Abstract
The molecule of the title compound, C25H24O, obtained by acid-catalysed 1,3-migration of an alcohol group, is T-shaped. The planes of the two phenyl rings are inclined to one another by 81.9 (2)°. In the crystal, molecules are linked by O—H⋯O hydrogen bonds, forming chains along [001].
Keywords: crystal structure; 1,3-migration; alcohol group; catalytic cyclization; styrylundecanene.
Related literature
For the 1,3-migration of an alcoholic group adjacent to a vinyl group in the presence of a Lewis acid, see: Piotti & Alper (1997 ▸); Poloukhtine & Popik (2005 ▸). For catalytic cyclization of alcohols containing a number of unsaturated groups, see: Teo et al. (2014 ▸).
Experimental
Crystal data
C25H24O
M r = 340.44
Trigonal,

a = 17.867 (2) Å
c = 5.3290 (6) Å
V = 1473.3 (4) Å3
Z = 3
Mo Kα radiation
μ = 0.07 mm−1
T = 103 K
0.34 × 0.04 × 0.04 mm
Data collection
Bruker Kappa APEXII CCD diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2013 ▸) T min = 0.74, T max = 1.00
14182 measured reflections
4846 independent reflections
2945 reflections with I > 2σ(I)
R int = 0.078
Refinement
R[F 2 > 2σ(F 2)] = 0.058
wR(F 2) = 0.129
S = 0.98
4846 reflections
235 parameters
1 restraint
H-atom parameters constrained
Δρmax = 0.39 e Å−3
Δρmin = −0.24 e Å−3
Data collection: APEX2 (Bruker, 2013 ▸); cell refinement: SAINT (Bruker, 2013 ▸); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick 2008 ▸); program(s) used to refine structure: SHELXL2014 (Sheldrick, 2008 ▸); molecular graphics: ORTEP-3 for Windows (Farrugia, 2012 ▸) and Mercury (Macrae et al., 2008 ▸); software used to prepare material for publication: SHELXL2014, PLATON (Spek, 2009 ▸) and publCIF (Westrip, 2010 ▸).
Supplementary Material
Crystal structure: contains datablock(s) I, New_Global_Publ_Block. DOI: 10.1107/S205698901402742X/su5038sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S205698901402742X/su5038Isup2.hkl
Supporting information file. DOI: 10.1107/S205698901402742X/su5038Isup3.cml
. DOI: 10.1107/S205698901402742X/su5038fig1.tif
A view of the molecular structure of the title compound, with atom labelling. Displacement ellipsoids are drawn at the 50% probability level.
c . DOI: 10.1107/S205698901402742X/su5038fig2.tif
A partial view along the c axis of the crystal packing of the title compound. Hydrogen bonds are shown as dashed lines (see Table 1 for details).
. DOI: 10.1107/S205698901402742X/su5038fig3.tif
Reaction scheme.
CCDC reference: 1009363
Additional supporting information: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (, ).
| DHA | DH | HA | D A | DHA |
|---|---|---|---|---|
| O1H1AO1i | 0.84 | 1.83 | 2.652(3) | 166 |
Symmetry code: (i) 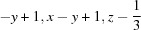 .
.
supplementary crystallographic information
S1. Comment
1,3-migration of an alcoholic group adjacent to a vinyl group in the presence of a Lewis-acid is widely known (Piotti & Alper, 1997). One such example was demonstrated recently in the preparation of 4-(α-hydroxybenzyl)-1-tert-butyldimethylsilyloxy-4-cyclodecene-2,6-diyne (Poloukhtine & Popik, 2005). In addition, alcohols containing many unsaturated groups provide an access to a myriad of types of functionalization such as catalytic cyclization (Teo et al., 2014). Herein, we report on the synthesis and crystal structure of the title compound, obtained by the acid-catalyzed 1,3-migration of an alcoholic group.
The molecular structure of the title compound is illustrated in Fig. 1. The molecule is T-shaped with the two phenyl ring inclined to one another by 81.9 (2) °.
In the crystal, molecules are linked by O—H···O hydrogen bonds forming chains along the c axis direction (Table 1 and Fig. 2).
S2. Synthesis and crystallization
The synthesis of the title compound is illustrated in Fig. 3. (Z)-1-phenyl-3-styrylundeca-1-en-4,10-diyn-3-ol (1mmol, 340.5 mg) was dissolved in 10 ml of CH2Cl2. DMAP [4-(dimethylamino)pyridine; 0.1 mmol, 12 mg], triethylamine (5 mmol, 0.70 ml) and acetic anhydride (5 mmol, 0.47 ml) were added sequentially and the reaction mixture was stirred overnight. It was then washed with saturated sodium bicarbonate and extracted twice with CH2Cl2. The organic layers were combined, dried with MgSO4 and the solvent was removed under reduced pressure. The resulting oil was purified by column chromatography with hexane/ethylacetate as eluent. The product was recrystallized with ethylacetate to give a colourless compound in 80% yield. Slow evaporation of a solution in ethylacetate gave needle-like crystals. 1H NMR (400 MHz, CDCl3) 1.71–1.83 (m, 4H), 1.97 (t, 1H), 2.20 (s, 1H), 2.26–2.30 (m, 2H), 2.55 (t, 2), 5.90 (d, 1H), 6.09 (d, 1H), 6.68 (d, 1H), 6.99 (d, 1H), 7.21–7.47 (m, 10H) 13 C NMR (100 MHz, CDCl3) 18.0, 19.2, 27.7, 27.7, 68.8, 72.5, 75.3, 84.0, 97.9, 124.1, 125.9, 126.8, 127.7, 127.9, 128.6, 128.6, 132.4, 136.8, 140.4, 142.7
S3. Refinement
The OH and C-bound H atoms were included in calculated positions and treated as riding atoms: O—H = 0.84 Å, C—H = 0.95 - 1.00 Å with Uiso(H) = 1.5Ueq(O) for the OH H atom and = 1.2Ueq(C) for other H atoms.
Figures
Fig. 1.

A view of the molecular structure of the title compound, with atom labelling. Displacement ellipsoids are drawn at the 50% probability level.
Fig. 2.

A partial view along the c axis of the crystal packing of the title compound. Hydrogen bonds are shown as dashed lines (see Table 1 for details).
Fig. 3.
Reaction scheme.
Crystal data
| C25H24O | Dx = 1.151 Mg m−3 |
| Mr = 340.44 | Mo Kα radiation, λ = 0.71073 Å |
| Trigonal, P32 | Cell parameters from 1285 reflections |
| a = 17.867 (2) Å | θ = 2.3–20.5° |
| c = 5.3290 (6) Å | µ = 0.07 mm−1 |
| V = 1473.3 (4) Å3 | T = 103 K |
| Z = 3 | Needle, colourless |
| F(000) = 546 | 0.34 × 0.04 × 0.04 mm |
Data collection
| Bruker Kappa APEXII CCD diffractometer | 4846 independent reflections |
| Radiation source: fine-focus sealed tube | 2945 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.078 |
| ω and φ scan | θmax = 28.3°, θmin = 2.6° |
| Absorption correction: multi-scan (SADABS; Bruker, 2013) | h = −23→14 |
| Tmin = 0.74, Tmax = 1.00 | k = −22→23 |
| 14182 measured reflections | l = −7→7 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.058 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.129 | H-atom parameters constrained |
| S = 0.98 | w = 1/[σ2(Fo2) + (0.0486P)2] where P = (Fo2 + 2Fc2)/3 |
| 4846 reflections | (Δ/σ)max < 0.001 |
| 235 parameters | Δρmax = 0.39 e Å−3 |
| 1 restraint | Δρmin = −0.24 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.30859 (17) | 0.59437 (17) | 0.3483 (5) | 0.0251 (7) | |
| H1A | 0.3463 | 0.6311 | 0.2521 | 0.038* | |
| C1 | 0.3069 (3) | 0.5136 (2) | 0.3226 (7) | 0.0199 (9) | |
| H1 | 0.3048 | 0.4898 | 0.4940 | 0.024* | |
| C2 | 0.2281 (2) | 0.4484 (3) | 0.1774 (7) | 0.0197 (9) | |
| C3 | 0.1897 (3) | 0.3614 (3) | 0.2407 (8) | 0.0281 (10) | |
| H3 | 0.2105 | 0.3441 | 0.3801 | 0.034* | |
| C4 | 0.1209 (3) | 0.2999 (3) | 0.1007 (9) | 0.0341 (12) | |
| H4 | 0.0948 | 0.2407 | 0.1461 | 0.041* | |
| C5 | 0.0899 (3) | 0.3235 (3) | −0.1032 (9) | 0.0333 (11) | |
| H5 | 0.0434 | 0.2809 | −0.1998 | 0.040* | |
| C6 | 0.1275 (3) | 0.4099 (3) | −0.1651 (8) | 0.0305 (11) | |
| H6 | 0.1060 | 0.4269 | −0.3038 | 0.037* | |
| C7 | 0.1962 (3) | 0.4721 (3) | −0.0270 (8) | 0.0272 (10) | |
| H7 | 0.2216 | 0.5313 | −0.0722 | 0.033* | |
| C8 | 0.3878 (2) | 0.5282 (2) | 0.1948 (7) | 0.0196 (9) | |
| H8 | 0.4050 | 0.5639 | 0.0497 | 0.024* | |
| C9 | 0.4386 (2) | 0.4964 (2) | 0.2634 (7) | 0.0160 (8) | |
| C10 | 0.5143 (2) | 0.5153 (2) | 0.1121 (7) | 0.0171 (8) | |
| H10 | 0.5253 | 0.5510 | −0.0311 | 0.021* | |
| C11 | 0.5691 (2) | 0.4869 (2) | 0.1575 (7) | 0.0171 (8) | |
| H11 | 0.5608 | 0.4552 | 0.3082 | 0.021* | |
| C12 | 0.6413 (2) | 0.5000 (2) | −0.0039 (7) | 0.0173 (9) | |
| C13 | 0.6850 (2) | 0.4550 (2) | 0.0458 (7) | 0.0184 (9) | |
| H13 | 0.6692 | 0.4182 | 0.1882 | 0.022* | |
| C14 | 0.7511 (3) | 0.4630 (3) | −0.1091 (8) | 0.0220 (9) | |
| H14 | 0.7805 | 0.4324 | −0.0718 | 0.026* | |
| C15 | 0.7739 (3) | 0.5160 (3) | −0.3182 (7) | 0.0245 (10) | |
| H15 | 0.8184 | 0.5209 | −0.4264 | 0.029* | |
| C16 | 0.7321 (2) | 0.5620 (3) | −0.3704 (7) | 0.0231 (9) | |
| H16 | 0.7482 | 0.5985 | −0.5134 | 0.028* | |
| C17 | 0.6665 (3) | 0.5545 (3) | −0.2137 (7) | 0.0204 (9) | |
| H17 | 0.6385 | 0.5867 | −0.2492 | 0.024* | |
| C18 | 0.4152 (2) | 0.4385 (3) | 0.4758 (8) | 0.0184 (8) | |
| C19 | 0.3892 (2) | 0.3880 (2) | 0.6457 (7) | 0.0201 (9) | |
| C20 | 0.3519 (3) | 0.3267 (3) | 0.8553 (8) | 0.0275 (10) | |
| H20A | 0.3052 | 0.3336 | 0.9313 | 0.033* | |
| H20B | 0.3973 | 0.3426 | 0.9844 | 0.033* | |
| C21 | 0.3157 (3) | 0.2328 (3) | 0.7913 (8) | 0.0310 (11) | |
| H21A | 0.2888 | 0.1977 | 0.9434 | 0.037* | |
| H21B | 0.3637 | 0.2235 | 0.7405 | 0.037* | |
| C22 | 0.2484 (3) | 0.2009 (3) | 0.5807 (8) | 0.0317 (11) | |
| H22A | 0.2021 | 0.2137 | 0.6248 | 0.038* | |
| H22B | 0.2760 | 0.2322 | 0.4237 | 0.038* | |
| C23 | 0.2091 (3) | 0.1044 (3) | 0.5383 (9) | 0.0467 (14) | |
| H23A | 0.1740 | 0.0881 | 0.3830 | 0.056* | |
| H23B | 0.2562 | 0.0913 | 0.5127 | 0.056* | |
| C24 | 0.1549 (3) | 0.0521 (3) | 0.7450 (9) | 0.0326 (11) | |
| C25 | 0.1097 (3) | 0.0105 (3) | 0.9069 (10) | 0.0431 (13) | |
| H25 | 0.0727 | −0.0235 | 1.0394 | 0.052* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0252 (16) | 0.0188 (15) | 0.0341 (17) | 0.0130 (13) | 0.0087 (14) | 0.0019 (13) |
| C1 | 0.024 (2) | 0.021 (2) | 0.021 (2) | 0.0159 (19) | 0.0076 (18) | 0.0067 (18) |
| C2 | 0.019 (2) | 0.021 (2) | 0.024 (2) | 0.0129 (18) | 0.0084 (18) | 0.0036 (18) |
| C3 | 0.022 (2) | 0.025 (2) | 0.036 (3) | 0.010 (2) | 0.002 (2) | 0.008 (2) |
| C4 | 0.026 (2) | 0.026 (3) | 0.047 (3) | 0.010 (2) | 0.003 (2) | 0.005 (2) |
| C5 | 0.024 (2) | 0.034 (3) | 0.038 (3) | 0.011 (2) | −0.001 (2) | −0.006 (2) |
| C6 | 0.031 (3) | 0.035 (3) | 0.030 (3) | 0.020 (2) | −0.005 (2) | −0.002 (2) |
| C7 | 0.028 (2) | 0.027 (2) | 0.029 (2) | 0.016 (2) | 0.003 (2) | 0.005 (2) |
| C8 | 0.020 (2) | 0.018 (2) | 0.021 (2) | 0.0093 (18) | 0.0026 (17) | 0.0022 (17) |
| C9 | 0.016 (2) | 0.0124 (19) | 0.019 (2) | 0.0071 (17) | −0.0012 (17) | −0.0009 (16) |
| C10 | 0.018 (2) | 0.0110 (19) | 0.019 (2) | 0.0051 (17) | 0.0007 (17) | 0.0011 (16) |
| C11 | 0.019 (2) | 0.015 (2) | 0.017 (2) | 0.0078 (18) | 0.0010 (17) | 0.0004 (16) |
| C12 | 0.014 (2) | 0.017 (2) | 0.020 (2) | 0.0066 (17) | −0.0008 (17) | −0.0034 (17) |
| C13 | 0.018 (2) | 0.019 (2) | 0.018 (2) | 0.0087 (18) | 0.0014 (17) | 0.0001 (17) |
| C14 | 0.021 (2) | 0.025 (2) | 0.024 (2) | 0.0145 (19) | −0.0020 (18) | −0.0037 (19) |
| C15 | 0.019 (2) | 0.031 (2) | 0.023 (2) | 0.012 (2) | 0.0035 (18) | −0.0035 (19) |
| C16 | 0.021 (2) | 0.025 (2) | 0.022 (2) | 0.0096 (19) | 0.0035 (18) | 0.0020 (19) |
| C17 | 0.017 (2) | 0.021 (2) | 0.025 (2) | 0.0119 (18) | −0.0006 (18) | 0.0014 (18) |
| C18 | 0.015 (2) | 0.020 (2) | 0.021 (2) | 0.0094 (17) | −0.0002 (17) | −0.0042 (18) |
| C19 | 0.017 (2) | 0.020 (2) | 0.021 (2) | 0.0073 (18) | 0.0002 (18) | 0.0011 (18) |
| C20 | 0.031 (3) | 0.024 (2) | 0.020 (2) | 0.008 (2) | −0.001 (2) | 0.0045 (19) |
| C21 | 0.033 (3) | 0.026 (2) | 0.027 (2) | 0.010 (2) | 0.002 (2) | 0.002 (2) |
| C22 | 0.032 (3) | 0.024 (2) | 0.029 (3) | 0.006 (2) | 0.003 (2) | 0.003 (2) |
| C23 | 0.055 (3) | 0.028 (3) | 0.039 (3) | 0.007 (2) | 0.006 (3) | −0.002 (2) |
| C24 | 0.033 (3) | 0.019 (2) | 0.043 (3) | 0.010 (2) | −0.004 (2) | −0.003 (2) |
| C25 | 0.037 (3) | 0.029 (3) | 0.054 (4) | 0.009 (2) | 0.001 (3) | 0.006 (3) |
Geometric parameters (Å, º)
| O1—C1 | 1.435 (4) | C13—C14 | 1.388 (5) |
| O1—H1A | 0.8400 | C13—H13 | 0.9500 |
| C1—C8 | 1.498 (5) | C14—C15 | 1.385 (6) |
| C1—C2 | 1.516 (6) | C14—H14 | 0.9500 |
| C1—H1 | 1.0000 | C15—C16 | 1.388 (6) |
| C2—C3 | 1.390 (5) | C15—H15 | 0.9500 |
| C2—C7 | 1.390 (5) | C16—C17 | 1.390 (5) |
| C3—C4 | 1.387 (6) | C16—H16 | 0.9500 |
| C3—H3 | 0.9500 | C17—H17 | 0.9500 |
| C4—C5 | 1.378 (6) | C18—C19 | 1.197 (5) |
| C4—H4 | 0.9500 | C19—C20 | 1.470 (5) |
| C5—C6 | 1.381 (6) | C20—C21 | 1.504 (6) |
| C5—H5 | 0.9500 | C20—H20A | 0.9900 |
| C6—C7 | 1.385 (6) | C20—H20B | 0.9900 |
| C6—H6 | 0.9500 | C21—C22 | 1.531 (6) |
| C7—H7 | 0.9500 | C21—H21A | 0.9900 |
| C8—C9 | 1.341 (5) | C21—H21B | 0.9900 |
| C8—H8 | 0.9500 | C22—C23 | 1.519 (6) |
| C9—C18 | 1.446 (5) | C22—H22A | 0.9900 |
| C9—C10 | 1.461 (5) | C22—H22B | 0.9900 |
| C10—C11 | 1.332 (5) | C23—C24 | 1.456 (7) |
| C10—H10 | 0.9500 | C23—H23A | 0.9900 |
| C11—C12 | 1.467 (5) | C23—H23B | 0.9900 |
| C11—H11 | 0.9500 | C24—C25 | 1.161 (6) |
| C12—C13 | 1.399 (5) | C25—H25 | 0.9500 |
| C12—C17 | 1.401 (5) | ||
| C1—O1—H1A | 109.5 | C12—C13—H13 | 119.4 |
| O1—C1—C8 | 109.5 (3) | C15—C14—C13 | 119.5 (4) |
| O1—C1—C2 | 111.5 (3) | C15—C14—H14 | 120.2 |
| C8—C1—C2 | 110.3 (3) | C13—C14—H14 | 120.2 |
| O1—C1—H1 | 108.5 | C14—C15—C16 | 120.3 (4) |
| C8—C1—H1 | 108.5 | C14—C15—H15 | 119.9 |
| C2—C1—H1 | 108.5 | C16—C15—H15 | 119.9 |
| C3—C2—C7 | 118.8 (4) | C15—C16—C17 | 120.1 (4) |
| C3—C2—C1 | 119.0 (4) | C15—C16—H16 | 120.0 |
| C7—C2—C1 | 122.0 (4) | C17—C16—H16 | 120.0 |
| C2—C3—C4 | 120.2 (4) | C16—C17—C12 | 120.6 (4) |
| C2—C3—H3 | 119.9 | C16—C17—H17 | 119.7 |
| C4—C3—H3 | 119.9 | C12—C17—H17 | 119.7 |
| C5—C4—C3 | 120.9 (4) | C19—C18—C9 | 174.8 (4) |
| C5—C4—H4 | 119.5 | C18—C19—C20 | 176.0 (4) |
| C3—C4—H4 | 119.5 | C19—C20—C21 | 116.2 (4) |
| C4—C5—C6 | 119.0 (4) | C19—C20—H20A | 108.2 |
| C4—C5—H5 | 120.5 | C21—C20—H20A | 108.2 |
| C6—C5—H5 | 120.5 | C19—C20—H20B | 108.2 |
| C5—C6—C7 | 120.7 (4) | C21—C20—H20B | 108.2 |
| C5—C6—H6 | 119.6 | H20A—C20—H20B | 107.4 |
| C7—C6—H6 | 119.6 | C20—C21—C22 | 113.6 (4) |
| C6—C7—C2 | 120.4 (4) | C20—C21—H21A | 108.8 |
| C6—C7—H7 | 119.8 | C22—C21—H21A | 108.8 |
| C2—C7—H7 | 119.8 | C20—C21—H21B | 108.8 |
| C9—C8—C1 | 126.9 (3) | C22—C21—H21B | 108.8 |
| C9—C8—H8 | 116.5 | H21A—C21—H21B | 107.7 |
| C1—C8—H8 | 116.5 | C23—C22—C21 | 111.3 (4) |
| C8—C9—C18 | 120.1 (3) | C23—C22—H22A | 109.4 |
| C8—C9—C10 | 119.7 (3) | C21—C22—H22A | 109.4 |
| C18—C9—C10 | 120.0 (3) | C23—C22—H22B | 109.4 |
| C11—C10—C9 | 125.7 (4) | C21—C22—H22B | 109.4 |
| C11—C10—H10 | 117.1 | H22A—C22—H22B | 108.0 |
| C9—C10—H10 | 117.1 | C24—C23—C22 | 113.4 (4) |
| C10—C11—C12 | 125.9 (4) | C24—C23—H23A | 108.9 |
| C10—C11—H11 | 117.0 | C22—C23—H23A | 108.9 |
| C12—C11—H11 | 117.0 | C24—C23—H23B | 108.9 |
| C13—C12—C17 | 118.2 (3) | C22—C23—H23B | 108.9 |
| C13—C12—C11 | 119.7 (4) | H23A—C23—H23B | 107.7 |
| C17—C12—C11 | 122.1 (3) | C25—C24—C23 | 178.0 (5) |
| C14—C13—C12 | 121.3 (4) | C24—C25—H25 | 180.0 |
| C14—C13—H13 | 119.4 | ||
| O1—C1—C2—C3 | −145.9 (3) | C8—C9—C10—C11 | −179.1 (4) |
| C8—C1—C2—C3 | 92.2 (4) | C18—C9—C10—C11 | −3.5 (6) |
| O1—C1—C2—C7 | 38.1 (5) | C9—C10—C11—C12 | 175.0 (3) |
| C8—C1—C2—C7 | −83.8 (4) | C10—C11—C12—C13 | −169.7 (4) |
| C7—C2—C3—C4 | 0.2 (6) | C10—C11—C12—C17 | 8.1 (6) |
| C1—C2—C3—C4 | −175.9 (4) | C17—C12—C13—C14 | −0.6 (5) |
| C2—C3—C4—C5 | 0.4 (6) | C11—C12—C13—C14 | 177.2 (3) |
| C3—C4—C5—C6 | −1.0 (7) | C12—C13—C14—C15 | −0.6 (6) |
| C4—C5—C6—C7 | 1.0 (7) | C13—C14—C15—C16 | 1.1 (6) |
| C5—C6—C7—C2 | −0.3 (7) | C14—C15—C16—C17 | −0.5 (6) |
| C3—C2—C7—C6 | −0.2 (6) | C15—C16—C17—C12 | −0.8 (6) |
| C1—C2—C7—C6 | 175.8 (4) | C13—C12—C17—C16 | 1.3 (5) |
| O1—C1—C8—C9 | 133.8 (4) | C11—C12—C17—C16 | −176.5 (4) |
| C2—C1—C8—C9 | −103.1 (4) | C19—C20—C21—C22 | −54.5 (5) |
| C1—C8—C9—C18 | 2.6 (6) | C20—C21—C22—C23 | −175.6 (4) |
| C1—C8—C9—C10 | 178.2 (3) | C21—C22—C23—C24 | 68.5 (6) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1A···O1i | 0.84 | 1.83 | 2.652 (3) | 166 |
Symmetry code: (i) −y+1, x−y+1, z−1/3.
Footnotes
Supporting information for this paper is available from the IUCr electronic archives (Reference: SU5038).
References
- Bruker (2013). APEX2, SAINT and SADABS. Bruker AXS Inc., Madison, Wisconsin, USA.
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Macrae, C. F., Bruno, I. J., Chisholm, J. A., Edgington, P. R., McCabe, P., Pidcock, E., Rodriguez-Monge, L., Taylor, R., van de Streek, J. & Wood, P. A. (2008). J. Appl. Cryst. 41, 466–470.
- Piotti, M. E. & Alper, H. (1997). J. Org. Chem. 62, 8484–8489. [DOI] [PubMed]
- Poloukhtine, A. & Popik, V. (2005). J. Org. Chem. 70, 1297–1305. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Teo, W. T., Rao, W., Ng, C. J. H., Koh, S. W. Y. & Chan, P. W. H. (2014). Org. Lett. 16, 1248–1251. [DOI] [PubMed]
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, New_Global_Publ_Block. DOI: 10.1107/S205698901402742X/su5038sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S205698901402742X/su5038Isup2.hkl
Supporting information file. DOI: 10.1107/S205698901402742X/su5038Isup3.cml
. DOI: 10.1107/S205698901402742X/su5038fig1.tif
A view of the molecular structure of the title compound, with atom labelling. Displacement ellipsoids are drawn at the 50% probability level.
c . DOI: 10.1107/S205698901402742X/su5038fig2.tif
A partial view along the c axis of the crystal packing of the title compound. Hydrogen bonds are shown as dashed lines (see Table 1 for details).
. DOI: 10.1107/S205698901402742X/su5038fig3.tif
Reaction scheme.
CCDC reference: 1009363
Additional supporting information: crystallographic information; 3D view; checkCIF report



