Abstract
Background
Several studies have reported an association between the A23G polymorphism (rs 1800975) in the xeroderma pigmentosum group A (XPA) gene and risk of digestive system cancers. However, the results are inconsistent. In this study, we performed a meta-analysis to assess the association between XPA A23G polymorphism and the risk of digestive system cancers.
Methods
Relevant studies were identified using the PubMed, Web of Science, China National Knowledge Infrastructure, WanFang, and VIP databases up to August 30, 2014. The pooled odds ratio (OR) with a 95% confidence interval (CI) was calculated using the fixed or random effects model.
Results
A total of 18 case-control studies from 16 publications with 4,170 patients and 6,929 controls were included. Overall, no significant association was found between XPA A23G polymorphism and the risk of digestive system cancers (dominant model: GA + AA versus GG, OR 0.89, 95% CI 0.74–1.08; recessive model: AA versus GA + GG, OR 0.94, 95% CI 0.74–1.20; GA versus GG, OR 0.89, 95% CI 0.77–1.03; and AA versus GG, OR 0.87, 95% CI 0.64–1.19). When the analysis was stratified by ethnicity, similar results were observed among Asians and Caucasians in all genetic models. In stratified analysis based on tumor type, we also failed to detect any association between XPA A23G polymorphism and the risk of esophageal, gastric, or colorectal cancers.
Conclusion
This meta-analysis indicates that the XPA A23G polymorphism is not associated with a risk of digestive system cancers.
Keywords: xeroderma pigmentosum group A, polymorphism, digestive system cancer, meta-analysis
Introduction
Gastrointestinal cancers, referring to a group of malignancies affecting the esophagus, stomach, liver, bowel, pancreas, gallbladder, and anus, are the most common cancers worldwide.1 There are an estimated 3.4 million new cases worldwide each year, and their mortality rates have increased gradually over the past decade.1 The exact mechanism of carcinogenesis is still not fully understood. It is well established that some risk factors (eg, dietary, racial, and socioeconomic) and interactions between genetic and environmental factors play important roles in the pathogenesis of cancer.2,3
Deregulation of DNA repair is a crucial factor in the multistep process of carcinogenesis. A variety of mechanisms for DNA repair have been developed to ensure integrity of the genome in humans, and the xeroderma pigmentosum group A (XPA) gene is a vital component of the DNA repair machinery. The XPA gene is located on chromosome 9q22.3 and encodes a zinc finger DNA-binding protein participating in DNA excision repair to maintain genomic integrity.4 The XPA protein plays a central role in nucleotide excision repair (NER) through its interaction with replication protein A, transcription factor II H, and the excision repair cross-complementing group 1-xeroderma pigmentosum group F protein complex.5,6 In addition, XPA is involved in both global genome and transcription-coupled repair pathways,7 and interacts with many core repair factors during the DNA repair process.8 In the XPA gene, a polymorphic site has been identified in the 5′ untranslated region and consists of an A to G substitution in the fourth nucleotide before the ATG start codon (XPA A23G, rs 1800975). It has been shown that the polymorphism could affect protein levels in the cell.9,10 To date, a large number of molecular epidemiologic studies have been conducted to assess the role of A23G polymorphism in XPA gene on various types of cancers, especially those affecting the digestive system.11–30 However, the results have been inconclusive or inconsistent. Individual studies might have been underpowered to detect the effect of this polymorphism on susceptibility to cancer. Therefore, we conducted a meta-analysis to evaluate the association between XPA A23G polymorphism and the susceptibility to digestive system cancers.
Methods
Search strategy
We searched the electronic literature in the PubMed, Web of Science, China National Knowledge Infrastructure, WanFang, and VIP databases for all relevant articles. The last search update was August 30, 2014, using the search terms: “xeroderma pigmentosum group A or XPA or DNA repair gene or NER”, “genetic polymorphism or polymorphisms or variant”, and “digestive system cancer or gastrointestinal cancers or gastric cancer or colorectal cancer or hepatocellular carcinoma or esophageal cancer or pancreatic cancer”. The search was restricted to humans without language restrictions. Additional studies were identified by a hand search of references of original or review articles on this topic. If more than one geographic or cancer type was reported in one report, each was extracted separately. If data or data subsets were published in more than one article, only the publication with the largest sample size was included.
Inclusion and exclusion criteria
Studies included in this meta-analysis had to meet the following criteria: studies that evaluated the association between XPA A23G polymorphism and digestive system cancers, in a case-control study design, and had detailed genotype frequency of cases and controls or could be calculated from the article text. We excluded case-only studies, case reports, review articles, studies without raw data for the XPA A23G genotype, and repetitive publications.
Data extraction
For each study, the following data were extracted independently by two investigators: the first author’s name, year of publication, country of origin, ethnicity, age, sex, source of controls, genotype methods, number of cases and controls (total and genotypes), and Hardy–Weinberg equilibrium (HWE) in controls (P-value). The results were compared, and disagreements were discussed among all authors and resolved with consensus. Different ethnicity was categorized as Asian and Caucasian.
Quality assessment
The quality of the eligible studies was assessed using Newcastle–Ottawa Scale (NOS), which is widely used for assessment of the quality of observational studies, including cohort or case–control studies.31 NOS, consisting of three parts (selection, comparability, and exposure), is a star-rewarded scale. A total of four, two, and three stars, respectively, will be rewarded if the criteria are met. A study with seven or more stars was categorized as high quality, otherwise, the study was categorized as low quality.
Statistical analysis
HWE was evaluated for each study using an Internet-based HWE calculator (http://ihg.gsf.de/cgi-bin/hw/hwa1.pl). The risk of digestive system cancers associated with XPA A23G polymorphism was estimated for each study by odds ratio (OR) and 95% confidence interval (CI). The most common G allele was considered the reference genotype and the rare A allele was examined as the variant in this analysis. Four different ORs were calculated: the dominant model (AG + AA versus GG), the recessive model (AA versus AG + GG), heterozygote comparison (AG versus GG), and homozygote comparison (AA versus GG). A χ2-test-based Q statistic test was performed to assess between-study heterogeneity.32 We also quantified the effect of heterogeneity by I2 test. When a significant Q test (P>0.1) or I2<50% indicated homogeneity across studies, the fixed effects model was used,33 or else the random effects model was used.34 We then performed stratification analyses on ethnicity, tumor type, and source of control. Analysis of sensitivity was performed to evaluate the stability of the results, namely, a single study in the meta-analysis was deleted each time to reflect the influence of the individual data set on the pooled OR. Finally, potential publication bias was investigated using Begg’s funnel plot and Egger’s regression test.35,36 P<0.05 was considered to be statistically significant. All analyses were performed using the Cochrane Collaboration RevMan 5.2 and Stata package version 12.0 (Stata Corp, College Station, TX, USA).
Results
Study characteristics
After an initial search, a total of 101 published articles relevant to the topic were identified. According to the inclusion criteria, 20 studies11–30 with full text were included in this meta-analysis and 81 studies were excluded. The flow chart for study selection is summarized in Figure 1. Because the study by Huang et al17 included two types of cancer, we treated them separately in this meta-analysis; three articles27–29 that had overlapped study data were also excluded. Moreover, we excluded one study30 because it did not present detailed genotyping information. Therefore, as shown in Table 1, there were 18 case-control studies11–26 with 4,170 cases and 6,929 controls concerning XPA A23G polymorphism. Of the 18 eligible studies, nine studies12,14,15,17,20,22,24–26 involved esophageal cancers, four studies11,17,21 involved gastric cancers, four13,16,18,19 involved colorectal cancers, and one23 involved hepatocellular carcinoma. Two ethnicities were addressed: eleven studies11,12,14,17,20,23–26 were conducted in Asian populations and seven studies13,15,16,18,19,21,22 in Caucasian populations. The distribution of genotypes in the controls was consistent with HWE for all selected studies. The quality of all eligible studies was categorized as high except for one study.20
Figure 1.
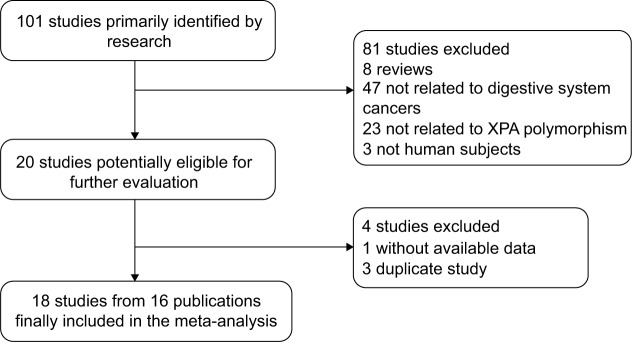
Flow chart showing study selection procedure.
Table 1.
Characteristics of studies included in the meta-analysis
| Study | Year | Country | Ethnicity | Age, years
|
Sex (male/female)
|
Cancer type | Source of controls | Genotype methods | Genotype (case/control)
|
P HWE | NOS | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | Case | Control | Total | GG | GA | AA | |||||||||
| Dong et al11 | 2008 | People’s Republic of China | Asian | 60.4±8.30 | 60.4±8.42 | 165/88 | 385/227 | Gastric | PB | PCR–RFLP | 253/612 | 47/128 | 120/322 | 86/162 | 0.169 | 8 |
| Feng et al12 | 2008 | People’s Republic of China | Asian | 59.09±8.43 | 58.81±8.58 | 125/71 | 130/71 | Esophageal | HB | PCR–RFLP | 196/201 | 28/56 | 83/91 | 85/54 | 0.181 | 7 |
| Gil et al13 | 2012 | Poland | Caucasian | 63.22±11.36 | 74.89±7.63 | NR | NR | Colorectal | HB | PCR–RFLP | 100/133 | 26/50 | 58/67 | 16/16 | 0.369 | 8 |
| Guo et al14 | 2008 | People’s Republic of China | Asian | 60.0±9.33 | 60.4±8.42 | 218/109 | 385/227 | Esophageal | PB | PCR–RFLP | 327/612 | 65/128 | 139/322 | 123/162 | 0.169 | 8 |
| Hall et al15 | 2007 | Europe | Caucasian | NR | NR | NR | NR | Esophageal | HB | TaqMan | 171/974 | 75/398 | 81/451 | 15/125 | 0.875 | 9 |
| Hansen et al16 | 2007 | Denmark | Caucasian | 59 (51–64) | 56 (50–63) | 219/178 | 434/366 | Colorectal | PB | NR | 394/788 | 176/339 | 187/359 | 31/90 | 0.731 | 8 |
| Huang et al17 | 2007a | People’s Republic of China | Asian | NR | NR | 112/38 | 301/101 | Esophageal | PB | PCR–RFLP | 150/402 | 22/32 | 69/160 | 59/210 | 0.843 | 7 |
| Huang et al17 | 2007b | People’s Republic of China | Asian | NR | NR | 116/29 | 130/50 | Cardiac | PB | PCR–RFLP | 145/180 | 20/13 | 60/55 | 65/112 | 0.097 | 7 |
| Huang et al17 | 2007c | People’s Republic of China | Asian | NR | NR | 111/35 | 130/50 | Gastric | PB | PCR–RFLP | 146/180 | 12/13 | 57/55 | 77/112 | 0.097 | 7 |
| Jelonek et al18 | 2010 | Poland | Caucasian | NR | NR | NR | NR | Colorectal | HB | PCR–RFLP | 66/133 | 29/46 | 33/70 | 4/17 | 0.225 | 8 |
| Joshi et al19 | 2009 | USA | Caucasian | 60.0±11.3 | 59.3±11.8 | NR | NR | Colorectal | PB | TaqMan | 302/349 | 136/149 | 133/170 | 33/30 | 0.056 | 8 |
| Liu et al20 | 2007 | People’s Republic of China | Asian | 63.67±9.58 | 62.50±9.39 | 56/41 | 56/41 | Esophageal | PB | PCR–RFLP | 96/96 | 11/11 | 35/47 | 50/38 | 0.535 | 6 |
| Palli et al21 | 2010 | Italy | Caucasian | 68.8±9.9 | 55.5±7.0 | 177/137 | 270/278 | Gastric | PB | TaqMan | 284/523 | 134/249 | 115/215 | 35/59 | 0.227 | 8 |
| Pan et al22 | 2009 | USA | Caucasian | 63.13±10.60 | 62.91±10.38 | 343/44 | 397/65 | Esophageal | HB | PCR–RFLP | 380/458 | 179/151 | 166/219 | 35/88 | 0.589 | 8 |
| Xie et al23 | 2007 | People’s Republic of China | Asian | 49.1 (17–80) | 48.6 (28–79) | 377/57 | 384/96 | Hepatocellular | PB | PCR–RFLP | 415/479 | 139/144 | 203/219 | 73/116 | 0.071 | 7 |
| Zhang et al24 | 2006 | People’s Republic of China | Asian | NR | NR | NR | NR | Esophageal | HB | PCR–RFLP | 206/206 | 33/44 | 82/96 | 91/66 | 0.412 | 7 |
| Zhen et al25 | 2012 | People’s Republic of China | Asian | NR | NR | 237/114 | 258/142 | Esophageal | PB | PCR–RFLP | 351/400 | 99/53 | 145/188 | 107/159 | 0.826 | 7 |
| Zhu et al26 | 2008 | People’s Republic of China | Asian | 61.03 | 60.77 | 105/83 | 126/77 | Esophageal | PB | PCR–SSCP | 188/203 | 50/52 | 69/88 | 69/63 | 0.063 | 7 |
Notes: Age is expressed either as mean ± standard deviation or median (interquartile range).
Esophageal study;
cardiac study;
gastric study.
Abbreviations: HWE, Hardy–Weinberg equilibrium; PCR–RFLP, polymerase chain reaction–restriction fragment length polymorphism; PCR–SSCP, polymerase chain reaction–single strand conformation polymorphism; PB, population-based; HB, hospital-based; NR, not reported; NOS, Newcastle–Ottawa Scale.
Quantitative data synthesis
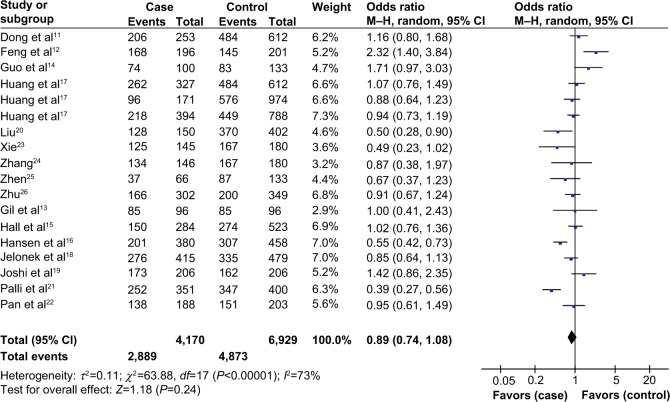
As shown in Table 2, overall no significant association was found between XPA A23G polymorphism and the risk of digestive system cancers (dominant model: OR 0.89, 95% CI 0.74–1.08; recessive model: OR 0.94, 95% CI 0.74–1.20; GA versus GG, OR 0.89, 95% CI 0.77–1.03; and AA versus GG, OR 0.87, 95% CI 0.64–1.19, Figure 2).
Table 2.
Summary of odds ratios for the risk of the XPG Asp1104His polymorphism and gastrointestinal cancers
| Variables | Na | Dominant model
|
Recessive model
|
GA versus GG
|
AA versus GG
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | Pb | I2 | OR (95% CI) | Pb | I2 | OR (95% CI) | Pb | I2 | OR (95% CI) | Pb | I2 | ||
| Total | 18 | 0.89 (0.74–1.08) | <0.00001 | 73 | 0.94 (0.74–1.20) | <0.00001 | 83 | 0.89 (0.77–1.03) | 0.005 | 52 | 0.87 (0.64–1.19) | <0.00001 | 83 |
| Ethnicity | |||||||||||||
| Asian | 11 | 0.90 (0.66–1.21) | <0.00001 | 78 | 1.04 (0.77–1.41) | <0.00001 | 86 | 0.87 (0.68–1.10) | 0.007 | 59 | 0.95 (0.62–1.44) | <0.00001 | 86 |
| Caucasian | 7 | 0.88 (0.71–1.10) | 0.007 | 66 | 0.78 (0.55–1.13) | 0.006 | 67 | 0.90 (0.79–1.03) | 0.09 | 45 | 0.76 (0.49–1.18) | 0.0004 | 76 |
| Tumor type | |||||||||||||
| Esophageal | 9 | 0.87 (0.61–1.23) | <0.00001 | 84 | 1.04 (0.71–1.52) | <0.00001 | 88 | 0.81 (0.62–1.06) | 0.002 | 68 | 0.89 (0.52–1.54) | <0.00001 | 90 |
| Gastric | 4 | 0.99 (0.80–1.22) | 0.22 | 32 | 0.86 (0.53–1.41) | 0.0006 | 83 | 0.98 (0.78–1.23) | 0.85 | 0 | 0.88 (0.52–1.50) | 0.02 | 69 |
| Colorectal | 4 | 0.96 (0.80–1.14) | 0.14 | 45 | 0.91 (0.56–1.47) | 0.08 | 56 | 0.98 (0.82–1.17) | 0.21 | 33 | 0.92 (0.52–1.62) | 0.04 | 63 |
| Hepatocellular | 1 | 0.85 (0.64–1.13) | NA | NA | 0.67 (0.48–0.93) | NA | NA | 0.96 (0.71–1.30) | NA | NA | 0.65 (0.45–0.95) | NA | NA |
| Source of control | |||||||||||||
| PB | 12 | 0.83 (0.69–1.00) | 0.002 | 63 | 0.93 (0.72–1.20) | <0.00001 | 81 | 0.86 (0.77–0.96) | 0.08 | 40 | 0.81 (0.60–1.11) | <0.00001 | 77 |
| HB | 6 | 1.09 (0.69–1.73) | <0.00001 | 85 | 0.96 (0.52–1.76) | <0.00001 | 87 | 1.04 (0.74–1.47) | 0.005 | 70 | 1.00 (0.44–2.27) | <0.00001 | 90 |
Notes:
Number of comparisons;
test for heterogeneity.
Abbreviations: CI, confidence interval; HB, hospital-based; NA, not applicable; OR, odds ratio; PB, population-based.
Figure 2.
Forest plots of odds ratios for the association of XPA A23G polymorphism and digestive system cancer risk (dominant model).
Abbreviations: CI, confidence interval; M–H, Mantel–Haenszel method.
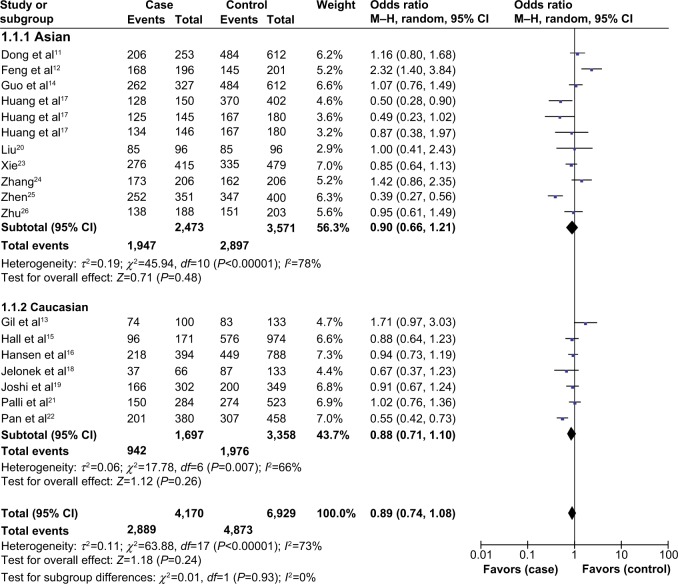
In subgroup analysis by ethnicity, there was no significant association between XPA A23G polymorphism and the risk of digestive system cancers in either Asians or Caucasians (Table 2, Figure 3).
Figure 3.
Subgroup analysis by ethnicity of odds ratios for the association of XPA A23G polymorphism and digestive system cancer risk (dominant model).
Abbreviations: CI, confidence interval; M–H, Mantel–Haenszel method.
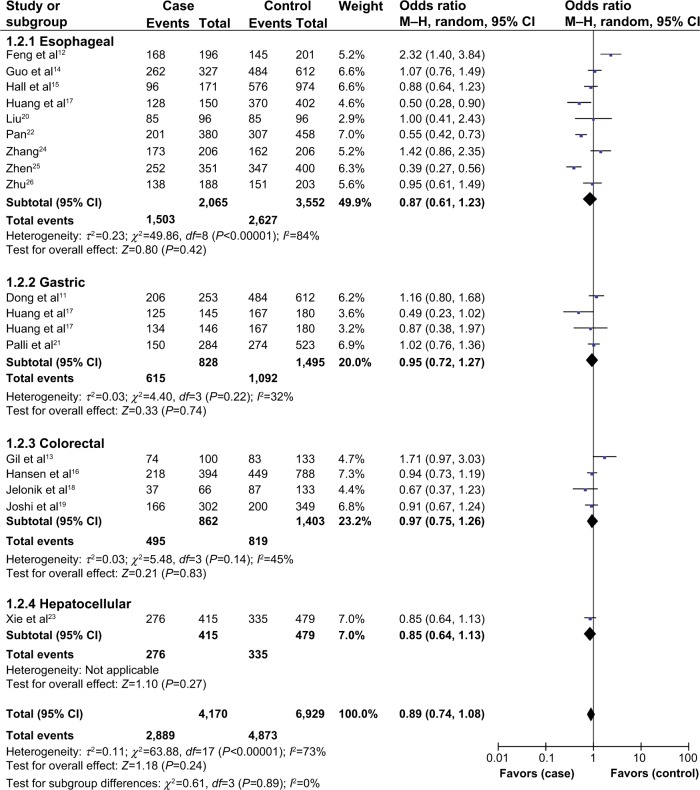
In stratified analysis based on tumor type, we also failed to detect any association between XPA A23G polymorphism and the risk of esophageal, gastric, or colorectal cancers. In addition, only one study focused on hepatocellular carcinoma, and the results showed no association between XPA A23G polymorphism and the risk of hepatocellular carcinoma (Table 2, Figure 4).
Figure 4.
Subgroup analysis by tumor type of odds ratios for the association of XPA A23G polymorphism and digestive system cancer risk (dominant model).
Abbreviations: CI, confidence interval; M–H, Mantel–Haenszel method.
When the analysis was stratified by source of control, we found that XPA A23G polymorphism was associated with a decreased risk of digestive system cancers in population-based models (GA versus GG, OR 0.86, 95% CI 0.77–0.96), but not in other genetic models or hospital-based populations (Table 2). However, it is worth noting that there was moderate heterogeneity (I2=40%) in the subgroup analysis; when the study by Zhen et al was excluded, the heterogeneity disappeared (I2=0%), and the pooled results showed no significant differences in genotype distribution between digestive system cancer cases and controls (OR 0.92, 95% CI 0.82–1.04). Therefore, the results that included the study by Zhen et al should be cautiously interpreted.
Heterogeneity and sensitivity analyses
Substantial heterogeneity was observed between studies for the association between XPA A23G polymorphism and digestive system cancer risk in all genetic models (dominant model: I2=73%, P<0.00001; recessive model: I2=83%, P<0.00001; GA versus GG, I2=52%, P=0.005; and AA versus GG, I2=83%, P<0.00001). We therefore assessed the source of heterogeneity by ethnicity, tumor type, and source of control. The heterogeneity was partly decreased or removed for gastric cancers, colorectal cancers, Caucasians, and population-based studies. However, there was still significant heterogeneity for esophageal cancer, Asians, and hospital-based populations. Sensitivity analysis was then performed to evaluate the stability of the results. The statistical significance of the results was not altered when any single study was omitted, confirming the stability of the findings.
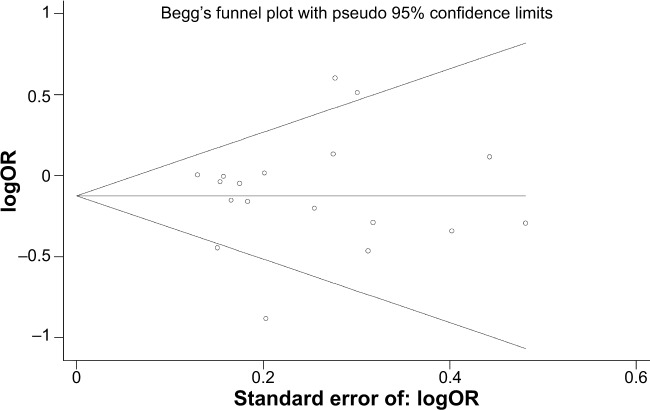
Publication bias
We used the Begg’s funnel plot and Egger’s test to address potential publication bias in the available literature. The shape of the funnel plots did not reveal any evidence of funnel plot asymmetry (Figure 5). Egger’s test also showed that there was no statistical significance for the evaluation of publication bias (dominant model: P=0.703; GA versus GG, P=0.792; AA versus GG, P=0.895; recessive model, P=0.678).
Figure 5.
Begg’s funnel plot for publication bias (GA versus GG).
Discussion
The evidence suggests that reduced DNA repair capacity may lead to genetic instability and carcinogenesis, genes involved in DNA repair have been proposed as candidate cancer susceptibility genes.37 The NER pathway may be important in modulating susceptibility to cancer, because it is the primary mechanism for repair of a wide variety of types of DNA damage.38–40 There are several core genes in the NER pathway (eg, ERCC1, XPA, XPB/ERCC3, XPC, XPD/ERCC2, XPE/DDB1, XPF/ERCC4, and XPG/ERCC5). Of these, the XPA gene is one of the central players, with a vital role in repairing DNA damage and maintaining the integrity of the genome.4 Recently, A23G polymorphism of the XPA gene was reported to confer a risk of digestive system cancers. Furthermore, a number of epidemiological studies have evaluated the association between this polymorphism and risk of digestive system cancers, but the results remain inconclusive. Dong et al11 and Guo et al14 reported that the XPA A23G polymorphism was associated with a decreased risk of esophageal squamous cell carcinoma and gastric cardiac adenocarcinoma in a high-incidence population in northern China; however, in a study from the USA, Pan et al22 suggested that the heterozygous AG genotype of the XPA 5′ untranslated region was associated with a 2.11-fold increased risk, and the increased risk reached 3.10-fold for the homozygous variant GG genotype in esophageal cancer. Furthermore, Gil et al13 found that the XPA A23G polymorphism may be unrelated to the risk of sporadic colorectal cancer; similarly, Hansen et al16 failed to detect an association between the polymorphism and risk of colorectal cancer in a Danish population. These inconsistent results may be attributed to differences in genetic backgrounds, environmental factors, and other factors.
A recent meta-analysis41 evaluated the association between XPA A23G polymorphism and cancer risk, and reported that this polymorphism is associated with an increased lung cancer risk and may be a low-penetrant risk factor for development of cancer in people of Asian ethnicity. Subsequently, Liu et al42 conducted another meta-analysis to assess the association between A23G polymorphism and risk of cancer, and suggested that the XPA A23G G allele is a low-penetrant risk factor for development of cancer. However, only few studies focusing on digestive system cancers (eight and nine studies, respectively) were included in the above meta-analysis, and due to the limited study number, further analyses was not conducted. Compared with those studies, we conducted a more comprehensive literature search in different databases (including Web of Science, China National Knowledge Infrastructure, WanFang, and VIP) and included several additional studies,17,19,20,23–26 which allowed for a larger number of subjects and more precise risk estimation. In this meta-analysis, we pooled 18 studies to explore the association between A23G polymorphism and risk of gastrointestinal cancers. The results demonstrated that XPA A23G polymorphism is not associated with digestive system cancer risk.
The outcomes from meta-analysis can be affected by several factors, such as ethnicity, cancer origin, and control selection. Therefore, subgroup analyses were conducted. In this study, stratification by ethnicity, tumor type, and source of control revealed no significant association. The results seem to contradict the previous meta-analyses. The discrepancies are probably due to the small size of the A23G polymorphism in determining susceptibility to digestive system cancers in the previous meta-analyses. Moreover, the biological mechanisms of the XPA gene in carcinogenesis are complicated, and may be mediated by the activities of multiple genes (such as ERCC1 and XPF) in the NER pathway, the function of which may be different in digestive system cancers than in other cancers. In addition, cancer is a multifactorial disease that results from complex interactions between many environmental and genetic factors. Therefore, when we only consider suspected gene polymorphism in digestive system cancers and ignore the role of other genes and environmental factors, we might fail to conclude a real association.
Two significant issues should be addressed in this study, ie, heterogeneity and publication bias, which may influence the results of a meta-analysis. We did not detect a significant publication bias in this meta-analysis, suggesting that our results are reliable. With regard to heterogeneity, in this meta-analysis, heterogeneity was found in overall comparison under all genetic models, when stratified by ethnicity, tumor type, and source of control, the heterogeneity was partly decreased or removed among gastric and colorectal cancers, Caucasians, and population-based subgroups. However, heterogeneity still existed for esophageal cancer, Asians, and hospital-based populations. In addition, when the study by Zhen et al was excluded, the heterogeneity decreased. Our results suggest that the ethnic background, different types of tumor, and the particular study might be the source of heterogeneity. Then sensitivity analyses were conducted by successively excluding one study, the estimated pooled odd ratio changed quite little, indicating that the results of this meta-analysis were stable.
This meta-analysis has limitations that must be acknowledged. First, all case-control studies included were done in Asians and Caucasians, so our results may be applicable only to these populations. More studies on Africans and other ethnic groups are needed. Second, the controls included in our analysis were selected variously from either population-based or hospitals. Therefore, misclassification bias was possible because these studies may have included control groups that have different risks for developing digestive system cancers. Third, our results were based on unadjusted estimates, without adjustment for family history or other risk factors, which may cause serious confounding bias.
Conclusion
In summary, this meta-analysis suggests that XPA A23G polymorphism is not associated with a risk of digestive system cancers. However, large and well-designed studies taking into consideration gene–gene and gene–environment interactions are warranted to validate our findings.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Pharoah PD, Dunning AM, Ponder BA, Easton DF. Association studies for finding cancer-susceptibility genetic variants. Nat Rev Cancer. 2004;4(11):850–860. doi: 10.1038/nrc1476. [DOI] [PubMed] [Google Scholar]
- 3.Risch N. The genetic epidemiology of cancer: interpreting family and twin studies and their implications for molecular genetic approaches. Cancer Epidemiol Biomarkers Prev. 2001;10(7):733–741. [PubMed] [Google Scholar]
- 4.Asahina H, Kuraoka I, Shirakawa M, et al. The XPA protein is a zinc metalloprotein with an ability to recognize various kinds of DNA damage. Mutat Res. 1994;315(3):229–237. doi: 10.1016/0921-8777(94)90034-5. [DOI] [PubMed] [Google Scholar]
- 5.Batty DP, Wood RD. Damage recognition in nucleotide excision repair of DNA. Gene. 2000;241(2):193–204. doi: 10.1016/s0378-1119(99)00489-8. [DOI] [PubMed] [Google Scholar]
- 6.Rademakers S, Volker M, Hoogstraten D, et al. Xeroderma pigmentosum group A protein loads as a separate factor onto DNA lesions. Mol Cell Biol. 2003;23(16):5755–5767. doi: 10.1128/MCB.23.16.5755-5767.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Volker M, Mone MJ, Karmakar P, et al. Sequential assembly of the nucleotide excision repair factors in vivo. Mol Cell. 2001;8(1):213–224. doi: 10.1016/s1097-2765(01)00281-7. [DOI] [PubMed] [Google Scholar]
- 8.de Laat WL, Jaspers NG, Hoeijmakers JH. Molecular mechanism of nucleotide excision repair. Genes Dev. 1999;13(7):768–785. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- 9.Kozak M. Interpreting cDNA sequences: some insights from studies on translation. Mamm Genome. 1996;7(8):563–574. doi: 10.1007/s003359900171. [DOI] [PubMed] [Google Scholar]
- 10.Wu X, Zhao H, Wei Q, et al. XPA polymorphism associated with reduced lung cancer risk and a modulating effect on nucleotide excision repair capacity. Carcinogenesis. 2003;24(3):505–509. doi: 10.1093/carcin/24.3.505. [DOI] [PubMed] [Google Scholar]
- 11.Dong Z, Guo W, Zhou R, et al. Polymorphisms of the DNA repair gene XPA and XPC and its correlation with gastric cardiac adenocarcinoma in a high incidence population in North China. J Clin Gastroenterol. 2008;42(8):910–915. doi: 10.1097/MCG.0b013e3180f6262c. [DOI] [PubMed] [Google Scholar]
- 12.Feng XX, Duan PF, Wang LB, Zhang JB, Lu ZX. Study on the relationship between polymorphisms of XPA gene and susceptibility of esophageal cancer. Zhonghua Liu Xing Bing Xue Za Zhi. 2008;29(9):930–933. Chinese. [PubMed] [Google Scholar]
- 13.Gil J, Ramsey D, Stembalska A, et al. The C/A polymorphism in intron 11 of the XPC gene plays a crucial role in the modulation of an individual’s susceptibility to sporadic colorectal cancer. Mol Biol Rep. 2012;39(1):527–534. doi: 10.1007/s11033-011-0767-5. [DOI] [PubMed] [Google Scholar]
- 14.Guo W, Zhou RM, Wan LL, et al. Polymorphisms of the DNA repair gene xeroderma pigmentosum groups A and C and risk of esophageal squamous cell carcinoma in a population of high incidence region of North China. J Cancer Res Clin Oncol. 2008;134(2):263–270. doi: 10.1007/s00432-007-0283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall J, Hashibe M, Boffetta P, et al. The association of sequence variants in DNA repair and cell cycle genes with cancers of the upper aerodigestive tract. Carcinogenesis. 2007;28(3):665–671. doi: 10.1093/carcin/bgl160. [DOI] [PubMed] [Google Scholar]
- 16.Hansen RD, Sorensen M, Tjonneland A, et al. XPA A23G, XPC Lys939Gln, XPD Lys751Gln and XPD Asp312Asn polymorphisms, interactions with smoking, alcohol and dietary factors, and risk of colorectal cancer. Mutat Res. 2007;619(1–2):68–80. doi: 10.1016/j.mrfmmm.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 17.Huang XX. Master’s thesis. Fujian, People’s Republic of China: Fujian Medical University; 2007. Polymorphisms of XPA and XRCC3, environmental agent and susceptibilities to esophageal, cardia and non-cardia gastric cancer. [Google Scholar]
- 18.Jelonek K, Gdowicz-Klosok A, Pietrowska M, et al. Association between single-nucleotide polymorphisms of selected genes involved in the response to DNA damage and risk of colon, head and neck, and breast cancers in a Polish population. J Appl Genet. 2010;51(3):343–352. doi: 10.1007/BF03208865. [DOI] [PubMed] [Google Scholar]
- 19.Joshi AD, Corral R, Siegmund KD, et al. Red meat and poultry intake, polymorphisms in the nucleotide excision repair and mismatch repair pathways and colorectal cancer risk. Carcinogenesis. 2009;30(3):472–479. doi: 10.1093/carcin/bgn260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu R. PhD thesis. Nanjing, People’s Republic of China: Southeast University; 2007. Study on expression characteristics of susceptibility genes and genetic susceptibility markers for esophageal carcinomas. [Google Scholar]
- 21.Palli D, Polidoro S, D’Errico M, et al. Polymorphic DNA repair and metabolic genes: a multigenic study on gastric cancer. Mutagenesis. 2010;25(6):569–575. doi: 10.1093/mutage/geq042. [DOI] [PubMed] [Google Scholar]
- 22.Pan J, Lin J, Izzo JG, et al. Genetic susceptibility to esophageal cancer: the role of the nucleotide excision repair pathway. Carcinogenesis. 2009;30(5):785–792. doi: 10.1093/carcin/bgp058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie WM. PhD thesis. Nanning, People’s Republic of China: Guangxi Medical University; 2007. Association between the nucleotide excision repair (NER) genes polymorphisms and genetic susceptibility and clinical phenotype of hepatocellular carcinoma. [Google Scholar]
- 24.Zhang WC. Master’s thesis. Nanjing, People’s Republic of China: Southeast University; 2006. Effects of genetic and environmental risk factors to esophageal cancer in Huaian population. [Google Scholar]
- 25.Zhen Q. Master’s thesis. Zhengzhou, People’s Republic of China: Zhengzhou University; 2012. XPA gene polymorphisms and susceptibility to developing esophageal cancer in Chinese Han population. [Google Scholar]
- 26.Zhu XL. Master’s thesis. Wuhan, People’s Republic of China: Huazhong University of Science and Technology; 2008. Study on relationship between DNA repair genes polymorphism of hOGG1, XPC, XPA and esophageal squamous cell carcinoma susceptibility. [Google Scholar]
- 27.Duan PF. Master’s thesis. Shanxi, People’s Republic of China: Shanxi Medical University; 2007. A case-control study on polymorphisms of XPA, XPC and esophageal carcinoma. [Google Scholar]
- 28.Wan LL, Zhou RM, Wang N, Li Y, Guo W. Correlation of XPA polymorphisms to the risk of squamous cell carcinoma and gastric cardia adenocarcinoma. Cancer Research of Prevention and Treatment. 2007;34(1):63–67. Chinese. [Google Scholar]
- 29.Zhang WC, Yin LH, Pu YP, et al. Relationship between polymorphisms of DNA repair genes and esophageal cancer susceptibility. Chin J Public Health. 2006;22(5):557–558. Chinese. [Google Scholar]
- 30.Ma WJ, Lv GD, Zheng ST, et al. DNA polymorphism and risk of esophageal squamous cell carcinoma in a population of North Xinjiang, China. World J Gastroenterol. 2010;16(5):641–647. doi: 10.3748/wjg.v16.i5.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25(9):603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 32.Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med. 1997;127(9):820–826. doi: 10.7326/0003-4819-127-9-199711010-00008. [DOI] [PubMed] [Google Scholar]
- 33.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22(4):719–748. [PubMed] [Google Scholar]
- 34.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 35.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–1101. [PubMed] [Google Scholar]
- 36.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wood RD, Mitchell M, Sgouros J, Lindahl T. Human DNA repair genes. Science. 2001;291(5507):1284–1289. doi: 10.1126/science.1056154. [DOI] [PubMed] [Google Scholar]
- 38.Goode EL, Ulrich CM, Potter JD. Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiol Biomarkers Prev. 2002;11(12):1513–1530. [PubMed] [Google Scholar]
- 39.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Ann Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 40.Wood RD. DNA damage recognition during nucleotide excision repair in mammalian cells. Biochimie. 1999;81(1–2):39–44. doi: 10.1016/s0300-9084(99)80036-4. [DOI] [PubMed] [Google Scholar]
- 41.Ding D, Zhang Y, Yu H, et al. Genetic variation of XPA gene and risk of cancer: a systematic review and pooled analysis. Int J Cancer. 2012;131(2):488–496. doi: 10.1002/ijc.26391. [DOI] [PubMed] [Google Scholar]
- 42.Liu J, Zhang Z, Cao XL, et al. XPA A23G polymorphism and susceptibility to cancer: a meta-analysis. Mol Biol Rep. 2012;39(6):6791–6799. doi: 10.1007/s11033-012-1504-4. [DOI] [PubMed] [Google Scholar]