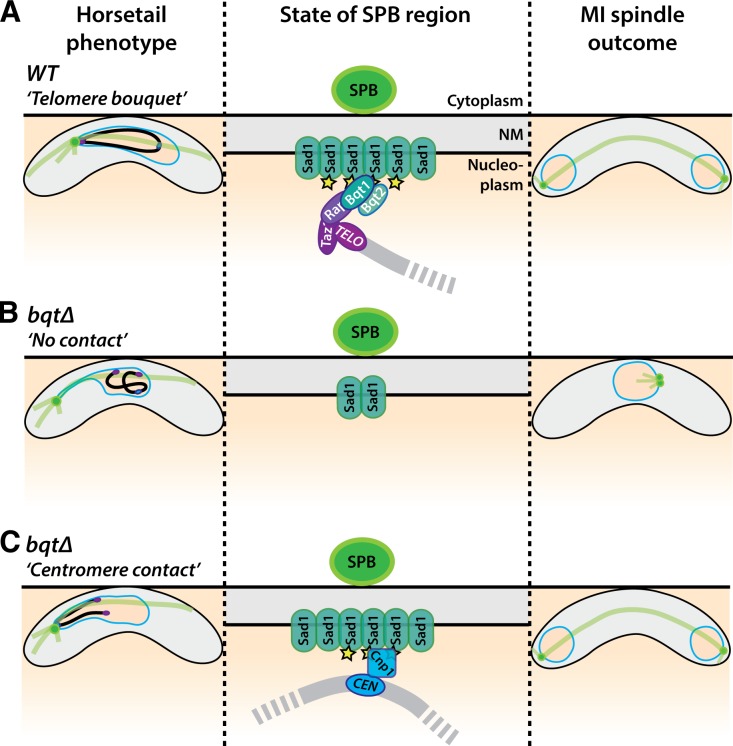
Both centromere–centrosome and telomere–centrosome contacts can promote spindle formation during meiosis.
Abstract
Telomeres and centromeres have traditionally been considered to perform distinct roles. During meiotic prophase, in a conserved chromosomal configuration called the bouquet, telomeres gather to the nuclear membrane (NM), often near centrosomes. We found previously that upon disruption of the fission yeast bouquet, centrosomes failed to insert into the NM at meiosis I and nucleate bipolar spindles. Hence, the trans-NM association of telomeres with centrosomes during prophase is crucial for efficient spindle formation. Nonetheless, in approximately half of bouquet-deficient meiocytes, spindles form properly. Here, we show that bouquet-deficient cells can successfully undergo meiosis using centromere–centrosome contact instead of telomere–centrosome contact to generate spindle formation. Accordingly, forced association between centromeres and centrosomes fully rescued the spindle defects incurred by bouquet disruption. Telomeres and centromeres both stimulate focal accumulation of the SUN domain protein Sad1 beneath the centrosome, suggesting a molecular underpinning for their shared spindle-generating ability. Our observations demonstrate an unanticipated level of interchangeability between the two most prominent chromosomal landmarks.
Introduction
Telomeres maintain the integrity of linear chromosomes by preventing the unsolicited recruitment of DNA repair machineries to chromosome ends (de Lange, 2009; Dehé and Cooper, 2010; Jain and Cooper, 2010) and engaging telomerase to solve the end replication problem (Greider and Blackburn, 1985; Artandi and Cooper, 2009). At a distinct site on the chromosome, centromeres mediate the attachment of chromosomes to spindle microtubules in specific orientations that allow accurate chromosome segregation (Cheeseman and Desai, 2008; Watanabe, 2012). Despite these separate functions, telomeres and centromeres share several similarities. Both direct the assembly of specific nucleoprotein complexes and both, as a consequence of their underlying repetitive DNA sequences, are packaged into heterochromatin (Karpen and Allshire, 1997; Stimpson and Sullivan, 2010).
Meiosis ensures the correct distribution of chromosomes from diploid progenitor cells to haploid gametes by incorporating two sequential nuclear divisions (meiosis I [MI] and II [MII]) after only a single round of DNA replication (Petronczki et al., 2003; Yanowitz, 2010). A widely conserved feature of meiotic prophase is formation of the telomere bouquet in which telomeres gather to a confined region of the nuclear membrane (NM), often near the centrosome (Chikashige et al., 1994; Chikashige et al., 1997; Scherthan, 2001). The bouquet stage is particularly well characterized in fission yeast. During mitotic interphase, telomeres localize to two or three clusters around the NM, whereas centromeres form a single cluster beneath the centrosome, which in fission yeast is called the spindle pole body (SPB) and is located on the cytoplasmic surface of the NM. This clustering requires interactions between centromeres and the SUN domain inner NM protein Sad1 (Hagan and Yanagida, 1995; Nabetani et al., 2001; Hou et al., 2012). Sad1 interacts with the outer NM KASH domain protein Kms1, which in turn contacts the SPB. Upon meiotic induction, the meiosis-specific proteins Bqt1 and Bqt2 interact with Rap1 (a partner of the telomeric dsDNA binding protein, Taz1) and recruit Sad1-Kms1 to the distally located telomeres (Chikashige et al., 2006). Kms1 interacts with cytoplasmic dynein, forming a telomere–Bqt–Sad1–Kms–dynein bridge dubbed the telocentrosome, as it nucleates cytoplasmic microtubules that provide tracks for the movement of telomeres to the SPB (Yoshida et al., 2013). As telomeres accumulate beneath the SPB during bouquet formation, the centromeres are released from this site (Asakawa et al., 2005; Klutstein et al., 2015). Hence, centromeres and telomeres sequentially interact with the SPB in a cell cycle–dependent manner.
Bouquet formation is followed by the onset of dramatic oscillatory movements of the SPB that pull the telomere-led chromosomes back and forth, generating the elongated horsetail nucleus (Ding et al., 1998). At the end of prophase, the SPB settles in the middle of the cell and the telomeres dissociate from the bouquet in a concerted fashion dubbed telomere fireworks (Tomita and Cooper, 2007). This marks a critical stage for the SPB: it must complete duplication while remaining anchored to the cytoplasmic surface of the NM; mother and daughter SPBs then insert into the NM to nucleate the spindles that orchestrate MI and MII. It is at this stage that SPB behavior fails in bouquet-deficient (taz1Δ, rap1Δ, or bqt1Δ) cells, 50% of which display SPBs that fail to nucleate spindles (Tomita and Cooper, 2007).
Here we explore the low penetrance of spindle defects in the absence of the telomere bouquet; how is spindle formation achieved without prior contact between telomeres and the SPB? Strikingly, we find that bouquet-deficient cells can successfully undergo meiosis using centromeres instead of telomeres to generate spindle formation. Our observations indicate a surprising level of telomere–centromere interchangeability.
Results
Sporadic centromere-mediated chromatin–SPB contacts during prophase rescue bqt1Δ spindle defect
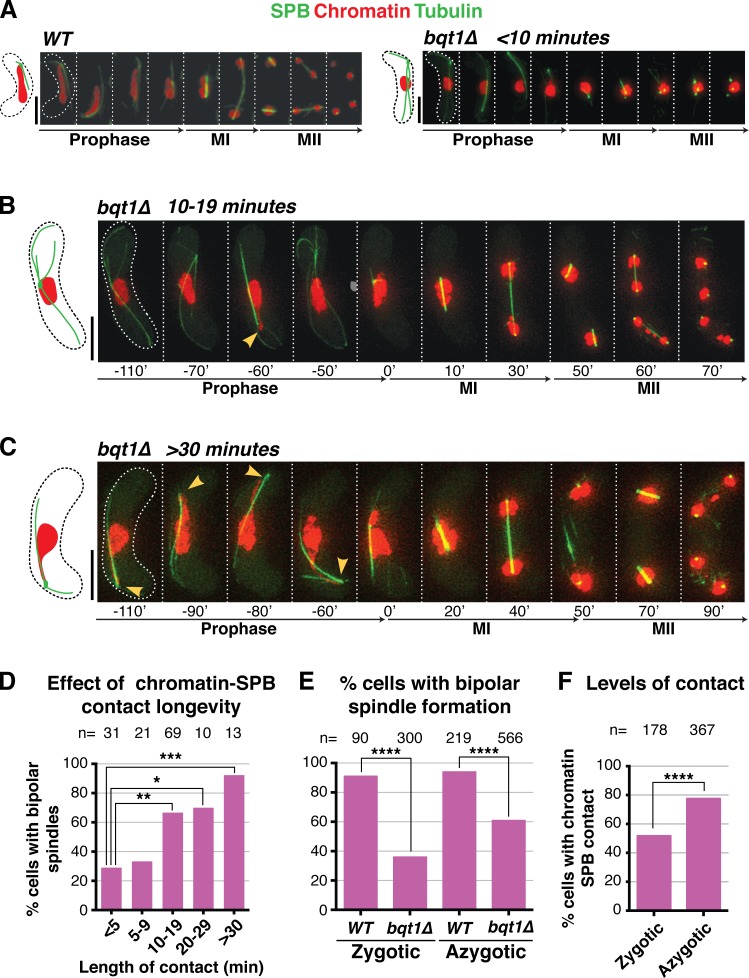
To address the low penetrance of spindle defects in bouquet-deficient meiosis, we examined the properties of those bqt1Δ meiocytes that succeed in meiotic spindle formation. In bouquet-deficient settings, the fluctuating SPB lacks stable contact with chromatin, preventing the chromosome oscillations that generate the horsetail nuclear shape (Fig. 1 A and Video 1; Chikashige et al., 2006; Tomita and Cooper, 2007). Nonetheless, we noticed that the SPB often appears to catch a chromatin segment as it passes through the static chromatin mass, generating a streak that emanates from the main bulk of chromatin (Fig. 1, B and C; and Video 2). Remarkably, although cells lacking visible chromatin–SPB contact tend to sustain spindle formation defects, those with visible chromatin–SPB contacts during meiotic prophase are significantly more likely to form robust bipolar spindles at MI despite the absence of the bouquet (Figs. 1 D and S1, A–D).
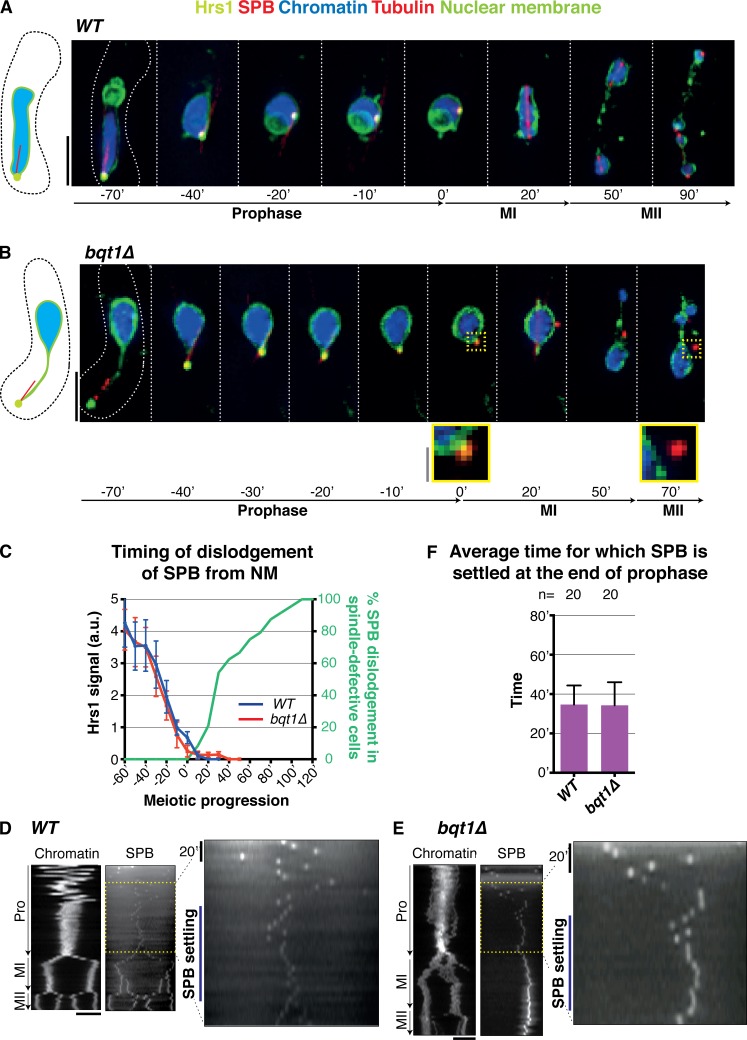
Figure 1.
Rescue of bqt1Δ spindle defect by prophase chromatin–SPB contacts. (A–C) Frames from films of meiocytes carrying Hht1-mRFP (histone H3 tagged at one of the two endogenous hht1+ loci; Chromatin), Sid4-GFP (endogenously tagged; SPB), and ectopically expressed GFP-Atb2 (nmt1 promoter controlled; Tubulin). Numbering indicates meiotic progression in minutes; t = 0 is just before spindle formation. Bars, 5 µm. (A) A bqt1Δ meiocyte with <10-min contact displays a monopolar MI spindle and unstable MII spindles. (B and C) bqt1Δ cells with 10–19-min and >30-min chromatin–SPB contact (yellow arrowheads) show proper spindles at MI and MII. (D) Quantitation of effect of chromatin–SPB contact duration on bipolar MI spindle formation. n is the total number of cells scored in eight independent experiments; data were subject to Fisher’s exact test: ***, 0.0001 < P < 0.001; **, 0.001 < P < 0.01; *, 0.01 < P < 0.05. All cells scored for A–D of this figure are more extensively analyzed in Fig. S1 (C and D). (E) Bipolar spindle formation is more frequent in azygotic than zygotic bqt1Δ meiosis. n is the total number of cells scored from at least two (WT) and more than eight (bqt1Δ) independent experiments in a range of strain backgrounds; ****, P < 0.0001. (F) Comparison of chromatin–SPB contact frequency in zygotic and azygotic meiosis. n is the total number of cells scored from >3 (WT) and >10 (bqt1Δ) independent experiments (for chromatin–SPB contact data) and ≥5 (WT and bqt1Δ) independent experiments (for centromere–SPB contact data).
To further define the correlation between bqt1Δ chromatin–SPB contacts and proper spindle formation, we allocated cells into categories according to the longevity of their longest contact (see Fig. S1 and Materials and methods), ranging from <5-min chromatin–SPB contact (i.e., “no apparent contact”) to “throughout” contacts that span the entirety of prophase. This analysis reveals a clear correlation between the longevity of prophase chromatin–SPB contacts and the recovery of bqt1Δ MI spindle formation (Fig. 1 D). Indeed, instances of bipolar spindle formation with no apparent contact are likely a result of contacts whose durations are shorter than the intervals between time-lapse images, as their frequency is reduced from 45% to 29% when the intervals between time-lapse images are reduced from 10 to 5 min.
During azygotic meiosis, in which stable diploids enter meiosis upon nitrogen starvation (as opposed to zygotic meiosis, in which haploids conjugate and immediately undergo meiosis, the scenario preferred by fission yeast and referred to throughout this paper unless specified), we observe a high number of cells with contacts throughout meiotic prophase. These long-lived contacts are associated with a bipolar spindle formation frequency of 100%. This explains the reduction in overall bipolar spindle formation in zygotic versus azygotic bqt1Δ meiosis (bipolar spindles are seen in 40% of the former and 60% of the latter; Fig. 1 E). As azygotic meiotic cells are smaller than zygotes, an intuitive expectation is that the likelihood of contacts between SPBs and chromatin is increased simply by crowding (Fig. 1 F). Correspondingly, we noticed a twofold increase in spindle defects in bqt1Δ meiocytes grown in liquid media, which are larger than those grown on solid media. An inverse relationship between cell size and proper spindle formation is also suggested by the results of bouquet loss in the cell size mutants wee1-50 and cdc25-22 (Cdc25-C532Y). Although loss of bqt1 in the smaller wee1-50 cells conferred only a slight reduction in sporulation efficiency (∼90% asci with four equally sized spores), bouquet loss in the larger cdc25-22 cells yields four healthy-looking spores in only 20% of asci. Thus, the rate of aberrant spindle formation in a bqt1Δ setting correlates with cell size, most likely because of the enhanced probability of prophase chromatin–SPB contacts in smaller cells.
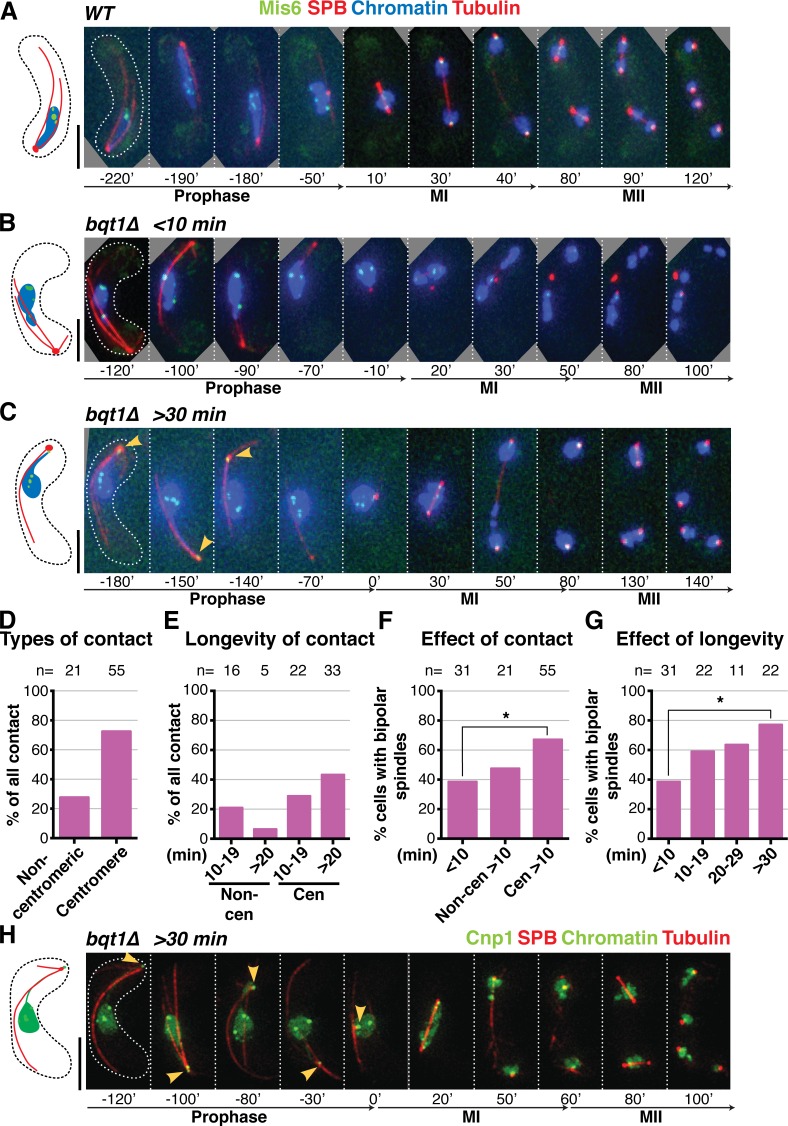
The foregoing observations implicate chromatin–SPB contacts in successful meiotic spindle formation. To define the nature of these contacts, we visualized the centromere via Mis6, an inner kinetochore component that remains at centromeres throughout meiotic prophase (Asakawa et al., 2005; Fig. 2, A–C; and Video 3). In a WT setting, centromeres dissociate from the SPB once the telomeres arrive at the onset of meiotic prophase (Fig. 2 A; Klutstein et al., 2015). The bulk of centromeres also dissociate from the SPB at the onset of prophase in a bouquet-deficient setting, leaving the fluctuating SPB to stray far from the chromatin mass, which remains static in the middle of the cell. Despite this separation of the SPB from the main chromatin mass, it always remains associated with the NM throughout prophase (see below). The SPB settles at the end of prophase in bqt1Δ cells as it does in WT cells, even though it subsequently fails to nucleate stable spindles (Fig. 2 B). As noted above, a considerable proportion of bqt1Δ meiocytes displays a segment of chromatin associated with the SPB during prophase along with proper bipolar spindles. Strikingly, in 72% of these cases the chromatin segment contains a clear Mis6 signal, indicating that the centromere itself contacts the SPB (Figs. 2, C and D). On the rare occasions where contact appears to be mediated by a noncentromeric region, the contact tends to be shorter (Fig. 2 E); moreover, as the Mis6 signal is often faint, the lack of a clear signal cannot be used to definitively rule out centromeric localization. In line with the correlation between chromatin–SPB contact duration and proper spindle formation (Fig. 1, B–D), we observe a correlation between Mis6–SPB contact duration and proper spindle formation (Fig. 2, F and G; and Fig. S1, A–F).
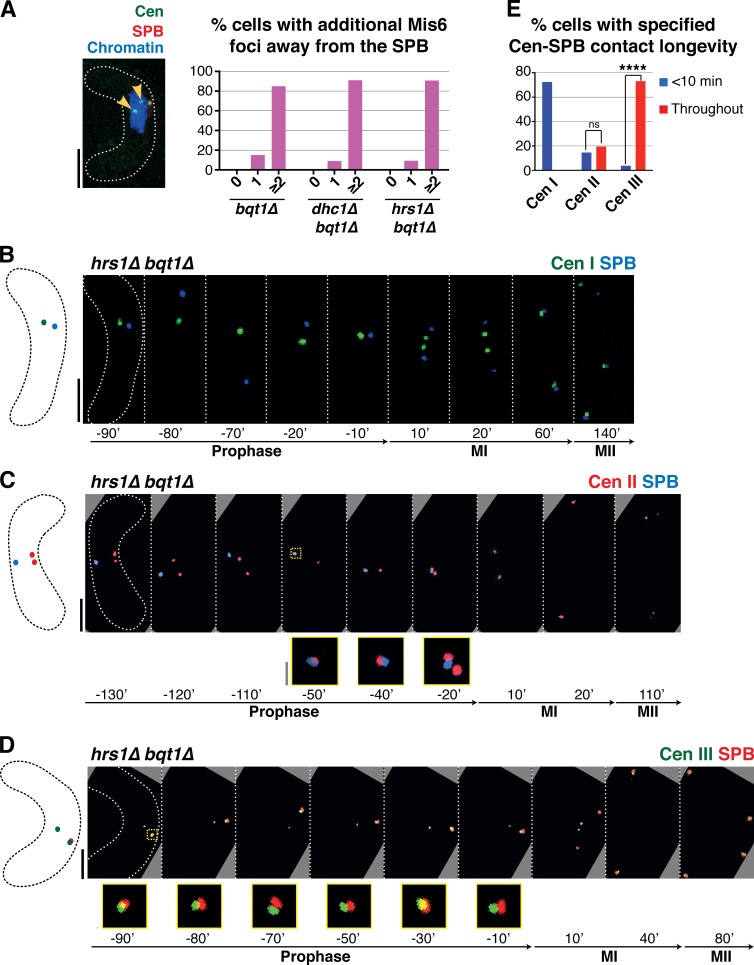
Figure 2.
Centromeres mediate the chromatin–SPB contacts that rescue bqt1Δ spindle defects. (A–C and H) Frames from films of meiocytes carrying Hht1-CFP (at a single endogenous locus as in Fig 1; Chromatin), Sid4-mCherry (SPB), ectopically expressed mCherry-Atb2 (Tubulin), and endogenously tagged Mis6-GFP (A–G) or Cnp1-GFP (ectopically expressed under control of endogenous promoter; H). Bars, 5 µm. (A) In WT cells, centromeres do not localize to the SPB during meiotic prophase. (B) bqt1Δ cell showing <10-min centromere–SPB contact during prophase followed by failed spindle formation. (C and H) A centromere–SPB contact lasting >30 min (indicated by yellow arrowheads) is followed by bipolar spindle formation. (D) Levels of centromeric versus noncentromeric chromatin–SPB contact during bqt1Δ meiotic prophase. (E) Longevity of centromeric versus noncentromeric contacts. (F) Levels of bqt1Δ bipolar MI spindle formation seen in cells with the specified types of chromatin–SPB contact. The percentage of bipolar spindle formation seen in noncentromeric >10-min contact (Non-cen >10 min) is likely an overestimate caused by the faintness of Mis6-GFP signals. (G) Proper spindle formation is quantified as a function of centromere–SPB contact duration. n is the number of cells scored in 14 independent experiments. All cells scored for this figure are more extensively analyzed in Fig. S1 (E and F). *, 0.01 < P < 0.05.
Observation of the centromeric histone H3 variant Cnp1 (Fig. 2 H) again reveals that centromeres mediate the chromatin–SPB contacts that rescue spindle formation in bouquet-deficient settings. Likewise, a higher frequency of centromere–SPB interactions is seen in azygotic than zygotic bqt1Δ meiosis. Therefore, prophase centromere–SPB contacts predict improved spindle formation in bqt1Δ meiosis.
Forced long-lived centromere–SPB contacts fully rescue bqt1Δ spindles
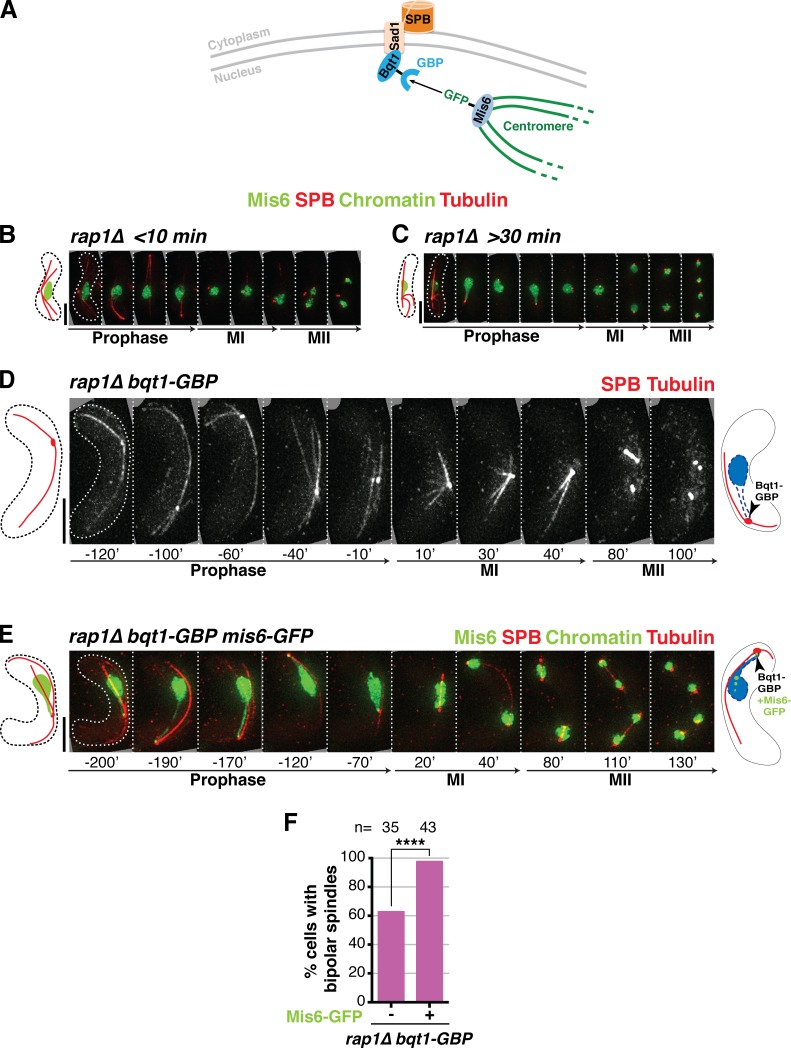
To further substantiate the idea that long-lived centromere–SPB contacts confer proper spindle formation, we sought to induce a complete rescue of the bqt1Δ spindle defect by maintaining centromeres at the SPB throughout meiotic prophase. To achieve this, we used the GFP-binding protein (GBP; Rothbauer et al., 2006) fused to the C terminus of endogenous Bqt1 to recruit GFP-tagged Mis6 (and hence, centromeres) to the SPB in a rap1Δ setting (Fig. 3 A). Indeed, coexpression of Mis6-GFP and Bqt1-GBP results in efficient recruitment such that at least one centromere is seen at the SPB throughout prophase in nearly all cells. In this forced centromere–SPB interaction scenario, nearly 100% bipolar MI spindle formation is achieved despite the absence of Rap1 (and therefore the absence of the bouquet; Fig. 3, B–F; and Video 4). Conversely, in those rare Mis6-GFP Bqt1-GBP cells lacking Mis6-Bqt1 contact, the spindle is defective. Hence, forced long-lived centromere–SPB contacts fully rescue spindle defects in bouquet-deficient settings.
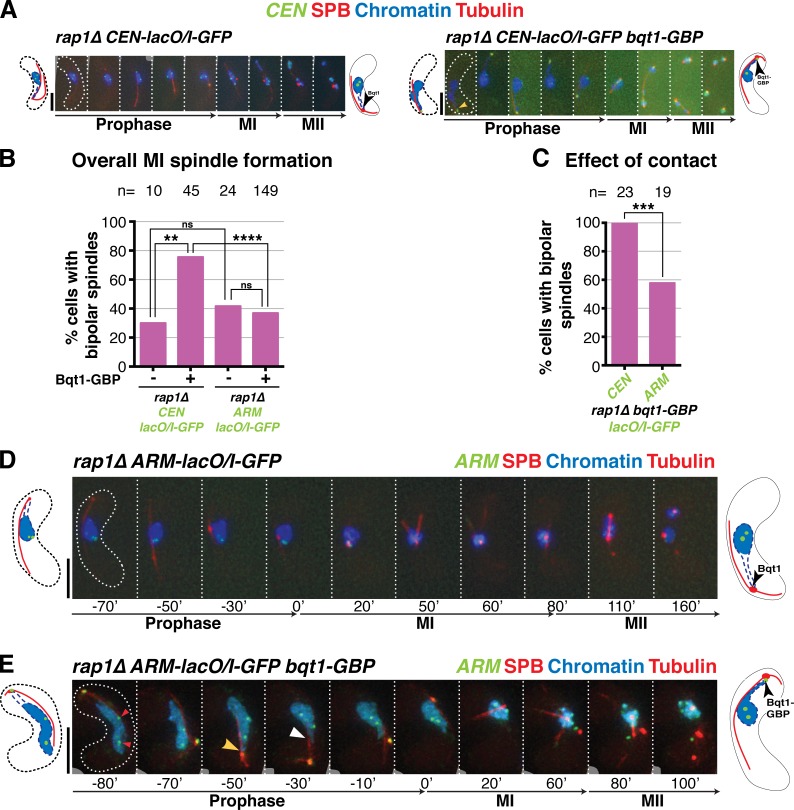
Figure 3.
Maintaining centromeres at the SPB for the entirety of prophase ensures spindle formation in the absence of the bouquet. (A) Schematic of the GBP-GFP system used to force centromere–SPB interactions. (B–E) Frames of films of meiocytes harboring the specified tags: SPB, Mis6, chromatin, and tubulin tagged as in Fig 2; Bqt1 is endogenously fused at its C terminus with GBP. Schematics on the right of each series show the expected prophase phenotype (nuclear membrane outlined with a dashed blue line). (B) In rap1Δ cells lacking centromere–SPB contacts, Bqt1 localizes to the nonchromatin-associated SPB and spindle formation is defective. (C) A rap1Δ meiocyte with >30-min centromere–SPB contact forms proper bipolar spindles. (D) Introduction of Bqt1-GBP in rap1Δ cells lacking a GFP tag on Mis6 fails to rescue abnormal spindle formation. (E) Introduction of Mis6-GFP in the Bqt1-GBP setting confers association of the SPB with a centromere, resulting in proper bipolar spindles. Bars, 5 µm. (F) Levels of bipolar MI spindle formation in rap1Δ bqt1-GBP background with and without Mis6-GFP. n is the total number of cells scored from more than two independent experiments. ****, P < 0.0001.
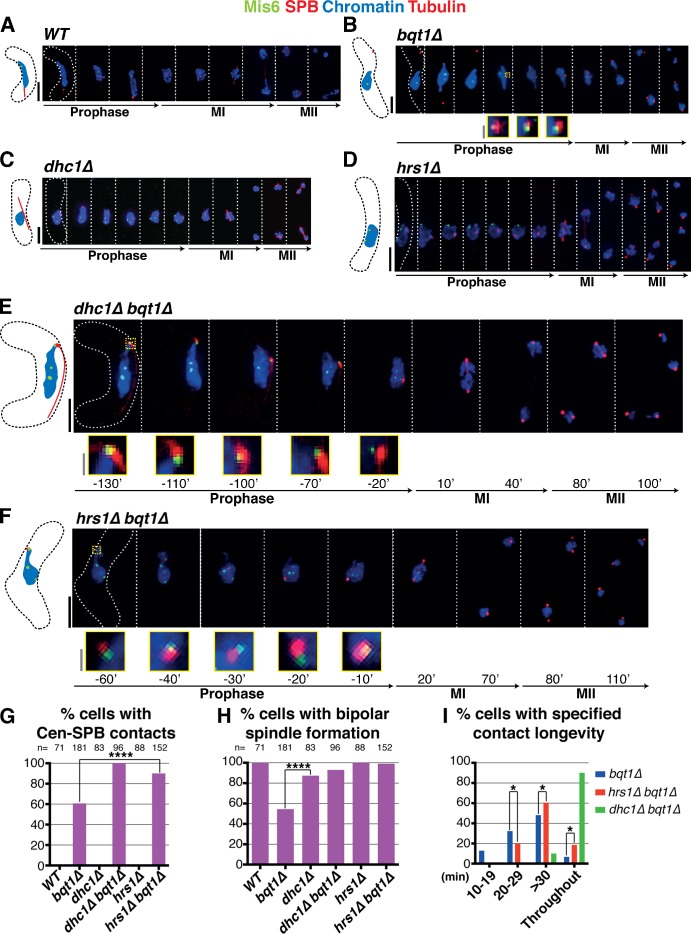
An independent approach to assessing the result of “throughout” centromere–SPB contacts in bouquet-deficient settings is provided by meiocytes in which horsetail movements are compromised via deletion of dhc1+ (which encodes the dynein heavy chain) or hrs1+ (which encodes a meiosis-specific SPB component required for horsetail movement; Yamamoto et al., 1999; Saito et al., 2005; Tanaka et al., 2005). We previously observed that dhc1Δ bqt1Δ meiocytes suffer defective spindle formation reminiscent of bqt1Δ single mutant meiocytes (Tomita and Cooper, 2007). However, if we revisit these analyses excluding all dhc1Δ bqt1Δ meiocytes displaying karyogamy defects, we find that the majority of dhc1Δ bqt1Δ meiocytes show proper bipolar spindle formation; this has also been observed by others (Chikashige et al., 2014). Hence, the loss of vigorous nuclear movement suppresses bqt1Δ spindle defects in those cells displaying robust karyogamy. Complete centromere release from the meiotic SPB occurs only upon the onset of horsetail nuclear movements (Klutstein et al., 2015), suggesting that disruption of such movement could confer maintenance of stable centromere–SPB contact in bqt1Δ mutants, thus explaining the restored bipolar spindle formation in dhc1Δ bqt1Δ cells. Hence, we investigated centromere–SPB association in these backgrounds. Whereas dhc1Δ and hrs1Δ single mutant meiocytes show normal centromere release during prophase (Fig. 4, C and D), dhc1Δ bqt1Δ and hrs1Δ bqt1Δ meiocytes show incomplete centromere release, with all cells displaying at least one centromere–SPB contact throughout prophase (Fig. 4, E–G; Fig. S1 G; and Video 5; Klutstein et al., 2015). These “throughout” centromere–SPB contacts can account for proper bipolar spindle formation (Fig. 4 H).
Figure 4.
Loss of Dhc1 or Hrs1 confers long-lived centromere–SPB contact in bqt1Δ zygotes. (A–F) All labels are as in Fig 2 with Mis6-GFP marking the centromere. Bars: (black) 5 µm; (gray) 1 µm. (A) Centromeres are absent from the SPB during horsetail movement in WT meiotic prophase. (B) Chromatin occasionally follows the SPB during prophase in bqt1Δ cells; sporadic centromere–SPB contacts (inset and magnified in yellow) confer bipolar spindle formation. (C and D) In dhc1Δ bqt1+ and hrs1Δ bqt1+ zygotes, vigorous SPB movement is abolished but centromeres are released from the SPB. (E and F) In contrast, dhc1Δ bqt1Δ and hrs1Δ bqt1Δ zygotes maintain at least one centromere at the SPB throughout prophase, ensuring normal spindle formation. (G) Although only around 60% of bqt1Δ cells show centromere–SPB contacts, all dhc1Δ bqt1Δ and the majority of hrs1Δ bqt1Δ cells show this interaction. No such contacts are observed in WT, dhc1Δ, or hrs1Δ cells. The analyses use Mis6-GFP as a centromere marker; the faintness of this marker explains the appearance of cells in which no centromere–SPB contact can be seen in a hrs1Δ bqt1Δ setting. Using the brighter Swi6-GFP as a marker for centromeres, we observe contact throughout in all cells (not depicted). (H) Quantitation shows complete restoration of bqt1Δ spindle formation by hrs1+ or dhc1+ deletion. n is the total number of cells scored from greater than six independent experiments. ****, P < 0.0001. (I) Categorization of zygotes according to longevity of their centromere–SPB contacts. The majority of dhc1Δ bqt1Δ cells show “throughout” (entire length of horsetail stage) prophase centromere–SPB contacts. All cells scored for this figure are more extensively analyzed in Fig. S1 G. The data shown are from three independent experiments analyzing >50 cells each. *, 0.01 < P < 0.05.
Although the persistence of centromere–SPB interactions upon cessation of prophase SPB movement can explain the suppression of bqt1Δ spindle defects by dhc1 and hrs1 deletion, the loss of movement by itself could also contribute to rescued spindle assembly; indeed, such spindle rescue in a related bouquet-deficient setting (bqt2Δ) has recently been interpreted in this context (Chikashige et al., 2014). For instance, one might envision that without associated telomeres (or centromeres) the SPB could be propelled away from the NM by the vigorous horsetail movements. However, dislodgement of the SPB from the NM occurs only after horsetail movements have ceased (Fig. 5, A–C). Indeed, the SPBs of bouquet-defective meiocytes dissociate from the NM only after Hrs1 signal has completely disappeared. Moreover, the SPB settles in the middle of the cell and remains fairly immobile at this position for ∼40 min before SPB duplication and insertion in both WT and bqt1Δ cells (Fig. 5, D–F). This observation argues against the idea that horsetail movements are responsible for defective SPB and spindle behavior in the absence of the bouquet. Instead, “throughout” centromere–SPB contacts most likely explain the rescue of spindle formation by dhc1 or hrs1 deletion in a bouquet-deficient setting.
Figure 5.
SPB settling occurs regardless of bouquet status. (A and B) Frames from films of meiocytes with endogenously GFP-tagged Hrs1 and nmt1-controlled Ish1-GFP (to visualize the NM) along with the indicated markers (as in Fig. 2). Bars: (black) 5 µm; (gray) 1 µm. (A) In WT meiocytes, the SPB colocalizes with the NM throughout meiosis. (B) In bqt1Δ meiocytes, the SPB detaches from the NM only after SPB movement ceases, i.e., upon disappearance of Hrs1-GFP (t = 0′). (C) Quantitation and timing of phenotypes shown in A and B from three independent experiments analyzing 30 cells for each strain. Note that dislodgement of the SPB from the NM is never observed in WT meiocytes. SPBs of bqt1Δ cells dissociate from the NM only after Hrs1 signal has disappeared. (D and E) Kymographs of Hht1-CFP (Chromatin) and Sid4-mCherry (SPB) during prophase, MI, and MII. The SPB settling phase (yellow) is shown magnified. The data shown are from a single representative experiment out of >10 repeats. (F) Time (in minutes) for which SPB (as visualized via Hrs1-GFP) settles after prophase. n is the total number of cells scored from greater than six independent experiments. Error bars show the mean SD.
CenIII preferentially interacts with the SPB in the absence of the bouquet
In bouquet-defective cells sustaining centromere signals at the SPB during meiotic prophase, additional centromeric foci are observed away from the SPB; indeed, the majority of centromere signals separate from the SPB in all backgrounds observed (Fig. 6 A). This suggests that successful spindle formation can be conferred by interaction between only one centromere and the SPB. To investigate whether these centromere–SPB interactions are chromosome specific, we used strains harboring individual centromeres tagged with fluorescent lacO or tetO arrays (Fig. 6). We engineered these centromeres in the bqt1Δ hrs1Δ background, as the absence of Hrs1 increases the probability of centromere–SPB contact (Fig. 4). Intriguingly, interactions between cenIII-lacO and the SPB are more frequent and of greater longevity than interactions with cenI-lacO or cenII-tetO; moreover, cenII-tetO interacts more frequently than cenI-lacO with the SPB (Fig. 6, B–E; and Fig. S1, H–J). Hence, centromere size (which is largest for chromosome III and smallest for chromosome I) may influence SPB interactions; for example, a larger centromere may persist longer at the SPB during its horsetail movements or may have a greater chance of reencountering the SPB once dissociated. Current studies aim to decipher the basis for these preferential interactions.
Figure 6.
Prophase centromere–SPB contacts in a bqt1Δ setting show a preference for cenIII. (A) In cells with centromere–SPB contacts, additional centromeric foci are observed away from the SPB. Arrowheads mark Mis6-GFP dots far from the SPB during dhc1Δ bqt1Δ prophase. Quantitation is shown to the right. More than 100 cells were scored in five independent experiments. (B–D) Series of frames of films of hrs1Δ bqt1Δ meiosis. Numbering as in Fig 1; the SPB is viewed via endogenously tagged Sad1-CFP. Bars: (black) 5 µm; (gray) 1 µm. (B) The centromere of chromosome I is visualized via a LacI-GFP–bound lys1+-lacO array. (C) The centromere of chromosome II is visualized via a TetR-bound cnt2-tetO array. (D) The centromere of chromosome III was followed via a LacI-bound ade6-lacO array. In these cells, Sad1 is endogenously tagged with mRFP. (E) Collated centromere–SPB interactions. SPB interactions with cenIII are more frequent and longer lasting than those with cenI or cenII; moreover, cenII–SPB interactions are more frequent and longer than cenI–SPB interactions. More than five independent experiments were performed. All cells scored for this figure are more extensively analyzed in Fig. S1 (H–J). ****, P < 0.0001.
Substitution of telomeres by centromeres is independent of Clr4, Dcr1, Nuf2, and Csi1
What feature of telomeres and centromeres allows both to promote meiotic spindle formation after prophase SPB contact? Their shared status as major heterochromatic regions prompted us to ask whether the histone methyltransferase Clr4 and/or the RNAi pathway component Dcr1 are required. Double mutant bqt1Δ clr4Δ and bqt1Δ dcr1Δ meiocytes show levels of spindle defects similar to those of bqt1Δ single mutants and, as in bqt1Δ meiocytes, the double mutants maintain a tight correlation between chromatin–SPB contacts and proper spindle formation (Fig. S2). Therefore, Clr4 and Dcr1 are dispensable for rescue of the bqt1Δ spindle defect by chromatin–SPB contact.
The control experiments assessing single clr4Δ and dcr1Δ mutants initially surprised us by revealing proper spindle formation (Fig. S2, A, C, and E) despite previous observations, using FISH to mark the telomere-adjacent rDNA region, that these single mutants sustain bouquet defects (Hall et al., 2003; Tuzon et al., 2004). Using live analysis to monitor telomeres via Taz1, we find that bouquet formation is largely intact in the absence of Clr4 or Dcr1. However, instances in which multiple Taz1 foci are seen during meiotic prophase are more frequent in some clr4Δ and dcr1Δ clones, suggesting that the bouquet is less stable in these settings and more prone to disruption by fixation and FISH (unpublished data). Moreover, as SPB contact with a single telomere stretch is sufficient to confer spindle formation (Tomita et al., 2013), cells suffering only a partial bouquet disruption would not suffer perturbed spindle formation.
Hence, the ability to control meiotic spindle formation may be independent of heterochromatinicity. However, the possibility remains that some self-propagating feature downstream of Clr4 or Dcr1 function is required for SPB control in a scenario analogous to centromere assembly itself; although adjacent heterochromatin is required for establishment of centromeres on naive sequences, this heterochromatin is dispensable for the inheritance of preformed centromeres (Karpen and Allshire, 1997; Folco et al., 2008).
To explore the roles of factors known to affect centromere–SPB interactions during mitotic cell cycles, we examined Nuf2, an outer kinetochore component whose meiotic prophase-specific disappearance correlates with disassociation of centromeres from the SPB, and Csi1, a protein that binds the SUN domain protein Sad1 and is required for full centromere–SPB association during mitotic interphase (Asakawa et al., 2005; Hou et al., 2012). As seen in WT meiosis (Asakawa et al., 2005), Nuf2 disappears from the kinetochore upon meiotic entry in bqt1Δ cells, regardless of their centromere–SPB contact status (unpublished data); hence, Nuf2 does not appear to mediate the ability of centromeres to contact the bqt1Δ SPB. Similarly, the ability of the centromere to bind the SPB in a bouquet-deficient background is independent of Csi1 (Fig. S3). Future work aims to identify the specific feature of the centromere that mediates stable interaction with the meiotic SPB.
Noncentromeric chromatin regions are less able than centromeres to restore bqt1Δ bipolar spindle formation
A key question is whether the centromere specificity of the chromatin contacts that rescue spindle formation stems from an ability uniquely shared by centromeres and telomeres to successfully modify SPB-associated factors or whether all chromatin possesses this SPB-modifying capacity but centromeres and telomeres have a higher propensity to associate with the SPB. Although contacts of >20 min between noncentromeric regions and the SPB are too rare to lend firm conclusions, we find that for a given SPB contact duration, the chances of bqt1Δ spindle rescue by noncentromeric chromatin are lower than that for centromeres (Fig. 2 F). To address this issue directly, we used Bqt1-GBP in combination with lacO/I-GFP arrays inserted at centromeric versus noncentromeric sites, theoretically conferring the respective recruitment of the two sites to the meiotic prophase SPB (Fig. S4). As expected, centromere-proximal lacO/I-GFP Bqt1-GBP ensures successful recruitment of centromeric chromatin to the SPB for long periods in rap1Δ meiocytes, and this contact confers vastly improved bipolar spindle formation (Fig. 7, A–C). In contrast, arm-proximal lacO/I-GFP Bqt1-GBP fails to confer improved rap1Δ bipolar spindle formation (Fig. 7, B–E), suggesting that contact between the SPB and a random chromatin site is insufficient to promote spindle formation. However, the efficacy of SPB recruitment by Bqt1-GBP differs between the two lacO/I-GFP–tagged loci. For CEN-proximal lacO/I-GFP, one bright focus remains associated with Bqt1-GBP at the prophase SPB, whereas another stays with the nuclear bulk; the two foci presumably represent the two homologues, which remain largely unpaired in bouquet-deficient settings (Ding et al., 2004; Fig. 7 A). In contrast, arm-associated cut3-lacO/I-GFP appears less tightly associated with Bqt1-GBP, as both bright GFP foci are often seen in the nuclear bulk with only a diffuse GFP halo appearing near the SPB, presumably representing excess free LacI molecules (Fig. 7 E). We presume that the enhanced SPB recruitment of centromere-proximal sites stems from reinforcement of GFP-GBP–based recruitment by the natural affinity of centromeric chromatin for the SPB. Nonetheless, despite the caveat that euchromatic sequences are recruited less efficiently than centromeric sequences to the SPB using this system, those instances of clear recruitment suggest that chromosome arm regions are less able than centromeres to confer proper spindle formation upon contact. Hence, centromeres and telomeres have both a greater propensity to interact with the SPB and a stronger ability to modify its behavior.
Figure 7.
CEN-proximal regions show greater affinity for the SPB, and greater ability to rescue spindle formation, than ARM-proximal regions. (A, D, and E) Series of frames of films of meiocytes harboring the tags detailed in Fig. S4; numbering as in Fig. 1. Bars, 5 µm. (A) rap1Δ meiotic spindle defect is rescued by forcing long-lived interaction between CEN-proximal region and SPB during meiotic prophase. (B) Quantitation of overall bipolar meiotic spindle formation in specified backgrounds. (C) Quantitation of effect of specified contact on bipolar spindle formation, scoring only those cells with contact >50 min. For meiocytes harboring cut3+-lacO/I-GFP, cells with one GFP focus (n = 4) or two foci in nucleoplasm were scored if and only if a clear GFP focus was present at the SPB. n is the total number of cells scored; data were subject to Fisher’s exact test: ****, P < 0.0001; ***, 0.0001 < P < 0.001; **, 0.001 < P < 0.01. (D) As expected, rap1Δ cells with no chromatin contact have defective meiotic spindles. (E) Example of a rap1Δ cut3+-lacO/I-GFP bqt1-GBP zygote. One GFP focus is seen at the SPB and two are seen within the bulk of the nucleus (left, red arrowheads), indicating incomplete recruitment of cut3 locus to the SPB. Yellow arrowhead indicates clear chromatin–SPB contacts; white arrowhead indicates occasions of less clear contact, in which the nucleus appears to poke out in the direction of the SPB but chromatin markers are not clearly visible.
Centromeres and telomeres share the ability to confer increased levels of Sad1 at the SPB
The foregoing results indicate that telomeres and centromeres share the ability to confer proper SPB behavior and spindle formation. To investigate the molecular underpinnings of this shared ability, we analyzed the behavior of Sad1, the SUN domain inner NM protein that connects centromeres and telomeres with the NM and is crucial for spindle formation (Hagan and Yanagida, 1995). We followed the dynamics of endogenously GFP-tagged Sad1 in live WT and bqt1Δ meiocytes and quantified focal Sad1 intensity throughout meiosis (see Materials and methods). Interestingly, the signal intensity of Sad1-GFP at the SPB is significantly reduced in those bqt1Δ meiocytes displaying defective spindle formation (Fig. 8, A and B). This reduction of Sad1 level is consistent and stable throughout prophase at all positions along the trajectory of horsetail movement, as well as at MI and MII.
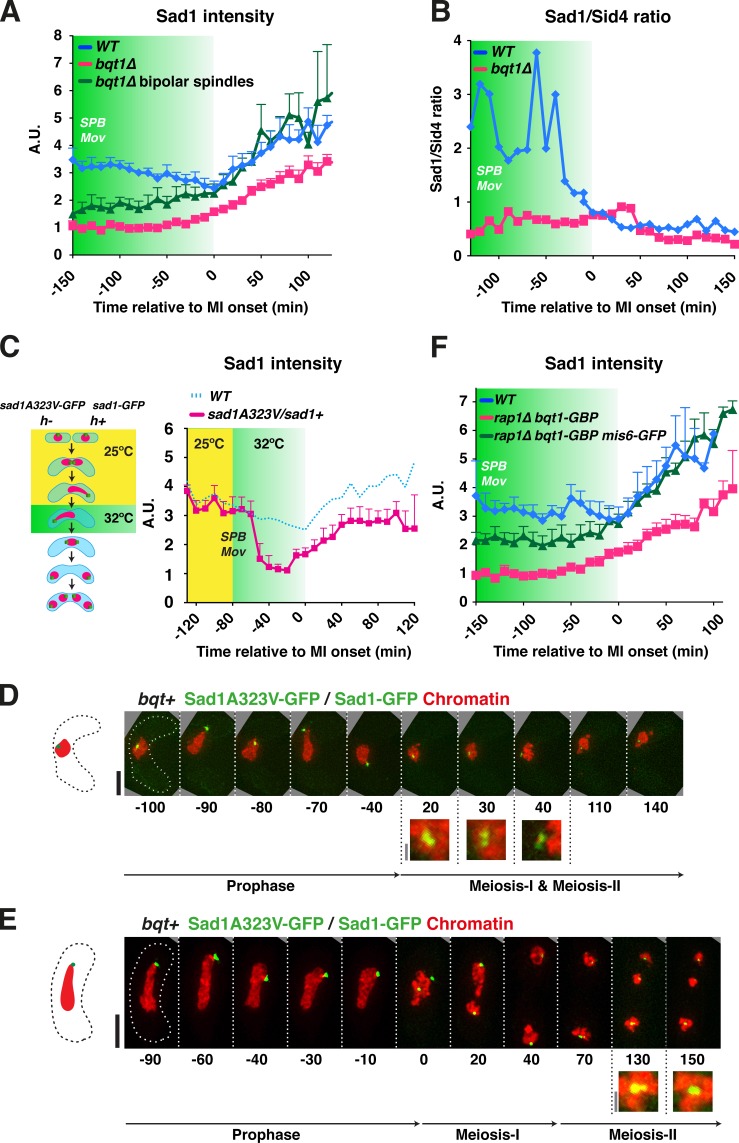
Figure 8.
Sad1 accumulation at the SPB is promoted by telomeres and centromeres. (A) Mean Sad1-GFP signal intensities at the SPB through meiosis were quantified in 19 WT meiocytes, 21 bqt1Δ meiocytes showing spindle defects, and 19 bqt1Δ meiocytes with proper spindle formation. The data shown are from >10 independent experiments. 0 min represents the onset of MI and error bars represent standard deviations. Green shading indicates the period of horsetail SPB movement. (B) Sad1-GFP/Sid4-mCherry intensity ratios are shown for the same cells quantified in A, as Sid4 intensity profiles through meiosis are identical in WT and bouquet-defective meiocytes (unpublished data). (C, left) Schematic of the strategy used to achieve an approximate halving of Sad1 level. A diploid constructed by crossing h+ sad1-GFP hht1-mRFP and h− sad1-A323V-GFP hht1-mRFP was meiotically induced. Once prophase horsetail movement commenced, the temperature was switched to 32°C to inactivate Sad1-A323V; the subsequent meiosis was filmed to assess spindle formation. (right) Sad1-GFP intensities, from 10 meiocytes for each genotype shown, are plotted over time relative to MI onset. Yellow shading indicates the time points taken at 25°C; the subsequent time points were taken 32°C. (D and E) Series of frames of Sad1-GFP/Sad1A323V-GFP meiocytes showing SPB problems when the temperature was switched to 32°C. Bars: (black) 5 µm; (gray) 1 µm. (F) Quantitation of Sad1-mCherry intensity levels in the strains indicated. Greater than 10 meiocytes are represented for each genotype.
To test the idea that Sad1 levels at the SPB are a relevant bouquet-controlled parameter for promoting spindle formation, we designed a system to test for meiotic haploinsufficiency of sad1+ using the ts allele sad1.1 (Hagan and Yanagida, 1995), which contains an alanine to valine substitution at position 323 in the N-terminal region of the SUN domain. To follow Sad1-A323V behavior by live microscopy, we inserted a C-terminal GFP tag and confirmed that growth is temperature sensitive (Fig. S5 A). In heterozygous sad1-GFP/sad1A323V-GFP strains, Sad1 levels are reduced to ∼50% of WT levels within an hour after shift to the nonpermissive temperature of 32°C (Fig. S5). We allowed sad1-GFP/sad1A323V-GFP meiocytes to undergo normal bouquet formation at 25°C and then switched the temperature to 32°C, filming throughout (Fig. 8 C). After the temperature shift–induced reduction in Sad1 levels at the SPB, chromosome separation fails in a manner reminiscent of that seen upon bouquet disruption (Fig. 8, D and E). These observations confirm that a threshold level of Sad1 at the SPB must be attained to ensure successful meiosis.
Notably, we found that in those bqt1Δ meiocytes that form proper bipolar spindles, Sad1 intensities at MI are similar to those in WT meiocytes (Fig. 8 A, green line). This observation not only reinforces the correlation between Sad1 accumulation at the SPB and proper spindle formation, but also suggests that centromeres and telomeres share the ability to promote enhanced Sad1 accumulation at the SPB. To explore this idea explicitly, we forced the interaction between centromeres and the SPB in the absence of the bouquet, using the Bqt1-GBP/Mis6-GFP system described above, and asked whether this forced interaction guarantees Sad1 accumulation. Remarkably, the GBP-GFP–induced centromere–SPB interaction resulted in levels of Sad1 that increased through meiotic prophase and achieved WT levels at MI and MII (Fig. 8 F). Hence, both accidental and forced centromere–SPB contacts rescue spindle formation in bouquet-deficient cells at least in part by conferring accumulation of Sad1 at the SPB.
Discussion
The 50% penetrance of spindle defects in bouquet-deficient meiosis indicated the existence of a redundant pathway by which spindle formation could be stimulated in the absence of telomere–SPB contact. Here we show that centromere–SPB interactions provide this alternative pathway. Hence, centromeres and telomeres are interchangeable in this respect, sharing an ability not possessed by other chromatin regions to contact the NM just beneath the SPB, in turn promoting Sad1 accumulation and spindle nucleation.
Meiotic spindle formation requires several compositional changes within the SPB during the preceding prophase, as several proteins that show constitutive SPB localization during mitotic cell cycles are evicted in early prophase and then actively stockpiled at the SPB before MI onset (Jin et al., 2002; Ohta et al., 2012). Remarkably, this SPB maturation process remains unaltered in a bouquet-deficient background, regardless of whether a given bqt1Δ meiocyte is destined to achieve successful spindle formation (unpublished data). In contrast, Sad1 accumulation and SPB insertion into the NM at MI onset are compromised in the absence of the bouquet (this work; unpublished data). Importantly, those long-lived prophase centromere–SPB contacts that rescue subsequent meiotic spindle formation also rescue Sad1 accumulation. Therefore, the functional interchangeability between centromeres and telomeres likely resides in their ability to interact with and accrue Sad1 at the SPB, thereby ensuring spindle nucleation (Fig. 9, A–C). Interestingly, simple overexpression of Sad1 is not sufficient to promote its accumulation at the SPB; instead, overexpressed Sad1 spreads around the NM (Hagan and Yanagida, 1995; unpublished data). Hence, the bouquet- or centromere-stimulated SPB accumulation of Sad1 must involve some conformational change or modification of Sad1 or of the local NM milieu that allows its retention beneath the SPB, rather than just the mobilization of non-SPB–associated Sad1 from distal regions of the NM. The idea of chromatin-stimulated SUN domain protein modification has precedent in studies of Caernohabditis elegans SUN1, whose phosphorylation during meiotic prophase is promoted by contact with the pairing centers, unique chromosome regions that localize near but not at telomeres and initiate homologue pairing. Pairing center–mediated SUN1 phosphorylation is in turn required for SUN1- and dynein-driven chromosome movements and full meiotic progression. These phospho-modifications depend not only on pairing center contact but also on the polo and Chk-2 kinases (Sato et al., 2009; Harper et al., 2011; Labella et al., 2011). In mouse, the meiosis-specific telomere binding protein TERB1 promotes association of telomeres with the NM and actively recruits the SUN–KASH complex to telomere–NM attachment sites (Shibuya et al., 2014), again highlighting a dynamic interplay between SUN domain protein function and the chromatin sites with which it interacts. However, it remains unknown whether worm or mouse chromosome–SUN interactions impact spindle assembly as they do in fission yeast.
Figure 9.
Centromere–telomere interchangeability in promoting meiotic spindle formation. Meiotic progression is represented from left to right. (A) In WT meiocytes, the movement of telomeres to the SPB promotes modification of Sad1 (yellow stars) or the NM surrounding Sad1, in turn promoting Sad1 accumulation and ensuring bipolar spindle formation. (B) In bqtΔ meiocytes with no chromatin–SKS contact, Sad1 accumulation fails, as does spindle nucleation. (C) In bqtΔ meiocytes with centromere–SPB contact, Sad1 concentrates beneath the SPB as in WT.
Although the factors that confer prophase interactions between telomeres and the SPB are known, the mechanism by which centromeres interact with Sad1 to afford its accumulation at the SPB in a bouquet-deficient setting remains enigmatic. Centromeres are generally released from the SUN–KASH–SPB (SKS) linkage regardless of whether the bouquet forms (Tomita and Cooper, 2007; this work); nonetheless, we observe a single centromere associating with the SKS for at least 10 min during prophase in over half of zygotic bqt1Δ meiocytes. Hence, we propose that although centromere release initiates upon dissolution of the outer kinetochore (Asakawa et al., 2005) and occurs in earnest upon the onset of horsetail nuclear movements (Klutstein et al., 2015), the absence of the telomere bouquet liberates the SKS from steric hindrance, providing centromeres with the opportunity to reassociate with the SKS at any point during prophase. The likelihood of this situation is increased dramatically when both bouquet formation and nuclear movement are disrupted (bqt1Δ dhc1Δ). Moreover, centromeres clearly have a higher propensity than noncentromeric regions to interact stably with the SPB. Our efforts to define the molecular underpinnings of this propensity have thus far ruled out essential roles for Clr4 and Dcr1, both of which are required to maintain pericentric heterochromatin; hence, we expect that elements of the centromere that can be maintained in the absence of methylated H3-K9, most likely the Cnp1-packaged centromeric central core, confer SKS interaction. Indeed, simultaneous disruption of clr4 and clr3 (which encodes a histone deacetylase) in the bqt1Δ setting leads to reduced levels of centromere–SPB contact and defective spindle formation (unpublished data), consistent with the effects of such double gene disruption on centromeric central core Cnp1 maintenance. Therefore, centromere identity by itself likely dictates the ability of centromeres to associate with the meiotic prophase SPB and substitute for telomeres in promoting spindle formation.
Although Csi1 is known to mediate central core–SPB interactions in mitotic interphase (Hou et al., 2012), the fact that some centromeres retain interphase SPB association in the absence of Csi1 (Hou et al., 2012) implies the existence of further uncharacterized components involved in this interaction; the dispensability of Csi1 for the rescuing centromere–SPB contacts we observe in bqt1Δ meiocytes further underscores the existence of such additional factors. In considering factors that might confer SKS-modifying ability, Rec8 and Moa1 (a meiosis-specific cohesin subunit and stimulator of monopolar attachment, respectively [Yokobayashi and Watanabe, 2005; Sakuno et al., 2009]) arose as viable candidates, as both are recruited to the centromeric central core in a H3-K9 methylation-independent manner; however, both are dispensable for centromeric rescue of bqt1Δ meiotic spindle formation (unpublished data). Hence, some other central core–associated (presumably Cnp1-dependent) feature imparts SKS modification ability. The supremacy of CenIII in terms of bqt1Δ spindle-rescuing ability could be explained by its longer central core domain.
Layered upon the issue of how meiotic centromeres associate stably with the SKS in the absence of the bouquet is the question of what feature shared between telomeres and centromeres endows these distinct chromosome sites with SPB regulatory capacity. The fact that a single telomere repeat stretch (Tomita et al., 2013) or a single centromere (Fig. 6) is sufficient to ensure bipolar spindle formation rules out the possibility that the relevant property involves the clustering of multiple such regions; moreover, our observation that forced interaction between lac arrays in euchromatin and the SKS fail to confer proper spindle formation argues against the possibility that repeats by themselves possess SKS-modifying ability. Conceivably, this ability is inherent to stretches of chromatin with some specific torsional or condensation-related property of centromeres and telomeres. It will be fascinating to determine the features of these regions that imparts their interchangeability.
The finding that centromeres can substitute for telomeres in controlling meiotic spindle formation may have important implications for mitotic cell cycles as well. Indeed, the well-established mitotic interphase contact between centromeres and the SKS suggests the tantalizing possibility that centromeres control mitotic SPB behavior in a manner reminiscent of the role of meiotic telomeres. Such mitotic chromatin–based control could provide an extra layer of cell cycle regulation, perhaps ensuring a dependency of spindle assembly upon the completion of specific chromosomal events (like DNA replication, chromatin assembly, or chromatin repair). Intriguingly, the origin recognition complex replication activation proteins have been shown to interact with human centrosomes and regulate the centrosome cycle, perhaps comprising a related mode by which chromosomal and centrosomal events are coupled (Prasanth et al., 2004; Hemerly et al., 2009).
Our discovery of an interchangeable role of centromeres and telomeres is consistent with the notion that centromeres derived from telomeres in parallel with the evolution of a complex cytoskeleton during the transition from a single circular genophore to multiple, linear, eukaryotic chromosomes (Méndez-Lago et al., 2009). Whereas the observation of a shared role for centromere and telomeres in promoting spindle formation is unprecedented, the use of centromeres in one organism to perform functions assumed by telomeres in another organism is prominently illustrated by the meiotic centromere clustering seen in Drosophila melanogaster oocytes that is thought to be analogous to the bouquet in facilitating homologue pairing (Takeo and Hawley, 2012). Centromere clustering is also thought to augment early meiotic chromosome sorting processes in polyploid plants (Wen et al., 2012). Moreover, a role for chromatin in controlling spindle formation is likely to be conserved in higher eukaryotes, especially as chromatin has been shown to promote spindle nucleation in Xenopus laevis extracts (Heald et al., 1997). Centromeres and telomeres may have coevolved to share features that enhance this spindle-promoting ability.
Materials and methods
Strains and media
Strains are listed in Table S1. Gene deletions/tag insertions were created as described previously (Grimm et al., 1988; Tomita and Cooper, 2007). Insertions of mCherry-Atb2 at the aur1 locus (Hashida-Okado et al., 1998) used pYC19-mCherry-atb2 (Nakamura et al., 2011) provided by T. Toda (Cancer Research UK, London, UK). The vector for GFP binding protein tagging (Rothbauer et al., 2006) was provided by M. Sato (Waseda University, Tokyo, Japan).
Media were as described previously (Moreno et al., 1991). Cells grown in YES at 32°C were plated on malt extract at 30°C to induce meiosis and analyzed 5–7 h later (live analysis).
Live analysis
Cells were adhered to 35-mm glass culture dishes (MatTek Corporation) using 0.2 mg/ml of soybean lectin (Sigma-Aldrich) and immersed in EMM-N (with required supplements ± 15 mM thiamine). Time-lapse imaging was performed at 27°C in an Environmental Chamber with a DeltaVision Spectris (Applied Precision) comprising a widefield inverted epifluorescence microscope (IX70; Olympus), a 100x NA 1.4 oil immersion objective (UPlanSapo; Olympus), and a charge coupled device CoolSnap HQ camera (Photometrics). Images were acquired over 26 focal planes at a 0.35-µm step size with frames taken every 10 min for 7 h. Images were deconvolved (enhanced ratio method) and combined into a 2D image using the maximum intensity projection setting for analysis using SoftWorx (Applied Precision). Sid4-GFP and Atb2-GFP were captured with 0.2-s exposures per plane, Hht1-mRFP for 0.06 s/plane, Atb2-mCherry and Sid4-mCherry for 0.4 s/plane, Hht1-CFP for 0.2 s/plane, Mis6-GFP for 0.35 s/plane, and Cnp1-GFP and Swi6-GFP for 0.3 s/plane. When three fluorescent channels were imaged, a fast acquisition mode was used in which each Z plane was exposed to each wavelength sequentially.
SPB (Sad1 or Sid4) signal quantitation was performed using Volocity software on images acquired over 26 focal planes at a 0.35-µm step size at each time point. Images were deconvolved (enhanced ratio method) and combined into a 2D image using SoftWorx. For each time point, the intensity of the area containing a given signal was quantified and that of an equivalent signal-free region within the same cell was subtracted; the resulting signal intensities were normalized to the mean intensity for one pixel of background outside the cell. Image processing and analysis were performed using Photoshop CS5.1 (Adobe).
Scoring criteria
To score contact categories (Fig. 1), only those cells observed for at least 40 min of meiotic prophase and through the meiotic divisions were included. For those zygotic meiotic cells in which karyogamy was observed, scoring began two frames after the completion of karyogamy. For all other zygotic cells, scoring began upon the start of live analysis. For azygotic cells, scoring began two frames after the first dramatic SPB fluctuation upon initiation of horsetail movement (once interphase microtubules depolymerized). To categorize contact duration, scoring was terminated two frames before MI spindle formation. Cells were categorized according to their longest single contact. In the rap1Δ mis6-GFP bqt1-GBP background (Fig. 3), only those meiocytes with at least 50 min of contact were scored. Cells with defects in karyogamy or chromosome condensation, which occasionally stem from UV exposure during karyogamy regardless of genotype, were discarded. In strains harboring tagged histone H3 but no tagged centromere protein, frames in which the SPB was within bulk chromatin were discarded, as it was impossible to assign direct chromatin–SPB contacts. Spindles were scored as bipolar if separation of duplicated SPBs and subsequent elongation of a spindle separating the chromatin masses was seen. Fischer’s exact test was performed as described previously (Agresti, 1992). The two-tail p-value was used.
A MATLAB script (see online supplemental material) was written to clearly display the range of contact lengths occurring in individual cells using vector diagrams (Fig. S1), enabling analysis of individual cells within the population according to their contact-to-spindle phenotypes.
Western blot analysis
Cells were collected by centrifugation, washed with Tris-HCl, pH 9.0, and resuspended in 20% TCA with PMSF (1 mM) and protease inhibitors mix (SetIII; EMD Millipore). After agitation with acid washed glass beads (Sigma-Aldrich), tubes were pierced at the bottom and samples were recovered by centrifugation. Beads were washed with 20% TCA with PMSF (1 mM) and protease inhibitors mix and centrifuged, and the final pellet was resuspended in LDS sample buffer (Invitrogen). All manipulations were performed at 4°C. Proteins were separated on NuPAGE 4–12% Bis-Tris acrylamide gradient gels with MOPS SDS running buffer (Invitrogen). Blots were probed anti-GFP antibody produced in rabbit (Sigma-Aldrich) or anti-Histone H2B antibody produced in rabbit (Sigma-Aldrich). Horseradish peroxidase–conjugated anti–rabbit IgG (GE Healthcare) were used as secondary antibodies. Visualization was performed using the ECL Plus Western Blotting Detection System (GE Healthcare).
Online supplemental material
Fig. S1 shows single cell analysis of chromatin/centromere–SPB contacts during prophase in bouquet-deficient meiocytes. Fig. S2 shows that the centromere–SPB contacts that rescue bqt1Δ spindle formation require neither Clr4 nor Dcr1. Fig. S3 shows that these centromere–SPB interactions are also independent of Csi1. Fig. S4 shows that forced interactions between the SPB and centromere- or chromosome arm–proximal loci have no effect on meiotic progression in a bouquet-proficient background. Fig. S5 shows the analysis of Sad1 protein levels in sad1+/sad1-A323V cells. Table S1 shows the genotypes of the strains used in this work. The MATLAB script is the source code for the vector diagram in Fig. S1. Video 1 shows a bqt1Δ meiocyte with no contact and a defective spindle. Video 2 shows a bqt1Δ meiocyte with persistent chromatin–SPB contact showing bipolar spindle formation. Video 3 shows that the rescuing chromatin–SPB contact is mediated by the centromere. Video 4 shows that forced centromere–SPB contact ensures bipolar spindle formation. Video 5 shows that loss of nuclear movement leads to centromere–SPB contact throughout and bipolar spindle formation. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201409058/DC1.
Supplementary Material
Acknowledgments
We thank Cécile Bez for advice and discussion throughout this work, Alex Tournier for invaluable help with MATLAB analysis, Michael Lichten for comments on the manuscript, and our laboratory members for discussions. We thank Yasushi Hiraoka and Yuji Chikashige for sharing unpublished data and strains and Masamitsu Sato for strains.
This work was supported by the European Research Council, Cancer Research UK, the National Institutes of Health, and a European Molecular Biology Organization long-term fellowship to A. Fernández-Álvarez.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- GBP
- GFP-binding protein
- MI
- meiosis I
- NM
- nuclear membrane
- SKS
- SUN–KASH–SPB
- SPB
- spindle pole body
References
- Agresti A.1992. A survey of exact inference for contingency tables. Stat. Sci. 7:131–153 10.1214/ss/1177011454 [DOI] [Google Scholar]
- Artandi S.E., and Cooper J.P.. 2009. Reverse transcribing the code for chromosome stability. Mol. Cell. 36:715–719 10.1016/j.molcel.2009.11.030 [DOI] [PubMed] [Google Scholar]
- Asakawa H., Hayashi A., Haraguchi T., and Hiraoka Y.. 2005. Dissociation of the Nuf2-Ndc80 complex releases centromeres from the spindle-pole body during meiotic prophase in fission yeast. Mol. Biol. Cell. 16:2325–2338 10.1091/mbc.E04-11-0996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., and Desai A.. 2008. Molecular architecture of the kinetochore–microtubule interface. Nat. Rev. Mol. Cell Biol. 9:33–46 10.1038/nrm2310 [DOI] [PubMed] [Google Scholar]
- Chikashige Y., Ding D.Q., Funabiki H., Haraguchi T., Mashiko S., Yanagida M., and Hiraoka Y.. 1994. Telomere-led premeiotic chromosome movement in fission yeast. Science. 264:270–273 10.1126/science.8146661 [DOI] [PubMed] [Google Scholar]
- Chikashige Y., Ding D.Q., Imai Y., Yamamoto M., Haraguchi T., and Hiraoka Y.. 1997. Meiotic nuclear reorganization: switching the position of centromeres and telomeres in the fission yeast Schizosaccharomyces pombe. EMBO J. 16:193–202 10.1093/emboj/16.1.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikashige Y., Tsutsumi C., Yamane M., Okamasa K., Haraguchi T., and Hiraoka Y.. 2006. Meiotic proteins bqt1 and bqt2 tether telomeres to form the bouquet arrangement of chromosomes. Cell. 125:59–69 10.1016/j.cell.2006.01.048 [DOI] [PubMed] [Google Scholar]
- Chikashige Y., Yamane M., Okamasa K., Mori C., Fukuta N., Matsuda A., Haraguchi T., and Hiraoka Y.. 2014. Chromosomes rein back the spindle pole body during horsetail movement in fission yeast meiosis. Cell Struct. Funct. 39:93–100 10.1247/csf.14007 [DOI] [PubMed] [Google Scholar]
- Dehé P.M., and Cooper J.P.. 2010. Fission yeast telomeres forecast the end of the crisis. FEBS Lett. 584:3725–3733 10.1016/j.febslet.2010.07.045 [DOI] [PubMed] [Google Scholar]
- de Lange T.2009. How telomeres solve the end-protection problem. Science. 326:948–952 10.1126/science.1170633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding D.Q., Chikashige Y., Haraguchi T., and Hiraoka Y.. 1998. Oscillatory nuclear movement in fission yeast meiotic prophase is driven by astral microtubules, as revealed by continuous observation of chromosomes and microtubules in living cells. J. Cell Sci. 111:701–712. [DOI] [PubMed] [Google Scholar]
- Ding D.Q., Yamamoto A., Haraguchi T., and Hiraoka Y.. 2004. Dynamics of homologous chromosome pairing during meiotic prophase in fission yeast. Dev. Cell. 6:329–341 10.1016/S1534-5807(04)00059-0 [DOI] [PubMed] [Google Scholar]
- Folco H.D., Pidoux A.L., Urano T., and Allshire R.C.. 2008. Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science. 319:94–97 10.1126/science.1150944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider C.W., and Blackburn E.H.. 1985. Identification of a specific telomere terminal transferase activity in tetrahymena extracts. Cell. 43:405–413 10.1016/0092-8674(85)90170-9 [DOI] [PubMed] [Google Scholar]
- Grimm C., Kohli J., Murray J., and Maundrell K.. 1988. Genetic engineering of Schizosaccharomyces pombe: A system for gene disruption and replacement using the ura4 gene as a selectable marker. Mol. Gen. Genet. 215:81–86 10.1007/BF00331307 [DOI] [PubMed] [Google Scholar]
- Hagan I., and Yanagida M.. 1995. The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J. Cell Biol. 129:1033–1047 10.1083/jcb.129.4.1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall I.M., Noma K., and Grewal S.I.. 2003. RNA interference machinery regulates chromosome dynamics during mitosis and meiosis in fission yeast. Proc. Natl. Acad. Sci. USA. 100:193–198 10.1073/pnas.232688099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper N.C., Rillo R., Jover-Gil S., Assaf Z.J., Bhalla N., and Dernburg A.F.. 2011. Pairing centers recruit a Polo-like kinase to orchestrate meiotic chromosome dynamics in C. elegans. Dev. Cell. 21:934–947 10.1016/j.devcel.2011.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashida-Okado T., Yasumoto R., Endo M., Takesako K., and Kato I.. 1998. Isolation and characterization of the aureobasidin A-resistant gene, aur1R, on Schizosaccharomyces pombe: roles of Aur1p+ in cell morphogenesis. Curr. Genet. 33:38–45 10.1007/s002940050306 [DOI] [PubMed] [Google Scholar]
- Heald R., Tournebize R., Habermann A., Karsenti E., and Hyman A.. 1997. Spindle assembly in Xenopus egg extracts: respective roles of centrosomes and microtubule self-organization. J. Cell Biol. 138:615–628 10.1083/jcb.138.3.615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemerly A.S., Prasanth S.G., Siddiqui K., and Stillman B.. 2009. Orc1 controls centriole and centrosome copy number in human cells. Science. 323:789–793 10.1126/science.1166745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou H., Zhou Z., Wang Y., Wang J., Kallgren S.P., Kurchuk T., Miller E.A., Chang F., and Jia S.. 2012. Csi1 links centromeres to the nuclear envelope for centromere clustering. J. Cell Biol. 199:735–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain D., and Cooper J.P.. 2010. Telomeric strategies: means to an end. Annu. Rev. Genet. 44:243–269 10.1146/annurev-genet-102108-134841 [DOI] [PubMed] [Google Scholar]
- Jin Y., Uzawa S., and Cande W.Z.. 2002. Fission yeast mutants affecting telomere clustering and meiosis-specific spindle pole body integrity. Genetics. 160:861–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen G.H., and Allshire R.C.. 1997. The case for epigenetic effects on centromere identity and function. Trends Genet. 13:489–496 10.1016/S0168-9525(97)01298-5 [DOI] [PubMed] [Google Scholar]
- Klutstein M., Fennell A., Fernández-Álvarez A., and Cooper J.P.. 2015. The telomere bouquet regulates meiotic centromere assembly. Nat. Cell Biol. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labella S., Woglar A., Jantsch V., and Zetka M.. 2011. Polo kinases establish links between meiotic chromosomes and cytoskeletal forces essential for homolog pairing. Dev. Cell. 21:948–958 10.1016/j.devcel.2011.07.011 [DOI] [PubMed] [Google Scholar]
- Méndez-Lago M., Wild J., Whitehead S.L., Tracey A., de Pablos B., Rogers J., Szybalski W., and Villasante A.. 2009. Novel sequencing strategy for repetitive DNA in a Drosophila BAC clone reveals that the centromeric region of the Y chromosome evolved from a telomere. Nucleic Acids Res. 37:2264–2273 10.1093/nar/gkp085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S., Klar A., and Nurse P.. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795–823 10.1016/0076-6879(91)94059-L [DOI] [PubMed] [Google Scholar]
- Nabetani A., Koujin T., Tsutsumi C., Haraguchi T., and Hiraoka Y.. 2001. A conserved protein, Nuf2, is implicated in connecting the centromere to the spindle during chromosome segregation: a link between the kinetochore function and the spindle checkpoint. Chromosoma. 110:322–334 10.1007/s004120100153 [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Arai A., Takebe Y., and Masuda M.. 2011. A chemical compound for controlled expression of nmt1-driven gene in the fission yeast Schizosaccharomyces pombe. Anal. Biochem. 412:159–164 10.1016/j.ab.2011.01.039 [DOI] [PubMed] [Google Scholar]
- Ohta M., Sato M., and Yamamoto M.. 2012. Spindle pole body components are reorganized during fission yeast meiosis. Mol. Biol. Cell. 23:1799–1811 10.1091/mbc.E11-11-0951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronczki M., Siomos M.F., and Nasmyth K.. 2003. Un ménage à quatre: the molecular biology of chromosome segregation in meiosis. Cell. 112:423–440 10.1016/S0092-8674(03)00083-7 [DOI] [PubMed] [Google Scholar]
- Prasanth S.G., Prasanth K.V., Siddiqui K., Spector D.L., and Stillman B.. 2004. Human Orc2 localizes to centrosomes, centromeres and heterochromatin during chromosome inheritance. EMBO J. 23:2651–2663 10.1038/sj.emboj.7600255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbauer U., Zolghadr K., Tillib S., Nowak D., Schermelleh L., Gahl A., Backmann N., Conrath K., Muyldermans S., Cardoso M.C., and Leonhardt H.. 2006. Targeting and tracing antigens in live cells with fluorescent nanobodies. Nat. Methods. 3:887–889 10.1038/nmeth953 [DOI] [PubMed] [Google Scholar]
- Saito T.T., Tougan T., Okuzaki D., Kasama T., and Nojima H.. 2005. Mcp6, a meiosis-specific coiled-coil protein of Schizosaccharomyces pombe, localizes to the spindle pole body and is required for horsetail movement and recombination. J. Cell Sci. 118:447–459 10.1242/jcs.01629 [DOI] [PubMed] [Google Scholar]
- Sakuno T., Tada K., and Watanabe Y.. 2009. Kinetochore geometry defined by cohesion within the centromere. Nature. 458:852–858 10.1038/nature07876 [DOI] [PubMed] [Google Scholar]
- Sato A., Isaac B., Phillips C.M., Rillo R., Carlton P.M., Wynne D.J., Kasad R.A., and Dernburg A.F.. 2009. Cytoskeletal forces span the nuclear envelope to coordinate meiotic chromosome pairing and synapsis. Cell. 139:907–919 10.1016/j.cell.2009.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherthan H.2001. A bouquet makes ends meet. Nat. Rev. Mol. Cell Biol. 2:621–627 10.1038/35085086 [DOI] [PubMed] [Google Scholar]
- Shibuya H., Ishiguro K., and Watanabe Y.. 2014. The TRF1-binding protein TERB1 promotes chromosome movement and telomere rigidity in meiosis. Nat. Cell Biol. 16:145–156 10.1038/ncb2896 [DOI] [PubMed] [Google Scholar]
- Stimpson K.M., and Sullivan B.A.. 2010. Epigenomics of centromere assembly and function. Curr. Opin. Cell Biol. 22:772–780 10.1016/j.ceb.2010.07.002 [DOI] [PubMed] [Google Scholar]
- Takeo S., and Hawley R.S.. 2012. Rumors of its disassembly have been greatly exaggerated: the secret life of the synaptonemal complex at the centromeres. PLoS Genet. 8:e1002807 10.1371/journal.pgen.1002807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Kohda T., Yamashita A., Nonaka N., and Yamamoto M.. 2005. Hrs1p/Mcp6p on the meiotic SPB organizes astral microtubule arrays for oscillatory nuclear movement. Curr. Biol. 15:1479–1486 10.1016/j.cub.2005.07.058 [DOI] [PubMed] [Google Scholar]
- Tomita K., and Cooper J.P.. 2007. The telomere bouquet controls the meiotic spindle. Cell. 130:113–126 10.1016/j.cell.2007.05.024 [DOI] [PubMed] [Google Scholar]
- Tomita K., Bez C., Fennell A., and Cooper J.P.. 2013. A single internal telomere tract ensures meiotic spindle formation. EMBO Rep. 14:252–260 10.1038/embor.2012.218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuzon C.T., Borgstrom B., Weilguny D., Egel R., Cooper J.P., and Nielsen O.. 2004. The fission yeast heterochromatin protein Rik1 is required for telomere clustering during meiosis. J. Cell Biol. 165:759–765 10.1083/jcb.200312061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y.2012. Geometry and force behind kinetochore orientation: lessons from meiosis. Nat. Rev. Mol. Cell Biol. 13:370–382 10.1038/nrm3349 [DOI] [PubMed] [Google Scholar]
- Wen R., Moore G., and Shaw P.J.. 2012. Centromeres cluster de novo at the beginning of meiosis in Brachypodium distachyon. PLoS ONE. 7:e44681 10.1371/journal.pone.0044681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A., West R.R., McIntosh J.R., and Hiraoka Y.. 1999. A cytoplasmic dynein heavy chain is required for oscillatory nuclear movement of meiotic prophase and efficient meiotic recombination in fission yeast. J. Cell Biol. 145:1233–1250 10.1083/jcb.145.6.1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanowitz J.2010. Meiosis: making a break for it. Curr. Opin. Cell Biol. 22:744–751 10.1016/j.ceb.2010.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokobayashi S., and Watanabe Y.. 2005. The kinetochore protein Moa1 enables cohesion-mediated monopolar attachment at meiosis I. Cell. 123:803–817 10.1016/j.cell.2005.09.013 [DOI] [PubMed] [Google Scholar]
- Yoshida M., Katsuyama S., Tateho K., Nakamura H., Miyoshi J., Ohba T., Matsuhara H., Miki F., Okazaki K., Haraguchi T., et al. 2013. Microtubule-organizing center formation at telomeres induces meiotic telomere clustering. J. Cell Biol. 200:385–395 10.1083/jcb.201207168 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.