Abstract
Background
Recent studies have proposed a unified genetic model for Facioscapulohumeral muscular dystrophy (FSHD), identifying potential therapeutic targets for future clinical trials. Serum biomarkers related to disease activity will be important for proof of concept or early phase clinical studies.
Objective
To identify potential serum biomarkers in FSHD for possible use in future clinical trials.
Methods
We performed a prospective cross-sectional study of serum biomarkers in 22 FSHD patients (19 FSHD1, 3 FSHD2) compared to 23 age and gender-matched healthy controls using a commercial multiplex, microsphere-based immune-fluorescent assay of 243 markers (Myriad, Human Discovery MAP 250, v2.0).
Results
169 markers had values sufficient for analysis. Correction for multiple testing identified 7 biomarkers below a 5% false discovery rate: creatine kinase MB fraction (CKMB, 6.52 fold change, P<0.0001), tissue-type plasminogen activator (PLAT, 1.64 fold change, P<0.0001), myoglobin (2.23 fold change, P=0.0001), epidermal growth factor (EGF, 2.33 fold change, P=0.0004), chemokine (C-C motif) ligand 2 (1.48 fold change, P=0.0004), CD 40 ligand (1.89 fold change, P=0.001), and vitronectin (VTN, 1.28 fold change, P=0.001). Moderate correlations to measures of FSHD disease were seen for CKMB, PLAT, and EGF. Markers in the plasminogen pathway (PLAT, serpin peptidase inhibitor, and VTN) were correlated with each other in FSHD but not healthy controls.
Conclusions
Commercial multiplex immune-fluorescent screening is a potentially powerful tool for identifying biomarkers for future FSHD therapeutic trials. Biomarkers identified in this study warrant further study in a larger prospective validation study.
Keywords: Muscle Disease, Facioscapulohumeral Muscular Dystrophy, DUX4, Biomarker, Proteomics
Introduction
Facioscapulohumeral muscular dystrophy is one of the most prevalent muscular dystrophies (prevalence 1:15,000–1:20,000), with a typically descending pattern of weakness, starting in the face and shoulders, followed later by the distal and proximal lower extremities [1–5]. The current genetic model proposes both FSHD types 1 and 2 are caused by de-repression of DUX4 expression, a retrogene believed to cause disease in toxic gain of function manner [6]. This model identifies potential therapeutic targets. Biomarkers serve two equally important roles for future FSHD clinical trials: 1) mechanistic biomarkers to serve as proof of concept for early phase studies of disease-modifying therapies; and 2) biomarkers that reliably track disease activity. Currently there are no validated serum biomarkers for future FSHD clinical trials, and identification of biomarkers was listed as a major goal of a 2012 FSHD clinical trial workshop held in the Netherlands [7].
DUX4 is a double homeobox transcription factor that alters regulation of a large number of genes – most notably genes involved in germline and early stem cell development, cancer testis antigens and genes involved in innate immunity [8]. Although DUX4 itself is difficult to measure directly in muscle tissue from FSHD patients, downstream targets of DUX4 expression are more reliably detected. A prominent feature of FSHD muscle is the presence of inflammatory infiltrates in up to 30% of muscle biopsies [9]. Since DUX4 is normally only expressed in testes, one avenue of thinking is that activation of genes normally expressed in immune-privileged environments could initiate an immune response in FSHD muscle. An Italian study identified STIR positive muscles on MRI which, when biopsied, demonstrated CD8+ immune infiltrations, and in half of these biopsies up-regulation of downstream DUX4 targets [10, 11]. The same study also demonstrated that patients with STIR positive muscles had elevations in a number of inflammatory markers from peripheral blood monocyte culture: including IL6, IL10, IL12, and TNF-alpha.
In addition to studies of direct targets of DUX4, genome screens identified several broad categories of genes with altered regulation in FSHD: including genes involved in angiogenesis, myogenic differentiation, oxidative stress, and inflammation [12–14]. However no study to date has identified downstream proteins from these targets increased in serum of FSHD patients.
Here we performed a prospective study of serum biomarkers in FSHD using a commercial multiplex, microsphere-based immune-fluorescent assay of 243 markers – with the goals of demonstrating the usefulness of this approach in FSHD, and identifying potential biomarkers warranting further study.
Materials and Methods
We performed a prospective cross-sectional study of 22 FSHD participants and 23 age and gender matched healthy volunteers at the University of Rochester Medical Center between 5/2012 and 12/2012. The study was approved by institutional review board and written and informed consent was obtained from all participants.
FSHD participants were recruited to match the most likely participants recruited in future clinical trials: between 18 and 75 years of age; symptomatic weakness in at least one limb; independently ambulatory (up to 30 feet without assistance); and genetically confirmed as previously described [15, 16]. Briefly, D4Z4 repeat array size on chromosome 4q35 was determined by Southern blot after double digestion with EcoRI/BlnI restriction enzymes. Normal individuals have fragments >38kb (≥11 units) while patients with FSHD have fragments between 10–38 kb (1–10 units). For contraction-independent FSHD2 CpG methylation measurements were taken after cleavage with the methylation-sensitive endonuclease FseI, using a methylation threshold of <25%[17]. For this investigational study participants were excluded if medical conditions precluded performing study procedures safely or participants had medical conditions or were on medications believed to influence biomarker discovery: this included participants with a recent history of infection; use of warfarin or other anticoagulation; history of malignancy with ongoing treatment with chemotherapeutic agents; history of autoimmune disorder; use of immunomodulating agents; any history of intermittent use of large doses of muscle anabolic or catabolic agents such as corticosteroids, oral testosterone or derivatives, or oral beta agonists; or pregnancy.
We collected serum samples from healthy adults (18–75 years of age) for comparison, equally divided by gender and age (in 10 year increments). To be included as normal controls, participants had a normal neuromuscular exam and had to meet the same exclusion criteria listed above for FSHD subjects.
Participants came for a 1 day study visit in the general clinical research center, where all subjects underwent a blood draw for biomarker testing, and bedside neurological exam and manual muscle testing. FSHD participants also had clinical severity scored on a scale of 1–10 (1 minimally affected, 10 severely affected) and filled out a medical history and symptom questionnaire [18]. The severity score takes into account the extent of weakness in various body regions and considers the descending spread of symptoms from face and shoulders to pelvic and leg muscles typical of FSHD. Higher scores are assigned to patients with involvement of pelvic and proximal lower limb muscles.
Multi-analyte profile
We collected 10cc of whole blood from all participants in red top Vacutainer (Becton Dickinson) tubes. The samples were centrifuged at 1,350 × g for 20 minutes. Following centrifugation, the liquid component (serum) was transferred into a clean polypropylene tube using a Pasteur pipette. The serum was apportioned into 0.5 ml aliquots and stored at –80°C. Samples were sent to Myriad Rule Based Medicine (RBM, Austin, TX) for analysis of 243 analytes using the Human Discovery Multi-Analyte Profile (MAP) 250, version 2.0 and a Luminex xMAP technology platform as previously described [19, 20]. The RBM Discovery MAP 250 is a commercial validated platform which measures a battery of analytes including markers of autoimmunity, infection, cancer-related, hormones, cytokines, cardiovascular risk, acute phase reactants, and others (for full list of analytes, see Supplementary Table 1). Samples were thawed and pipetted into a 96 microtiter plate. The rest of the process was fully automated per RBM procedures (for white paper http://rbm.myriad.com/scientific-literature/data-quality/). Samples from each plate were added to reaction wells containing fluorescently labeled microspheres conjugated to antibodies encoded with unique fluorescent signatures. The beads were incubated with the sample and antigens of interest allowed to bind to their targets. Multiplexed cocktails of biotinylated detection reagents were added to the sample followed by the addition of fluorescent reporter molecules. RBM determined analyte concentrations using The Luminex machine, which functions similarly to a flow cytometer, using hydrodynamic focusing to pass the microspheres between lasers, and fluorescent sensors to measure the median fluorescent intensity. RBM used a unique set of controls with known quantities of the analyte of interest to create two sets of 8 point calibrators. Concentrations were determined by fitting to standardized concentration curves using RBM curve fitting routines. This provided a dynamic concentration range between fg/ml to mg/ml and intra-assay coefficient of variation < 10%. Data was reported back as concentrations (average of two independent measures) and lower assay limits (lowest level of quantification, LLOQ). RBM MAP technology meets Clinical Laboratory Standard Institute standards.
Statistics
Population characteristics were investigated using medians, quartiles, and confidence limits. We tested for differences in gender between groups using Fisher exact test. We evaluated the age matching of healthy controls by decade using the Wald test from a logistic regression. Multi-analyte profile analysis: only markers with protein levels in the detectible range were used for analysis. 243 markers are included on the panel (Supplemental Table 1). 74 markers had undetectable or > 30% missing values and were excluded from analysis. Values below the lower limit of quantification (LLOQ) were imputed by LLOQ/2, and represented no more than 1.3% of values. Distributions were tested for normality using the Shapiro-Wilk test and visually inspected for linearity using QQ plots, and non-normal distributions were log transformed for analysis, and verified by checking linearity of QQ plot. If normality was not achieved non-parametric testing was performed using the Kruskal-Wallis Test. Differences between FSHD and healthy control populations were evaluated using two sample t-test, assuming equal variance in the two groups. All results were checked for residual gender or age effects by analysis of variance using the SAS GLM procedure and adjusting for gender and age. If a significant interaction was found adjusted estimates using least means squares test with associated confidence limits were determined from the GLM model. Fold change was calculated by dividing the larger mean by the smaller (positive if FSHD>healthy control, negative if healthy control > FSHD). P-value of 0.05 was used for a cut-off, and all tests were two-sided. Multiple testing was addressed utilizing the false discovery rate (FDR) as described by Benjamini and Hochberg [21]. FDR adjusted p-values and FDR q values were calculated utilizing an online published FDR calculator [22]. As this is an investigational study with limited power FDR values were presented for all markers with a P-value <0.10. Associations between markers and measures of FSHD clinical severity and between markers in the same metabolic pathway were calculated using Spearman correlation coefficients, and Fisher’s ζ transformation. Statistical analysis was performed using SAS version 9.3 (SAS Institute Inc., Cary, NC), and STATA version 11.2 (StataCorp, College Station, TX).
Results
We found no differences between FSHD participants and healthy controls in gender frequency or age by decade, with equal numbers of men and women, with most participants between 30 and 69 years of age (77.27% for FSHD, and 73.91% of healthy control, Table 1), but all decades represented. FSHD participants were mostly FSHD type 1 (19, or 86.36%). The full range of D4Z4 contraction and clinical severity was represented, with a median clinical severity score of 5.5 (range 0 to 8), and median D4Z4 contraction size of 25 kb (or 6 repeats, range for FSHD type 1 of 13–35 kb, or 2–10 repeats).
Table 1.
Patient Characteristics.
| Category | FSHD | Control | P-value |
|---|---|---|---|
| n | 22 | 23 | - |
| Male (%) | 12 (54.55) | 11 (47.83) | 0.77 |
| Age Years (%) | 1.0* | ||
| 18–19 | 1 (4.55) | 1 (4.35) | |
| 20–29 | 3 (13.64) | 4 (17.39) | |
| 30–39 | 4 (18.18) | 4 (17.39) | |
| 40–49 | 5 (22.73) | 5 (21.74) | |
| 50–59 | 2 (9.09) | 2 (8.70) | |
| 60–69 | 6 (27.27) | 6 (26.09) | |
| 70–79 | 1 (4.55) | 1 (4.35) | |
| FSHD1 (%) | 19 (86.36) | n/a | - |
| median CSS (Q1, Q3) | 5.5 (2, 6) | n/a | - |
| median D4Z4 kb (Q1, Q3) | 25 (23, 32) | n/a | - |
P-value from logistic regression Wald statistic for interaction between age category and disease.
FSHD = Facioscapulohumeral; n=number; CSS = clinical severity score; kb = kilobase
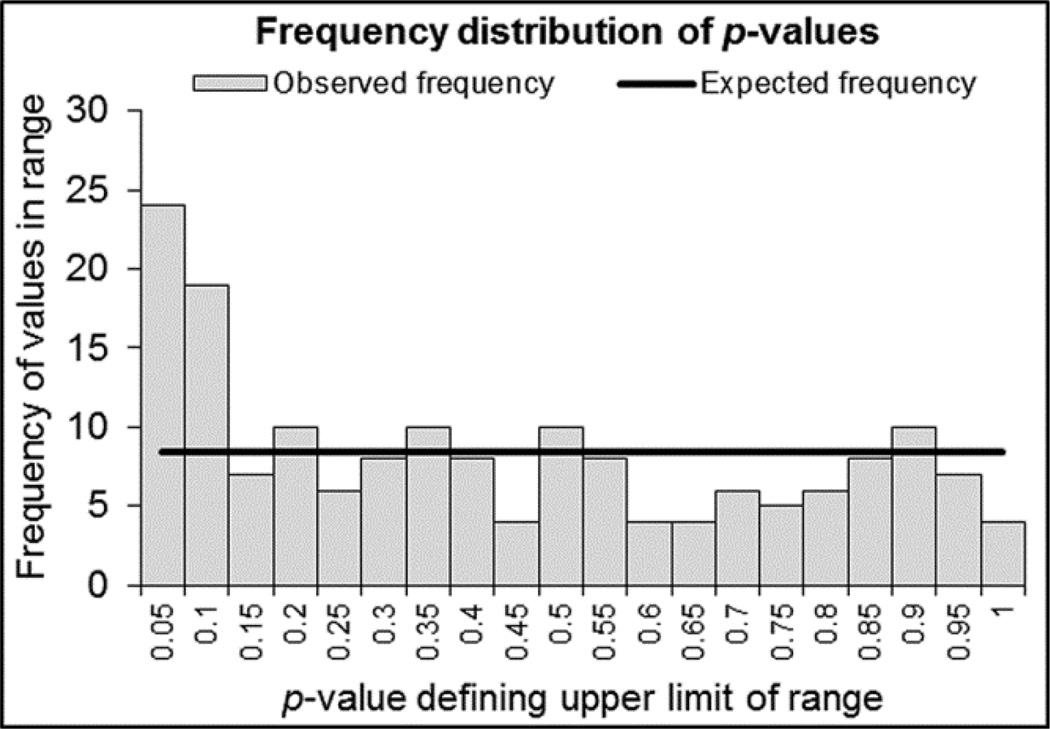
169 analytes had > 30% of values present and were included in the analysis. Twenty-four analytes (14.20%) were significantly increased or decreased in FSHD compared to healthy control and 43 (25.44%) had a P-value <0.10 (Supplemental Table 2). By comparing the distribution of actual P-values to the expected P-value frequency given a normal distribution we identified a peak for markers with P-values <0.10 for FSHD participants (Figure 1), suggesting the Myriad Discovery MAP panel included markers abnormally increased or decreased in FSHD.
Figure 1.
Observed P-values versus expected. Frequency plot showing the number of markers within a given p-value range. The frequency below P<0.10 is above that expected by chance given a normal distribution of p-values, suggested the RBM MAP panel is picking up a signal in analytes different in FSHD participants compared to normal controls. Expected frequency = total markers/20 P-value divisions, or 8.5 (solid line). Frequency plot created using the FDR calculator [22].
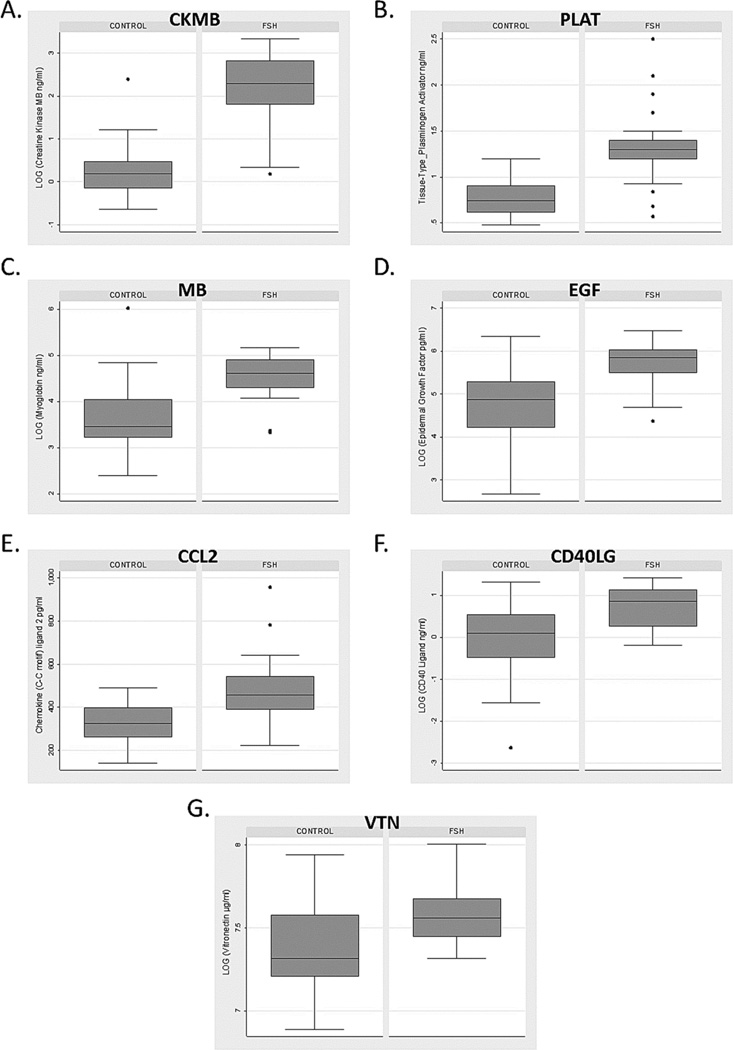
We identified 7 markers with a false discovery rate q-value less than 5% (Table 2, Figure 2). All were increased in FSHD compared to healthy controls and included muscle-related proteins (creatine kinase MB [CKMB], 6.52 fold change, P<0.0001; myoglobin [MB], 2.23 fold change, P=0.006), plasminogen pathway markers (tissue-type plasminogen activator [PLAT], 1.64 fold change, P<0.0001; vitronectin [VTN], 1.28 fold change, P=0.03), inflammatory markers (chemokine (C-C motif) ligand 2 [CCL2], 1.48 fold change, P=0.0004; CD40 ligand [CD40LG], 1.89 fold change, P=0.03), and epidermal growth factor (EGF, 2.33 fold change, P=0.01). No differences were detected between FSHD types 1 and 2 for these markers.
Table 2.
Markers with FDR <0.05
| Marker | FC | P-value | FDRq | Description& |
|---|---|---|---|---|
| Creatine Kinase MB*#$ | 6.52 | <0.0001 | <0.0001 | cytoplasmic enzyme involved in energy homeostasis – cardiac isoenzyme |
| Tissue-type Plasminogen Activator*% | 1.64 | <0.0001 | 0.0002 | serine protease involved in fibrinolysis, tumor angiogenesis, and myogenesis [29] |
| Myoglobin*# | 2.23 | 0.0001 | 0.006 | Iron-binding muscle protein |
| Epidermal Growth Factor*# | 2.33 | 0.0004 | 0.01 | Potent mitogenic factor, important role in the growth, proliferation and differentiation of numerous cell types. |
| Chemokine (C-C motif) ligand 2 | 1.48 | 0.0004 | 0.01 | Recruits monocytes to sites of injury and infection. |
| CD40 Ligand*# | 1.89 | 0.001 | 0.03 | Expressed on the surface of T cells, regulates B cell function by engaging CD40 |
| Vitronectin*# | 1.28 | 0.001 | 0.03 | Cell adhesion factor. Stabilizes PAI-1*. Inhibitor of the membrane-damaging effect of the terminal cytolytic complement pathway. |
Descriptions taken from the Human Protein Reference Data Base, and NCBI gene reference
Markers with false discovery rate <5%
Log transformed for analysis.
Group estimates and statistical test from the GLM model adjusted for gender
Group estimates and statistical test from the GLM model adjusted for age
FC = fold change (positive FSH > control; negative control > FSH); FDR = false discovery rate
Figure 2.
Box plots of markers positive after adjustment for multiple comparisons. A) creatine kinase MB (CKMB); B) tissue-type plasminogen activator (PLAT); C) myoglobin (MB); D) epidermal growth factor (EGF); E) chemokine (C-C motif) ligand 2 (CCL2); F) CD40 ligand (CD40LG); and G) vitronectin (VN). Box and whisker plots show distribution of concentrations for each analyte. The box represents 50% of population; dark line is the median; whiskers are adjacent upper/lower values; and dots represent outliers.
When evaluating categories of markers markers with a P-value <0.1 might be of interest for comparison to future studies, and help identify classes of biomarkers for future investigations. Two muscle related proteins (CKMB and MB) were increased, as might be expected in a muscular dystrophy with leaky or degenerative muscle fibers, and served as a positive control for the panel. Inflammatory markers increased included CCL2 (synonym: monocyte chemotactic protein 1, MCP1), CD40LG, complement C3 (C3), and CD40 antigen (CD40); and decreased included S100 calcium binding protein A12 (S100A12, synonym: advanced glycation end products receptor ligand, EN-RAGE), tumor necrosis factor receptor superfamily, member 10c, decoy without an intracellular domain (TNFRSF10C, synonym: TNF-related apoptosis inducing ligand receptor 3, TRAIL-R3) and chemokine (C-C motif) ligand 23 (CCL23, synonym: myeloid progenitor inhibitory factor 1, MPIF-1). Cancer-related markers increased or decreased included cancer antigen 15-3 (CA 15-3), mesothelin (MSLN), phosphoserine aminotransferase (PSAT1), and v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 (ERBB2, synonym: human epidermal growth factor receptor 2, HER-2). Metabolic factors in glucose metabolism included leptin (LEP) and insulin-like growth factor binding protein 1 (IGFBP1). Interestingly, a number of markers related to the plasminogen pathway were increased in FSHD: including PLAT, serpin peptidase inhibitor (SERPINE1, synonym: plasminogen activator inhibitor type 1, PA-1), and vitronectin (VTN). Notable markers which were undetectable in serum from FSHD participants were interleukins 6, 10, and 12, and tumor necrosis factor alpha, which had previously been reported to be increased in the supernatant of cultured peripheral blood monocytes in patients with FSHD [10].
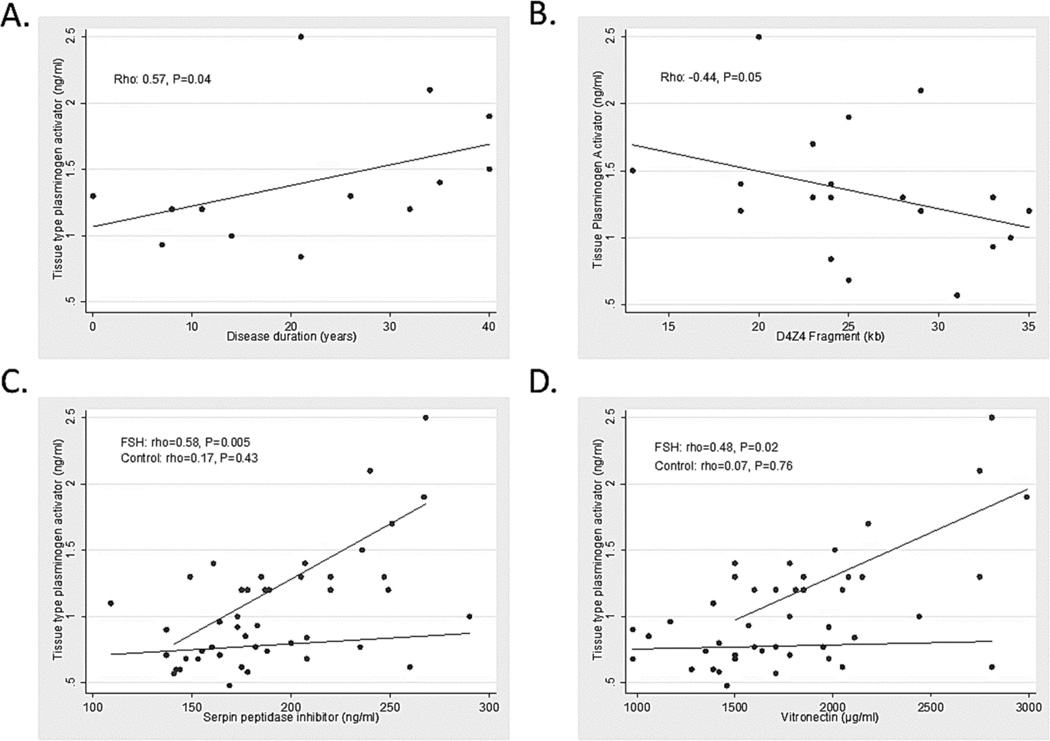
Multiple biomarkers were correlated to measures of disease severity or D4Z4 contraction size despite the small sample size (Table 3). CKMB showed a moderate correlation to clinical severity (rho 0.44, P=0.04). EGF showed a moderate inverse correlation to age of symptom onset (rho -0.54, P=0.03). The plasminogen pathway markers showed inverse correlations to both age of symptom onset and D4Z4 contraction size, and positive correlation to disease duration (PLAT versus age of symptom onset, rho −0.67, P=0.003, versus disease duration, rho=0.57, P=0.04, and versus D4Z4 contraction size, rho −0.45, P=0.05, Figure 3A–B).
Table 3.
Correlations for markers positive after adjusting for multiple comparisons. Spearman rho values with P-values in parenthesis.
| Marker | Clinical Severity Score* (P-value) |
Age of Symptom Onset (P-value) |
Disease Duration# (P- value) |
D4Z4 Contraction (P- value) |
|---|---|---|---|---|
| Creatine kinase MB | 0.44 (0.04) | −0.24 (0.36) | 0.14 (0.64) | 0.17 (0.49) |
| Tissue-type plasminogen activator | 0.21 (0.43) | −0.67 (0.003) | 0.57 (0.04) | −0.45 (0.05) |
| Myoglobin | 0.47 (0.06) | 0.01 (0.96) | −0.05 (0.77) | 0.11 (0.65) |
| Epidermal growth factor | 0.44 (0.07) | −0.54 (0.03) | 0.01 (0.98) | −0.36 (0.13) |
| Chemokine (C-C motif) ligand 2 | 0.03 (0.90) | 0.15 (0.57) | −0.05 (0.87) | 0.08 (0.75) |
| CD40 ligand | −0.09 (0.73) | −0.02 (0.93) | −0.52 (0.07) | −0.20 (0.41) |
| Vitronectin | −0.07 (0.78) | −0.47 (0.06) | 0.17 (0.59) | −0.41 (0.07) |
CSS adjusted for age = 1000*CSS/age
Disease duration = age-age of symptom onset
Figure 3.
Association of tissue-type plasminogen activator to measures of disease progression, and other markers in the plasminogen pathway. A) Disease duration; B) D4Z4 fragment size; C) serpin peptidase inhibitor; and D) vitronectin.
A number of markers showed expected correlations within a metabolic pathway for both FSHD participants and healthy controls. Insulin compared to LEP or IGFBP1were significantly positively correlated (rho-values between 0.39–0.56, and P-values between 0.006–0.07). EGF and CD40LG were positively correlated (rho-values 0.63–0.74 and P-values <0.01). Proteins in the plasminogen pathway were only correlated in FSHD (PLAT and SERPINE1 rho=0.58, p=0.005; PLAT and VTN rho=0.48, P=0.02), suggesting this pathway may be selectively activated in FSHD compared to healthy controls (Figure 3C–D).
Discussion
This is the first study of its kind utilizing a broad automated multi-analyte array platform to identify serum biomarkers in FSHD. We found 24 markers with concentrations significantly different from healthy controls, and seven remained positive after adjustment for multiple testing and a false discovery rate of 5%. Markers of interest included proteins in the plasminogen, inflammatory, and wound healing pathways. Markers in the plasminogen pathway had cross-sectional correlations to FSHD measures of disease and appeared to be selectively metabolically activated in FSHD compared to healthy controls, suggesting they might serve as useful markers of muscle regeneration.
Since the publication of the unified genetic hypothesis for FSHD there has been considerable interest in identifying DUX4-related mechanistic biomarkers [7, 23]. The obvious target, DUX4, has been difficult to measure directly in muscle tissue of FSHD patients, possibly because DUX4 demonstrates a stochastic burst-like expression pattern and is present a very low abundance [24]. While DUX4 itself is in very low abundance and hard to detect, the downstream targets of the DUX4 transcription factor have been more reliably identified in in vitro models and muscle tissue from FSHD patients [8]. Genes up-regulated by DUX4 include cancer-testis antigens. One model of disease activity posits that genes normally only expressed in an immune-privileged environment, such as in the testis or during development, can trigger an immune response in somatic cells [8, 24]. This model is attractive for FSHD as it explains one observation which has never been completely understood: a high frequency of endomysial and perivascular inflammatory cell infiltrates in up to a third of FSHD biopsies [9, 25]. The inflammatory infiltrates are predominantly CD8+ with more prominent CD4+ T cells in the perivascular infiltrates –and while inflammatory infiltrates can be seen in other dystrophies, the predilection of the inflammatory infiltrates for the perivascular regions is unique to FSHD [9, 10]. In support of this model a study identified MRI STIR positivity in FSHD patient muscles which corresponded on muscle biopsy to CD8+ inflammatory infiltrates, and on half of the sampled muscle biopsies, elevation of downstream DUX4 targets were identified [10, 11]. In addition a variety of inflammatory markers were increased in FSHD patients with MRI STIR positive muscles [10]. Although we were unable to detect circulating levels of the same inflammatory mediators (IL6, IL10, IL12, and TNF-alpha), we did detect several inflammatory mediators significantly increased in FSHD compared to healthy volunteers (CCL2, CD40LG, and CD40 antigen). In addition several markers important in regulation of T-cell pathways were decreased in FSHD participants (S100A12 and CCL23). Muscle biopsies from children with Duchenne muscular dystrophy (DMD) showed increased CCL2 in the endothelium, and CCL2 has been implicated in macrophage and t-cell recruitment in the DMD mdx mouse model.[26, 27] It is possible these changes reflect physiological responses to inflammatory infiltrates in FSHD participant muscles, but further studies will be required to elucidate the exact relationship of circulating proteins to muscle inflammation.
This is the first study in FSHD to find overlap between serum proteins, and genes with altered regulation identified in previous studies of FSHD patient muscle [11–14]. C3, CKMB, angiogenin (ANG), matrix metalloproteinase 9, v-erb-b2 avian erythroblastic leukemia viral oncogene homolog, and LEP levels were altered or showed trends towards being increased or decreased in FSHD serum. A broad genome screen of muscle from FSHD patients identified up-regulation of genes in the complement pathway [12]. Another study identified up-regulation of genes for C3, ANG, and LEP in FSHD muscle samples, but not in samples from other muscle diseases characterized by prominent inflammatory infiltrates (dysferlinopathy or inflammatory myopathies)[11].
Although the Myriad Discovery MAP panel was not specifically designed for FSHD the panel still had a strong signal for positive markers in this patient population, so may indeed serve as an effective approach for biomarker screening. The Discovery MAP panel identified a number of potential biomarkers for disease activity in FSHD. The plasminogen pathway is classically involved in the fibrinolytic pathway; however animal studies have suggested a role for the activated plasminogen pathway in myogenesis [28, 29]. The alpha-enolase-type plasminogen receptor was shown to be induced in mouse C2Cl2 myoblasts during differentiation. Blocking this pathway prevented myoblast fusion. More importantly both plasmin activity and plasminogen receptor expression were induced in in vivo mouse models of either experimentally-induced muscle injury or muscular dystrophy (mdx mouse)[29]. Proteolysis mediated by plasmin has a key role in controlling inflammation and satellite-cell dependent myogenesis. Blocking the plasminogen pathway impaired muscle regeneration in both injured wild type mouse models and the mdx mouse [28]. To the best of our knowledge this is the first study showing activation of the plasminogen pathway in FSHD participants. We propose this activation may serve as a useful measure of disease activity in FSHD, but is likely not a disease-specific measure of muscle turnover, consistent with the observed correlation between PLAT levels and disease progression. A prior study in myotonic dystrophy type 1 (DM1) identified increases in the PLAT antigen and the PLAT/SERPINE1[30]. The authors concluded this represented a metabolic pathway altered in myotonic dystrophy, in the same way that insulin pathways are altered, but did not attempt to correlate this to other measures of disease activity.
Insulin resistance and alterations in insulin-related pathways have been described previously in DM1. Insulin and its regulatory proteins are up-regulated in DM1, and are associated with similar elevations in LEP. In DM1 elevations in LEP are not correlated to differences in body fat or other measures of body composition, and are believed to be due to the broad metabolic alterations seen in DM1[30, 31]. Disordered insulin regulation has not been described previously in FSHD. Here we see trends towards elevation of insulin, LEP, and ILGFBP1, which in FSHD may represent a non-specific alteration in insulin pathways due to muscle injury, as opposed to the clinical insulin resistance seen in DM1.
Limitations to this study include the small sample size, and insufficient power to determine disease-related correlations. Correction for a false discovery rate of 5% in such a small sample will likely discard many biomarkers which may ultimately prove useful in FSHD. However such smaller studies are essential prior to devoting the resources and time for larger multi-center validation studies of potential biomarkers, and have a valuable role for hypothesis generation and as references for future studies. No differences were seen between FSHD types 1 and 2. FSHD types 1 and 2 converge at an early point in the DUX4 pathway, and are clinically indistinguishable, so we would not expect to see differences in biomarkers related to the progression of the muscle manifestations of FSHD. However, broad biomarker screens might still be important for identifying other targets of altered SMCHD1 expression in FSHD2.[17] An important next step will be determining which, if any, biomarkers discovered here are specific for FSHD disease mechanisms. This study did identify several markers which appear to be good candidates for non-specific markers of disease activity – and may prove valuable in the setting of a clinical trial. Ultimately a larger validation study will be required to confirm the findings reported here, to investigate the differences between FSHD types 1 and 2, and to further stratify the patient population based on disease severity to confirm the relationship of biomarkers to disease mechanism or progression.
Supplementary Material
Acknowledgements
The authors would like to thank Peter A.C. 't Hoen, PhD for his valuable comments during the preparation of this manuscript. JS effort was supported by the MDA Clinical Research Training Grant, and a CTSA grant from NCATS awarded to the University of Kansas Medical Center for Frontiers: The Heartland Institute for Clinical and Translational Research # KL2TR000119. RT received research support from the NIH NINDS 1P01NS069539-01 (co-PI), NIH NCRR UL1RR024160, and the Fields Center for FSHD and Neuromuscular Research.
Footnotes
Conflicts of Interest
Both JS and RT are consultants for Cytokinetics and Regeneron.
ST has no disclosures
SM and PH has no disclosures
CMD has no disclosures
Author contributions
Jeffrey Statland and Rabi Tawil: drafting/revising the manuscript for content; study concept or design; analysis or interpretation of data; in addition acquisition of data and statistical analysis.
Stephen J Tapscott and Silvere van der Maarel: drafting/revising the manuscript for content; analysis or interpretation of data; in addition acquisition of data.
Colleen Donlin-Smith: study concept or design; drafting/revising the manuscript for content; in addition study coordination.
Contributor Information
Colleen M Donlin-Smith, Email: Colleen_Donlin-Smith@URMC.Rochester.edu.
Stephen J Tapscott, Email: stapscot@fhcrc.org.
Silvere van der Maarel, Email: S.M.van_der_Maarel@lumc.nl.
Rabi Tawil, Email: Rabi_Tawil@URMC.Rochester.edu.
References
- 1.Flanigan KM, Coffeen CM, Sexton L, Stauffer D, Brunner S, Leppert MF. Genetic characterization of a large, historically significant Utah kindred with facioscapulohumeral dystrophy. Neuromuscul Disord. 2001;11:525–529. doi: 10.1016/s0960-8966(01)00201-2. [DOI] [PubMed] [Google Scholar]
- 2.Mostacciuolo ML, Pastorello E, Vazza G, et al. Facioscapulohumeral muscular dystrophy: epidemiological and molecular study in a north-east Italian population sample. Clin Genet. 2009;75:550–555. doi: 10.1111/j.1399-0004.2009.01158.x. [DOI] [PubMed] [Google Scholar]
- 3.Norwood FL, Harling C, Chinnery PF, Eagle M, Bushby K, Straub V. Prevalence of genetic muscle disease in Northern England: in-depth analysis of a muscle clinic population. Brain. 2009;132:3175–3186. doi: 10.1093/brain/awp236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Padberg GW, Frants RR, Brouwer OF, Wijmenga C, Bakker E, Sandkuijl LA. Facioscapulohumeral muscular dystrophy in the Dutch population. Muscle Nerve. 1995;2:S81–S84. [PubMed] [Google Scholar]
- 5.Tawil R, van der Maarel S, Padberg GW, van Engelen BG. 171st ENMC international workshop: Standards of care and management of facioscapulohumeral muscular dystrophy. Neuromuscul Disord. 2010;20:471–475. doi: 10.1016/j.nmd.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 6.van der Maarel SM, Tawil R, Tapscott SJ. Facioscapulohumeral muscular dystrophy and DUX4: breaking the silence. Trends Mol Med. 2011;17:252–258. doi: 10.1016/j.molmed.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tawil R, Shaw DW, van der Maarel SM, Tapscott SJ. Clinical trial preparedness in facioscapulohumeral dystrophy: outcome measures and patient access: 8–9 April 2013, Leiden, The Netherlands. Neuromuscul Disord. 2014;24:79–85. doi: 10.1016/j.nmd.2013.07.009. [DOI] [PubMed] [Google Scholar]
- 8.Geng LN, Yao Z, Snider L, et al. DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Dev Cell. 2012;22:38–51. doi: 10.1016/j.devcel.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arahata K, Ishihara T, Fukunaga H, et al. Inflammatory response in facioscapulohumeral muscular dystrophy (FSHD): immunocytochemical and genetic analyses. Muscle Nerve. 1995;2:S56–S66. [PubMed] [Google Scholar]
- 10.Frisullo G, Frusciante R, Nociti V, et al. CD8(+) T cells in facioscapulohumeral muscular dystrophy patients with inflammatory features at muscle MRI. J Clin Immunol. 2011;31:155–166. doi: 10.1007/s10875-010-9474-6. [DOI] [PubMed] [Google Scholar]
- 11.Tasca G, Pescatori M, Monforte M, et al. Different molecular signatures in magnetic resonance imaging-staged facioscapulohumeral muscular dystrophy muscles. PLoS One. 2012;7:e38779. doi: 10.1371/journal.pone.0038779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osborne RJ, Welle S, Venance SL, Thornton CA, Tawil R. Expression profile of FSHD supports a link between retinal vasculopathy and muscular dystrophy. Neurology. 2007;68:569–577. doi: 10.1212/01.wnl.0000251269.31442.d9. [DOI] [PubMed] [Google Scholar]
- 13.Rahimov F, King OD, Leung DG, et al. Transcriptional profiling in facioscapulohumeral muscular dystrophy to identify candidate biomarkers. Proc Natl Acad Sci U S A. 2012;109:16234–16239. doi: 10.1073/pnas.1209508109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Winokur ST, Chen YW, Masny PS, et al. Expression profiling of FSHD muscle supports a defect in specific stages of myogenic differentiation. Hum Mol Genet. 2003;12:2895–2907. doi: 10.1093/hmg/ddg327. [DOI] [PubMed] [Google Scholar]
- 15.de Greef JC, Lemmers RJ, Camano P, et al. Clinical features of facioscapulohumeral muscular dystrophy 2. Neurology. 2010;75:1548–1554. doi: 10.1212/WNL.0b013e3181f96175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orrell RW, Tawil R, Forrester J, Kissel JT, Mendell JR, Figlewicz DA. Definitive molecular diagnosis of facioscapulohumeral dystrophy. Neurology. 1999;52:1822–1826. doi: 10.1212/wnl.52.9.1822. [DOI] [PubMed] [Google Scholar]
- 17.Lemmers RJ, Tawil R, Petek LM, et al. Digenic inheritance of an SMCHD1 mutation and an FSHD-permissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2. Nat Genet. 2012;44:1370–1374. doi: 10.1038/ng.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ricci E, Galluzzi G, Deidda G, et al. Progress in the molecular diagnosis of facioscapulohumeral muscular dystrophy and correlation between the number of KpnI repeats at the 4q35 locus and clinical phenotype. Ann Neurol. 1999;45:751–757. doi: 10.1002/1531-8249(199906)45:6<751::aid-ana9>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 19.Chowdhury F, Williams A, Johnson P. Validation and comparison of two multiplex technologies, Luminex and Mesoscale Discovery, for human cytokine profiling. J Immunol Methods. 2009;340:55–64. doi: 10.1016/j.jim.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 20.Krishhan VV, Khan IH, Luciw PA. Multiplexed microbead immunoassays by flow cytometry for molecular profiling: Basic concepts and proteomics applications. Crit Rev Biotechnol. 2009;29:29–43. doi: 10.1080/07388550802688847. [DOI] [PubMed] [Google Scholar]
- 21.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and pwerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 22.Pike N. Using false discovery rates for multiple comparisons in ecology and evolution. Ecology and Evolution. 2011;2:278–282. [Google Scholar]
- 23.Lemmers RJ, van der Vliet PJ, Klooster R, et al. A unifying genetic model for facioscapulohumeral muscular dystrophy. Science. 2010;329:1650–1653. doi: 10.1126/science.1189044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Snider L, Geng LN, Lemmers RJ, et al. Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene. PLoS Genet. 2010;6:e1001181. doi: 10.1371/journal.pgen.1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carpenter SKG. Pathology of Skeletal Muscle. Second Edition. New York: Oxford University Press; 2001. [Google Scholar]
- 26.De Paepe B, Creus KK, Martin JJ, De Bleecker JL. Upregulation of chemokines and their receptors in Duchenne muscular dystrophy: potential for attenuation of myofiber necrosis. Muscle Nerve. 2012;46:917–925. doi: 10.1002/mus.23481. [DOI] [PubMed] [Google Scholar]
- 27.Porter JD, Guo W, Merriam AP, et al. Persistent over-expression of specific CC class chemokines correlates with macrophage and T-cell recruitment in mdx skeletal muscle. Neuromuscul Disord. 2003;13:223–235. doi: 10.1016/s0960-8966(02)00242-0. [DOI] [PubMed] [Google Scholar]
- 28.Diaz-Ramos A, Roig-Borrellas A, Garcia-Melero A, Llorens A, Lopez-Alemany R. Requirement of plasminogen binding to its cell-surface receptor alpha-enolase for efficient regeneration of normal and dystrophic skeletal muscle. PLoS One. 2012;7:e50477. doi: 10.1371/journal.pone.0050477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lopez-Alemany R, Suelves M, Munoz-Canoves P. Plasmin generation dependent on alpha-enolase-type plasminogen receptor is required for myogenesis. Thromb Haemost. 2003;90:724–733. doi: 10.1160/TH03-04-0291. [DOI] [PubMed] [Google Scholar]
- 30.Johansson A, Boman K, Cederquist K, Forsberg H, Olsson T. Increased levels of tPA antigen and tPA/PAI-1 complex in myotonic dystrophy. J Intern Med. 2001;249:503–510. doi: 10.1046/j.1365-2796.2001.00832.x. [DOI] [PubMed] [Google Scholar]
- 31.Gomez JM, Molina A, Fernandez-Castaner M, Casamitjana R, Martinez-Matos JA, Soler J. Insulin regulation of leptin synthesis and secretion in humans: the model of myotonic dystrophy. Clin Endocrinol (Oxf) 1999;50:569–575. doi: 10.1046/j.1365-2265.1999.00675.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.