The Asterless N terminus promotes Plk4 homodimerization and autophosphorylation during interphase, whereas its C terminus stabilizes Plk4 during mitosis.
Abstract
Plk4 (Polo-like kinase 4) and its binding partner Asterless (Asl) are essential, conserved centriole assembly factors that induce centriole amplification when overexpressed. Previous studies found that Asl acts as a scaffolding protein; its N terminus binds Plk4’s tandem Polo box cassette (PB1-PB2) and targets Plk4 to centrioles to initiate centriole duplication. However, how Asl overexpression drives centriole amplification is unknown. In this paper, we investigated the Asl–Plk4 interaction in Drosophila melanogaster cells. Surprisingly, the N-terminal region of Asl is not required for centriole duplication, but a previously unidentified Plk4-binding domain in the C terminus is required. Mechanistic analyses of the different Asl regions revealed that they act uniquely during the cell cycle: the Asl N terminus promotes Plk4 homodimerization and autophosphorylation during interphase, whereas the Asl C terminus stabilizes Plk4 during mitosis. Therefore, Asl affects Plk4 in multiple ways to regulate centriole duplication. Asl not only targets Plk4 to centrioles but also modulates Plk4 stability and activity, explaining the ability of overexpressed Asl to drive centriole amplification.
Introduction
Centrosomes serve as microtubule-organizing centers and facilitate chromosome segregation and spindle orientation during cell division (Bornens, 2012; Tang and Marshall, 2012). Centrosomes are also the precursors to basal bodies of cilia and are involved in regulation of cell cycle transitions and responses to cell stress and DNA damage (Krämer et al., 2004; Shimada and Komatsu, 2009; Kim and Dynlacht, 2013; Nakamura et al., 2013). Cells exert tight control over centrosome number by regulating assembly of the centriole pair, the core duplicating elements of the organelle (Brito et al., 2012). Centriole duplication occurs in a cell cycle–dependent manner and is restricted to only one iteration during S phase when a single nascent procentriole emerges orthogonally from each centriole within the pair (Nigg and Stearns, 2011). Errors in this process result in abnormal centrosome numbers that may perturb spindle orientation and chromosome segregation (Vitre and Cleveland, 2012). Centriole amplification—the overduplication and subsequent overabundance of centrioles within cells—can drive tumorigenic chromosomal instability and is often observed in cancer cells (Nigg and Raff, 2009). Conversely, too few centrioles can lead to a variety of ciliopathies (Bettencourt-Dias et al., 2011).
Plk4 (Polo-like kinase 4) is a conserved master regulator of centriole duplication, and its overexpression induces centriole amplification as well as de novo centriole assembly (Avidor-Reiss and Gopalakrishnan, 2013). Plk4 is primarily regulated by protein turnover and efficiently promotes its own destruction to suppress centriole overduplication (Cunha-Ferreira et al., 2013; Klebba et al., 2013). Unlike other Polo kinase family members, Plk4 forms a homodimer, mediated through an interaction between its first two Polo boxes (PB1 and PB2; formerly known as the cryptic Polo box; Slevin et al., 2012). Upon dimerization, Plk4 extensively trans-autophosphorylates a region near the kinase domain, which then recruits the SCFSlimb/β-TrCP ubiquitin (Ubi) ligase, resulting in its ubiquitination and proteasomal degradation (Cunha-Ferreira et al., 2009; Rogers et al., 2009; Holland et al., 2010; Guderian et al., 2010; Cunha-Ferreira et al., 2013; Klebba et al., 2013). However, during mitosis in flies, autophosphorylation is counteracted by Protein Phosphatase 2A (PP2A) and, consequently, Plk4 protein levels rise (Brownlee et al., 2011). Plk4 then targets mitotic centrioles, appearing as a single asymmetric spot on each centriole (Rogers et al., 2009). Plk4 within each spot is thought to modify this site on a centriole, making each centriole competent to spawn a single daughter centriole during the next S phase (Kleylein-Sohn et al., 2007).
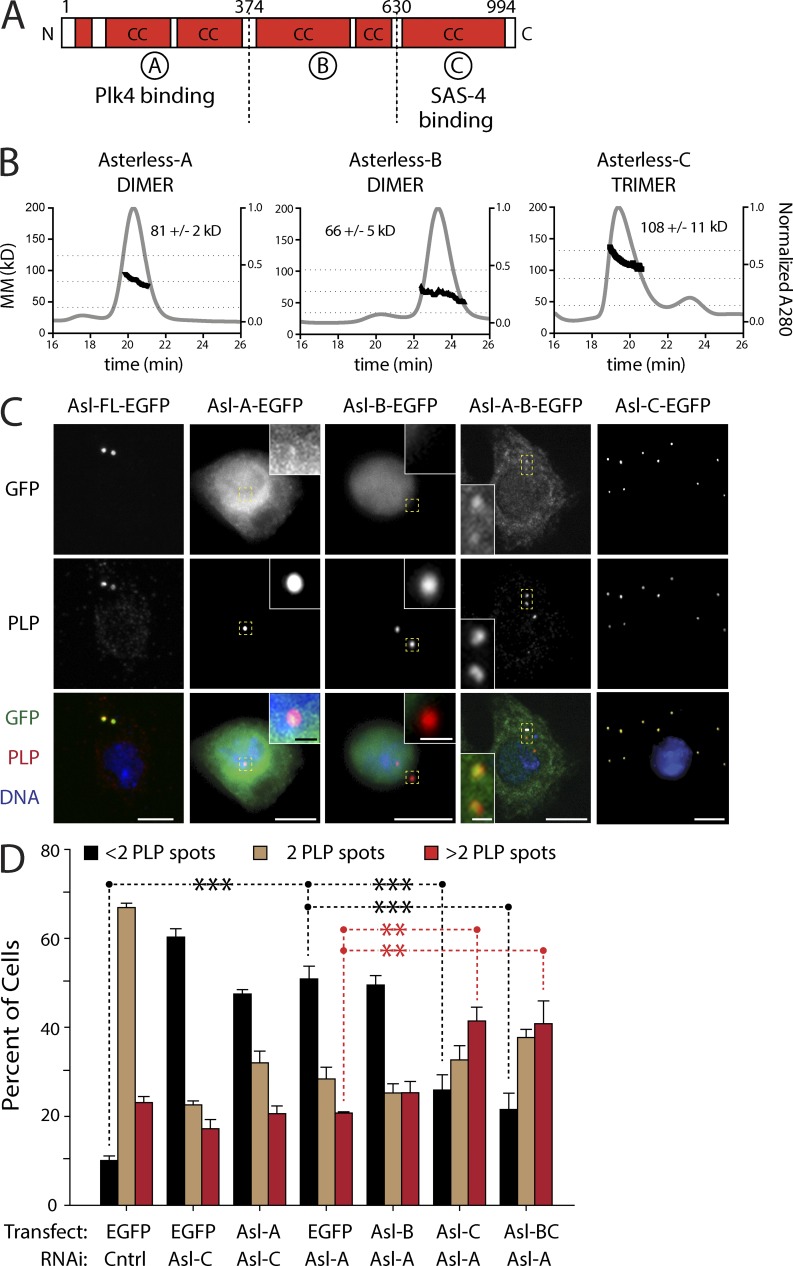
The Drosophila melanogaster protein Asterless (Asl) is required for centriole duplication and its overexpression also induces centriole overduplication and de novo centriole assembly (Varmark et al., 2007; Blachon et al., 2008; Dzhindzhev et al., 2010; Stevens et al., 2010). Notably, the Asl human orthologue, Cep152, is linked to microcephaly (MCPH9) and Seckel syndrome (SCKL5; Guernsey et al., 2010; Kalay et al., 2011). Asl/Cep152 is a large protein containing extensive coiled-coil regions and acts as a platform for procentriole assembly by binding several centrosomal proteins including SAS-4/centrosomal P4.1-associated protein (CPAP), Cep63, Cep192, and Plk4 (Dzhindzhev et al., 2010; Hatch et al., 2010; Cizmecioglu et al., 2010; Sir et al., 2011; Sonnen et al., 2013). Previous studies identified three distinct scaffolding domains within Asl, which we refer to as Asl-A, -B, and -C (Fig. 1 A; Cizmecioglu et al., 2010; Dzhindzhev et al., 2010; Hatch et al., 2010). The C-terminal Asl-C region associates with the centriolar outer-surface protein SAS-4/CPAP, whereas a large central region in Cep152 binds Cep192 (Spd-2 in Drosophila and Caenorhabditis elegans). The N-terminal Asl-A region directly binds the central tandem Polo Box cassette (PB1-PB2) within Plk4. Asl is thought to bind Plk4 and target it to the centriole surface via its association with SAS-4/CPAP to initiate procentriole assembly (Dzhindzhev et al., 2010; Cizmecioglu et al., 2010). Thus, in Asl-depleted Drosophila cells, Plk4 fails to localize to centrioles and centrioles do not duplicate (Dzhindzhev et al., 2010).
Figure 1.
The Asl-C region is sufficient for centriole duplication, whereas the Asl-A region is not. (A) Linear map of the Drosophila Asl polypeptide showing functional and structural domains. Red boxes indicate regions of predicted coiled coils (CC). Asl is divided into three previously identified scaffolding domains (A, B, and C). Asl-A binds Plk4 and Asl-C binds SAS-4. (B) Asl fragments oligomerize. Three consecutive His6-tagged Asl fragments: Asl-A (aa 1–357), Asl-B (aa 358–625), and Asl-C (aa 626–994) were analyzed using SEC-MALS. Normalized A280 is shown in gray (y axis at right), and elution time is indicated in minutes (x axis). Calculated MM was determined for each elution peak and is represented by the dark trace (y axis at left, n = 3 experiments). Mean molecular masses (±SD) for the traces are indicated. Horizontal dashed lines indicate the molecular masses of hypothetical monomeric, dimeric, and trimeric species for each fragment. (C) Asl-C contains the centriole-targeting domain. S2 cells were depleted of endogenous Asl for 3 d using RNAi targeting the 5′–3′UTR and transfected with the indicated inducible Asl-EGFP construct. While under continuous RNAi treatment, transfected cells were allowed to recover for 24 h, induced to express for an additional 24 h, and then immunostained for centrioles using the anti-PLP antibody. Representative images of localization patterns of the Asl fragments are shown (n = 100 cells scored/construct). Insets show centrioles (yellow boxes) at higher magnification. Bars: (main images) 5 µm; (insets) 0.5 µm. (D) Replacement of endogenous Asl with Asl-BC or Asl-C rescues centriole duplication and promotes centriole amplification. S2 cells were depleted of endogenous Asl using the indicated exon-targeting RNAi for 3 d and transfected with the indicated inducible Asl-EGFP construct (or control EGFP). While under continuous RNAi treatment, transfected cells were allowed to recover for 24 h and induced to express for an additional 3 d, and centrioles were visualized by anti-PLP immunostaining. Each bar shows the mean percentage of cells containing the indicated number of centrioles (n = 3 experiments; 300 cells counted per treatment, per experiment). Asterisks mark significant differences (relative to control) for selected comparisons. **, 0.01 > P ≥ 0.001; ***, P < 0.001. Error bars indicate SEM. Cell lysates were immunoblotted to verify expression of the Asl-EGFP fragments and knockdown of endogenous Asl (Fig. S1 D). Cntrl, control.
Endogenous Plk4 protein is nearly undetectable in cells because it efficiently promotes its own destruction (Cunha-Ferreira et al., 2013; Klebba et al., 2013). This observation raises a perplexing question: if the role of Asl is only to target Plk4 to centrioles, how does Asl overexpression induce centriole amplification? Inspired by this question, we sought to identify new Asl functions by performing a comprehensive analysis of the interaction between Plk4 and Asl. We have discovered that a second Plk4-binding domain in the Asl C-terminal region (Asl-C) is sufficient to induce centriole amplification. Furthermore, mechanistic experiments demonstrate that Asl is an important regulator of Plk4 levels; Asl-A primarily promotes Plk4 dimerization and facilitates its degradation, whereas Asl-C stabilizes Plk4 to promote centriole amplification. Collectively, Asl possesses two functionally distinct Plk4-binding domains with opposing activities and acts not only to shuttle Plk4 to centrioles but forms a stabilizing complex during mitosis to prevent Plk4 degradation.
Results
Asl forms a trimer and its C-terminal region targets centrioles
The oligomeric state of the scaffolding protein Asl is unknown. To better understand the functional interactions between Asl and its binding partners, we first characterized its ability to oligomerize. First, full-length (FL) Asl-EGFP from S2 cell lysates retrieved endogenous Asl by coimmunoprecipitation (Fig. S1 A). Second, yeast two-hybrid (Y2H) analysis of the three Asl functional domains (A, B, and C; Fig. 1 A) revealed that Asl-B and Asl-C self-interact (Fig. S1 B). Because Asl-A autoactivates in the Y2H system, we were unable to determine whether it self-associates. Interestingly, Asl-B can associate with Asl-C (Fig. S1 B), suggesting that Asl may adopt a folded conformation as different regions form intramolecular interactions. Third, size-exclusion chromatography with multiangle laser light scattering (SEC-MALS) was used to determine the oligomeric states of the Asl domains (Fig. 1 B). Results suggested that both Asl-A and Asl-B form homodimers (predicted Asl-A monomer molecular mass [MM], 43 kD; dimer MM, 86 kD; predicted Asl-B monomer MM, 31 kD; dimer MM, 62 kD), whereas Asl-C assembles as a homotrimer (predicted monomer MM, 43 kD; trimer MM, 129 kD). Indeed, computer analysis using MultiCoil identified several distinct regions in Asl-C likely to form a trimeric structure (Fig. S1 C). Thus, our findings suggest that Asl homo-oligomerizes via spatially distinct two- and three-stranded coiled-coil regions.
Next, we determined the intracellular localization of each Asl fragment. Because Asl oligomerizes, we eliminated the influence of endogenous Asl on the binding and localization of the Asl fragments by first depleting cells of Asl (Fig. S1 D). After 3 d of RNAi knockdown, we transiently expressed each fragment as well as FL Asl (Fig. 1 C). Asl-A and Asl-AB were largely diffuse and cytoplasmic and weakly localized to centrioles. Surprisingly, Asl-B localization was restricted to the nucleus. However, Asl-C efficiently colocalized with the centriole marker pericentrin-like protein (PLP), as did Asl-FL and an Asl-BC fragment (unpublished data). Thus, the C terminus of Asl is required to target centrioles, as reported for its orthologue Cep152 (Cizmecioglu et al., 2010).
Replacement of endogenous Asl with Asl-C rescues centriole duplication
Asl overexpression triggers centriole amplification (Dzhindzhev et al., 2010; Stevens et al., 2010), although the mechanism for this is unclear. Therefore, we next addressed whether a specific region in Asl is sufficient for this activity by expressing Asl fragments (in the presence of endogenous Asl) and measuring centriole numbers (Fig. S2 A). Because of their small size, mother and daughter centrioles cannot be distinguished within an engaged pair using standard light microscopy in most fly cells. However, by immunostaining for PLP, a protein recruited to the outer surface of mature centrioles (Fu and Glover, 2012; Mennella et al., 2012), we can accurately measure centriole loss (less than two spots) and amplification (greater than two spots) in these cells (Dzhindzhev et al., 2010; Brownlee et al., 2011). Consistent with a previous study, expression of FL Asl induced centriole amplification (Dzhindzhev et al., 2010) compared with controls. Asl-A and Asl-B expression did not have this effect. Strikingly, Asl-C expression induced centriole amplification at levels similar to FL Asl.
It was surprising that Asl-C expression induced centriole amplification because the only known Plk4-binding domain is in Asl-A. However, because transgenic Asl binds endogenous Asl (Fig. S1 A), Asl-C could amplify centrioles by influencing Plk4 through its association with endogenous Asl. Therefore, we performed “replacement” experiments by depleting endogenous Asl (Fig. S1 D), transfecting with inducible Asl-EGFP fragments, and then measuring centriole numbers after an additional 3 d of expression. (Levels of transgenic proteins are compared with endogenous Asl levels in Fig. S1, E and F.) As expected, Asl depletion caused significant centriole loss (less than two centrioles) compared with controls (Fig. 1 D). Replacement with either Asl-A or Asl-B could not rescue this defect. Strikingly, Asl replacement with Asl-BC or Asl-C not only significantly rescued centriole loss but also lead to significant overduplication (Fig. 1, C and D, last two sets of bars). Thus, our findings indicate that Asl-C expression alone is sufficient to rescue centriole duplication and induce centriole amplification in Asl-depleted cells. Moreover, Asl-A is expendable with regard to centriole duplication, a remarkable result given that Asl-A contains the known Plk4-binding domain thought to position Plk4 at centrioles (Dzhindzhev et al., 2010).
Asl stabilizes Plk4 through Asl-C
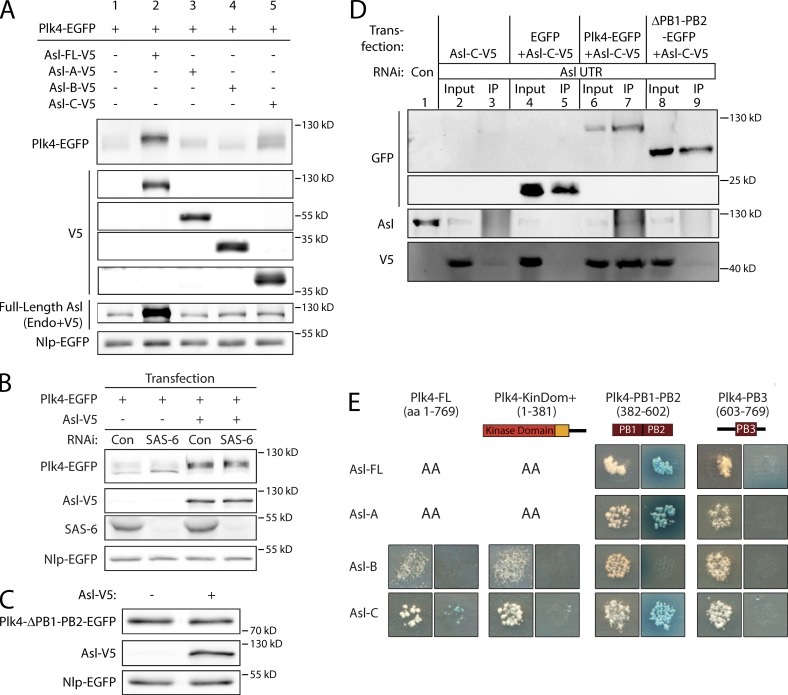
Given these results, we hypothesized that Asl possesses multiple mechanisms to regulate Plk4 activity. Possibly, Asl not only shuttles Plk4 to centrioles but also stabilizes Plk4 by suppressing its degradation. To test these hypotheses, we expressed Plk4-EGFP with either FL Asl or the Asl fragments and analyzed Plk4 levels. As expected, Plk4-EGFP was not easily detectable as a result of its ability to autodestruct (Fig. 2 A, lane 1). However, coexpression with Asl-FL resulted in a marked rise in Plk4 level as well as an upward shift in its electrophoretic mobility (Fig. 2 A, lane 2). This stabilizing effect was not caused simply by an increase in centrioles, as Plk4-EGFP stability was also observed in Asl-FL-V5–expressing cells that were depleted of the essential centriole protein SAS-6 (Fig. 2 B, lane 4), which efficiently reduced centriole numbers in these cells (Fig. S2 B). The reduced electrophoretic mobility of Plk4 raises the interesting possibility that Asl stabilizes Plk4 in a phosphorylated state, perhaps by stimulating Plk4 autophosphorylation. Indeed, λ phosphatase treatment of Plk4 immunoprecipitated from cells coexpressing Asl shifted Plk4 to faster migrating species, suggesting that Asl may promote Plk4 autophosphorylation (Fig. S2 C). Coexpression of Asl-A or Asl-B did not alter Plk4 levels (Fig. 2 A, lanes 3 and 4). To eliminate the possibility that endogenous Asl is necessary for Asl-C to stabilize Plk4, we depleted endogenous Asl in cells expressing Plk4-EGFP alone or Plk4-EGFP with Asl-C. Asl-C significantly stabilized Plk4 in the presence or absence of endogenous Asl relative to Plk4 expression alone (Fig. S2 D). These findings demonstrate a new role for Asl in stabilizing Plk4 and provide a mechanism explaining how Asl expression promotes centriole amplification. Moreover, Asl-C is largely responsible for this activity.
Figure 2.
Asl contains a second Plk4 PB1-PB2–binding domain in its C terminus that is sufficient for stabilizing Plk4. (A) Asl stabilizes Plk4 primarily through its C terminus. The relative stability of Plk4-EGFP protein was analyzed by immunoblotting lysates of S2 cells transiently coexpressing the indicated inducible Asl-V5 constructs. Cotransfected Nlp-EGFP was expressed under its endogenous promoter and served as a loading control. Blots were probed with anti-GFP, V5, and Asl. In lane 2, FL Asl-V5 and endogenous (Endo) Asl are not resolvable, so the band contains both species. (B) Asl stabilizes Plk4 in cells lacking centrioles. S2 cells were control or SAS-6 RNAi treated for 6 d. On day 4, cells were transfected with inducible Plk4-EGFP alone or with Asl-V5 and then induced to express the next day for 24 h. Proteins were detected by immunoblotting cell lysates with anti-GFP, V5, and SAS-6 antibodies. Cotransfected Nlp-EGFP was used as a loading control. Con, control. (C) Asl stabilization of Plk4 requires PB1-PB2. The relative protein stability of a Plk4-EGFP mutant lacking PB1-PB2 was analyzed by immunoblotting lysates of S2 cells that were or were not transiently coexpressing Asl-V5. Cotransfected Nlp-EGFP was used as a loading control. (D) PB1-PB2 is necessary for Plk4 to associate with Asl-C. S2 cells were control or Asl 5′–3′UTR RNAi treated for 7 d. On day 5, cells were cotransfected with inducible Asl-C–V5 and the indicated inducible Plk4-EGFP construct (or control EGFP) and induced to express the next day for 24 h, and then lysates were prepared for anti-GFP immunoprecipitation. Immunoblots were probed for V5, GFP, and endogenous Asl. IP, immunoprecipitation. (E) Asl-A and Asl-C interact specifically with PB1-PB2 in Plk4 by Y2H analysis. FL and fragments of Asl and Plk4 were screened by Y2H. In each image, colonies from replica plating are shown. (left image) Growth indicates the presence of both bait and prey. (right image) Growth on DDOXA, and color indicates an interaction. AA indicates that one or both protein fragments autoactivated the Y2H reporters on their own and could not be tested. Both Asl-A and Asl-C strongly interacted with PB1-PB2 but not PB3. KinDom, kinase domain.
Asl contains a second Plk4-binding domain in Asl-C
Our finding that Asl-C stabilizes Plk4 and induces centriole amplification suggests that Asl contains a previously unidentified Plk4-binding domain in the C-terminal region. First, we tested whether Plk4 stabilization requires direct interaction with Asl by coexpressing Asl with a Plk4 mutant lacking the critical Asl-binding domain, Polo boxes 1 and 2 (Plk4-ΔPB1-PB2-EGFP). Asl expression had no effect on the level of the ΔPB1-PB2 mutant (Fig. 2 C). Therefore, the Plk4 Asl-binding domain (PB1-PB2) is required for Asl to stabilize Plk4, suggesting that Asl directly interacts with Plk4.
To test whether Asl-C binds PB1-PB2, we used immunoprecipitations, in vitro protein binding assays, and Y2H analysis. S2 cells depleted of endogenous Asl were induced to express Asl-C alone or Asl-C plus either control EGFP or different Plk4-EGFP constructs (Fig. 2 D). Western blots of the immunoprecipitated GFP-tagged proteins (Fig. 2 D, lanes 5 and 7) demonstrated that Asl-C readily bound FL Plk4, whereas deletion of PB1-PB2 abolished the interaction (Fig. 2 D, lanes 7 and 9). Therefore, Asl-C binds Plk4 and PB1-PB2 is necessary for this interaction. Furthermore, GST pull-down assays showed that, similar to Asl-A, Asl-C bound to PB1-PB2 in vitro (Fig. S2, E and F).
Lastly, interactions between Asl and Plk4 were examined by Y2H, which included both FL proteins, Asl fragments, and Plk4 functional domains (Fig. 2 E). As expected, both Asl-FL and Asl-A strongly interacted with PB1-PB2 but not with the last, C-terminal Plk4 Polo box (PB3). Similarly, Asl-C associated with Plk4-FL and strongly interacted with PB1-PB2 but not PB3. No binding was detected between Asl-B and Plk4. Collectively, our results demonstrate that Asl contains two distinct domains at either end of the protein that directly bind Plk4.
The Plk4-binding domain in Asl-C is important for centriole duplication
Because an Asl-BC construct can effectively replace endogenous Asl and promote centriole duplication, we next asked whether the Plk4-binding domain in Asl-C is necessary for this function. Using random PCR-based mutagenesis, we generated five Asl-C mutants that fail to interact with PB1-PB2 but that still bind Asl-B (Fig. S3 A). One of these mutants, J11F10 containing three missense mutations (E698G, E789D, and S906P), was chosen for further characterization. The J11F10 mutations specifically blocked the interaction of Asl with Plk4 but did not interfere with its binding to SAS-4 (Fig. S3 A). Moreover, we examined its ability to interact with Spd-2 because Cep152 binds Cep192 (the human orthologues of Asl and Spd-2, respectively; Sonnen et al., 2013). We found that Asl-C also strongly interacts with the C-terminal half of Spd-2 (Fig. S3 A) and that the J11F10 point mutations do not inhibit this association (Fig. S3 A). In addition, the J11F10 mutations do not prevent centriole targeting because a construct of EGFP–Asl-BC containing the J11F10 mutations still localized to centrioles (Fig. S3 B).
Lastly, recent work in human cells has shown that Plk4 PB1-PB2 also binds Cep192, which cooperates with Cep152 to target Plk4 to centrioles (Kim et al., 2013; Sonnen et al., 2013). Using a Y2H assay, we found that Spd-2 did not interact with any fly Plk4 domain (Fig. S3 C). The absence of a Plk4–Spd-2 interaction potentially explains why Plk4 does not localize to centrioles in Asl-depleted Drosophila cells, unlike the situation in Cep152-depleted human cells (Cizmecioglu et al., 2010; Dzhindzhev et al., 2010; Hatch et al., 2010). Thus, Plk4 targeting to centrioles in fly cells appears to be exclusively mediated by two Plk4 binding sites in Asl but not by Spd-2.
We now tested the functional importance of the Plk4-binding domain within Asl-C by performing replacement experiments using an Asl-C construct unable to bind Plk4. Specifically, centrioles were counted in cells depleted of endogenous Asl and expressing either wild-type (WT) or J11F10-mutated Asl-BC. As before, centriole number was significantly decreased after Asl depletion, and this phenotype was rescued by expression of EGFP–Asl-BC (Fig. S3 D). However, expression of EGFP–Asl-BC containing the J11F10 mutations failed to rescue centriole duplication (Fig. S3 D). Thus, the Plk4-binding activity within Asl-C is necessary and sufficient for centriole duplication.
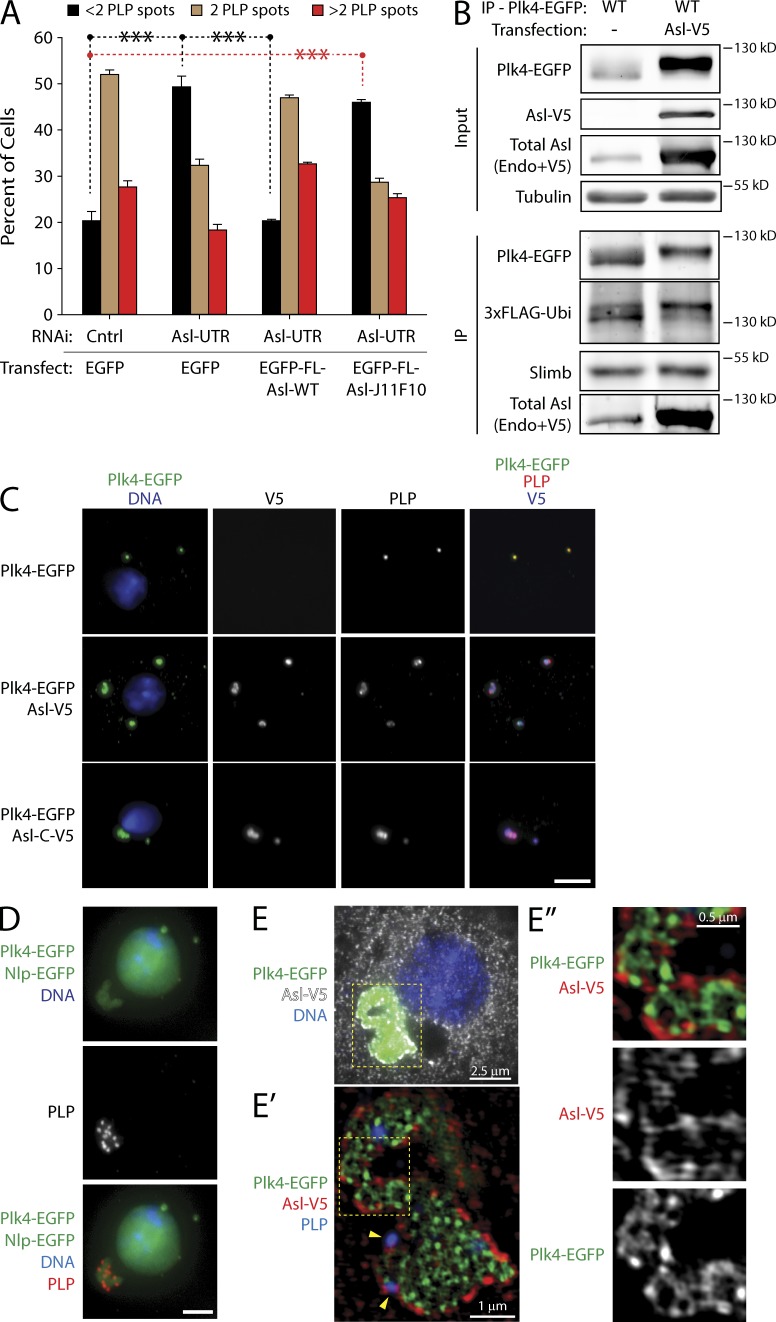
To test whether the two Plk4 binding sites of Asl are functionally redundant, we expressed WT or J11F10-mutated FL Asl in cells that had been depleted of endogenous Asl. As expected, Asl-FL-WT expression rescued centriole loss, but Asl-FL-J11F10 expression did not (Fig. 3 A). Thus, to the extent demonstrable by this assay, the Plk4-binding activity within the Asl C-terminal region is not redundant to the activity within the N-terminal region. In order for centriole duplication to occur properly, Asl requires the C-terminal Plk4-binding domain.
Figure 3.
Asl stabilizes Plk4 in aggregates with supernumerary centrioles. (A) Disruption of the C-terminal Plk4-binding domain with the J11F10 mutation compromises centriole duplication. S2 cells were transfected with inducible EGFP-FL-Asl, J11F10 mutant, or EGFP and either control or Asl depleted using Asl UTR RNAi for 6 d. Cells were anti-PLP immunostained, and centriole numbers were measured. Each bar shows the mean percentage of cells containing the indicated number of centrioles (n = 5 experiments; 200 cells counted per treatment, per experiment). Asterisks mark significant differences between treatments. ***, P < 0.001. Error bars indicate SEM. Cntrl, control. (B) Asl stabilizes Plk4 but does not prevent its ubiquitination. Anti-GFP immunoprecipitates (IPs) were prepared from lysates of S2 cells transiently co-overexpressing Plk4-EGFP and 3×FLAG-ubiquitin (Ubi) and either with or without Asl-V5. Volumes of the IPs were adjusted to load an equivalent amount of Plk4 for each immunoblot. Blots were probed for α-tubulin, GFP, V5, and Asl. Endo, endogenous. (C) Plk4-EGFP forms aggregates with Asl or Asl-C that recruit PLP. S2 cells were transfected with inducible Plk4-EGFP either alone, with Asl-V5, or with Asl-C–V5, and then induced to express the next day for 24 h. Cells were immunostained for PLP. Bar, 5 µm. (D) S2 cell coexpressing Plk4-EGFP and Asl-V5 was immunostained for PLP to mark centrioles. Coexpressing Nlp-EGFP (a nuclear protein) was used to identify transfected cells. Bar, 2.5 µm. (E–E″) An S2 cell coexpressing Plk4-EGFP and Asl-V5 (white in E) was imaged using superresolution microscopy. Cells were immunostained for V5 to mark Asl-V5. (E′ and E″) Higher magnifications of yellow boxed regions show PLP-labeled centrioles (indicated with yellow arrowheads) and ring-like Asl and Plk4 structures.
Asl stabilizes Plk4 by aggregation and suppresses Plk4 turnover
Our results demonstrate that Asl stabilizes Plk4 but the mechanism is unclear. One possibility is that Asl binding to PB1-PB2 could prevent recognition by the SCFSlimb Ubi ligase and, thus, ubiquitination of Plk4. To test this, we expressed Plk4-EGFP with Asl-V5 in cells cotransfected with 3×FLAG-Ubi. As shown previously (Rogers et al., 2009), immunoprecipitation of Plk4 retrieves endogenous Slimb, and Plk4 is robustly labeled with FLAG-Ubi (Fig. 3 B, lane 1). Coexpression with Asl dramatically increased Plk4 protein levels (Fig. 3 B, lane 2, input) but, surprisingly, did not affect Slimb binding or ubiquitination (Fig. 3 B, lane 2, IP). Thus, Asl does not inhibit Slimb-mediated ubiquitination of Plk4. Moreover, the association with Slimb suggests that stabilized Plk4 is catalytically active, capable of trans-autophosphorylating to recruit Slimb.
Transiently expressed Plk4-EGFP is normally observed at centrioles (Fig. 3 C). However, when Asl is coexpressed with Plk4, cells contained large aggregates of both proteins and PLP (Fig. 3 C). Indeed, 80% of coexpressing cells contained Plk4 aggregates with PLP (Fig. S3 E). Aggregates were not simply the result of Plk4 overabundance because expression of stable, nondegradable Plk4 did not generate such aggregates (Klebba et al., 2013). Moreover, Asl-C, which is sufficient to stabilize Plk4 (Fig. 2 A), also induced formation of identical Plk4 aggregates (Fig. 3 C). Notably, these complexes contained numerous PLP spots (Fig. 3 D). Although both Plk4 and Asl form a partially overlapping ring–link network within the aggregate, Asl also concentrates at the periphery of the structure (Fig. 3 E). We note that centrioles in cells with Asl–Plk4 aggregates display distorted PLP structures (Fig. S4, A and B). Possibly, Plk4–Asl aggregates act as sinks for centriolar proteins, reducing their availability and promoting defects in centriolar structure. Therefore, coexpression of Asl and Plk4 promotes the formation of complexes containing centriolar proteins and, at least in the case of Plk4, can stabilize the ubiquitinated protein. Though we acknowledge that these aggregates are not physiological, the arrangement of Asl as a shell enclosing a bulk of Plk4 suggests that Asl has the ability to sequester Plk4 into a stable complex.
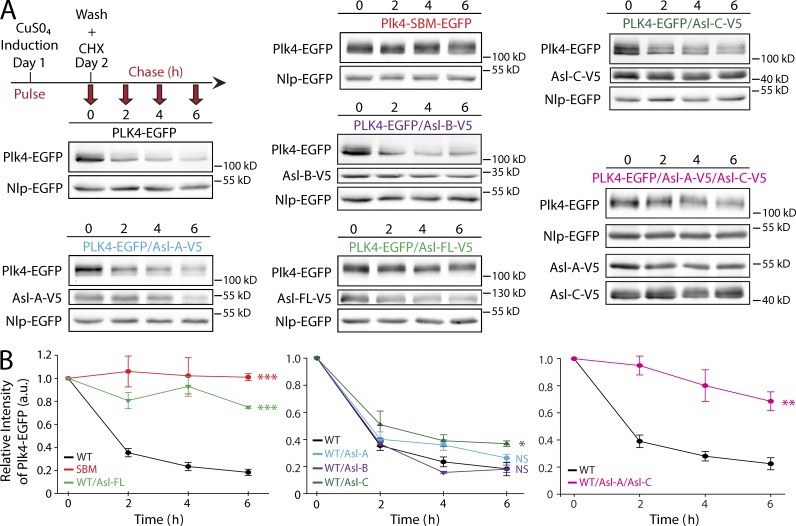
If the Asl–Plk4 complexes protect Plk4 from proteasome-mediated degradation, this might explain the observed accumulation of Plk4 (Fig. 2 A). To test this, Plk4-EGFP expression was induced overnight, and then, cycloheximide (CHX) was introduced to block further protein synthesis. Plk4 protein levels were then assayed every 2 h (Fig. 4 A). Plk4 is relatively short lived: 50% of Plk4 was eliminated <2 h after CHX addition, and only ∼20% remained at the 6 h time point (Fig. 4 B). Protein levels of the control (nondegradable Plk4–Slimb-binding mutant [SBM]–EGFP) were constant throughout the time course (Fig. 4, A and B). Strikingly, coexpression of Asl-FL significantly stabilized Plk4. By 6 h after CHX addition, ∼80% of the Plk4 protein remained. To determine which Asl region decreased Plk4 turnover, we repeated the assay with the three Asl fragments (Fig. 4 A). Whereas Asl-A and Asl-B did not affect the kinetics of Plk4 degradation, Asl-C expression significantly slowed Plk4 turnover rate but not nearly as dramatically as Asl-FL (Fig. 4 B). Neither Asl-A nor Asl-C were nearly as efficient as Asl-FL in stabilizing Plk4, suggesting that Asl-C has a role in forming an Asl–Plk4 stabilizing complex but that the Plk4 binding site within Asl-A may work together with Asl-C for maximal suppression of Plk4 turnover. To test this, we coexpressed both Asl-A and Asl-C together with Plk4. The two fragments suppressed Plk4 turnover to a similar extent as Asl-FL (Fig. 4, A and B). Thus, Asl-A and Asl-C act synergistically to stabilize Plk4 without residing on the same polypeptide.
Figure 4.
FL Asl or Asl-C suppresses Plk4 turnover. (A) S2 cells cotransfected with Plk4-EGFP or nondegradable Plk4 Slimb-binding mutant (SBM) and the indicated Asl-V5 constructs were induced to express for 24 h and then treated with fresh media and cycloheximide (CHX) to inhibit protein translation. Immunoblots of cell lysates were probed for GFP and V5. Plk4-EGFP protein turnover over a 6-h time course was analyzed by densitometry of the anti-GFP immunoblots. (Cotransfected Nlp-EGFP was used as a loading control.) (B) Graphs show relative Plk4-EGFP levels for each treatment normalized against Nlp-EGFP over the 6-h time course for three experiments. Error bars show SEM. Expression of Asl-V5 or Asl-C–V5 suppresses Plk4-EGFP degradation. Asterisks indicate significant differences between a treatment and the WT control at 6 h. *, 0.05 > P ≥ 0.01; **, 0.01 > P ≥ 0.001; ***, P < 0.001; NS, not significant. a.u., arbitrary unit.
Asl promotes Plk4 autophosphorylation
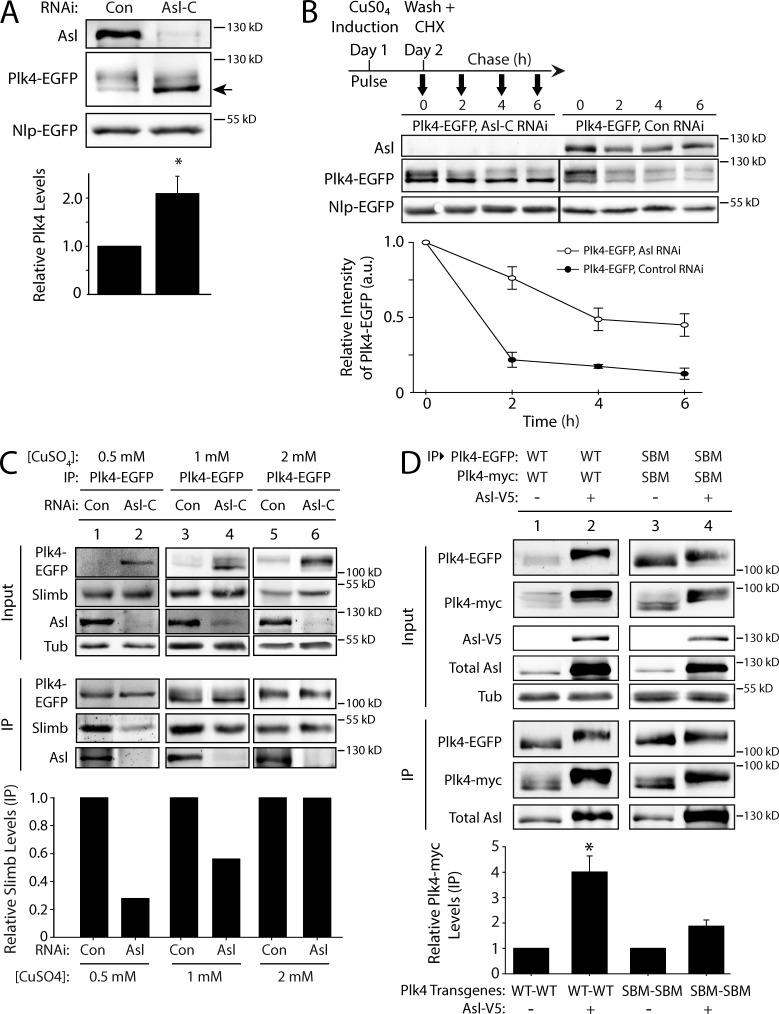
Because Asl expression stabilizes Plk4, we predicted that Asl depletion would decrease Plk4 protein levels. Surprisingly, Asl depletion doubled Plk4 levels (Figs. 5 A and S2 D). The explanation for this unexpected result may lie in the change of Plk4’s electrophoretic mobility after Asl depletion. Plk4 normally runs as a multibanded smear on SDS-PAGE (Fig. 5 A, lane 1) but collapses to a single band if mutated to kinase dead (Klebba et al., 2013) or if treated with a phosphatase (Fig. S2 C). Therefore, the multiple bands likely correspond to multiple phosphorylation states. Depletion of Asl altered the electrophoretic mobility of Plk4, increasing the proportion of Plk4 present in a focused, fast-migrating band, which likely corresponds to a low- or nonphosphorylated isoform (Fig. 5 A, lane 2, arrow). Since previous studies have shown that Plk4 phosphorylation is a prerequisite for Slimb-mediated degradation (Cunha-Ferreira et al., 2009; Rogers et al., 2009), it follows that Plk4 levels would increase if its autophosphorylation is suppressed by Asl depletion.
Figure 5.
Asl functions to promote Plk4 dimerization and autophosphorylation. (A) Asl increases the phosphorylation state of Plk4. S2 cells were control or Asl depleted for 5 d. On day 3, cells were transfected with Plk4-EGFP, and the next day, they were induced to express for 24 h. Immunoblots of cell lysates were probed for Asl and GFP. Cotransfected Nlp-EGFP was used as a loading control. Asl depletion increases the proportion of Plk4-EGFP with higher electrophoretic mobility (arrow), indicating decreased Plk4 phosphorylation. The graph shows relative amounts of Plk4-EGFP as determined by densitometry of the anti-GFP immunoblots, normalized to Nlp-EGFP, and plotted relative to control (lane 1). n = 6 experiments. (B) Plk4 turnover is suppressed in the absence of Asl. S2 cells were control or Asl depleted for 7 d. On day 5, cells were cotransfected with Plk4-EGFP and Nlp-EGFP (loading control) and induced the next day to express for 24 h. Cycloheximide (CHX) treatment was performed as indicated in the schematic and as described in Fig. 4 A. Immunoblots of cell lysates were probed for GFP and Asl. Plk4-EGFP protein levels were analyzed by densitometry of the anti-GFP immunoblots. Graphs show relative Plk4-EGFP levels for each treatment normalized against Nlp-EGFP for two experiments. (C) The impact of Asl on Plk4 autophosphorylation and Slimb binding can be masked by high levels of Plk4 expression. S2 cells were control or Asl-RNAi treated for 7 d. On day 5, cells were transfected with Plk4-EGFP, and the next day, they were induced to express using three different concentrations of CuSO4 for 24 h. Anti-GFP immunoprecipitates (IPs) were then prepared from lysates, and immunoblots were probed for GFP, Slimb, Asl, and α-tubulin. Graph shows relative amounts of Slimb as determined by densitometry of the anti-Slimb immunoblots, normalized to Plk4-EGFP. Data shown are from a single representative experiment (n = 2 independent experiments). (D) Asl increases Plk4 dimerization. Anti-GFP immunoprecipitates were prepared from lysates of S2 cells transiently coexpressing Plk4-EGFP and Plk4-myc and with or without Asl-V5. Blots of the input lysates and immunoprecipitates were probed for α-tubulin, GFP, V5, Asl, and myc. The graph shows relative amounts of Plk4-myc bound to Plk4-EGFP. For each treatment, levels of tagged Plk4 in the immunoprecipitates were determined by densitometry of the anti-GFP and myc immunoblots, normalized to measure Plk4-EGFP, and then plotted relative to control (lane 1). Asterisks mark significant difference compared with control. *, 0.05 > P ≥ 0.01. n = 3–4 experiments per treatment. Error bars indicate SEM. a.u., arbitrary unit; Con, control; Tub, tubulin.
The previous result was further supported by measuring Plk4 turnover in Asl-depleted cells. Compared with controls, Asl depletion significantly slowed Plk4 turnover (P = 0.0236 at 6 h), increasing Plk4 stability (Fig. 5 B). Strikingly, not all of the Plk4-phosphorylated isoforms are equally stabilized by Asl depletion: the low-phosphorylated Plk4 isoform preferentially accumulated in the absence of Asl. Thus, Asl normally increases Plk4 turnover, probably by facilitating its phosphorylation.
Asl facilitates Plk4 homodimerization
Homodimers of Plk4 trans-autophosphorylate to generate a Slimb-binding phosphodegron, thereby promoting its own destruction (Holland et al., 2010; Guderian et al., 2010; Cunha-Ferreira et al., 2013; Klebba et al., 2013). Our findings suggest that Asl increases Plk4 autophosphorylation and degradation. In contrast, previous studies found that Asl does not regulate Plk4 levels (Cizmecioglu et al., 2010; Dzhindzhev et al., 2010). However, it is possible that high levels of exogenous Plk4 expression might have obscured the effect of Asl on Plk4 regulation. To test this, we induced Plk4-EGFP expression at a range of concentrations in control and Asl-depleted S2 cells (Fig. 5 C). Plk4-EGFP protein was immunoprecipitated from cell lysates, and quantitative immunoblotting was used to measure the amount of associated Slimb, which only binds trans-autophosphorylated Plk4 (Guderian et al., 2010; Cunha-Ferreira et al., 2013; Klebba et al., 2013). At low expression, Plk4 was nearly undetectable in control lysates (Fig. 5 C, lane 1, Input) but readily pulled down Slimb (Fig. 5 C, lane 1, IP). However, at the same level of induction, Slimb binding was sharply decreased (and Plk4 protein in the lysate increased) by depletion of Asl (Fig. 5 C, lane 2). Increasing Plk4 expression moderately (Fig. 5 C, lanes 3 and 4) or to higher levels (Fig. 5 C, lanes 5 and 6) diminished the effect of Asl on Plk4 autophosphorylation (as indicated by the increasing levels of bound Slimb in Asl-depleted cells). Thus, at low Plk4 levels, Asl plays an important role in facilitating the Plk4–Slimb interaction, revealing a new layer of complexity to Plk4 regulation and Asl function in cells.
Plk4 homodimerization is mediated by interactions between the PB1-PB2 domains (Slevin et al., 2012), which also bind Asl. We hypothesized that Asl facilitates Plk4 autophosphorylation by interacting with multiple Plk4 monomers, thus increasing the probability of dimerization. Alternatively, Asl may stabilize existing dimers, thereby promoting Plk4 trans-autophosphorylation and degradation. To test the effect of Asl on Plk4 dimerization, we coexpressed Plk4-EGFP and Plk4-myc in S2 cells and measured the amount of Plk4-myc in the anti-GFP immunoprecipitate. As expected, Plk4-myc binds Plk4-EGFP and is readily recovered (Fig. 5 D, lane 1). However, coexpression of Asl in these cells significantly increased the amount of bound Plk4-myc by approximately fourfold (Fig. 5 D, lane 2). To eliminate the possibility that the increased dimerization was simply a result of increased Plk4 levels as a result of Asl expression, we performed the same assay using nondegradable (SBM) Plk4 mutants. A modest increase in dimerization was obtained with Plk4-SBM (Fig. 5 D, lanes 3 and 4), likely because high Plk4 levels are not as reliant on Asl for dimerization (Fig. 5 C). Thus, these results support the hypothesis that Asl stabilizes Plk4 dimers.
Asl-A promotes Plk4 homodimerization and autophosphorylation
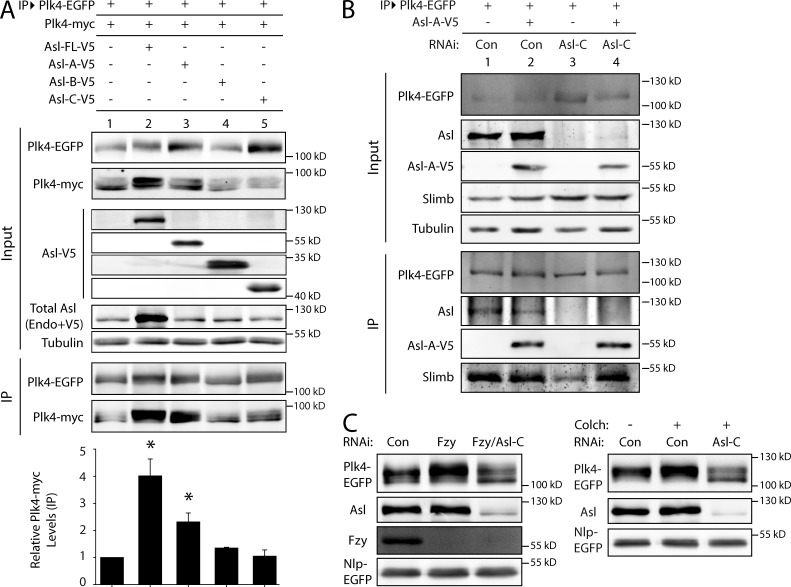
To identify which Asl region increases Plk4 dimerization, we performed dimerization assays using Asl fragments or Asl-FL. Expression of Asl-A significantly increased dimerization, as did Asl-FL expression (Fig. 6 A, lanes 2 and 3). Expression of either Asl-B or Asl-C did not affect Plk4 dimerization (Fig. 6 A, lanes 4 and 5). Therefore, Asl-A increases Plk4 dimerization.
Figure 6.
Asl-A is sufficient to promote Plk4 dimerization and autophosphorylation, and Asl is required for stabilizing Plk4 during mitosis. (A) Asl-A is sufficient to induce Plk4 dimerization. Anti-GFP immunoprecipitates (IPs) were prepared from lysates of S2 cells transiently coexpressing Plk4-EGFP, Plk4-myc, and full-length (FL) Asl-V5 or the indicated Asl-V5 fragment. Blots of the input lysates and IPs were probed for α-tubulin, GFP, V5, Asl, and myc. The graph shows relative amounts of Plk4-myc bound to Plk4-EGFP. For each treatment, levels of tagged Plk4 in the IPs were determined by densitometry of the anti-GFP and myc immunoblot and then normalized to measure Plk4-EGFP, and the results were plotted relative to control (lane 1). Asterisks mark significant differences compared with control. *, 0.05 > P ≥ 0.01. n = 2–4 experiments per treatment. Error bars indicate SEM. Endo, endogenous. (B) Asl-A is sufficient to facilitate Plk4 autophosphorylation and Slimb binding in the absence of endogenous Asl. S2 cells were control or Asl depleted for 7 d. On day 5, cells were transfected with Plk4-EGFP with or without Asl-A–V5, and the next day, they were induced to express for 24 h. Anti-GFP immunoprecipitates were then prepared from lysates, and immunoblots of the inputs and IPs were probed for GFP, Slimb, Asl, V5, and α-tubulin. (C) Asl is required for stabilizing Plk4 levels during mitosis. S2 cells were arrested in mitosis by Fizzy (Fzy) depletion (left) or 24 h of colchicine (Colch) treatment (right). Cells were control, Asl, Fizzy, or Fizzy/Asl RNAi treated for 6 d. On day 4, cells were transfected with Plk4-EGFP and Nlp-GFP (loading control) and induced to express for 24 h the next day. Cells were treated with colchicine on day 6. Immunoblots of cell lysates were probed for Asl, Fzy, and GFP. Con, control.
If Asl-A promotes Plk4 dimerization, this predicts that it should also promote Plk4 autophosphorylation. We tested whether Asl-A is sufficient to induce Plk4 trans-autophosphorylation using Slimb binding as a readout for Plk4 autophosphorylation in cells. Asl levels were depleted in S2 cells followed by Plk4-EGFP expression with or without Asl-A. (Induction was moderated to generate an expressed Plk4-EGFP level that requires Asl for efficient Slimb recruitment; Fig. 5 C, lanes 1 and 2.) Plk4-EGFP was then immunoprecipitated from cell lysates and probed for associated Slimb. Asl-A did not alter the amount of Slimb bound to Plk4-EGFP in control cells containing endogenous Asl (Fig. 6 B, lanes 1 and 2), inferring that Plk4 autophosphorylation was unchanged. However, when the same assay was performed in a background of depleted endogenous Asl, the decrease in Slimb binding that resulted from Asl depletion was rescued by Asl-A (Fig. 6 B, lanes 3 and 4). These findings suggest that Asl-A promotes Plk4 dimerization and autophosphorylation.
Asl is required to stabilize Plk4 during mitosis
Thus far, our results indicate that Asl contains two separate Plk4-binding domains that exert opposite influences on Plk4 stability. Whereas Asl-C decreases Plk4 turnover, Asl-A facilitates Plk4 dimerization, autophosphorylation, and Slimb binding. These opposing Asl activities could function differentially throughout the cell cycle. Asl-A activity could predominate during interphase to stimulate Plk4 degradation, whereas Asl-C activity might stabilize Plk4 during mitosis (Fode et al., 1996; Rogers et al., 2009). To test whether Asl stabilizes mitotic Plk4, we examined Plk4-EGFP levels in asynchronous (i.e., almost entirely interphase) and mitotic cells that were either Asl depleted or control treated. Plk4-EGFP–expressing cells were arrested in mitosis either by depletion of Fizzy (Pesin and Orr-Weaver, 2008) or by treatment with colchicine. Although both treatments only elevated the mitotic index to ∼25%, Plk4-EGFP levels were markedly increased in mitotic cells as compared with asynchronous culture and were shifted to a higher phosphorylated form (Fig. 6 C), as previously described (Brownlee et al., 2011). Strikingly, Asl depletion decreased Plk4 levels and reduced apparent phosphorylation levels of Plk4. Thus, our findings reveal a new role for Asl in stabilizing Plk4 during mitosis.
Discussion
Our investigations of the interactions between the conserved proteins Asl and Plk4 have revealed new features of this centriole-assembly complex. Previous studies have shown that the Asl/Cep152 N terminus (Asl-A) binds Plk4 on its central tandem Polo Box cassette (PB1-PB2) and shuttles it to centrioles to promote centriole duplication (Cizmecioglu et al., 2010; Dzhindzhev et al., 2010; Hatch et al., 2010). In human cells, Plk4 targeting is more complex because Cep152 cooperates with Cep192 in this function (Kim et al., 2013; Sonnen et al., 2013). Plk4 targeting to centrioles appears to be simpler in Drosophila. We found that Plk4 does not interact with the Cep192 orthologue, Spd-2, and so its targeting relies primarily on Asl. Surprisingly, removal of the Asl-A region, which contains the previously characterized Plk4 binding domain, did not prevent centriole overduplication by overexpressed Asl. This is caused by a second Plk4-binding domain in the Asl C terminus (Asl-C), which also associates with SAS-4 and Spd-2, localizes to centrioles, and is sufficient for their overduplication when overexpressed. It would be intriguing to know whether Cep152 also contains a second Plk4 binding site. If not, then perhaps the multiple functions of Asl have been divided between Cep152 and Cep192, with Cep192 performing the functions allocated to Asl-C.
Asl overexpression induces centriole amplification (Dzhindzhev et al., 2010; Stevens et al., 2010). Our experiments have revealed several new properties of Asl that better explain the control Asl exerts on centriole numbers. Structurally it appears that different regions of Asl collaborate when interacting with Plk4. Our experiments indicate that Asl-C and -B bind, suggesting that Asl adopts a folded conformation with potential important functional consequences. Future studies should be aimed at solving the atomic structure of Asl in complex with PB1-PB2 to determine whether Asl-A and Asl-C compete for the same binding site in Plk4 as observed with Cep152 and Cep192. Intriguingly, Asl expression does not prevent Plk4 ubiquitination. Although it is not clear how Plk4 escapes proteasomal degradation, it is possible that formation of higher-order Asl–Plk4 complexes hinder Plk4 shuttling to proteasomes. Indeed, Plk4 and Asl form aggregates in cells, which recruit PLP. At present, it is not known whether these Plk4–Asl aggregates sequester Plk4 and suppress its turnover or whether aggregation is simply a consequence of Plk4 stabilization.
This study highlights an odd functional duality of Asl. Although Asl overexpression induces centriole amplification by stabilizing Plk4, Asl depletion does not have the opposite effect in asynchronous cells but, instead, increases the levels of a low or nonphosphorylated Plk4 species. We found that Asl-A binds and promotes Plk4 dimerization, thereby facilitating its autophosphorylation and recruitment of Slimb. These opposing Asl activities are likely cell cycle dependent. Most of our experiments examined Asl activity during interphase, when Asl primarily assists in promoting Plk4 dimerization and degradation. However, during mitosis, Plk4 protein levels rise, and it decorates centrioles as an asymmetric spot where it has been proposed to act as a platform to initiate centriole duplication (Rodrigues-Martins et al., 2007; Rogers et al., 2009). Previously, we have shown that in Drosophila PP2A, in complex with its regulatory subunit Twins, counteracts Plk4 autophosphorylation of the Slimb-binding domain to promote Plk4 stabilization during mitosis (Brownlee et al., 2011). Remarkably, the impact of Asl on Plk4 switches during mitosis, and Asl stabilizes mitotic Plk4. An important goal will be to understand the cell cycle–dependent changes in Asl that regulate its ability to either stabilize Plk4 or facilitate its degradation. Possibly, Asl and PP2ATwins collaborate in a functional complex to promote mitotic Plk4 stability. Notably, the N terminus of Cep152 is also a Plk4 substrate (Hatch et al., 2010), and, when Plk4 levels rise during mitosis, it may phosphorylate Asl to modulate this region’s Plk4 stabilization activity. Future studies will be necessary to determine the physiological significance of Asl as a Plk4 substrate.
Lastly, our analysis suggests that Asl forms a complex oligomer with Asl-A and -B regions each homodimerizing and Asl-C forming a homotrimer. Likely, Asl molecules form parallel coiled coils as their N and C termini spatially map to distinct, independent locations on the centriole surface (Mennella et al., 2012). Based on its ability to both dimerize and trimerize, we propose that Asl may exist as a homohexamer, capable of forming higher-order structures that could be enhanced with Plk4 binding (Fig. 7). Accordingly, during interphase, Asl-A dimers bind and promote/maintain Plk4 homodimer status, thereby facilitating Plk4 trans-autophosphorylation and Slimb recruitment. During mitosis, Plk4 is stabilized by PP2ATwins and shuttled to the centriole via Asl where it may be transferred to multiple Asl-C regions, thereby cross-linking the kinase into a stable complex. Interestingly, Asl-C expression alone does not stabilize Plk4 to the same extent as FL Asl. In fact, expression of Asl-A and Asl-C work synergistically to suppress Plk4 turnover.
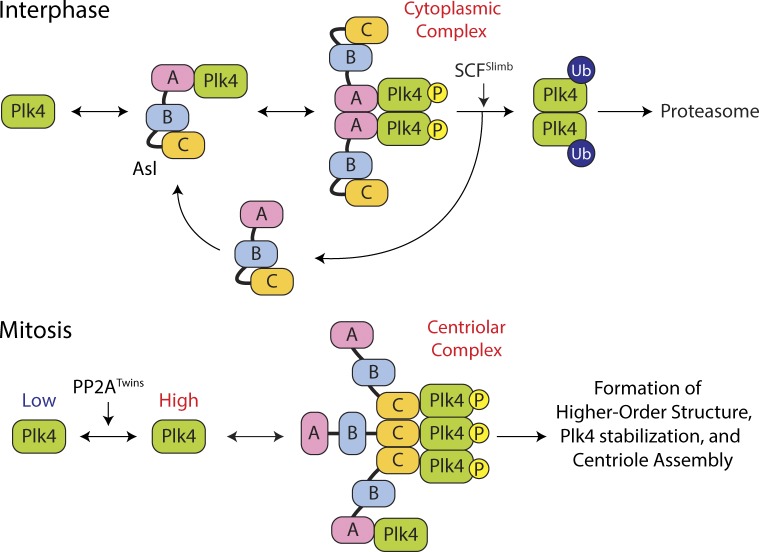
Figure 7.
Model of Plk4 turnover regulation by domain-specific and cell cycle–dependent activities of Asl. (top) A cytoplasmic Asl–Plk4 complex forms during interphase to facilitate Plk4 degradation. Asl-A binding to Plk4 is dominant and facilitates Plk4 homodimerization and autophosphorylation, triggering its ubiquitination by SCFSlimb and degradation. (bottom) A centriolar Asl–Plk4 complex forms during mitosis to stabilize Plk4 and initiate centriole duplication. PP2ATwins counteracts Plk4 autophosphorylation, allowing Plk4 protein levels to rise. Asl-C trimers bind Plk4 dimers, forming high-order structures that protect Plk4 from proteasome-mediated degradation. Asl-A also participates in stabilizing mitotic Plk4, but the mechanism by which this occurs is unclear. Plk4 aggregates into an asymmetric spot on the centriole surface, which modifies the centriole, making it competent to assemble a single daughter during S phase. P, phosphorylation.
It is tempting to speculate that the asymmetric Plk4 spot on mitotic centrioles consists of an organized higher-order Asl–Plk4 assembly, possibly similar to the aggregates observed in Asl–Plk4-overexpressing interphase cells, albeit on a smaller scale. Understanding how Plk4 centriolar spots form and then disassemble during mitotic exit is another important question. Modulating Asl activity may play a crucial role in the event because Plk4 ubiquitination alone may not efficiently remove the spot. We found that the second Plk4-binding domain within the C terminus enlarges the functional repertoire of Asl to include not only Plk4 centriole targeting but also opposing, cell cycle–dependent effects that regulate Plk4 stability and activity. Future studies of the Asl–Plk4 interaction will provide a better understanding of how a mother centriole is mechanistically restricted to the assembly of a single daughter centriole.
Materials and methods
Cell culture and double-stranded RNAi
Drosophila S2 cell culture, in vitro double-stranded RNA (dsRNA) synthesis, and RNAi treatments were performed as previously described (Rogers and Rogers, 2008). In brief, cells were cultured in Sf-900 II serum-free media (Life Technologies). RNAi was performed in 6-well plates. Cells (50–90% confluency) were treated with 5 µg of dsRNA in 1 ml of media and replenished with fresh media/dsRNA every day for 4–7 d. A ∼550-bp control dsRNA was synthesized from DNA template amplified from a non-GFP sequence of the pEGFP-N1 vector (Takara Bio Inc.) using the primers 5′-CGCTTTTCTGGATTCATCGAC-3′ and 5′-TGAGTAACCTGAGGCTATGG-3′ (all primers used for dsRNA synthesis begin with the T7 promoter sequence 5′-TAATACGACTCACTATAGGG-3′). dsRNA was synthesized from cDNA using the primers against the following genes: Slimb, 5′-GGCCGCCACATGCTGCG-3′ and 5′-CGGTCTTGTTCTCATTGGG-3′; SAS-6, 5′-ATGTGGCCTCCAGGGAGC-3′ and 5′-TGATGTTGGCCACATCCCC-3′; Fizzy, 5′-AAACTGCCTTCTTGGACGC-3′ and 5′-ACTCATTCCTGGTTTCCTCTGG-3′; Asl-A targeting exon, 5′-GGAGGAGGAAGAGGCGC-3′ and 5′-GGCGTTCCGCTCCTCCC-3′; and Asl-C targeting exon, 5′-CGTCTGATCCATCGCCC-3′ and 5′-CATCGCCTCTTCGTGGG-3′. dsRNA targeting the Asl UTR was synthesized from an EST template by first removing the Asl cDNA, joining 76 bp of 5′UTR with 114 bp of 3′UTR, and amplified using the primers 5′-GTTGCCTACGAAAATAGCGCC-3′ and 5′-TTTTGTTAGGAATGTACAGCG-3′. Immunoblots confirmed that Asl UTR RNAi depleted endogenous Asl by ∼70–80%, whereas Asl-A and Asl-C RNAi depleted endogenous Asl by over 90%. UTR RNAi was primarily used in replacement experiments, in which Asl-FL was expressed, otherwise Asl-A or Asl-C RNAi was used as indicated.
Immunofluorescence microscopy
For immunostaining, S2 cells were fixed and processed as previously described (Rogers and Rogers, 2008) by spreading S2 cells on concanavalin A–coated, glass-bottom dishes and fixing with 10% formaldehyde. Primary antibodies were diluted to concentrations ranging from 1 to 20 µg/ml. They included rabbit anti-PLP (Rogers et al., 2009), guinea pig anti-Asl (Klebba et al., 2013), and mouse anti-V5 (Life Technologies) antibodies. Goat secondary antibodies (conjugated with Cy2, Rhodamine red-X, or Cy5; Jackson ImmunoResearch Laboratories, Inc.) were used at manufacturer-recommended dilutions. Hoechst 33342 (Life Technologies) was used at a final dilution of 3.2 µM. Cells were mounted in 0.1 M n-propyl gallate, 90% (by volume) glycerol, and 10% PBS solution. Specimens were imaged using a DeltaVision Core system (Applied Precision) equipped with a microscope (IX71; Olympus), a 100× objective (NA 1.4), and a cooled charge-coupled device camera (CoolSNAP HQ2; Photometrics). Images were acquired with softWoRx v1.2 software (Applied Precision). Superresolution microscopy was performed using a superresolution microscopy system for structured illumination (ELYRA S1; Carl Zeiss) equipped with an inverted microscope stand (Axio Observer.Z1; Carl Zeiss) with transmitted, UV, and solid-state (405/488/561 nm) laser illumination sources, a 60× objective (NA 1.4), and an electron-multiplying charge-coupled device camera (iXon; Andor Technology). Images were acquired with ZEN 2011 software (Carl Zeiss).
Immunoblotting
S2 cell extracts were produced by lysing cells in cold PBS and 0.1% Triton X-100. Laemmli sample buffer was then added and boiled for 5 min. Samples of equal total protein were resolved by SDS-PAGE, blotted, probed with primary and secondary antibodies, and scanned on an Odyssey imager (LI-COR Biosciences). Care was taken to avoid saturating the scans of blots. Transfected Nlp-EGFP (a constitutively expressed nuclear protein; Rogers et al., 2009) was used as a loading control and transfection marker. Antibodies used for Western blotting include guinea pig anti-Slimb (Brownlee et al., 2011), guinea pig anti–SAS-6 (Rogers laboratory), rabbit anti-Fizzy (Rogers laboratory), guinea pig anti-Asl (Klebba et al., 2013), mouse anti-V5 monoclonal (Life Technologies), mouse anti-GFP monoclonal JL8 (Takara Bio Inc.), mouse anti-myc (Cell Signaling Technology), mouse anti–α-tubulin monoclonal DM1A (Sigma-Aldrich), and mouse anti-FLAG monoclonal (Sigma-Aldrich) used at 1:1,000 dilutions. IRDye 800CW secondary antibodies (LI-COR Biosciences) were prepared according to the manufacturer’s instructions and used at 1:1,500 dilutions.
Constructs and transfection
FL cDNAs of Drosophila Asl and Plk4 were subcloned into a pMT vector containing in-frame coding sequence for EGFP, V5, or myc and the inducible metallothionein promoter. Phusion polymerase (Thermo Fisher Scientific) was used according to manufacturer’s instructions to generate the various Asl and Plk4 deletion and point mutants. Transient transfections of S2 cells were performed using a Nucleofector II and Nucleofector kit V (Lonza) according to manufacturer’s instructions. In most experiments, 1.8 µg of Asl or Plk4 expression plasmid was mixed with 0.2 µg of Nlp-EGFP expression plasmid to either identify transfected cells or to serve as a loading control. Expression of all Plk4 constructs (and GFP control) was induced by the addition of 50 µM–2 mM copper sulfate to the culture medium. Colchicine and CHX (Sigma-Aldrich) were used at final concentrations of 30 and 100 µM, respectively.
Protein binding assays
Asl fragments (A, aa 1–357; B, aa 358–625; and C, aa 626–994) were PCR amplified from a FL Asl cDNA, and directionally subcloned into pET28b (Life Technologies) using BamHI and EcoRI restriction enzymes (Promega) to generate N-terminal His6-tagged constructs. The PB1-PB2 domain was PCR amplified from a Plk4 cDNA and directionally subcloned into pGEX-6p2 (GE Healthcare) using BamHI and EcoRI restriction enzymes. BL21 DE3 Escherichia coli were grown at 37°C to an OD600 of 0.6 and induced with 0.1 mM IPTG, and cells were shifted to 16°C for 18 h. Cells were centrifuged for 10 min at 2,100 g, and then, the pellets stored in buffer A (PBS, 10 mM imidazole, and 0.1% β-mercaptoethanol) at −80°C. Cells were lysed in buffer A by either sonication or using a cell disruptor (Avestin) and centrifuged at 23,000 g for 20 min at 4°C, and the supernatant was mixed with either Ni2+-NTA resin (QIAGEN), TALON cobalt resin (Takara Bio Inc.), or glutathione resin (GE Healthcare). Resins were washed in buffer A and eluted with either buffer A + 10 mM glutathione or a linear gradient of buffer A to buffer B (25 mM Tris, pH 8.0, 300 mM NaCl, 300 mM imidazole, and 0.1% β-mercaptoethanol). Protein-containing fractions were pooled and dialyzed overnight in either buffer C (25 mM Hepes, pH 7.5, 300 mM NaCl, and 0.1% β-mercaptoethanol) for SEC-MALS analysis or buffer D (25 mM Hepes, pH 7.4, 150 mM NaCl, and 1 mM DTT) for GST pull-down assays. Purified proteins were also concentrated using spin concentrators (Amicon Ultra; EMD Millipore). For in vitro binding assays, GST or GST–PB1-PB2 was immobilized on glutathione, mixed with His6-Asl-A or Asl-C, rocked at 25°C for 35 min, and pelleted at 500 g for 1 min. For Asl-C pull-down assays, GST or GST–PB1-PB2 were first chemically crossed linked to glutathione beads in buffer D by incubating with 20 mM dimethyl pimelimidate dihydrochloride, pH 8.3, for 2 h at 22°C and then quenching the coupling reaction by incubating with 0.2 M ethanolamine, pH 8.3, for 1 h at 22°C. Supernatant and pellets were analyzed by SDS-PAGE.
SEC-MALS
Asl fragments were individually injected (50 µM; 100 µl) onto a Superdex 200 10/300 GL size exclusion column (GE Healthcare) at 0.5 ml/min in buffer D (25 mM, Hepes, pH 7.5, 300 mM NaCl, 0.1% β-mercaptoethanol, and 0.2 g/liter sodium azide) and then passed consecutively through a UV detector, a light scattering instrument (DAWN HELEOS II; Wyatt Technology), and a refractometer (Optilab rEX; Wyatt Technology). The light scattering and refractive index data were used to calculate the weight-averaged molar mass using Wyatt Astra V software (Wyatt Technology). Data were processed with ASTRA software and plotted using SigmaPlot v11.
Y2H assay
Y2H experiments were performed using the Matchmaker Gold Y2H system (Takara Bio Inc.) with significant modifications. pDEST-GADT7 and pDEST-GBKT7, modified versions of Matchmaker vectors compatible with the Gateway cloning system (Life Technologies), were used (Rossignol et al., 2007). pDEST-GADT7 and pDEST-GBKT7 contain the 2µ and pUC origins of replication for growth in yeast and bacteria. Both contain the Gateway cassette and use the ADH1 promoter to drive expression in Saccharomyces cerevisiae. pDEST-GADT7 fuses the SV40 nuclear localization signal, the GAL4 activation domain, and the HA epitope tag to the N terminus of the protein encoded by DNA inserted into the Gateway cassette. pDEST-GBKT7 fuses the SV40 nuclear localization signal, the GAL4 DNA binding domain, and the c-Myc epitope tag to the N terminus of the protein encoded by DNA inserted into the Gateway cassette. pDest-pGBKT7 was modified by yeast-mediated recombination to confer resistance to ampicillin instead of kanamycin. pDEST-GADT7 and pDEST-GBKT7 plasmids containing fragments encoding the protein regions to be tested for interaction were transformed into Y187 and Y2HGold yeast strains, respectively, using standard techniques. Liquid cultures of yeast carrying these plasmids were grown at 30°C, with shaking, to an OD600 of ∼0.5 in synthetic defined (SD)/–Leu or SD/–Trp media, as appropriate to maintain plasmid selection. Interactions were tested by mating, mixing 20 µl each of a Y187 strain and a Y2HGold strain in 100 µl of 2× YPD media in a 96-well plate. Mating cultures were grown for 20–24 h at 30°C with shaking. Cells were pinned onto SD/–Leu/–Trp (minimal media double dropout [DDO]) plates, to select for diploids carrying both plasmids, using a Multi-Blot Replicator (VP 407AH; V&P Scientific), and grown for 5 d at 30°C. These plates were then replica plated onto DDO, SD/–Ade/–Leu/–Trp/–Ura (minimal media quadruple dropout [QDO]), SD/–Leu/–Trp + aureobasidin A (Takara Bio Inc.) + X-α-Gal (Takara Bio Inc. and Gold Biotechnology; supplemented DDO media [DDOXA]), and/or SD/–Ade/–Leu/–Trp/–Ura + aureobasidin A + X-α-Gal (5-bromo-4-chloro-3-indolyl-α-d-galactopyranoside; supplemented QDO media [QDOXA]). Replica plates were grown for 5 d at 30°C. Interactions were scored based on growth and/or blue color, as appropriate. All clones used were confirmed to not activate Y2H reporters in isolation.
To generate a mutant version of Asl (aa 626–994) no longer able to bind Plk4, a reverse two-hybrid screen was conducted as described by Bennett et al. (2004) with significant modifications. Low-fidelity PCR was performed using Taq polymerase (New England Biolabs, Inc.) in the supplied buffer with the addition of 0.05 mM MnCl2. Deoxy-CTP, deoxy-GTP, and deoxy-TTP were added to 0.25 mM. Deoxy-ATP was added to 0.06 mM. asl (aa 626–994) in pGBKT7 was used as a template. Primers were designed to complement sequence in the vector, allowing for homologous recombination-mediated reconstruction of mutagenized asl (aa 626–994) in pGBKT7 to be performed in yeast. The primers used were 5′-TAATACGACTCACTATAGGGCG-3′ and 5′-CGGAATTAGCTTGGCTGC-3′. pGBKT7 was linearized by digestion with EcoRI and PstI, and amplified PCR products and the linearized vector both were separated by agarose gel electrophoresis and purified from the gel using DEAE cellulose paper. Linearized pGBKT7 and the mutagenized Asl (aa 626–994) PCR product were cotransformed into Y2HGold yeast using standard methods. Yeast carrying the reconstituted plasmid were selected by growth on SD/–Trp plates. Approximately 1,200 independent clones containing randomly mutagenized versions of asl (aa 626–994) were isolated. These clones were individually crossed to a Y187 strain carrying plk4 (aa 382–602) in PGADT7. Diploids carrying both plasmids were selected by growth on media selective for both plasmids (DDO) for 5 d at 30°C. Yeast were then replica plated to DDO and QDOXA. Clones that grew on DDO but failed to grow on QDOXA were selected for further study. The failure to grow when crossed to the plk4 (aa 382–602) strain was confirmed. These clones were then tested to identify one that retained the ability to interact with Asl (aa 358–625), Sas-4 (aa 1–347), Sas-4 (aa 348–707), and Spd-2 (aa 664–1,146).
GFP immunoprecipitation assays
GFP-binding protein (GBP; Rothbauer et al., 2008) was fused to the fragment crystallizable domain of human IgG (cloned from the L1-Fc plasmid; Addgene), tagged with His6 in pET28a (EMD), expressed in E. coli, and purified on HisPur resin (Thermo Fisher Scientific) according to the manufacturer’s instructions (Buster et al., 2013). Purified GBP was bound to protein A–coupled Sepharose and then cross-linked to the resin by incubating with 20 mM dimethyl pimelimidate dihydrochloride in PBS, pH 8.3, 2 h at 22°C, and then, the coupling reaction was quenched by incubating with 0.2 M ethanolamine, pH 8.3, for 1 h at 22°C. Antibody-coated beads were washed three times with 1.5 ml of cell lysis buffer (CLB; 50 mM Tris, pH 7.2, 125 mM NaCl, 2 mM DTT, 0.1% Triton X-100, and 0.1 mM PMSF). Transfected cells expressing recombinant proteins were lysed in CLB, and the lysates were clarified by centrifugation at 20,000 g for 10 min at 4°C. 10% of the inputs were used for immunoblots. GBP-coated beads were rocked with lysate for 1 h at 4°C, washed two times with 1 ml CLB, and then boiled in Laemmli sample buffer. In vivo ubiquitination assays were performed by coexpressing Plk4-EGFP constructs with triple FLAG-tagged Drosophila Ubi (CG32744; also under the metallothionein promoter and Cu induced; Buster et al., 2013) and then probing the immunoblot of the cell lysate with mouse anti-FLAG antibody (Sigma-Aldrich).
Statistical analysis
Means of measurements were analyzed for significant differences by one-way analysis of variance (followed by Tukey’s post-test to evaluate differences between treatment pairs) using Prism 6 (GraphPad Software) software. Means are taken to be significantly different if P < 0.05. P-values shown for pairwise comparisons of Tukey’s post-test are adjusted for multiplicity. In the figures, a single asterisk indicates 0.05 > P ≥ 0.01, a double asterisk indicates 0.01 > P ≥ 0.001, a trouble asterisk indicates P < 0.001, and NS indicates P > 0.05 for the indicated pairwise comparison. The Plk4 levels in Fig. 5 A were evaluated with the Wilcoxon signed rank test (Prism 6). Error bars in all figures indicate SEM.
Online supplemental material
Fig. S1 shows that transgenic Asl coimmunoprecipitates with endogenous Asl and self-interacts via Y2H, that Asl-C is predicted to form trimeric coiled coils, and that immunoblots of endogenous Asl replaced with Asl fragments. Fig. S2 shows that Asl-C binds Plk4 PB1-PB2 and is sufficient to induce centriole amplification and stabilize Plk4. Fig. S3 shows that the J11F10 mutant of Asl-C localizes to centrioles but fails to rescue centriole duplication in Asl-depleted cells. Fig. S4 shows images of centrioles in Asl- and Plk4-coexpressing cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201410105/DC1.
Supplementary Material
Acknowledgments
We thank D. Elliott for structured illumination microscopy assistance, C. Fagerstrom and J. Ortega for help with cloning and Y2H, D. Morgan for use of his facilities, and P. Krieg for editing.
K.C. Slep is supported by the National Institutes of Health (R01GM094415) and the March of Dimes (FY11-434). N.M. Rusan is supported by the Division of Intramural Research National Institutes of Health/National Heart, Lung, and Blood Institute (1ZIAHL006104). G.C. Rogers is grateful for support from National Cancer Institute (30 CA23074), National Science Foundation (1158151), Arizona Biomedical Research Commission (1210), and the Phoenix Friends.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- Asl
- Asterless
- CHX
- cycloheximide
- CLB
- cell lysis buffer
- CPAP
- centrosomal P4.1-associated protein
- DDO
- double dropout
- dsRNA
- double-stranded RNA
- FL
- full length
- GBP
- GFP-binding protein
- MM
- molecular mass
- PLP
- pericentrin-like protein
- PP2A
- protein phosphatase 2A
- QDO
- quadruple dropout
- SBM
- Slimb-binding mutant
- SD
- synthetic defined
- SEC-MALS
- size-exclusion chromatography with multiangle laser light scattering
- Ubi
- ubiquitin
- WT
- wild type
- Y2H
- yeast two hybrid
References
- Avidor-Reiss T., and Gopalakrishnan J.. 2013. Building a centriole. Curr. Opin. Cell Biol. 25:72–77 10.1016/j.ceb.2012.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M.A., Shern J.F., and Kahn R.A.. 2004. Reverse two-hybrid techniques in the yeast Saccharomyces cerevisiae. Methods Mol. Biol. 261:313–326. [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M., Hildebrandt F., Pellman D., Woods G., and Godinho S.A.. 2011. Centrosomes and cilia in human disease. Trends Genet. 27:307–315 10.1016/j.tig.2011.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blachon S., Gopalakrishnan J., Omori Y., Polyanovsky A., Church A., Nicastro D., Malicki J., and Avidor-Reiss T.. 2008. Drosophila asterless and vertebrate Cep152 Are orthologs essential for centriole duplication. Genetics. 180:2081–2094 10.1534/genetics.108.095141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornens M.2012. The centrosome in cells and organisms. Science. 335:422–426 10.1126/science.1209037 [DOI] [PubMed] [Google Scholar]
- Brito D.A., Gouveia S.M., and Bettencourt-Dias M.. 2012. Deconstructing the centriole: structure and number control. Curr. Opin. Cell Biol. 24:4–13 10.1016/j.ceb.2012.01.003 [DOI] [PubMed] [Google Scholar]
- Brownlee C.W., Klebba J.E., Buster D.W., and Rogers G.C.. 2011. The Protein Phosphatase 2A regulatory subunit Twins stabilizes Plk4 to induce centriole amplification. J. Cell Biol. 195:231–243 10.1083/jcb.201107086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buster D.W., Daniel S.G., Nguyen H.Q., Windler S.L., Skwarek L.C., Peterson M., Roberts M., Meserve J.H., Hartl T., Klebba J.E., et al. 2013. SCFSlimb ubiquitin ligase suppresses condensin II–mediated nuclear reorganization by degrading Cap-H2. J. Cell Biol. 201:49–63 10.1083/jcb.201207183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cizmecioglu O., Arnold M., Bahtz R., Settele F., Ehret L., Haselmann-Weiss U., Antony C., and Hoffmann I.. 2010. Cep152 acts as a scaffold for recruitment of Plk4 and CPAP to the centrosome. J. Cell Biol. 191:731–739 10.1083/jcb.201007107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha-Ferreira I., Rodrigues-Martins A., Bento I., Riparbelli M., Zhang W., Laue E., Callaini G., Glover D.M., and Bettencourt-Dias M.. 2009. The SCF/Slimb ubiquitin ligase limits centrosome amplification through degradation of SAK/PLK4. Curr. Biol. 19:43–49 10.1016/j.cub.2008.11.037 [DOI] [PubMed] [Google Scholar]
- Cunha-Ferreira I., Bento I., Pimenta-Marques A., Jana S.C., Lince-Faria M., Duarte P., Borrego-Pinto J., Gilberto S., Amado T., Brito D., et al. 2013. Regulation of autophosphorylation controls PLK4 self-destruction and centriole number. Curr. Biol. 23:2245–2254 10.1016/j.cub.2013.09.037 [DOI] [PubMed] [Google Scholar]
- Dzhindzhev N.S., Yu Q.D., Weiskopf K., Tzolovsky G., Cunha-Ferreira I., Riparbelli M., Rodrigues-Martins A., Bettencourt-Dias M., Callaini G., and Glover D.M.. 2010. Asterless is a scaffold for the onset of centriole assembly. Nature. 467:714–718 10.1038/nature09445 [DOI] [PubMed] [Google Scholar]
- Fode C., Binkert C., and Dennis J.W.. 1996. Constitutive expression of murine Sak-a suppresses cell growth and induces multinucleation. Mol. Cell. Biol. 16:4665–4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J., and Glover D.M.. 2012. Structured illumination of the interface between centriole and peri-centriolar material. Open Biol. 2:120104 10.1098/rsob.120104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guderian G., Westendorf J., Uldschmid A., and Nigg E.A.. 2010. Plk4 trans-autophosphorylation regulates centriole number by controlling βTrCP-mediated degradation. J. Cell Sci. 123:2163–2169 10.1242/jcs.068502 [DOI] [PubMed] [Google Scholar]
- Guernsey D.L., Jiang H., Hussin J., Arnold M., Bouyakdan K., Perry S., Babineau-Sturk T., Beis J., Dumas N., Evans S.C., et al. 2010. Mutations in centrosomal protein CEP152 in primary microcephaly families linked to MCPH4. Am. J. Hum. Genet. 87:40–51 10.1016/j.ajhg.2010.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch E.M., Kulukian A., Holland A.J., Cleveland D.W., and Stearns T.. 2010. Cep152 interacts with Plk4 and is required for centriole duplication. J. Cell Biol. 191:721–729 10.1083/jcb.201006049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland A.J., Lan W., Niessen S., Hoover H., and Cleveland D.W.. 2010. Polo-like kinase 4 kinase activity limits centrosome overduplication by autoregulating its own stability. J. Cell Biol. 188:191–198 10.1083/jcb.200911102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalay E., Yigit G., Aslan Y., Brown K.E., Pohl E., Bicknell L.S., Kayserili H., Li Y., Tüysüz B., Nürnberg G., et al. 2011. CEP152 is a genome maintenance protein disrupted in Seckel syndrome. Nat. Genet. 43:23–26 10.1038/ng.725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., and Dynlacht B.D.. 2013. Assembling a primary cilium. Curr. Opin. Cell Biol. 25:506–511 10.1016/j.ceb.2013.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.S., Park J.E., Shukla A., Choi S., Murugan R.N., Lee J.H., Ahn M., Rhee K., Bang J.K., Kim B.Y., et al. 2013. Hierarchical recruitment of Plk4 and regulation of centriole biogenesis by two centrosomal scaffolds, Cep192 and Cep152. Proc. Natl. Acad. Sci. USA. 110:E4849–E4857 10.1073/pnas.1319656110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebba J.E., Buster D.W., Nguyen A.L., Swatkoski S., Gucek M., Rusan N.M., and Rogers G.C.. 2013. Polo-like kinase 4 autodestructs by generating its Slimb-binding phosphodegron. Curr. Biol. 23:2255–2261 10.1016/j.cub.2013.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleylein-Sohn J., Westendorf J., Le Clech M., Habedanck R., Stierhof Y.D., and Nigg E.A.. 2007. Plk4-induced centriole biogenesis in human cells. Dev. Cell. 13:190–202 10.1016/j.devcel.2007.07.002 [DOI] [PubMed] [Google Scholar]
- Krämer A., Lukas J., and Bartek J.. 2004. Checking out the centrosome. Cell Cycle. 3:1390–1393 10.4161/cc.3.11.1252 [DOI] [PubMed] [Google Scholar]
- Mennella V., Keszthelyi B., McDonald K.L., Chhun B., Kan F., Rogers G.C., Huang B., and Agard D.A.. 2012. Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nat. Cell Biol. 14:1159–1168 10.1038/ncb2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Saito H., and Takekawa M.. 2013. SAPK pathways and p53 cooperatively regulate PLK4 activity and centrosome integrity under stress. Nat. Commun. 4:1775 10.1038/ncomms2752 [DOI] [PubMed] [Google Scholar]
- Nigg E.A., and Raff J.W.. 2009. Centrioles, centrosomes, and cilia in health and disease. Cell. 139:663–678 10.1016/j.cell.2009.10.036 [DOI] [PubMed] [Google Scholar]
- Nigg E.A., and Stearns T.. 2011. The centrosome cycle: Centriole biogenesis, duplication and inherent asymmetries. Nat. Cell Biol. 13:1154–1160 10.1038/ncb2345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesin J.A., and Orr-Weaver T.L.. 2008. Regulation of APC/C activators in mitosis and meiosis. Annu. Rev. Cell Dev. Biol. 24:475–499 10.1146/annurev.cellbio.041408.115949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues-Martins A., Riparbelli M., Callaini G., Glover D.M., and Bettencourt-Dias M.. 2007. Revisiting the role of the mother centriole in centriole biogenesis. Science. 316:1046–1050 10.1126/science.1142950 [DOI] [PubMed] [Google Scholar]
- Rogers G.C., Rusan N.M., Roberts D.M., Peifer M., and Rogers S.L.. 2009. The SCFSlimb ubiquitin ligase regulates Plk4/Sak levels to block centriole reduplication. J. Cell Biol. 184:225–239 http://jcb.rupress.org/content/184/2/225.long [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers S.L., and Rogers G.C.. 2008. Culture of Drosophila S2 cells and their use for RNAi-mediated loss-of-function studies and immunofluorescence microscopy. Nat. Protoc. 3:606–611 10.1038/nprot.2008.18 [DOI] [PubMed] [Google Scholar]
- Rossignol P., Collier S., Bush M., Shaw P., and Doonan J.H.. 2007. Arabidopsis POT1A interacts with TERT-V(I8), an N-terminal splicing variant of telomerase. J. Cell Sci. 120:3678–3687 10.1242/jcs.004119 [DOI] [PubMed] [Google Scholar]
- Rothbauer U., Zolghadr K., Muyldermans S., Schepers A., Cardoso M.C., and Leonhardt H.. 2008. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol. Cell. Proteomics. 7:282–289 10.1074/mcp.M700342-MCP200 [DOI] [PubMed] [Google Scholar]
- Shimada M., and Komatsu K.. 2009. Emerging connection between centrosome and DNA repair machinery. J. Radiat. Res. (Tokyo). 50:295–301 10.1269/jrr.09039 [DOI] [PubMed] [Google Scholar]
- Slevin L.K., Nye J., Pinkerton D.C., Buster D.W., Rogers G.C., and Slep K.C.. 2012. The structure of the plk4 cryptic polo box reveals two tandem polo boxes required for centriole duplication. Structure. 20:1905–1917 10.1016/j.str.2012.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sir J.H., Barr A.R., Nicholas A.K., Carvalho O.P., Khurshid M., Sossick A., Reichelt S., D’Santos C., Woods C.G., and Gergely F.. 2011. A primary microcephaly protein complex forms a ring around parental centrioles. Nat. Genet. 43:1147–1153 10.1038/ng.971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnen K.F., Gabryjonczyk A.M., Anselm E., Stierhof Y.D., and Nigg E.A.. 2013. Human Cep192 and Cep152 cooperate in Plk4 recruitment and centriole duplication. J. Cell Sci. 126:3223–3233 10.1242/jcs.129502 [DOI] [PubMed] [Google Scholar]
- Stevens N.R., Dobbelaere J., Brunk K., Franz A., and Raff J.W.. 2010. Drosophila Ana2 is a conserved centriole duplication factor. J. Cell Biol. 188:313–323 10.1083/jcb.200910016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang N., and Marshall W.F.. 2012. Centrosome positioning in vertebrate development. J. Cell Sci. 125:4951–4961 10.1242/jcs.038083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varmark H., Llamazares S., Rebollo E., Lange B., Reina J., Schwarz H., and Gonzalez C.. 2007. Asterless is a centriolar protein required for centrosome function and embryo development in Drosophila. Curr. Biol. 17:1735–1745 10.1016/j.cub.2007.09.031 [DOI] [PubMed] [Google Scholar]
- Vitre B.D., and Cleveland D.W.. 2012. Centrosomes, chromosome instability (CIN) and aneuploidy. Curr. Opin. Cell Biol. 24:809–815 10.1016/j.ceb.2012.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.