Abstract
Coq5 catalyzes the only C-methylation involved in the biosynthesis of coenzyme Q (Q or ubiquinone) in humans and yeast Saccharomyces cerevisiae. As one of eleven polypeptides required for Q production in yeast, Coq5 has also been shown to assemble with the multi-subunit complex termed the CoQ-synthome. In humans, mutations in several COQ genes cause primary Q deficiency, and a decrease in Q biosynthesis is associated with mitochondrial, cardiovascular, kidney and neurodegenerative diseases. In this study, we characterize the human COQ5 polypeptide and examine its complementation of yeast coq5 point and null mutants. We show that human COQ5 RNA is expressed in all tissues and that the COQ5 polypeptide is associated with the mitochondrial inner membrane on the matrix side. Previous work in yeast has shown that point mutations within or adjacent to conserved COQ5 methyltransferase motifs result in a loss of Coq5 function but not Coq5 steady state levels. Here, we show that stabilization of the CoQ-synthome within coq5 point mutants or by over-expression of COQ8 in coq5 null mutants permits the human COQ5 homolog to partially restore coq5 mutant growth on respiratory media and Q6 content. Immunoblotting against the human COQ5 polypeptide in isolated yeast mitochondria shows that the human Coq5 polypeptide migrates in two-dimensional blue-native/SDS-PAGE at the same high molecular mass as other yeast Coq proteins. The results presented suggest that human and Escherichia coli Coq5 homologs expressed in yeast retain C-methyltransferase activity but are capable of rescuing the coq5 yeast mutants only when the CoQ-synthome is assembled.
Abbreviations: 2D-BN-SDS/PAGE, two-dimensional blue-native-sodium dodecyl sulfate/polyacrylamide gel electrophoresis; DDMQ, demethyl-demethoxy-Q; DDMQH2, demethyl-demethoxy-QH2; DMQ, demethoxy-Q; DOD, drop-out growth medium with dextrose; 4HB, 4-hydroxybenzoic acid; HPLC, high performance liquid chromatography; IDMQ, 4-imino-demethoxy-Q; MRM, multiple reaction monitoring; MTase, methyltransferase; pABA, para-aminobenzoic acid; PK, proteinase K; Q, coenzyme Q or ubiquinone; QH2, coenzyme QH2, ubiquinol, or ubihydroquinone; SD, minimal synthetic media with dextrose; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis; TCA, trichloroacetic acid; YPD, rich growth medium with dextrose; YPG, rich growth medium with glycerol; YPGal, rich growth medium with galactose
Keywords: Human COQ gene, Mitochondrial metabolism, Protein complex, Q-biosynthetic intermediate, Saccharomyces cerevisiae, Ubiquinone
Graphical abstract
Highlights
-
•
Yeast Coq5 functions as a C-methyltransferase in coenzyme Q biosynthesis.
-
•
Human COQ5 is located within the mitochondria matrix of human cells.
-
•
Expression of human COQ5 partially rescues yeast coq5 point but not null mutants.
-
•
Human COQ5 rescues yeast coq5 null mutants provided that Coq8 is over-expressed.
-
•
Coq5 homologs rescue the coq5 yeast mutants when the CoQ-synthome is assembled.
1. Introduction
Coenzyme Q (ubiquinone or Q) is an essential lipophilic electron carrier found within the mitochondrial inner membrane of eukaryotes and the plasma membrane of many prokaryotes. Q is composed of a polyisoprenoid “tail” that anchors it to the lipid membrane, and a benzoquinone “head” that confers the abilities to shuttle electrons from Complexes I and II to Complex III [1], [2]. Q also acts as a cofactor of uncoupling proteins and several mitochondrial dehydrogenases, and in its reduced or hydroquinone form can quench lipid radical species as an antioxidant [3], [4].
The biosynthesis of the isoprenoid tail derives from either the mevalonate or 1-deoxy-d-xylulose-5-phosphate pathways [5], with the number of isoprene units varying in different species: six in Saccharomyces cerevisiae, eight in Escherichia coli, and ten in humans. In S. cerevisiae, the hexaprenyl diphosphate tail is attached to either 4-hydroxybenzoic acid (4HB) or para-aminobenzoic acid (pABA), both of which have been shown to serve as aromatic ring precursors in S. cerevisiae Q6 biosynthesis [6], [7].
Eleven S. cerevisiae genes (COQ1-9, ARH1, YAH1) are required for Q6 biosynthesis, several of which encode proteins associated with the mitochondrial inner membrane in a multi-subunit complex termed the CoQ-synthome [8]. In the absence or deficiency of any one of these genes, Q6 is not made and growth on medium containing a non-fermentable carbon source is not possible; the presence of each Coq polypeptide is essential for the assembly of the Coq polypeptide complex and for the proper function of each individual enzyme [9]. The over-expression of COQ8, which encodes a putative regulatory kinase, has been shown to restore steady state levels of several Coq polypeptides [10] and the assembly of the high molecular mass CoQ-synthome [8].
In humans, decreased levels of Q10 are associated with mitochondrial, cardiovascular, kidney and neurodegenerative diseases [3], [11]. Mutations in COQ genes cause primary Q10 deficiency (OMIM #607426), one of the few treatable mitochondrial disorders; in fact some affected patients respond well to oral Q10 supplementation [12]. COQ genes are highly conserved throughout evolution, and several human COQ genes have complemented the corresponding yeast coq null mutant [11], [13]. Previously, we have shown that expression of human ADCK3 (a yeast Coq8 ortholog) fused with an N-terminal yeast mitochondrial leader sequence rescued the growth of yeast coq8 null mutants and restored de novo Q biosynthesis [14].
In this study, we report the cloning and functional characterization of the human ortholog of yeast COQ5 and test its ability to complement yeast coq5 point and null mutants. Coq5 catalyzes the only C-methylation involved in the synthesis of Q6 in yeast [15]. Previous work in yeast has shown that certain point mutations within or adjacent to conserved COQ5 methyltransferase motifs result in a loss of Coq5 methyltransferase function, but mutants harboring these alleles (coq5-2, coq5-5) still retain steady state levels of Coq5 protein [15], [16]. Complementation of these yeast point mutants with ubiE, an E. coli COQ5 homolog, restored respiration and C-methyltransferase activity [16]. Here, we examine the function of human COQ5, and show that expression of human COQ5 in yeast mutants identifies the functional conservation of the yeast and human Q biosynthetic pathways, with implications for the diagnosis and treatment of Q10 deficiencies in patients.
2. Material and methods
2.1. Yeast strains and growth media
S. cerevisiae strains used in this study are listed in Table 1. Media were prepared as described [17], and included: YPD (2% glucose, 1% yeast extract, 2% peptone), YPGal (2% galactose, 1% yeast extract, 2% peptone, 0.1% glucose), and YPG (3% glycerol, 1% yeast extract, 2% peptone). Synthetic dextrose/minimal medium (SD, SD–Ura, and SD–Ura–Leu) consisted of 0.18% yeast nitrogen base without amino acids, 2% dextrose, 0.14% NaH2PO4, and 0.5% (NH4)2SO4, and amino acids (minus uracil or leucine for selective media) were added at final concentrations as described in [15]. Drop-out media with dextrose (DOD) lacking folate and para-amino benzoic acid and with proper amino acid selection were prepared as described in [18]. Plate media contained 2% bacto agar.
Table 1.
Genotype and source of yeast strains.
| Strain | Genotype | Source or reference |
|---|---|---|
| W303-1A | MAT aade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 | R. Rothsteina |
| W303ΔCOQ5 | MAT aade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 coq5::HIS3 | [15] |
| CH83-B3 | MAT α ade2-1 coq5-2 his3-1,15 ura3-1 | [16] |
| CH316-6B | MAT α coq5-5 trp1-1 ura3-1 | [16] |
Dr. Rodney Rothstein, Department of Human Genetics, Columbia University.
2.2. Identification of human COQ5
The human COQ5 gene (hCOQ5) cDNA sequence was identified by screening the Expressed Sequence Tag (EST) database with the tBLASTn algorithm (www.ncbi.nlm.nih.gov/blast) and the yeast protein sequence as bait [19].
2.3. Plasmid construction
Plasmids used in this study are listed in Table 2. Human COQ5 was amplified from cDNA obtained from human skin fibroblasts [20] using primers COQ5F and COQ5-1077R. PCR products were cloned in pCRII TOPO (Invitrogen), and the high copy pYES2.1V5His yeast expression vectors (Invitrogen). The COQ5 insert from pCRIITOPO-COQ5 was then cloned into the centromeric pCM189 vector (EUROSCARF) [21]. Yeast COQ5 (yCOQ5) was amplified from genomic DNA obtained from a wild-type BY4741 strain using standard protocols and cloned into the same vectors. Amplification primers and conditions for all reactions are shown in Table S1. The hybrid yeast–human COQ5 (yhCOQ5) gene was obtained by amplifying a 5′ segment of yCOQ5, corresponding to the mitochondrial targeting region (encoding aa 1–54; with primers yCOQ5F and hybridCOQ5R) and the 3′ of human COQ5 (encoding aa 56–327; with primers hybridCOQ5F and COQ5-1037R). The two PCR products were joined with a sequential PCR protocol [22].
Table 2.
Plasmids.
| Plasmids | Relevant genes/markers | Source or reference |
|---|---|---|
| Yeast vectors | ||
| pyCOQ5 | pCM189 with S. cerevisiae COQ5, low copy | This work |
| phCOQ5 | pCM189 with human COQ5, low copy | This work |
| pyhCOQ5 | pCM189 with the first 55 amino acids of human COQ5 replaced with the first 54 amino acids of S. cerevisiae Coq5 | This work |
| pYES-hCOQ5 | pYES2.1V5His TOPOwith human COQ5 high copy | This work |
| pUE3 | pQM with E. coli ubiE; low copy | [29] |
| p4HN5 (hcCOQ8) | pRS426 with yeast ABC1/COQ8; high copy | C. He a |
| pRS316 (vector) | pRS316; low copy | Addgene |
| Human vectors | ||
| pCOQ4-V5 | Human COQ4-V5 fusion cloned in pCDNA3.1TOPO | [20] |
| pCOQ5-V5 | Human COQ5-V5 fusion cloned in pCDNA3.1TOPO | This work |
| pCOQ5-myc | Human COQ5-myc fusion cloned in pCDNA3.1TOPO | This work |
| pCOQ5-GFP | Human COQ5-GFP fusion cloned in pEGFPN1 (Clontech) | This work |
Cuiwen H. He, Department of Chemistry and Biochemistry, UCLA.
To generate the COQ5-GFP fusion gene, the human COQ5 coding region (devoid of the termination codon) was amplified from pCRIITOPO-COQ5 with primers containing HindIII and PstI restriction sites, digested with HindIII and Pst1, and cloned into the pEGFP-N1 vector (Clontech) digested with the same enzymes. Finally a myc-tag was added to the C-terminus of hCOQ5 by PCR with primers COQ5F and COQ5mycR (Table S1) and cloned into the pCDNA3.1V5HisTOPO vector (Invitrogen) to yield pCOQ5-myc. All plasmid-constructs were sequenced to confirm the presence of the inserted DNA segments and to ascertain they were free of errors.
2.4. Northern blot and RACE
The probe for Northern blot analysis was obtained by EcoRI digestion of pCRII TOPO-COQ5 and was labeled and purified as described in [19] and hybridized to a commercial membrane (FirstChoice Human Blot 1 membrane–Ambion) containing 2 μg/lane of poly(A) + RNA from ten human tissues, previously used as described [23] and stripped. Radioactivity was detected with a Storm PhosphoImager (Molecular Dynamics, Sunnyvale, CA, USA) after an overnight exposure. The 5′ and 3′ termini of COQ5 transcripts were characterized by RACE (Rapid Amplification of cDNA Ends), a protocol that has been detailed elsewhere [19]. RNA was extracted from cultured skin fibroblasts and from HeLa cells. Primers are reported in Table S1.
2.5. Localization of human COQ5 polypeptide in HeLa and HEK cells
The COQ5-GFP construct was used to transfect HeLa cells stably expressing mitochondrial Red Florescent Protein (mtRFP) as previously reported [20]. Procedures for mitochondrial isolation, proteinase K protection assays, and carbonate extraction were performed as described [24]. Antibodies used are described in Table 3. Tagged versions of COQ5 were employed since our anti-COQ5 antibody detected only very faint signals in cultured cells.
Table 3.
Description and source of antibodies.
| Antibody | Source |
|---|---|
| Atp2 | Carla. M. Koehlera |
| Coq5 (yeast) | [16] |
| hCOQ5 (human) | Santa Cruz Biotechnology |
| Coq9 | [9] |
| GRP75 | Santa Cruz |
| OPA1 | BD Biosciences |
| OAT | Abcam |
| Porin | Mitoscience |
| SDHA | Molecular Probes |
| TOM20 | Santa Cruz |
| myc | Sigma |
| V5 | Invitrogen |
Dr. Carla. M. Koehler, Department of Chemistry and Biochemistry, UCLA.
2.6. Immunoprecipitation of hCOQ5 from cultured cells
HEK 293 cells were co-transfected with the pCOQ5-myc and pCOQ4-V5, and were harvested after 48 h and mitochondria were isolated as above. About 3–4 mg of mitochondria were resuspended in 1 ml of lysis buffer (150 mM NaCl; 10 mM Tris/HCl pH 7,4; 1 mM EDTA; 0.5% Triton X-100; protease and phosphatase inhibitors) and incubated with 400 μl of protein A-Sepharose (GE Lifesciences) 50% (v/v) for 1 h at 4 °C. An antibody-protein A-Sepharose 50% slurry was obtained by crosslinking anti-V5, anti-myc antibodies (or the preimmune serum), and incubated with 500 μl of the mitochondrial lysate overnight at 4 °C with rotation. The immune-complex was washed with lysis buffer 0.1% Triton X-100 and PBS (Life Technologies). The immune-precipitated proteins were eluted with SDS-Laemmli buffer without β-mercaptoethanol and heated for 10 min at 95 °C. The supernatant was collected, β-mercaptoethanol was added to a final concentration of 4%, heated again, and subjected to SDS-PAGE. Proteins were transferred to membranes, which were then probed with either anti-V5 or anti-myc antibodies.
2.7. Functional complementation of the yeast coq5 mutant
Yeast transformations with the plasmids described in Table 2 were performed with the PEG-lithium acetate method [25]. Transformed yeast strains were selected and maintained in SD–Ura or SD–Ura–Leu medium. For plate dilution assays, each strain was cultured overnight in synthetic dextrose medium with proper amino acid selection. Optical densities (A600 nm) of harvested cells were adjusted to 0.2, and 2 μl of 1:5 serial dilutions were spotted onto YPG, YPD, and SD plate media, corresponding to a final A600 nm of 0.2, 0.04, 0.008, 0.0016, and 0.00032.
2.8. Lipid extraction and quantification of Q6 by HPLC and tandem mass spectrometry
To analyze Q6 content, yeast cells were seeded in SD with proper amino acid selection at 0.2 OD/ml and collected after 3.5 to 4 h during log phase (a total of 15–30 A600 nm were collected). Lipid extraction and quantification of Q6 in yeast cell lipid extracts were determined by HPLC/MS–MS as described [6], [10]. Q4 was used as internal standard (expected final concentration, 1 pmol/μl upon analysis). For de novo Q6 labeling experiments, yeast were grown to log phase in SD medium with proper selection and then seeded at 0.07 OD/ml into DOD medium with proper amino acid selection supplemented with 8 μg/ml 13C6-pABA or 13C6-4HB (Cambridge Isotopes). Cultures were grown for 10 h to late log phase, and lipid extraction and quantification of 13C6–Q6 was determined by HPLC/MS–MS as described [6], [10].
2.9. Preparation of mitochondria from yeast
The yeast strains were grown in selective media to log phase. An aliquot of the log phase culture was seeded into 600 ml YPGal + 0.1% dextrose to a final density of 0.05 OD600 nm and incubated with shaking (250 rpm, 30 °C). The cells were harvested at OD600 nm between 2 and 3. The purified mitochondria were isolated as described [18], [26] in the presence of 1× EDTA-free protease inhibitor (Roche) and 1:100 phosphatase inhibitor cocktail sets I and II (Calbiochem). Pure mitochondria were flash frozen in liquid nitrogen and stored at − 80 °C. The protein concentration was measured with a bicinchoninic acid assay with bovine serum albumin as standard (Thermo Scientific).
2.10. Two-dimensional blue-native SDS/PAGE
Aliquots of purified mitochondria (200 μg protein) were solubilized in 50 μl of 1.2% digitonin (Biosynth AG), 1 × protease inhibitor (Roche), 1:100 phosphatase inhibitor cocktail sets I and II (Calbiochem) and 1 × NativePAGE sample buffer (Invitrogen). Samples were incubated on ice for 1 h and mixed by pipetting up and down every 10 min. The soluble supernatant fraction was separated from the insoluble pellet by centrifugation in a Beckman Airfuge (100,000 ×g, 10 min, chilled rotor). NativePAGE 5% G-250 sample additive (Invitrogen) was added to the supernatant from 200 μg of digitonin-solubilized mitochondria (50 μl) to a final concentration of 2.5%. BN-PAGE was performed as described in NativePAGE user manual with NativePAGE 4–16% Bis–Tris gel 1.0 mm × 10 wells (Invitrogen). First dimension gel slices were soaked in heated 2× SDS sample buffer for 20 min before loading onto pre-cast 10% SDS-polyacrylamide gels. Proteins were transferred to Immobilon-P transfer membrane (Millipore), and blocked in 1% skim milk, phosphate-buffered saline, and 0.1% Tween-20 (phosphate buffered saline, pH 7.4, is composed of 0.141 M NaCl, 1.2 mM NaH2PO4, and 8.1 mM Na2HPO4). Membranes were treated with the following primary antibodies (Table 3): human Coq5, 1:2500, Coq5, 1:5000, Coq9, 1:1000; Atp2, 1:4000. Goat anti-rabbit secondary antibodies conjugated to horseradish peroxidase (Calbiochem) were used at a 1:10,000 dilution and visualized using the Alpha Innotech FluorChem FC2 Imaging System.
3. Results
3.1. Identification and genetic characterization of human COQ5
The gene encoding human COQ5 spans to 26 kb on chromosome 12q24.31 and is comprised of seven exons. The open reading frame is 981 nucleotides long encoding a predicted 327 amino acid protein which contains a putative mitochondrial targeting sequence and shares 45% identity and 60% similarity with the yeast protein (excluding the putative targeting region). Fig. 1B shows the amino acid alignment of COQ5 from different species. Northern blot analysis detected a single human COQ5 transcript of about 1.5 kb expressed in all tissues tested (Fig. 2A). Human COQ5 steady-state RNA levels were higher in liver, lung, placenta, and skeletal muscle, similar to what was detected for COQ6[23] (Fig.S1). RACE analysis on mRNA extracted from primary skin fibroblast detected a single transcription initiation site located 14 bp upstream of the ATG initiation codon, while the 3′ untranslated region (UTR) extends to 501 nucleotides downstream of the termination signal, and contains a non-canonical AGTAAA polyadenylation signal (Fig. 2B). The size of the transcript corresponds to the hCOQ5 RNA observed by Northern blot. There was no bioinformatics evidence of COQ5 pseudogenes, or other genes with significant similarity to hCOQ5. A search of the human EST database showed evidence of a second transcript that includes an additional 90 bp exon between exon 1 and exon 2 (Fig. 2C). This exon contains a termination codon so the predicted protein encoded by this transcript (which we have termed COQ5 isoform 2) is only 70 aa. However, we did not detect this transcript in RNA extracted from cultured skin fibroblasts or skeletal muscle.
Fig. 1.
Biosynthesis of Q in S. cerevisiae and amino acid alignment of COQ5 homologs. (A) The benzoquinone head group of Q derives from either the 4HB or pABA aromatic ring precursor, is prenylated by Coq2 and further modified by Coq polypeptides to make the Q6-intermediate demethyl-demethoxy-hydroquinone (DDMQ6H2). Coq5 catalyzes the transfer of a methyl group from AdoMet to the C2 position of the hydroquinone ring to make demethoxy-Q6 (DMQ6H2), which is ultimately converted into the fully substituted Q6H2. (B) Amino acid alignments of COQ5 homologs were generated by MultAlin. In red are highly conserved residues, in blue, moderately conserved residues. Methyltransferase motifs [38], [39] are designated by the dashed-line boxes. The aligned sequences include: Homo sapiens COQ5 (84274), Mus musculus Coq5 (52064), Gallus gallus COQ5 (416975), Drosophila melanogaster CG2453 (32272), Arabidopsis thaliana AT5G57300 (835835), Caenorhabditis elegans COQ-5 (176099), Saccharomyces cerevisiae Coq5 (854930), and Escherichia coli UbiE (12932088). Single unique nucleotide substitutions in coq5 mutant alleles producing amino acid substitutions are designated by arrowheads (▼) on the figure as G596A (G199D) for coq5-2, G911A (G304D) for coq5-3, G279A (W93STOP) for coq5-4, and G361A (G121R) for coq5-5. S. cerevisiae Coq5 residues making contacts with S-adenosyl-l-methionine [35] are identified (red triangles). The yeast DNA segment encoding the amino terminal 54 residues (underlined) replaced the corresponding segment of human COQ5 DNA to generate the hybrid yeast/human gene.
Fig. 2.
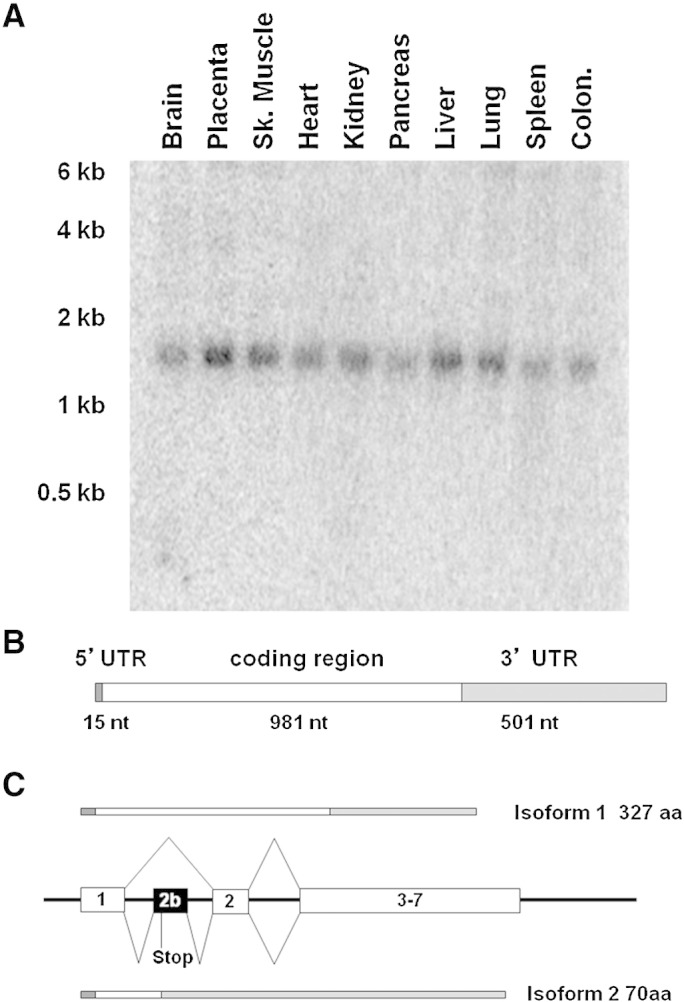
Characterization of RNA steady-state levels, transcript isoforms, and gene structure of human COQ5. (A) Northern blot analysis of COQ5 expression in different issues. Each lane contains 200 ng of polyA + mRNA. (B) Schematic structure of the principal transcript of human COQ5 as determined by RACE experiments. (C) Genomic structure of the 5 ′ region of COQ5 showing the alternatively spliced exon 2b and the resulting proteins.
3.2. hCOQ5 encodes a mitochondrial matrix protein associated with hCOQ4
To determine the subcellular localization of human COQ5, we transfected HeLa cells stably expressing mitochondrial-targeted RFP [20] with a construct expressing a hCOQ5-GFP fusion protein. As seen in Fig. 3A, the green and red fluorescence signals coincide, indicating a mitochondrial localization of hCOQ5. To determine the precise sub-mitochondrial localization of hCOQ5, mitochondria prepared from cultured HEK293 cells transfected with pCOQ5-myc were subjected to treatment with proteinase K under different conditions (hypotonic buffer to disrupt outer membrane, or triton X-100 to disrupt both outer and inner membranes). Extracts were separated by SDS-PAGE and immunoblot analyses were performed with antibodies recognizing TOM20 (outer membrane protein), OPA1 (intermembrane space), GRP75 (matrix enzyme) and COQ5Myc (Fig. 3B, Table 3). The hCOQ5 signal disappears only after treatment with triton X-100 indicating that hCOQ5 is localized to the mitochondrial matrix, similar to the GRP75 matrix marker.
Fig. 3.
Human COQ5 is located within the mitochondrial matrix and is peripherally associated with the inner membrane. (A) Co-localization of COQ5-GFP fusion proteins transiently expressed from pCOQ5-GFP in HeLa cells stably expressing mitochondria targeted RFP (mtRFP). (B) Proteinase K (PK) protection assays. Mitochondria were subjected to PK treatment directly after isolation and only outer membrane proteins are degraded (e.g. TOM20). Alternatively, mitochondria were treated with PK after treatment with hypotonic solution, which causes swelling (SW), and renders proteins of the inter-membrane space (e.g. OPA1) accessible to PK; or after treatment with a detergent (Triton X-100) which disrupts all mitochondrial membranes and allows PK degradation of all compartments including matrix proteins (e.g. GRP75). The susceptibility of COQ5 to PK follows the same pattern as GRP75, a mitochondrial matrix protein. (C) Membrane association assays. Mitochondrial membranes were prepared by sonication followed by centrifugation (12,000 g for 10 min) to generate pellet and supernatant fractions: isotonic buffer conditions (SON), NaCO3 treatment (which solubilizes peripheral membrane proteins), or Triton X-100 (which solubilizes integral membrane proteins). Aliquots of the supernatants (S) and pellets (P) were separated by SDS-PAGE and proteins were detected with the designated antibodies. A sample of the total membrane preparation (T) was included as control. (D) Mitochondrial extracts of HEK293 cells co-expressing COQ5-myc and COQ4-V5 were immune-precipitated with anti-myc, anti-V5 or with pre-immune sera. Immune-precipitates were separated by SDS-PAGE, transferred to membranes and the tagged-polypeptides were detected with either anti-V5 or anti-myc antibodies.
To determine whether hCOQ5 is associated with the mitochondrial inner membrane, mitochondrial extracts were treated with sonication, sodium carbonate, or Triton X-100 (Fig. 3C). After treatment samples were precipitated with TCA and centrifuged. Aliquots of the supernatant and of the resuspended pellets were then separated with SDS-PAGE and polypeptides were detected with immune-blot analyses. Membranes were assayed with antibodies recognizing porin (an integral membrane protein), SDHA (a peripheral membrane protein), and COQ5Myc. The association of hCOQ5 to the membrane is looser than SDHA, since sonication itself partially released the protein (Fig. 3C). Overall these results indicate that hCOQ5 is peripherally associated with the mitochondrial inner membrane on the matrix side, and confirm the findings of Chen et al. [27].
In yeast, Coq4 is a crucial organizer of the CoQ-synthome, a multi-subunit complex required for Q biosynthesis [8]. To determine the association of COQ4 with COQ5 in human cells, we performed immune-precipitation assays. Tagged versions of COQ4-V5 and COQ5-myc were co-expressed in HEK293 cells. After cell harvesting, mitochondrial lysates were immune-precipitated with either anti-V5 or anti-myc antibodies. Immune-precipitates were separated by SDS-PAGE, and the proteins transferred to a membrane that was then probed with antibodies to the V5- or myc-epitopes. Fig. 3D shows that immune-precipitation of COQ4-V5 captures COQ5-myc and vice versa, indicating that these COQ proteins physically associate in a complex in human cells.
3.3. Expression of human or E. coli COQ5 homologs rescues yeast coq5Δ mutants over-expressing COQ8
We examined if expression of hCOQ5 could complement the Q6 biosynthetic defect in coq5Δ mutant yeast. Human COQ5 expressed from either low- or high-copy expression plasmids failed to rescue growth of coq5Δ mutant yeast on medium containing glycerol as the sole carbon source (Fig. 4 and data not shown). Human proteins are sometimes not properly imported into yeast mitochondria [14], [23]. Thus, a hybrid gene construct was prepared in which amino acids 1–55 of hCOQ5 were replaced with the first 54 amino acids of S. cerevisiae Coq5. This construct is referred to as yeast–human hybrid COQ5 or yhCOQ5 (Table 2). As shown by a plate dilution assay, neither hCOQ5 nor yhCOQ5 rescues the growth of yeast coq5Δ on non-fermentable YPG medium (Fig. 4A). This result corroborates previous studies demonstrating that expression of the E. coli Coq5 homolog, UbiE, also failed to rescue growth of the yeast coq5Δ mutant on YPG medium [16].
Fig. 4.
COQ5 homologs partially restore growth on respiratory medium and Q6-content in yeast coq5 null mutants over-expressing COQ8. Yeast coq5 null mutants (W303ΔCOQ5) were transformed with the designated plasmids without (A) or with (B) a high copy COQ8 plasmid (hcCOQ8). Each mutant strain was cultured overnight in SD-selective media, adjusted to 0.2 optical density (OD), and 2 μl of 1:5 serial dilutions was spotted onto plate media. Plates are depicted after 3 days of incubation at 30 °C. (C) Yeast were seeded in SD–Ura at 0.2 OD and collected after 4.5 h. Q6 content in cell lipid extracts was determined by HPLC/MS–MS as described [10] with a limit of detection of 10 fmol/μl. Each bar represents a total of four measurements from two independent samples each with two injections. Error bars represent standard deviations.
We therefore asked whether the problem could be related to a failure of the human COQ5 protein to stabilize the Coq polypeptide complex in yeast coq5Δ cells. Recent work has shown that the over-expression of the putative Coq8 kinase in certain coq null mutants allows synthesis of late-stage Q-intermediates, restores steady-state levels of Coq polypeptides, and their association in a high molecular mass complex known as the CoQ-synthome [8], [10], [28]. To examine rescue of coq5Δ in the presence of the CoQ-synthome, yeast Coq8 was over-expressed on a high copy plasmid (hcCOQ8) and strains were grown on selective media (Fig. 4B). Co-expression of yeast Coq8 with either yhCOQ5 or E. coli UbiE, both of which contain an amino-terminal yeast mitochondrial leader sequence, rescues growth of coq5Δ on YPG medium (Fig. 4B). The rescue by hCOQ5 is less robust, suggesting that a yeast mitochondrial leader sequence is critical for efficient import of hCOQ5 into yeast mitochondria.
To examine Q6 levels, lipids were extracted from yeast cells and subjected to HPLC and tandem mass spectrometry [10]. In the absence of yeast Coq8 over-expression, Q6 was not detected in any of the coq5Δ mutant yeast strains (Fig. 4C). However, in the presence of yeast Coq8 over-expression, the Q6 content is partially restored in strains expressing E. coli UbiE, yhCOQ5, or hCOQ5 (Fig. 4C). These results indicate that human and E. coli Coq5 homologs can rescue the defect in the yeast coq5Δ mutant only when other yeast Coq polypeptides are stabilized by Coq8 over-expression.
3.4. Expression of human and E. coli COQ5 homologs partially restores Q6-content in coq5-2 and coq5-5 point mutants
To further examine whether expression of human COQ5 homologs is able to rescue coq5 mutant strains retaining a stable CoQ-synthome, yeast coq5 strains with point mutations within or adjacent to one of the methyltransferase motifs were employed (Fig. 1B). Two point mutants, coq5-2 (CH83-B3) and coq5-5 (CH316-6B) lack C-methyltransferase activity and Q6 biosynthesis but retain steady state levels of Coq5 and other Coq polypeptides [15], [16], [29]. Expression of E. coli UbiE, hCOQ5 or yhCOQ5 in these yeast coq5 point mutants rescued growth on rich glycerol (YPG) medium and partially restored Q6 content (Fig. 5).
Fig. 5.
COQ5 homologs partially restore growth on respiratory medium and Q6-content in yeast coq5 point mutants. (A and B) Yeast were seeded in SD–Ura at 0.2 OD and collected after 3 h for Q6 analysis and plate dilution assays. Plates are depicted after 3 days of incubation at 30 °C. (C) Q6 content in cell lipid extracts was determined by HPLC/MS–MS as described [10]. Limit of detection was 50 amol/μl. Each bar represents a total of four measurements from two independent samples each with two injections. Error bars represent standard deviations.
Expression of hCOQ5 results in detectable growth in the coq5-5 mutant on respiratory media even though the content of Q6 ranges from 0.4% to 3% of wild-type Q6 content (Fig. 5C). Serial plate dilution assays show that growth on respiratory medium requires only a small amount of Q6, as restoration of a low Q6 content is sufficient to rescue plate growth of the coq5-2 and coq5-5 point mutants (Fig. 5).
3.5. Yeast coq5-5 mutants rescued with human COQ5 homologs show low de novo Q6 biosynthesis and accumulation of Q6-intermediates
The Q6 levels measured in Figs. 4C and 5C account for total content of Q6 present at the time of harvest; to assay the ability of mutants to synthesize new Q6 de novo, coq5-5 point mutants harboring hCOQ5 or yhCOQ5 were grown in medium containing either 13C6-pABA or 13C6-4HB. pABA and 4HB are two aromatic ring precursors of yeast Q6 biosynthesis, and inclusion of 13C6-pABA or 13C6-4HB will result in the new synthesis of 13C6–Q6 detectable by its additional mass of + 6 indicating the incorporation of a 13C6-label. The yeast coq5-5 point mutant rescued with either hCOQ5 or yhCOQ5 was able to synthesize similar amounts of 13C6–Q6 from either 13C6-pABA (Fig. 6A) or 13C6-4HB (Fig. 6B). However, the Q6 synthesis was much less efficient as compared to either wild-type yeast or to the coq5-5 mutant rescued by yeast COQ5.
Fig. 6.
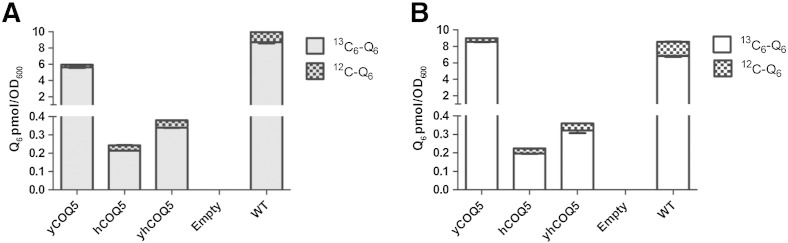
COQ5 homologs partially restore de novo Q6 content of yeast coq5-5 mutants. Yeast cells were seeded at 0.07 OD into DOD medium supplemented with (A) 13C6-pABA or (B) 13C6-4HB. After a 10-h incubation with label at 30 °C, 13C6–Q6 lipid extracts were determined by HPLC/MS–MS as in [10] with a limit of detection of 10 fmol/μl. Each bar represents two measurements from one sample. Error bars represent standard deviations.
The low content of newly synthesized 13C6–Q6 in the yeast coq5-5 point mutants expressing hCOQ5 or yhCOQ5 suggested that Q6-intermediates might accumulate in these strains. To determine this, lipid extracts were examined for the presence of 13C6–Q6 intermediates. Of particular interest is the substrate of Coq5, demethyl-demethoxy-Q6 (DDMQ6) (Fig. 1A). DDMQ6 was first detected in coq5Δ strains over-expressing Coq8 [10]. DDMQ6 is not detected in wild-type yeast, presumably because it is quickly methylated in the production of Q6. We therefore determined whether DDMQ6 could be detected in strains with less efficient Q6 biosynthesis, for example in yeast coq5-5 mutants expressing hCOQ5 or yhCOQ5.
The yeast coq5-5 mutant harboring yhCOQ5 was cultured in the presence of 13C6-pABA. Lipid extracts were prepared and subjected to reversed phase HPLC–MS/MS as described (Section 2.8). In extracts prepared from 13C6-pABA-labeled yeast cells, a predicted precursor-to-product ion transition for 13C6-labeled DDMQ6 eluted at 4.69 min. The fragmentation spectrum (Fig. 7) revealed the 13C6-DDMQ6 [M + H]+ precursor ion, the 13C6-DDMQ6 tropylium product ion [M]+, and the 13C6-DDMQ6 chromenylium product ion [M]+.
Fig. 7.
Identification of de novo demethyl-demethoxy-Q6 (DDMQ6) in lipid extracts of coq5-5:yhCOQ5 yeast cells. 13C6-DDMQ6 was detected in the lipid extracts of coq5-5:yhCOQ5 cells grown in the presence of 13C6-pABA as described in Materials and methods (Section 2.8). The 13C6-DDMQ6 [M + H]+ precursor ion (13C612C31H55O3+; monoisotopic mass 553.4), the 13C6-DDMQ6 tropylium product ion [M]+ (13C612C2H9O3+; monoisotopic mass 159.05), and the 13C6-DDMQ6 chromenylium product ion [M]+ (13C612C5H13O3+; monoisotopic mass 199.1) are consistent with the presence of 13C6-aromatic ring label as denoted by red asterisks.
The levels of the DDMQ6 and demethoxy-Q6 (DMQ6) were next examined in yeast coq5 mutants expressing yhCOQ5 or E. coli UbiE (Fig. 8). DMQ6 is detected in Q6-producing yeast strains characterized so far and is a reflection of the Q6-content [30]. In mutants rescued by yeast COQ5 (blue trace in Fig. 8), DMQ6 is detected, but not DDMQ6, suggesting that yeast Coq5 efficiently methylates DDMQ6 to make DMQ6. In contrast, the yeast coq5-5 mutant harboring the empty vector control accumulates DDMQ6 (gray trace in Fig. 8A and C) but DMQ6 is not detected (gray trace, Fig. 8B and D). Notably, coq5-5 mutants harboring either yhCOQ5 or UbiE accumulate DDMQ6 but also make small amounts of DMQ6 (green and red traces, Fig. 8). We reason that the expression of yhCOQ5 or UbiE results in slower, less efficient methylation of DDMQ6, leading to an accumulation of DDMQ6 and lower levels DMQ6. These data indicate that in this system yhCOQ5 is less efficient than yCOQ5 as a C-methyltransferase.
Fig. 8.
Yeast coq5 mutants rescued with COQ5 homologs accumulate Q6-intermediate DDMQ6. The designated yeast coq5 mutants were seeded in selective SD medium at 0.2 OD, collected after 3 h, and lipids were extracted for Q6 analysis. MRM detected precursor-to-product ion transition 547.4/153.1 (DDMQ6) in panels (A) and (C); and precursor-to-product ion transition 561.4/167.0 (DMQ6) in panels (B) and (D). Red trace indicates the presence of the yeast-human hybrid COQ5, blue trace indicates yeast COQ5, green indicates E. coli UbiE, and gray indicates the empty vector control.
3.6. Human Coq5 polypeptide co-migrates with Coq9 in a high molecular mass complex
The lower methyltransferase efficiency of the human and yeast–human COQ5 orthologs elicits an interesting question: what interaction, if any, do COQ5 orthologs have with the yeast CoQ-synthome? We probed this question in a preliminary way by determining whether the human COQ5 polypeptide co-migrated with yeast Coq polypeptides at high molecular mass. Previous studies have shown that the Coq9 polypeptide co-purifies with other yeast Coq polypeptides and migrates at high molecular mass in solubilized extracts of mitochondria [9]. Here, the antibody against yeast Coq9 was used as a benchmark for the presence of high molecular mass CoQ-synthome [8], [9], [10].
Mitochondria were purified from wild-type yeast and from coq5Δ mutants over-expressing yeast Coq8 in combination with yhCOQ5, yeast Coq5, or empty vector, and the digitonin extracts were separated by 2D-BN-SDS/PAGE. We used a polyclonal antibody against hCOQ5 that identified yhCOQ5 expressed in the yeast coq5Δ mutant (Fig. S2). The yhCOQ5 polypeptide was detected at high molecular mass (~ 650 kDa), although most of the signal was observed in a broad region around 66 kDa (Fig. 9A). Yeast Coq9 was also found to migrate with a pattern similar to the yhCOQ5 polypeptide on the same immune-blot membrane (Fig. 9A). In the coq5Δ strain co-expressing high copy yeast Coq8 and yeast Coq5, the yeast Coq5 polypeptide was detected at high molecular mass complex together with yeast Coq9 (~ 300 kDa) (Fig. 9B). Both yeast Coq5 and Coq9 were also present at lower mass (Fig. 9B). In the coq5Δ strain co-expressing high copy yeast Coq8 and vector control, no Coq5 is detected, while Coq9 is observed at both low and high molecular weight (Fig. 9C). In wild-type yeast Coq9 is present across a wide range of high molecular mass complexes, while Coq5 is present mainly in a broad region around 66 kDa, perhaps indicating the presence of Coq5 and Coq9 in sub-complexes.
Fig. 9.
Human COQ5 polypeptide co-migrates with yeast Coq9 at high molecular mass. Purified mitochondria (200 μg of protein) from the designated yeast strains were separated by 2D-BN-SDS/PAGE, and the membranes containing transferred proteins were probed with antibodies against human COQ5, yeast Coq9 or yeast Coq5. Mitochondria were isolated from coq5 null with co-expression of Coq8 (hcCOQ8) and (A) yhCOQ5, (B) yCOQ5 and (C) empty vector. (D), Mitochondria were purified from wild-type yeast, W3031A. A sample of wild-type mitochondria separated just in the SDS-second dimension served as a positive control and is designated as WT Mito. The positions of molecular mass standards in the first dimension are indicated at the top of each panel.
4. Discussion
COQ5 encodes an essential protein involved in the biosynthetic pathway of Q6 in S. cerevisiae. It is the only known C-methyltransferase enzyme involved in Q6 synthesis, and catalyzes the methylation of demethyl-demethoxy-Q6H2 (DDMQ6H2) to form demethoxy-Q6H2 (DMQ6H2). In yeast, Coq5 and several COQ gene products are associated in the CoQ-synthome, a high molecular mass complex peripherally associated with the matrix side of the mitochondrial inner membrane [8]. In this study, we confirmed that the human protein is localized to the mitochondrial matrix and is peripherally associated with the inner membrane. We also showed that hCOQ5 physically associates with hCOQ4, indicating that a COQ multi-enzyme complex is also present in human cells.
Human COQ5 RNA was expressed ubiquitously in human organ samples as assayed by Northern blotting (Fig. 3). Steady state RNA levels for COQ2, COQ4, and COQ6 have been determined for a variety of human tissues [20], [23], [31]. In comparing the expression levels of these COQ genes, there does not seem to be a common pattern: COQ5 had the highest expression levels in the placenta, liver, skeletal muscle, and lung, COQ6 in skeletal muscle, kidney, and placenta, COQ2 in skeletal muscle and heart, while COQ4 RNA abundance was highest in lung, colon and pancreas.
A search of the EST database revealed a possible second transcript encoding a truncated form of COQ5. The presence of transcripts encoding non-functional proteins is not unusual for COQ genes and has been reported also for PDSS2, COQ4, and COQ6[20], [23], [32]. The functional significance of the truncated proteins is still unclear, although they could play a regulatory role [33].
Despite a relatively high degree of homology to its yeast counterpart, the human COQ5 protein failed to complement yeast mutants with a deletion in COQ5, as determined by a lack of growth on non-fermentable carbon source and absence of Q6. Rescue by human COQ5 was more efficient when the protein contained an appended S. cerevisiae Coq5 mitochondrial leader sequence. However, even under this condition, rescue was observed only in (1) yeast coq5 point mutants harboring stable but catalytically inactive Coq5 polypeptide, or (2) yeast coq5 null mutants over-expressing COQ8. We note that both conditions permit the stabilization of CoQ-synthome required for Q6 biosynthesis in yeast. The results here suggest that COQ5 orthologs can serve as functional C-methyltransferases and restore Q biosynthesis in the yeast coq5 mutants only when the other yeast Coq polypeptides are preserved.
Interestingly, the expression of human COQ5 was recently shown to complement a Schizosaccharomyces pombe coq5Δ mutant for growth in medium containing a nonfermentable carbon source [34]. Unlike the S. cerevisiae coq5Δ mutant, the S. pombe coq5Δ mutant contained peaks tentatively identified as late-stage Q-intermediates, suggesting that the S. pombe coq deletion mutants do not have the same drastic “early intermediate” phenotype as observed in the S. cerevisiae coq deletion mutants. Indeed, human COQ homologs were shown to rescue each of the S. pombe coq deletion mutants, with the exception of Coq9 [34]. For both S. pombe and S. cerevisiae, expression of human COQ5 results in production of a very small amount of Q, as compared to wild-type cells. Several interspecific complementation studies suggest that such rescue of yeast growth on nonfermentable carbon sources can be accomplished with less than 0.2% of the normal levels of Q [14], [23].
Yeast coq5-5 mutants rescued by yeast Coq5 are able to efficiently methylate DDMQ6H2 to make DMQ6H2 and high amounts of Q6H2. Due to autoxidation, the DDMQ6 and DMQ6 quinones are the predominant forms recovered in lipid extracts. In contrast, the accumulation of the Coq5 substrate DDMQ6 in yeast coq5-5 mutants expressing either human COQ5 or E. coli UbiE indicates a slower or less efficient methylation of DDMQ6H2, and is also consistent with the lower levels of both DMQ6 and Q6. A recent crystal structure of the S. cerevisiae Coq5 polypeptide showed the structure to be a dimer, and identified amino acid residues at the dimer interface domain [35]. The residues at the yeast Coq5 dimer interface are not as highly conserved between Coq5 homologs as the active-site residues that contact S-adenosyl-l-methionine [35]. We speculate that the interaction of the yeast Coq5 dimer with the yeast CoQ-synthome enhances the catalytic efficiency of the C-methylation, and that the partial rescue observed by the heterologous COQ5 polypeptides results from a less efficient or absence of interaction with the yeast CoQ-synthome.
To explore this idea we examined whether the human COQ5 homolog might interact with the yeast CoQ-synthome as determined by separation of digitonin solubilized mitochondrial extracts with 2D-BN-SDS/PAGE. Small amounts of the hCOQ5 polypeptide were observed at high molecular mass in the coq5 null mutant harboring yhCOQ5 with Coq8 over-expression. Similarly, small amounts of the yeast Coq5 polypeptide were observed at high molecular mass in the coq5 null mutant harboring yeast COQ5 with Coq8 over-expression. This may be due to the presence of over-expressed Coq8. However, we did not detect Coq5 co-migrating with other Coq polypeptides at a high molecular mass in mitochondria prepared from wild-type yeast cells (Fig. 9D and unpublished results). It is very challenging to recover the intact CoQ-synthome. The complex must be first solubilized from the inner mitochondrial membrane with detergent, and depending on the separation conditions, may behave very differently. For example, although we readily detect the presence of the Coq5 polypeptide in complexes captured with tagged forms of Coq9 [9], only very small amounts of Coq5 are detected at high molecular mass by 2D-blue native-SDS PAGE [8]. Tauche et al. observed a Coq3–Coq5–Coq9 complex with myc-tagged Coq polypeptides [36]. We do not always observe the Coq5 polypeptide at high molecular mass when separated by size exclusion chromatography [37]. Based on these observations we think that it is very likely that Coq5 readily dissociates from the CoQ synthome. Obviously, the absence of co-migration does not rule out the possibility of a transient or weak physical association.
A physical interaction between the human COQ5 and the yeast Coq polypeptide complex may not be necessary for partial rescue of Q6 content. The CoQ-synthome needs only to provide human COQ5 with its substrate DDMQ6H2, which may be accomplished with human COQ5 peripherally associated with the complex. Low Q6 content and the presence of Q6-intermediates suggest either inefficient or weak interaction with the Coq complex. The results presented here suggest that human and E. coli Coq5 homologs expressed in yeast retain C-methyltransferase function, but rescue the yeast coq5 mutants only in the presence of the assembled yeast CoQ-synthome.
The following are the Supplementary data related to this article.
Densitometric analyses of COQ5 RNA in human tissues. The Northern blot image was analyzed as described previously [23]. Densitometric signals for COQ5 RNA were normalized to beta actin expression (reported previously) [23] and expressed in arbitrary units (highest expression = 100).
Determination of antibody specificity to hCOQ5 and yeast Coq5 polypeptides. Purified mitochondria (35 μg protein) from W303ΔCOQ5 harboring hcCOQ8 and the designated COQ5 plasmids were subjected to immune-blot analysis. The membrane was first probed with antibodies to human COQ5, and stripped and re-probed with antisera to yeast Coq5. The membrane was then probed for Coq9 and Atp2 (beta subunit of the F1 sector of mitochondrial F1F0 ATP synthase).
Acknowledgements
We thank the members of the CF Clarke lab for their advice and input on this manuscript. We thank Dr. C. M. Koehler (UCLA) for the antibodies to yeast Atp2.
Footnotes
This work was supported in part by grants from Telthon Italy (GGP13222), the University of Padova (CPDA123573/12), and Fondazione CARIPARO (to L.S.); the Italian Ministry of Health (GR-2009-1578914) (to E.T.); and the National Science Foundation Grant MCB-1330803 (to C.F.C.); and by the National Institutes of HealthS10RR024605 from the National Center for Research Resources for the purchase of the LC–MS/MS system.
Contributor Information
Catherine F. Clarke, Email: cathy@chem.ucla.edu.
Leonardo Salviati, Email: leonardo.salviati@unipd.it.
References
- 1.Crane F.L., Hatefi Y., Lester R.L., Widmer C. Isolation of a quinone from beef heart mitochondria. Biochim. Biophys. Acta. 1957;25:220–221. doi: 10.1016/0006-3002(57)90457-2. [DOI] [PubMed] [Google Scholar]
- 2.Bentinger M., Tekle M., Dallner G. Coenzyme Q-biosynthesis and functions. Biochem. Biophys. Res. Commun. 2010;396:74–79. doi: 10.1016/j.bbrc.2010.02.147. [DOI] [PubMed] [Google Scholar]
- 3.Rotig A., Appelkvist E.L., Geromel V., Chretien D., Kadhom N., Edery P., Lebideau M., Dallner G., Munnich A., Ernster L., Rustin P. Quinone-responsive multiple respiratory-chain dysfunction due to widespread coenzyme Q10 deficiency. Lancet. 2000;356:391–395. doi: 10.1016/S0140-6736(00)02531-9. [DOI] [PubMed] [Google Scholar]
- 4.Bentinger M., Brismar K., Dallner G. The antioxidant role of coenzyme Q. Mitochondrion. 2007;7(Suppl.):S41–S50. doi: 10.1016/j.mito.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Nowicka B., Kruk J. Occurrence, biosynthesis and function of isoprenoid quinones. Biochim. Biophys. Acta. 2010;1797:1587–1605. doi: 10.1016/j.bbabio.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 6.Marbois B., Xie L.X., Choi S., Hirano K., Hyman K., Clarke C.F. Para-aminobenzoic acid is a precursor in coenzyme Q6 biosynthesis in Saccharomyces cerevisiae. 2010;285:27827–27838. doi: 10.1074/jbc.M110.151894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pierrel F., Hamelin O., Douki T., Kieffer-Jaquinod S., Mühlenhoff U., Ozeir M., Lill R., Fontecave M. Involvement of mitochondrial ferredoxin and para-aminobenzoic acid in yeast coenzyme Q biosynthesis. Chem. Biol. 2010;17:449–459. doi: 10.1016/j.chembiol.2010.03.014. [DOI] [PubMed] [Google Scholar]
- 8.He C.H., Xie L.X., Allan C.M., Tran U.C., Clarke C.F. Coenzyme Q supplementation or over-expression of the yeast Coq8 putative kinase stabilizes multi-subunit Coq polypeptide complexes in yeast coq null mutants. Biochim. Biophys. Acta. 2014;1841:630–644. doi: 10.1016/j.bbalip.2013.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsieh E.J., Gin P., Gulmezian M., Tran U.C., Saiki R., Marbois B.N., Clarke C.F. Saccharomyces cerevisiae Coq9 polypeptide is a subunit of the mitochondrial coenzyme Q biosynthetic complex. Arch. Biochem. Biophys. 2007;463:19–26. doi: 10.1016/j.abb.2007.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie L.X., Ozeir M., Tang J.Y., Chen J.Y., Jaquinod S.K., Fontecave M., Clarke C.F., Pierrel F. Overexpression of the Coq8 kinase in Saccharomyces cerevisiae coq null mutants allows for accumulation of diagnostic intermediates of the coenzyme Q6 biosynthetic pathway. J. Biol. Chem. 2012;287:23571–23581. doi: 10.1074/jbc.M112.360354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y., Hekimi S. Molecular genetics of ubiquinone biosynthesis in animals. Crit. Rev. Biochem. Mol. Biol. 2013;48:69–88. doi: 10.3109/10409238.2012.741564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salviati L., Sacconi S., Murer L., Zacchello G., Franceschini L., Laverda A.M., Basso G., Quinzii C., Angelini C., Hirano M., Naini A.B., Navas P., DiMauro S., Montini G. Infantile encephalomyopathy and nephropathy with CoQ10 deficiency: a CoQ10-responsive condition. Neurology. 2005;65:606–608. doi: 10.1212/01.wnl.0000172859.55579.a7. [DOI] [PubMed] [Google Scholar]
- 13.Tran U.C., Clarke C.F. Endogenous synthesis of coenzyme Q in eukaryotes. Mitochondrion 7. Supplement. 2007:S62–S71. doi: 10.1016/j.mito.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie L.X., Hsieh E.J., Watanabe S., Allan C.M., Chen J.Y., Tran U.C., Clarke C.F. Expression of the human atypical kinase ADCK3 rescues coenzyme Q biosynthesis and phosphorylation of Coq polypeptides in yeast coq8 mutants. Biochim. Biophys. Acta. 2011;1811:348–360. doi: 10.1016/j.bbalip.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barkovich R.J., Shtanko A., Shepherd J.A., Lee P.T., Myles D.C., Tzagoloff A., Clarke C.F. Characterization of the COQ5 gene from Saccharomyces cerevisiae. Evidence for a C-methyltransferase in ubiquinone biosynthesis. J. Biol. Chem. 1997;272:9182–9188. doi: 10.1074/jbc.272.14.9182. [DOI] [PubMed] [Google Scholar]
- 16.Baba S.W., Belogrudov G.I., Lee J.C., Lee P.T., Strahan J., Shepherd J.N., Clarke C.F. Yeast Coq5 C-methyltransferase is required for stability of other polypeptides involved in coenzyme Q biosynthesis. J. Biol. Chem. 2004;279:10052–10059. doi: 10.1074/jbc.M313712200. [DOI] [PubMed] [Google Scholar]
- 17.Burke D., Dawson D., Stearns T. Cold Spring Harbor Laboratory Press; Plainview, NY: 2000. Methods in Yeast Genetics. [Google Scholar]
- 18.Allan C.M., Hill S., Morvaridi S., Saiki R., Johnson J.S., Liau W.S., Hirano K., Kawashima T., Ji Z., Loo J.A., Shepherd J.N., Clarke C.F. A conserved START domain coenzyme Q-binding polypeptide is required for efficient Q biosynthesis, respiratory electron transport, and antioxidant function in Saccharomyces cerevisiae. Biochim. Biophys. Acta. 2013;1831:776–791. doi: 10.1016/j.bbalip.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sacconi S., Trevisson E., Pistollato F., Baldoin M.C., Rezzonico R., Bourget I., Desnuelle C., Tenconi R., Basso G., DiMauro S., Salviati L. hCOX18 and hCOX19: two human genes involved in cytochrome c oxidase assembly. Biochem. Biophys. Res. Commun. 2005;337:832–839. doi: 10.1016/j.bbrc.2005.09.127. [DOI] [PubMed] [Google Scholar]
- 20.Casarin A., Jimenez-Ortega J.C., Trevisson E., Pertegato V., Doimo M., Ferrero-Gomez M.L., Abbadi S., Artuch R., Quinzii C., Hirano M., Basso G., Ocana C.S., Navas P., Salviati L. Functional characterization of human COQ4, a gene required for coenzyme Q10 biosynthesis. Biochem. Biophys. Res. Commun. 2008;372:35–39. doi: 10.1016/j.bbrc.2008.04.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gari E., Piedrafita L., Aldea M., Herrero E. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast. 1997;13:837–848. doi: 10.1002/(SICI)1097-0061(199707)13:9<837::AID-YEA145>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 22.Paret C., Ostermann K., Krause-Buchholz U., Rentzsch A., Rodel G. Human members of the SCO1 gene family: complementation analysis in yeast and intracellular localization. FEBS Lett. 1999;447:65–70. doi: 10.1016/s0014-5793(99)00266-5. [DOI] [PubMed] [Google Scholar]
- 23.Heeringa S.F., Chernin G., Chaki M., Zhou W., Sloan A.J., Ji Z., Xie L.X., Salviati L., Hurd T.W., Vega-Warner V., Killen P.D., Raphael Y., Ashraf S., Ovunc B., Schoeb D.S., McLaughlin H.M., Airik R., Vlangos C.N., Gbadegesin R., Hinkes B., Saisawat P., Trevisson E., Doimo M., Casarin A., Pertegato V., Giorgi G., Prokisch H., Rotig A., Nurnberg G., Becker C., Wang S., Ozaltin F., Topaloglu R., Bakkaloglu A., Bakkaloglu S.A., Muller D., Beissert A., Mir S., Berdeli A., Varpizen S., Zenker M., Matejas V., Santos-Ocana C., Navas P., Kusakabe T., Kispert A., Akman S., Soliman N.A., Krick S., Mundel P., Reiser J., Nurnberg P., Clarke C.F., Wiggins R.C., Faul C., Hildebrandt F. COQ6 mutations in human patients produce nephrotic syndrome with sensorineural deafness. J. Clin. Invest. 2011;121:2013–2024. doi: 10.1172/JCI45693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doimo M., Desbats M.A., Baldoin M.C., Lenzini E., Basso G., Murphy E., Graziano C., Seri M., Burlina A., Sartori G., Trevisson E., Salviati L. Functional analysis of missense mutations of OAT, causing gyrate atrophy of choroid and retina. Hum. Mutat. 2013;34:229–236. doi: 10.1002/humu.22233. [DOI] [PubMed] [Google Scholar]
- 25.Gietz R.D., Woods R.A. Yeast transformation by the LiAc/SS carrier DNA/PEG method. Methods Mol. Biol. 2006;313:107–120. doi: 10.1385/1-59259-958-3:107. [DOI] [PubMed] [Google Scholar]
- 26.Glick B.S., Pon L.A. Isolation of highly purified mitochondria from Saccharomyces cerevisiae. Methods Enzymol. 1995;260:213–223. doi: 10.1016/0076-6879(95)60139-2. [DOI] [PubMed] [Google Scholar]
- 27.Chen S.W., Liu C.C., Yen H.C. Detection of suppressed maturation of the human COQ5 protein in the mitochondria following mitochondrial uncoupling by an antibody recognizing both precursor and mature forms of COQ5. Mitochondrion. 2013;13:143–152. doi: 10.1016/j.mito.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 28.Padilla S., Tran U.C., Jimenez-Hidalgo M., Lopez-Martin J.M., Martin-Montalvo A., Clarke C.F., Navas P., Santos-Ocana C. Hydroxylation of demethoxy-Q6 constitutes a control point in yeast coenzyme Q6 biosynthesis. Cell. Mol. Life Sci. 2009;66:173–186. doi: 10.1007/s00018-008-8547-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee P.T., Hsu A.Y., Ha H.T., Clarke C.F. A C-methyltransferase involved in both ubiquinone and menaquinone biosynthesis: isolation and identification of the Escherichia coli ubiE gene. J. Bacteriol. 1997;179:1748–1754. doi: 10.1128/jb.179.5.1748-1754.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Padilla S., Jonassen T., Jimenez-Hidalgo M.A., Fernandez-Ayala D.J., Lopez-Lluch G., Marbois B., Navas P., Clarke C.F., Santos-Ocana C. Demethoxy-Q, an intermediate of coenzyme Q biosynthesis, fails to support respiration in Saccharomyces cerevisiae and lacks antioxidant activity. J. Biol. Chem. 2004;279:25995–26004. doi: 10.1074/jbc.M400001200. [DOI] [PubMed] [Google Scholar]
- 31.Forsgren M., Attersand A., Lake S., Grunler J., Swiezewska E., Dallner G., Climent I. Isolation and functional expression of human COQ2, a gene encoding a polyprenyl transferase involved in the synthesis of CoQ. Biochem. J. 2004;382:519–526. doi: 10.1042/BJ20040261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saiki R., Lunceford A.L., Shi Y., Marbois B., King R., Pachuski J., Kawamukai M., Gasser D.L., Clarke C.F. Coenzyme Q10 supplementation rescues renal disease in Pdss2kd/kd mice with mutations in prenyl diphosphate synthase subunit 2. Am. J. Physiol. Renal Physiol. 2008;295:F1535–F1544. doi: 10.1152/ajprenal.90445.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doimo M., Trevisson E., Airik R., Bergdoll M., Santos-Ocana C., Hildebrandt F., Navas P., Pierrel F., Salviati L. Effect of vanillic acid on COQ6 mutants identified in patients with coenzyme Q10 deficiency. Biochim. Biophys. Acta. 2014;1842:1–6. doi: 10.1016/j.bbadis.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hayashi K., Ogiyama Y., Yokomi K., Nakagawa T., Kaino T., Kawamukai M. Functional conservation of coenzyme Q biosynthetic genes among yeasts, plants, and humans. PLoS One. 2014;9:e99038. doi: 10.1371/journal.pone.0099038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dai Y.N., Zhou K., Cao D.D., Jiang Y.L., Meng F., Chi C.B., Ren Y.M., Chen Y., Zhou C.Z. Crystal structures and catalytic mechanism of the C-methyltransferase Coq5 provide insights into a key step of the yeast coenzyme Q synthesis pathway. Acta Crystallogr. D Biol. Crystallogr. 2014;70:2085–2092. doi: 10.1107/S1399004714011559. [DOI] [PubMed] [Google Scholar]
- 36.Tauche A., Krause-Buchholz U., Rodel G. Ubiquinone biosynthesis in Saccharomyces cerevisiae: the molecular organization of O-methylase Coq3p depends on Abc1p/Coq8p. FEMS Yeast Res. 2008;8:1263–1275. doi: 10.1111/j.1567-1364.2008.00436.x. [DOI] [PubMed] [Google Scholar]
- 37.Marbois B., Gin P., Faull K.F., Poon W.W., Lee P.T., Strahan J., Shepherd J.N., Clarke C.F. Coq3 and Coq4 define a polypeptide complex in yeast mitochondria for the biosynthesis of coenzyme Q. J. Biol. Chem. 2005;280:20231–20238. doi: 10.1074/jbc.M501315200. [DOI] [PubMed] [Google Scholar]
- 38.Petrossian T.C., Clarke S.G. Multiple motif scanning to identify methyltransferases from the yeast proteome. Mol. Cell. Proteomics. 2009;8:1516–1526. doi: 10.1074/mcp.M900025-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petrossian S.G. Clarke. Uncovering the human methyltransferasome. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M110.000976. (M110 000976) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Densitometric analyses of COQ5 RNA in human tissues. The Northern blot image was analyzed as described previously [23]. Densitometric signals for COQ5 RNA were normalized to beta actin expression (reported previously) [23] and expressed in arbitrary units (highest expression = 100).
Determination of antibody specificity to hCOQ5 and yeast Coq5 polypeptides. Purified mitochondria (35 μg protein) from W303ΔCOQ5 harboring hcCOQ8 and the designated COQ5 plasmids were subjected to immune-blot analysis. The membrane was first probed with antibodies to human COQ5, and stripped and re-probed with antisera to yeast Coq5. The membrane was then probed for Coq9 and Atp2 (beta subunit of the F1 sector of mitochondrial F1F0 ATP synthase).










