Abstract
MicroRNAs (miRNAs) are small, endogenous RNAs that play important gene-regulatory roles by binding to the imperfectly complementary sequences at the 3′-UTR of mRNAs and directing their gene expression. Here, we first discovered that miR-576-3p was down-regulated in human bladder cancer cell lines compared with the non-malignant cell line. To better characterize the role of miR-576-3p in bladder cancer cells, we over-expressed or down-regulated miR-576-3p in bladder cancer cells by transfecting with chemically synthesized mimic or inhibitor. The overexpression of miR-576-3p remarkably inhibited cell proliferation via G1-phase arrest, and decreased both mRNA and protein levels of cyclin D1 which played a key role in G1/S phase transition. The knock-down of miR-576-3p significantly promoted the proliferation of bladder cancer cells by accelerating the progression of cell cycle and increased the expression of cyclin D1. Moreover, the dual-luciferase reporter assays indicated that miR-576-3p could directly target cyclin D1 through binding its 3′-UTR. All the results demonstrated that miR-576-3p might be a novel suppressor of bladder cancer cell proliferation through targeting cyclin D1.
Keywords: bladder cancer, cyclin D1, microRNA-576-3p, proliferation
INTRODUCTION
Bladder cancer is the most common malignant tumor in urogenital tract and the ninth common cancer in worldwide cancer incidence, with an estimated 386,300 new cases and 150, 200 deaths in 2008 (Jemal et al., 2011; Ploeg et al., 2009). Although localized bladder cancers could be managed by surgical resection, the recurrence and progression rates are still high. In patients with advanced bladder cancer who receive radiotherapy or chemotherapy, the therapeutic outcomes remain unsatisfactory. The absence of more effective therapies for bladder cancer requires research into the underlying molecular mechanisms of bladder tumorigenesis and the development of new treatments aimed at specific molecular targets.
MicroRNAs (miRNAs) are small (~23 nucleotides), endogenous RNAs that play important gene-regulatory roles by binding to the imperfectly complementary sequences at the 3′ untranslated regions (3′-UTR) of mRNAs and directing their posttranscriptional repression (Bartel, 2009). It is estimated that over 1000 miRNAs are encoded in the human genome, and they target approximately 60% of mammalian genes (Friedman et al., 2009). The deregulation of miRNAs has been identified in various types of cancer including bladder cancer, which is involved in tumor cell proliferation, apoptosis, differentiation, angiogenesis, metastasis, and other functions (Catto et al., 2011; Fendler et al., 2011). Some microRNAs, such as miR-23b, miR-26a, miR-143, miR-195 and miR-490-5p have been proved as tumor suppressors by suppressing the proliferation of bladder cancer (Li et al., 2013; Lin et al., 2009; 2012; 2013; Majid et al., 2013). Recently several studies (Coskun et al., 2013; Tatarano et al., 2011; Zhang et al., 2013) have reported a series of aberrant expressed miRNAs in bladder cancer and leukemia, among which miR-576-3p was one of the significantly down-regulated miRNAs. However, there has been no relevant study on miR-576-3p before and its functions and targets remain unclear.
In our study, we confirmed the down regulation of miR-576-3p in human bladder cancer cells. Furthermore, for the first time, we revealed that the overexpression of miR-576-3p could inhibit the proliferation of bladder cancer cells in vitro via targeting cyclinD1 (CCND1).
MATERIALS AND METHODS
Cell lines and cell culture
Four human bladder cancer cell lines (UM-UC-3, 5637, J82 and T24) and one non-tumor urothelial cell line (SV-HUC-1) were purchased from the Shanghai Institute of Cell Biology (China). All the cell lines were cultured in RPMI-1640 medium supplemented with 10% heat-inactivated fetal bovine serum under a humidified atmosphere of 5% CO2 at 37°C.
Transient transfection of miRNA mimic, inhibitor and small interfering RNA
The miR-576-3p mimic (named as miR-576-3p) and the negative control duplex (named as NC) which was nonhomologous to all human gene sequences were used for transient gain of function study. The mir-576-3p inhibitor oligo (named as miR-576-3p inhibitor) and inhibitor negative control oligo (named as inhibitor NC) were used for transient loss of function study. A small interfering RNA duplex (siRNA) targeting human CCND1 mRNA was used for RNAi study (named as siCCND1). All the RNA duplexes and RNA oligos were synthesized by Gene Pharma (China). T24 and UM-UC-3 cells were seeded into 6-well plates 24 h before transfection to ensure 60–70% confluence at the time of transfection. The Lipofectamine 2000 Reagent (Invitrogen, USA) was used for transfections following the manufacturer’s instructions. The sequences of RNA duplexes and RNA oligos used in transfection are listed in Table 1.
Table 1.
The oligonucleotides used in this study
| Namea | Sequence (5′ → 3′)b |
|---|---|
| miR-576-3p mimic (sense) | AAGAUGUGGAAAAAUUGGAAUC |
| miR-576-3p inhibitorc | GAUUCCAAUUUUUCCACAUCUU |
| NC (sense) | ACUACUGAGUGACAGUAGA |
| inhibitor NCc | CAGUACUUUUGUGUAGUACAA |
| siCCND1(sense) | GGAGAACAAACAGAUCAUCTT |
| U6 F | TGCGGGTGCTCGCTTCGGCAGC |
| miR-576-3p F | AAGATGTGGAAAAATTGGAATC |
| miR-576-5p F | ATTCTAATTTCTCCACGTCTTT |
| CCND1 F | GCTGCGAAGTGGAAACCATC |
| CCND1 R | CCTCCTTCTGCACACATTTGAA |
| GAPDH F | AAGGTGAAGGTCGGAGTCA |
| GAPDH R | GGAAGATGGTGATGGGATTT |
| CCND1-utr F | CGACAAAATAGACAATTTGCACATCTTGGCTATGTAATTCTTGTAAG |
| CCND1-utr R | TCGACTTACAAGAATTACATAGCCAAGATGTGCAAATTGTCTATTTTGTCGAGCT |
| CCND1-mut F | CGACAAAATAGACAATTTGGTGTAGATGGCTATGTAATTCTTGTAAG |
| CCND1-mut R | TCGACTTACAAGAATTACATAGCCATCTACACCAAATTGTCTATTTTGTCGAGCT |
F, forward primer; R, reverse primer.
Restriction sites are in bold; Mutated sites are underlined.
Oligonucleotides with 2′-O-methyl modification.
RNA isolation and quantitative real-time PCR
MircoRNAs were extracted from cultured cell lines with the RNAiso kit for small RNA (Takara, China) and reversely transcribed into cDNA with the One Step PrimeScript miRNA cDNA Synthesis Kit (Takara, China). Total RNA was isolated with TRIzol reagent (Takara, China) and reversely transcribed into cDNAs with the PrimeScript RT reagent Kit (Takara, China). The resulting cDNAs were quantified with SYBR Green (Takara, China) using an ABI 7500 fast real-time PCR System (Applied Biosystems, USA). The relative expression levels of miRNAs (miR-576-3p or miR-576-5p) and CCND1 normalized by small nuclear RNA U6 and GAPDH mRNA respectively were calculated with the 2−ΔΔCt method. All the qPCR primers were provided by Sango Biotech (China). All primers used are listed in Table 1.
Cell cycle analysis by flow cytometry
The 48 h after the transfection of RNA duplexes (50 nM of NC, miR-576-3p or siCCND1) or the co-transfection of miR-576-3p (50 nM) and RNA oligos (mir-576-3p inhibitor or inhibitor NC, 100 nM), bladder cancer cells were harvested and washed with PBS and fixed with 75% ethanol at −20°C. After 24 h fixation, the cells were washed with PBS again and stained with propidium iodide using the cell cycle and apoptosis analysis kit (Beyotime, China) for 30 min. Cell cycle features were analyzed by BD LSRII Flow cytometry system with FACSDiva software (BD Bioscience, USA). The data were analyzed by ModFit LT 3.2 software (Verity Software House, USA).
Colony formation assay
T24 and UM-UC-3 cells were collected 24 h after transfected with RNA duplexes (50 nM of NC, miR-576-3p or siCCND1) or co-transfected with both 50 nM of miR-576-3p or NC and 1 μg of pReceiver-CCND1 or empty pReceiver vector. The cells were then resuspended in RPMI-1640 medium supplemented with 10% FBS and seeded in 6-well plates at a density of 400 cells per well. After 10 days of culture under standard conditions, the colonies on the plates were fixed with absolute methanol for 15 min and stained with 0.1% crystal violet for 20 min. The colonies with a diameter over 2 mm were counted. The rate of colony formation was calculated by the following formula: colony formation rate = (number of colonies/number of seeded cells) × 100%.
Cell growth and cell viability assay
Bladder cancer cells were plated in 96-well plates at the density of 5000 cells per well. After an overnight cultivation, all the cells were incubated with RNA duplexes (NC, miR-576-3p or siCCND1) for 48–72 h. The concentration of RNA duplexes ranged from 25 to 75 nM. As soon as reached the time limit of incubation, the medium were removed and a Cell Counting solution (WST-8, Dojindo Laboratories, Japan) was added to each well and incubated at 37°C for another 2 h. The absorbance of the solution was measured at 450 nm with MRX II absorbance reader (Dynex Technologies, USA).
Western blotting analysis
All the cells were gathered and lysed in cell lysis buffer 48 h after the transfection. The BCA protein assay kit (Thermo Scientific, USA) was used to calculate the total protein concentration in every lysate. The same amounts of protein samples were separated by 10% SDS-polyacrylamide gels and transferred to polyvinylidene fluoride (PVDF) membranes. The membranes were blocked with 5% non-fat milk for 1 h, and incubated overnight with primary antibodies including rabbit polyclonal anti-CCND1 and anti-GAPDH (Epitomics, USA) at dilutions instructed by the manufacturer. After being washed in TBS-T three times, PVDF membranes were incubated with horseradish peroxidase (HRP)-conjugated goat anti-rabbit secondary antibody at a 1:5000 dilution for 1 h. The binding secondary antibody was detected by the enhanced chemiluminescence (ECL) system (Pierce Biotechnology, USA).
Vector construction and dual-luciferase reporter assay
A bioinformatic analysis through the microRNA target gene prediction database TargetScan (http://www.targetscan.org) revealed that CCND1 is most likely a direct binding target of miR-576-3p. Based on the 3′-UTR sequence of human CCND1 mRNA, two pairs of oligonucleotides which contained the desired (wild type) miRNA target region and mutant miRNA target region (primers set in Table 1) were designed and ordered from Sangon Biotech (China). After the annealing procedure, the duplexes of annealed oligonucleotides were inserted into the downstream of the firefly luciferase reporter in the pmirGLO Dual-luciferase miRNA target expression vector (Promega, USA) between the SacI and SalI sites. The insertions were verified by sequencing. T24 and UM-UC-3 cells were plated in 24-well plates for 24 h, then co-transfected with both 50 nM of miR-576-3p or NC and 200 ng of pmirGLO vector carrying either wild type or mutant 3′-UTR sequence. The relative luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega, USA) 48 h after co-transfection.
CCND1 rescue experiments
The human CCND1 coding sequence excluded 3′-UTR was inserted into the pReceiver vector (Genecopoeia, USA), which formed the pReceiver-CCND1 vector. The bladder cancer cells were co-transfected with both miR-576-3p or NC and pReceiver-CCND1 or empty pReceiver vector. All the cells were harvested 48 h after the transfection. The cell cycle was analyzed by flow cytometry and the CCND1 expression was determined by Western blotting as mentioned above.
Statistical analysis
All the data were expressed as the mean ± deviation (SD) for three independent experiments. Student’s t-test was used to contrast quantitative data between two test groups, while two-way ANOVA was used to compare the differences of quantitative data among three or more experimental groups. Statistical analysis was performed using GraphPad Prism, version 5.0 for Windows. Values of P < 0.05 were considered to be statistically significant and indicated by asterisks in the figures.
RESULTS
miR-576-3p is down-regulated in bladder cancer cells
It has been reported that miR-576-3p was down-regulated in a patient with bladder urothelial carcinoma (Tatarano et al., 2011; Zhang et al., 2013). We questioned whether miR-576-3p acts as a tumor suppressor in bladder cancer. By using the online database, TargetScan, we determined that CCND1, which was well known as a regulator of cell proliferation, was predicted to be a direct target of miR-576-3p. In order to further verify the expression pattern of miR-576-3p in human bladder cancer, we used real-time RT-PCR to quantify and compare the expression levels of miR-576-3p in four bladder cancer cell lines versus SV-HUC-1 cell (a normal transitional epithelial cell line). It showed that miR-576-3p expression levels in bladder cancer cell lines were down-regulated significantly compared with SV-HUC-1 cell (Fig. 1A). In the same manner, we found that the levels of miR-576-5p were remarkably down-regulated in three bladder cancer cell lines (Fig. 1B), which confirmed the low expression levels of miR-576 family in bladder cancer cells. Furthermore, we tested the expression levels of CCND1 in T24 and UM-UC-3 bladder cancer cell lines versus SV-HUC-1 cell. The results revealed that the mRNA and protein levels of CCND1 in T24 and UM-UC-3 cells were dramatically increased in contrast with SV-HUC-1 cell (Figs. 1C and 1D).
Fig. 1.

The altered expression of miR-576-3p, miR-576-5p and CCND1 in bladder cancer cell lines. (A, B) Relative miR-576-3p and miR-576-5p levels analyzed by qRT-PCR in four bladder cancer cell lines (UM-UC-3, 5637, J82 and T24) and one non-tumor urothelial cell line (SV-HUC-1) were normalized with U6 snRNA. (C, D) Relative CCND1 mRNA and protein levels in T24 and UM-UC-3 cell lines compared with SV-HUC-1 cell line were detected by qRT-PCR (*P < 0.05). GAPDH was used as internal control.
miR-576-3p suppresses bladder cancer cell proliferation, inhibits colony formation and induces G1-phase arrest
Based on the low expression levels of miR-576-3p and the overexpression of its predicted target gene CCND1 in human bladder cancer cells, we believed that miR-576-3p could function as a suppressor of cell proliferation. T24 and UM-UC-3 cells were therefore transfected with miR-576-3p mimic for the gain-of-function analysis. The cell viability test showed that miR-576-3p could sharply suppress the growth of bladder cancer cells at a concentration of 25 nM, which reduced cell viability by 38% and 47%, respectively, at 48 or 72 h after transfection in T24 cells. The corresponding results of UM-UC-3 were 31% and 43% (Fig. 2A). In like manner, the colony formation ability was inhibited in both of the bladder cancer lines. The colony formation rates of miR-576-3p transfected cells were much lower in contrast with those transfected with NC (Fig. 2B).
Fig. 2.
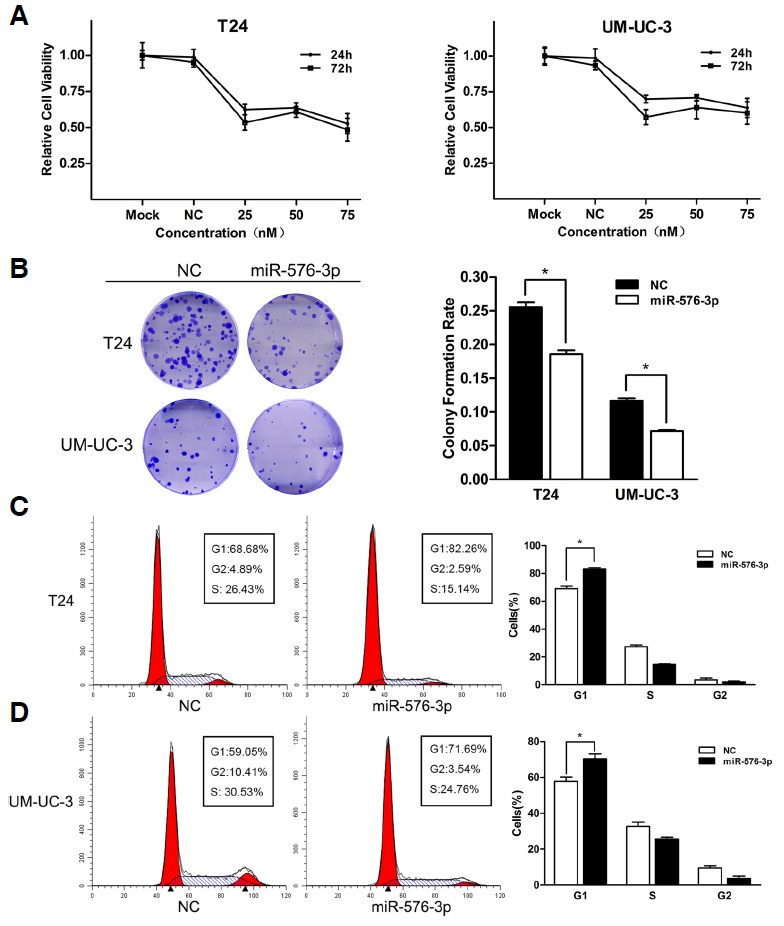
The overexpression of miR-576-3p suppressed the bladder cancer cell proliferation. (A) Cell growth/viability assay. The relative cell viability of T24 and UM-UC-3 cells treated with miR-576-3p mimic was lower than that of treated with NC. Cell viability of 0 Nm (mock) was regarded as 1.0. (B) The colony formation rates of miR-576-3p mimic transfected groups were lower in contrast with NC transfected groups (*P < 0.05). (C, D) Over-expression of miR-576-3p induced significant G1-phase arrest (representative histograms were shown above. The indicated percentages were means of triplicate experiments) (*P < 0.05).
In order to uncover the underlying mechanisms of cell proliferation suppression and colony formation inhibition mediated by miR-576-3p, we used flow cytometry to observe the distribution of cell cycle after the transfection of miR-576-3P mimic or the co-transfection of miR-576-3p mimic and inhibitor. The outcomes showed that the overexpression of miR-576-3P induced a significant G1-phase arrest in both T24 and UM-UC-3 cells (Figs. 2C and 2D), and correspondingly, according to the method of a previous researcher’s (Place et al., 2008), the down-regulation of miR-576-3p simulated by the co-transfection of miR-576-3p mimic and inhibitor (Fig. 4A) significantly promoted bladder cancer cell proliferation by accelerating the progression of cell cycle (Figs. 4D and 4E), which supported that miR-576-3P was a suppressor of cell proliferation in bladder cancer cells.
Fig. 4.
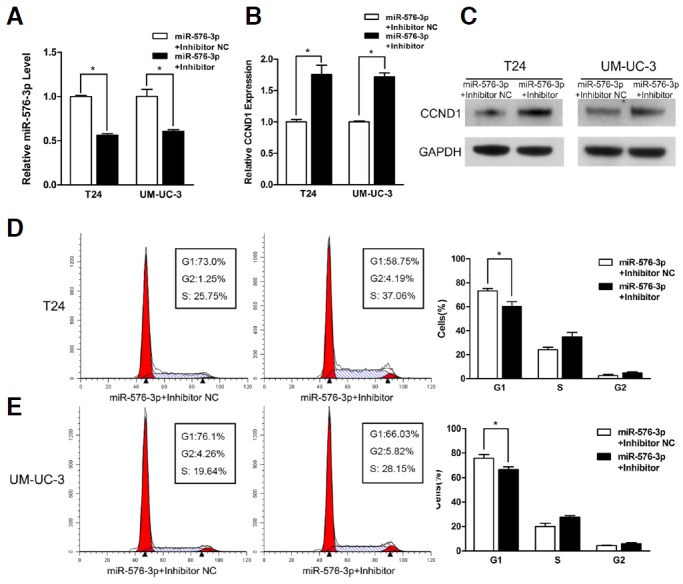
The miR-576-3p inhibitor significantly increased the expression of CCND1 and promoted the proliferation of bladder cancer cells by accelerating the progression of cell cycle. (A, B) T24 and UM-UC-3 cells were transfected with 50nM of miR-576-3p mimic in combination with 100 nM of miR-576-3p inhibitor or inhibitor NC for 48 h. Relative miR-576-3p and CCND1 expression levels were analyzed by qRT-PCR (*P < 0.05). (C) Relative CCND1 protein levels were assessed by Western blotting. (D, E) miR-576-3p inhibitor promoted bladder cancer cells proliferation by accelerating the progression of cell cycle (Representative histograms were shown above. The indicated percentages were means of triplicate experiments) (*P < 0.05).
miR-576-3P down-regulates the expression of CCND1 via its 3′-UTR
As CCND1 was a binding target of miR-576-3p predicted by the online database, TargetScan, we performed quantitative real-time PCR and Western blotting to observe the expression of CCND1 on mRNA and protein levels in T24 and UM-UC-3 cells transfected with miR-576-3P mimic or co-transfected with miR-576-3p mimic and inhibitor. The results indicated that both of the two levels were remarkably decreased after the overexpression of miR-576-3p (Figs. 3A and 3B), and significantly increased by the knock-down of miR-576-3p after the co-transfection of miR-576-3p mimic and inhibitor (Figs. 4B and 4C), which proved that the inhibition of CCND1 was specific to the sequence of miR-576-3p.
Fig. 3.
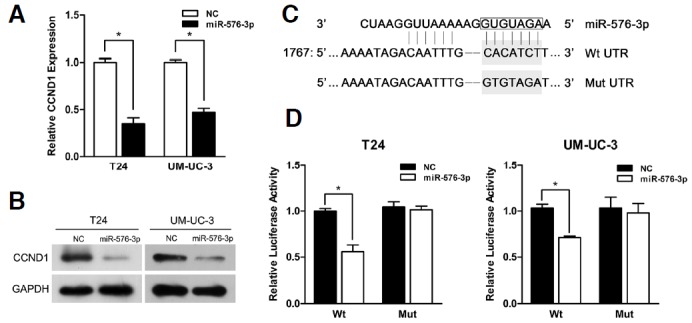
CCND1 was a direct target of miR-576-3p. (A) The miR-576-3p mimic significantly reduced the levels of CCND1 mRNA in T24 and UM-UC-3 cells (*P < 0.05). (B) Western blotting analysis was performed to detect the expression of CCND1 and GAPDH in T24 and UM-UC-3 cells transfected with miR-576-3p mimic. (C) A predicted seed region in the 3′-UTR of CCND1 (Wt) and mutated sequence (Mut) were highlighted in grey respectively. (D) T24 and UM-UC-3 cells were co-transfected with both 50 nM of miR-576-3p mimic or NC and 200 ng pmirGLO Dual-luciferase miRNA Target Expression vector containing the Wt or Mut 3′-UTR of CCND1. The relative firefly luciferase activity normalized with renilla luciferase was measured 48 h after transfection (*P < 0.05).
Likewise, the knock-down of CCND1 by transfecting with siCCND1 caused the inhibition of cell proliferation and G1-phase arrest in T24 and UM-UC-3 cells (Figs. 5A–5D). Moreover, siCCND1 also demonstrated a similar effect on the mRNA and protein levels of CCND1 treated with miR-576-3p (Figs. 5E and 5F). All the outcomes above showed that the knock-down of CCND1 could copy the effect of miR-576-3p in bladder cancer cells.
Fig. 5.
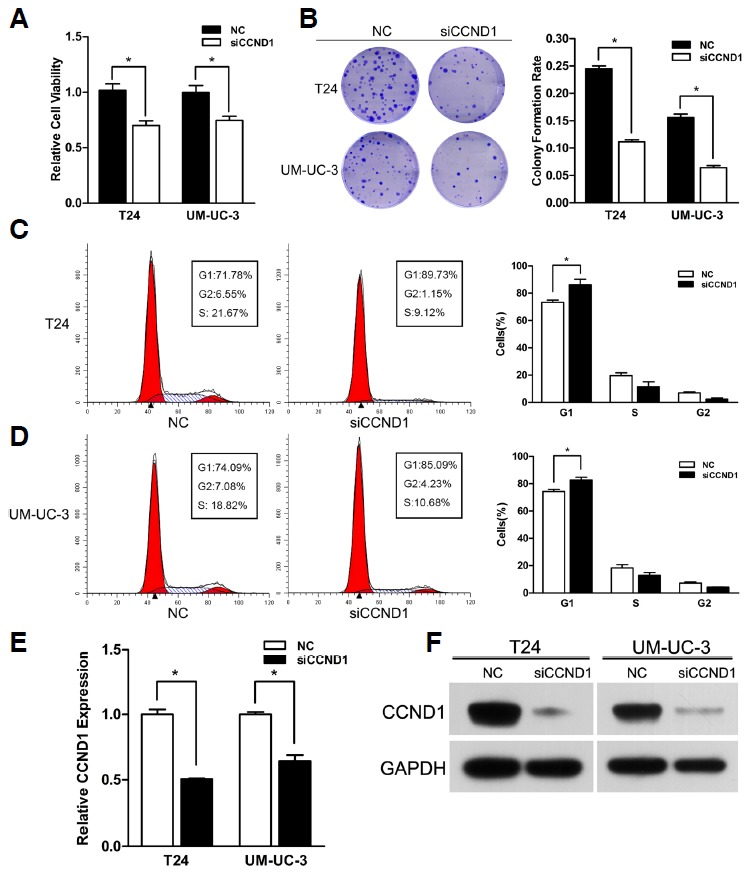
Knock-down of CCND1 phenoco- pied the effect of miR-576-3p. (A) Knockdown of miR-576-3p suppressed bladder cancer cell growth. The 50 nM of SiCCND1 caused significant cell viability inhibition in T24 and UM-UC-3 cells 48 h after transfection (*P < 0.05). (B) SiCCND1 reduced the colony formation rate in both of the cells (Representative wells were presented) (*P < 0.05). (C, D) Knock-down of CCND1 induced significant accumulation of cells in G1 phase (Representative histograms were shown above. The indicated percentages were means of triplicate experiments) (*P < 0.05). (E, F) Knock-down of CCND1 reduced the expression of CCND1 at the mRNA and protein levels in both two cell lines (*P < 0.05).
For the purpose of identifying CCND1 as a direct target of miR-576-3p, a fragment of CCND1 3′-UTR including the putative miR-576-3P binding site was inserted into the down-stream of the firefly luciferase of the pmirGLO Dual-Luciferase miRNA target expression vector. Another vector with a mutated putative binding site was also constructed (Fig. 3C). As expected, the relative luciferase activities in T24 and UM-UC-3 cells co-transfected with miR-576-3p and wild type (Wt) 3′-UTR vector were reduced significantly in contrast with NC, while in the same cells co-transfected with miR-576-3p and mutant (Mut) 3′-UTR vector the luciferase activities were unaffected (Fig. 3D). Therefore we came to the conclusion that CCND1 was a direct target of miR-576-3p.
Restoration of CCND1 could partially rescue miR-576-3P-induced G1-phase arrest
From the former results, we confirmed that miR-576-3p could induce G1-phase arrest. Then we questioned whether the overexpression of CCND1 could restore the inhibition of cell cycle caused by miR-576-3p. For this purpose, we inserted the human CCND1 coding sequence excluded 3′-UTR into the pReceiver vector, and it showed that CCND1 expression was regained in T24 cells after treated with pReceiver-CCND1 vector (Fig. 6A). The cell cycle was evaluated by flow cytometry and the colony formation rates were calculated in T24 cells co-transfected with both miR-576-3P mimic or NC and pReceiver-CCND1 or empty pReceiver vector. The result revealed that forced expression of CCND1 could partially rescue miR-576-3p induced G1-phase arrest and inhibition of colony formation (Figs. 6B–6D).
Fig. 6.
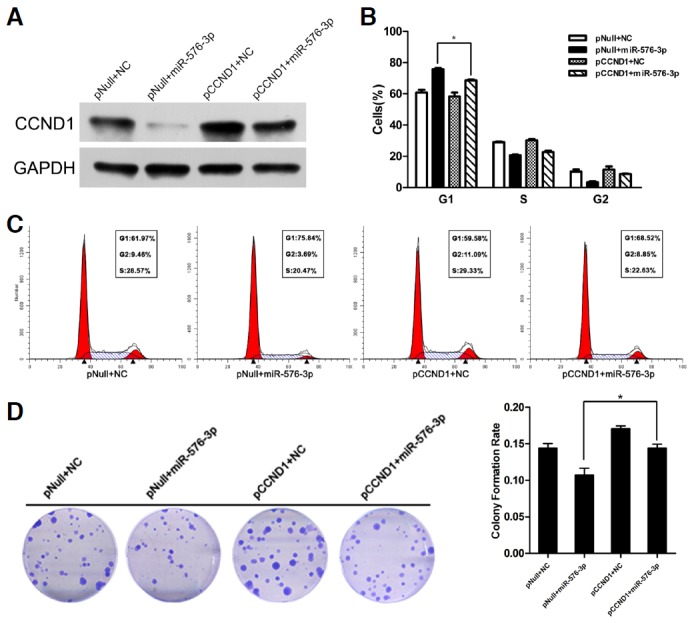
Forced expression of CCND1 rescued miR-576-3p induced G1 phase arrest and inhibition of colony formation. (A) T24 cells were co-transfected with both miR-576-3P mimic or NC and pReceiver-CCND1 or empty pReceiver vector. The expression of CCND1 or GAPDH was detected by Western blotting analysis. (B, C, and D) Forced expression of CCND1 partly restored cell cycle arrest effect and inhibition of colony formation rate of miR-576-3p in T24 cells (*P < 0.05).
DISCUSSION
In recent years, an increasing number of studies demonstrated the potential of microRNAs as promising diagnostic or prognostic biomarkers and novel therapeutic targets in the treatment of malignant tumors (Allegra et al., 2012; Cho, 2010; Nana-Sinkam and Croce, 2010; Osaki et al., 2008; Piva et al., 2013). Moreover, the aberrant expressions of miRNAs in bladder cancer have been identified as well, which play crucial roles in tumorigenesis, progression and metastasis (Ratert et al., 2013; Shi et al., 2014). MiR-576-3p is a recently discovered miRNA with a decreased expression in bladder cancer (Tatarano et al., 2011; Zhang et al., 2013) and leukemia (Coskun et al., 2013). Our quantification analyses showed the similar outcomes that miR-576-3p and miR-576-5p were both down-regulated in most bladder cancer cell lines, suggesting miR-576-3p might be implicated in tumorigenesis. Since the expression pattern of miR-576-3p in other tumors has not been reported and its functions in bladder cancer or other tumors are still unknown, this is the first study to explore miR-576-3p about its mechanism of action in tumors.
In order to discover the function of miR-576-3p in bladder cancer, we conducted both gain-of-function and loss-of-function studies in bladder cancer lines, T24 and UM-UC-3. After transfected with miR-576-3p, T24 and UM-UM-3 showed significant decreases in cell viability and colony formation rate, which indicated that miR-576-3p was able to inhibit the proliferation in bladder cancer cells. Further analysis by FACS demonstrated that the overexpression miR-576-3p could induce G1-phase arrest, and on the contrary, the loss of miR-576-3p expression could promote bladder cancer cells proliferation by accelerating the progression of cell cycle. All these results suggested that G1-phase arrest specifically induced by miR-576-3p was the potential cause of the growth suppression.
Based on the growth suppression function of miR-576-3p discovered in our study, we chose one of the predicted target genes of miR-576-3p, CCND1, which plays an important role in cell cycle and cell growth, to investigate the mechanism of miR-576-3p in regulating the growth of bladder cancer cells. CCND1 is a crucial regulator of cell cycle, and related to the development of many cancers. It binds and activates cyclin-dependent kinases (CDK), CDK4 and CDK6, which subsequently phosphorylates tumor suppressor protein Rb and allows the cell cycle to progress through G1 into S phase (Malumbres and Barbacid, 2009; Satyanarayana and Kaldis, 2009; Tashiro et al., 2007). Given its important role in cell cycle, the expression of CCND1 needs to be strictly ruled. It is subject to all aspects of regulation, including gene transcription, RNA processing, splicing, modification, protein stability and cellular localization (Witzel et al., 2010). Deregulation of any of these procedures could induce overexpression of CCND1, and subsequently promote tumorigenesis in many kinds of cells (Fu et al., 2004). Recently, lots of studies demonstrate that CCND1 is frequently up-regulated in cancers and its overexpression might be due to many factors including increased transcription, translation, and protein stability (Kim and Diehl, 2009). In bladder cancers, the overexpression of CCND1 has already been identified (Kopparapu et al., 2013), and our experiments also revealed the same up-regulation of CCND1 in T24 and UM-UC-3 cell lines. Furthermore, numerous evidences have proved that the variation of CCND1 expression could remarkably affect the proliferation of bladder cancer cells (Fang et al., 2013; Liu et al., 2013). And microRNAs, on the other hand, such as miR-15b, miR-34a and miR-193a have also been reported to inhibit proliferation through down-regulation of CCND1 expression in various cancers (Chen et al., 2010; Sun et al., 2008; Xia et al., 2009).
In our study, Western blotting and real-time PCR analyses demonstrated that overexpression of miR-576-3p was able to down-regulate the expression of CCND1, and it could also inhibit the cell viability, colony formation as well as induce G1-phase arrest in T24 and UM-UC-3 cells. Correspondingly, the knockdown of miR-576-3p increased the expression of CCND1 and promoted the proliferation of bladder cancer cells by accelerating the progression of cell cycle. Meanwhile, the knock-down of CCND1 could copy the effects of over-expressed miR-576-3p on cell growth and the distribution of cell cycle. All these evidences implied that miR-576-3p might act as a tumor suppressor or a negative regulator in bladder cancer cell lines by inhibiting the expression of CCND1. And we then supposed that the down-regulation of miR-576-3p might oppositely enhance the growth of bladder cancer cells and subsequently facilitate the development of bladder cancer through up-regulating the expression of CCND1. Moreover, luciferase assay further proved that miR-576-3p could directly target the 3′-UTR of CCND1. The restoration of CCND1 expression could partly reverse the cell cycle arrest induced by miR-576-3p. Therefore, our study finally revealed that miR-576-3p was able to suppress the proliferation of bladder cancer cells by directly targeting the 3′-UTR of CCND1.
In conclusion, our study identified the down-regulation of miR-576-3p in T24 and UM-UC-3 cell lines and first reported that miR-576-3p, as a potential tumor suppressor in bladder cancer cells, could suppress cell proliferation by targeting CCND1. Despite the absence of further studies to identify the other targets and investigate the expression of miR-576-3p in bladder cancer tissues or other cell lines, our experiment indicates that restoration of miR-576-3p might be a promising therapeutic option for inhibiting cell proliferation in bladder cancers.
Acknowledgments
This study was supported by Grants from the National Key Clinical Specialty Construction Project of China, Key medical disciplines of Zhejiang province, National Natural Science Foundation of China (Grant No. 30900552), the Public Welfare Technology Application Research project of Zhejiang Provincial Science Technology Department of China (Grant No. 2014C33198) and the Zhejiang Provincial Medical Science Foundation of China (Grant No. 2014KYB084) and Zhejiang Provincial Natural Science Foundation of China (LQ14H160012, Y2110120 and LY12H05006).
REFERENCES
- Allegra A, Alonci A, Campo S, Penna G, Petrungaro A, Gerace D, Musolino C. Circulating microRNAs: new biomarkers in diagnosis, prognosis and treatment of cancer (review) Int J Oncol. 2012;41:1897–1912. doi: 10.3892/ijo.2012.1647. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catto JW, Alcaraz A, Bjartell AS, De Vere White R, Evans CP, Fussel S, Hamdy FC, Kallioniemi O, Mengual L, Schlomm T, et al. MicroRNA in prostate, bladder, and kidney cancer: a systematic review. Eur Urol. 2011;59:671–681. doi: 10.1016/j.eururo.2011.01.044. [DOI] [PubMed] [Google Scholar]
- Chen J, Feilotter HE, Pare GC, Zhang X, Pemberton JG, Garady C, Lai D, Yang X, Tron VA. MicroRNA-193b represses cell proliferation and regulates cyclin D1 in melanoma. Am J Pathol. 2010;176:2520–2529. doi: 10.2353/ajpath.2010.091061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho WC. MicroRNAs: potential biomarkers for cancer diagnosis, prognosis and targets for therapy. Int J Biochem Cell Biol. 2010;42:1273–1281. doi: 10.1016/j.biocel.2009.12.014. [DOI] [PubMed] [Google Scholar]
- Coskun E, Neumann M, Schlee C, Liebertz F, Heesch S, Goekbuget N, Hoelzer D, Baldus CD. MicroRNA profiling reveals aberrant microRNA expression in adult ETP-ALL and functional studies implicate a role for miR-222 in acute leukemia. Leuk Res. 2013;37:647–656. doi: 10.1016/j.leukres.2013.02.019. [DOI] [PubMed] [Google Scholar]
- Fang Y, Cao Z, Hou Q, Ma C, Yao C, Li J, Wu XR, Huang C. Cyclin d1 downregulation contributes to anticancer effect of isorhapontigenin on human bladder cancer cells. Mol Cancer Ther. 2013;12:1492–1503. doi: 10.1158/1535-7163.MCT-12-0922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendler A, Stephan C, Yousef GM, Jung K. MicroRNAs as regulators of signal transduction in urological tumors. Clin Chem. 2011;57:954–968. doi: 10.1373/clinchem.2010.157727. [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu M, Wang C, Li Z, Sakamaki T, Pestell RG. Minireview: Cyclin D1: normal and abnormal functions. Endocrinology. 2004;145:5439–5447. doi: 10.1210/en.2004-0959. [DOI] [PubMed] [Google Scholar]
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- Kim JK, Diehl JA. Nuclear cyclin D1: an oncogenic driver in human cancer. J Cell Physiol. 2009;220:292–296. doi: 10.1002/jcp.21791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopparapu PK, Boorjian SA, Robinson BD, Downes M, Gudas LJ, Mongan NP, Persson JL. Expression of cyclin d1 and its association with disease characteristics in bladder cancer. Anticancer Res. 2013;33:5235–5242. [PMC free article] [PubMed] [Google Scholar]
- Li S, Xu X, Xu X, Hu Z, Wu J, Zhu Y, Chen H, Mao Y, Lin Y, Luo J, et al. MicroRNA-490-5p inhibits proliferation of bladder cancer by targeting c-Fos. Biochem Biophys Res Commun. 2013;441:976–981. doi: 10.1016/j.bbrc.2013.11.006. [DOI] [PubMed] [Google Scholar]
- Lin T, Dong W, Huang J, Pan Q, Fan X, Zhang C, Huang L. MicroRNA-143 as a tumor suppressor for bladder cancer. J Urol. 2009;181:1372–1380. doi: 10.1016/j.juro.2008.10.149. [DOI] [PubMed] [Google Scholar]
- Lin Y, Wu J, Chen H, Mao Y, Liu Y, Mao Q, Yang K, Zheng X, Xie L. Cyclin-dependent kinase 4 is a novel target in micoRNA-195-mediated cell cycle arrest in bladder cancer cells. FEBS Lett. 2012;586:442–447. doi: 10.1016/j.febslet.2012.01.027. [DOI] [PubMed] [Google Scholar]
- Lin Y, Chen H, Hu Z, Mao Y, Xu X, Zhu Y, Xu X, Wu J, Li S, Mao Q, et al. miR-26a inhibits proliferation and motility in bladder cancer by targeting HMGA1. FEBS Lett. 2013;587:2467–2473. doi: 10.1016/j.febslet.2013.06.021. [DOI] [PubMed] [Google Scholar]
- Liu JY, Qian D, He LR, Li YH, Liao YJ, Mai SJ, Tian XP, Liu YH, Zhang JX, Kung HF, et al. PinX1 suppresses bladder urothelial carcinoma cell proliferation via the inhibition of telomerase activity and p16/cyclin D1 pathway. Mol Cancer. 2013;12:148. doi: 10.1186/1476-4598-12-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majid S, Dar AA, Saini S, Deng G, Chang I, Greene K, Tanaka Y, Dahiya R, Yamamura S. MicroRNA-23b functions as a tumor suppressor by regulating Zeb1 in bladder cancer. PLoS One. 2013;8:e67686. doi: 10.1371/journal.pone.0067686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9:153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- Nana-Sinkam SP, Croce CM. MicroRNA dysregulation in cancer: opportunities for the development of microRNA-based drugs. Invest Drugs J. 2010;13:843–846. [PubMed] [Google Scholar]
- Osaki M, Takeshita F, Ochiya T. MicroRNAs as biomarkers and therapeutic drugs in human cancer. Biomarkers. 2008;13:658–670. doi: 10.1080/13547500802646572. [DOI] [PubMed] [Google Scholar]
- Piva R, Spandidos DA, Gambari R. From microRNA functions to microRNA therapeutics: novel targets and novel drugs in breast cancer research and treatment (Review) Int J Oncol. 2013;43:985–994. doi: 10.3892/ijo.2013.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci USA. 2008;105:1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploeg M, Aben KK, Kiemeney LA. The present and future burden of urinary bladder cancer in the world. World J Urol. 2009;27:289–293. doi: 10.1007/s00345-009-0383-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratert N, Meyer HA, Jung M, Lioudmer P, Mollenkopf HJ, Wagner I, Miller K, Kilic E, Erbersdobler A, Weikert S, et al. miRNA profiling identifies candidate mirnas for bladder cancer diagnosis and clinical outcome. J Mol Diagn. 2013;15:695–705. doi: 10.1016/j.jmoldx.2013.05.008. [DOI] [PubMed] [Google Scholar]
- Satyanarayana A, Kaldis P. Mammalian cell-cycle regulation: several Cdks, numerous cyclins and diverse compensatory mechanisms. Oncogene. 2009;28:2925–2939. doi: 10.1038/onc.2009.170. [DOI] [PubMed] [Google Scholar]
- Shi Z, Wei Q, Zhang M, She J. MicroRNAs in bladder cancer: expression profiles, biological functions, regulation, and clinical implications. Crit Rev Eukaryot Gene Expr. 2014;24:55–75. doi: 10.1615/critreveukaryotgeneexpr.2014007798. [DOI] [PubMed] [Google Scholar]
- Sun F, Fu H, Liu Q, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. Downregulation of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Lett. 2008;582:1564–1568. doi: 10.1016/j.febslet.2008.03.057. [DOI] [PubMed] [Google Scholar]
- Tashiro E, Tsuchiya A, Imoto M. Functions of cyclin D1 as an oncogene and regulation of cyclin D1 expression. Cancer Sci. 2007;98:629–635. doi: 10.1111/j.1349-7006.2007.00449.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatarano S, Chiyomaru T, Kawakami K, Enokida H, Yoshino H, Hidaka H, Yamasaki T, Kawahara K, Nishiyama K, Seki N, et al. miR-218 on the genomic loss region of chromosome 4p15.31 functions as a tumor suppressor in bladder cancer. Int J Oncol. 2011;39:13–21. doi: 10.3892/ijo.2011.1012. [DOI] [PubMed] [Google Scholar]
- Witzel II, Koh LF, Perkins ND. Regulation of cyclin D1 gene expression. Biochem Soc Trans. 2010;38:217–222. doi: 10.1042/BST0380217. [DOI] [PubMed] [Google Scholar]
- Xia H, Qi Y, Ng SS, Chen X, Chen S, Fang M, Li D, Zhao Y, Ge R, Li G, et al. MicroRNA-15b regulates cell cycle progression by targeting cyclins in glioma cells. Biochem Biophys Res Commun. 2009;380:205–210. doi: 10.1016/j.bbrc.2008.12.169. [DOI] [PubMed] [Google Scholar]
- Zhang ZC, Huang Y, Wang XJ, Wang M, Ma LL. Expression of circulating microRNAs in patients with bladder urothelial carcinoma. Beijing Da Xue Xue Bao. 2013;45:532–536. [PubMed] [Google Scholar]


