Abstract
Background
Protein-protein interactions (PPIs) are key to understanding diverse cellular processes and disease mechanisms. However, current PPI databases only provide low-resolution knowledge of PPIs, in the sense that "proteins" of currently known PPIs generally refer to "genes." It is known that alternative splicing often impacts PPI by either directly affecting protein interacting domains, or by indirectly impacting other domains, which, in turn, impacts the PPI binding. Thus, proteins translated from different isoforms of the same gene can have different interaction partners.
Results
Due to the limitations of current experimental capacities, little data is available for PPIs at the resolution of isoforms, although such high-resolution data is crucial to map pathways and to understand protein functions. In fact, alternative splicing can often change the internal structure of a pathway by rearranging specific PPIs. To fill the gap, we systematically predicted genome-wide isoform-isoform interactions (IIIs) using RNA-seq datasets, domain-domain interaction and PPIs. Furthermore, we constructed an III database (IIIDB) that is a resource for studying PPIs at isoform resolution. To discover functional modules in the III network, we performed III network clustering, and then obtained 1025 isoform modules. To evaluate the module functionality, we performed the GO/pathway enrichment analysis for each isoform module.
Conclusions
The IIIDB provides predictions of human protein-protein interactions at the high resolution of transcript isoforms that can facilitate detailed understanding of protein functions and biological pathways. The web interface allows users to search for IIIs or III network modules. The IIIDB is freely available at http://syslab.nchu.edu.tw/IIIDB.
Background
Protein-protein interactions (PPIs) perform and regulate fundamental cellular processes. As a consequence, identifying interacting partners for a protein is essential to understand its functions. In recent years, remarkable progress has been made in the annotation of all functional interactions among proteins in the cell. However, in both experimentally derived and computationally predicted protein-protein interactions, a "protein" generally refers to "all isoforms of the respective gene." Yet, it is known that alternative splicing often impacts PPI by either directly affecting protein interacting domains, or by indirectly impacting other domains, which, in turn, impact the PPI binding [1]. That is, alternative splicing can modulate the PPIs by altering the protein structures and the domain compositions, leading to the gain or loss of specific molecular interactions that could be key links of pathways (reviewed in reference [2]). It is very likely that different isoforms of the same protein interact with different proteins, thus exerting different functional roles. For example, the protein BCL2L1 is alternatively spliced into two isoforms: Bcl-xL (long form) and Bcl-xS (short form) [3], in which Bcl-xL inhibits apoptosis whereas Bcl-xS promotes apoptosis [4]. Vogler et al. reported that the interaction of Bcl-xL and BAK1 in platelets ensures platelet survival [5]. Therefore, comprehensively identifying protein-protein interactions at the isoform level is important to systematically dissect cellular roles of proteins, to elucidate the exact composition of protein complexes, and to gain insights into metabolic pathways and a wide range of direct and indirect regulatory interactions.
Thus far, a series of studies have systematically predicted PPIs [6-10] and established PPI databases, e.g., OPHID [11], POINT [12], STRING [10] and PIPs [7]. With the exception that the IntAct database [13] contains 116 human PPIs with isoform specification, currently, none of those PPI databases has isoform-level PPI data. This is a huge knowledge gap yet to be filled. The rapid accumulation of RNA-seq data provides unprecedented opportunities to study the structures and topological dynamics of PPI networks at the isoform resolution. RNA-seq data provides two unique informative sources for Isoform-Isoform Interaction (III) reconstruction: the absence or presence of specific isoforms under specific conditions, and the co-expression of two isoforms that may contribute to their interaction propensity. In this study, we seize this opportunity to comprehensively predict the possible interactions between splicing isoforms by integrating a series of RNA-seq data with domain-domain interaction data and PPI database. The resulting III network presents a high-resolution map of PPIs, which could be invaluable in studying biological processes and understanding cellular functions.
In this report, we described a database, IIIDB, for accessing and managing predicted human IIIs. In the IIIDB, users can differentiate access high-confidence and low-confidence predictions of human IIIs (see detailed description in Result section), and then see the full evidence values for each predicted III. Figure 1 shows the IIIDB web interface screen-shots of the III search and isoform module search function in the IIIDB. Users can upload their won gene expression data for III prediction, and then the users can download the predicted result. The searching function has three major parts: high-confidence interaction prediction search, low-confidence interaction prediction search, and isoform module search. The IIIDB provides auto-complete function in all search functions. Users can input a gene symbol or gene ID in the auto-complete field which provides an interface to quickly find and select matched values.
Figure 1.
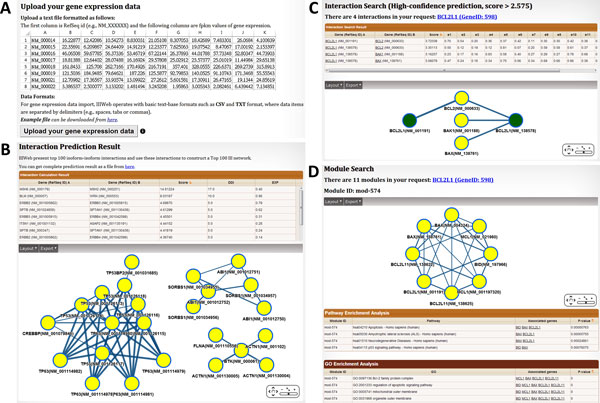
The screenshots of the isoform interaction and module search function in the IIIDB. (A) Users can upload their own gene expression data for III prediction. (B) IIIDB shows top 100 IIIs and uses these interactions to construct an III network. Users can download complete prediction result as a file. (C) In interaction search function, users can input a gene symbol or gene ID to search associated IIIs and isoform modules. The interaction search section provides interface on searching high-confidence (score > 2.575) or low-confidence (score > 1.692) prediction. The resulting III table for interaction search shows full evidence values including Pearson correlations for 19 RNA-seq datasets and domain interaction score. (D) The resulting page for module search includes the user friendly network graph and pathway/GO enrichment analysis result.
The IIIDB allows users to easily search IIIs and isoform modules, and then provides the evidence that led to each III prediction. To visualize the interactions with the isoforms of the input gene, we integrated CytoscapeWeb [14] to generate the interactive web-based III network (Figure 1B). Interestingly, the different isoforms within the same gene can be involved with different isoform modules that may open a new door to study differential functionality of isoforms of the gene. The IIIDB also provided GO/pathway enrichment analysis results for each isoform module, which helps biologists to study the biological insights of network modules at isoform level.
Results
To obtain the isoform annotations in the human genome, we used the NCBI Reference Sequences (RefSeq) mRNAs as transcriptome annotation [15]. Figure 2 shows the framework of III prediction. We employed the logistic regression approach with 19 RNA-seq datasets (Table 1) and the domain-domain interaction database to infer IIIs. To confirm with PPI, given an III prediction I1 and I2, we only keep this prediction if the gene symbols of I1 and I2 have PPI in IntAct database.
Figure 2.
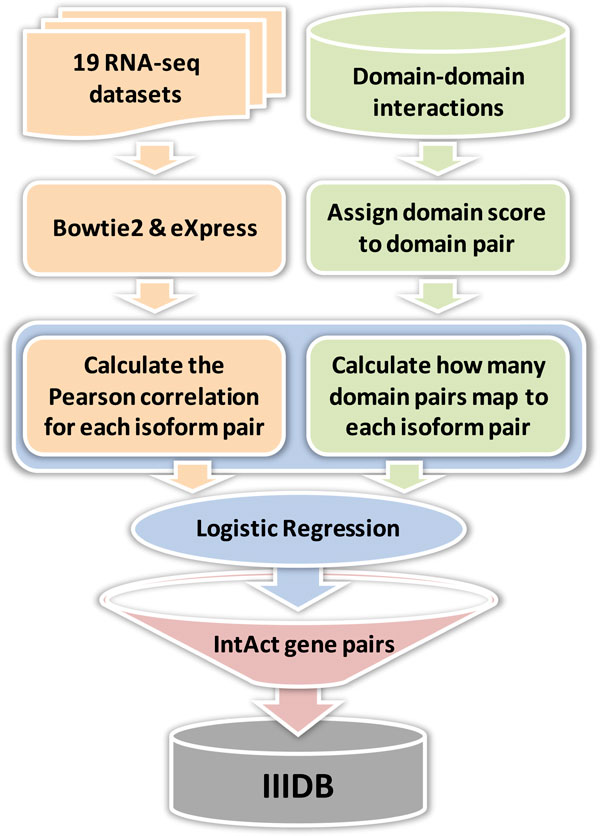
We performed logistic regression with 19 RNA-seq datasets and the domain-domain interaction database to construct IIIDB. In RNA-seq data processing, Bowtie2 and eXpress were used to calculate isoform expressions. To confirm with PPI, given an III prediction I1 and I2, we only keep this isoform interaction if the gene symbols of I1 and I2 have PPI in IntAct database.
Table 1.
19 RNA-seq datasets from SRA
| ID | SRA ID | # Exp | Title |
|---|---|---|---|
| d1 | ERP000546 | 48 | Illumina bodyMap2 transcriptome |
| d2 | SRP005169 | 41 | Widespread splicing changes in human brain development and aging |
| d3 | SRP005408 | 31 | Gene expression profile in postmortem hippocampus using RNAseq for addicted human samples |
| d4 | SRP010280 | 31 | Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins |
| d5 | SRP002628 | 30 | Comparative transcriptomic analysis of prostate cancer and matched normal tissue using RNA-seq |
| d6 | ERP000550 | 29 | Complete transcriptomic landscape of prostate cancer in the Chinese population using RNA-seq |
| d7 | SRP005242 | 21 | A Comparison of Single Molecule and Amplification Based Sequencing of Cancer Transcriptomes: RNA-Seq Comparison |
| d8 | SRP002079 | 20 | GSE20301: Dynamic transcriptomes during neural differentiation of human embryonic stem cells |
| d9 | ERP000992 | 18 | The effect of estrogen and progesterone and their antagonists in Ishikawa cell line compared to MCF7 and T47D cells |
| d10 | SRP000727 | 16 | Alternative Isoform Regulation in Human Tissue Transcriptomes |
| d11 | SRP007338 | 16 | GSE30017: Widespread regulated alternative splicing of single codons accelerates proteome evolution |
| d12 | SRP010166 | 16 | GSE34914: Deep Sequence Analysis of non-small cell lung cancer: Integrated analysis of gene expression, alternative splicing, and single nucleotide variations in lung adenocarcinomas with and without oncogenic KRAS mutations |
| d13 | ERP000710 | 12 | Transciptome profiling of ovarian cancer cell lines |
| d14 | SRP005411 | 11 | RNA-Seq Quantification of the Complete Transcriptome of Genes Expressed in the Small Airway Epithelium of Nonsmokers and Smokers |
| d15 | SRP006731 | 11 | GSE29155: RNA-Seq anlalysis of prostate cancer cell lines using Next Generation Sequencing |
| d16 | SRP013224 | 11 | GSE38006: Next-generation sequencing reveals HIV-1-mediated suppression of T cell activation and RNA processing and the regulation of non-coding RNA expression in a CD4+ T cell line |
| d17 | ERP000418 | 10 | Gene expression profiles between normal and breast tumor genomes |
| d18 | ERP000573 | 10 | RNA and chromatin structure |
| d19 | SRP010483 | 10 | GSE35296: The human pancreatic islet transcriptome: impact of pro-inflammatory cytokines |
High-confidence and low-confidence prediction of IIIs
In the IIIDB, we provided two III prediction sets using the logistic regression model: (a) high-confidence prediction: logit score > 2.575 (precision 60% and recall 15%), it resulted in 4,476 IIIs; and (b) low-confidence prediction: logit score > 1.692 (precision 40% and recall 68%). In addition, given a known PPI, the isoform pair with the best logit score among this PPI will be selected as low-confidence prediction. Thus, a PPI has at least one isoform-isoform interaction. It resulted in 54,605 IIIs (Figure 3).
Figure 3.
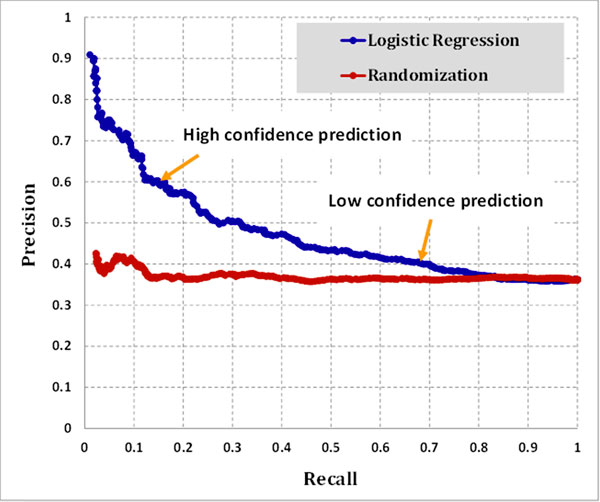
Precision and recall curve for logistic regression model. At recall 15%, logistic regression model achieve 60% precision (High-confidence prediction); at recall 68%, logistic regression model achieve 40% precision (Low-confidence prediction).
Isoform module discovery
To discover functional modules in the III network, we applied MODES network clustering method [16] on low-confidence III network to discover isoform modules with the given parameters (minimum module size 3, maximum module size 30, and density cutoff 0.7). An important feature of MODES is that it can discover overlapping dense isoform modules which allows one isoform to belong to multiple modules. We obtained 1025 modules with size of 5.08 isoforms on average. To provide functional annotation and evaluate the module functional enrichment, we performed the enrichment analyses with GO [17] and KEGG pathway [18] databases. These databases are protein-level annotation which provides approximately functional annotation of isoforms.
To evaluate the significance, we randomly generated the same number and the same size of MODES isoform modules (i.e. 1025 randomized isoform modules). Table 2 shows the results of enrichment analyses for MODES and randomization modules for comparison. The MODES isoform modules have significantly higher functional enrichment rate than those of random cases, showing strong biological relevance of the predicted modules. The MODES isoform modules were further used to build the isoform module database in the IIIDB, and all isoform modules were listed in Additional file 1. We also stored the GO and KEGG enrichment results for all isoform modules in the IIIDB to provide potential functional annotation. In the IIIDB web interface, Figure 1C shows the result page of the isoform module search including the network graph and the enrichment analysis results. In the module result page, users can click any gene symbol to iteratively search isoform modules, and sort the module result table by clicking the column header.
Table 2.
The enrichment rate of isoform modules based on GO and pathway enrichments
| Isoform modules | # modules | % modules enriched with GOa | % modules enriched with pathwayb |
|---|---|---|---|
| MODES modules | 1025 | 88.7% | 36.1% |
| Randomization | 1025 | 49.7% | 10.1% |
a The modules enriched with GO term (P-value < 0.001).
b The modules enriched with pathway (P-value < 0.01).
Integrating diverse data sources for III prediction
Currently, most PPI databases do not provide information at the level of isoforms, which thus presents challenges for constructing a gold standard positive set (GSP) for the III prediction. Fortunately, in the June 2013 version of the IntAct database http://www.ebi.ac.uk/intact/[13], we identified 116 human PPIs with isoform specification (out of the total 43,508 distinct human PPIs). For example, IntAct has III between P29590-5 and P03243-1, which correspond to the 5th isoform of the protein P29590 and the 1st isoform of P03243. In addition, to obtain more IIIs for the GSP, we applied the following rule: given a PPI between protein P1 and P2, if both P1 and P2 only have single isoforms we also take it as the GSP. It resulted in 11,356 IIIs in the GSP set.
GSP covered 5,503 RefSeq IDs, and we used these RefSeq IDs to construct gold standard negative set (GSN). The GSN was defined as isoform pairs in which one isoform was assigned to the plasma membrane cellular component, and the other was assigned to the nuclear cellular component by the isoform-specific sub-cellular localization, in which we performed sequence-based predictions using the CELLO (subCELlular LOcalization predictor) [19]. To obtain the accurate isoform-specific annotation, we only used the cellular localization prediction results that consist of UniProt GO annotations. It resulted in 36 RefSeq IDs for plasma membrane cellular component and 739 RefSeq IDs for nuclear cellular component. In addition, isoforms that are assigned to both the plasma membrane and the nuclear cellular component are excluded in GSN.
To calculate precision and recall, we used timestamp to divided GSP into training and test GSP sets, in which if an interaction with timestamp after 1st Jan 2012, it will be assigned to test GSP set (10,408 IIIs); otherwise, it will be assign to training GSP set (948 IIIs). When the GSP is decided, we used the RefSeq IDs covered in GSP to build GSN. Figure 3 shows the precision and recall curve for the logistic regression model.
Case studies
To demonstrate the biological importance of the IIIDB, we searched for isoform-associated reports in the literature. Although isoform-specific protein function studies are very rare, we found isoform-specific biological evidences with BCL2L1, which validated our III prediction of BCL2L1. In addition, we also found diverse biological functions with Ras association domain family in our isoform modules.
BCL2L1
BCL2L1 has two isoforms (Table 3), in which BCL2L1 isoform 1 is called Bcl-xL (long form) and BCL2L1 isoform 2 is called Bcl-xS (short form) [3]. These isoforms play important roles in apoptosis as follows: Bcl-xL inhibits apoptosis whereas Bcl-xS promotes apoptosis [4]. In our high-confidence prediction, BCL2L1 isoform 1 (Bcl-xL) interacts with BAK1, BAX and NLRP1 to inhibit apoptosis, but BCL2L1 isoform 2 (Bcl-xS) doesn't (Table 3). In previous reports, the fluorescence anisotropy, analytical ultracentrifugation, and NMR assays confirmed a direct interaction between Bcl-xL and BAK1 [5,20]. Vogler et al. also reported that the interaction of Bcl-xL and BAK1 in platelets ensures cell survival [5]. Edlich et al. reported that an interaction between Bcl-xL and BAX not only inhibits BAX activity but also maintains BAX in the cytosol [21]. On the other hand, Chang et al. demonstrated that Bcl-xL interacted with endogenous BAX in 293 cells. However, no significant amount of BAX was detectable in the Bcl-xS immunoprecipitation [21], suggesting that Bcl-xS does not interact with BAX. In addition, Bruey et al. reported that Bcl-xL interacts with NALP1 to suppress apoptosis [22]. Thus, these previous biological studies validated our III prediction of BCL2L1.
Table 3.
BCL2L1 isoform interaction partners.
RASSF1
RASSF1 is RAS association domain family member 1. RASSF1 has four isoforms, of which two isoforms belong to isoform modules (Mod-162 and Mod-384). The Mod-162 consists of RASSF1 isoform A, RASSF5 and STK4, whereas the Mod-384 consists of RASSF1 isoform C, RASSF3, RASSF4, SAV1, STK3 and STK4 (Figure 4). Interestingly, several studies reported that RASSF5 may interact with RASSF1 isoform A to suppress tumors [23-25], suggesting that the Mod-162 is validated. On the other hand, RASSF1 isoform C may play a completely different role as an oncogene in high-grade tumors [26]. Although the function of RASSF1 isoform C is still not clear, the RASSF1 isoform A and C should have distinct functions. The IIIDB assigned the RASSF1 isoform A and C into the Mod-162 and the Mod-384, respectively, suggesting a new hypothesis of the isoform-level modules.
Figure 4.
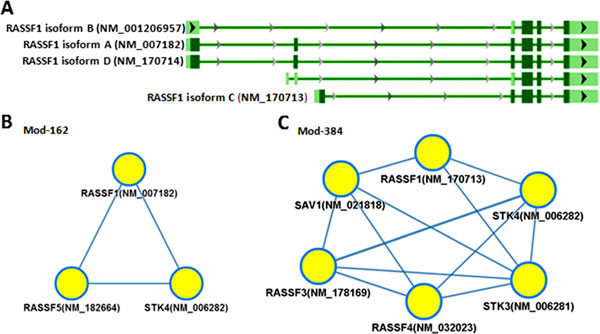
Two isoform modules of RASSF1. (A) RASSF1 has four isoforms (isoform A, B, C and D), of which two isoforms belong to module Mod-162 and Mod-384. (B) The isoform module Mod-162 includes RASSF1 isoform A (NM_007182), RASSF5 and STK4. (C) The isoform module Mod-384 includes RASSF1 isoform C (NM_170713), RASSF3, RASSF4, SAV1, STK3 and STK4.
Methods
Isoform coexpression network construction
To construct the Isoform coexpression networks, we selected 19 RNA-seq datasets with at least 10 experiments from Sequence Read Archive (SRA) database (Table 1) [27]. We performed the eXpress [28] with Bowtie2 aligner [29] to obtain isoform expression values. NCBI Reference Sequences (RefSeq) mRNAs with protein sequences was used as transcriptome annotation, which included 31,454 RefSeq IDs (Jan 2013 version) [15].
Logistic regression model
The logistic regression model has been applied to PPI prediction [30,31], as it is suitable to describe the relationship between a binary response variable and a set of explanatory variables. We used logistic regression approach to build the prediction model as follows:
where α0,α1,...,α20 are regression coefficients and yij is the probability of the isoform interaction between isoform i and isoform j. DDI, E1, E2, ..., E19 are described as follows: (a) Domain-domain interaction (DDI) score: For all combinations of human isoform pairs, if a isoform pair has domain-domain interaction in the DOMINE database [32,33], then we assign the DDI score by the confidence level in the DOMINE database as follows: high-confidence prediction: 3, medium-confidence prediction: 2, and low-confidence prediction: 1. If the isoform pair has several DDI scores, we take the highest score. (b) 19 RNA-seq datasets (E1, E2, ..., E19): the absolute values of Pearson correlations for all isoform pairs derived from RNA-seq datasets. Since each RNA-seq dataset may have different quality and data type, we used the logistic regression model to integrate 19 RNA-seq datasets, and then the coefficients of datasets reflected quality of the RNA-seq datasets.
Competing interests
The authors declare that they have no competing interests.
Supplementary Material
The isoform modules. There are 1025 isoform modules with gene symbols and RefSeq IDs.
Contributor Information
Yu-Ting Tseng, Email: ting0514@gmail.com.
Wenyuan Li, Email: wel@usc.edu.
Ching-Hsien Chen, Email: chchencmc@gmail.com.
Shihua Zhang, Email: zsh@amss.ac.cn.
Jeremy JW Chen, Email: jwchen@dragon.nchu.edu.tw.
Xianghong Jasmine Zhou, Email: xjzhou@usc.edu.
Chun-Chi Liu, Email: jimliu@nchu.edu.tw.
Acknowledgements
The National Institutes of Health (NIH) [NHLBI MAPGEN U01HL108634 and NIGMS R01GM105431] and the National Science Foundation [0747475 to X.J.Z.]; the Taiwan Ministry of Science and Technology grant [MOST 103-2320-B-005-004] and the Ministry of Education, Taiwan, R.O.C. under the ATU plan (to C.C.L.).
This article has been published as part of BMC Genomics Volume 16 Supplement 2, 2015: Selected articles from the Thirteenth Asia Pacific Bioinformatics Conference (APBC 2015): Genomics. The full contents of the supplement are available online at http://www.biomedcentral.com/bmcgenomics/supplements/16/S2
References
- Ellis JD, Barrios-Rodiles M, Colak R, Irimia M, Kim T, Calarco JA, Wang X, Pan Q, O'Hanlon D, Kim PM. et al. Tissue-specific alternative splicing remodels protein-protein interaction networks. Molecular cell. 2012;46(6):884–892. doi: 10.1016/j.molcel.2012.05.037. [DOI] [PubMed] [Google Scholar]
- Leeman JR, Gilmore TD. Alternative splicing in the NF-kappaB signaling pathway. Gene. 2008;423(2):97–107. doi: 10.1016/j.gene.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perumalsamy A, Fernandes R, Lai I, Detmar J, Varmuza S, Casper RF, Jurisicova A. Developmental consequences of alternative Bcl-x splicing during preimplantation embryo development. FEBS J. 2010;277(5):1219–1233. doi: 10.1111/j.1742-4658.2010.07554.x. [DOI] [PubMed] [Google Scholar]
- Rajan P, Elliott DJ, Robson CN, Leung HY. Alternative splicing and biological heterogeneity in prostate cancer. Nat Rev Urol. 2009;6(8):454–460. doi: 10.1038/nrurol.2009.125. [DOI] [PubMed] [Google Scholar]
- Vogler M, Hamali HA, Sun XM, Bampton ET, Dinsdale D, Snowden RT, Dyer MJ, Goodall AH, Cohen GM. BCL2/BCL-XL inhibition induces apoptosis, disrupts cellular calcium homeostasis, and prevents platelet activation. Blood. 2011;117(26):7145–7154. doi: 10.1182/blood-2011-03-344812. [DOI] [PubMed] [Google Scholar]
- Cho YR, Shi L, Ramanathan M, Zhang A. A probabilistic framework to predict protein function from interaction data integrated with semantic knowledge. BMC Bioinformatics. 2008;9:382. doi: 10.1186/1471-2105-9-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowall MD, Scott MS, Barton GJ. PIPs: human protein-protein interaction prediction database. Nucleic Acids Res. 2009. pp. D651–656. [DOI] [PMC free article] [PubMed]
- Rhodes DR, Tomlins SA, Varambally S, Mahavisno V, Barrette T, Kalyana-Sundaram S, Ghosh D, Pandey A, Chinnaiyan AM. Probabilistic model of the human protein-protein interaction network. Nat Biotechnol. 2005;23(8):951–959. doi: 10.1038/nbt1103. [DOI] [PubMed] [Google Scholar]
- Scott MS, Barton GJ. Probabilistic prediction and ranking of human protein-protein interactions. BMC Bioinformatics. 2007;8:239. doi: 10.1186/1471-2105-8-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011. pp. D561–568. [DOI] [PMC free article] [PubMed]
- Brown KR, Jurisica I. Online predicted human interaction database. Bioinformatics. 2005;21(9):2076–2082. doi: 10.1093/bioinformatics/bti273. [DOI] [PubMed] [Google Scholar]
- Huang TW, Tien AC, Huang WS, Lee YC, Peng CL, Tseng HH, Kao CY, Huang CY. POINT: a database for the prediction of protein-protein interactions based on the orthologous interactome. Bioinformatics. 2004;20(17):3273–3276. doi: 10.1093/bioinformatics/bth366. [DOI] [PubMed] [Google Scholar]
- Aranda B, Achuthan P, Alam-Faruque Y, Armean I, Bridge A, Derow C, Feuermann M, Ghanbarian AT, Kerrien S, Khadake J, The IntAct molecular interaction database in 2010. Nucleic Acids Res. 2010. pp. D525–531. [DOI] [PMC free article] [PubMed]
- Lopes CT, Franz M, Kazi F, Donaldson SL, Morris Q, Bader GD. Cytoscape Web: an interactive web-based network browser. Bioinformatics. 2010;26(18):2347–2348. doi: 10.1093/bioinformatics/btq430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Brown GR, Maglott DR. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 2012. pp. D130–135. [DOI] [PMC free article] [PubMed]
- Hu H, Yan X, Huang Y, Han J, Zhou XJ. Mining coherent dense subgraphs across massive biological networks for functional discovery. Bioinformatics. 2005;21(Suppl 1):i213–221. doi: 10.1093/bioinformatics/bti1049. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CS, Chen YC, Lu CH, Hwang JK. Prediction of protein subcellular localization. Proteins. 2006;64(3):643–651. doi: 10.1002/prot.21018. [DOI] [PubMed] [Google Scholar]
- Sot B, Freund SM, Fersht AR. Comparative biophysical characterization of p53 with the pro-apoptotic BAK and the anti-apoptotic BCL-xL. J Biol Chem. 2007;282(40):29193–29200. doi: 10.1074/jbc.M705544200. [DOI] [PubMed] [Google Scholar]
- Edlich F, Banerjee S, Suzuki M, Cleland MM, Arnoult D, Wang C, Neutzner A, Tjandra N, Youle RJ. Bcl-x(L) retrotranslocates Bax from the mitochondria into the cytosol. Cell. 2011;145(1):104–116. doi: 10.1016/j.cell.2011.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruey JM, Bruey-Sedano N, Luciano F, Zhai D, Balpai R, Xu C, Kress CL, Bailly-Maitre B, Li X, Osterman A. et al. Bcl-2 and Bcl-XL regulate proinflammatory caspase-1 activation by interaction with NALP1. Cell. 2007;129(1):45–56. doi: 10.1016/j.cell.2007.01.045. [DOI] [PubMed] [Google Scholar]
- Park J, Kang SI, Lee SY, Zhang XF, Kim MS, Beers LF, Lim DS, Avruch J, Kim HS, Lee SB. Tumor suppressor ras association domain family 5 (RASSF5/NORE1) mediates death receptor ligand-induced apoptosis. J Biol Chem. 2010;285(45):35029–35038. doi: 10.1074/jbc.M110.165506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes MS, Carneiro F, Oliveira C, Seruca R. Colorectal cancer and RASSF family--a special emphasis on RASSF1A. Int J Cancer. 2013;132(2):251–258. doi: 10.1002/ijc.27696. [DOI] [PubMed] [Google Scholar]
- Richter AM, Pfeifer GP, Dammann RH. The RASSF proteins in cancer; from epigenetic silencing to functional characterization. Biochim Biophys Acta. 2009;1796(2):114–128. doi: 10.1016/j.bbcan.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Pelosi G, Fumagalli C, Trubia M, Sonzogni A, Rekhtman N, Maisonneuve P, Galetta D, Spaggiari L, Veronesi G, Scarpa A. et al. Dual role of RASSF1 as a tumor suppressor and an oncogene in neuroendocrine tumors of the lung. Anticancer Res. 2010;30(10):4269–4281. [PubMed] [Google Scholar]
- Kodama Y, Shumway M, Leinonen R, International Nucleotide Sequence Database C. The Sequence Read Archive: explosive growth of sequencing data. Nucleic Acids Res. 2012. pp. D54–56. [DOI] [PMC free article] [PubMed]
- Roberts A, Pachter L. Streaming fragment assignment for real-time analysis of sequencing experiments. Nat Methods. 2013;10(1):71–73. doi: 10.1038/nmeth.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Schmidt B. Long read alignment based on maximal exact match seeds. Bioinformatics. 2012;28(18):i318–i324. doi: 10.1093/bioinformatics/bts414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Klein-Seetharaman J, Bar-Joseph Z. A mixture of feature experts approach for protein-protein interaction prediction. BMC Bioinformatics. 2007;8(Suppl 10):S6. doi: 10.1186/1471-2105-8-S10-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzak E, Altuvia Y, Margalit H. Characterization and prediction of protein-protein interactions within and between complexes. Proc Natl Acad Sci USA. 2006;103(40):14718–14723. doi: 10.1073/pnas.0603352103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavachari B, Tasneem A, Przytycka TM, Jothi R. DOMINE: a database of protein domain interactions. Nucleic Acids Res. 2008. pp. D656–661. [DOI] [PMC free article] [PubMed]
- Yellaboina S, Tasneem A, Zaykin DV, Raghavachari B, Jothi R. DOMINE: a comprehensive collection of known and predicted domain-domain interactions. Nucleic Acids Res. 2011. pp. D730–735. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The isoform modules. There are 1025 isoform modules with gene symbols and RefSeq IDs.


