band was manually excised and cut in pieces before being washed by six successive incubations of 15 min in 25 mm NH4HCO3 and in 25 mm NH4HCO3 containing 50% (v/v) acetonitrile. Gel pieces were then dehydrated with 100% acetonitrile and incubated for 45 min at 53°C with 10 mm DTT in 25 mm NH4HCO3 and for 35 min in the dark with 55 mm iodoacetamide in 25 mm NH4HCO3. Alkylation was stopped by adding 10 mm DTT in 25 mm NH4HCO3 and mixing for 10 min. Gel pieces were then washed again by incubation in 25 mm NH4HCO3 before dehydration with 100% acetonitrile. A total of 0.15 μg of modified trypsin (sequencing grade; Promega) in 25 mm NH4HCO3 was added to the dehydrated gel pieces for an overnight incubation at 37°C. Peptides were then extracted from gel pieces in three 15 min sequential extraction steps in 30 μl of 50% acetonitrile; 30 μl of 5% formic acid; and, finally, 30 μl of 100% acetonitrile. The pooled supernatants were then dried under vacuum.
For nano-LC-MS/MS analyses, the dried extracted peptides were resuspended in 5% acetonitrile and 0.1% trifluoroacetic acid and analyzed by on-line nano-LC-MS/MS [Ultimate 3000 (Dionex) and LTQ-Orbitrap Velos pro (Thermo Fisher Scientific)]. Peptides were sampled on a 300 μm × 5 mm PepMap C18 precolumn and separated on a 75 μm × 250 mm C18 column (PepMap; Dionex). The nano-LC method consisted of a 120 min gradient at a flow rate of 300 nl/min, ranging from 5 to 37% acetronitrile in 0.1% formic acid for 114 min before reaching 72% for the last 6 min. MS and MS/MS data were acquired using Xcalibur (Thermo Fisher Scientific). Spray voltage and heated capillary were, respectively, set at 1.4 kV and 200°C. Survey full-scan MS spectra (m/z = 400–1600) were acquired in the Orbitrap with a resolution of 60,000 after accumulation of 106 ions (maximum filling time, 500 ms). The 20 most intense ions from the preview survey scan delivered by the Orbitrap were fragmented by collision-induced dissociation (collision energy, 35%) in the LTQ after accumulation of 104 ions (maximum filling time, 100 ms).
For bioinformatic analyses, data were processed automatically using Mascot Daemon software (version 2.3.2; Matrix Science). Concomitant searches against the Uniprot database (August 2012 version, Rattus taxonomy, 42,469 sequences), the classical contaminants database (67 sequences, homemade), and the corresponding reversed databases were performed using Mascot (version 2.4). ESI-TRAP was chosen as the instrument and trypsin/P as the enzyme, and two missed cleavage were allowed. Precursor and fragment mass error tolerances were set, respectively, at 10 ppm and 0.6 Da. Peptide modifications allowed during the search were carbamidomethyl (C, fixes) acetyl (N-ter, variable), oxidation (M, variable), and deamidation (NQ, variable). The IRMa software (version 1.30.4) was used to filter the results: conservation of rank 1 peptides, peptide identification False Discovery Rate < 1% (as calculated by using the reverse database strategy), and minimum of one specific peptide per identified protein group. The filtered results were uploaded into a relational mass spectrometry identification database (MSIdb), and a homemade tool (hEIDI; Milbradt et al., 2014) was used for the compilation, grouping, and comparison of the protein groups from the different samples. hEIDI provides modules to help exploring validated search results that have been compiled into a MSIdb. The user creates contexts, which represent sets of identifications, and runs peptides and proteins grouping algorithms to cluster peptides identifying a family of proteins in these contexts. The grouping algorithm will first identify identical peptides based on their mass and sequence. Groups of proteins matching the same set of peptides are then created as is done in the IRMa toolbox for a single identification. This is typically useful to consider results at the sample level instead of the replicate level. To compare several contexts, hEIDI aligns each protein group from each context to be compared. This alignment is based on protein and peptide set similarity metrics.
To be considered as specifically associated with the Chmp2b immunoprecipitate, the proteins identified by this analysis were filtered according to the following inclusion criteria: two or more unique peptides (specific spectral counts); two or more distinct protein fragments [spectral counts (SC)]; and ratio of SC in specific to control immunoprecipitate (SC 2B/SC control) ≥5.
Electrophysiological recordings in cultured neurons.
Mature [18–21 d in vitro (DIV)] hippocampal neurons expressing the transfection marker ZsGreen were used for whole-cell patch-clamp recording at room temperature (20–24°C). The neurons were continuously perfused with an extracellular solution containing 140 mm NaCl, 2 mm CaCl2, 3 mm KCl, 10 mm HEPES, 10 mm glucose, 0.5 μm tetrodotoxin, 50 μm bicuculline methiodide, and 1 μm strychnine, adjusted to pH 7.4 with NaOH and to 330 mOsm with Na-acetate. To induce LTP, glycine (200 μm in extracellular solution) was perfused for 3 min onto the neurons (Lu et al., 2001). For the experiments in which NMDA current amplitudes were measured, NMDA (100 μm) was added to this solution and perfused with a fast-exchange perfusion system. Recording pipettes were pulled from borosilicate glass and had a resistance of 3–5 ΩM when filled with a solution containing (in mm) 115 CsMeSO3, 20 CsCl, 10 HEPES, 0.6 EGTA, 4 Na2-ATP, 0.4 Na-GTP, 10 Na-phosphocreatine, and 2.5 MgCl2, adjusted to pH 7.2 with CsOH and to 300 mOsm with Cs-acetate. Currents were recorded from a holding potential of −60 mV using an Axon 200B amplifier (Molecular Devices). Cell capacitance and series resistance compensation were applied electronically. Signals were filtered at 1 kHz, digitized at 5 kHz, and recorded using the pClamp software. Miniature EPSCs (mEPSCs) and NMDA-evoked currents were analyzed using ClampFit (Molecular Devices).
Live imaging.
Image acquisition and analysis were performed blind to transfected plasmids. For live imaging during chemical LTP, neurons were transferred to a perfusion chamber (Pocon), washed, and kept in an extracellular solution (125 mm NaCl, 2.5 mm KCl, 2 mm CaCl2, 1 mm MgCl2, 5 mm HEPES, 33 mm glucose, 50 μm bicuculline, and 0.5 μm TTX, 1 μm strychnine, pH 7.4) at 37°C on a heated stage. Neurons that simultaneously expressed GFP and mCherry (within the same fluorescence range throughout experiments) and had a healthy appearance were identified by epifluorescence, and dendritic segments were selected so that 30–100 spines could be imaged per neuron while minimizing exposure to light. mCherry fluorescence was excited with 75–150 μW nominal laser power at 561 nm. The pinhole diameter was set to 1 Airy unit. Z-stacks of 20–30 optical sections (voxel size, 0.11 × 0.11 × 0.2 μm3) per dendritic segment were acquired at 15 min intervals. Signal per pixel was averaged over two scans. After acquiring baseline images, an extracellular solution containing 200 μm glycine (warmed to 37°C by in-line heating) was delivered. Stimulation was stopped after 4 min by perfusing a warm glycine-free extracellular solution. Only modest bleaching was observed during the experiment. Cells showing signs of damage (pearling or blebbing) were discarded.
To measure changes in spine volume, image stacks were subjected to blind (adaptive) deconvolution with AutoQuant as above. The deconvolved images (scaled to 8 bit) were used as input for 3D reconstruction of dendritic morphology by the software package NeuronStudio (http://www.mssm.edu/cnic/tools.html), which allocates voxels to spines through a specific algorithm (Rodriguez et al., 2006, 2008). Individual spines labeled by the software and that were clearly identifiable at both initial and final time points were manually selected. The volume of the spines before and after stimulation was estimated by counting spine voxels. The fractional increase in spine volume was calculated for each spine as follows:
with Vi the volume at 5 min before the start of the stimulation and Vf the volume at 45 min after the end of the stimulation. The median values of ΔV/V were calculated for each neuron. Differences between cultures did not significantly contribute to the observed effects (two-way ANOVA, p = 0.015 for factor plasmid, p = 0.93 for factor culture, p = 0.56 for interaction). Therefore, the results from seven independent cultures were pooled.
Statistical analysis.
The statistical significance of differences between sample means or sample distributions was calculated as described in the figure legends. The normality of samples was systematically checked by the Shapiro-Wilks test, and nonparametric tests were used where appropriate. Statistical tests were performed with R (R Core Team, 2013). Error bars indicate SEM in all figures.
Results
A pool of Chmp2b localizes in dendritic shafts and spines
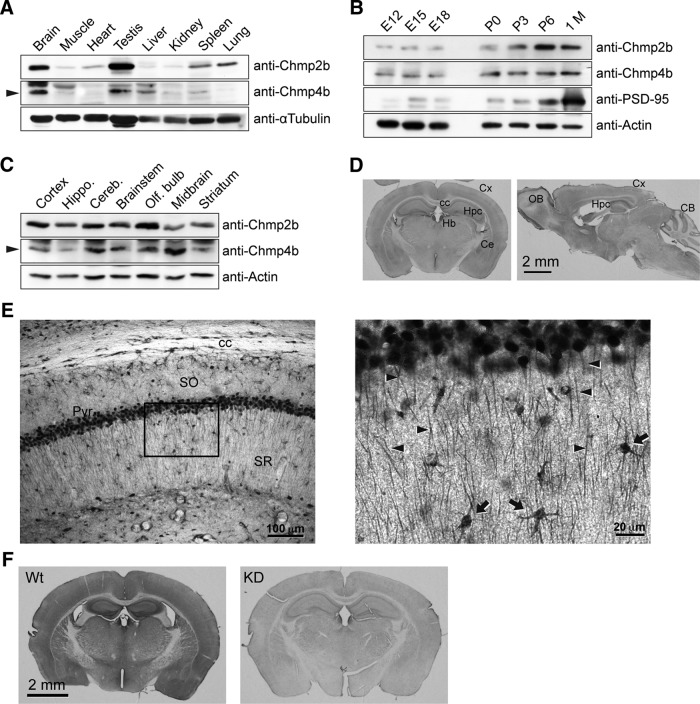
To address the physiological function of Chmp2b in neurons, we first evaluated the relative level of Chmp2b protein in the brain compared with that in other organs. Mouse tissue extracts were analyzed by Western immunoblotting with an anti-Chmp2b antibody. The same blots were also probed with an antibody raised against the core ESCRT-III subunit Chmp4b. For both proteins, expression was strikingly higher in the brain than in the other organs, except for testes (Fig. 1A). Analysis of mouse brain at various stages of development indicated that Chmp2b expression increased during the first postnatal week (Fig. 1B). Both Chmp2b and Chmp4b were detected in extracts from various regions of the adult brain, with the highest Chmp2b levels found in olfactory bulb and cerebral cortex (Fig. 1C).
Figure 1.
Chmp2b expression in the mouse brain. A, An adult male mouse was dissected, and equal amounts of total protein from each tissue were analyzed by SDS-PAGE and immunoblotting with the indicated antibodies. The arrowhead indicates the Chmp4b-specific band. α-Tubulin was used as the loading control. B, Mouse brains at increasing developmental ages were analyzed by immunoblotting with the indicated antibodies. PSD-95 was used as a marker of synaptogenesis. P, Postnatal age (days); 1 M, 1 month. C, Equal protein amounts from the indicated regions of an adult mouse brain were analyzed by immunoblotting as above. D, Frontal (left) or sagittal (right) sections from adult mouse brain were stained by immunoperoxidase with anti-Chmp2b antibody. Cx, Cortex; cc, corpus callosum; Hpc, hippocampus; Hb, habenula; Ce, central amygdala; OB, olfactory bulb; CB, cerebellum. E, Left, Immunostained CA1 layer in a frontal section of the hippocampus. Note the strong staining of pyramidal cell bodies and the weaker but definite immunoreactivity of the neuropil. SO, Stratum oriens; Pyr, stratum pyramidale; SR, stratum radiatum. Right, Higher magnification of the boxed region. Immunoreactivity is observed in apical dendrites (arrowheads) and interneurons (arrows). F, Specificity of Chmp2b immunostaining. As a control, we used brain sections from Chmp2b KO mice (Ghazi-Noori et al., 2012), here called Chmp2b KD because of the presence of residual Chmp2b protein in these mice (Fig. 7G,H). Chmp2b immunoreactivity was revealed by immunoperoxidase in a frontal section of the brain of a 5-week-old wild-type (Wt) mouse (left) or in an equivalent section of a sibling KD mouse (right). Note that development of DAB staining was performed identically for the two sections, but for a longer time than in D, which therefore appears lighter than F (Wt).
To define the topographical pattern of Chmp2b protein expression, we performed immunohistochemistry on mouse brain sections (Fig. 1D). Chmp2b immunoreactivity was particularly strong in habenula, olfactory bulb, deep layers of the neocortex, amygdala, and hippocampus. This pattern generally coincided with in situ hybridization data from the Allen Brain Atlas (http://mouse.brain-map.org/gene/show/44784). In higher-magnification views, immunostaining was evident in the somatodendritic compartment of numerous neurons (e.g., in the CA1 region of the hippocampus; Fig. 1E). Specificity of labeling was confirmed by showing reduced staining in the brain of Chmp2b-deficient (KD) mice (Fig. 1F).
Chmp2b was also readily detectable in primary cultures of rat neurons. Consistent with the developmental profile shown in Figure 1B, immunoblotting analysis of cultured hippocampal (Belly et al., 2010) or cortical (Fig. 2A) neurons showed that endogenous Chmp2b expression steadily increased during neuronal maturation, reaching a plateau after 9 DIV, thus overlapping the period of synapse formation. As already observed in non-neuronal cells, immunofluorescent staining of mature hippocampal neurons revealed Chmp2b immunoreactivity throughout the entire cytoplasm. However, focusing on dendrites, we noted that Chmp2b staining formed numerous small clusters, many of which were found at the base, neck, and head of dendritic spines. Within spine heads, Chmp2b clusters were often juxtaposed to PSDs labeled with anti-PSD-95 antibody (Fig. 2B). The overlap of Chmp2b with PSD-95 puncta was significantly more frequent than expected by random superposition (Fig. 2C). The specificity of synaptic staining with the anti-Chmp2b antibody was confirmed by extinction with a well validated shRNA construct (Belly et al., 2010; Fig. 2D).
Chmp2b regulates the stability of dendritic branches
After these observations, we used cultured hippocampal neurons to examine whether Chmp2b played a role in the maintenance of dendrites and synapses. For this, neurons were transfected with the same Chmp2b-depleting shRNA as above. Control transfections were performed with a GFP-specific shRNA with no effect on neurons (Alvarez et al., 2006). The shRNA vectors also expressed mCherry, allowing visualization of the cellular morphology. To assess the effect of Chmp2b depletion on dendrite branching, neurons were transfected at 10 DIV and imaged at 14 DIV by confocal microscopy. As shown in Figure 3, A and B, dendritic branching was reduced in Chmp2b-depleted neurons compared with neurons transfected with control shRNA. Coexpression of an RNAi-resistant version of the Chmp2b mRNA together with the shRNA almost fully rescued dendritic complexity. These data indicate that Chmp2b is required for maintaining the integrity of dendritic trees.
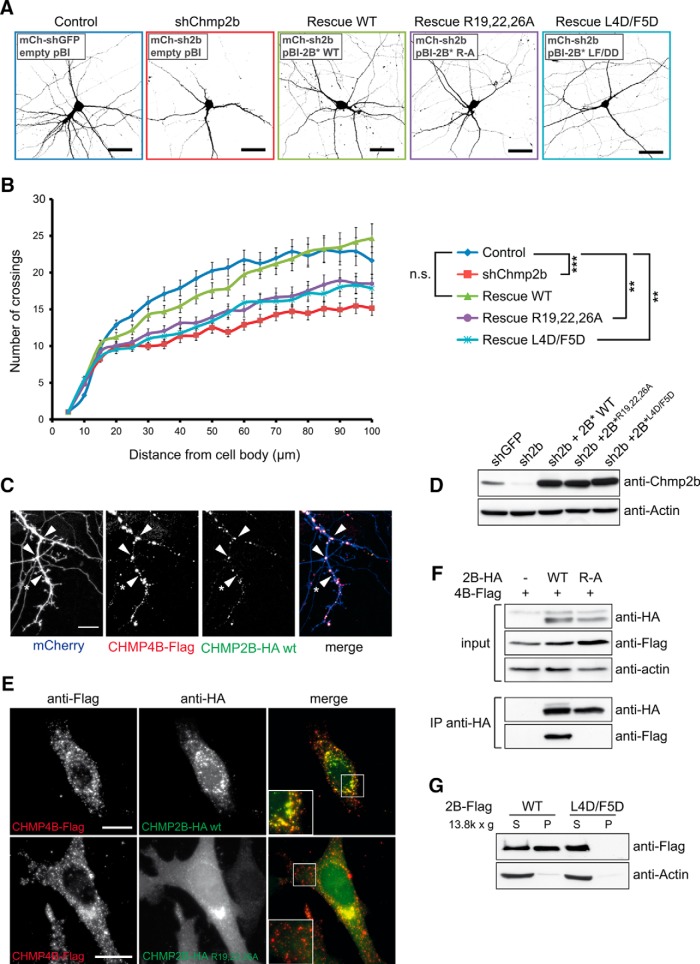
Figure 3.
Chmp2b regulates dendritic morphology in an ESCRT-III-dependent manner. A, Hippocampal neurons were cotransfected at 10 DIV with the indicated mixtures of pSuper-based and rescue (pBI-based) plasmids. pSuper vectors expressed mCherry together with the indicated shRNA [GFP shRNA (shGFP); Chmp2b shRNA (shChmp2b or sh2b)]. Rescue plasmids expressed ZsGreen, alone or together with the indicated version of RNAi-resistant Chmp2b (2B*). Images of doubly transfected neurons at 14 DIV were acquired by confocal microscopy. Depicted is the morphology of representative neurons in each condition, as visualized by mCherry fluorescence (negative contrast, maximum intensity projections). B, Sholl analysis of dendritic arborization in neurons transfected as in A. For each plasmid mix, the graph shows mean numbers of dendritic branches as a function of radial distance (n = 40 neurons per condition). Chmp2b knockdown strongly reduced the distance-dependent increase in branch number (***p = 0.00014 compared with control curve, Kolmogorov–Smirnov test with Bonferroni's correction). Expression of wild-type Chmp2b* restored the control phenotype (p = 1). Chmp4b-binding-defective (Chmp2b*R19/22/26A) and membrane-binding-defective (Chmp2b* L4D/F5D) mutants of Chmp2b were unable to fully restore the control phenotype (**p = 0.004 and p = 0.006, respectively). C, Hippocampal neurons were triply cotransfected at 10 DIV with HA-tagged Chmp2bWT, Flag-tagged Chmp4b, and mCherry plasmids (1 μg of each plasmid). The cells were fixed 24 h later and stained by dual immunofluorescence with anti-HA and anti-Flag antibodies. Confocal microscopy images (z-projections) of dendritic fluorescence are shown for a representative neuron. Chmp2b was recruited to Chmp4b puncta, some of which were found at dendritic branch points (arrowheads) or at the base of spine-looking protrusions (asterisk). Scale bar, 10 μm. D, N2A cells were transfected with the indicated combinations of RNAi-inducing plasmids and rescue plasmids. The cells were lysed and analyzed by immunoblotting with anti-Chmp2b antibody. E, HeLa cells were cotransfected with HA-tagged Chmp2bWT or Chmp2bR19/22/26A together with Chmp4b-Flag. Cells were stained by dual immunofluorescence with anti-HA and anti-Flag antibodies and imaged by confocal microscopy. Insets, High magnification of the boxed regions. Scale bars, 20 μm. Colocalization of Chmp2b and Chmp4b was 80% for Chmp2bWT and 12% for Chmp2bR19/22/26A (quantified as the percentage of Chmp2b-HA-positive pixels overlapping Chmp4b-Flag puncta). F, HEK-293T cells were transfected with Flag-tagged Chmp4b plasmid, alone or together with HA-tagged Chmp2bWT or Chmp2bR19/22/26A. Anti-HA immunoprecipitates prepared from the transfected cells were analyzed by immunoblotting with anti-Flag or anti-HA antibodies. G, HEK-293T cells were transfected with Flag-tagged Chmp2bWT or Chmp2bL4D/F5D. The transfected cells were homogenized, the membrane (P) and cytosolic (S) fractions were separated by centrifugation, and equivalent amounts of each fraction were analyzed by immunoblotting with anti-Flag antibody.
In non-neuronal settings, Chmp2 proteins are known to interact with Chmp4 for formation of biologically active ESCRT-III (Morita et al., 2011). Consistent with such an interaction, we observed that when coexpressed in neurons at 10 DIV, epitope-tagged Chmp2b and Chmp4b colocalized into puncta distributed along the dendrites (Fig. 3C). Chmp2b/Chmp4b puncta were repeatedly found at dendritic branch points or at the base of dendritic protrusions. We then tested whether the effect of Chmp2b on dendrite branching depended on its ability to bind to endogenous Chmp4b. For this, we designed a Chmp2b point mutant deficient for binding to Chmp4b. Morita et al. (2011) showed that binding of Chmp4b to the close Chmp2b homolog Chmp2a depends on three arginine residues in the Chmp2a N terminus. Based on sequence alignment with Chmp2a, we replaced arginine residues 19, 22, and 26 of Chmp2b by alanine. We demonstrated that, unlike Chmp2bWT, Chmp2bR19/22/26A failed to colocalize with Chmp4b in the cell cytoplasm and to coimmunoprecipitate with Chmp4b in cell lysates (Fig. 3D–F). Thus, Chmp2bR19/22/26A is unable to interact with Chmp4b.
In Chmp2b-depleted neurons, Chmp2bR19/22/26A was much less efficient than Chmp2bWT for restoring dendritic complexity (Fig. 3B), suggesting that Chmp2b activity in dendrite morphogenesis depends on its capacity to bind to Chmp4b. A second point mutant, Chmp2bL4D/F5D, was also tested in dendrite branching assays (Fig. 3B). The L4D/F5D mutation prevents recruitment of the protein to the plasma membrane (Fig. 3G) and abolishes the ability of Chmp2b to deform the plasma membrane when overexpressed (Bodon et al., 2011). We found that Chmp2bL4D/F5D was severely deficient in the dendrite rescue assay, suggesting that the contribution of Chmp2b to dendrite branching correlates with its membrane-deforming potential. Together, the results show that Chmp2b regulates dendrite morphogenesis in a way correlated with its capacity to bind to Chmp4b, thereby suggesting a role for ESCRT-III complexes in this process.
Chmp2b is required for maintenance of synapse density in vitro and in vivo
We previously showed that Chmp2b depletion impaired the morphological maturation of dendritic spines in young cultured neurons (transfected at 10 DIV; Belly et al., 2010). To investigate the contribution of Chmp2b to the maintenance of mature postsynaptic spines, neurons were transfected at 16 DIV with control or Chmp2b-targeting shRNA plasmid, together with control or Chmp2b rescue vector. Two days later, the neurons were fixed, and spines were labeled by immunofluorescence with anti-PSD-95 antibody. Transfected neurons were then imaged by confocal microscopy, and postsynaptic spines were counted (Fig. 4A). In neurons transfected with the Chmp2b-depleting construct, the density of postsynaptic spines along the dendrites was reduced to 53% of that in control neurons (Fig. 4B,C). Coexpression of RNAi-resistant Chmp2bWT restored spine density to 89% of the control value. In contrast, replacing Chmp2bWT with either the Chmp2bR19/22/26A or the Chmp2bL4D/F5D mutant resulted in a complete lack of spine restoration. These results indicate that Chmp2b is necessary for spine maintenance and suggest that this function requires interaction of Chmp2b with both Chmp4b and the plasma membrane.
Figure 4.
Chmp2b is involved in the maintenance of excitatory synapses in cultured neurons. A, Hippocampal neurons were transfected at 16 DIV with pSuper-mCherry encoding control (GFP) shRNA. The neurons were fixed at 18 DIV, stained by dual immunofluorescence with anti-PSD-95 and anti-synapsin I antibodies, and dendrites were imaged by confocal microscopy. Postsynaptic spines (arrowheads) were identified as dendritic protrusions (top) containing PSD-95 puncta (middle). Nearly all of the spines colocalized with the presynaptic marker Synapsin (bottom). mCherry fluorescence was used as a mask to facilitate visualization of puncta belonging to the transfected neuron. Scale bar, 15 μm. B, Neurons were transfected at 16 DIV with the same plasmid combinations as in Figure 3A. Images of dendritic morphology were acquired, and postsynaptic spines were identified as in A and counted. A representative field is shown for each condition. Scale bar, 5 μm. C, Mean density of postsynaptic spines along the dendrites of neurons transfected as in B. Compared with the control (GFP) shRNA, the Chmp2b-depleting shRNA induced a large drop in spine density (***p = 2.4 × 10−11, two-sided Wilcoxon test with Holm correction). Coexpressing wild-type Chmp2b* completely restored density (p = 1 vs control, ***p = 6.3 × 10−14 vs Chmp2b shRNA alone). Coexpressing the Chmp4b binding-defective mutant Chmp2b*R19/22/26A did not rescue spine density at all (***p = 3.8 × 10−6 vs Chmp2b*WT, p = 0.14 vs Chmp2b shRNA alone); neither did the membrane binding-defective mutant Chmp2b* L4D/F5D (***p = 2.9 × 10−7 vs Chmp2bWT, p = 1 vs Chmp2b shRNA alone). Neurons per condition: control, n = 34; Chmp2b shRNA, n = 33; Chmp2b*WT, n = 30; Chmp2b*R19/22/26A, n = 16; Chmp2b*L4D/F5D, n = 14. D–F, Hippocampal neurons were transfected at 18 DIV with the indicated plasmids. Transfected neurons were identified by fluorescence at 21 DIV, and their miniature EPSCs were measured by patch-clamp recordings in whole-cell configuration. D, Sample traces from neurons transfected as indicated. E, Histogram of mean mEPSC amplitudes in neurons transfected as indicated (n = 5 cells, >100 minis per cell in each condition). The amplitude of mEPSCs was markedly reduced in Chmp2b-deficient neurons (**p = 0.005, two-sided t test). F, Histogram of mean mEPSC frequencies in neurons transfected as indicated. Frequencies were lower in Chmp2b-deprived neurons (*p = 0.012, two-sided t test). G, Neurons transfected as above were stimulated at 21 DIV by bath-applied NMDA (control, n = 6 cells; Chmp2b shRNA, n = 5 cells). The density of NMDA-evoked currents (calculated as current normalized by capacitance) was strongly decreased in Chmp2b-deprived neurons (*p = 0.031, two-sided t test).
To determine whether the loss of morphologically identified spines correlated with changes in synaptic transmission, we performed whole-cell patch-clamp recordings of spontaneous mEPSCs in cultured hippocampal neurons transfected with control or shChmp2b constructs (Fig. 4D–F). Transfection was done at 18 DIV, and recordings were performed on mCherry fluorescent neurons at 21 DIV. In Chmp2b-depleted neurons, the mean amplitude of mEPSCs was reduced by 43% and the mean frequency by 30% (Fig. 4E,F). We also recorded excitatory currents evoked by direct activation with bath-applied NMDA. Transfection of the Chmp2b-depleting plasmid resulted in a 63% drop in NMDA receptor-mediated current density (Fig. 4G). Cell capacitance was not significantly affected (GFP shRNA, 16.2 ± 0.8 pF; Chmp2b shRNA, 14.0 ± 1.0 pF; p = 0.3037, t test). Thus, Chmp2b knockdown induced a loss of functional NMDA receptors. Together, these data indicate that Chmp2b is required for maintenance of functional excitatory synapses in cultured hippocampal neurons.
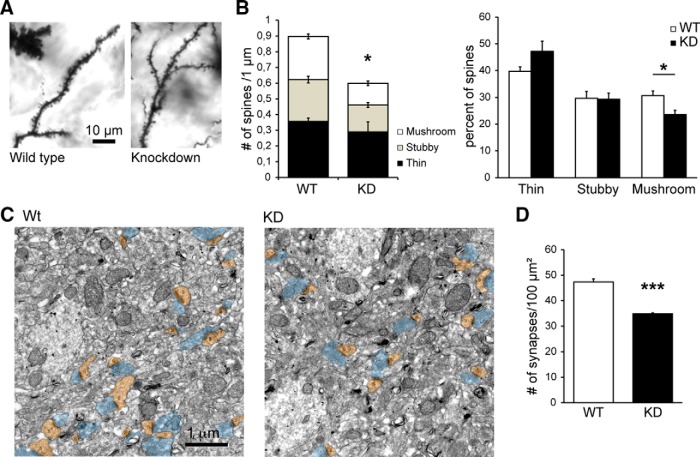
To confirm the specificity of the shRNA effects and to determine whether these results extended to in vivo conditions, we investigated the dendritic morphology of neurons stained by Golgi impregnation in hippocampal CA1 sections from wild-type and Chmp2b KD mice (Fig. 5A; Ghazi-Noori et al., 2012). We found that the total density of spines was reduced by 33% in KD mice compared with age-matched control siblings. This included a 50% loss in the density of mushroom spines (Fig. 5B). We also evaluated the density of excitatory synapses in wild-type and KD hippocampus by counting asymmetric synaptic profiles in electron microscopic images of hippocampal stratum radiatum (Fig. 5C). KD hippocampus displayed a 33% drop in excitatory synapse density (Fig. 5D). Together, the data strongly support the conclusion that Chmp2b plays an important role in formation and maintenance of postsynaptic spines.
Figure 5.
Chmp2b is involved in the maintenance of excitatory synapses in vivo. A, Brain sections from 5-week-old Chmp2b knockdown mice (KD) or sibling controls (Wild type) were stained by the Golgi impregnation technique. Stained dendritic segments were visualized by bright-field microscopy of the stratum radiatum. Shown are representative images of dendritic shafts and spines in both genotypes. B, Spines were classified into thin, stubby, or mushroom types, and their density per unit dendritic length was measured for each type (n = 4 animals, 36 dendrites per genotype). Left, Histogram showing the mean density of each spine type in KD and WT mice. The total spine density was significantly lower in KD mice than in controls (*p = 0.039, two-sided t test). Right, Histogram showing the mean proportion of the three spine types. The percentage of mushroom spines was significantly reduced in the KD mice (*p = 0.02). C, Representative electron micrographs of the CA1 stratum radiatum from Wt and KD siblings. Presynaptic profiles are highlighted in blue, and postsynaptic spines (s) are in brown. D, Quantification of synapse density in the electron micrographs. WT: n = 3 animals, 851 axo-spinous synapses from 30 micrographs; KD: n = 3 animals, 627 axo-spinous synapses from 30 micrographs. The density of postsynaptic spines per unit surface was significantly lower in the KD (***p = 0.0008).
Chmp2b is required for stable spine expansion and synaptic potentiation during chemical LTP
We then examined whether Chmp2b was required for activity-induced changes in spine structure and synaptic function. We first measured the change in spine volume in cultured hippocampal neurons subjected to a chemical LTP (cLTP) protocol, after transfection with control or Chmp2b-depleting vectors. Hippocampal neurons were transfected at 16–18 DIV and imaged 3 d later during cLTP induced by brief stimulation of synaptic NMDA receptors with glycine. The volume of individual spines 5 min before and 45 min after cLTP induction was estimated by 3D reconstruction and voxel counting (Fig. 6A). As expected (Park et al., 2006), in control neurons, spine volumes were increased at 45 min by ≈15% (median value of the relative increment). In contrast, no such increase was seen in Chmp2b-depleted neurons (Fig. 6B). Coexpression of RNAi-resistant Chmp2b fully restored the increase in spine volume (Fig. 6B). These data indicated that Chmp2b was required for stable expansion of spine heads during cLTP.
We also determined the effect of the cLTP protocol on mEPSCs in control and Chmp2b-depleted neurons. As expected, glycine induced a stable increase in the amplitude of mEPSCs in control neurons, thus demonstrating effective LTP (Fig. 6C,D). Knockdown of Chmp2b impaired the increase in synaptic currents, and LTP could be rescued by expression of RNAi-resistant Chmp2bWT (Fig. 6D). These data indicated that Chmp2b was required for long-term synaptic plasticity.
CHMP2B is concentrated at the PSD and close to the spine membrane
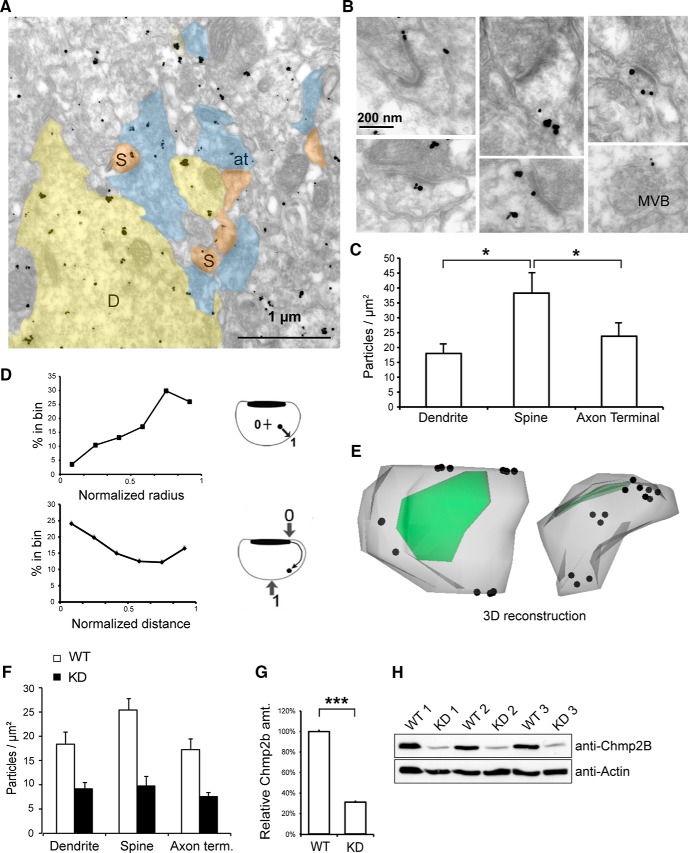
Chmp2b might regulate synaptic function by acting within synapses. To investigate this point, we examined the localization of the protein in neuronal processes in situ at ultrastructural resolution, using the same antibody as above for pre-embedding immunogold labeling of the adult mouse hippocampus. EM of the stratum radiatum revealed Chmp2b immunoreactivity in axons, dendrites, and spines (Fig. 7A,B). Importantly, immunogold labeling was greatly reduced in Chmp2b KD brain, confirming the specificity of labeling (Fig. 7F–H). In the dendritic shafts, as expected based on the known endosomal function of ESCRT-III, particles sometimes decorated multivesicular bodies (MVBs; Fig. 7B, bottom right). Spines (which are usually devoid of MVB; Cooney et al., 2002) displayed a density of immunogold particles that was twofold higher than in dendritic shafts or axonal boutons (Fig. 7C). The particles found in spines showed no obvious association with internal vesicular compartments.
Figure 7.
Ultrastructural localization of Chmp2b in dendritic spines. A, Electron micrograph of Chmp2b immunoreactivity in the mouse stratum radiatum, detected by pre-embedding immunogold labeling. D, Dendrites; S, spines; at, axon terminals. B, High-magnification views of typical dendritic spines. Chmp2b usually appears on the periphery of the PSD or on the neck of the spine, beneath the plasma membrane. Bottom right, Labeled MVB in a dendritic shaft. C, Quantitative analysis of immunogold labeling in the stratum radiatum. The graph shows mean density of gold particles in three neuronal compartments. The ultrastructural localization of 2100 gold particles was analyzed in 30 fields similar to that shown in A. Chmp2b density was significantly higher in spines than in shafts or axonal boutons (p = 0.0498, n = 3 animals, 3 compartments and 10 fields per animal, Friedman test). D, Distribution of gold particle positions inside dendritic spines (n = 335 particles, 3 animals). Top, Histogram of normalized positions along the spine radius. Inset, Coordinate 0 corresponds to the spine center, and 1.0 corresponds to the plasma membrane. Particles concentrate in the subplasmalemmal cortex. Bottom, Histogram of normalized particle distances from the PSD. Inset, Coordinate 0 corresponds to the PSD edge, and 1.0 corresponds to the point equidistant from both edges of the PSD. Particles concentrate in the proximity of the PSD and become scarcer further away, with a slight rebound in frequency at the usual position of the neck. E, 3D reconstruction of a typical spine using immuno-electron micrographs of serial sections. The plasma membrane is in gray, PSD in green, and particles are marked by dots. Particles appear beneath the perisynaptic membrane around the PSD and also in the neck. Left, View from above the PSD; right, lateral view. An animated version of the 3D model appears in Movie 1 (see Notes). F, Quantitation of immunogold labeling in the stratum radiatum of WT and KD siblings. The density of gold particles in the three neuronal compartments is much lower in the KD animals (p < 10−5, n = 3 animals per genotype, Kruskal–Wallis test, density vs genotype). G, Comparison of Chmp2b amounts in WT and KD brains. Total brain extract was obtained from three animals of each genotype and analyzed by SDS-PAGE and Western immunoblotting with anti-Chmp2b antibody. Actin was used as a loading control. Quantitation of the Chmp2b band in each lane was obtained by densitometry and normalized to the respective actin control. KD brain still expressed 30% of the wild-type amount. H, Western blot used for the quantitation in G.
We quantitated the distribution of Chmp2b within dendritic spines by measuring the normalized distance of the gold particles relative to either the center of the spine (radial distance) or the edge of the PSD (tangential distance), following the method of Rácz and Weinberg (2008). Chmp2b immunoreactivity peaked at radial positions corresponding to submembrane cortex and at tangential positions immediately adjacent to the PSD (Fig. 7D). Correcting the radial distribution for distance-dependent increase in the sampling area did not change the submembrane location of the density maximum (see Materials and Methods). Along the tangential coordinate, in addition to the PSD-proximal peak, a second peak was observed at large tangential distances, corresponding to a distinct accumulation in the spine neck. These results were confirmed by reconstructing the three-dimensional distribution of Chmp2b immunoreactivity within individual spines (Fig. 7E; Movie 1). Most particles were arrayed just beneath the spine membrane around the PSD. Particles were also found next to the limiting membrane of the spine neck. The concentration of Chmp2b molecules in the perisynaptic domain is consistent with the possibility that Chmp2b-containing assemblies might associate with the exterior fringe of the PSD.
Animated view of a reconstructed spine, showing under various angles the distribution of Chmp2b labeling (black dots) around the PSD (green) and in the neck.
Synaptic Chmp2b is part of a stable synaptic ESCRT-III complex
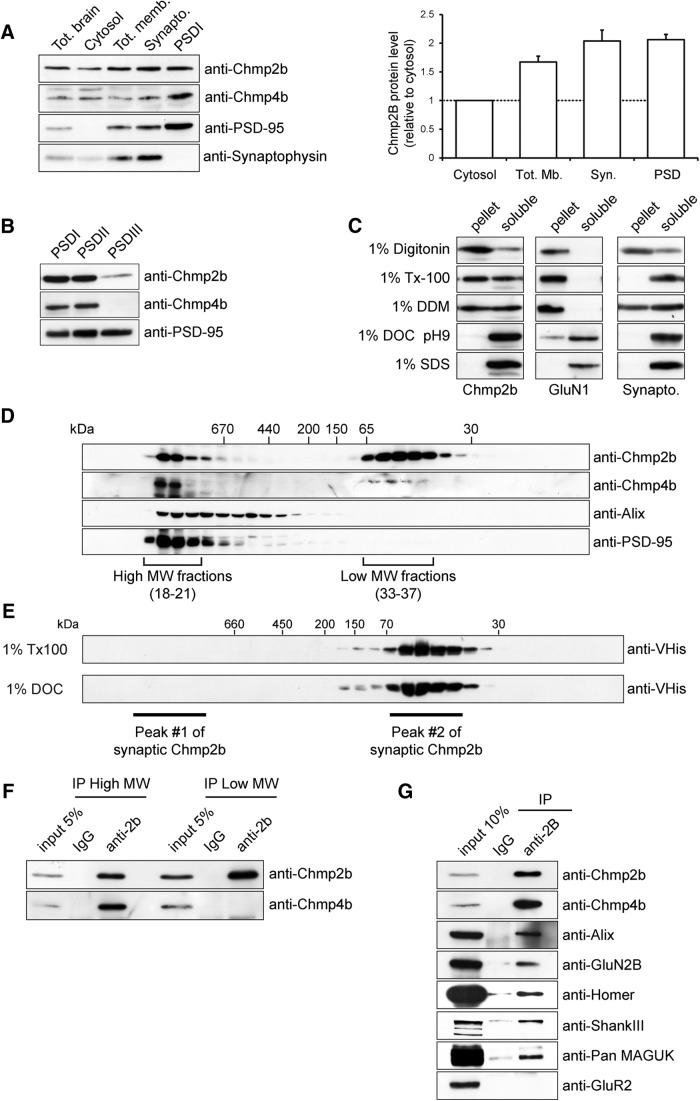
We used immunoblotting to analyze the distribution of Chmp2b and its direct interactor Chmp4b among subcellular fractions derived from rat forebrain. Soluble protein and membranous organelles were recovered from cortical lysates, and synaptoneurosomes were isolated from the total membranes by sedimentation on sucrose gradients. Antibodies against synaptophysin and PSD-95 were used as markers of the presynaptic and postsynaptic compartments, respectively. Chmp2b was more concentrated in the membranes and in the synaptoneurosomes than in the soluble fraction (Fig. 8A). By comparing the intensity of the Chmp2b band with recombinant protein standards, we estimated that Chmp2b represented ∼0.03% (w/w) of total synaptosomal protein (data not shown). The relative level of Chmp2b remained elevated in the Triton X-100-insoluble material extracted from synaptoneurosomes (PSDI fraction). Chmp4b was mostly Triton insoluble and highly enriched in the PSDI fraction. Subsequent solubilization of PSDI revealed that unlike PSD-95, Chmp2b and Chmp4b could be solubilized from the PSD fraction using the more stringent detergent Sarkosyl (Fig. 8B, fraction PSDIII). Solubilization of Chmp2b from synaptoneurosomes was completely ineffective with 1% digitonin, only partial with the mild nonionic detergent dodecylmaltoside, and very efficient with 1% DOC (Fig. 8C). These data indicated that although Chmp2b and Chmp4b were associated with Triton-insoluble fractions containing postsynaptic scaffolding proteins, they did not behave as “core” proteins of the PSD, but rather as peripherally attached PSD proteins.
Figure 8.
Chmp2b is part of a stable ESCRT-III-type complex. A, Rat forebrain homogenates were fractionated as described in Materials and Methods. Equivalent amounts of each fraction were analyzed by Western immunoblotting with the indicated antibodies. Both Chmp2b and Chmp4b were enriched in the PSD fraction. B, PSDI was re-extracted with either 0.5% Triton X-100 or 3% Sarkosyl to yield insoluble fractions PSDII or PSDIII, respectively. Equivalent amounts of each fraction were analyzed by Western immunoblotting. C, Synaptoneurosomes were solubilized with the indicated detergents and centrifuged at high speed. The resulting supernatants and pellets were analyzed by immunoblotting with the indicated antibodies. D, Gradient-purified synaptoneurosomes were lysed in 1% DOC, insoluble material was removed, and the extract was fractionated by size-exclusion chromatography on a Sephacryl S-300 column. Column fractions were analyzed by immunoblotting with the indicated antibodies. Apparent molecular weights (MW) were calibrated with protein markers. A subset of Chmp2b displayed the same high MW as the PSD-95 complex. E, Monomeric recombinant His-tagged Chmp2b was incubated with either 1% Triton X-100 or 1% DOC and fractionated by size-exclusion chromatography on a Sephacryl S-300 column. Fractions were analyzed by immunoblotting with an anti-His tag antibody. DOC did not cause detectable aggregation of monomeric Chmp2b. The elution peaks of synaptic Chmp2b-containing fractions resolved on the same column are indicated for reference. F, Fractions corresponding to HMW and LMW Chmp2b populations were separately pooled and immunoprecipitated with anti-Chmp2b antibody or control IgG. The immunoprecipitates were analyzed by immunoblotting with anti-Chmp2b or anti-Chmp4b antibody. Chmp4b coprecipitated with Chmp2b in the HMW peak. G, Synaptoneurosome extracts were prepared as in D and immunoprecipitated with anti-Chmp2b antibody or control IgG. The immunoprecipitates were analyzed by immunoblotting with the indicated antibodies.
To determine the oligomeric state of Chmp2b at synapses, gradient-purified synaptoneurosomes were lysed in DOC (Husi and Grant, 2001). The lysates were clarified by high-speed centrifugation, and the solubilized proteins were then analyzed by gel filtration and immunoblotting of fractions. This revealed that 30% of synaptic Chmp2b was associated with large molecular assemblies (>1.5 MDa), coeluting with the expected peak of postsynaptic receptor/scaffold complexes (Fig. 8D). The remaining Chmp2b molecules were eluted in low-molecular-weight (LMW) fractions (peaking at ≅50 kDa apparent molecular weight) corresponding to monomeric Chmp2b, as indicated by comparison with the purified recombinant protein (Fig. 8E). Importantly, DOC buffer did not affect the apparent size of monomeric recombinant Chmp2b (Fig. 8E), demonstrating that Chmp2b polymerization is not activated by the detergent during solubilization. The vast majority of synaptic Chmp4b, as well as a fraction of the Chmp4b-binding protein Alix, coeluted with the HMW pool of Chmp2b and PSD-95 (Fig. 8D). These data show that at synapse Chmp2b, Chmp4b and Alix are part of stable polymers, possibly representing ESCRT-III complexes associated with synaptic structures. To investigate this point further, we performed coimmunoprecipitation experiments. The HMW and monomeric entities resolved by gel filtration were separately immunoprecipitated with anti-Chmp2b, and the immunoprecipitates were probed with anti-Chmp4b antibody. A large proportion of the Chmp4b present in the HMW peak coprecipitated along with Chmp2b (Fig. 8F, lanes 1–3). In contrast, Chmp4b molecules and Chmp2b present in the LMW fraction were not coprecipitated (Fig. 8F, lanes 4–6). Thus, in synaptoneurosomes solubilized by DOC, the polymeric pool of Chmp4b corresponds to molecules associated with Chmp2b, supporting the conclusion that synapses contain stable ESCRT-III complexes.
To further define the composition of synaptic ESCRT-III, Chmp2b immunoprecipitates derived from the purified HMW peak or from total synaptoneurosomal lysates were analyzed by mass spectrometry. In agreement with the above data, Chmp4b was detected as one of the most prominent proteins among the numerous Chmp2b-associated species (Table 3). Alix and the ESCRT-III subunits Chmp1A, Chmp2A, Chmp3, Chmp5, and Chmp6 were also identified, confirming that synaptic Chmp2b is part of an ESCRT-III-type complex. On the other hand, proteomic analysis also identified GluN2B, Homer, and Shank peptides among Chmp2b-bound species. The presence of these postsynaptic proteins was confirmed by immunoblotting (Fig. 8G). In addition to Chmp4b and Alix, Chmp2b immunoprecipitates derived from synaptosomal DOC lysates also contained detectable amounts of NMDA receptors but no GluA2-containing AMPA receptors. Furthermore, mass spectrometry also revealed a set of proteins regulating or belonging to the cortical actin cytoskeleton (Table 4). Together with the intraspine distribution of Chmp2b immunoreactivity, these biochemical data suggest that a subset of Chmp2b molecules exist within stable ESCRT-III polymers attached to the spinoskeleton, around the PSD and in the spine shell.
Table 3.
ESCRT-III subunits and postsynaptic scaffold proteins detected by mass spectrometry of the synaptic Chmp2b-containing complex
| Protein name | Accession number | Unique peptides (IP HMW complex) |
Spectral counts (IP HMW complex) |
Spectral counts (IP lysate) |
|||
|---|---|---|---|---|---|---|---|
| Chmp2b IgG | Control IgG | Chmp2b IgG | Control IgG | Chmp2b IgG | Control IgG | ||
| CHMP2B | F1M8B7_RAT | 2 | 0 | 2 | 0 | 2 | 0 |
| CHMP2A | B2RZB5_RAT | 8 | 0 | 8 | 0 | 4 | 0 |
| CHMP4B | D4A9Z8_RAT | 12 | 0 | 12 | 0 | 14 | 0 |
| CHMP6 | D3ZDR2_RAT | 3 | 0 | 3 | 0 | 3 | 0 |
| CHMP5 | CHMP5_RAT | 3 | 0 | 3 | 0 | 4 | 0 |
| CHMP1A | D4AE79_RAT | 4 | 0 | 4 | 0 | 4 | 0 |
| CHMP3 | CHMP3_RAT | 8 | 0 | 8 | 0 | 3 | 0 |
| ALIX | PDC6I_RAT | 10 | 0 | 10 | 0 | 2 | 0 |
| GLUN2B | NMDE2_RAT | 2 | 0 | 2 | 0 | 10 | 0 |
| PSD-95 | DLG4_RAT | 3 | 0 | 3 | 0 | 26 | 7 |
| SHANK1 | SHAN1_RAT | 5 | 0 | 5 | 0 | 7 | 0 |
| HOMER1 | HOME1_RAT | 6 | 0 | 6 | 0 | 11 | 3 |
Gradient-purified synaptoneurosomes were lysed in DOC buffer and fractionated by gel filtration. The HMW fractions were pooled and immunoprecipitated with anti-Chmp2b or control IgG. Immunoprecipitates were analyzed by LC-MS/MS. For each protein, the total number of protein-derived peptides (spectral counts) is shown as a semiquantitative estimate of relative protein abundance. The number of unique peptides was equal to the spectral count for all proteins shown. Spectral counts are also given for the same proteins identified in an independent immunoprecipitate prepared from total synaptosomal lysate (right column). Note the consistency of Chmp spectral counts between experiments. Proteins were detected in a total of three immunoprecipitation (IP) experiments.
Table 4.
Complete list of proteins detected by mass spectrometry of the synaptic Chmp2b-containing HMW complex
| Protein name | Accession number | Note | SC 2B/SC Control | Chmp2b IgG SC | Control IgG |
||
|---|---|---|---|---|---|---|---|
| SSC | SC | SSC | |||||
| Bag6 | BAG6_RAT | 2B only | 35 | 35 | |||
| Gja1 | CXA1_RAT | 2B only | 29 | 29 | |||
| Get4 | F1LXF5_RAT | 2B only | 23 | 23 | |||
| Camk2g | KCC2G_RAT | PSD | 2B only | 14 | 4 | ||
| Chmp4b | D4A9Z8_RAT | ESCRT-III | 2B only | 12 | 12 | ||
| Fasn | FAS_RAT | 2B only | 11 | 11 | |||
| Ptpn23 | F1M951_RAT | ESCRT-III | 2B only | 10 | 10 | ||
| Alix | PDC6I_RAT | ESCRT-III | 2B only | 10 | 10 | ||
| Ubl4a | UBL4A_RAT | 2B only | 10 | 10 | |||
| Wdr7 | WDR7_RAT | 2B only | 10 | 10 | |||
| Myo6 | D4A5I9_RAT | CSK | 2B only | 9 | 9 | ||
| Hk1 | HXK1_RAT | 2B only | 9 | 9 | |||
| Sptbn2 | SPTN2_RAT | CSK | 2B only | 9 | 7 | ||
| Chmp2a | B2RZB5_RAT | ESCRT-III | 2B only | 8 | 8 | ||
| Chmp3 | CHMP3_RAT | ESCRT-III | 2B only | 8 | 8 | ||
| Pfkp | K6PP_RAT | 2B only | 8 | 7 | |||
| Ogt | OGT1_RAT | 2B only | 8 | 8 | |||
| Pfkm | Q52KS1_RAT | 2B only | 8 | 7 | |||
| Srcin1 | SRCN1_RAT | 2B only | 8 | 8 | |||
| Myo1d | MYO1D_RAT | CSK | 2B only | 7 | 7 | ||
| Capzb | CAPZB_RAT | CSK | 2B only | 6 | 6 | ||
| Myh14 | F1LNF0_RAT | CSK | 2B only | 6 | 2 | ||
| Ank2 | F1M9N9_RAT | CSK | 2B only | 6 | 6 | ||
| Sfxn | G3V804_RAT | 2B only | 6 | 2 | |||
| Homer1 | HOME1_RAT | PSD | 2B only | 6 | 6 | ||
| Ppp1ca | PP1A_RAT | 2B only | 6 | 2 | |||
| Ppp1cc | PP1G_RAT | 2B only | 6 | 2 | |||
| Hspa12a | D3ZC55_RAT | 2B only | 5 | 5 | |||
| Dbn1 | DREB_RAT | PSD | 2B only | 5 | 5 | ||
| Gphn | GEPH_RAT | 2B only | 5 | 5 | |||
| Idh2 | IDHP_RAT | 2B only | 5 | 5 | |||
| Shank1 | SHAN1_RAT | PSD | 2B only | 5 | 5 | ||
| Tpi1 | TPIS_RAT | 2B only | 5 | 5 | |||
| Sptbn1 | G3V6S0_RAT | CSK | 35, 0 | 35 | 33 | (1) | (1) |
| Myh10 | F1LQ02_RAT | CSK | 24, 0 | 24 | 19 | (1) | (1) |
| Dync1h1 | DYHC1_RAT | 22, 0 | 22 | 5 | (1) | (1) | |
| Ap3d1 | B5DFK6_RAT | 12, 0 | 12 | 12 | (1) | (1) | |
| Myh9 | G3V6P7_RAT | CSK | 11, 0 | 11 | 6 | (1) | (1) |
| Cyfip2 | D3ZX82_RAT | CSK | 2B only | 4 | 4 | ||
| Chmp1a | D4AE79_RAT | ESCRT-III | 2B only | 4 | 4 | ||
| Glud1 | DHE3_RAT | 2B only | 4 | 4 | |||
| Sucla2 | F1LM47_RAT | 2B only | 4 | 4 | |||
| Epb41l3 | G3V874_RAT | CSK | 2B only | 4 | 4 | ||
| Kif2a | KIF2A_RAT | 2B only | 4 | 4 | |||
| Pclo | PCLO_RAT | 2B only | 4 | 4 | |||
| Snf8 | SNF8_RAT | 2B only | 4 | 4 | |||
| Snap25 | SNP25_RAT | 2B only | 4 | 4 | |||
| Actn1 | ACTN1_RAT | CSK | 2B only | 3 | 3 | ||
| Agap2 | AGAP2_RAT | 2B only | 3 | 3 | |||
| Snap91 | AP180_RAT | 2B only | 3 | 3 | |||
| Atp5f1 | AT5F1_RAT | 2B only | 3 | 3 | |||
| LOC681996 | B0BN63_RAT | 2B only | 3 | 3 | |||
| Arpc4 | B2RZ72_RAT | CSK | 2B only | 3 | 3 | ||
| Ndufa4 | B2RZD6_RAT | 2B only | 3 | 3 | |||
| Capza2 | CAZA2_RAT | CSK | 2B only | 3 | 3 | ||
| Chmp5 | CHMP5_RAT | ESCRT-III | 2B only | 3 | 3 | ||
| Rapgef4 | D3KR63_RAT | 2B only | 3 | 3 | |||
| Chmp6 | D3ZDR2_RAT | ESCRT-III | 2B only | 3 | 3 | ||
| Dclk1 | DCLK1_RAT | 2B only | 3 | 3 | |||
| Dlg4 | DLG4_RAT | PSD | 2B only | 3 | 3 | ||
| Dynll1 | DYL1_RAT | 2B only | 3 | 3 | |||
| H2a1c | H2A1C_RAT | 2B only | 3 | 3 | |||
| Prkar2b | KAP3_RAT | 2B only | 3 | 3 | |||
| Prkcg | KPCG_RAT | 2B only | 3 | 3 | |||
| Nckap1 | NCKP1_RAT | CSK | 2B only | 3 | 3 | ||
| Ndufs2 | NDUS2_RAT | 2B only | 3 | 3 | |||
| Ndufv2 | NDUV2_RAT | 2B only | 3 | 3 | |||
| Plec | PLEC_RAT | 2B only | 3 | 3 | |||
| Pura | PURA_RAT | 2B only | 3 | 3 | |||
| Pygb | PYGB_RAT | 2B only | 3 | 3 | |||
| Syt7 | Q99P34_RAT | 2B only | 3 | 3 | |||
| Rab2a | RAB2A_RAT | 2B only | 3 | 3 | |||
| Ran | RAN_RAT | 2B only | 3 | 3 | |||
| Dnm1 | DYN1_RAT | 8, 7 | 26 | 26 | 3 | 3 | |
| Atp2b1 | AT2B1_RAT | 8, 0 | 8 | 3 | (1) | (1) | |
| Nsf | NSF_RAT | 8, 0 | 16 | 16 | 2 | 2 | |
| Rab3c | RAB3C_RAT | 8, 0 | 8 | 4 | (1) | (1) | |
| Atp2b2 | AT2B2_RAT | 7, 0 | 7 | 3 | (1) | (1) | |
| Rab14 | RAB14_RAT | 7, 0 | 7 | 3 | (1) | (1) | |
| Rims1 | RIMS1_RAT | 7, 0 | 7 | 7 | (1) | (1) | |
| Syn2 | SYN2_RAT | 7, 0 | 7 | 4 | (1) | (1) | |
| Add2 | ADDB_RAT | CSK | 5, 0 | 5 | 4 | (1) | (1) |
| Atp5o | ATPO_RAT | 5, 0 | 5 | 5 | (1) | (1) | |
| RGD1307615 | D3ZKQ4_RAT | 5, 0 | 5 | 5 | (1) | (1) | |
| Sptan1 | E9PSZ3_RAT | CSK | 5, 0 | 35 | 35 | 7 | 7 |
| Immt | IMMT_RAT | 5, 0 | 5 | 5 | (1) | (1) | |
| Camk2b | KCC2B_RAT | PSD | 5, 0 | 30 | 13 | 6 | (1) |
| Aco2 | ACON_RAT | 2B only | 2 | 2 | |||
| Maoa | AOFA_RAT | 2B only | 2 | 2 | |||
| Ap3m2 | AP3M2_RAT | 2B only | 2 | 2 | |||
| Actr2 | ARP2_RAT | CSK | 2B only | 2 | 2 | ||
| Arpc5l | ARP5L_RAT | CSK | 2B only | 2 | 2 | ||
| Atad3 | ATAD3_RAT | 2B only | 2 | 2 | |||
| Cacna1a | CAC1A_RAT | 2B only | 2 | 2 | |||
| Cand1 | CAND1_RAT | 2B only | 2 | 2 | |||
| Crym | CRYM_RAT | 2B only | 2 | 2 | |||
| Caskin1 | CSKI1_RAT | 2B only | 2 | 2 | |||
| Atp5j2 | D3ZAF6_RAT | 2B only | 2 | 2 | |||
| Dsg1b | D3ZQ45_RAT | 2B only | 2 | 2 | |||
| Atp6v1a | D4A133_RAT | 2B only | 2 | 2 | |||
| Add3 | D4A3Q7_RAT | CSK | 2B only | 2 | 2 | ||
| Epb41l1 | E41L1_RAT | CSK | 2B only | 2 | 2 | ||
| Hadha | ECHA_RAT | 2B only | 2 | 2 | |||
| Ank3 | F1LP19_RAT | CSK | 2B only | 2 | 2 | ||
| Mrpl48 | F1LWB8_RAT | 2B only | 2 | 2 | |||
| Chmp2b | F1M8B7_RAT | ESCRT-III | 2B only | 2 | 2 | ||
| Gnb1 | GBB1_RAT | 2B only | 2 | 2 | |||
| Slc2a1 | GTR1_RAT | 2B only | 2 | 2 | |||
| Eif4a2 | IF4A2_RAT | 2B only | 2 | 2 | |||
| Kcnh1 | KCNH1_RAT | 2B only | 2 | 2 | |||
| Prkcb | KPCB_RAT | 2B only | 2 | 2 | |||
| Ldha | LDHA_RAT | 2B only | 2 | 2 | |||
| Mink1 | MINK1_RAT | 2B only | 2 | 2 | |||
| Grin2b | NMDE2_RAT | PSD | 2B only | 2 | 2 | ||
| Pdhb | ODPB_RAT | 2B only | 2 | 2 | |||
| Pde2a | PDE2A_RAT | 2B only | 2 | 2 | |||
| Mrps9 | Q5I0K4_RAT | 2B only | 2 | 2 | |||
| Ndufs7 | Q5RJN0_RAT | 2B only | 2 | 2 | |||
| Ndufv1 | Q5XIH3_RAT | 2B only | 2 | 2 | |||
| Uqcrc2 | QCR2_RAT | 2B only | 2 | 2 | |||
| Rab18 | RAB18_RAT | 2B only | 2 | 2 | |||
| Rab7a | RAB7A_RAT | 2B only | 2 | 2 | |||
| Rac1 | RAC1_RAT | 2B only | 2 | 2 | |||
| Mras | RASM_RAT | 2B only | 2 | 2 | |||
| Rpl23a | RL23A_RAT | 2B only | 2 | 2 | |||
| Rtn1 | RTN1_RAT | 2B only | 2 | 2 | |||
| Sept5 | SEPT5_RAT | CSK | 2B only | 2 | 2 | ||
| Sipa1l1 | SI1L1_RAT | 2B only | 2 | 2 | |||
| Ssbp1 | SSBP_RAT | 2B only | 2 | 2 | |||
| Stx1b | STX1B_RAT | 2B only | 2 | 2 | |||
Four hundred twenty-eight candidate proteins detected in the sample were filtered as described in Materials and Methods, yielding a list of 134 proteins, shown by order of decreasing spectral counts. The Note column points out ESCRT-III proteins, PSDs, and components of the spine actin cytoskeleton (CSK). SC, Spectral counts (cut- off 2); SSC, specific spectral counts (unique peptides); SC 2B/SC Control, ratio of spectral counts between specific and control immunoprecipitation (cut-off 5); 2B only, no peptides in control immunoprecipitation; (1), SSC = 1, insufficient for definite identification.
Discussion
Our previous work indicated that Chmp2b depletion, as well as expression of FTD-linked Chmp2b mutants, impaired the morphological maturation of dendritic spines in cultured hippocampal neurons, suggesting that this extremely conserved protein might play a specific role at synapses (Belly et al., 2010). In the present study, we confirmed and extended these results. We found that, compared with other tissues, Chmp2b was highly expressed in the brain and that its expression was upregulated during synaptogenesis. In cultured hippocampal neurons, depleting Chmp2b by RNAi decreased the stability of dendritic branches and the density, efficacy, and plasticity of excitatory synapses. Chmp2b-deficient mouse hippocampus displayed a striking reduction in spine and synapse density, supporting the specificity and in vivo relevance of the RNAi effects. Furthermore, EM observations showed that the protein preferentially accumulated close to the spine plasma membrane and near the PSD. Finally, biochemical data demonstrated that the protein was present inside ESCRT-III polymers physically associated with the PSD. Thus, Chmp2b-containing ESCRT-III complexes play a previously unsuspected role in the regulation of long-term synaptic plasticity. The circular geometry and membrane-scaffolding properties of ESCRT-III polymers suggest that they may directly contribute to shaping domains of the spine membrane.
The importance of Chmp2b for synaptic stability was supported by the concomitant drop in synaptic markers, mEPSC frequency, and NMDA-evoked current density in mature cultured neurons in which Chmp2b had been depleted. In these neurons, remaining synapses were less efficient than in control neurons and less apt to undergo potentiation. We also showed that Chmp2b is necessary for synapse integrity in vivo, based on the observed decrease in both Golgi-stained spines and EM synaptic profiles in the stratum radiatum of “leaky KO” mice expressing 15–30% of the normal Chmp2b amount (Ghazi-Noori et al., 2012).
Given the established function of Chmp proteins in the transfer of material to the endo-lysosomal apparatus, the neuronal effects of Chmp2b deficiency might arise from impaired endosomal traffic. Endosomes have been shown to play a crucial role in postsynaptic function and plasticity, e.g., for receptor delivery and as membrane reservoirs for spine expansion (Park et al., 2006). Chmp2b might conceivably regulate some aspect of endosomal dynamics or function in spines. Consistent with this possibility, in non-neuronal cells, ESCRT complexes seem to be involved in trafficking endosomal cargo to multiple cellular localization, including the plasma membrane and associated cytoskeletal structures (Baldys and Raymond, 2009; Lobert and Stenmark, 2012).
However, native endosomal ESCRT-III complexes are thought to be very transient, in contrast with the synaptic complex described here. An alternative hypothesis, which we favor, rests on the finding that ESCRT-III assemblies made of Alix, Chmp4, and Chmp2 can locally reshape the plasma membrane, as during virus egress, cytokinesis, and membrane repair (Henne et al., 2013; Jimenez et al., 2014). That Chmp2b might have such a membrane-scaffolding activity in neurons was suggested by the finding that Chmp2bL4D/F5D was not able to restore dendritic complexity in Chmp2B-depleted neurons. Since we had previously characterized this mutant for its inability to bind to and scaffold the plasma membrane (Bodon et al., 2011), these results suggested that Chmp2b locally acts as a morphoregulatory protein. The effect of Chmp2B on dendritic branches also correlated with its ability to bind its direct ESCRT-III partner Chmp4b, in agreement with previous findings that Chmp4b was required for dendritic growth and maintenance in Drosophila and mouse neurons (Sweeney et al., 2006; Lee et al., 2007). However, since Chmp2b depletion did not cause the rapid lethality induced by Chmp4b RNAi, the morphogenetic function mediated by Chmp2b appears to be distinct from other vital ESCRT-III activities. Interestingly, mutation of Chmp2b arginine 22 (one of the three residues required for binding to Chmp4b) was recently detected in a sporadic case of amyotrophic lateral sclerosis (van Blitterswijk et al., 2012), suggesting that dysfunction of the Chmp2b–Chmp4b interaction might have disease relevance.
Gel filtration of synaptosomal lysates showed that at least 30% of synaptic Chmp2b was bound into a large supramolecular complex that comigrated with postsynaptic scaffolds. Coimmunoprecipitation indicated that within this complex, Chmp2b was associated with the Shank–Homer network and GluN2B-containing NMDA receptors but not with the more mobile GluA2-containing AMPA receptors. In accordance with the strong concentration of Chmp4b in the PSD fraction, ∼70% of synaptosomal Chmp4b comigrated with the Chmp2b/PSD-95 peak and coprecipitated with Chmp2b. Unbiased analysis by mass spectrometry confirmed the presence of Chmp4b in the Chmp2b immunoprecipitates. Using mass spectral counts as a semiquantitative criterion for abundance, Chmp4b; ESCRT-III components Chmp2A, Chmp3, Chmp1A, and Chmp5; and the Chmp4b-binding platform Alix were all strongly represented. This composition corresponds to that expected for ESCRT-III. The simplest explanation of these data is that Chmp2b, Chmp4b, and other subunits were all bound together in stable ESCRT-III polymers attached to postsynaptic scaffolds. The size of scaffolds would account for the apparent molecular weight of the complex, which was >1 MDa larger than the 450 kDa measured in yeast ESCRT-III (Teis et al., 2008). The difference in solubility between Chmp4b and Chmp2b is consistent with multimerization of these molecules in filaments with a high Chmp4/Chmp2 ratio, similar to that determined in yeast.
In view of the known features of ESCRT-III in yeast and cell lines, the apparent stability of synaptic ESCRT-III was unexpected. The transient nature of ESCRT-III assembly in vivo is considered one of its cardinal features, as it arises from tight coupling between subunit polymerization, recruitment of the depolymerizing ATPase Vps4, and membrane scission (Lata et al., 2008; Henne et al., 2013). Detection of wild-type yeast ESCRT-III polymers required protein cross-linking (Teis et al., 2008), and to our knowledge, the biochemical recovery of physiologically assembled ESCRT-III from metazoan cells has not been previously reported. In contrast, in synaptoneurosomes, a large fraction of Chmp2b and Chmp4b was polymerized at steady state, and the polymer survived solubilization in deoxycholate. One possible explanation for this stability is that Vps4 may not have access to synaptic ESCRT-III because of steric hindrance by synaptic proteins. In line with this possibility, proteomic analysis failed to detect Vps4 among Chmp2b-associated proteins, even though Vps4 was present in total synaptosomal lysates.
That a subset of Chmp2b-containing ESCRT-III assembles under the spine membrane is supported by congruent immuno-EM and biochemical data. First, the preferential localization of Chmp2b around the PSD and in the neck indicated that Chmp2b was recruited to the submembrane scaffold of spines. Moreover, isolation and differential detergent extraction of the PSD fraction showed that Chmp2b was anchored to but not embedded within the PSD. Our biochemical data suggest that most of the Chmp2b molecules lying under the spine membrane are part of ESCRT-III polymers. Because Chmp2b exists in axonal boutons as well as in spines, it is possible that some of the complexes originated in the presynaptic compartment. However, since Chmp2b was significantly more concentrated in spines than in boutons, and given that postsynaptic entities predominate in the PSD-95 peak and among Chmp2b-bound proteins, it is likely that most of the ESCRT-III identified in synaptoneurosome extracts was postsynaptic.
We speculate that the intrinsic geometry and rigidity of ESCRT-III filaments may be responsible for imparting spines with some aspects of their typical architecture, thus promoting synaptic stability and plasticity. Chmp4 and Chmp2 polymers reconstituted in vitro from recombinant subunits or via overexpression in transfected cells, consistently form 100- to 300-nm-wide circular or helical arrays strongly adhering to the plasma membrane. Together with the ultrastructural distribution of Chmp2b within dendritic spines, these dimensions are compatible with circular ESCRT-III filaments lining the cytoplasmic side of the spine plasma membrane in the neck and around the PSD. The filaments might conceivably help shape the neck or set up boundaries for PSD growth. Because of the high affinity of the filaments for lipids, they could also act as diffusion barriers, regulating exchange of molecules between the PSD and the extrasynaptic membrane or between spine and shaft. Given the numerous actin-regulating proteins detected by LC-MS in Chmp2b immunoprecipitates, ESCRT-III filaments might also provide an anchor for assembly of actin-based structures or a fulcrum for application of mechanical force by actin.
The Chmp2bR19/22/26A and Chmp2bL4D/F5D mutants were defective for both dendrite and spine maintenance, suggesting that both processes share a requirement for ESCRT-III assembly and binding to the plasma membrane. In cultured neurons, overexpressed ESCRT-III assemblies were frequently localized at dendritic branch points. Chmp2b-containing ESCRT-III might control local cytoskeletal rearrangements, or stabilize membrane curvature at the base of dendritic branches, perhaps by assembling into ≈1-μm-wide rings, akin to those observed in the intercellular bridge of dividing cells (Guizetti et al. 2011). This diameter would correspond to that of distal dendrites, known by EM measurements to be no larger than 0.5–1 μm (Bartlett and Banker, 1984). More studies are necessary to identify ESCRT-III structures in neurons and to elucidate the mechanisms whereby they help maintain dendrites and spines.
Our work showing that CHMP2B resides at synapses and is involved in synaptic plasticity opens new avenues for understanding how Chmp2b deficiency contributes to the cognitive effects of Chmp2b mutations in FTD patients.
Footnotes
This work was supported by a grant from the Agence Nationale de la Recherche (ANR), MALZ program, France. R.C. and M.L. were supported by the Ministère de l'Enseignement Supérieur et de la Recherche, France. J.M.-H. and N.B. were supported by ANR. A.M.I. was supported by the United Kingdom Medical Research Council. We thank Y. Saoudi for help with microscopy and N. Jaafari, A. Petiot, and W. Weissenhorn for critically reading this manuscript.
The authors declare no competing financial interests.
This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/3.0), which permits unrestricted use, distribution and reproduction in any medium provided that the original work is properly attributed.
References
- Alvarez VA, Ridenour DA, Sabatini BL. Retraction of synapses and dendritic spines induced by off-target effects of RNA interference. J Neurosci. 2006;26:7820–7825. doi: 10.1523/JNEUROSCI.1957-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajorek M, Schubert HL, McCullough J, Langelier C, Eckert DM, Stubblefield WM, Uter NT, Myszka DG, Hill CP, Sundquist WI. Structural basis for ESCRT-III protein autoinhibition. Nat Struct Mol Biol. 2009;16:754–762. doi: 10.1038/nsmb.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldys A, Raymond JR. Critical role of ESCRT machinery in EGFR recycling. Biochemistry. 2009;48:9321–9323. doi: 10.1021/bi900865u. [DOI] [PubMed] [Google Scholar]
- Bartlett WP, Banker GA. An electron microscopic study of the development of axons and dendrites by hippocampal neurons in culture. J Neurosci. 1984;4:1944–1953. doi: 10.1523/JNEUROSCI.04-08-01944.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belly A, Bodon G, Blot B, Bouron A, Sadoul R, Goldberg Y. CHMP2B mutants linked to frontotemporal dementia impair maturation of dendritic spines. J Cell Sci. 2010;123:2943–2954. doi: 10.1242/jcs.068817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodon G, Chassefeyre R, Pernet-Gallay K, Martinelli N, Effantin G, Hulsik DL, Belly A, Goldberg Y, Chatellard-Causse C, Blot B, Schoehn G, Weissenhorn W, Sadoul R. Charged multivesicular body protein 2B (CHMP2B) of the endosomal sorting complex required for transport-III (ESCRT-III) polymerizes into helical structures deforming the plasma membrane. J Biol Chem. 2011;286:40276–40286. doi: 10.1074/jbc.M111.283671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S, Cordelières FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- Buchkovich NJ, Henne WM, Tang S, Emr SD. Essential N-terminal insertion motif anchors the ESCRT-III filament during MVB vesicle formation. Dev Cell. 2013;27:201–214. doi: 10.1016/j.devcel.2013.09.009. [DOI] [PubMed] [Google Scholar]
- Carlin RK, Grab DJ, Cohen RS, Siekevitz P. Isolation and characterization of postsynaptic densities from various brain regions: enrichment of different types of postsynaptic densities. J Cell Biol. 1980;86:831–845. doi: 10.1083/jcb.86.3.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooney JR, Hurlburt JL, Selig DK, Harris KM, Fiala JC. Endosomal compartments serve multiple hippocampal dendritic spines from a widespread rather than a local store of recycling membrane. J Neurosci. 2002;22:2215–2224. doi: 10.1523/JNEUROSCI.22-06-02215.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filimonenko M, Stuffers S, Raiborg C, Yamamoto A, Malerød L, Fisher EM, Isaacs A, Brech A, Stenmark H, Simonsen A. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J Cell Biol. 2007;179:485–500. doi: 10.1083/jcb.200702115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazi-Noori S, Froud KE, Mizielinska S, Powell C, Smidak M, Fernandez de Marco M, O'Malley C, Farmer M, Parkinson N, Fisher EM, Asante EA, Brandner S, Collinge J, Isaacs AM. Progressive neuronal inclusion formation and axonal degeneration in CHMP2B mutant transgenic mice. Brain. 2012;135:819–832. doi: 10.1093/brain/aws006. [DOI] [PubMed] [Google Scholar]
- Goetze B, Grunewald B, Baldassa S, Kiebler M. Chemically controlled formation of a DNA/calcium phosphate coprecipitate: application for transfection of mature hippocampal neurons. J Neurobiol. 2004;60:517–525. doi: 10.1002/neu.20073. [DOI] [PubMed] [Google Scholar]
- Guizetti J, Schermelleh L, Mäntler J, Maar S, Poser I, Leonhardt H, Müller-Reichert T, Gerlich DW. Cortical constriction during abscission involves helices of ESCRT- III-dependent filaments. Science. 2011;331:1616–1620. doi: 10.1126/science.1201847. [DOI] [PubMed] [Google Scholar]
- Hanson PI, Roth R, Lin Y, Heuser JE. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J Cell Biol. 2008;180:389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E, Lane DP. Antibodies: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- Henne WM, Buchkovich NJ, Zhao Y, Emr SD. The endosomal sorting complex ESCRT-II mediates the assembly and architecture of ESCRT-III helices. Cell. 2012;151:356–371. doi: 10.1016/j.cell.2012.08.039. [DOI] [PubMed] [Google Scholar]
- Henne WM, Stenmark H, Emr SD. Molecular mechanisms of the membrane sculpting ESCRT pathway. Cold Spring Harb Perspect Biol. 2013;5:a016766. doi: 10.1101/cshperspect.a016766. pii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husi H, Grant SG. Isolation of 2000-kDa complexes of N-methyl-D-aspartate receptor and postsynaptic density 95 from mouse brain. J Neurochem. 2001;77:281–291. doi: 10.1046/j.1471-4159.2001.t01-1-00248.x. [DOI] [PubMed] [Google Scholar]
- Isaacs AM, Johannsen P, Holm I, Nielsen JE. Frontotemporal dementia caused by CHMP2B mutations. Curr Alzheimer Res. 2011;8:246–251. doi: 10.2174/156720511795563764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Chen G. High Ca2+-phosphate transfection efficiency in low-density neuronal cultures. Nat Protoc. 2006;1:695–700. doi: 10.1038/nprot.2006.86. [DOI] [PubMed] [Google Scholar]
- Jimenez AJ, Maiuri P, Lafaurie-Janvore J, Divoux S, Piel M, Perez F. ESCRT machinery is required for plasma membrane repair. Science. 2014;343:1247136. doi: 10.1126/science.1247136. [DOI] [PubMed] [Google Scholar]
- Lata S, Schoehn G, Jain A, Pires R, Piehler J, Gottlinger HG, Weissenhorn W. Helical structures of ESCRT-III are disassembled by VPS4. Science. 2008;321:1354–1357. doi: 10.1126/science.1161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA, Beigneux A, Ahmad ST, Young SG, Gao FB. ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr Biol. 2007;17:1561–1567. doi: 10.1016/j.cub.2007.07.029. [DOI] [PubMed] [Google Scholar]
- Lobert VH, Stenmark H. The ESCRT machinery mediates polarization of fibroblasts through regulation of myosin light chain. J Cell Sci. 2012;125:29–36. doi: 10.1242/jcs.088310. [DOI] [PubMed] [Google Scholar]
- Lu W, Man H, Ju W, Trimble WS, MacDonald JF, Wang YT. Activation of synaptic NMDA receptors induces membrane insertion of new AMPA receptors and LTP in cultured hippocampal neurons. Neuron. 2001;29:243–254. doi: 10.1016/S0896-6273(01)00194-5. [DOI] [PubMed] [Google Scholar]
- McCullough J, Colf LA, Sundquist WI. Membrane fission reactions of the mammalian ESCRT pathway. Annu Rev Biochem. 2013;82:663–692. doi: 10.1146/annurev-biochem-072909-101058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milbradt J, Kraut A, Hutterer C, Sonntag E, Schmeiser C, Ferro M, Wagner S, Lenac T, Claus C, Pinkert S, Hamilton ST, Rawlinson WD, Sticht H, Couté Y, Marschall M. Proteomic analysis of the multimeric nuclear egress complex of human cytomegalovirus. Mol Cell Proteomics. 2014;13:2132–2146. doi: 10.1074/mcp.M113.035782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E, Sandrin V, McCullough J, Katsuyama A, Baci Hamilton I, Sundquist WI. ESCRT-III protein requirements for HIV-1 budding. Cell Host Microbe. 2011;9:235–242. doi: 10.1016/j.chom.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M, Salgado JM, Ostroff L, Helton TD, Robinson CG, Harris KM, Ehlers MD. Plasticity-induced growth of dendritic spines by exocytic trafficking from recycling endosomes. Neuron. 2006;52:817–830. doi: 10.1016/j.neuron.2006.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rácz B, Weinberg RJ. Organization of the Arp2/3 complex in hippocampal spines. J Neurosci. 2008;28:5654–5659. doi: 10.1523/JNEUROSCI.0756-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. Vienna: R Foundation for Statistical Computing; 2013. R: a language and environment for statistical computing. http://www.R-project.org/ [Google Scholar]
- Rodriguez A, Ehlenberger DB, Hof PR, Wearne SL. Rayburst sampling, an algorithm for automated three-dimensional shape analysis from laser scanning microscopy images. Nat Protoc. 2006;1:2152–2161. doi: 10.1038/nprot.2006.313. [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Ehlenberger DB, Dickstein DL, Hof PR, Wearne SL. Automated three-dimensional detection and shape classification of dendritic spines from fluorescence microscopy images. PLoS One. 2008;3:e1997. doi: 10.1371/journal.pone.0001997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim S, Kimpler LA, Hanson PI. Structure/function analysis of four core ESCRT-III proteins reveals common regulatory role for extreme C-terminal domain. Traffic. 2007;8:1068–1079. doi: 10.1111/j.1600-0854.2007.00584.x. [DOI] [PubMed] [Google Scholar]
- Skibinski G, Parkinson NJ, Brown JM, Chakrabarti L, Lloyd SL, Hummerich H, Nielsen JE, Hodges JR, Spillantini MG, Thusgaard T, Brandner S, Brun A, Rossor MN, Gade A, Johannsen P, Sørensen SA, Gydesen S, Fisher EM, Collinge J. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat Genet. 2005;37:806–808. doi: 10.1038/ng1609. [DOI] [PubMed] [Google Scholar]
- Sweeney NT, Brenman JE, Jan YN, Gao FB. The coiled-coil protein shrub controls neuronal morphogenesis in Drosophila. Curr Biol. 2006;16:1006–1011. doi: 10.1016/j.cub.2006.03.067. [DOI] [PubMed] [Google Scholar]
- Teis D, Saksena S, Emr SD. Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev Cell. 2008;15:578–589. doi: 10.1016/j.devcel.2008.08.013. [DOI] [PubMed] [Google Scholar]
- Urwin H, Authier A, Nielsen JE, Metcalf D, Powell C, Froud K, Malcolm DS, Holm I, Johannsen P, Brown J, Fisher EM, van der Zee J, Bruyland M, Van Broeckhoven C, Collinge J, Brandner S, Futter C, Isaacs AM. Disruption of endocytic trafficking in frontotemporal dementia with CHMP2B mutations. Hum Mol Genet. 2010;19:2228–2238. doi: 10.1093/hmg/ddq100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Blitterswijk M, Vlam L, van Es MA, van der Pol WL, Hennekam EA, Dooijes D, Schelhaas HJ, van der Kooi AJ, de Visser M, Veldink JH, van den Berg LH. Genetic overlap between apparently sporadic motor neuron diseases. PLoS One. 2012;7:e48983. doi: 10.1371/journal.pone.0048983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Nonogaki M, Madhira R, Ma HY, Hermanson O, Kioussi C, Gross MK. Population-specific regulation of Chmp2b by Lbx1 during onset of synaptogenesis in lateral association interneurons. PLoS One. 2012;7:e48573. doi: 10.1371/journal.pone.0048573. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Animated view of a reconstructed spine, showing under various angles the distribution of Chmp2b labeling (black dots) around the PSD (green) and in the neck.