Abstract
• Premise of the study: While numerous software packages enable scientists to evaluate molecular data and transform them for phylogenetic analyses, few such tools exist for phenomic data. We introduce MatrixConverter, a program that helps expedite and facilitate the transformation of raw phenomic character data into discrete character matrices that can be used in most evolutionary inference programs.
• Methods and Results: MatrixConverter is an open source program written in Java; a platform-independent binary executable, as well as sample data sets and a user’s manual, are available at https://github.com/gburleigh/MatrixConverter/tree/master/distribution. MatrixConverter has a simple, intuitive user interface that enables the user to immediately begin scoring phenomic characters. We demonstrate the performance of MatrixConverter on a phenomic data set from cycads.
• Conclusions: New technologies and software make it possible to obtain phenomic data from species across the tree of life, and MatrixConverter helps to transform these new data for evolutionary or ecological inference.
Keywords: MatrixConverter, morphology, phenomics, phylogenetic matrix, software
The study of phenomics (i.e., morphology, physiology, behavior, or other phenotypic traits) is among the oldest disciplines in biology, and it is important for the study of plant form and function and for building and interpreting phylogenetic trees. In recent years, new technologies such as transmission electron microscopy, scanning electron microscopy, and X-ray computed tomography enable scientists to observe and describe morphologies in unprecedented detail (e.g., Dhondt et al., 2010), and new efforts in crowd-sourcing, computer vision, and developing ontologies can help describe and report phenomic characters across many taxa (Burleigh et al., 2013; Deans et al., 2015). In addition, CharaParser, a semantic parser for taxonomic literature, enables scientists to identify phenomic characters from the wealth of existing species descriptions (Cui, 2012). Although these new technologies and software make it possible to obtain phenomic data from species across the tree of life, the resulting data—species with character state information—are not easily usable for evolutionary inference.
In this paper, we introduce MatrixConverter, a program that helps expedite and facilitate the transformation of raw phenomic character data into discrete character matrices that can be used in most popular evolutionary inference programs. Specifically, it takes as input a table of phenomic characters and enables scientists to quickly and easily evaluate the underlying character data and translate them into a matrix of discrete, numerical character states. Previously, scoring and discretizing phenomic data and formatting the data for phylogenetic analyses have largely been done by hand. We are not aware of any existing software to automate and ease the processing of phenomic data for evolutionary inference.
METHODS AND RESULTS
MatrixConverter is an open source program written in Java. The source code is available at https://github.com/gburleigh/MatrixConverter/. A platform-independent binary executable, as well as sample data sets and a user’s manual, are also available at https://github.com/gburleigh/MatrixConverter/tree/master/distribution. To install the binary executable, the user simply needs to download and unzip the file on their computer. On a Mac or Windows computer, to open the user interface (Fig. 1), the user just has to click the executable JAR file. On a Linux machine, the user must open the terminal shell, move to the directory with the executable, and then type “java –jar MatrixConverter.jar”. The executable has been successfully tested on Windows 7 and Mac OS X version 10.9. MatrixConverter requires Java Runtime Environment (JRE) 6.0 or later to be installed on the computer.
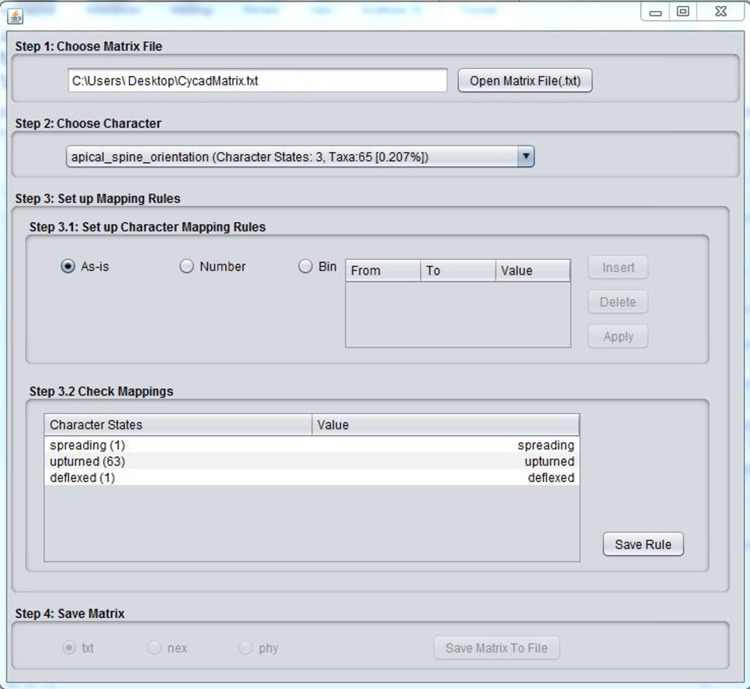
Fig. 1.
User interface for MatrixConverter. The Step 1 section is where users upload a phenomic character matrix formatted as a tab-delimited text or CSV file. The Step 2 tab allows the user to choose a phenomic character, defined in a column heading in the input file; it also provides the number and percentage of taxa with data for the character and the total number of character states. Step 3 enables the user to code the character states as integers. Step 3.1 provides simple ways to automate the character scoring, while the user can evaluate and manually score characters in Step 3.2. In Step 4, the user can save the newly scored characters along with scoring rules in a variety of formats.
MatrixConverter has a simple and intuitive user interface that should enable the user to immediately begin scoring phenomic characters with little or no prior instruction on running the program (Fig. 1). The input for this MatrixConverter can be either a tab-delimited text file (*.txt) or a CSV file of morphological characters (although note that the CSV format may be problematic if commas are used to separate multiple character states for a polymorphic character). The first line of the input file is a header line with a description of the columns. The first column of the file lists the taxon names, and the rest of the columns include the character states for phenomic characters. The character states can be any combination of integers, real numbers, or words. With phenomic data, it is also likely that many of the cells will have no values (i.e., the character is missing or character state unknown). MatrixConverter can easily handle missing data, which can either be coded as empty cells or as “NA” in the input files, or cases where a taxon has multiple character states. See the sample input files (e.g., Cycadtest.txt, available at https://github.com/gburleigh/MatrixConverter/tree/master/distribution) for examples.
Once the file is uploaded, the user must select the informative characters that will be used to build the matrix and review the character states. In the user interface under “Step 2: Choose a Character,” the user can select a character from the dropdown list. The name of the character is followed by the number of character states and the number and total percentage of taxa that have information for that character. Step 3 is designed to facilitate character coding. Characters can be coded in Step 3.1 using the default coding (As-is), converted to numerical values, or binned. The As-is feature may be useful if the character states are already digits (e.g., 0, 1, 2). If you choose the “Number” option, the character states listed under Step 3.2 will be converted into digits (0, 1, 2, etc.) in the order in which they appear in the list. The “Bin” option enables the user to categorize numerical characters based on their numerical values. The options in Step 3 automate the process of coding phenomic characters into discrete character states. However, the user can manually modify the values of any character state in Step 3.2. After the character states have been defined, the character is incorporated into the growing matrix by clicking on the button “Save Rule.” The final matrix is saved by clicking the “Save Matrix” button.
MatrixConverter can output discrete character matrices for phylogenetic or other evolutionary analyses, such as tests for phylogenetic signal, ancestral state reconstructions, or tests to relate phenotypic characters with diversification, in NEXUS (Maddison et al., 1997), PHYLIP, or NeXML format (Vos et al., 2012), or as a text file. These file formats enable the output files to be used as input in many evolutionary analysis programs including PAUP* (Swofford, 2003), TNT (Goloboff et al., 2008), MacClade (Maddison and Maddison, 2005), Mesquite (Maddison and Maddison, 2014), MrBayes (Ronquist et al., 2012), GARLI (Zwickl, 2006), PHYLIP (Felstenstein, 1989), RAxML (Stamatakis, 2014), or packages within R (R Core Team, 2014). The saved file will include not only the discrete phenomic character matrix, but also information describing how the character states for each character were coded.
We tested the performance of MatrixConverter on a set of characters from cycads. The sample input file (Cycadtest.txt) is a subset of the original phenomic data set. The original cycad phenomic character set, the input for MatrixConverter, was obtained by parsing the taxonomic descriptions available at the cycad pages (http://plantnet.rbgsyd.nsw.gov.au/PlantNet/cycad/) with CharaParser beta version 0.1 (Cui, 2012). Such a character data set can be generated using the “Text Capture” and “Matrix Generation” features on the ETC website (Explorer of Taxonomic Concepts; http://etc.cs.umb.edu/etcsite/). The original data set contained 647 characters. However, only 307 of these characters had data (i.e., defined character states) from at least four taxa, and only 174 of these characters contained at least two character states. With the character frequency data listed in the “Step 2: Choose Character” tab of MatrixConverter (Fig. 1), we filtered out characters with fewer data points, which would provide little or no information for most evolutionary inference analyses. Of the remaining 174 characters, we determined that 52 of them contained character states that were not homologous. Examples of nonhomologous character states include a combination of qualitative and numerical character states (e.g., character “Lamina width” had values “thickened” and “0.04”), or the character states describing different aspects of a character (e.g., “Leaflet shape” had the values “entire,” “divided,” “serrate,” “lacerate,” “lanceolate,” “falcate,” “elliptic,” “with 1–2 lobes,” “dentate,” and “rounded”). It was easy to identify and ultimately exclude characters with nonhomologous character states using the “Step 3.2: Check Mappings” window (Fig. 1). Of the remaining characters, 67 were in natural discrete categories that were simple to automatically discretize, and 55 were continuous, numerical characters that we could bin and discretize using Step 3. Thus, the phylogenetic matrix in NEXUS format that was output from MatrixConverter contained a total of 122 variable, discrete numerical characters ready for evolutionary inference. Coding these cycad characters with MatrixConverter took only a few hours, while attempting to code them without MatrixConverter doubtlessly would have taken many days.
CONCLUSIONS
While numerous software packages enable scientists to evaluate molecular data and transform them for phylogenetic analyses, few such tools exist for phenomic data. Thus, despite recent developments in methods to identify, observe, and collect phenomic data, the work of transforming these data into a usable format for evolutionary or ecological inference can still be extremely time-consuming and tedious. MatrixConverter facilitates the process of evaluating, scoring, and formatting raw phenomic data so that the data can be used for evolutionary inference. Its intuitive and simple user interface and platform-free implementation make it easy for any scientist to use without training. Although the development of MatrixConverter was motivated by the large phenomic character matrices produced by CharaParser, it can be used to evaluate and transform any data sets of phenomic characters into discrete character matrices, as long as they are in a tab-delimited text or CSV file.
MatrixConverter makes phenomic character coding easy, but character coding decisions may be contentious and ideally should be done with much expert consideration (e.g., Brazeau, 2011). In designing MatrixConverter, we sought to expedite the process of character coding while remaining agnostic with respect to character coding decisions. Thus, it is easy to make extremely bad character coding decisions using MatrixConverter. For example, if you are using MatrixConverter with data generated from CharaParser, it is important to keep in mind that processing nonparallel taxonomic descriptions can erroneously inflate the number of characters extracted from the text, and it is important to be vigilant about other possible errors in character state designations. Furthermore, although we have tried to make the output of MatrixConverter compatible with many phylogenetic software programs, some character codings are not allowed by all phylogenetic programs. For example, many programs do not allow polymorphic characters (i.e., multiple character states from a character in a taxon), and many programs have limits on the number of character states allowed for a single character. MatrixConverter is meant to enhance the process of evaluating and coding phenomic characters for scientists, but it does not replace the need for thoughtful consideration of the characters.
LITERATURE CITED
- Brazeau M. D. 2011. Problematic character coding methods in morphology and their effects. Biological Journal of the Linnean Society 104: 489–498. [Google Scholar]
- Burleigh J. G., Alphonse K., Alverson A. J., Bik H. M., Blank C., Cirranello A. L., Cui H., et al. 2013. Next-generation phenomics for the Tree of Life. PLoS Currents 5: ecurrents.tol.085c713acafc8711b2ff7010a4b03733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H. 2012. CharaParser for fine-grained semantic annotation of organism morphological descriptions. Journal of the American Society for Information Science and Technology 63: 738–754. [Google Scholar]
- Deans A. R., Lewis S. E., Huala E., Anzaldo S. S., Ashburner M., Balhoff J. P., Blackburn D. C., Blake J. A., et al. 2015. Finding our way through phenotypes. PLoS Biology 13: e1002033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhondt S., Vanhaeren H., Van Loo D., Cnudde V., Inzé D. 2010. Plant structure visualization by high-resolution X-ray computed tomography. Trends in Plant Science 15: 419–422. [DOI] [PubMed] [Google Scholar]
- Felstenstein J. 1989. PHYLIP—Phylogeny Inference Package. Cladistics 5: 164–166. [Google Scholar]
- Goloboff P. A., Farris J. S., Nixon K. C. 2008. TNT, a free program for phylogenetic analysis. Cladistics 24: 774–786. [Google Scholar]
- Maddison D. R., Swofford D. L., Maddison W. P. 1997. NEXUS: An extensible file format for systematic information. Systematic Biology 46: 590–621. [DOI] [PubMed] [Google Scholar]
- Maddison D. R., Maddison W. P. 2005. MacClade 4: Analysis of phylogeny and character evolution. Website http://macclade.org [accessed 20 January 2015].
- Maddison W. P., Maddison D. R. 2014. Mesquite: A modular system for evolutionary analysis. Version 3.01. Website http://mesquiteproject.org [accessed 20 January 2015].
- R Core Team. 2014. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Website http://www.R-project.org [accessed 20 January 2015].
- Ronquist F., Teslenko M., Van Der Mark P., Ayers D. L., Darling A., Höhna S., Larget B., et al. 2012. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large phylogeny. Systematic Biology 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics (Oxford, England) 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford D. L. 2003. PAUP*. Phylogenetic analysis using parsimony (*and Other Methods). Version 4. Sinauer, Sunderland, Massachusetts, USA. [Google Scholar]
- Vos R. A., Balhoff J. P., Caravas J. A., Holder M. T., Lapp H., Maddison W. P., Midford P. E., et al. 2012. NeXML: Rich, extensible, and verifiable representation of comparative data and metadata. Systematic Biology 61: 675–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwickl D. J. 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. Ph.D. dissertation, The University of Texas at Austin, Austin, Texas, USA.