Mitofusin 2 plays an unexpected role in maintaining the terpenoid biosynthesis pathway and is necessary for mitochondrial coenzyme Q biosynthesis.
Abstract
Mitochondria form a dynamic network within the cell as a result of balanced fusion and fission. Despite the established role of mitofusins (MFN1 and MFN2) in mitochondrial fusion, only MFN2 has been associated with metabolic and neurodegenerative diseases, which suggests that MFN2 is needed to maintain mitochondrial energy metabolism. The molecular basis for the mitochondrial dysfunction encountered in the absence of MFN2 is not understood. Here we show that loss of MFN2 leads to impaired mitochondrial respiration and reduced ATP production, and that this defective oxidative phosphorylation process unexpectedly originates from a depletion of the mitochondrial coenzyme Q pool. Our study unravels an unexpected and novel role for MFN2 in maintenance of the terpenoid biosynthesis pathway, which is necessary for mitochondrial coenzyme Q biosynthesis. The reduced respiratory chain function in cells lacking MFN2 can be partially rescued by coenzyme Q10 supplementation, which suggests a possible therapeutic strategy for patients with diseases caused by mutations in the Mfn2 gene.
Introduction
Mammalian Mitofusin 1 (MFN1) and Mitofusin 2 (MFN2) belong to the GTPase family of proteins (Bourne et al., 1990) and are required for mitochondrial outer membrane fusion (Rojo et al., 2002). Ubiquitous knockout of the Mfn1 or Mfn2 gene results in embryonic lethality during midgestation (Chen et al., 2003) due to placental dysfunction (Chen et al., 2003, 2007). Remarkably, by using a conditional knockout allele in conjunction with cre-recombinase expression only in the embryo, Mfn1 or Mfn2 knockout mice are born alive (Chen et al., 2007). Mice ubiquitously lacking MFN1 are apparently healthy, whereas mice without MFN2 die in the early postnatal period and show severe defects in movement and balance (Chen et al., 2007). Experiments performed on immortalized Mfn1 and Mfn2 knockout mouse embryonic fibroblasts (MEFs) have shown that hetero-oligomeric complexes between MFN1 and MFN2 are important for control of mitochondrial fusion (Detmer and Chan, 2007). Despite its well-established role in mitochondrial fusion, a growing body of evidence suggests that MFN2 has additional functions, such as ER–mitochondria tethering (de Brito and Scorrano, 2008) and control of mitochondrial transport in axons (Misko et al., 2010), which may explain some of the disease manifestations in humans with Mfn2 mutations (Züchner et al., 2004; Detmer et al., 2008). The different functions of MFN2 are supported by in vivo studies in mice, showing that MFN2 is required for normal glucose homeostasis (Sebastián et al., 2012), and is essential for axonal projections of dopaminergic neurons (Lee et al., 2012; Pham et al., 2012). However, the molecular basis for the mitochondrial dysfunction, caused by loss of MFN2, remains to be established. Here we report the unexpected finding that loss of MFN2 causes depletion of mitochondrial coenzyme Q, which, in turn, leads to respiratory chain dysfunction. A global reduction in the terpenoid synthesis pathway is present in both Mfn2 and Mfn1 conditional knockout mice, but the defect is more severe and impacts respiratory chain function only in Mfn2 knockout mice. Moreover, addition of coenzyme Q to immortalized Mfn2 knockout MEFs or isolated mitochondria partially rescues the respiratory chain dysfunction, which suggests a therapeutic strategy for intervention in affected patients.
Results
MFN2 is required to maintain mitochondrial morphology and function in cardiomyocytes
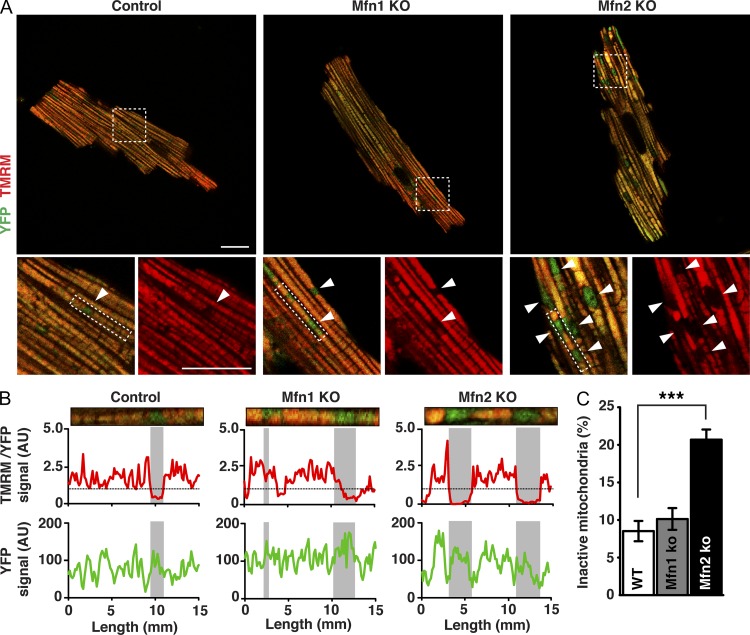
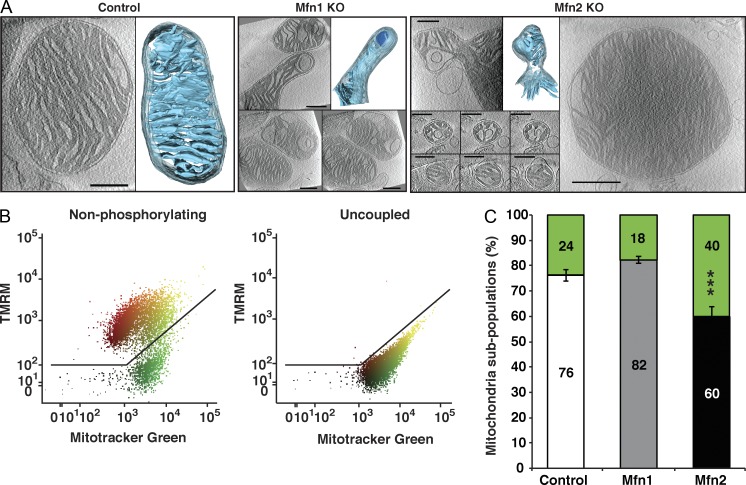
To further investigate the role of mitofusins in energy metabolism, we performed a detailed characterization of how loss of MFN1 and MFN2 affects mitochondrial morphology and oxidative phosphorylation (OXPHOS) in heart, a typical highly energy-demanding tissue. Heart conditional knockouts for Mfn1 or Mfn2 were born at Mendelian proportions and had no enlargement of the heart when assessed at the age of 20 wk (Fig. S1, A and B), which is consistent with results from independent knockout strains (Papanicolaou et al., 2011, 2012). Furthermore, mitochondrial DNA (mtDNA) levels (Fig. S1 C) and mitochondrial mass were not affected in Mfn1 and Mfn2 knockout hearts, as determined by electron microscopy analysis (Fig. S1 D). The absence of changes in mitochondrial content prompted us to analyze mitochondrial activity at the single-cell level. To this end, we performed retrograde perfusion to isolate adult cardiomyocytes from control and Mfn1 and Mfn2 knockout hearts expressing a mitochondrial targeted YFP. Confocal microscopy analysis of cardiomyocytes isolated from Mfn1 knockout hearts showed that the mitochondria were homogeneous in shape and organized in a classical linear arrangement between myofibrils, as observed in controls (Fig. 1 A). In contrast, the mitochondrial organization was disrupted in Mfn2 knockout cardiomyocytes with the appearance of enlarged mitochondria (Fig. 1 A). To gain functional insights into the observed mitochondrial morphological alterations, we assessed the mitochondrial membrane potential (Δψ) by loading the cells with the potentiometric probe tetramethylrhodamine methyl ester (TMRM). Interestingly, the membrane potential varied greatly among individual mitochondria in control cardiomyocytes, as indicated by the strong variations in TMRM fluorescence in line-scan analysis (Figs. 1 B and S1 E). This finding is in agreement with previous reports suggesting that cardiac mitochondria are electrically and functionally independent (Kuznetsov et al., 2006; Beraud et al., 2009). Strikingly, Δψ analysis disclosed the presence of inactive mitochondria that were almost twice as abundant in Mfn2 knockout cardiomyocytes in comparison with Mfn1 knockout or control cardiomyocytes (Fig. 1, A–C; and Fig. S1, E and F). Consistent with the light microscopy results, mitochondrial morphological heterogeneity of Mfn2 knockout heart mitochondria was also found by electron cryo-tomography analysis of fully hydrated mitochondria at high resolution (Fig. 2 A). Interestingly, a small fraction of mitochondria isolated from Mfn1 and Mfn2 knockout hearts exhibited atypical morphology with bud formation (Fig. 2 A). In addition, significant fractions of abnormally small (diameter <300 nm) or large (diameter >1.5 µm) mitochondria were observed in Mfn2 knockout hearts (Figs. 2 A and S2 A). The functional defects observed in single cells were further quantified by FACS analysis of isolated heart mitochondria after double staining with MitoTracker green and TMRM, which revealed a twofold increase of Δψ-negative mitochondria in Mfn2 knockout in comparison with Mfn1 knockout or control hearts (Fig. 2, B and C).
Figure 1.
Abnormal mitochondrial morphology and function in Mfn2 knockout cardiomyocytes. (A) Representative confocal microscopy images of adult cardiomyocytes isolated from control, Mfn1 knockout, and Mfn2 knockout mouse hearts that also express mitochondria-targeted YFP. Live cardiomyocytes were loaded with TMRM to evaluate Δψ. Magnifications of the boxed areas below show merged channels of TMRM and mito-YFP (left) and single channels of TMRM (right). Arrowheads point to TMRM negative mitochondria. Bars, 20 µm. (B) Representative single-cell line-scan profiles of the rectangular boxed areas in A showing the intensity levels (arbitrary units) of TMRM normalized to YFP (top) and YFP (bottom) in individual mitochondria. Gray boxes in the graphs highlight the tracks corresponding to the TMRM-negative mitochondria. (C) Quantification of inactive mitochondria in adult cardiomyocytes isolated from control, Mfn1 conditional knockout, and Mfn2 conditional knockout animals (n ≥ 25 cardiomyocytes from three independent experiments. Error bars indicate ±SEM; ***, P < 0.001. (No error bar applies in B, as this specific analysis is from one cell and representative of the cell population analyzed in C.)
Figure 2.
Loss of MFN2 results in mitochondrial heterogeneity and dysfunction. (A) Electron cryo-tomography analysis of isolated mitochondria. Slices through typical tomographic volumes and example segmented 3D representations from control, Mfn1, and Mfn2 knockout heart mitochondria are shown (blue, cristae; transparent gray, IMS; outer membrane omitted for clarity). Bars, 250 nm. (B) Representative analysis out of 14 FACS analyses of isolated control heart mitochondria incubated with succinate and rotenone, and stained with MitoTracker green FM and TMRM. FACS analyses were performed using the FlowJo software, applying a biexponential transformation to digital data. (C) FACS analysis of control (white bars, n = 5 independent experiments), Mfn1 knockout (gray bars, n = 4 independent experiments), and Mfn2 knockout (black bars, n = 5 independent experiments) heart mitochondria to determine the percentage of membrane potential–negative (green) mitochondria in the presence of succinate and rotenone. Error bars indicate ±SEM. ***, P < 0.001.
Loss of MFN2 impairs the mitochondrial respiratory chain capacity
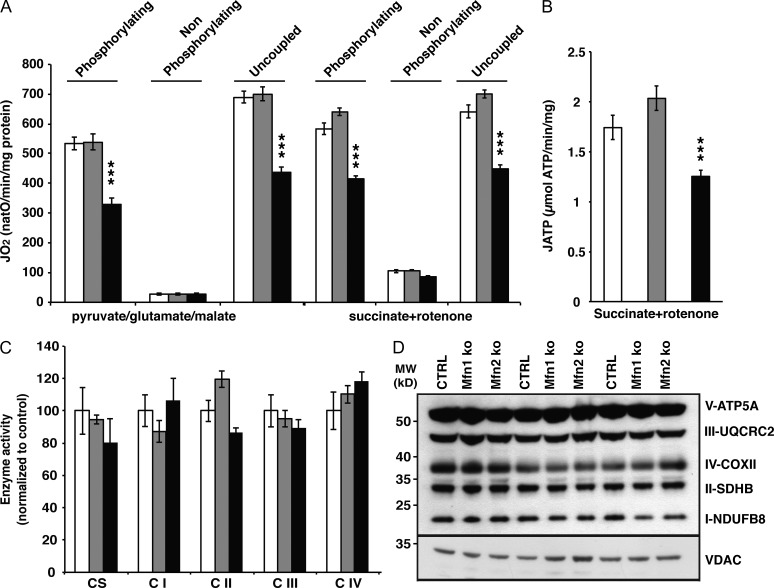
The increase of Δψ-negative mitochondria in hearts lacking MFN2 prompted us to characterize the OXPHOS system properties. To this end, isolated heart mitochondria were incubated with respiratory substrates whose metabolism results in delivery of electrons at the level of complex I (pyruvate, glutamate, malate) or complex II (succinate + rotenone), and the oxygen consumption rate was recorded in the phosphorylating (ADP and Pi), nonphosphorylating (oligomycin), and uncoupled state (carbonyl cyanide m-chlorophenyl hydrazone [CCCP]). We observed a clear decrease in respiration capacity under phosphorylating and uncoupled conditions in Mfn2 knockout heart mitochondria at 20 (Fig. 3 A) and 60 wk of age (Fig. S2 B). In agreement with the reduced respiratory chain capacity, the ATP production rate was also affected in the absence of MFN2 (Fig. 3 B). To decipher the molecular origin of the respiratory chain dysfunction, we assessed the enzyme activities and steady-state levels of the respiratory chain complexes. Surprisingly, the respiratory chain complex enzyme activities were not decreased (Fig. 3 C) and the levels of different cytochromes, such as a-a3 (complex IV), bH-bL (complex III), and c, were normal in Mfn2 knockout heart mitochondria (Fig. S2 C). Also, the levels of different subunits of the OXPHOS system complexes as determined by Western blot analyses were unaffected (Fig. 3 D).
Figure 3.
Loss of MFN2 affects mitochondrial respiration without affecting the levels of OXPHOS components. (A) Oxygen consumption rates in heart mitochondria from control (white bars, n = 14 independent experiments), Mfn1 (gray bars, n = 13 independent experiments), and Mfn2 conditional knockout (black bars, n = 16 independent experiments) animals at age 20 wk. (B) ATP synthesis flux assessed in heart mitochondria from control (white bars, n = 7 independent experiments), Mfn1 (gray bars, n = 9 independent experiments), and Mfn2 conditional knockout (black bars, n = 8 independent experiments) animals at age 20 wk, in the presence of succinate and rotenone. (C) Citrate synthase (CS) and respiratory chain enzyme activity (CI–IV) measurements in isolated mitochondria from control (white bars, n = 4 independent experiments), Mfn1 (gray bars, n = 4 independent experiments), and Mfn2 conditional knockout (black bars, n = 4 independent experiments) animals. (D) Steady-state levels of different OXPHOS system subunits determined by Western blot analysis of heart mitochondria from control, Mfn1, and Mfn2 conditional knockout animals at age 20 wk. Error bars indicate ±SEM. ***, P < 0.001.
Mfn2 heart knockout mitochondria display a severe coenzyme Q deficiency
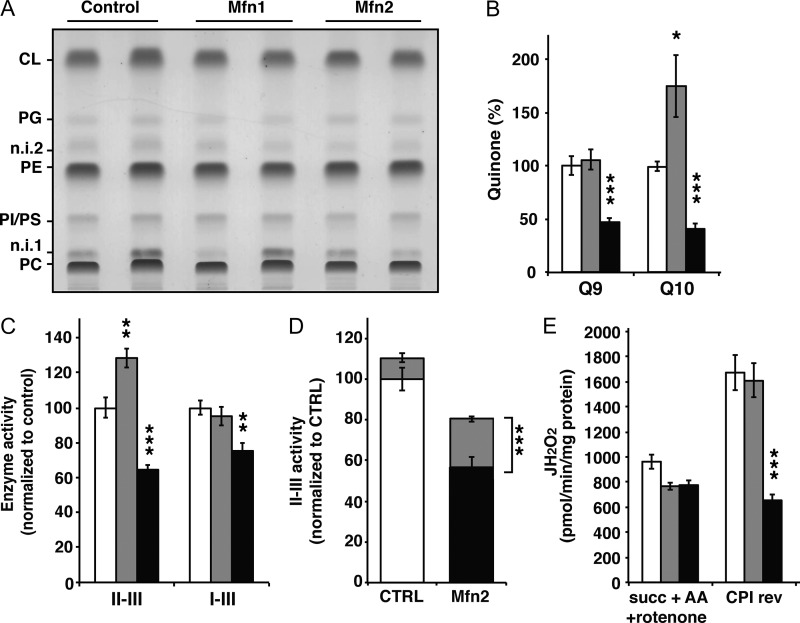
Because the lipid environment has been shown to impact the respiratory activity, we determined the mitochondrial lipid composition and found no alterations in phospholipid content (Fig. 4 A and S2 D). However, the levels of coenzyme Q9 and Q10 were severely decreased in Mfn2 knockout heart mitochondria in comparison with controls (Fig. 4 B). The coenzyme Q isoforms are organic antioxidant compounds present in all cellular membranes and are specifically required in the mitochondrial inner membrane to transfer electrons from complex I and II to complex III of the respiratory chain. To gain further insight into the potential consequences of the observed coenzyme Q deficiency, we measured the NADH–cytochrome c reductase (complex I-III) and succinate-cytochrome c reductase (complex II-III) activities, as they are dependent on the endogenous coenzyme Q pool. Interestingly, both of these respiratory chain enzyme activities were impaired in Mfn2 knockout heart mitochondria (Fig. 4 C). Moreover, the succinate cytochrome c reductase deficiency observed in isolated mitochondria from Mfn2 knockout hearts could be partially rescued by the addition of the artificial quinone decylubiquinone (Fig. 4 D). The rescue of the succinate cytochrome c reductase deficiency was comparable to the rescue seen after decylubiquinone treatment of cell lines from patients with genetic defects impairing the synthesis of coenzyme Q10 (Mollet et al., 2007). Reactive oxygen species (ROS) production was assessed by measuring H2O2 production at the level of complex III (succinate + rotenone) in the presence (Fig. 4 E) or absence of antimycin A (Fig. S2 E), and no difference between Mfn2 knockout and control mitochondria was observed. In contrast, ROS production induced by reversed electron flow at the level of complex I (succinate, rotenone sensitive) was clearly reduced in Mfn2 knockout heart mitochondria (Fig. 4 E), which suggests an important role for coenzyme Q levels in this process.
Figure 4.
Mfn2 knockout heart mitochondria are severely coenzyme Q deficient. (A) Mitochondrial phospholipid composition in hearts from control, Mfn1, and Mfn2 conditional knockout animals at age 20 wk analyzed by TLC. CL, cardiolipin; PG, phosphatidylglycerol; PE, phosphatidylethanolamine; PI/PS, phosphatidylinositol and phosphatidylserine; PC, phosphatidylcholine; n.i. 1 and n.i. 2, nonidentified 1 and 2. (B) Coenzyme Q content of isolated cardiac mitochondria from control (white bars, n = 10 independent experiments), Mfn1 (gray bars, n = 10 independent experiments), and Mfn2 conditional knockout (black bars, n = 10 independent experiments) animals. (C) Succinate-cytochrome c reductase (complex II-III) and NADH-cytochrome c reductase (complex I-III) enzyme activities determined in isolated heart mitochondria from control (white bars, n = 5 independent experiments), Mfn1 (gray bars, n = 5 independent experiments), and Mfn2 conditional knockout (black bars, n = 5 independent experiments) animals. (D) Succinate-cytochrome c reductase activity determined in the absence (white and black bars) or presence (gray bars) of an exogenous coenzyme Q analogue (decylubiquinone). The experiment was performed on mitochondria from control (white bars, n = 5 independent experiments) and Mfn2 knockout (black bars, n = 5 independent experiments) hearts. (E) Assessment of the hydrogen peroxide production rate in nonphosphorylating conditions in the presence of succinate, antimycin A, and rotenone (Succ+AA+rot) and succinate alone (CPI rev). The latter condition assesses the rotenone-sensitive H2O2 production caused by reverse electron flow through complex I. This experiment was performed on heart mitochondria from control (white bars, n = 5 independent experiments), Mfn1 (gray bars, n = 5 independent experiments), and Mfn2 conditional knockout (black bars, n = 5 independent experiments) animals. Error bars indicate ±SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
The coenzyme Q deficiency is caused by multiple terpenoid synthesis defects
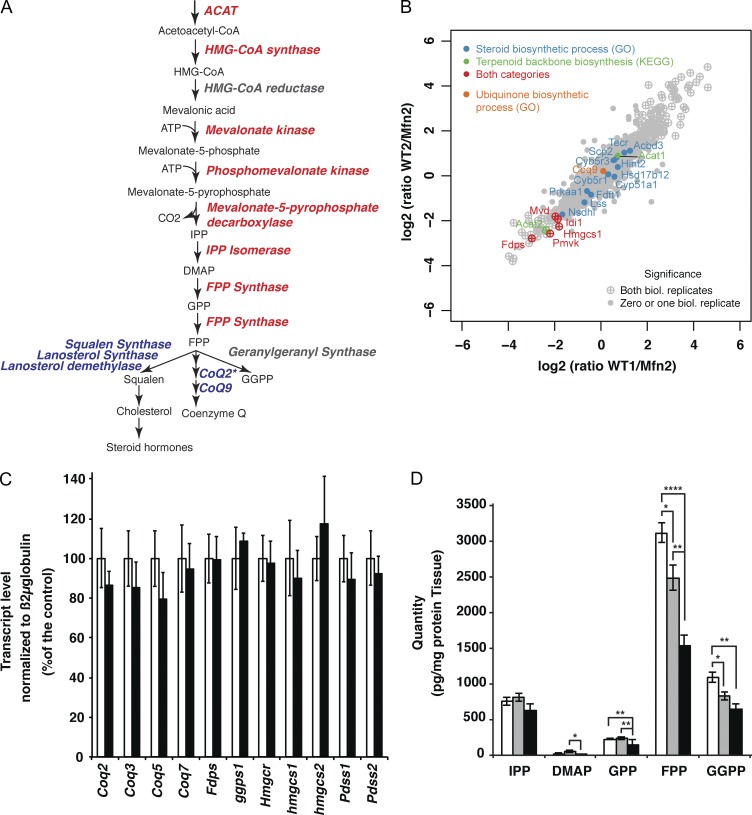
To decipher the molecular origin of the coenzyme Q deficiency, we characterized the proteome changes associated with loss of the MFN1 or MFN2 proteins, respectively. To this end, we performed stable isotope labeling by amino acids in cell culture (SILAC) experiments by quantitatively comparing the proteomes of Mfn1 and Mfn2 knockout MEFs (Fig. 5, A and B; and Fig. S3, B–D) with control MEFs, whose proteome had been saturated with heavy isotope labeled arginine and lysine (13C6 15N4 l-arginine, 13C6 15N4 l-lysine). Surprisingly, the terpenoid biosynthesis pathway clearly stood out as the unique metabolic process significantly down-regulated in both Mfn1 and Mfn2 knockout MEFs. The levels of all detected enzymes in the terpenoid biosynthesis pathway, except one of the two acetyl-CoA acetyltransferase isoforms (Acat 1), were decreased in comparison with controls (Fig. 5, A and B; and Fig. S3, B–D). In contrast, none of the downstream enzymes acting in the coenzyme Q biosynthesis pathway (Coq2, Coq9) or in the cholesterol and steroidogenesis pathways were significantly affected (Fig. 5, A and B; and Fig. S3 D). Furthermore, qRT-PCR analyses showed that the levels of transcripts encoding enzymes involved in the terpenoid (Fdps, ggps1, Hmgcr, Hmgcs1, Hmgcs2) and coenzyme Q synthesis pathways (coq2, coq3, coq5, coq7, pdss1, pdss2) were not affected in Mfn2 knockout hearts (Fig. 5 C). Thus, the decreased levels of enzymes involved in the terpenoid biosynthesis pathway could not be attributed to changes in their mRNA expression.
Figure 5.
Terpenoid synthesis pathway defects in mitofusin knockouts. (A) Scheme of the mevalonate pathway in mammals. Red, proteins down-regulated according to SILAC-based protein quantification performed with Mfn1 and Mfn2 knockout MEFs; blue, not affected; gray, not detected. *, COQ2 protein level was determined by Western blot analysis. (B) Quantitative proteomic analysis of two independent batches of control and Mfn2 knockout MEFs using SILAC analysis. Negative values indicate down-regulation of the protein in Mfn2 knockout relative to control MEFs. (C) Transcript levels of different enzymes involved in the terpenoid and coenzyme Q synthesis pathways determined by qRT-PCR with total RNA extracted from control (white bars) and Mfn2 knockout (black bars) hearts (n = 5 pairs of mice). (D) Terpenoid metabolites content of whole heart extracts isolated from control (white bars, n = 12 samples), Mfn1 knockout (gray bars, n = 7 samples), and Mfn2 knockout (black bars, n = 6 samples). Error bars indicate ±SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To investigate how the global down-regulation of terpenoid synthesis enzyme levels impacted terpenoid biosynthesis, we quantified terpenoid metabolites in Mfn1 and Mfn2 knockout hearts. The levels of isopentenyl-pyrophosphate (IPP) and p-HBA (unpublished data) were unaltered in Mfn1 and Mfn2 knockout hearts. However, the levels of dimethylallyl pyrophosphate (DMAP), geranyl diphosphate (GPP), farnesyl diphosphate (FPP), and geranyl-geranyl-diphosphate (GGPP) were severely reduced in the Mfn2 knockout hearts (Fig. 5 D) and were normal (IPP; DMAP and GPP) or slightly reduced (FPP and GGPP) in the Mfn1 knockout hearts. The proteomic and metabolomic analyses (Fig. 5, A–D; and Fig. S3, B–D) thus show that the coenzyme Q deficiency in Mfn2 knockout hearts is likely to be explained by a severe global defect of the terpenoid biosynthesis pathway. Similar to coenzyme Q, the synthesis of squalene and cholesterol is also dependent on the terpenoid pathway (Fig. 5 A). Surprisingly, the cholesterol content was unchanged in both Mfn1 and Mfn2 knockout hearts (Fig. S3 A), which indicates that the severe down-regulation of the terpenoid biosynthesis pathway in Mfn2 knockout hearts (Fig. 5 and Fig. S3) has differential effects on the multiple downstream pathways.
Coenzyme Q10 supplementation partially rescues mitochondrial morphology and function in Mfn2 knockout MEFs
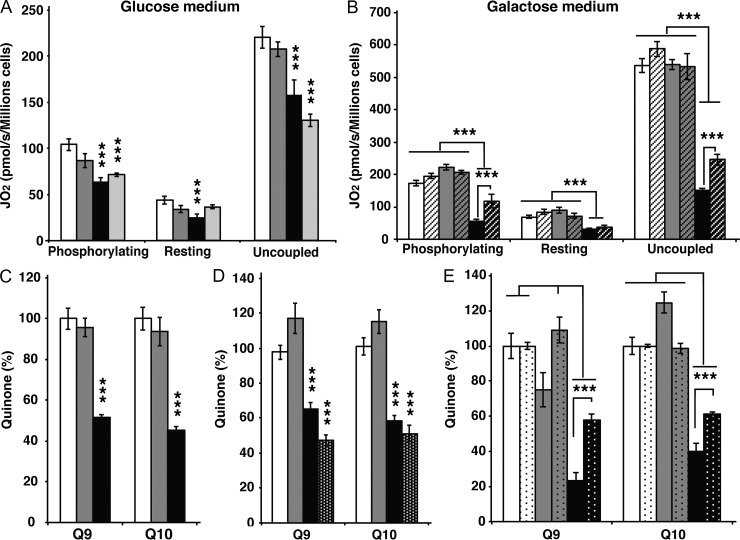
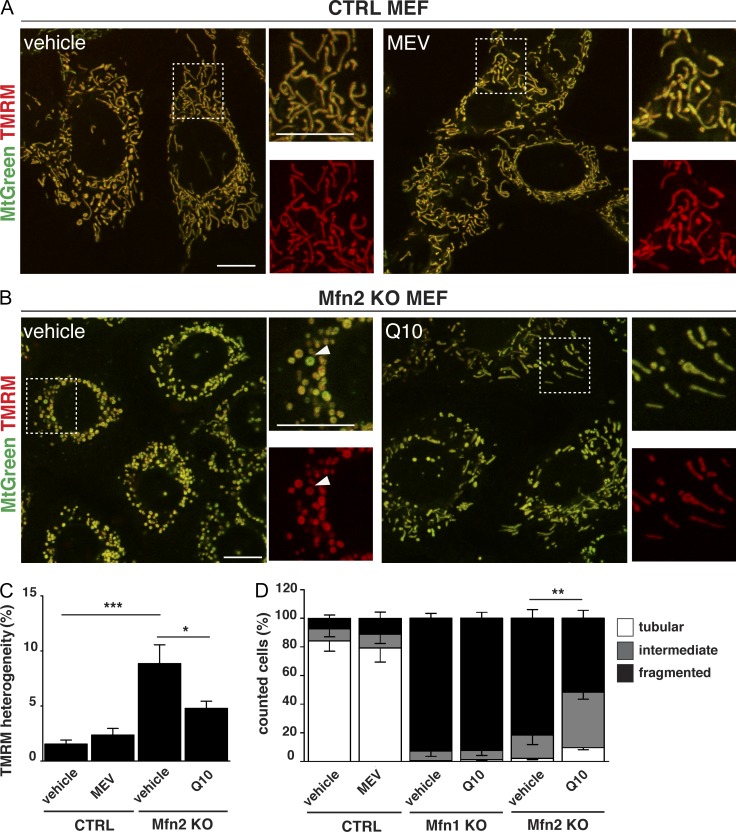
The loss of MFN2 in cardiomyocytes leads to impaired respiratory chain activity that is caused by CoQ deficiency, which, in turn, originates from a global defect of the terpenoid synthesis pathway, as determined by a combined proteomic and metabolomic approach. It should be noted that cultured cells lacking MFN1 or MFN2 typically have a fragmented mitochondrial network. It is thus formally possible that the coenzyme Q deficiency originates from impaired mitochondrial fusion as such rather than from a specific role for MFN2. To distinguish between these possibilities, we investigated the connection between coenzyme Q homeostasis and mitochondrial network morphology in Mfn1 and Mfn2 knockout MEFs. We first grew MEFs in glycolytic (glucose) and oxidative (galactose) medium and observed a growth defect of Mfn2 knockout MEFs, but not control or Mfn1 knockout MEFs, in galactose medium (Fig. S4, A–D). The cell growth phenotype suggests that loss of MFN2 causes a respiratory chain deficiency and, consistent with this observation, we observed a pronounced respiration defect in Mfn2 knockout MEFs grown on galactose (Fig. 6, A and B). The defective mitochondrial respiration in permeabilized Mfn2 knockout MEFs was associated with a deficiency of coenzyme Q9 and Q10 in both isolated mitochondria (Fig. 6 C) and whole cells (Fig. 6 D). To investigate how impaired terpenoid biosynthesis impacts respiratory chain function and mitochondrial morphology, we treated control MEFs with mevastatin (MEV), a well-known inhibitor of the terpenoid biosynthetic pathway. MEV treatment of wild-type MEFs lead to a profound reduction of coenzyme Q levels and impaired mitochondrial respiration, thus closely mimicking the bioenergetic phenotype of Mfn2 knockout MEFs (Fig. 6, A and D). However, the MEV treatment did not affect the mitochondrial network morphology of control cells (Fig. 7, A, C, and D), thus showing that the coenzyme Q deficiency as such does not induce profound alterations of mitochondrial dynamics. As the opposite might still be true, we investigated whether the coenzyme Q deficiency could be rescued by restoring a tubular mitochondrial network in MEFs lacking MFN2. To this end, we knocked down Drp1, which is involved in mitochondrial fission, in control MEFs and found a slight increase in mitochondrial elongation in comparison with MEFs transfected with a scrambled siRNA (Fig. S5 A), whereas the fragmented mitochondrial network of Mfn2 knockout MEFs was rescued (Fig. S5 C), in agreement with previous reports (Smirnova et al., 2001; Motori et al., 2013). However, the rescue of mitochondrial network morphology induced by Drp1 knockdown in Mfn2 knockout MEFs causes no changes of coenzyme Q content (Fig. S5, B and D), thus demonstrating that the coenzyme Q deficiency is not directly related to the mitochondrial network morphology. Interestingly, supplementation of the culture medium with coenzyme Q10 partially rescued the cellular respiration (Fig. 6 B) and the quinone deficiency (Fig. 6 E) in MEFs lacking MFN2. As previously reported (Chen et al., 2003), the absence of MFN2 causes a stochastic loss of Δψ, as depicted by the strong variation in the TMRM staining (Fig. 7 B). Strikingly, we observed that supplementation of the culture medium with coenzyme Q could partially rescue the TMRM heterogeneity (Fig. 7, B and C; and Fig. S5, E and F). This partial rescue in Δψ was also accompanied by a partial rescue of the mitochondrial network morphology, as indicated by the appearance of more elongated mitochondria (Fig. 7, B and D). Importantly, this rescue in mitochondrial morphology was not observed when Mfn1 knockout MEFs were supplemented with coenzyme Q10, which additionally supports the finding that MFN2 has a distinct role in coenzyme Q pool maintenance.
Figure 6.
OXPHOS dysfunction in the absence of Mfn2 is due to coenzyme Q deficiency. (A) Assessment of cellular respiration in control (white bars, n = 5 independent experiments), Mfn1 knockout (gray bars, n = 5 independent experiments), Mfn2 knockout (black bars, n = 5 independent experiments), and MEV-treated control (light gray bars, n = 5 independent experiments) MEFs that were grown in DMEM glucose, permeabilized, and analyzed in the presence of succinate and glycerol-3-phosphate. (B) Assessment of cellular respiration in control (white bars, n = 5 independent experiments), Mfn1 knockout (gray bars, n = 5 independent experiments), and Mfn2 knockout (black bars, n = 5 independent experiments) MEFs that were grown in DMEM galactose, permeabilized, and analyzed in the presence of succinate and glycerol-3-phosphate. Dashed bars indicate cells of the same genotypes grown in the presence of coenzyme Q10 (n = 5 per genotype). (C) Quinone quantification performed on mitochondria isolated from control (white bars, n = 5 independent experiments), Mfn1 knockout (gray bars, n = 5 independent experiments), and Mfn2 knockout (black bars, n = 5 independent experiments) MEFs. (D) Quinone quantification performed on whole cell extracts isolated from control (white bars, n = 5 independent experiments), Mfn1 knockout (gray bars, n = 5 independent experiments), Mfn2 knockout (black bars, n = 5 independent experiments), and MEV-treated control (black bars with white dots, n = 5 independent experiments) MEFs grown in DMEM glucose. (E) Quinone quantification performed on whole cell extracts isolated from control (white bars, n = 5 independent experiments), Mfn1 knockout (gray bars, n = 5 independent experiments), and Mfn2 knockout (black bars, n = 5 independent experiments) MEFs grown in DMEM galactose. Dotted bars indicate cells of the same genotypes grown in the presence of coenzyme Q10 (n = 5 independent experiments). Error bars indicate ±SEM. ***, P < 0.001.
Figure 7.
Morphological and functional incidence of terpenoid and coenzyme Q deficiency in Mfn2 knockout MEFs. (A and B) Representative confocal microscopy images of control (A) and Mfn2 knockout (B) MEFs, untreated (left) and supplemented with MEV or Q10 (right). MEFs were loaded with TMRM and MitoTracker green to evaluate Δψ. Magnifications of the boxed areas (on the right) show merged channels of TMRM and MitoTracker green (top) and single channels of TMRM (bottom). Arrowheads point to a mitochondrion presenting a low TMRM/MitoTracker green ratio. Bars, 10 µm. (C) Analysis of TMRM heterogeneity in control and Mfn2 knockout MEFs in the presence of MEV or Q10 (from independent experiments, n = 9). *, P < 0.05; **, P < 0.01; ***, P < 0.001. (D) Quantification of mitochondrial morphology in CTRL, Mfn1 knockout, and Mfn2 knockout MEFs upon treatment with MEV or Q10 (from independent experiments, n ≥ 300 cells analyzed). Error bars indicate ±SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Discussion
Mammalian MFN1 and MFN2 have originally been described as playing a central role in mitochondrial outer membrane fusion to control mitochondrial network morphology and allow the exchange of content between adjacent mitochondria. The role for MFN2 in mammalian mitochondria has attracted much interest because mutations in Mfn2 cause the Charcot Marie Tooth type 2A (CMT2A) axonopathy (Züchner et al., 2004; Detmer et al., 2008). Recently, MFN2 has been shown to be essential for axonal mitochondrial transport (Misko et al., 2010) and to maintain the axonal projections of dopaminergic neurons (Lee et al., 2012; Pham et al., 2012). Beyond its role in mitochondrial fusion, MFN2 has also been implicated in ER–mitochondria tethering (de Brito and Scorrano, 2008) and in ER stress caused by mitochondrial dysfunction (Sebastián et al., 2012). However, the molecular basis for the mitochondrial dysfunction caused by loss of Mfn2 has been unclear. We report here that mouse hearts or MEFs lacking MFN2 are respiratory chain deficient despite having normal levels and normal enzyme activities of individual respiratory chain complexes, which is consistent with previous studies of fibroblasts from CMT2A patients (Loiseau et al., 2007; Guillet et al., 2010). Unexpectedly, we show here that the deficient respiratory chain function in the absence of MFN2 is explained by a deficiency of coenzyme Q. Loss of MFN2 is associated with a fragmentation of the mitochondrial network, but this alteration is likely not the cause of the coenzyme Q deficiency because normalization of the mitochondrial network morphology in Mfn2 knockout MEFS does not normalize coenzyme Q levels (Fig. S5 G).
A continuous cellular supply of coenzyme Q requires de novo synthesis from the terpenoid pathway, a multistep process that has been reported to involve many subcellular compartments (Szkopińska, 2000). Whereas the condensation of the long isoprenoid side chain to the para-hydroxybenzoic acid (PHB) can occur in mitochondria (Tran and Clarke, 2007; Wang and Hekimi, 2013), the exact subcellular localization of the terpenoid and coenzyme Q biosynthetic pathways as well as the mechanisms that govern the intracellular transport of their metabolites remain to be defined (Szkopińska, 2000). Our analyses shed important new light on these molecular pathways, and we show here that loss of MFN1 and MFN2 is associated with a down-regulation of both levels of enzymes and metabolites of the terpenoid biosynthesis pathway. The enzymes of the terpenoid biosynthesis pathway have been found in different organelles such as mitochondria, ER, and peroxisomes (Keller et al., 1986; Olivier and Krisans, 2000; Kovacs et al., 2001). Interestingly, MFN2 was previously reported to be key in high-level steroid production of trophoblast and Leydig cells (Duarte et al., 2012, 2014; Wasilewski et al., 2012). Our results suggest that the previously observed steroidogenesis defects of trophoblast and Leydig cells lacking MFN2 are likely to originate from a deficiency in the terpenoid synthesis pathway.
In summary, we report here the novel and unexpected finding that MFN2, an outer mitochondrial membrane protein, has an essential role in regulating the terpenoid biosynthesis pathway, which, in turn, is required to maintain mitochondrial coenzyme Q levels for optimal function of the respiratory chain. We show that the coenzyme Q deficiency and the respiratory chain deficiency can be partially rescued by coenzyme Q10 supplementation. This finding may well be of direct clinical importance because mutations in MFN2 are known to cause human disease. Indeed, a recently published case study indicates that coenzyme Q supplementation may have a positive effect in patients with pathogenic Mfn2 mutations (Takahashi et al., 2012). Our results suggest that clinical trials to stringently evaluate the effects of coenzyme Q10 supplementation in patients with diseases caused by Mfn2 mutations may be warranted.
Materials and methods
Creation of Mfn1 and Mfn2 knockout mice
Mice with a loxP-flanked exon 3 of the Mfn1 gene or a loxP-flanked exon 5 of the Mfn2 gene were generated in the C57BL/6N background as described previously (Lee et al., 2012) and crossed with mice expressing cre-recombinase in heart and skeletal muscle (+/Ckmm-cre). The resulting double heterozygous offspring (+/Mfn1loxP, +/Ckmm-cre and +/Mfn2loxP, +/Ckmm-cre) were crossed with mice having homozygous loxP-flanked alleles (Mfn1loxP/Mfn1loxP and Mfn2loxP/Mfn2loxP) to generate tissue-specific knockouts for Mfn1 (Mfn1loxP/Mfn1loxP; +/Ckmm-cre) and Mfn2 (Mfn2loxP/Mfn2loxP; +/Ckmm-cre). In some crosses, an allele allowing cell-type-specific expression of mitochondria-targeted YFP from the ROSA26 locus (Gt(ROSA26Sor+/lox-Stop-lox-mito-YFP; Sterky et al., 2011) was introduced.
Electron cryo-microscopy
For analysis by electron cryo-microscopy, mitochondria were washed twice with 320 mM trehalose, 20 mM Tris, pH 7.3, and 1 mM EGTA. Samples were mixed 1:1 with fiducial gold markers (10 nm gold particles conjugated to protein A; Aurion), blotted, and immediately plunge-frozen in liquid ethane on holey carbon grids (Quantifoil Micro Tools). Single tilt series (±60°, step size 1.5°) were collected on a Polara microscope (300 kV; FEI) using an Ultrascan 4 × 4k CCD (Gatan, Inc.) and a post-column Quantum energy filter (Gatan, Inc.) at −9 µm defocus. The nominal magnification was 34,000×, resulting in a pixel size of 6 Å. A total dose of ∼130 e−/Å2 was used. Tilt series alignment using the gold fiducial markers and tomogram reconstruction by back-projection were performed using the IMOD software package (Kremer et al., 1996). To increase contrast, a final filtering step applying nonlinear anisotropic diffusion (Frangakis and Hegerl, 2001) was performed. Manual segmentation was performed with the program Amira (FEI).
Isolation of cardiomyocytes
To isolate adult cardiomyocytes, 15-wk-old mice were sacrificed by cervical dislocation, and the aorta was quickly clamped to the perfusion cannula. Using a Radnotti heart perfusion apparatus, the heart was first washed (1.5 ml/min) for 3 min with the perfusion buffer (120 mM NaCl, 4.8 mM KCl, 1.25 mM KH2PO4, 1.25 mM MgSO4, 11 mM glucose, 20 mM Hepes, and 30 mM taurine, pH 7.2). Then the heart was perfused for 3 min with the calcium-free perfusion buffer, and 240 U/ml collagenase type II (EMD Millipore) was added for ∼30 min digestion. During digestion, the calcium concentration was progressively increased to 250 µM. To stop the digestion, the heart, which was flaccid and looked pale, was disrupted in perfusion buffer (250 µM calcium, 0.5 mg/ml BSA). After several rounds of washing by sedimentation, the isolated cardiomyocytes, which had a mostly rod-shaped morphology, were seeded at 70–80% density in culture medium (DMEM F-12 without phenol red [Invitrogen], 1 µg/ml insulin, 0.55 µg/ml transferrin, 0.65 µg/ml selenium, 2 mM glutamine, 10 mM Hepes, 1% penicillin/streptomycin, and 0.05% BSA). Plating was performed on 23-mm glass-bottomed dishes (World Precision Instruments) and cardiomyocytes were left to attach for 2 h at room temperature before confocal imaging. To avoid spontaneous contraction of cells, 25 µM blebbistatin was added.
Live imaging of adult cardiomyocytes and MEFs
Δψ was assessed by live imaging using a laser-scanning confocal microscope (TCS SP5-X; Leica) equipped with a white light laser. Adult cardiomyocytes derived from mice expressing a mitochondrially targeted YFP were first equilibrated 30 min at 37°C, 5% CO2, then loaded with 50 nM TMRM (Invitrogen) in culture medium for 30 min at 37°C. Fluorescence images were acquired with a 100× objective lens (HCX Plan-Apochromat CS 100× oil, 1.46 NA), using the LAS AF software. The following parameters in sequential line scan were used: YFP (excitation, 488 nm; emission, 512–552 nm; image size 1,024 × 1,024 pixels, scan speed 200 Hz) and TMRM (excitation, 550 nm; emission, 595–670 nm; image size 1,024 × 1,024 pixels, scan speed 200 Hz). For TMRM labeling in MEFs, cells were first loaded with 100 nM MitoTracker Green (Invitrogen) for 15 min at 37°C, 5% CO2, then washed twice with culture medium to remove the probe in excess. Finally, MEFs were loaded with 20 nM TMRM for 30 min at 37°C before imaging. Images were acquired using the following parameters: MitoTracker Green (excitation, 488 nm; emission, 504–543 nm; image size 1,024 × 1,024 pixels, scan speed 400 Hz) and TMRM (excitation, 550 nm; emission, 590–668 nm; image size 1,024 × 1,024 pixels, scan speed 400 Hz). During acquisition, the laser power was kept as low as possible to reduce photo-toxicity; a 2× averaging of the optical plane was also applied to increase the signal-to-noise ratio. Transmission-light images were acquired with a scan-BF detector and a 488-nm excitation laser line.
Imaging analysis and processing
Analysis of mitochondrial potential in adult cardiomyocytes.
The fraction of inactive mitochondria was quantified using the JACoP plugin (Bolte and Cordelières, 2006) from ImageJ by colocalization analysis of YFP signal (corresponding to the overall mitochondrial population) and TMRM signal (corresponding to the active mitochondria). The fraction of YFP signal overlapping the TMRM signal was expressed with the Mander’s coefficient. The intensity profile analysis was obtained by measuring the maximum fluorescence intensity of a segmented line drawn along mitochondria using ImageJ.
Analysis of mitochondrial potential in MEFs.
The heterogeneity in TMRM signal was assessed at a single-cell level using the Image Calculator plugin from ImageJ. In brief, single-stack confocal images corresponding to the TMRM and MitoTracker Green channels were calculated for their difference by using the equation signal = |TMRM-MitoTracker Green|. The pixel area in the resulting difference mask was then quantified and subsequently normalized over the corresponding thresholded single-stack image of the MitoTracker Green channel in order to obtain the fraction of mitochondria displaying heterogeneous membrane potential.
Analysis of mitochondrial morphology in MEFs.
Classification of the mitochondrial morphology was performed according to the following criteria: cells displaying an intact tubular mitochondrial network were classified as tubular; cells with globular or rod-like mitochondria were classified as fragmented; cells showing equal presence of both tubular and fragmented mitochondria were instead classified as intermediate.
Image processing.
Image panels were assembled with Photoshop (Adobe); no digital manipulation was applied except for adjustment of brightness and contrast.
Isolation of mitochondria
To isolate intact heart mitochondria, mice were sacrificed by cervical dislocation and hearts were quickly collected in ice-cold DPBS (Gibco), minced, and homogenized with a few strokes of a Potter S homogenizer (Sartorius) in 5 ml of ice-cold mitochondria isolation buffer (MIB; 310 mM sucrose, 20 mM Tris-HCl, and 1 mM EGTA, pH 7.2). Mitochondria were purified by differential centrifugation (1,200 g for 10 min) and supernatants were then centrifuged at 10,000 g for 10 min. The crude mitochondrial pellet was resuspended in an appropriate volume of MIB. The mitochondrial protein concentration was determined using the Protein DC Lowry based assay (Bio-Rad Laboratories, Inc.).
Transmission electron microscopy
Electron micrographs of mitochondria were obtained as previously described (Hansson et al., 2004). In brief, small pieces from the left myocardium were fixed in 2% glutaraldehyde, 0.5% paraformaldehyde, 0.1 M sodium cacodylate, 0.1 M sucrose, and 3 mM CaCl2, pH 7.4, at room temperature for 30 min, followed by 24 h at 4°C. Specimens were rinsed in a buffer containing 0.15 M sodium cacodylate and 3 mM CaCl2, pH 7.4; postfixed in 2% osmium tetroxide, 0.07 M sodium cacodylate, and 1.5 mM CaCl2, pH 7.4, at 4°C for 2 h; dehydrated in ethanol followed by acetone; and embedded in LX-112 (Ladd Research Industries). Ultra-thin sections (40–50 nm) from longitudinal parts were cut and examined in a transmission electron microscope (Tecnai 10; FEI) at 80 kV. Digital images at a final magnification of 8,200× were randomly taken on myofibrils from sections of the myocardium. Printed digital images were used and the volume density (Vv) of mitochondria was calculated by point counting using a 2-cm square lattice (Weibel, 1979). A pilot study was performed to determine the number of blocks and images needed for an appropriate sample using a cumulative mean plot for evaluation (Weibel, 1979). Two different blocks from one animal were sectioned and 10 random images were collected from each block.
Measurement of mitochondrial respiration
The mitochondrial oxygen consumption flux was measured as described previously (Mourier et al., 2014) at 37°C using 65–125 µg of crude mitochondria diluted in 2.1 ml of mitochondrial respiration buffer (120 mM sucrose, 50 mM KCl, 20 mM Tris-HCl, 4 mM KH2PO4, 2 mM MgCl2, and 1 mM EGTA, pH 7.2) in an Oxygraph-2k (Oroboros Instruments). The oxygen consumption rate was measured using either 10 mM pyruvate, 5 mM glutamate, and 5 mM malate or 10 mM succinate and 10 nM rotenone. Oxygen consumption was assessed in the phosphorylating state with 1 mM ADP or in the nonphosphorylating state by adding 2.5 µg/ml oligomycin. In the control mitochondria, the respiratory control ratio (RCR) values were >10 with pyruvate/glutamate/malate and >5 with succinate/rotenone. Respiration was uncoupled by successive addition of CCCP up to 3 µM to reach maximal respiration.
Measurement of respiratory chain enzyme activities
Mitochondrial proteins (15–50 µg) were diluted in phosphate buffer (50 mM KH2PO4, pH 7.4) followed by spectrophotometric analysis of isolated respiratory chain complex activities at 37°C using a spectrophotometer (UV-3600; Hitachi). Citrate synthase activity was measured at 412 nm (ε = 13,600 M−1cm−1) after the addition of 0.1 mM acetyl-CoA, 0.5 mM oxaloacetate, and 0.1 mM 5,5′-dithiobis-2-nitrobenzoic acid (DTNB). NADH dehydrogenase activity was determined at 340 nm (ε = 6220M−1cm-1) after the addition of 0.25 mM NADH, 0.25 mM decylubiquinone, and 1 mM KCN, controlling for rotenone sensitivity. Succinate dehydrogenase (SDH) activity was measured at 600 nm (ε = 21,000 M−1cm−1) after the addition of 40 mM succinate, 35 µM dichlorophenolindophenol (DCPIP), and 1 mM KCN. The complex III activity was measured after the reduction of cytochrome c at 540 nm (ε =18,000 M−1cm−1) in the presence of 1 µg/ml decylubiquinol, 80 µg/ml cytochrome c, and 1 mM of sodium azide. The specific complex III activity was defined as the flux difference before and after the addition of 1 µM antimycin A. The NADH and succinate cytochrome c reductase activity was also followed at 540 nm (ε = 18,000 M−1cm−1) in the presence of 40 mM succinate or 5 mM NADH, 80 µg/ml cytochrome c, and 1 mM of sodium azide. The specific II-III or I-III activities were defined as the flux difference before and after the addition of 1 µM antimycin A. When used, decylubiquinone was added at 0.02 mg/ml. The COX activity was assessed using a classical TMPD/ascorbate assay. In brief, isolated mitochondria (65 µg/ml) were suspended in mitochondrial respiration buffer (see “Measurement of mitochondrial respiration”). Oxygen consumption was assessed in the presence of TMPD (0.2 mM), ascorbate (1 mM), and antimycin A (0.5 µM). After a few minutes of stationary respiration, 2 mM KCN was injected into the chamber. The COX activity corresponds to the KCN-sensitive respiration. All chemicals were obtained from Sigma-Aldrich.
Measurement of mitochondrial ATP production
To assess the ATP production rate, isolated mitochondria (65 µg/ml) were suspended in the mitochondrial respiration buffer (see “Measurement of mitochondrial respiration”). After addition of ADP (1 mM), succinate (2 mM), and rotenone (10 nM) or addition of ADP (1 mM), pyruvate (10 mM), glutamate (5 mM), and malate (5 mM), the oxygen consumption and ATP synthesis rates were both measured. Aliquots were collected every 20 s and precipitated in 7% HClO4/25 mM EDTA, centrifuged at 16,000 g for 10 min, and then neutralized with 2 M KOH and 0.3 M MOPS. The ATP content in these samples was determined with the ATPlite 1step (PerkinElmer). In a parallel experiment, oligomycin (2.5 µg/ml protein) was added to the mitochondrial suspension to determine the nonoxidative ATP synthesis rate.
Measurement of hydrogen peroxide production
The rate of H2O2 production was determined by monitoring the oxidation of the fluorogenic indicator Amplex red (Life Technologies) in the presence of horseradish peroxidase. The concentrations of horseradish peroxidase and Amplex red in the incubation medium were 5 U/ml and 1 µM, respectively. Fluorescence was recorded at the following wavelengths: excitation, 560 nm; emission, 590 nm. A standard curve was obtained by adding known amounts of H2O2 to the assay medium in the presence of the reactants. Mitochondria (65 µg protein/ml) were incubated in the respiratory medium (see “Measurement of mitochondrial respiration”) at 37°C and the H2O2 production rate measurements were initiated by substrate addition. The H2O2 production rate was determined from the slope of a plot of the fluorogenic indicator versus time.
FACS analysis of mitochondria
FACS analyses were performed using freshly isolated mitochondria (10–20 µg/ml) incubated in respiration buffer supplemented with 10 mM pyruvate, 5 mM glutamate, and 5 mM malate or 10 mM succinate and 10 nM rotenone. After a few minutes of preincubation at 37°C with 250 nM Mitotracker Green FM and 400 nM TMRM, the membrane potential positive and negative populations were analyzed by quadrant and polygonal region analysis. The cytofluorometric analysis was performed by using a FACSAria III cell sorter (BD) equipped with 488 nm and 633 nm lasers, and fluorescence was measured using the FL1 and FL2 channels. Data were acquired using DIVA 6.0 software (BD). To analyze the membrane potential, optimum concentration of oligomycin and CCCP defined in the respiratory assay were successively added. FACS analyses were performed using the FlowJo software applying a biexponential transformation to digital data.
Determination of coenzyme Q and terpenoids intermediates content
Mitochondria were additionally purified on a 20% Percoll gradient to separate mitochondria from mitochondrial-associated membranes (MAM). Quinones were extracted by suspending pellets of isolated mitochondria or MEFs in 200 µl of 1 mM cupric sulfate followed by addition of 200 µl ethanol. After sonication, the homogenate was vortexed in the presence of 400 µl hexane. After collection of the upper phase containing hexane, it was dried using a SpeedVac apparatus (Thermo Fisher Scientific). Next, samples were suspended in 50–200 µl ethanol/methanol (9:1), vortexed, incubated for 2 min in an ultrasonication bath, and filtered through a 0.2-µm modified nylon centrifugal filter (VWR) using a centrifuge (5424R; Eppendorf) set to 6°C and 15,000 rpm.
For absolute quantification of Q9 and Q10 in positive ESI multiple reaction monitoring (MRM) mode, an Acquity UPLC system (Waters) was connected to a Xevo TQ (Waters). An Acquity UPLC (Waters) BEH C18 1.7 µm, 2.1 × 50 mm column was used at 40°C. Solvent A was 90% methanol + 10% propanol + 0.1% FA, and solvent B was 45% acetonitrile/acetone + 10% propanol + 0.1% FA. A linear gradient of solvent A ranging from 100% to 0% in 3.5 min at a flow rate of 0.45 ml/min was used. Between 2 and 6 µl of the samples were injected. The sample manager was set to 6°C for the standards and the samples were defrosted and directly injected. The MRM transitions used for quantification are presented in the Fig. S5 H. Terpenoid intermediates were extracted as described previously (Henneman et al., 2008). In brief, half of a heart was homogenized in 2 ml of 2-propanol:100 mM NH4HCO3, pH 7.8 (1:1 vol/vol), and sonicated for 5 min. After 10 min of centrifugation (20,000 g at 4°C), supernatant was collected and evaporated for 30 min using a SpeedVac apparatus. Samples were then suspended in 100 µl ethanol/methanol (9:1), sonicated for 1 min, and filtrated successively through a 0.45 µm and 0.2 µm modified nylon centrifugal filter (VWR). The absolute quantification of DMAP, FPP, GGPP, GPP, and IPP in negative ESI MRM was determined using an Acuity UPLC (Waters) connected to a Xevo TQ (Waters). An Acquity UPLC BEH C18 1.7 µm, 2.1 × 50 mm column was used at 40°C. Solvent A was 20 mM ammonium bicarbonate + 0.1% FA and B acetonitrile/water (3:1) + 0.1% FA. A linear gradient from 95% solvent A to 0% in 5 min at a flow rate of 0.35 ml/min was used. 7.5 µl was injected. The sample manager was set to 8°C for the standards and the samples.
For both quinone and terpenoid quantification, the source temperature was set to 150°C, desolvation temperature was 650°C, desolvation gas was set to 800 liter/h, and cone gas was set to 50 liter/h. The software used for data management was MassLynx (Waters) and the software for data evaluation and absolute quantification was TargetLynx (Waters). All compounds were freshly prepared daily and dissolved in ethanol/methanol (9:1). Quality control standards of each standard were used during sample analysis and showed between 0.5% and 40% deviation for Q9 and Q10, and between 0.5% and 25% deviation for terpenoids. Blanks after the standards, quality control, and sample batch proved to be sufficient. No carry-over was detected. The quinone quantification performed by UPLC-LC/MS/MS was validated by using an HPLC-EC quinone measurement performed in the STROMALab (Galinier et al., 2004).
Cell culture
Immortalized MEFs from control, Mfn1 (Exon 3) knockout, or Mfn2 (Exon 3) knockout mice from the C57BL/6J background were a gift from D. Chan at California Institute of Technology, Pasadena, CA. Cells were cultivated in DMEM containing 4.5 g/liter glucose, supplemented with 1% FBS, 1% NEAA, and 1% penicillin/streptomycin, and maintained at 37°C in 5% CO2 or in galactose medium (glucose-free DMEM supplemented with 1% FBS, 15 mM galactose, 1 mM pyruvate, 1% NEAA, and 1% penicillin/streptomycin). For some experiments, MEV (0.1 µM) or coenzyme Q10 (0.5 mM) were added to the culture media 24 h before cell collection. To assess the growth curve, MEFs were plated at the density of 8,000 cells per well in a six-well dish and collected at 1–4 d after seeding. Cell count was performed with an automated system by using the Vi-Cell XR cell analyzer (Beckman Coulter).
RNAi
For the knockdown of Drp1, previously published constructs expressing either a GFP-encoding scramble or a GFP-encoding Drp1 microRNA were used (Motori et al., 2013). Cloning of constructs was performed by using the BLOCK-iT Lentiviral Pol II miR RNAi Expression System with EmGFP (Invitrogen). The following sequences were inserted into a pcDNA 6.2-GW/EmGFP-miR vector: 5′-GAAATGTACTGCGCGTGGAGACGTTTTGGCCACTGACTGACGTCTCCACGCAGTACATTT-3′ (scramble), and 5′-CCTGTAAACTT CTACAGGTAGGCGTCAGTCAGTGGCCAAAACGCCTACCTGAGTAGAAGTTTAC-3′ (Drp1 microRNA). 106 cells were electroporated with 1.5 µg of plasmid by using the Amaxa 4D Nucleofector (Lonza) according to the manufacturer’s instructions. Cells were then harvested at day 2 after electroporation and transfection efficiency was ≥60% in all analyzed samples.
Immunocytochemistry and confocal microscopy
Cells were fixed with 4% PFA in PBS and processed for immunocytochemistry as described previously (Motori et al., 2013). In brief, cells were fixed for 10 min at room temperature with 4% PFA in PBS, then permeabilized for 5 min at room temperature with 0.1% Triton X-100 in PBS. Blocking of unspecific sites as well as incubation with primary and secondary antibodies was performed in 3% BSA in PBS. Incubation with primary antibodies was carried at +4°C overnight by using chicken anti-GFP (1:1,000; Aves Labs) and rabbit anti-TOM20 (1:1,000; Santa Cruz Biotechnology, Inc.). After washing in blocking buffer, the secondary antibodies donkey anti–chicken conjugated to Alexa Fluor 488 and donkey anti–rabbit conjugated to Alexa Fluor 549 (1:1,000; Jackson ImmunoResearch Laboratories, Inc.) were applied for 2 h at room temperature. Slides were extensively washed in PBS and nuclei were counterstained with DAPI before mounting.
Fluorescence images were acquired using a laser-scanning confocal microscope (TCS SP5-X; Leica), equipped with a white light laser, a 405-diode UV laser, and a 100× objective lens (HCX Plan-Apochromat CS 100× oil, 1.46 NA). Individual stacks were taken in sequential mode by applying the following parameters: DAPI (excitation, 405 nm; emission, 428–467 nm; image size 1,024 × 1,024 pixels, scan speed 200 Hz), Alexa Fluor 488 (excitation, 488 nm; emission, 501–542 nm; image size 1,024 × 1,024 pixels, scan speed 200 Hz), Alexa Fluor 549 (excitation, 549 nm; emission, 563–638 nm; image size 1,024 × 1,024 pixels, scan speed 200 Hz).
Proteome analysis by SILAC
Stable isotope labeling by amino acids in culture was performed as described previously (Ong and Mann, 2006). In brief, control MEFs were grown in DMEM medium supplemented with dialyzed serum in the presence of either light or heavy arginine and lysine (13C6 15N4-l-arginine, 13C6 15N4-l-lysine; Thermo Fisher Scientific) to allow a full incorporation of the amino acids (>98%). After 7 d of culture, labeled control MEFs and nonlabeled Mfn1, Mfn2 knockout MEFs were collected and suspended in lysis buffer (6 M guanidinium chloride, 10 mM Tris(2-carboxyethyl)phosphine hydrochloride, 40 mM chloroacetamide, and 100 mM Tris-HCl; Kulak et al., 2014). After complete lysis, 100 µg of protein sample (50 µg labeled control [heavy] mixed with 50 µg nonlabeled samples [light]) was mixed with 2 µg of Trypsin gold (Promega) and incubated overnight at 37°C to achieve complete digestion. Peptides were cleaned with home-made StageTip (Empore Octadecyl C18; 3M) and eluted in 80% acetonitrile/0.1% formic acid buffer. After being dried in SpeedVac, the peptides were resuspended with 0.1% formic acid. 2 µg of peptides was analyzed with LC-MS/MS. Peptides were resolved with an Easy-nLC UPLC equipped with a 50 cm × 75 µm Easyspray column (Thermo Fisher Scientific) at 50°C using a 200 min linear gradient from 5 to 25% ACN in 0.1% formic acid, followed by a steeper final elution phase. The eluting flow rate was 250 nl/min. Data were collected on a mass spectrometer (QExactive Plus Orbitrap; Thermo Fisher Scientific) using a Top10 method. For MS mode, the following parameters were used: resolution 70,000, maximum injection time 100 ms, scan range 300–1,800 m/z. For MSMS mode: resolution 17,500, maximum injection time 120 ms, scan range 100–2,000 m/z.
Mass spectrometry data analysis
The raw data were analyzed with MaxQuant (Cox and Mann, 2008) version 1.4.1.2 using the integrated Andromeda search engine (Cox et al., 2011). The data were searched against the canonical and isoform sequences of the mouse reference proteome (proteome ID UP000000589, downloaded June 2014) from UniProt. The database was automatically complemented with 247 sequences of contaminating proteins using the “include contaminants” option of MaxQuant. For data analysis, cysteine carbamidomethylation was set as “fixed modification” and methionine oxidation and protein N-terminal acetylation as “variable modification.” The digestion parameters were set to “specific” and “Trypsin/P,” allowing for cleavage after lysine and also arginine when followed by proline. The minimum number of peptides and razor peptides for protein identification was 1; the minimum number of unique peptides was 0. The minimum peptide length was 7 amino acids. Protein identification was performed at a peptide spectrum matches and protein false discovery rate (FDR) of 0.01. The “second peptide” option was on in order to identify co-fragmented peptides. For protein quantification, the heavy labels were set to Arg10 and Lys8. The “requantify” option was on and a minimum of two ratio counts was required for reporting the protein ratio. Unique plus razor peptides were used for protein quantification.
Protein quantification significance analysis
Downstream data analysis was performed using the Perseus statistical framework (http://www.perseus-framework.org/) version 1.5.0.0. Protein contaminants and reverse identifications were removed and protein ratios were reversed. Next, protein ratios and intensities were transformed (log 2 and log 10, respectively). Proteins that were significantly down-regulated in the knockout versus the wild type were identified using significance B (Cox and Mann, 2008). A p-value of 0.05 was used as a threshold for significance. Only proteins that were down-regulated and significant in both knockout versus WT1 and WT2 experiments were considered for further analysis. Finally, protein annotation (“GOBP name,” “KEGG name,” “GOBP,” and “KEGG”) was added, based on the UniProt identifiers in the “Majority protein IDs” column.
Functional annotation enrichment analysis
Enrichment of functional annotation terms was performed using the DAVID bioinformatics resources (Huang et al., 2009). The official gene names of the down-regulated proteins were used while “Mus musculus” was applied as a species and background. Functional annotation clustering of the gene list was performed using the KEGG (Kanehisa and Goto, 2000) pathway (“KEGG_PATHWAY”) and Gene Ontology (GO; Ashburner et al., 2000). The “FAT” subsets of the three major GO domains: biological process (“GOTERM_BP_FAT”), cellular compartment (“GOTERM_CC_FAT”), and molecular function (“GOTERM_MF_FAT”) were used. The results of the enrichment analysis were filtered based on an FDR adjusted p-value (“Benjamini”) threshold. Only categories showing adjusted p-values for enrichment of 0.05 or lower were used for analysis.
Determination of tissue and mitochondrial cholesterol content
Cholesterol levels were determined by liquid chromatography coupled to electrospray ionization tandem mass spectrometry (LC-ESI-MS/MS). To tissue or mitochondria aliquots containing 100 µg protein, 100 µl water, 500 µl methanol, 250 µl chloroform, and 1.32 nmol deuterated cholesterol-D7 as an internal standard were added. The mixture was sonicated for 5 min, and lipids were extracted overnight at 48°C. After adding 2 ml of chloroform and 4 ml of water, the mixture was vortexed vigorously for 30 s and then centrifuged (4,000 g, 5 min, 4°C) to separate layers. The lower (organic) phase was transferred to a new tube, and the upper phase was extracted with an additional 2 ml of chloroform. The combined organic phases were dried under a stream of nitrogen. The residues were resolved in 200 µl of 5 mM ammonium acetate in methanol and sonicated for 5 min. After centrifugation (12,000 g, 5 min, 4°C), 40 µl of the clear supernatants was transferred to autoinjector vials. LC-MS/MS analysis was performed using a Core-Shell Kinetex C18 column (150 mm × 2.1 mm inner diameter, 2.6 µm particle size, 100 Å pore size; Phenomenex) with detection using a QTRAP 6500 triple quadrupole/linear ion trap mass spectrometer (AB SCIEX). The liquid chromatography device (1260 Infinity Binary LC System; Agilent Technologies) was operated isocratically at a flow rate of 300 µl/min with a mobile phase of 5 mM ammonium acetate in methanol. 20 µl of sample was injected. The total run time was 6 min. Cholesterol was monitored in the positive ion mode using its specific MRM transitions m/z 404.4 to 369.3 (quantifier) and 387.4 to 369.3 (qualifier). For the detection of the internal standard cholesterol-D7, the MRM transitions m/z 411.5 to 376.3 (quantifier) and 394.5 to 376.3 (qualifier) were used. The instrument settings for nebulizer gas (gas 1), turbogas (gas 2), curtain gas, and collision gas were 50 psi, 20 psi, 30 psi, and medium, respectively. The Turbo V ESI source temperature was 50°C, and the ion spray voltage was 5,500 V. For all MRM transitions, the values for declustering potential, entrance potential, collision energy, and cell exit potential were 100 V, 10 V, 15 V, and 10 V, respectively. The LC chromatogram quantifier peaks of endogenous cholesterol and cholesterol-D7 were integrated using the Analyst 1.6.2 software (AB SCIEX). Endogenous cholesterol was quantified on the basis of an external calibration curve, which was calculated from LC-MS/MS measurements of serially diluted synthetic cholesterol in the range between 0.0 and 250 pmol on column. The cholesterol standard dilutions were prepared in 5 mM ammonium acetate in methanol. To each dilution a fixed amount of the internal standard cholesterol-D7 was added. The standard calibration curve was plotted based on molar concentration versus peak area ratio of cholesterol to cholesterol-D7. Linearity and the correlation coefficient (R2) of the calibration curve were obtained via linear regression analysis. R2 of the calibration curve was 0.9958. The calculated amounts of endogenous cholesterol were normalized to the protein content of the sample.
Determination of mitochondrial phospholipid composition
The phospholipid composition was determined as described previously (Belgardt et al., 2010). In brief, 1 mg of mitochondria was homogenized in 1 ml of water using the Precellys 24 Homogenisator (Peqlab) at 6,500 rpm for 30 s. The protein content of the homogenate was routinely determined using bicinchoninic acid. Lipids were extracted as described previously (Belgardt et al., 2010). After addition of 2 ml methanol and 1 ml chloroform, lipids were extracted for 24 h at 37°C. The liquid phase was separated by filtration, and the solvent was evaporated in a stream of nitrogen. The residues were purified using a modification of the Bligh-Dyer procedure as described previously (Signorelli and Hannun, 2002). For the determination of the mitochondrial lipid content, the lipid extract was applied to 20 × 10 cm high-performance thin-layer chromatography (HPTLC) Silica Gel 60 plates (Merck), which were prewashed twice with chloroform/methanol 1:1 (vol/vol) and air-dried for 30 min. Each lane of the thin-layer chromatography (TLC) plate was loaded with the equivalent of 90 µg of protein. Glycerophospholipids were separated using chloroform/methanol/glacial acetic acid 65:28:8 (vol/vol/vol) as a solvent system. For quantitative analytical TLC determination, increasing amounts of standard lipids (Sigma-Aldrich) were applied to the TLC plates in addition to the lipid samples. For detection of lipid bands, the TLC plates were sprayed with a phosphoric acid/copper sulfate reagent (15.6 g of CuSO4(H2O)5 and 9.4 ml of H3PO4 [85%, wt/vol] in 100 ml of water) and charred at 180°C for 10 min (Yao and Rastetter, 1985). Lipid bands were then quantified by densitometry using the TLC-Scanner 3 (CAMAG) at a wavelength of 595 nm.
Western blot analyses
Proteins from mitochondrial and whole cell lysates were separated by SDS-PAGE and blotted onto PVDF membranes (GE Healthcare). The following primary antibodies were used: mouse anti–β-actin (1:5,000, A5316; Sigma-Aldrich), rabbit anti-Mitofusin1 (1:1,000, ab104274; Abcam), mouse anti-MFN2 (1:1,000, ab56889; Abcam), mouse anti-VDAC (1:5,000, MABN504; EMD Millipore), and mouse anti-OXPHOS antibody cocktail (1:1,000, ab110413; Abcam). The following secondary antibodies were used: donkey anti–rabbit IgG (NA9340V; GE Healthcare) and sheep anti–mouse (NXA931; GE Healthcare). Detection was done by incubation with HRP-conjugated secondary antibodies and conversion to chemiluminescence with ECL (GE Healthcare).
mtDNA quantification
DNA was extracted using the DNeasy Tissue and Blood kit (QIAGEN) according to the manufacturer’s instructions. 5 ng of total DNA from the heart was used for detection of mtDNA and genomic DNA by qPCR with the following Taqman probes: 16S (Mm03975671_s1), ATP6 (Mm03649417-g1), b-actin (Mm01205647-g1), and 18S (Hs99999901_s1), from Life Technologies.
qRT-PCR
RNA was isolated with TRIzol Reagent (Invitrogen) and resuspended in nuclease-free water (Ambion), and 2 µg of RNAs were reverse transcribed using the High-Capacity cDNA reverse transcription kit (Applied Biosystems). qPCR was performed on the obtained cDNAs using the Taqman Universal PCR Master Mix, No AmpErase UNG (Applied Biosystems), and the following Taqman probes: Mfn1 (Mm01289372_m1), Mfn2 (Mm00500120_m1), b2µglobulin (Mm00437762_m1), Ggps1 (Mm00656129_mH), HmgCR (Mm01282499_m1), HmgCS1 (Mm01304569_m1), HmgCS2 (Mm00550050_m1), Pdss1 (Mm00450958_m1), Pdss2 (Mm01190168_m1), Fdps (Mm00836315_g1), Coq2 (Mm01203260_g1), Coq3 (Mm01220614_m1), Coq5 (Mm00518239_m1), and Coq7 (Mm00501588_m1), from Life Technologies.
Ethics statement
This study was performed in strict accordance with the recommendations and guidelines of the Federation of European Laboratory Animal Science Associations (FELASA). The protocol was approved by the Landesamt für Natur, Umwelt und Verbraucherschutz in Nordrhein-Westfalen, in Germany.
Online supplemental material
Fig. S1 shows the mouse heart weight/body weight ratio, mtDNA quantification, mitochondrial mass analyzed by electron cryo-microscopy, and present microscopy analyses of isolated cardiomyocytes. Fig. S2 shows the size distribution of isolated mitochondria analyzed by electron cryo-tomography, the oxygen consumption rate of heart mitochondria isolated from Mfn1 and Mfn2 knockout animals at 60 wk of age, and mitochondrial cytochromes and phospholipids quantification analyses. Fig. S3 shows cholesterol and SILAC proteomic quantifications performed in Mfn1 and Mfn2 knockouts. Fig. S4 shows the characterization of Mfn1 and Mfn2 knockout MEFs and their respective growth curves on glucose and galactose media. Fig. S5 shows the mitochondrial network morphology and coenzyme Q quantification performed on Mfn2 knockout MEFs subjected to Drp1 knockdown and presents the method used to quantify the TMRM mitochondrial heterogeneity in MEFs. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201411100/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201411100.dv.
Supplementary Material
Acknowledgments
We thank Lysann Schmitz for technical assistance with the bioenergetic characterization of heart mitochondria. We also thank Yvonne Hinze for technical assistance for the coenzyme Q quantification and Dr. Xinping Li for technical assistance with the mass spectrometry. We thank Werner Kühlbrandt for support and critical reading of the manuscript. We also thank Prof. Thomas Langer and Prof. David Chan for the gift of MEFs.
N.G. Larsson was supported by a European Research Council Advanced Investigator grant and by grants from CECAD and Deutsche Forschungsgemeinschaft (SFB 829). Mouse colonies were maintained in the Animal Core Facility at the Max Planck Institute for Biology of Ageing. Imaging analyses were performed in the FACS & Imaging Core Facility at the Max Planck Institute for Biology of Ageing. Proteomics analyses were performed in the Proteomics and Bioinformatics Core Facility at the Max Planck Institute for Biology of Ageing.
The authors declare no competing financial interests.
Author contributions: E. Motori performed and analyzed the microscopic experiments. T. Brandt, M. Lagouge, I. Atanassov, A. Galinier, G. Rappl, K. Hultenby, S. Brodesser, and C. Dieterich helped with experimental work. A. Mourier performed experimental work, project planning, and data analysis, and wrote the manuscript together with N.-G. Larsson.
Footnotes
Abbreviations used in this paper:
- CCCP
- carbonyl cyanide m-chlorophenyl hydrazone
- Δψ
- mitochondrial membrane potential
- DMAP
- dimethylallyl pyrophosphate
- FPP
- farnesyl diphosphate
- GGPP
- geranyl-geranyl-diphosphate
- GPP
- geranyl diphosphate
- IPP
- isopentenyl-pyrophosphate
- MEF
- mouse embryonic fibroblast
- MEV
- mevastatin
- MRM
- multiple reaction monitoring
- mtDNA
- mitochondrial DNA
- OXPHOS
- oxidative phosphorylation
- SILAC
- stable isotope labeling by amino acids in cell culture
- TLC
- thin-layer chromatography
- TMRM
- tetramethylrhodamine methyl ester
References
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. 2000. Gene ontology: tool for the unification of biology. Nat. Genet. 25:25–29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgardt B.F., Mauer J., Wunderlich F.T., Ernst M.B., Pal M., Spohn G., Brönneke H.S., Brodesser S., Hampel B., Schauss A.C., and Brüning J.C.. 2010. Hypothalamic and pituitary c-Jun N-terminal kinase 1 signaling coordinately regulates glucose metabolism. Proc. Natl. Acad. Sci. USA. 107:6028–6033 10.1073/pnas.1001796107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beraud N., Pelloux S., Usson Y., Kuznetsov A.V., Ronot X., Tourneur Y., and Saks V.. 2009. Mitochondrial dynamics in heart cells: very low amplitude high frequency fluctuations in adult cardiomyocytes and flow motion in non beating Hl-1 cells. J. Bioenerg. Biomembr. 41:195–214 10.1007/s10863-009-9214-x [DOI] [PubMed] [Google Scholar]
- Bolte S., and Cordelières F.P.. 2006. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224:213–232 10.1111/j.1365-2818.2006.01706.x [DOI] [PubMed] [Google Scholar]
- Bourne H.R., Sanders D.A., and McCormick F.. 1990. The GTPase superfamily: a conserved switch for diverse cell functions. Nature. 348:125–132 10.1038/348125a0 [DOI] [PubMed] [Google Scholar]
- Chen H., Detmer S.A., Ewald A.J., Griffin E.E., Fraser S.E., and Chan D.C.. 2003. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 160:189–200 10.1083/jcb.200211046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., McCaffery J.M., and Chan D.C.. 2007. Mitochondrial fusion protects against neurodegeneration in the cerebellum. Cell. 130:548–562 10.1016/j.cell.2007.06.026 [DOI] [PubMed] [Google Scholar]
- Cox J., and Mann M.. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26:1367–1372 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., and Mann M.. 2011. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10:1794–1805 10.1021/pr101065j [DOI] [PubMed] [Google Scholar]
- de Brito O.M., and Scorrano L.. 2008. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 456:605–610 10.1038/nature07534 [DOI] [PubMed] [Google Scholar]
- Detmer S.A., and Chan D.C.. 2007. Complementation between mouse Mfn1 and Mfn2 protects mitochondrial fusion defects caused by CMT2A disease mutations. J. Cell Biol. 176:405–414 10.1083/jcb.200611080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detmer S.A., Vande Velde C., Cleveland D.W., and Chan D.C.. 2008. Hindlimb gait defects due to motor axon loss and reduced distal muscles in a transgenic mouse model of Charcot-Marie-Tooth type 2A. Hum. Mol. Genet. 17:367–375 10.1093/hmg/ddm314 [DOI] [PubMed] [Google Scholar]
- Duarte A., Poderoso C., Cooke M., Soria G., Cornejo Maciel F., Gottifredi V., and Podestá E.J.. 2012. Mitochondrial fusion is essential for steroid biosynthesis. PLoS ONE. 7:e45829 10.1371/journal.pone.0045829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte A., Castillo A.F., Podestá E.J., and Poderoso C.. 2014. Mitochondrial fusion and ERK activity regulate steroidogenic acute regulatory protein localization in mitochondria. PLoS ONE. 9:e100387 10.1371/journal.pone.0100387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangakis A.S., and Hegerl R.. 2001. Noise reduction in electron tomographic reconstructions using nonlinear anisotropic diffusion. J. Struct. Biol. 135:239–250 10.1006/jsbi.2001.4406 [DOI] [PubMed] [Google Scholar]
- Galinier A., Carrière A., Fernandez Y., Bessac A.M., Caspar-Bauguil S., Periquet B., Comtat M., Thouvenot J.P., and Casteilla L.. 2004. Biological validation of coenzyme Q redox state by HPLC-EC measurement: relationship between coenzyme Q redox state and coenzyme Q content in rat tissues. FEBS Lett. 578:53–57 10.1016/j.febslet.2004.10.067 [DOI] [PubMed] [Google Scholar]
- Guillet V., Gueguen N., Verny C., Ferre M., Homedan C., Loiseau D., Procaccio V., Amati-Bonneau P., Bonneau D., Reynier P., and Chevrollier A.. 2010. Adenine nucleotide translocase is involved in a mitochondrial coupling defect in MFN2-related Charcot-Marie-Tooth type 2A disease. Neurogenetics. 11:127–133 10.1007/s10048-009-0207-z [DOI] [PubMed] [Google Scholar]
- Hansson A., Hance N., Dufour E., Rantanen A., Hultenby K., Clayton D.A., Wibom R., and Larsson N.-G.. 2004. A switch in metabolism precedes increased mitochondrial biogenesis in respiratory chain-deficient mouse hearts. Proc. Natl. Acad. Sci. USA. 101:3136–3141 10.1073/pnas.0308710100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henneman L., van Cruchten A.G., Denis S.W., Amolins M.W., Placzek A.T., Gibbs R.A., Kulik W., and Waterham H.R.. 2008. Detection of nonsterol isoprenoids by HPLC-MS/MS. Anal. Biochem. 383:18–24 10.1016/j.ab.2008.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Sherman B.T., and Lempicki R.A.. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4:44–57 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Kanehisa M., and Goto S.. 2000. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller G.A., Pazirandeh M., and Krisans S.. 1986. 3-Hydroxy-3-methylglutaryl coenzyme A reductase localization in rat liver peroxisomes and microsomes of control and cholestyramine-treated animals: quantitative biochemical and immunoelectron microscopical analyses. J. Cell Biol. 103:875–886 10.1083/jcb.103.3.875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs W.J., Faust P.L., Keller G.A., and Krisans S.K.. 2001. Purification of brain peroxisomes and localization of 3-hydroxy-3-methylglutaryl coenzyme A reductase. Eur. J. Biochem. 268:4850–4859 10.1046/j.0014-2956.2001.02409.x [DOI] [PubMed] [Google Scholar]
- Kremer J.R., Mastronarde D.N., and McIntosh J.R.. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71–76 10.1006/jsbi.1996.0013 [DOI] [PubMed] [Google Scholar]
- Kulak N.A., Pichler G., Paron I., Nagaraj N., and Mann M.. 2014. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods. 11:319–324 10.1038/nmeth.2834 [DOI] [PubMed] [Google Scholar]
- Kuznetsov A.V., Troppmair J., Sucher R., Hermann M., Saks V., and Margreiter R.. 2006. Mitochondrial subpopulations and heterogeneity revealed by confocal imaging: possible physiological role? Biochim. Biophys. Acta. 1757:686–691 10.1016/j.bbabio.2006.03.014 [DOI] [PubMed] [Google Scholar]
- Lee S., Sterky F.H., Mourier A., Terzioglu M., Cullheim S., Olson L., and Larsson N.-G.. 2012. Mitofusin 2 is necessary for striatal axonal projections of midbrain dopamine neurons. Hum. Mol. Genet. 21:4827–4835 10.1093/hmg/dds352 [DOI] [PubMed] [Google Scholar]
- Loiseau D., Chevrollier A., Verny C., Guillet V., Gueguen N., Pou de Crescenzo M.-A., Ferré M., Malinge M.-C., Guichet A., Nicolas G., et al. 2007. Mitochondrial coupling defect in Charcot-Marie-Tooth type 2A disease. Ann. Neurol. 61:315–323 10.1002/ana.21086 [DOI] [PubMed] [Google Scholar]
- Misko A., Jiang S., Wegorzewska I., Milbrandt J., and Baloh R.H.. 2010. Mitofusin 2 is necessary for transport of axonal mitochondria and interacts with the Miro/Milton complex. J. Neurosci. 30:4232–4240 10.1523/JNEUROSCI.6248-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollet J., Giurgea I., Schlemmer D., Dallner G., Chretien D., Delahodde A., Bacq D., de Lonlay P., Munnich A., and Rötig A.. 2007. Prenyldiphosphate synthase, subunit 1 (PDSS1) and OH-benzoate polyprenyltransferase (COQ2) mutations in ubiquinone deficiency and oxidative phosphorylation disorders. J. Clin. Invest. 117:765–772 10.1172/JCI29089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motori E., Puyal J., Toni N., Ghanem A., Angeloni C., Malaguti M., Cantelli-Forti G., Berninger B., Conzelmann K.-K., Götz M., et al. 2013. Inflammation-induced alteration of astrocyte mitochondrial dynamics requires autophagy for mitochondrial network maintenance. Cell Metab. 18:844–859 10.1016/j.cmet.2013.11.005 [DOI] [PubMed] [Google Scholar]
- Mourier A., Ruzzenente B., Brandt T., Kühlbrandt W., and Larsson N.-G.. 2014. Loss of LRPPRC causes ATP synthase deficiency. Hum. Mol. Genet. 23:2580–2592 10.1093/hmg/ddt652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier L.M., and Krisans S.K.. 2000. Peroxisomal protein targeting and identification of peroxisomal targeting signals in cholesterol biosynthetic enzymes. Biochim. Biophys. Acta. 1529:89–102 10.1016/S1388-1981(00)00139-6 [DOI] [PubMed] [Google Scholar]
- Ong S.-E., and Mann M.. 2006. A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC). Nat. Protoc. 1:2650–2660 10.1038/nprot.2006.427 [DOI] [PubMed] [Google Scholar]
- Papanicolaou K.N., Khairallah R.J., Ngoh G.A., Chikando A., Luptak I., O’Shea K.M., Riley D.D., Lugus J.J., Colucci W.S., Lederer W.J., et al. 2011. Mitofusin-2 maintains mitochondrial structure and contributes to stress-induced permeability transition in cardiac myocytes. Mol. Cell. Biol. 31:1309–1328 10.1128/MCB.00911-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papanicolaou K.N., Ngoh G.A., Dabkowski E.R., O’Connell K.A., Ribeiro R.F., Stanley W.C., and Walsh K.. 2012. Cardiomyocyte deletion of mitofusin-1 leads to mitochondrial fragmentation and improves tolerance to ROS-induced mitochondrial dysfunction and cell death. Am. J. Physiol. Heart Circ. Physiol. 302:H167–H179 10.1152/ajpheart.00833.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham A.H., Meng S., Chu Q.N., and Chan D.C.. 2012. Loss of Mfn2 results in progressive, retrograde degeneration of dopaminergic neurons in the nigrostriatal circuit. Hum. Mol. Genet. 21:4817–4826 10.1093/hmg/dds311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo M., Legros F., Chateau D., and Lombès A.. 2002. Membrane topology and mitochondrial targeting of mitofusins, ubiquitous mammalian homologs of the transmembrane GTPase Fzo. J. Cell Sci. 115:1663–1674. [DOI] [PubMed] [Google Scholar]
- Sebastián D., Hernández-Alvarez M.I., Segalés J., Sorianello E., Muñoz J.P., Sala D., Waget A., Liesa M., Paz J.C., Gopalacharyulu P., et al. 2012. Mitofusin 2 (Mfn2) links mitochondrial and endoplasmic reticulum function with insulin signaling and is essential for normal glucose homeostasis. Proc. Natl. Acad. Sci. USA. 109:5523–5528 10.1073/pnas.1108220109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signorelli P., and Hannun Y.A.. 2002. Analysis and quantitation of ceramide. Methods Enzymol. 345:275–294 10.1016/S0076-6879(02)45023-9 [DOI] [PubMed] [Google Scholar]
- Smirnova E., Griparic L., Shurland D.L., and van der Bliek A.M.. 2001. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol. Biol. Cell. 12:2245–2256 10.1091/mbc.12.8.2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterky F.H., Lee S., Wibom R., Olson L., and Larsson N.-G.. 2011. Impaired mitochondrial transport and Parkin-independent degeneration of respiratory chain-deficient dopamine neurons in vivo. Proc. Natl. Acad. Sci. USA. 108:12937–12942 10.1073/pnas.1103295108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szkopińska A.2000. Ubiquinone. Biosynthesis of quinone ring and its isoprenoid side chain. Intracellular localization. Acta Biochim. Pol. 47:469–480. [PubMed] [Google Scholar]
- Takahashi R., Ikeda T., Hamaguchi A., Iwasa K., and Yamada M.. 2012. Coenzyme Q10 therapy in hereditary motor sensory neuropathy type VI with novel mitofusin 2 mutation. Intern. Med. 51:791–793 10.2169/internalmedicine.51.6676 [DOI] [PubMed] [Google Scholar]
- Tran U.C., and Clarke C.F.. 2007. Endogenous synthesis of coenzyme Q in eukaryotes. Mitochondrion. 7:S62–S71 10.1016/j.mito.2007.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., and Hekimi S.. 2013. Molecular genetics of ubiquinone biosynthesis in animals. Crit. Rev. Biochem. Mol. Biol. 48:69–88 10.3109/10409238.2012.741564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasilewski M., Semenzato M., Rafelski S.M., Robbins J., Bakardjiev A.I., and Scorrano L.. 2012. Optic atrophy 1-dependent mitochondrial remodeling controls steroidogenesis in trophoblasts. Curr. Biol. 22:1228–1234 10.1016/j.cub.2012.04.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weibel E.R.1979. Stereological Methods: Practical Methods of Biological Morphometry. Academic Press, London. 630 pp. [Google Scholar]
- Yao J.K., and Rastetter G.M.. 1985. Microanalysis of complex tissue lipids by high-performance thin-layer chromatography. Anal. Biochem. 150:111–116 10.1016/0003-2697(85)90447-6 [DOI] [PubMed] [Google Scholar]
- Züchner S., Mersiyanova I.V.I., Muglia M., Bissar-Tadmouri N., Rochelle J., Dadali E.L.E., Zappia M., Nelis E., Patitucci A., Senderek J., et al. 2004. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat. Genet. 36:449–451 (published erratum in Nat. Genet. 2004. 36:660) 10.1038/ng1341 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.