In the title compound, the ring conformations of the tricycles are in an envelope, a half-chair and a chair. In the crystal, intermolecular O—H⋯O and C—H⋯O hydrogen bonds and C—H⋯π interactions link the molecules into a three-dimensional architecture.
Keywords: crystal structure, hydrogen bonds, taxane skeleton, paclitaxel
Abstract
In the title compound, C36H44O10·C6H6, the dioxolane ring adopts an envelope conformation with the C atom bonded to the H atom as the flap, while the cyclohexene and cyclohexane rings are in half-chair and chair conformations, respectively. In the crystal, a pair of O—H⋯O hydrogen bonds with an R 2 2(26) graph-set motif connect the benzoate molecules into an inversion dimer. The dimers are linked by a weak C—H⋯O interaction into a tape structure along [01-1]. The benzene molecule links the tapes through C—H⋯O and C—H⋯π interactions, forming a sheet parallel to (100).
Chemical context
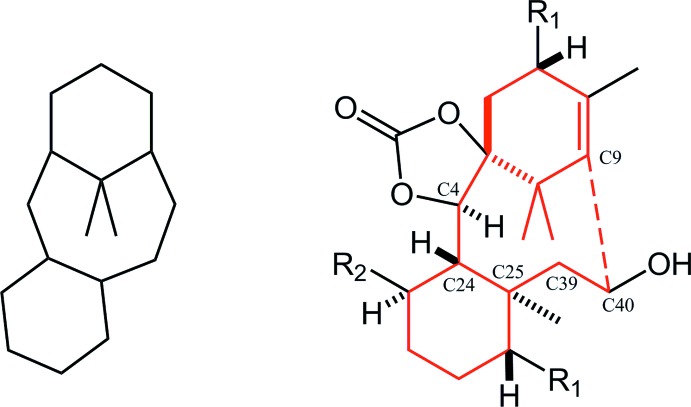
Paclitaxel is a well-known natural diterpenoid containing a taxane framework (tricyclo[9.3.1.03,8]pentadecane; Fig. 1 ▸), with potent antitumor activity (Wall & Wani, 1995 ▸). This unique and complicated structure has attracted significant interest, and a large number of synthetic studies have been reported. In these researches, whereas some structure data after cyclization into taxane or taxoid derivatives are available (§ 4), precursors just before cyclization are very few. The title compound has been obtained in our synthetic study of paclitaxel as a cyclization precursor to build the taxane skeleton (Fukaya et al., 2014 ▸).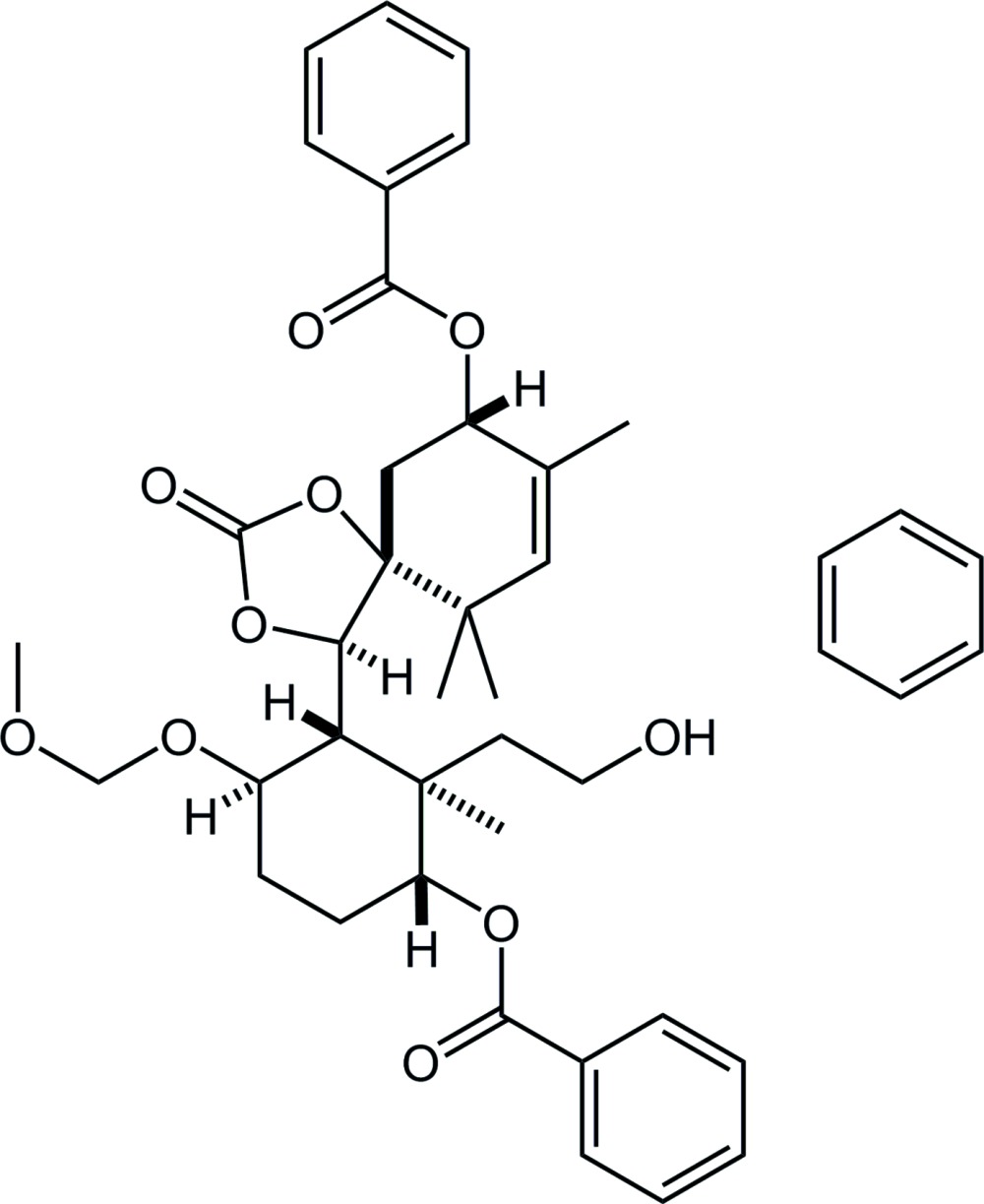
Figure 1.
Left: the structure of the tricyclo[9.3.1.03,8]pentadecane (taxane) skeleton. Right: the title compound. Red lines indicate the taxane skeleton with the expected bond (red dashed line). R 1 = –OC(=O)Ph, R 2 = –OCH2OCH3.
Structural commentary
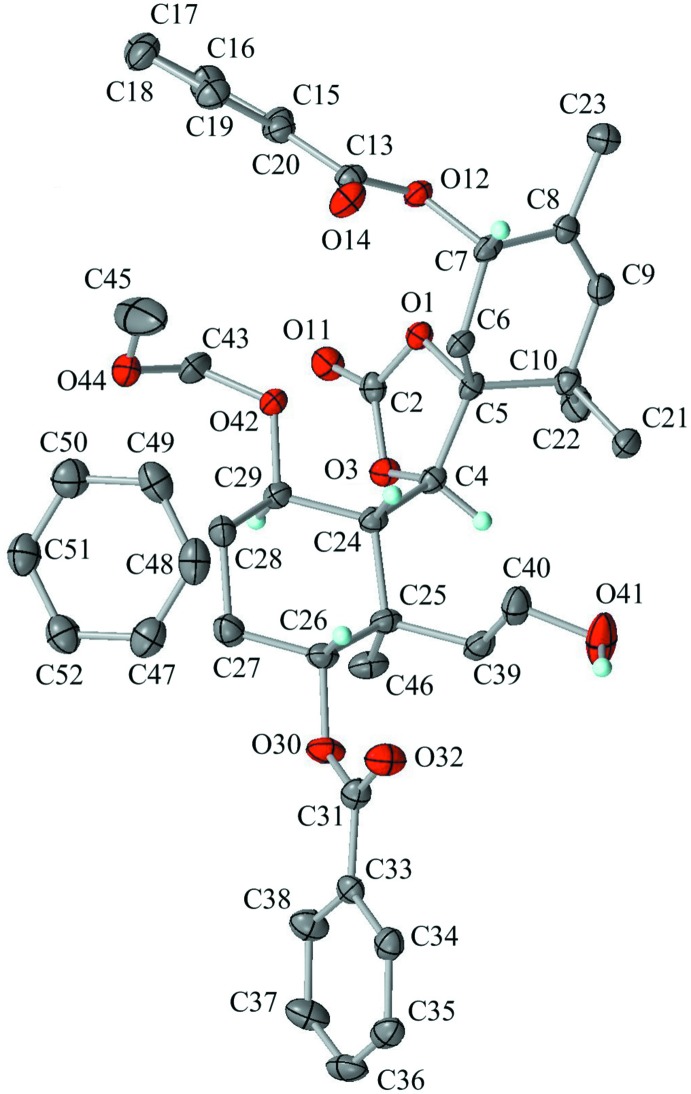
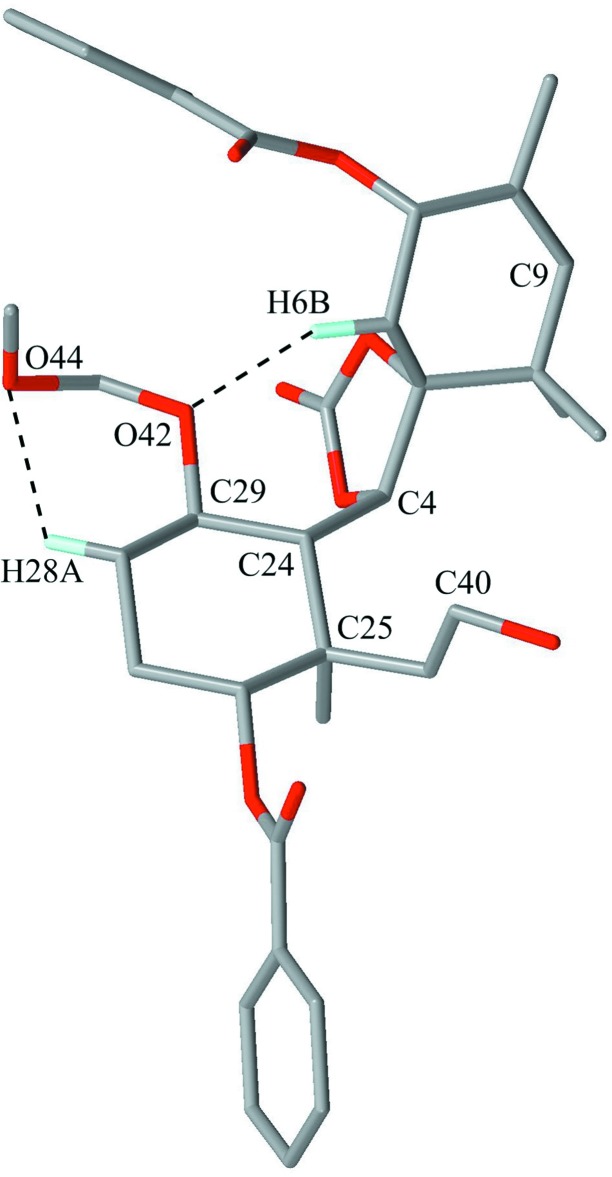
The molecular structure of the title compound is shown in Fig. 2 ▸. The dioxolane ring (O1/C2/O3/C4/C5) is in an envelope conformation with puckering parameters of Q(2) = 0.165 (2) Å and ϕ(2) = 114.5 (6)°. The flap atom C4 deviates from the mean plane of other atoms by 0.270 (3) Å. The cyclohexene ring (C5–C10), which is spiro-fused to the dioxolane ring, is in a half-chair conformation with puckering parameters of Q = 0.469 (2) Å, θ = 127.5 (2)°, ϕ(2) = 197.2 (3)°, Q(2) = 0.372 (2) Å and Q(3) = −0.285 (2) Å. Atoms C5 and C6 deviate from the mean plane of the other atoms by −0.493 (4) and 0.212 (4) Å, respectively. The cyclohexane ring (C24–C29) is in a chair conformation with puckering parameters Q = 0.587 (2) Å, θ = 4.6 (2)°, ϕ = 246 (3)°, Q(2) = 0.042 (2) Å and Q(3) = 0.585 (2) Å. The large substituents (C24—C4, C25—C39, C26—O30 and C29—O42) are in the equatorial positions. The methoxymethoxy group (O42/C43/O44/C45) shows a helical form with torsion angles of 76.5 (3)° for C29—O42—C43—O44 and 64.8 (3)° for O42—C43—O44—C45 held by weak intramolecular C—H⋯O interactions (Fig. 3 ▸, Table 1 ▸). The atom pairs which may be connected by cyclization into a taxane framework are C9 and C40 (Figs. 1 ▸ and 3 ▸) with their distance being 5.831 (3) Å in the present conformation. They are expected to approach each other by rotation about the C4–C24, C25–C39 and C39–C40 bonds.
Figure 2.
The asymmetric unit of the title compound with atom labelling. Displacement ellipsoids are drawn at the 50% probability level. Only H atoms connected to O and chiral C atoms are shown for clarity.
Figure 3.
The molecular conformation indicating the intramolecular C—H⋯O interactions with dashed lines. Only H atoms involved in hydrogen bonds are shown for clarity. The benzene solvent molecule has been omitted.
Table 1. Hydrogen-bond geometry (, ).
Cg is the centroid of the C47C52 ring.
| DHA | DH | HA | D A | DHA |
|---|---|---|---|---|
| C6H6BO42 | 0.99 | 2.32 | 3.095(2) | 135 |
| C28H28AO44 | 0.99 | 2.37 | 2.989(3) | 120 |
| O41H41O14i | 0.84 | 2.06 | 2.888(2) | 170 |
| C7H7O32i | 1.00 | 2.34 | 3.269(2) | 155 |
| C18H18O11ii | 0.95 | 2.53 | 3.465(2) | 168 |
| C49H49O11 | 0.95 | 2.46 | 3.300(3) | 147 |
| C27H27A Cg iii | 0.95 | 2.64 | 3.514(2) | 147 |
Symmetry codes: (i) 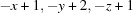 ; (ii)
; (ii) 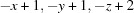 ; (iii)
; (iii) 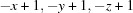 .
.
Supramolecular features
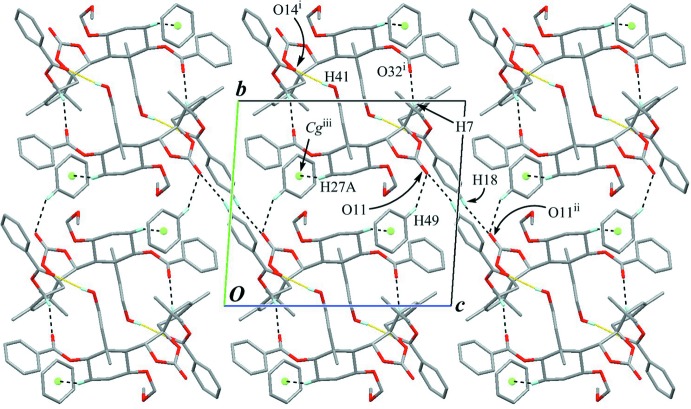
The crystal packing is stabilized by a pair of intermolecular O—H⋯O hydrogen bonds (O41—H41⋯O14i; Table 1 ▸) with an 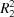 (26) graph-set motif, forming an inversion dimer (Fig. 4 ▸). In the dimer, a pair of C—H⋯O hydrogen bonds (C7—H7⋯O32i; Table 1 ▸) are also observed. The dimers are further linked by a weak intermolecular C—H⋯O hydrogen bond (C18—H18⋯O11ii; Table 1 ▸) into a tape along [01
(26) graph-set motif, forming an inversion dimer (Fig. 4 ▸). In the dimer, a pair of C—H⋯O hydrogen bonds (C7—H7⋯O32i; Table 1 ▸) are also observed. The dimers are further linked by a weak intermolecular C—H⋯O hydrogen bond (C18—H18⋯O11ii; Table 1 ▸) into a tape along [01 ]. The benzene molecule links adjacent tapes through C—H⋯O and C—H⋯π interactions (C49—H49⋯O11 and C27—H27A⋯Cg
iii; Table 1 ▸), forming a sheet parallel to (100).
]. The benzene molecule links adjacent tapes through C—H⋯O and C—H⋯π interactions (C49—H49⋯O11 and C27—H27A⋯Cg
iii; Table 1 ▸), forming a sheet parallel to (100).
Figure 4.
The crystal packing viewed along the a axis. Dotted yellow lines indicate the intermolecular O—H⋯O hydrogen bonds which form the inversion dimers. Black dashed lines indicate the intermolecular C—H⋯O and C—H⋯π interactions. Cg is the centroid of the benzene solvent molecule. Only H atoms involved in hydrogen bonds are shown for clarity. [Symmetry codes: (i) −x + 1, −y + 2, −z + 1; (ii) −x + 1, −y + 1, −z + 2; (iii) −x + 1, −y + 1, −z + 1.]
Database survey
In the Cambridge Structural Database (CSD, Version 5.35, November 2013; Groom & Allen, 2014 ▸), four compounds possessing a core of 6,6,8-trimethyl-1,3-dioxaspiro[4.5]dec-7-ene are found (Fig. 5 ▸). These include its derivatives with 2-one (PUQLAO; Nishizawa et al., 1998 ▸) and 2,2-dimethyl (NEGBOQ; Poujol et al., 1997 ▸) substitutes. Another tetracyclic taxoid (ILIQUP; Ohba et al., 2003 ▸) with a core of 6,6,8-trimethyl-1,3-dioxaspiro[4.5]decan-2-one, obtained in our previous study, is closely related to the title compound. Only one crystalline compound just before cyclization is found in the literature (Nicolaou et al., 1995 ▸), however it is not registered in the CSD.
Figure 5.
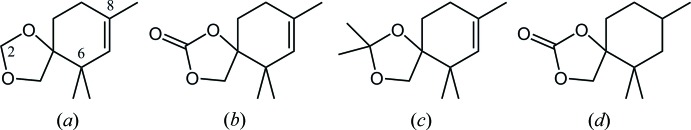
Core substructures for database survey; (a) 6,6,8-trimethyl-1,3-dioxaspiro[4.5]dec-7-ene, (b) its 2-one derivative, (c) the 2,2-dimethyl derivative and (d) 6,6,8-trimethyl-1,3-dioxaspiro[4.5]decan-2-one.
Synthesis and crystallization
The title compound was obtained in a synthetic study on paclitaxel. The cyclohexene unit (C5–C10) was provided according to the reported procedure (Nicolaou et al., 1995 ▸), and coupled with the substituted cyclohexane unit (C24–C29) synthesized from 3-methylanisole (Fukaya et al., 2014 ▸) by a Shapiro reaction (Nicolaou et al., 1995 ▸). Further manipulation of the functional groups afforded the title compound, which was purified by silica gel column chromatography. Colorless crystals were grown from a benzene solution under a pentane-saturated atmosphere by slow evaporation at ambient temperature.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. C-bound H atoms were positioned geometrically with C—H = 0.95–1.00 Å, and constrained to ride on their parent atoms with U iso(H) = 1.2U eq(C) or 1.5U eq(methyl C). The H atom of hydroxy group (O41) was placed guided by difference maps and then treated as riding, with O—H = 0.84 Å and with U iso(H) = 1.5U eq(O). 13 problematic reflections were omitted from the final refinement.
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C36H44O10C6H6 |
| M r | 714.82 |
| Crystal system, space group | Triclinic, P
|
| Temperature (K) | 90 |
| a, b, c () | 9.6397(6), 13.6008(8), 15.0461(10) |
| , , () | 83.6966(19), 77.488(2), 77.9768(18) |
| V (3) | 1879.2(2) |
| Z | 2 |
| Radiation type | Mo K |
| (mm1) | 0.09 |
| Crystal size (mm) | 0.50 0.37 0.19 |
| Data collection | |
| Diffractometer | Bruker D8 Venture |
| Absorption correction | Multi-scan (SADABS; Bruker, 2013 ▸) |
| T min, T max | 0.96, 0.98 |
| No. of measured, independent and observed [I > 2(I)] reflections | 25389, 6526, 5180 |
| R int | 0.043 |
| (sin /)max (1) | 0.595 |
| Refinement | |
| R[F 2 > 2(F 2)], wR(F 2), S | 0.042, 0.151, 0.93 |
| No. of reflections | 6526 |
| No. of parameters | 475 |
| H-atom treatment | H-atom parameters constrained |
| max, min (e 3) | 0.25, 0.25 |
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S2056989014026048/is5382sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989014026048/is5382Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989014026048/is5382Isup3.cml
CCDC reference: 1036428
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
We thank Professor S. Ohba (Keio University, Japan) and Dr K. Yoza (Bruker AXS Inc.) for providing valuable advice.
supplementary crystallographic information
Crystal data
| C36H44O10·C6H6 | F(000) = 764 |
| Mr = 714.82 | Dx = 1.263 Mg m−3 |
| Triclinic, P1 | Melting point: 459.2 K |
| a = 9.6397 (6) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 13.6008 (8) Å | Cell parameters from 9980 reflections |
| c = 15.0461 (10) Å | θ = 2.2–25.1° |
| α = 83.6966 (19)° | µ = 0.09 mm−1 |
| β = 77.488 (2)° | T = 90 K |
| γ = 77.9768 (18)° | Plate, colorless |
| V = 1879.2 (2) Å3 | 0.50 × 0.37 × 0.19 mm |
| Z = 2 |
Data collection
| Bruker D8 Venture diffractometer | 6526 independent reflections |
| Radiation source: fine-focus sealed tube | 5180 reflections with I > 2σ(I) |
| Multilayered confocal mirror monochromator | Rint = 0.043 |
| Detector resolution: 8.333 pixels mm-1 | θmax = 25.0°, θmin = 2.2° |
| φ and ω scans | h = −11→11 |
| Absorption correction: multi-scan (SADABS; Bruker, 2013) | k = −16→16 |
| Tmin = 0.96, Tmax = 0.98 | l = −17→17 |
| 25389 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.042 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.151 | H-atom parameters constrained |
| S = 0.93 | w = 1/[σ2(Fo2) + (0.0996P)2 + 1.0739P] where P = (Fo2 + 2Fc2)/3 |
| 6526 reflections | (Δ/σ)max = 0.023 |
| 475 parameters | Δρmax = 0.25 e Å−3 |
| 0 restraints | Δρmin = −0.25 e Å−3 |
Special details
| Experimental. Recrystallization from benzene, toluene, chloroform, dichloromethane, diethyl ether, tetrahydrofuran, ethyl acetate, acetonitrile and methanol solutions under the air were failed. These solutions under hexane atmosphere also gave unsatisfactory results. Only the condition mentioned above has been quite effective to afford the single crystals suitable for X-ray analysis.; M.p. 458.7–459.2 K (not corrected); IR (film): 3524, 2945, 2890, 1790, 1715, 1274, 714 cm-1; 1H NMR (500 MHz, CDCl3): δ (p.p.m.) 8.13–8.10 (m, 2H), 8.03–7.99 (m, 2H), 7.60–7.53 (m, 2H), 7.48–7.42 (m, 4H), 5.56 (d, J = 6.0 Hz, 1H), 5.41 (t, J = 0.9 Hz, 1H), 5.09 (dd, J = 11.5, 4.6 Hz, 1H), 4.84 (s, 1H), 4.49 (d, J = 7.7 Hz, 1H), 4.13 (d, J = 7.7 Hz, 1H), 3.76–3.68 (m, 2H), 3.63 (ddd, J = 10.6, 7.2, 7.2 Hz, 1H), 3.08 (d, J = 14.9 Hz, 1H), 2.66 (s, 3H), 2.40 (dddd, J = 12.9, 4.3, 4.0, 4.0 Hz, 1H), 2.34 (d, J = 10.3 Hz, 1H), 2.29 (dd, J = 15.5, 6.3 Hz, 1H), 1.93 (dddd, J = 12.9, 4.3, 4.3, 4.0 Hz, 1H), 1.75 (d, J = 0.9 Hz, 3H), 1.73–1.60 (m, 3H), 1.59–1.45 (m, 2H), 1.22 (s, 3H), 1.20 (s, 3H), 1.11 (s, 3H); 13C NMR (125 MHz, CDCl3): δ (p.p.m.) 166.4 (C), 166.0 (C), 155.2 (C), 135.1 (CH), 133.4 (CH), 133.3 (CH), 130.5 (C), 130.2 (C), 130.0 (CH), 129.7 (CH), 129.0 (C), 128.7 (CH), 128.7 (CH), 97.9 (CH2), 87.1 (C), 76.9 (CH), 76.5 (CH), 75.1 (CH), 68.7 (CH), 58.3 (CH2), 54.7 (CH3), 45.9 (CH), 42.0 (C), 41.2 (C), 38.1 (CH2), 31.5 (CH2), 30.6 (CH2), 25.7 (CH3), 25.1 (CH2), 22.3 (CH3), 20.2 (CH3), 16.8 (CH3); HRMS (ESI): m/z calcd for C36H44O10Na+ [M+Na]+ 659.2832, found 659.2836. |
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger.Problematic 13 reflections with |I(obs)-I(calc)|/σW(I) greater than 10 (–8 –2 1, –8 –2 2, –8 –1 2, –7 –4 3, –8 –3 6, 3 11 7, 3 10 8, 0 9 9, 2 10 9, 2 8 10, 4 9 10, 2 7 11, 3 8 11) have been omitted in the final refinement. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.25606 (14) | 0.79849 (9) | 0.81366 (8) | 0.0191 (3) | |
| C2 | 0.2032 (2) | 0.71963 (14) | 0.79948 (13) | 0.0201 (4) | |
| O3 | 0.16170 (14) | 0.73096 (9) | 0.71879 (9) | 0.0206 (3) | |
| C4 | 0.2098 (2) | 0.81686 (14) | 0.66496 (13) | 0.0185 (4) | |
| H4 | 0.1247 | 0.8597 | 0.6428 | 0.022* | |
| C5 | 0.2481 (2) | 0.87501 (14) | 0.73783 (12) | 0.0181 (4) | |
| C6 | 0.3931 (2) | 0.90852 (14) | 0.71464 (13) | 0.0194 (4) | |
| H6A | 0.3918 | 0.9613 | 0.6638 | 0.023* | |
| H6B | 0.4698 | 0.8506 | 0.6934 | 0.023* | |
| C7 | 0.4298 (2) | 0.94956 (14) | 0.79519 (13) | 0.0204 (4) | |
| H7 | 0.5042 | 0.9925 | 0.7716 | 0.024* | |
| C8 | 0.3023 (2) | 1.01014 (14) | 0.85459 (13) | 0.0204 (4) | |
| C9 | 0.1692 (2) | 1.01625 (14) | 0.84154 (13) | 0.0219 (4) | |
| H9 | 0.094 | 1.0582 | 0.8799 | 0.026* | |
| C10 | 0.1244 (2) | 0.96338 (14) | 0.77197 (13) | 0.0206 (4) | |
| O11 | 0.19139 (15) | 0.64864 (10) | 0.85222 (9) | 0.0258 (3) | |
| O12 | 0.48877 (14) | 0.86671 (9) | 0.85533 (9) | 0.0206 (3) | |
| C13 | 0.6265 (2) | 0.82130 (14) | 0.82802 (13) | 0.0204 (4) | |
| O14 | 0.70642 (15) | 0.84907 (11) | 0.76027 (10) | 0.0279 (3) | |
| C15 | 0.6684 (2) | 0.73187 (15) | 0.88941 (13) | 0.0221 (4) | |
| C16 | 0.8142 (2) | 0.68716 (16) | 0.88007 (14) | 0.0264 (5) | |
| H16 | 0.8853 | 0.7165 | 0.8378 | 0.032* | |
| C17 | 0.8556 (2) | 0.60012 (16) | 0.93234 (16) | 0.0324 (5) | |
| H17 | 0.955 | 0.5699 | 0.9263 | 0.039* | |
| C18 | 0.7518 (3) | 0.55705 (16) | 0.99346 (15) | 0.0328 (5) | |
| H18 | 0.7799 | 0.4965 | 1.0285 | 0.039* | |
| C19 | 0.6074 (3) | 0.60200 (16) | 1.00363 (15) | 0.0321 (5) | |
| H19 | 0.5367 | 0.5729 | 1.0465 | 0.039* | |
| C20 | 0.5651 (2) | 0.68926 (15) | 0.95176 (14) | 0.0269 (5) | |
| H20 | 0.4656 | 0.7198 | 0.9588 | 0.032* | |
| C21 | 0.0916 (2) | 1.04115 (15) | 0.69334 (15) | 0.0282 (5) | |
| H21A | 0.0245 | 1.1008 | 0.7182 | 0.042* | |
| H21B | 0.0476 | 1.0116 | 0.6524 | 0.042* | |
| H21C | 0.1818 | 1.0606 | 0.6593 | 0.042* | |
| C22 | −0.0155 (2) | 0.92602 (16) | 0.81716 (16) | 0.0299 (5) | |
| H22A | 0.0011 | 0.8803 | 0.8705 | 0.045* | |
| H22B | −0.045 | 0.8902 | 0.7734 | 0.045* | |
| H22C | −0.0921 | 0.9837 | 0.8365 | 0.045* | |
| C23 | 0.3361 (2) | 1.06479 (16) | 0.92679 (14) | 0.0274 (5) | |
| H23A | 0.2466 | 1.1054 | 0.9587 | 0.041* | |
| H23B | 0.4042 | 1.1089 | 0.8981 | 0.041* | |
| H23C | 0.3795 | 1.0157 | 0.9705 | 0.041* | |
| C24 | 0.3249 (2) | 0.78089 (14) | 0.58056 (13) | 0.0191 (4) | |
| H24 | 0.3926 | 0.8297 | 0.5659 | 0.023* | |
| C25 | 0.2548 (2) | 0.78407 (15) | 0.49501 (13) | 0.0213 (4) | |
| C26 | 0.3765 (2) | 0.74420 (15) | 0.41478 (13) | 0.0224 (4) | |
| H26 | 0.4423 | 0.7939 | 0.3956 | 0.027* | |
| C27 | 0.4645 (2) | 0.64260 (15) | 0.43479 (14) | 0.0269 (5) | |
| H27A | 0.5411 | 0.623 | 0.3809 | 0.032* | |
| H27B | 0.4018 | 0.5915 | 0.4485 | 0.032* | |
| C28 | 0.5328 (2) | 0.64693 (15) | 0.51624 (14) | 0.0248 (4) | |
| H28A | 0.5916 | 0.5802 | 0.5296 | 0.03* | |
| H28B | 0.5975 | 0.6967 | 0.5017 | 0.03* | |
| C29 | 0.4158 (2) | 0.67659 (14) | 0.59900 (13) | 0.0208 (4) | |
| H29 | 0.3513 | 0.6257 | 0.6133 | 0.025* | |
| O30 | 0.30900 (16) | 0.73471 (10) | 0.33928 (9) | 0.0268 (3) | |
| C31 | 0.3358 (2) | 0.79233 (15) | 0.26237 (13) | 0.0223 (4) | |
| O32 | 0.40856 (17) | 0.85680 (11) | 0.25168 (10) | 0.0316 (4) | |
| C33 | 0.2631 (2) | 0.76877 (15) | 0.19191 (13) | 0.0208 (4) | |
| C34 | 0.2657 (2) | 0.83032 (15) | 0.11194 (13) | 0.0247 (5) | |
| H34 | 0.3158 | 0.8849 | 0.1024 | 0.03* | |
| C35 | 0.1955 (2) | 0.81243 (17) | 0.04616 (14) | 0.0286 (5) | |
| H35 | 0.1968 | 0.855 | −0.0084 | 0.034* | |
| C36 | 0.1237 (2) | 0.73291 (18) | 0.05975 (14) | 0.0330 (5) | |
| H36 | 0.075 | 0.7209 | 0.0147 | 0.04* | |
| C37 | 0.1222 (3) | 0.67046 (18) | 0.13876 (15) | 0.0344 (5) | |
| H37 | 0.0732 | 0.6153 | 0.1476 | 0.041* | |
| C38 | 0.1919 (2) | 0.68816 (16) | 0.20503 (14) | 0.0296 (5) | |
| H38 | 0.1909 | 0.6452 | 0.2593 | 0.036* | |
| C39 | 0.1840 (2) | 0.89252 (15) | 0.46698 (14) | 0.0253 (5) | |
| H39A | 0.1477 | 0.8901 | 0.4107 | 0.03* | |
| H39B | 0.0986 | 0.9141 | 0.5153 | 0.03* | |
| C40 | 0.2753 (2) | 0.97374 (15) | 0.44990 (15) | 0.0303 (5) | |
| H40A | 0.3069 | 0.982 | 0.5066 | 0.036* | |
| H40B | 0.3627 | 0.954 | 0.4023 | 0.036* | |
| O41 | 0.1917 (2) | 1.06636 (12) | 0.42091 (11) | 0.0461 (5) | |
| H41 | 0.2305 | 1.0848 | 0.3679 | 0.069* | |
| O42 | 0.47536 (14) | 0.68229 (10) | 0.67742 (9) | 0.0223 (3) | |
| C43 | 0.5313 (3) | 0.58881 (17) | 0.71882 (16) | 0.0364 (6) | |
| H43A | 0.5402 | 0.599 | 0.7814 | 0.044* | |
| H43B | 0.4618 | 0.5432 | 0.724 | 0.044* | |
| O44 | 0.6660 (2) | 0.54241 (13) | 0.67142 (12) | 0.0538 (5) | |
| C45 | 0.7765 (3) | 0.5978 (3) | 0.6691 (3) | 0.0826 (13) | |
| H45A | 0.7436 | 0.6683 | 0.6492 | 0.124* | |
| H45B | 0.8639 | 0.5691 | 0.6262 | 0.124* | |
| H45C | 0.7983 | 0.5943 | 0.7301 | 0.124* | |
| C46 | 0.1359 (2) | 0.72061 (16) | 0.51442 (14) | 0.0273 (5) | |
| H46A | 0.096 | 0.7225 | 0.4595 | 0.041* | |
| H46B | 0.0588 | 0.748 | 0.5644 | 0.041* | |
| H46C | 0.1771 | 0.6508 | 0.5318 | 0.041* | |
| C47 | 0.1268 (2) | 0.40365 (17) | 0.63928 (16) | 0.0345 (5) | |
| H47 | 0.0799 | 0.4303 | 0.59 | 0.041* | |
| C48 | 0.1481 (2) | 0.46772 (17) | 0.69805 (16) | 0.0343 (5) | |
| H48 | 0.1157 | 0.5383 | 0.6892 | 0.041* | |
| C49 | 0.2161 (2) | 0.42938 (16) | 0.76949 (16) | 0.0335 (5) | |
| H49 | 0.2298 | 0.4736 | 0.8102 | 0.04* | |
| C50 | 0.2644 (3) | 0.32710 (17) | 0.78217 (16) | 0.0356 (5) | |
| H50 | 0.3119 | 0.3008 | 0.8312 | 0.043* | |
| C51 | 0.2433 (3) | 0.26278 (17) | 0.72304 (16) | 0.0350 (5) | |
| H51 | 0.2769 | 0.1923 | 0.7314 | 0.042* | |
| C52 | 0.1735 (2) | 0.30103 (17) | 0.65192 (16) | 0.0329 (5) | |
| H52 | 0.1578 | 0.2569 | 0.6119 | 0.04* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0219 (7) | 0.0173 (7) | 0.0177 (7) | −0.0039 (5) | −0.0048 (5) | 0.0020 (5) |
| C2 | 0.0178 (10) | 0.0194 (10) | 0.0209 (10) | −0.0022 (8) | −0.0001 (8) | −0.0021 (8) |
| O3 | 0.0242 (7) | 0.0203 (7) | 0.0186 (7) | −0.0082 (6) | −0.0052 (6) | 0.0023 (5) |
| C4 | 0.0169 (10) | 0.0187 (10) | 0.0202 (10) | −0.0046 (7) | −0.0050 (8) | 0.0029 (8) |
| C5 | 0.0185 (10) | 0.0174 (9) | 0.0168 (9) | −0.0019 (7) | −0.0045 (8) | 0.0045 (8) |
| C6 | 0.0198 (10) | 0.0191 (10) | 0.0190 (10) | −0.0024 (8) | −0.0063 (8) | 0.0030 (8) |
| C7 | 0.0214 (10) | 0.0179 (10) | 0.0223 (10) | −0.0037 (8) | −0.0089 (8) | 0.0056 (8) |
| C8 | 0.0252 (11) | 0.0160 (9) | 0.0202 (10) | −0.0040 (8) | −0.0065 (8) | 0.0021 (8) |
| C9 | 0.0240 (11) | 0.0171 (10) | 0.0229 (10) | −0.0012 (8) | −0.0040 (8) | −0.0005 (8) |
| C10 | 0.0165 (10) | 0.0198 (10) | 0.0255 (10) | −0.0021 (8) | −0.0060 (8) | −0.0011 (8) |
| O11 | 0.0350 (8) | 0.0217 (7) | 0.0201 (7) | −0.0099 (6) | −0.0021 (6) | 0.0025 (6) |
| O12 | 0.0216 (7) | 0.0199 (7) | 0.0212 (7) | −0.0036 (6) | −0.0088 (6) | 0.0026 (6) |
| C13 | 0.0185 (10) | 0.0228 (10) | 0.0232 (10) | −0.0073 (8) | −0.0083 (8) | −0.0012 (8) |
| O14 | 0.0221 (8) | 0.0314 (8) | 0.0292 (8) | −0.0074 (6) | −0.0062 (6) | 0.0090 (6) |
| C15 | 0.0234 (11) | 0.0225 (10) | 0.0220 (10) | −0.0046 (8) | −0.0083 (8) | 0.0000 (8) |
| C16 | 0.0227 (11) | 0.0294 (11) | 0.0286 (11) | −0.0074 (9) | −0.0074 (9) | 0.0006 (9) |
| C17 | 0.0279 (12) | 0.0309 (12) | 0.0389 (13) | −0.0003 (9) | −0.0163 (10) | 0.0041 (10) |
| C18 | 0.0432 (14) | 0.0257 (11) | 0.0294 (12) | −0.0017 (10) | −0.0157 (10) | 0.0064 (9) |
| C19 | 0.0397 (13) | 0.0277 (12) | 0.0258 (11) | −0.0066 (10) | −0.0023 (10) | 0.0034 (9) |
| C20 | 0.0264 (11) | 0.0275 (11) | 0.0248 (11) | −0.0033 (9) | −0.0046 (9) | 0.0022 (9) |
| C21 | 0.0295 (11) | 0.0220 (11) | 0.0359 (12) | 0.0001 (9) | −0.0170 (10) | −0.0024 (9) |
| C22 | 0.0181 (11) | 0.0296 (12) | 0.0413 (13) | −0.0026 (9) | −0.0032 (9) | −0.0092 (10) |
| C23 | 0.0284 (11) | 0.0287 (11) | 0.0265 (11) | −0.0050 (9) | −0.0080 (9) | −0.0042 (9) |
| C24 | 0.0206 (10) | 0.0190 (10) | 0.0188 (10) | −0.0053 (8) | −0.0060 (8) | 0.0013 (8) |
| C25 | 0.0249 (11) | 0.0238 (10) | 0.0171 (10) | −0.0059 (8) | −0.0087 (8) | 0.0022 (8) |
| C26 | 0.0293 (11) | 0.0230 (10) | 0.0180 (10) | −0.0079 (8) | −0.0093 (8) | 0.0001 (8) |
| C27 | 0.0346 (12) | 0.0238 (11) | 0.0212 (10) | −0.0040 (9) | −0.0041 (9) | −0.0024 (9) |
| C28 | 0.0275 (11) | 0.0201 (10) | 0.0248 (11) | 0.0016 (8) | −0.0056 (9) | −0.0038 (8) |
| C29 | 0.0228 (10) | 0.0209 (10) | 0.0195 (10) | −0.0027 (8) | −0.0083 (8) | 0.0003 (8) |
| O30 | 0.0391 (9) | 0.0297 (8) | 0.0164 (7) | −0.0147 (7) | −0.0100 (6) | 0.0027 (6) |
| C31 | 0.0229 (10) | 0.0226 (10) | 0.0191 (10) | −0.0027 (8) | −0.0018 (8) | 0.0011 (8) |
| O32 | 0.0425 (9) | 0.0319 (8) | 0.0260 (8) | −0.0188 (7) | −0.0114 (7) | 0.0054 (6) |
| C33 | 0.0200 (10) | 0.0236 (10) | 0.0165 (10) | −0.0013 (8) | −0.0003 (8) | −0.0039 (8) |
| C34 | 0.0237 (11) | 0.0251 (11) | 0.0227 (10) | −0.0014 (8) | −0.0022 (8) | −0.0013 (9) |
| C35 | 0.0271 (11) | 0.0364 (12) | 0.0180 (10) | 0.0023 (9) | −0.0036 (9) | −0.0017 (9) |
| C36 | 0.0323 (12) | 0.0495 (14) | 0.0196 (11) | −0.0069 (10) | −0.0068 (9) | −0.0109 (10) |
| C37 | 0.0412 (13) | 0.0407 (13) | 0.0272 (12) | −0.0188 (11) | −0.0063 (10) | −0.0072 (10) |
| C38 | 0.0351 (12) | 0.0334 (12) | 0.0218 (11) | −0.0120 (10) | −0.0039 (9) | −0.0012 (9) |
| C39 | 0.0263 (11) | 0.0296 (11) | 0.0181 (10) | 0.0001 (9) | −0.0077 (8) | 0.0029 (8) |
| C40 | 0.0350 (12) | 0.0222 (11) | 0.0282 (11) | 0.0008 (9) | −0.0038 (9) | 0.0043 (9) |
| O41 | 0.0580 (11) | 0.0276 (9) | 0.0340 (9) | 0.0095 (8) | 0.0067 (8) | 0.0132 (7) |
| O42 | 0.0253 (7) | 0.0208 (7) | 0.0203 (7) | 0.0018 (6) | −0.0096 (6) | −0.0009 (6) |
| C43 | 0.0510 (15) | 0.0277 (12) | 0.0318 (12) | 0.0042 (11) | −0.0239 (11) | 0.0017 (10) |
| O44 | 0.0635 (12) | 0.0453 (10) | 0.0521 (11) | 0.0305 (9) | −0.0381 (10) | −0.0268 (9) |
| C45 | 0.0367 (17) | 0.125 (3) | 0.092 (3) | 0.0260 (18) | −0.0341 (17) | −0.067 (2) |
| C46 | 0.0288 (11) | 0.0358 (12) | 0.0211 (10) | −0.0119 (9) | −0.0099 (9) | 0.0026 (9) |
| C47 | 0.0288 (12) | 0.0347 (13) | 0.0369 (13) | −0.0037 (10) | −0.0053 (10) | 0.0033 (10) |
| C48 | 0.0286 (12) | 0.0248 (11) | 0.0432 (14) | −0.0022 (9) | 0.0024 (10) | 0.0006 (10) |
| C49 | 0.0366 (13) | 0.0272 (12) | 0.0351 (13) | −0.0088 (10) | 0.0008 (10) | −0.0063 (10) |
| C50 | 0.0390 (13) | 0.0334 (13) | 0.0345 (13) | −0.0102 (10) | −0.0070 (10) | 0.0028 (10) |
| C51 | 0.0368 (13) | 0.0249 (12) | 0.0415 (14) | −0.0055 (10) | −0.0063 (11) | 0.0020 (10) |
| C52 | 0.0337 (13) | 0.0287 (12) | 0.0365 (13) | −0.0090 (10) | −0.0034 (10) | −0.0043 (10) |
Geometric parameters (Å, º)
| O1—C2 | 1.334 (2) | C27—C28 | 1.523 (3) |
| O1—C5 | 1.461 (2) | C27—H27A | 0.99 |
| C2—O11 | 1.189 (2) | C27—H27B | 0.99 |
| C2—O3 | 1.342 (2) | C28—C29 | 1.516 (3) |
| O3—C4 | 1.446 (2) | C28—H28A | 0.99 |
| C4—C24 | 1.544 (3) | C28—H28B | 0.99 |
| C4—C5 | 1.566 (3) | C29—O42 | 1.435 (2) |
| C4—H4 | 1.0 | C29—H29 | 1.0 |
| C5—C6 | 1.517 (3) | O30—C31 | 1.333 (2) |
| C5—C10 | 1.550 (3) | C31—O32 | 1.209 (2) |
| C6—C7 | 1.524 (3) | C31—C33 | 1.486 (3) |
| C6—H6A | 0.99 | C33—C38 | 1.385 (3) |
| C6—H6B | 0.99 | C33—C34 | 1.387 (3) |
| C7—O12 | 1.464 (2) | C34—C35 | 1.381 (3) |
| C7—C8 | 1.501 (3) | C34—H34 | 0.95 |
| C7—H7 | 1.0 | C35—C36 | 1.375 (3) |
| C8—C9 | 1.324 (3) | C35—H35 | 0.95 |
| C8—C23 | 1.507 (3) | C36—C37 | 1.382 (3) |
| C9—C10 | 1.513 (3) | C36—H36 | 0.95 |
| C9—H9 | 0.95 | C37—C38 | 1.383 (3) |
| C10—C22 | 1.534 (3) | C37—H37 | 0.95 |
| C10—C21 | 1.538 (3) | C38—H38 | 0.95 |
| O12—C13 | 1.338 (2) | C39—C40 | 1.519 (3) |
| C13—O14 | 1.213 (2) | C39—H39A | 0.99 |
| C13—C15 | 1.485 (3) | C39—H39B | 0.99 |
| C15—C20 | 1.388 (3) | C40—O41 | 1.427 (3) |
| C15—C16 | 1.393 (3) | C40—H40A | 0.99 |
| C16—C17 | 1.383 (3) | C40—H40B | 0.99 |
| C16—H16 | 0.95 | O41—H41 | 0.84 |
| C17—C18 | 1.385 (3) | O42—C43 | 1.408 (2) |
| C17—H17 | 0.95 | C43—O44 | 1.396 (3) |
| C18—C19 | 1.381 (3) | C43—H43A | 0.99 |
| C18—H18 | 0.95 | C43—H43B | 0.99 |
| C19—C20 | 1.385 (3) | O44—C45 | 1.420 (4) |
| C19—H19 | 0.95 | C45—H45A | 0.98 |
| C20—H20 | 0.95 | C45—H45B | 0.98 |
| C21—H21A | 0.98 | C45—H45C | 0.98 |
| C21—H21B | 0.98 | C46—H46A | 0.98 |
| C21—H21C | 0.98 | C46—H46B | 0.98 |
| C22—H22A | 0.98 | C46—H46C | 0.98 |
| C22—H22B | 0.98 | C47—C52 | 1.380 (3) |
| C22—H22C | 0.98 | C47—C48 | 1.380 (3) |
| C23—H23A | 0.98 | C47—H47 | 0.95 |
| C23—H23B | 0.98 | C48—C49 | 1.378 (3) |
| C23—H23C | 0.98 | C48—H48 | 0.95 |
| C24—C29 | 1.536 (3) | C49—C50 | 1.378 (3) |
| C24—C25 | 1.571 (2) | C49—H49 | 0.95 |
| C24—H24 | 1.0 | C50—C51 | 1.386 (3) |
| C25—C46 | 1.535 (3) | C50—H50 | 0.95 |
| C25—C26 | 1.547 (3) | C51—C52 | 1.383 (3) |
| C25—C39 | 1.545 (3) | C51—H51 | 0.95 |
| C26—O30 | 1.455 (2) | C52—H52 | 0.95 |
| C26—C27 | 1.500 (3) | C9—C40 | 5.831 (3) |
| C26—H26 | 1.0 | ||
| C2—O1—C5 | 110.64 (14) | C27—C26—C25 | 114.64 (16) |
| O11—C2—O1 | 124.53 (18) | O30—C26—H26 | 109.1 |
| O11—C2—O3 | 123.74 (17) | C27—C26—H26 | 109.1 |
| O1—C2—O3 | 111.71 (16) | C25—C26—H26 | 109.1 |
| C2—O3—C4 | 110.32 (14) | C26—C27—C28 | 109.18 (16) |
| O3—C4—C24 | 109.71 (15) | C26—C27—H27A | 109.8 |
| O3—C4—C5 | 102.47 (14) | C28—C27—H27A | 109.8 |
| C24—C4—C5 | 120.64 (15) | C26—C27—H27B | 109.8 |
| O3—C4—H4 | 107.8 | C28—C27—H27B | 109.8 |
| C24—C4—H4 | 107.8 | H27A—C27—H27B | 108.3 |
| C5—C4—H4 | 107.8 | C29—C28—C27 | 110.00 (16) |
| O1—C5—C6 | 106.35 (14) | C29—C28—H28A | 109.7 |
| O1—C5—C10 | 107.29 (14) | C27—C28—H28A | 109.7 |
| C6—C5—C10 | 110.80 (15) | C29—C28—H28B | 109.7 |
| O1—C5—C4 | 101.92 (14) | C27—C28—H28B | 109.7 |
| C6—C5—C4 | 117.25 (15) | H28A—C28—H28B | 108.2 |
| C10—C5—C4 | 112.21 (15) | O42—C29—C28 | 111.95 (15) |
| C5—C6—C7 | 112.96 (16) | O42—C29—C24 | 106.93 (15) |
| C5—C6—H6A | 109.0 | C28—C29—C24 | 110.66 (16) |
| C7—C6—H6A | 109.0 | O42—C29—H29 | 109.1 |
| C5—C6—H6B | 109.0 | C28—C29—H29 | 109.1 |
| C7—C6—H6B | 109.0 | C24—C29—H29 | 109.1 |
| H6A—C6—H6B | 107.8 | C31—O30—C26 | 119.33 (15) |
| O12—C7—C8 | 105.62 (15) | O32—C31—O30 | 124.21 (18) |
| O12—C7—C6 | 110.35 (14) | O32—C31—C33 | 124.19 (18) |
| C8—C7—C6 | 114.15 (16) | O30—C31—C33 | 111.59 (16) |
| O12—C7—H7 | 108.9 | C38—C33—C34 | 119.78 (19) |
| C8—C7—H7 | 108.9 | C38—C33—C31 | 121.56 (18) |
| C6—C7—H7 | 108.9 | C34—C33—C31 | 118.65 (18) |
| C9—C8—C7 | 120.99 (18) | C35—C34—C33 | 120.20 (19) |
| C9—C8—C23 | 122.92 (19) | C35—C34—H34 | 119.9 |
| C7—C8—C23 | 116.07 (17) | C33—C34—H34 | 119.9 |
| C8—C9—C10 | 126.79 (18) | C36—C35—C34 | 119.9 (2) |
| C8—C9—H9 | 116.6 | C36—C35—H35 | 120.1 |
| C10—C9—H9 | 116.6 | C34—C35—H35 | 120.1 |
| C9—C10—C22 | 108.36 (17) | C35—C36—C37 | 120.3 (2) |
| C9—C10—C21 | 108.05 (16) | C35—C36—H36 | 119.9 |
| C22—C10—C21 | 108.13 (16) | C37—C36—H36 | 119.9 |
| C9—C10—C5 | 109.25 (15) | C38—C37—C36 | 120.1 (2) |
| C22—C10—C5 | 111.47 (16) | C38—C37—H37 | 119.9 |
| C21—C10—C5 | 111.47 (16) | C36—C37—H37 | 119.9 |
| C13—O12—C7 | 117.11 (14) | C37—C38—C33 | 119.7 (2) |
| O14—C13—O12 | 123.85 (17) | C37—C38—H38 | 120.1 |
| O14—C13—C15 | 124.22 (18) | C33—C38—H38 | 120.1 |
| O12—C13—C15 | 111.92 (16) | C40—C39—C25 | 118.35 (17) |
| C20—C15—C16 | 119.80 (18) | C40—C39—H39A | 107.7 |
| C20—C15—C13 | 121.31 (18) | C25—C39—H39A | 107.7 |
| C16—C15—C13 | 118.79 (18) | C40—C39—H39B | 107.7 |
| C17—C16—C15 | 120.06 (19) | C25—C39—H39B | 107.7 |
| C17—C16—H16 | 120.0 | H39A—C39—H39B | 107.1 |
| C15—C16—H16 | 120.0 | O41—C40—C39 | 109.16 (18) |
| C18—C17—C16 | 119.9 (2) | O41—C40—H40A | 109.8 |
| C18—C17—H17 | 120.0 | C39—C40—H40A | 109.8 |
| C16—C17—H17 | 120.0 | O41—C40—H40B | 109.8 |
| C19—C18—C17 | 120.1 (2) | C39—C40—H40B | 109.8 |
| C19—C18—H18 | 120.0 | H40A—C40—H40B | 108.3 |
| C17—C18—H18 | 120.0 | C40—O41—H41 | 109.5 |
| C18—C19—C20 | 120.4 (2) | C43—O42—C29 | 115.15 (15) |
| C18—C19—H19 | 119.8 | O44—C43—O42 | 113.8 (2) |
| C20—C19—H19 | 119.8 | O44—C43—H43A | 108.8 |
| C15—C20—C19 | 119.7 (2) | O42—C43—H43A | 108.8 |
| C15—C20—H20 | 120.1 | O44—C43—H43B | 108.8 |
| C19—C20—H20 | 120.1 | O42—C43—H43B | 108.8 |
| C10—C21—H21A | 109.5 | H43A—C43—H43B | 107.7 |
| C10—C21—H21B | 109.5 | C43—O44—C45 | 112.71 (19) |
| H21A—C21—H21B | 109.5 | O44—C45—H45A | 109.5 |
| C10—C21—H21C | 109.5 | O44—C45—H45B | 109.5 |
| H21A—C21—H21C | 109.5 | H45A—C45—H45B | 109.5 |
| H21B—C21—H21C | 109.5 | O44—C45—H45C | 109.5 |
| C10—C22—H22A | 109.5 | H45A—C45—H45C | 109.5 |
| C10—C22—H22B | 109.5 | H45B—C45—H45C | 109.5 |
| H22A—C22—H22B | 109.5 | C25—C46—H46A | 109.5 |
| C10—C22—H22C | 109.5 | C25—C46—H46B | 109.5 |
| H22A—C22—H22C | 109.5 | H46A—C46—H46B | 109.5 |
| H22B—C22—H22C | 109.5 | C25—C46—H46C | 109.5 |
| C8—C23—H23A | 109.5 | H46A—C46—H46C | 109.5 |
| C8—C23—H23B | 109.5 | H46B—C46—H46C | 109.5 |
| H23A—C23—H23B | 109.5 | C52—C47—C48 | 120.1 (2) |
| C8—C23—H23C | 109.5 | C52—C47—H47 | 119.9 |
| H23A—C23—H23C | 109.5 | C48—C47—H47 | 119.9 |
| H23B—C23—H23C | 109.5 | C49—C48—C47 | 120.1 (2) |
| C29—C24—C4 | 112.45 (15) | C49—C48—H48 | 120.0 |
| C29—C24—C25 | 111.56 (15) | C47—C48—H48 | 120.0 |
| C4—C24—C25 | 111.32 (15) | C48—C49—C50 | 120.2 (2) |
| C29—C24—H24 | 107.1 | C48—C49—H49 | 119.9 |
| C4—C24—H24 | 107.1 | C50—C49—H49 | 119.9 |
| C25—C24—H24 | 107.1 | C49—C50—C51 | 119.7 (2) |
| C46—C25—C26 | 110.71 (16) | C49—C50—H50 | 120.2 |
| C46—C25—C39 | 107.03 (16) | C51—C50—H50 | 120.2 |
| C26—C25—C39 | 108.36 (15) | C52—C51—C50 | 120.2 (2) |
| C46—C25—C24 | 111.22 (15) | C52—C51—H51 | 119.9 |
| C26—C25—C24 | 107.84 (15) | C50—C51—H51 | 119.9 |
| C39—C25—C24 | 111.66 (15) | C47—C52—C51 | 119.7 (2) |
| O30—C26—C27 | 106.89 (15) | C47—C52—H52 | 120.2 |
| O30—C26—C25 | 107.73 (15) | C51—C52—H52 | 120.2 |
| C5—O1—C2—O11 | −176.77 (18) | C5—C4—C24—C29 | 87.2 (2) |
| C5—O1—C2—O3 | 1.9 (2) | O3—C4—C24—C25 | 94.64 (17) |
| O11—C2—O3—C4 | −171.43 (18) | C5—C4—C24—C25 | −146.76 (16) |
| O1—C2—O3—C4 | 9.9 (2) | C29—C24—C25—C46 | 69.7 (2) |
| C2—O3—C4—C24 | 113.11 (16) | C4—C24—C25—C46 | −56.9 (2) |
| C2—O3—C4—C5 | −16.21 (18) | C29—C24—C25—C26 | −51.9 (2) |
| C2—O1—C5—C6 | −134.83 (16) | C4—C24—C25—C26 | −178.43 (15) |
| C2—O1—C5—C10 | 106.55 (16) | C29—C24—C25—C39 | −170.86 (16) |
| C2—O1—C5—C4 | −11.50 (18) | C4—C24—C25—C39 | 62.6 (2) |
| O3—C4—C5—O1 | 16.00 (16) | C46—C25—C26—O30 | 50.78 (19) |
| C24—C4—C5—O1 | −106.16 (17) | C39—C25—C26—O30 | −66.31 (18) |
| O3—C4—C5—C6 | 131.61 (16) | C24—C25—C26—O30 | 172.66 (14) |
| C24—C4—C5—C6 | 9.4 (2) | C46—C25—C26—C27 | −68.1 (2) |
| O3—C4—C5—C10 | −98.47 (16) | C39—C25—C26—C27 | 174.86 (16) |
| C24—C4—C5—C10 | 139.36 (17) | C24—C25—C26—C27 | 53.8 (2) |
| O1—C5—C6—C7 | −57.91 (19) | O30—C26—C27—C28 | −177.59 (15) |
| C10—C5—C6—C7 | 58.4 (2) | C25—C26—C27—C28 | −58.3 (2) |
| C4—C5—C6—C7 | −171.06 (15) | C26—C27—C28—C29 | 59.7 (2) |
| C5—C6—C7—O12 | 81.26 (19) | C27—C28—C29—O42 | −179.61 (15) |
| C5—C6—C7—C8 | −37.5 (2) | C27—C28—C29—C24 | −60.4 (2) |
| O12—C7—C8—C9 | −114.50 (19) | C4—C24—C29—O42 | −54.6 (2) |
| C6—C7—C8—C9 | 6.9 (3) | C25—C24—C29—O42 | 179.52 (14) |
| O12—C7—C8—C23 | 67.2 (2) | C4—C24—C29—C28 | −176.75 (15) |
| C6—C7—C8—C23 | −171.41 (16) | C25—C24—C29—C28 | 57.4 (2) |
| C7—C8—C9—C10 | 2.7 (3) | C27—C26—O30—C31 | −120.95 (19) |
| C23—C8—C9—C10 | −179.15 (18) | C25—C26—O30—C31 | 115.37 (18) |
| C8—C9—C10—C22 | 139.3 (2) | C26—O30—C31—O32 | −3.6 (3) |
| C8—C9—C10—C21 | −103.8 (2) | C26—O30—C31—C33 | 177.38 (16) |
| C8—C9—C10—C5 | 17.7 (3) | O32—C31—C33—C38 | 174.2 (2) |
| O1—C5—C10—C9 | 69.18 (18) | O30—C31—C33—C38 | −6.9 (3) |
| C6—C5—C10—C9 | −46.5 (2) | O32—C31—C33—C34 | −6.7 (3) |
| C4—C5—C10—C9 | −179.68 (15) | O30—C31—C33—C34 | 172.23 (17) |
| O1—C5—C10—C22 | −50.6 (2) | C38—C33—C34—C35 | 1.1 (3) |
| C6—C5—C10—C22 | −166.25 (16) | C31—C33—C34—C35 | −178.07 (18) |
| C4—C5—C10—C22 | 60.6 (2) | C33—C34—C35—C36 | −0.5 (3) |
| O1—C5—C10—C21 | −171.49 (14) | C34—C35—C36—C37 | −0.4 (3) |
| C6—C5—C10—C21 | 72.82 (19) | C35—C36—C37—C38 | 0.6 (3) |
| C4—C5—C10—C21 | −60.3 (2) | C36—C37—C38—C33 | 0.0 (3) |
| C8—C7—O12—C13 | −158.97 (15) | C34—C33—C38—C37 | −0.8 (3) |
| C6—C7—O12—C13 | 77.20 (19) | C31—C33—C38—C37 | 178.25 (19) |
| C7—O12—C13—O14 | 5.0 (3) | C46—C25—C39—C40 | 175.91 (17) |
| C7—O12—C13—C15 | −173.77 (15) | C26—C25—C39—C40 | −64.7 (2) |
| O14—C13—C15—C20 | −163.81 (19) | C24—C25—C39—C40 | 54.0 (2) |
| O12—C13—C15—C20 | 15.0 (3) | C25—C39—C40—O41 | 177.32 (17) |
| O14—C13—C15—C16 | 12.6 (3) | C28—C29—O42—C43 | −74.0 (2) |
| O12—C13—C15—C16 | −168.65 (17) | C24—C29—O42—C43 | 164.63 (17) |
| C20—C15—C16—C17 | 0.5 (3) | C29—O42—C43—O44 | 76.5 (2) |
| C13—C15—C16—C17 | −175.95 (19) | O42—C43—O44—C45 | 64.8 (3) |
| C15—C16—C17—C18 | 0.5 (3) | C52—C47—C48—C49 | 0.1 (3) |
| C16—C17—C18—C19 | −1.3 (3) | C47—C48—C49—C50 | 0.6 (3) |
| C17—C18—C19—C20 | 1.2 (3) | C48—C49—C50—C51 | −0.4 (3) |
| C16—C15—C20—C19 | −0.6 (3) | C49—C50—C51—C52 | −0.3 (3) |
| C13—C15—C20—C19 | 175.70 (19) | C48—C47—C52—C51 | −0.8 (3) |
| C18—C19—C20—C15 | −0.2 (3) | C50—C51—C52—C47 | 0.9 (3) |
| O3—C4—C24—C29 | −31.4 (2) |
Hydrogen-bond geometry (Å, º)
Cg is the centroid of the C47–C52 ring.
| D—H···A | D—H | H···A | D···A | D—H···A |
| C6—H6B···O42 | 0.99 | 2.32 | 3.095 (2) | 135 |
| C28—H28A···O44 | 0.99 | 2.37 | 2.989 (3) | 120 |
| O41—H41···O14i | 0.84 | 2.06 | 2.888 (2) | 170 |
| C7—H7···O32i | 1.00 | 2.34 | 3.269 (2) | 155 |
| C18—H18···O11ii | 0.95 | 2.53 | 3.465 (2) | 168 |
| C49—H49···O11 | 0.95 | 2.46 | 3.300 (3) | 147 |
| C27—H27A···Cgiii | 0.95 | 2.64 | 3.514 (2) | 147 |
Symmetry codes: (i) −x+1, −y+2, −z+1; (ii) −x+1, −y+1, −z+2; (iii) −x+1, −y+1, −z+1.
References
- Bruker (2013). APEX2, SAINT and SADABS. Bruker AXS Inc., Madison, Wisconsin, USA.
- Fukaya, K., Sugai, T., Yamaguchi, Y., Watanabe, A., Sato, T. & Chida, N. (2014). In preparation.
- Groom, C. R. & Allen, F. H. (2014). Angew. Chem. Int. Ed. 53, 662–671. [DOI] [PubMed]
- Macrae, C. F., Edgington, P. R., McCabe, P., Pidcock, E., Shields, G. P., Taylor, R., Towler, M. & van de Streek, J. (2006). J. Appl. Cryst. 39, 453–457.
- Nicolaou, K. C., Liu, J.-J., Yang, Z., Ueno, H., Sorensen, E. J., Claiborne, C. F., Guy, R. K., Hwang, C.-K., Nakada, M. & Nantermet, P. G. (1995). J. Am. Chem. Soc. 117, 634–644.
- Nishizawa, M., Imagawa, H., Hyodo, I., Takeji, M., Morikuni, E., Asoh, K. & Yamada, H. (1998). Tetrahedron Lett. 39, 389–392.
- Ohba, S., Chinen, A., Matsumoto, Y. & Chida, N. (2003). Acta Cryst. E59, o1476–o1477.
- Poujol, H., Ahond, A., Al Mourabit, A., Chiaroni, A., Poupat, C., Riche, C. & Potier, P. (1997). Tetrahedron, 53, 5169–5184.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Wall, M. E. & Wani, M. C. (1995). ACS Symp. Ser. 583, 18–30.
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S2056989014026048/is5382sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989014026048/is5382Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989014026048/is5382Isup3.cml
CCDC reference: 1036428
Additional supporting information: crystallographic information; 3D view; checkCIF report