Abstract
Small RNAs (sRNAs) are ~20 to 24 nucleotide single-stranded RNAs that play crucial roles in regulation of gene expression. In plants, sRNAs are classified into microRNAs (miRNAs), repeat-associated siRNAs (ra-siRNAs), phased siRNAs (pha-siRNAs), cis and trans natural antisense transcript siRNAs (cis- and trans-nat siRNAs). Pima (Gossypium barbadense L.) is one of the most economically important fiber crops, producing the best and longest spinnable fiber. Although some miRNAs are profiled in Pima, little is known about siRNAs, the largest subclass of plant sRNAs. In order to profile these gene regulators in Pima, a comprehensive analysis of sRNAs was conducted by mining publicly available sRNA data, leading to identification of 678 miRNAs, 3,559,126 ra-siRNAs, 627 pha-siRNAs, 136,600 cis-nat siRNAs and 79,994 trans-nat siRNAs. The 678 miRNAs, belonging to 98 conserved and 402 lineage-specific families, were produced from 2,138 precursors, of which 297 arose from introns, exons, or intron/UTR-exon junctions of protein-coding genes. Ra-siRNAs were produced from various repeat loci, while most (97%) were yielded from retrotransposons, especially LTRs (long terminal repeats). The genes encoding auxin-signaling-related proteins, NBS-LRRs and transcription factors were major sources of pha-siRNAs, while two conserved TAS3 homologs were found as well. Most cis-NATs in Pima overlapped in enclosed and convergent orientations, while a few hybridized in divergent and coincided orientations. Most cis- and trans-nat siRNAs were produced from overlapping regions. Additionally, characteristics of length and the 5’-first nucleotide of each sRNA class were analyzed as well. Results in this study created a valuable molecular resource that would facilitate studies on mechanism of controlling gene expression.
Introduction
Small RNAs (sRNAs) are short, single-stranded RNAs that silence gene expression transcriptionally or post-transcriptionally [1–3], which is also known as RNA-induced silencing. sRNAs are produced from longer, double stranded RNA (dsRNA) precursors by DICER-LIKE proteins (DCLs) in plants [4]. Mature sRNAs are loaded into Argonaute (AGO) proteins to form RNA inducing complex in which sRNAs serve as guides to complementarily bind target RNA molecules, leading to degradation or translation repression of the targets [5,6]. In addition, some sRNAs are able to induce methylation or histone modification of target genomic loci as well [7–9].
Generally, sRNAs are classified into microRNAs (miRNAs) and small interfering RNAs (siRNAs), according to their origin and biogenesis [10]. miRNAs are processed from hairpin-shaped RNA precursors (pre-miRNAs) that are transcribed by RNA polymerase II [11]. In plants, most miRNAs are derived from non-coding genes, whose transcripts are further processed by DCLs with presence of the zinc finger protein SERRATE [12], HYPONASTIC LEAVES1 [13,14], the G-patch domain protein TOUGH [15] and hnRNP-like protein [16]. However, recent studies show that miRNAs can also be produced from introns or exon-intron junctions of protein-coding genes [17,18], which are regulated through alternative splicing (AS) [19]. In plants, canonical miRNAs are 21 nt in length and regulate gene expression transcriptionally [20,21]. Many miRNAs target transcription factors, such as SQUAMOSA Promoter Binding Protein-Like, MYB and HD-ZIP [22,23], which are crucial for plant growth, development and stress response [21,24,25], but the so-called long miRNAs (24-nt in length) function to direct DNA methylation [8]. In plants, siRNAs are produced from double stranded RNAs (dsRNAs), whose synthesis requires activities of RNA polymerase IV/V (Pol IV/V) [26,27] and RNA-dependent RNA polymerases (RDRs) [28,29]. siRNAs can be sub-divided into four groups, repeat-associated siRNAs (ra-siRNAs), phased siRNAs (pha-siRNAs), cis- and trans natural antisense siRNAs (cis- and trans-nat siRNAs) [10]. Ra-siRNAs, typically 24 nt in length, are also known as heterochromatic siRNAs (hc-siRNAs) transcribed from repeat DNA loci by RNA polymerase IV (Pol IV) [30] with assistance of certain proteins, such as CLASSY1 [31], SHH1[32] and DTF1[33]. Ra-siRNAs are the biggest subclass of plant sRNAs [34] and mediate epigenetic modification through DNA methylation [7]. Pha-siRNAs are plant-specific sRNAs cleaved from pha-siRNA-yielding transcripts (PYTs) in phased manner, by which sRNAs are produced precisely in a head-to-tail arrangement [35,36]. A subset of pha-siRNAs that can function in trans to repress gene expression at transcriptional level are termed as trans-acting siRNAs (ta-siRNAs), and the transcripts producing ta-siRNAs are called TASs [10,35,37]. To date, TAS3 homologs are widely found in various plants, and their ta-siRNAs are shown to play crucial roles in regulation of plant developmental timing and stress response [36–38]. Both cis- and trans-nat siRNAs are processed from natural antisense transcripts (NATs) by DCL proteins in plants [10,39] but differ by their origin. Generally, cis-NATs are transcribed from the same genomic loci with opposite polarities, whereas trans-NATs are generated from different genomic loci. Nat-derived siRNAs are shown to regulate diverse physiological processes, such as fertilization and stress response to environmental cues [7,39–42].
Pima (Gossypium barbadense L.) is one of the most important fiber crops worldwide, belonging to allotetraploid cotton that is evolved through hybridization and subsequent polyploidization events between the A-genome and D-genome cottons, closely related to Gossypium arboreum L. (A2) and Gossypium raimondii L. (D5), respectively [43,44]. The merger of the A and D genome species into allotetraploid species led to superior fiber length and quality, and sRNAs are shown to play crucial roles in regulation of fiber development [45–50]. Pang et al. discover a total of 31 miRNA families as well as their targets expressed in vegetative and reproductive tissues [51]. Analysis of deep sequencing data shows that a large number of known and novel miRNA families are expressed in the Upland cotton ovule and fiber, and a subset of them displays differential expression patterns [49–52]. For instance, miR166 and miR172 are expressed abundantly in the fiber, but miR828, miR475 and miR1023 are expressed at extremely low levels [50]. Comparative analysis of sRNAs reveals significant differences in miRNAs expression between the wild type Upland cotton and its fuzzless-lintless mutant [53]. Additionally, miR828 and miR858 targeting GhMYB2D mRNA leads to generation of ta-siRNAs, mediating fiber development [47]. In addition to miRNAs, TAS3-derived ta-siRNAs triggered by miR390 are found in Upland cotton as well [54]. The suppression of miR156/157 leads to the reduction of mature fiber length in Pima [48]. Compared to Upland cotton, apparently fewer miRNAs are identified in Pima [48,55], which produces the best and longest textile fiber, and siRNAs being the most abundant sRNA class in plants [34] still remain largely unknown. To profile these gene regulators in Pima, we analyzed two publicly available sRNA datasets from the root and fiber, resulting in identification of 5 major sRNA species. This is the first comprehensive analysis of Pima sRNAs and created a valuable molecular resource, which largely expands the scope of sRNAs in cotton and is very crucial for future studies on mechanism of regulating plant growth and development.
Methods and Materials
Small RNA datasets of G. barbadense
Two sRNA datasets of Pima prepared from the root (GSM699076) and 10 DPA (days post anthesis) fiber (GSM634227) were downloaded from NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo). The two datasets contain 15,599,325 and 7,764,225 reads (S1 Table), respectively. For further analysis, sequences 18 to 26 nt in length were retained, leaving a total of 9,518,811 unique sRNAs, and the total reads of each sRNA from the two datasets were recorded for expression-based filtration, using in-house Perl scripts.
The cotton nucleotide sequence dataset
Due to unavailability of the allopolyploid cotton genome, a comprehensive cotton nucleotide dataset was collected and assembled for this study, including cotton A2 [44] and D5 genomes [43], and cotton-derived transcripts, downloaded from NCBI (ftp://ftp.ncbi.nlm.nih.gov/repository/UniGene/), PlantDB (http://www.plantgdb.org), CottonGen (http://www.cottongen.org), Comparative Evolution of Genomics of Cotton (http://128.192.141.98/CottonFiber), and Cotton Gene Index (version 11, http://compbio.dfci.harvard.edu).
Identification of conserved and lineage-specific miRNAs
To identify miRNAs, sRNAs with a total abundance ≥10 reads were submitted to a modified miREAP (http://sourceforge.net/projects/mireap/), in which 3 nucleotide shifts from putative miRNA loci are allowed, using the following parameters: the maximal distance between miRNAs and miRNA*s (-d 450), the maximal copies on the genome (-u 5000), and defaults for others. The putative miRNAs obtained from miREAP were further filtered by strand bias (≥0.8), which was calculated by dividing reads from sense strand by total reads, and abundant bias (≥0.6), which was calculated by dividing total reads of top 3 sRNAs from miRNA loci by total reads [56,57]. Additionally, the remaining putative miRNAs were filtered by the secondary structures of their pre-miRNAs in which maximal four mismatches are allowed between miRNAs and miRNA*s [57], using CentroidFold [58]. The identified miRNAs that are homologous to known plant miRNAs in miRBase version 21 (http://www.mirbase.org) with three or less substitutions are classified into conserved miRNAs (cs-miRNAs), while the others are considered as candidates of lineage-specific miRNAs (ls-miRNAs) [10].
To determine miRNAs yielded from protein-coding genes, cotton-derived transcripts were mapped to the cotton A2 and D5 genomes, using Blastn [59,60] with e-value < 10-6. Those transcripts, which were mapped to upstream and/or downstream of miRNA loci within 5000 nt with ≥96% identity, ≤3 mismatches and no indel (insertion-deletion), were selected for further analysis. The transcripts mapped to miRNA loci were annotated using Blastx [59,60] against plant-derived proteins deposited in NCBI (http://www.ncbi.nlm.nih.gov/) with a stringent e-value <10-6. miRNAs derived from protein-coding genes were further classified into 3 classes based on the location of their pre-miRNAs, intron-derived miRNAs processed from introns of protein-coding genes [17], exon-derived miRNAs cleaved from exons of protein coding genes, and junction-derived miRNAs yielded from exon-intron/UTR junctions [18].
Identification of ra-siRNAs
To determine ra-siRNAs, we extracted repeat DNAs from the cotton A2 and D5 genomes [43,44]. Given that the information of repeat loci in the cotton A2 genome is not publicly available, we identified repeat DNA loci in the A2 genome using RepeatMasker [61] with default parameters. All sRNAs matching repeat DNAs were considered as ra-siRNAs.
Identification of pha-siRNAs
In order to identify pha-siRNAs, sRNAs (excluding miRNAs and ra-siRNAs) were subjected to pha-siRNA analysis, using UEA Small RNA WorkBench [62] with p-value <10-3 that employs the algorithm described previously [63]. The putative PYTs obtained from UEA Small RNA WorkBench [62] were further filtered by three extra characteristics: 1). the PYTs produced at least two pha-siRNAs; 2). at least a pha-siRNA had a total read count ≥5; 3). the ratio of pha-siRNAs/non-pha-siRNAs was ≥0.6. Analysis of miRNA initiators triggering the synthesis of pha-siRNAs was conducted, referring to the methodology described in early study [64] with minor modification. Briefly, putative initiators were predicted using psRNATarget with length for complementarity scoring (18) and expectation value (4.0). Then, the miRNAs that were predicted to flank the upstream or downstream of pha-siRNAs within 148 nt were selected for this analysis, allowing one nucleotide shift from the classic cleavage site [64].
Identification of cis- and trans-nat siRNAs
The cis-nat siRNAs were determined, referring to the methods described previously [65]. Briefly, cotton-derived transcripts were mapped to both cotton A2 and D5 genomes, and then pairs of transcripts derived from the same genomic loci with at least 50 nt overlap but from different orientations were chosen as cis-NATs. sRNAs produced from cis-NATs were considered as cis-nat siRNA candidates.
After excluding cis-NATs, the remaining transcripts were used for analysis of trans-NATs. Putative trans-NATs were determined, using Blastn search for pairs of sequences that were complementary to each other with at least 100 nt overlap. The putative trans-NATs were then filtered by secondary structure using RNAcoFold in Vienna RNA package [66]. Those putative trans-NATs that can form at least 50 nt dsRNAs, in which 90% nucleotides were base-paired, were considered as trans-NATs, and sRNAs matched to trans-NATs were trans-nat siRNA candidates.
Results and Discussion
Conserved and lineage-specific miRNAs in Pima
An analysis of the Pima sRNA datasets (S1 Table), which contained 9,518,811sRNAs, was performed. This led to identification of a total of 678 miRNAs from 2,138 pre-miRNAs (S2 Table), of which 222 and 456 were assigned to 98 cs-miRNA and 402 ls-miRNA families, respectively.
A comparison of the 98 Pima cs-miRNA families across plants was conducted (Table 1). Twenty three (23%) cs-miRNA families were present in both Eudicotyledon and Monocotyledon, with a subset also found in Embryophyta and/or Coniferophyta, suggesting that they have common ancient origins [67]. Notably, miR477 was a special family that was present in both Embryophyta and Eudicotyledons, but absent in Monocotyledon. Ten (10%) families were detected in only Eudicotyledons, indicating that they are Eudicotyledon-specific, and the other 64 families (65%) were restricted to Malvaceae, reflecting that they are poorly conserved.
Table 1. Conserved miRNA families of Pima across plants.
| miRNAs | Coniferophyta | Embryophyta | Magnoliophyta | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Eudicotyledons | Monocotyledons | |||||||||||||||||||
| Araliaceae | Asteraceae | Brassicaceae | Caricaceae | Cucurbitaceae | Euphorbiaceae | Fabaceae | Lamiales | Linaceae | Malvaceae | Ranunculaceae | Rhizophoraceae | Rosaceae | Rutaceae | Salicaceae | Solanaceae | Vitaceae | ||||
| miR156 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| miR396 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| miR160 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||
| miR166 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||
| miR171 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||
| miR395 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||
| miR159 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |||
| miR167 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |||
| miR169 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |||
| miR164 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||||
| miR172 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||||
| miR390 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||||
| miR394 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |||||
| miR162 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |||||
| miR168 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||||||
| miR393 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||||||
| miR397 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||||||
| miR399 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |||||
| miR482 | + | + | + | + | + | + | + | + | + | + | + | + | ||||||||
| miR530 | + | + | + | + | + | + | + | + | + | + | + | + | ||||||||
| miR535 | + | + | + | + | + | + | + | + | + | + | ||||||||||
| miR827 | + | + | + | + | + | + | + | + | ||||||||||||
| miR1877 | + | + | ||||||||||||||||||
| miR477 | + | + | + | + | + | + | + | + | + | + | + | |||||||||
| miR403 | + | + | + | + | + | + | + | + | + | + | ||||||||||
| miR2111 | + | + | + | + | + | + | + | + | ||||||||||||
| miR3627 | + | + | + | + | + | |||||||||||||||
| miR1446 | + | + | + | + | ||||||||||||||||
| miR2950 | + | + | + | |||||||||||||||||
| miR479 | + | + | + | + | ||||||||||||||||
| miR170 | + | + | ||||||||||||||||||
| miR3954 | + | + | ||||||||||||||||||
| miR6140 | + | + | ||||||||||||||||||
| miR8148 | + | + | ||||||||||||||||||
| miR2947 | + | |||||||||||||||||||
| miR2948 | + | |||||||||||||||||||
| miR2949 | + | |||||||||||||||||||
| miR3267 | + | |||||||||||||||||||
| miR3476 | + | |||||||||||||||||||
| miR7484 | + | |||||||||||||||||||
| miR7486 | + | |||||||||||||||||||
| miR7489 | + | |||||||||||||||||||
| miR7491 | + | |||||||||||||||||||
| miR7492 | + | |||||||||||||||||||
| miR7494 | + | |||||||||||||||||||
| miR7495 | + | |||||||||||||||||||
| miR7498 | + | |||||||||||||||||||
| miR7499 | + | |||||||||||||||||||
| miR7500 | + | |||||||||||||||||||
| miR7501 | + | |||||||||||||||||||
| miR7502 | + | |||||||||||||||||||
| miR7504 | + | |||||||||||||||||||
| miR7505 | + | |||||||||||||||||||
| miR7506 | + | |||||||||||||||||||
| miR7507 | + | |||||||||||||||||||
| miR7508 | + | |||||||||||||||||||
| miR7510 | + | |||||||||||||||||||
| miR7512 | + | |||||||||||||||||||
| miR7513 | + | |||||||||||||||||||
| miR8632 | + | |||||||||||||||||||
| miR8633 | + | |||||||||||||||||||
| miR8634 | + | |||||||||||||||||||
| miR8635 | + | |||||||||||||||||||
| miR8636 | + | |||||||||||||||||||
| miR8637 | + | |||||||||||||||||||
| miR8638 | + | |||||||||||||||||||
| miR8639 | + | |||||||||||||||||||
| miR8641 | + | |||||||||||||||||||
| miR8642 | + | |||||||||||||||||||
| miR8644 | + | |||||||||||||||||||
| miR8646 | + | |||||||||||||||||||
| miR8675 | + | |||||||||||||||||||
| miR8677 | + | |||||||||||||||||||
| miR8680 | + | |||||||||||||||||||
| miR8695 | + | |||||||||||||||||||
| miR8701 | + | |||||||||||||||||||
| miR8702 | + | |||||||||||||||||||
| miR8706 | + | |||||||||||||||||||
| miR8709 | + | |||||||||||||||||||
| miR8721 | + | |||||||||||||||||||
| miR8722 | + | |||||||||||||||||||
| miR8727 | + | |||||||||||||||||||
| miR8729 | + | |||||||||||||||||||
| miR8732 | + | |||||||||||||||||||
| miR8733 | + | |||||||||||||||||||
| miR8740 | + | |||||||||||||||||||
| miR8746 | + | |||||||||||||||||||
| miR8752 | + | |||||||||||||||||||
| miR8753 | + | |||||||||||||||||||
| miR8758 | + | |||||||||||||||||||
| miR8762 | + | |||||||||||||||||||
| miR8764 | + | |||||||||||||||||||
| miR8766 | + | |||||||||||||||||||
| miR8767 | + | |||||||||||||||||||
| miR8768 | + | |||||||||||||||||||
| miR8775 | + | |||||||||||||||||||
| miR8783 | + | |||||||||||||||||||
| miR8785 | + | |||||||||||||||||||
*: Each plus sign “+” represents the presence of miRNAs in corresponding plants.
Among the 98 cs-miRNA families, 25 (26%) were detected from all three reference resources (Fig. 1A), 40 (41%) were shared between A2 and D5 genomes but not detected from cotton transcripts, while two were shared between either of two genomes and cotton transcripts. In addition, 5, 25 and 1 cs-miRNA families were uniquely detected from A2, D5 genome, and cotton transcripts, respectively. By contrast, only two ls-miRNA families (0.5%) were detected in all three reference resources (Fig. 1B), 80 (20%) were shared between A2 and D5 genome but not found in cotton transcripts, while 168 (42%), 144 (36%) and 4 (1%) were uniquely detected from A2, D5 genome, and cotton transcripts, respectively.
Figure 1. Venn diagram shows the number of miRNAs derived from different originations.
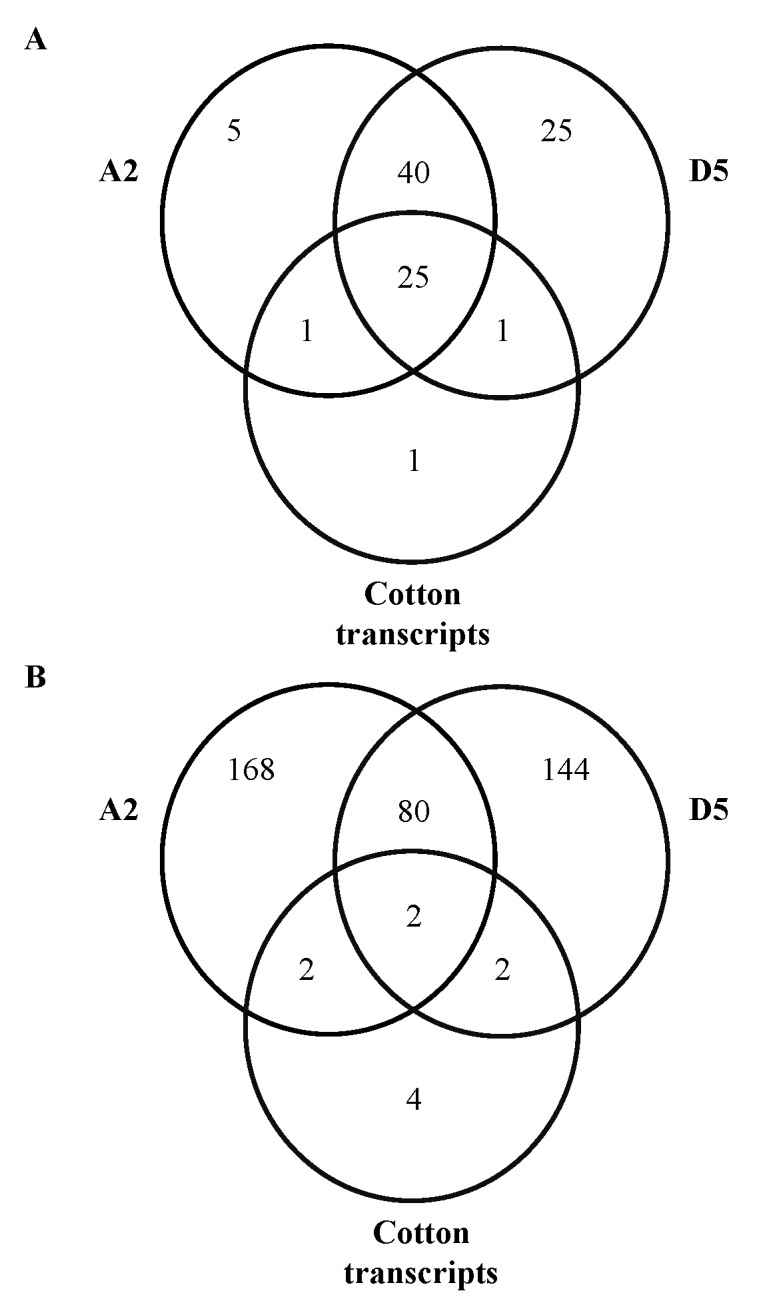
The numbers of miRNAs of Pima derived from different originations are shown in A (conserved miRNAs) and B (lineage-specific miRNAs), respectively. A2, D5 and cotton transcripts represent the number of miRNA family detected from A2 genome (G. arbroeum), D5 genome (G. raimondii) and cotton transcripts, respectively.
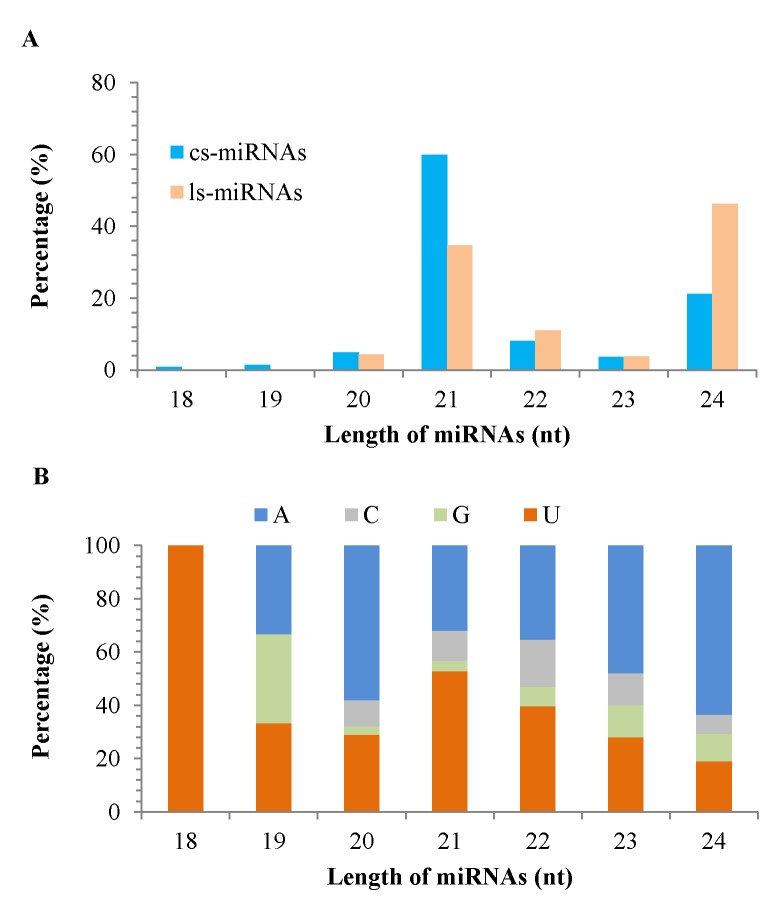
Pima miRNAs varied from 18 to 24 nt in length, with almost all miRNAs (99%) being 20 to 24 nt in length (Fig. 2A). Most cs-miRNAs (60%) were 21 nt in length, whereas nearly half ls-miRNAs (46%) were 24 nt in length. In plants, miRNAs are predominantly 21 nt in length and regulate gene expression post-transcriptionally [10,68], but long miRNAs (24 nt in length) function to direct chromatin modification and consequently result in transcriptional repression of target loci [8]. Cs-miRNAs and ls-miRNAs displayed different length characteristics, which might be associated with their regulatory roles [8,69].
Figure 2. Characteristics of Pima miRNAs.
The length distribution of Pima miRNAs is shown in A, and the 5’end nucleotide distribution of Pima miRNAs is shown in B.
Analysis of the 5’first nucleotide revealed a higher frequency of A or U and a less frequency of C or G at the 5’end of Pima miRNAs (Fig. 2B). The 5’-first nucleotide is important for sorting sRNAs into different AGO clades, and changing 5’-first nucleotide of a miRNA can redirects it to a different AGO complex and alter its biological activity [5]. Thus, it could be inferred that the characteristic of the 5’-first nucleotide is also functionally important for miRNAs in Pima.
Compared to previous studies in Pima and other cotton species [48–50,55], more conserved and lineage-specific miRNAs along with their pre-miRNAs are identified in Pima in this study, which largely expands cotton miRNA information. Consistent with previous study [67], most miRNAs are not conserved among plants, which are proposed to play species-specific roles in plant growth and development [70].
miRNAs derived from protein-coding genes
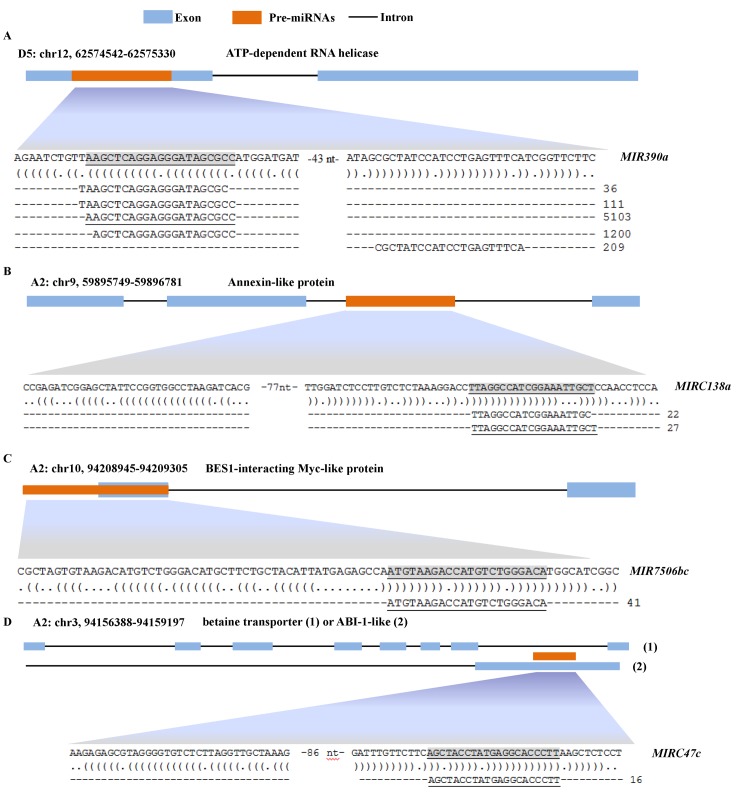
Typically, miRNAs in plants are produced from non-protein-coding genes [10,57], while recent studies show that miRNAs can also be produced from protein-coding genes in Arabidopsis and rice [71,72]. In the present study, 297 pre-miRNAs were identified to arise from introns, exons, or intron-exon junctions of protein-coding genes (S3 Table). These pre-miRNAs were able to form well-defined hairpin structures of plant miRNAs (Fig. 3A to 3D) [56,57]. Notably, a special pre-miRNA (MIRC47c) was observed, which could be processed from either an intron or exon of two different protein-coding genes from the same genomic loci (Fig. 3D), indicating a potential AS site.
Figure 3. Examples of four classes of protein-gene-derived pre-miRNAs.
Four classes of protein-gene-derived pre-miRNAs are shown in A (exon-derived), B (intron-derived), C (junction-derived), and D (intron/exon-derived). The nucleotide sequences represent the pre-miRNAs, in which the shaded and underlined nucleotides represent miRNA sequences. The secondary structure of pre-miRNAs is shown below each pre-miRNA. The sRNAs mapped to each pre-miRNAs are shown below the secondary structure and the reads of each sRNAs is shown on the right.
In animals, the biogenesis of miRNAs derived from exon-intron junctions is negatively regulated through AS, which is competitive with Drosha cleavage [19]. In Arabidopsis, an AS event that occurs in the intron where MIR400 is located is specifically induced by heat stress, providing the evidence that AS acts as a regulatory mechanism linking miRNAs and environmental stress in plants [72].
Identification of ra-siRNAs in Pima
To identify ra-siRNAs, sRNAs (excluding miRNAs) were mapped to repeat DNA loci in both cotton A2 [43] and D5 [44] genomes. Consequently, 3,559,126 sRNAs were mapped to repeat DNA loci, which were considered as ra-siRNA candidates (S4 Table). These ra-siRNAs were further classified into 5 different types, according to their origin (Table 2). A total of 3,447,580 ra-siRNAs arose from retrotransposons, of which 79% were derived from LTRs (long terminal repeats). Except for rRNA-derived ra-siRNAs, approximately 75% ra-siRNAs were uniquely detected in either A2 or D5 genome, indicating that ra-siRNAs are not well conserved between the two genomes.
Table 2. Component of ra-siRNAs of G. barbadense.
| Type | Cotton Sub-genome | |||
|---|---|---|---|---|
| A2 | D5 | Both A2 and D5 | Total | |
| Transposon | 31,102 | 49,934 | 14,973 | 66,063 |
| DNA/MuDR | 21,050 | 30,748 | 9,794 | 42,004 |
| DNA/En-Spm | 6,144 | 12,550 | 3,305 | 15,389 |
| DNA | 2,636 | 4,830 | 1,304 | 6,162 |
| DNA/hAT | 1,150 | 1,549 | 486 | 2,213 |
| DNA/Harbinger | 42 | 152 | 32 | 162 |
| DNA/TcMar | 80 | 96 | 52 | 124 |
| DNA/Mite | 0 | 6 | 0 | 6 |
| RC/Helitron | 0 | 3 | 0 | 3 |
| Retrotransposon | 2,091,427 | 2,213,611 | 857,458 | 3,447,580 |
| LTR | 796,849 | 922,373 | 343,074 | 1,376,148 |
| LTR/Gypsy | 737,565 | 536,980 | 282,674 | 991,871 |
| LTR/Copia | 216,186 | 227,997 | 89,959 | 354,224 |
| Non LTR | 37,724 | 56,264 | 18,520 | 75,468 |
| Retroelement | 301,759 | 467,958 | 122,859 | 646,858 |
| LINE | 1,344 | 2,037 | 372 | 3,009 |
| SINE | 0 | 2 | 0 | 2 |
| rRNA | 25,763 | 22,994 | 20,772 | 27,985 |
| Simple repeat | 15,852 | 7,133 | 5,641 | 17,344 |
| Others | 14 | 148 | 8 | 154 |
| Total | 2,164,158 | 2,293,820 | 898,852 | 3,559,126 |
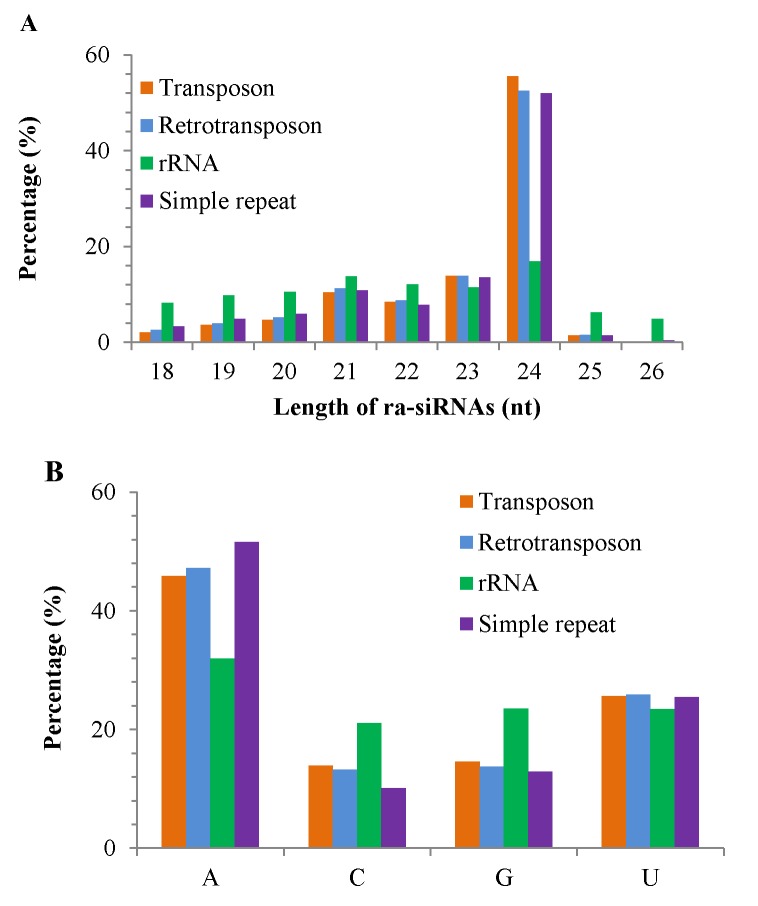
The length distribution of major classes of ra-siRNAs was shown in Fig. 4A. The 24 nt ra-siRNAs were predominant, corresponding to 52–56% ra-siRNAs from DNA transposons, retrotransposons or simple repeats. By contrast, no obvious length bias was observed among rRNA-derived ra-siRNAs. Moreover, analysis of the 5’ first nucleotide revealed a higher frequency of A than other residues at the 5’end of ra-siRNAs (Fig. 4B). These characteristics are consistent with that found in other plants [34,73].
Figure 4. Characteristics of ra-siRNAs in G. barbadense.
The length distribution of ra-siRNAs is shown in A, and the 5’first nucleotide of ra-siRNAs is shown in B. Transposon, retrotransposon, rRNA and simple repeat represent different derivations of ra-siRNAs.
PYTs and pha-siRNAs in Pima
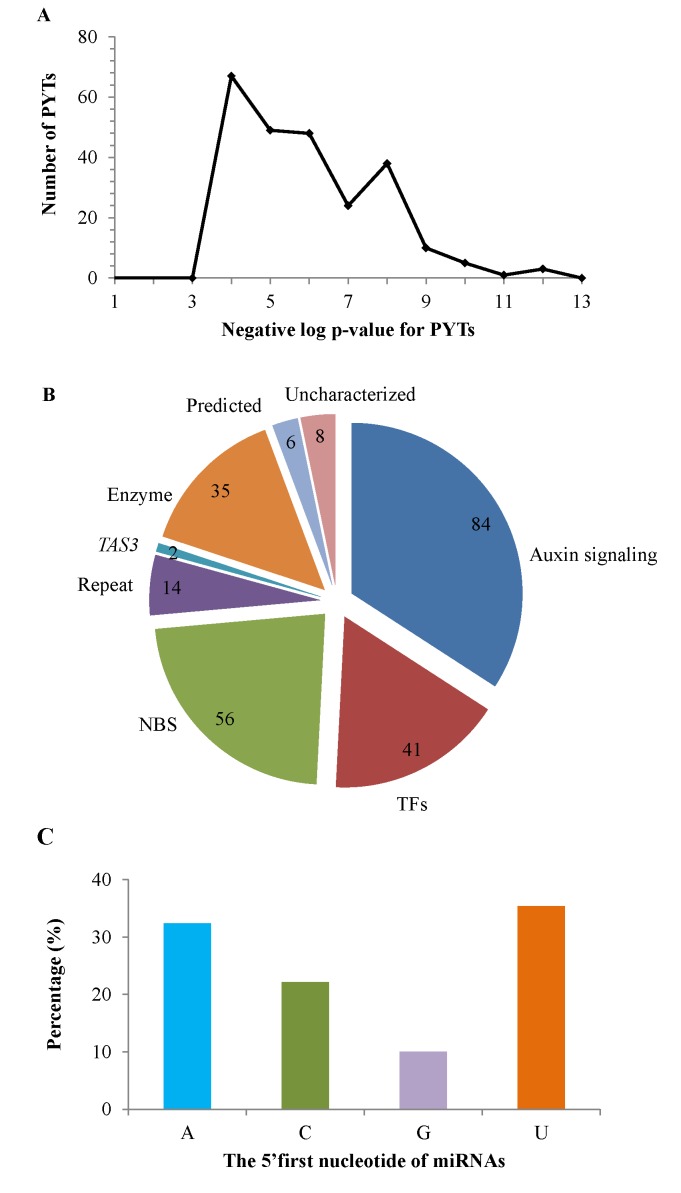
Analysis of pha-siRNA was conducted using a combination of UEA Small RNA Workbench [62] and in-house Perl scripts as described in the Method section. As a result, 246 PYTs that produced 627 pha-siRNAs were obtained with significant p-values ranging from 0 to 10-3 (Fig. 5A), indicating reliable capture of phasing phenomenon [63].
Figure 5. Characteristics of Pima pha-siRNAs.
The distribution of p-value for Pima PYTs is shown in A, the classification of PYTs is shown in B, and the distribution of the 5’first nucleotide is shown in C. Auxin: auxin-related transcription factors; Other TF: other transcription factors; NBS: NBS-LRR disease resistance proteins; Repeat: repeat proteins; TAS3: Arabidopsis TAS3 homologs; Enzymes: various enzymes; Predicted: predicted proteins; Uncharacterized: uncharacterized proteins.
The PYTs were further annotated using either Blastx against NCBI nr protein database or Blastn against TAIR gene database. Consequently, 236 PYTs were derived from protein coding genes (S5 Table and Fig. 5B), 2 were from non-coding TAS3 homologs, and 8 had no hit in the databases. Genes encoding auxin response factors (ARFs) and auxin signaling F-box were the richest source of Pima pha-siRNAs, followed by those encoding NBS-LRR disease resistance proteins and transcription factors. In addition, PYTs were also produced from genes encoding various proteins or enzymes, such as pentatricopeptide repeats, Ca2+ ATPase, Helicase, and peroxidase. Strikingly, a DCL1 isoform was able to produce pha-siRNAs (S6 Table), suggesting that the sRNA biogenesis machinery is subject to pha-siRNA regulation [38]. A subset of those PYT genes, which encode proteins, such as auxin signaling F-boxs, pentatricopeptide repeats, NBS-LRR and MYBs, et al., has been previously reported in other plants [23,38,74], indicating that these pha-siRNA pathways are likely conserved across plants. Furthermore, analysis of the 5’first nucleotide revealed a higher frequency of A and U and a lower occurrence of C and G at the 5’ end of pha-siRNAs (Fig. 5C). This is consistent with characteristics of DCL products, such as miRNAs and ra-siRNAs.
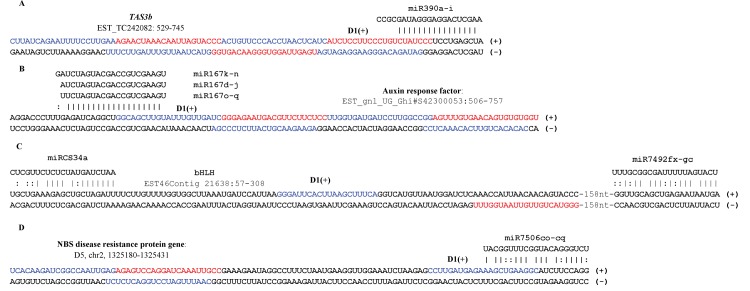
The biogenesis of most known pha-siRNAs is dependent on miRNA cleavage [35,75]. To better understand regulatory pathways of Pima pha-siRNAs, an analysis of phase-initiators was performed, referring to the methodology described previously [64]. As a consequence, 58 miRNA initiators were predicted to target 103 PYTs (S6 Table), including known miRNA-target pairs, such as miR390-TAS3 (Fig. 6A) [75,76] and miR167-ARF (Fig. 6B) [77]. However, other miRNA-target pairs, such as miRCS34/miR7492-bHLH (Fig. 6C) and miR7506-NBS-LRR (Fig. 6D), were firstly reported in plants, indicating that they are likely novel pha-siRNA regulatory pathways in plants. Except for the miRCS34/miR7492-bHLH pathway, most PYTs in Pima were triggered by “one-hit” model [35].
Figure 6. miRNA-mediated pha-siRNA yielding pathways.
Four miRNA-mediated pha-siRNAs yielding pathways are shown in A, B, C and D. The red and blue letters represent pha-siRNAs. The vertical line “|” and colon “:” represent Watson-Crick base-pairs and G-U pairs, respectively.
Cis-nat siRNAs in Pima
Cis-nat siRNAs have been identified in various plants and are shown to regulate plant development, disease resistance and stress response [40,41,78]. However, no cis-nat siRNA is reported in Pima to date, although some cotton trans-NATs are deposited in plant NAT database, PlantNATsDB (http://bis.zju.edu.cn/pnatdb/) [79]. In this study, 42,737 and 24,178 cis-NATs (S7 Table) were identified from cotton A2 and D5 genome, respectively. These cis-NATs yielded a total of 136,600 cis-nat siRNAs (S8 Table), representing 229,435 reads.
The cis-NATs were further classified into 4 types in accordance to overlapping orientations: convergent (3’end overlap), divergent (5’end overlap), enclosed (one transcript encompassed the other transcript), and coincided (two transcripts of cis-NATs completely overlapped). Most cis-NATs hybridized in enclosed and convergent orientations, corresponding to 42% and 30% of the total cis-NATs, respectively, while only 10–16% overlapped in the divergent or coincided way (Fig. 7A). Compared to earlier studies [41,78], cis-NATs in Pima can form dsRNAs in the coincided orientation, a new overlapping means found in plants. In addition, nearly half cis-NATs in Pima overlapped in the enclosed way, differing from that the convergent orientation is the major overlapping means found previously [41,78], which reflects the overlapping means of cis-NATs might vary in different plant species or tissue types.
Figure 7. Characteristics of cis-NATs and cis-nat siRNAs in Pima.
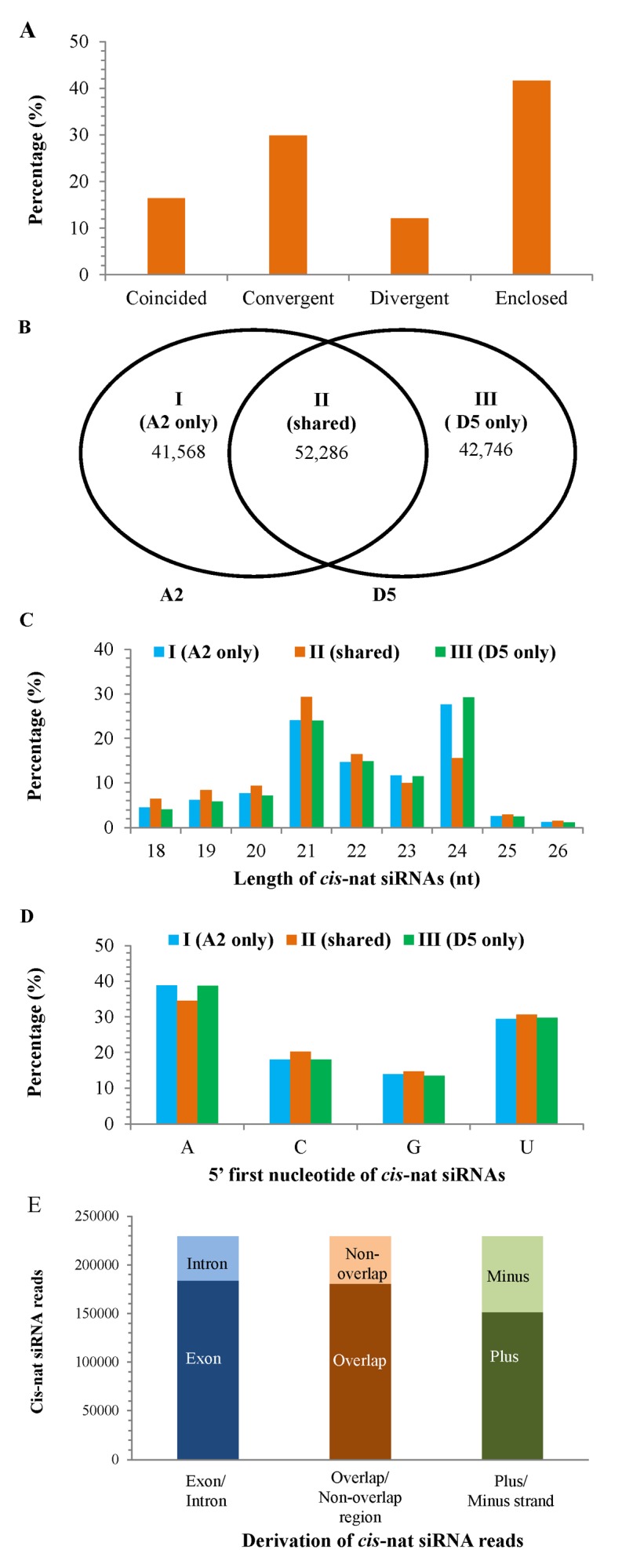
The numbers of each type of cis-NAT orientations are shown in A. The number of cis-nat siRNAs detected from the A2 and D5 genome is shown in B. The length distribution of cis-nat siRNAs is shown in C. The 5’first nucleotide of cis-nat siRNAs is shown in D. The enrichment analysis of cis-nat siRNAs is shown in E; exon/intron, overlap/non-overlap region, and plus/minus stand represent different derivations of cis-nat siRNAs.
Of the 136,600 cis-nat siRNAs, 38% (52,286) were expressed in both A2 and D5 genomes (Fig. 7B), while 41,568 (31%) and 42,746 (31%) were uniquely detected in A2 and D5 genome (Fig. 7B), respectively. Interestingly, cis-nat siRNAs uniquely detected in the A2 and D5 genome shared a similar length distribution, with the 24 nt cis-nat siRNAs (~29%) being the most abundant (Fig. 7C). However, those shared between the two genomes displayed a different length characteristic with the 21 nt cis-nat siRNAs (29%) being predominant (Fig. 7C). Moreover, the cis-nat siRNAs exhibited a 5’first nucleotide bias for A and U (Fig. 7D). These are consistent with previous studies in plant cis-nat siRNAs [65,73,78].
Furthermore, analysis of enrichment of cis-nat siRNAs was conducted by mapping sRNAs to the cis-NAT genomic loci, using Bowtie Mapping Utility [80]. The results revealed that most cis-nat siRNA reads were produced from exons (80%) or overlapping regions (79%) (Fig. 7E). In addition, 66% cis-nat siRNA reads were produced from plus strands (Fig. 7E), approximately two folds of that from minus strands, reflecting that cis-nat siRNAs exhibited strand-biased expression [41,73,78].
Trans-nat siRNAs in Pima
In this study, 1,250,581 trans-NATs were identified in Pima, of which 895,809 (72%) were detected to yield 79,994 trans-nat siRNAs (S9 Table). These trans-NATs overlapped in a wide length range from 50 to more than 1000 nt (Fig. 8A), while most (68%) had an overlapping region between 300 and 800 nt. The trans-NATs can form dsRNAs with a minimum free energy ranging from -1500 to -100 kcal.mol-1 (Fig. 8B), of which 85% ranged from -1200 to -400 kcal.mol-1, indicating that these trans-NATs can form stable dsRNAs.
Figure 8. Characteristics of trans-NATs and trans-nat siRNAs.
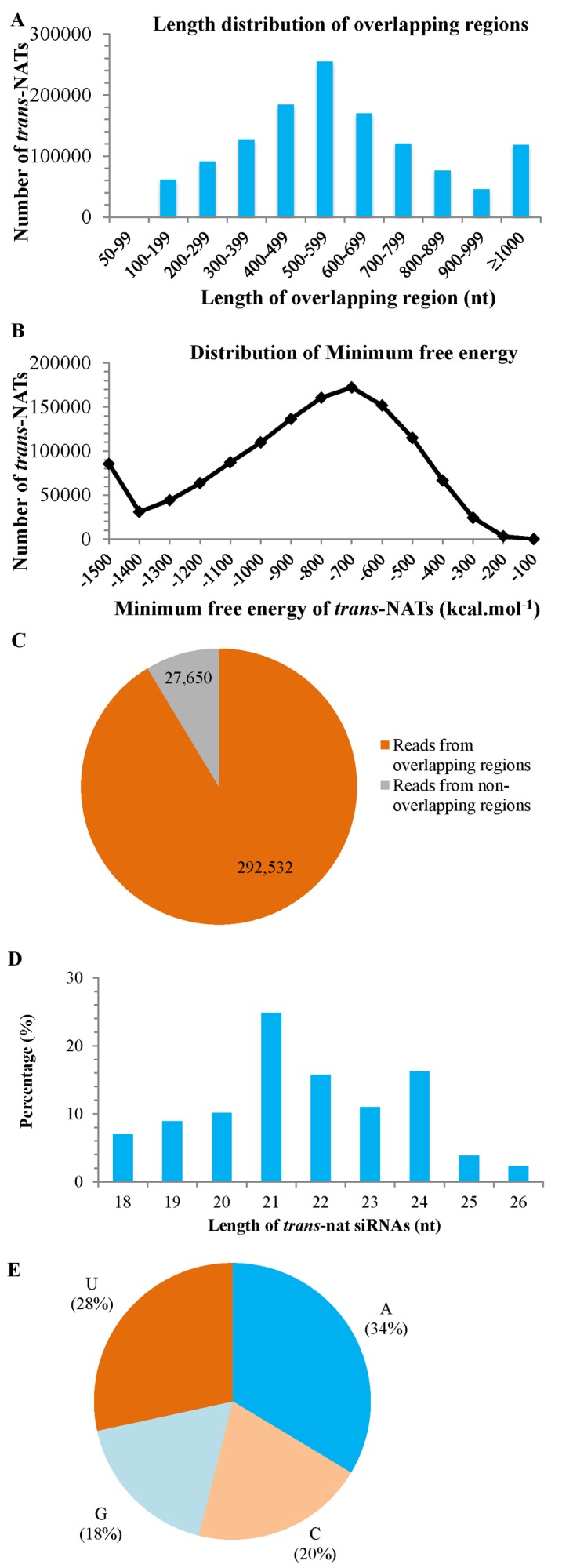
The length of overlapping regions of trans-NATs is shown in A. The distribution of minimal free energy of trans-NATs is shown in B. The enrichment analysis of trans-nat siRNAs along trans-NATs is shown in C. The distribution of length of trans-nat siRNAs is shown D. The 5’first nucleotide of trans-nat siRNAs is shown in E.
The 79,994 trans-nat siRNAs represented a total of 320,182 reads (Fig. 8C), and 91% reads were derived from overlapping regions. Trans-nat siRNAs ranged from 18 to 26 nt in length, with the 21 nt siRNAs being the most abundant (Fig. 8D). Most trans-nat siRNAs (62%) had an A or U as the 5’first nucleotide (Fig. 8E), and relatively fewer G and C appeared at the most 5’end.
To date, trans-NATs have been identified in a broad range of plants [79], such as Upland cotton and G. raimondii, while no trans-nat siRNA has been previously reported in Pima. In this study, we firstly identified a large number of trans-NATs in Pima, producing a significant number of trans-nat siRNAs. Consistent with the major length range of DCL products [10,81], most trans-nat siRNAs were 21 to 24 nt in length and had the characteristic of the 5’first nucleotide similar to other sRNAs in Pima, indicating that they are likely products of DCL cleavage. Like cis-nat siRNAs and in agreement with previous study [73], most trans-nat siRNA reads were produced from the overlapping regions, suggesting that a similar mechanism might govern the synthesis of NAT-derived siRNAs.
Conclusion
Through analysis of deep sequencing sRNA data of Pima, 5 major classes of sRNAs including miRNAs, ra-siRNAs, pha-siRNAs, cis-nat and trans-nat siRNAs along with their precursors were bioinformatically identified. This is the first comprehensive analysis of sRNAs in Pima and creates an important molecular resource for future studies on mechanism of regulating plant growth and development.
Supporting Information
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
Authors would also like to thank Dr. Narendra K. Singh at Auburn University for reviewing the manuscript.
Data Availability
Data are available from NCBI, accession No: GSM699076 (root) and GSM634227 (fiber).
Funding Statement
This project was supported by a grant from Fundamental Research Funds for the Central University (No. 2014PY042). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Ameres SL, Zamore PD (2013) Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol 14: 475–488. 10.1038/nrm3611 [DOI] [PubMed] [Google Scholar]
- 2. Meng Y, Shao C, Wang H, Chen M (2011) The regulatory activities of plant microRNAs: a more dynamic perspective. Plant Physiol 157: 1583–1595. 10.1104/pp.111.187088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pumplin N, Voinnet O (2013) RNA silencing suppression by plant pathogens: defence, counter-defence and counter-counter-defence. Nat Rev Microbiol 11: 745–760. 10.1038/nrmicro3120 [DOI] [PubMed] [Google Scholar]
- 4. Voinnet O (2009) Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687. 10.1016/j.cell.2009.01.046 [DOI] [PubMed] [Google Scholar]
- 5. Mi S, Cai T, Hu Y, Chen Y, Hodges E, et al. (2008) Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5' terminal nucleotide. Cell 133: 116–127. 10.1016/j.cell.2008.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mallory A, Vaucheret H (2010) Form, function, and regulation of ARGONAUTE proteins. Plant Cell 22: 3879–3889. 10.1105/tpc.110.080671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xu C, Tian J, Mo B (2013) siRNA-mediated DNA methylation and H3K9 dimethylation in plants. Protein Cell 4: 656–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu L, Zhou H, Zhang Q, Zhang J, Ni F, et al. (2010) DNA methylation mediated by a microRNA pathway. Mol Cell 38: 465–475. 10.1016/j.molcel.2010.03.008 [DOI] [PubMed] [Google Scholar]
- 9. Eun C, Lorkovic ZJ, Naumann U, Long Q, Havecker ER, et al. (2011) AGO6 functions in RNA-mediated transcriptional gene silencing in shoot and root meristems in Arabidopsis thaliana. PLoS One 6: e25730 10.1371/journal.pone.0025730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Axtell MJ (2013) Classification and comparison of small RNAs from plants. Annu Rev Plant Biol 64: 137–159. 10.1146/annurev-arplant-050312-120043 [DOI] [PubMed] [Google Scholar]
- 11. Lee Y, Kim M, Han J, Yeom KH, Lee S, et al. (2004) MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23: 4051–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yang L, Liu Z, Lu F, Dong A, Huang H (2006) SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J 47: 841–850. [DOI] [PubMed] [Google Scholar]
- 13. Yang SW, Chen HY, Yang J, Machida S, Chua NH, et al. (2010) Structure of Arabidopsis HYPONASTIC LEAVES1 and its molecular implications for miRNA processing. Structure 18: 594–605. 10.1016/j.str.2010.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wu F, Yu L, Cao W, Mao Y, Liu Z, et al. (2007) The N-terminal double-stranded RNA binding domains of Arabidopsis HYPONASTIC LEAVES1 are sufficient for pre-microRNA processing. Plant Cell 19: 914–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ren G, Xie M, Dou Y, Zhang S, Zhang C, et al. (2012) Regulation of miRNA abundance by RNA binding protein TOUGH in Arabidopsis. Proc Natl Acad Sci U S A 109: 12817–12821. 10.1073/pnas.1204915109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Koster T, Meyer K, Weinholdt C, Smith LM, Lummer M, et al. (2014) Regulation of pri-miRNA processing by the hnRNP-like protein AtGRP7 in Arabidopsis. Nucleic Acids Res 42: 9925–9936. 10.1093/nar/gku716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yang GD, Yan K, Wu BJ, Wang YH, Gao YX, et al. (2012) Genomewide analysis of intronic microRNAs in rice and Arabidopsis. J Genet 91: 313–324. [DOI] [PubMed] [Google Scholar]
- 18. Ramalingam P, Palanichamy JK, Singh A, Das P, Bhagat M, et al. (2014) Biogenesis of intronic miRNAs located in clusters by independent transcription and alternative splicing. RNA 20: 76–87. 10.1261/rna.041814.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Melamed Z, Levy A, Ashwal-Fluss R, Lev-Maor G, Mekahel K, et al. (2013) Alternative splicing regulates biogenesis of miRNAs located across exon-intron junctions. Mol Cell 50: 869–881. 10.1016/j.molcel.2013.05.007 [DOI] [PubMed] [Google Scholar]
- 20. Guo HS, Xie Q, Fei JF, Chua NH (2005) MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell 17: 1376–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang JW (2014) Regulation of flowering time by the miR156-mediated age pathway. J Exp Bot 65: 4723–4730. 10.1093/jxb/eru246 [DOI] [PubMed] [Google Scholar]
- 22. Kim JJ, Lee JH, Kim W, Jung HS, Huijser P, et al. (2012) The microRNA156-SQUAMOSA PROMOTER BINDING PROTEIN-LIKE3 Module Regulates Ambient Temperature-Responsive Flowering via FLOWERING LOCUS T in Arabidopsis. Plant Physiol 159: 461–478. 10.1104/pp.111.192369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xia R, Zhu H, An YQ, Beers EP, Liu Z (2012) Apple miRNAs and tasiRNAs with novel regulatory networks. Genome Biol 13: R47 10.1186/gb-2012-13-6-r47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Khraiwesh B, Zhu JK, Zhu J (2012) Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim Biophys Acta 1819: 137–148. 10.1016/j.bbagrm.2011.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rubio-Somoza I, Weigel D (2011) MicroRNA networks and developmental plasticity in plants. Trends Plant Sci 16: 258–264. 10.1016/j.tplants.2011.03.001 [DOI] [PubMed] [Google Scholar]
- 26. Herr AJ, Jensen MB, Dalmay T, Baulcombe DC (2005) RNA polymerase IV directs silencing of endogenous DNA. Science 308: 118–120. [DOI] [PubMed] [Google Scholar]
- 27. Lee TF, Gurazada SG, Zhai J, Li S, Simon SA, et al. (2012) RNA polymerase V-dependent small RNAs in Arabidopsis originate from small, intergenic loci including most SINE repeats. Epigenetics 7: 781–795. 10.4161/epi.20290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Marker S, Le Mouel A, Meyer E, Simon M (2010) Distinct RNA-dependent RNA polymerases are required for RNAi triggered by double-stranded RNA versus truncated transgenes in Paramecium tetraurelia. Nucleic Acids Res 38: 4092–4107. 10.1093/nar/gkq131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sugiyama T, Cam H, Verdel A, Moazed D, Grewal SI (2005) RNA-dependent RNA polymerase is an essential component of a self-enforcing loop coupling heterochromatin assembly to siRNA production. Proc Natl Acad Sci U S A 102: 152–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Onodera Y, Haag JR, Ream T, Costa Nunes P, Pontes O, et al. (2005) Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 120: 613–622. [DOI] [PubMed] [Google Scholar]
- 31. Smith LM, Pontes O, Searle I, Yelina N, Yousafzai FK, et al. (2007) An SNF2 protein associated with nuclear RNA silencing and the spread of a silencing signal between cells in Arabidopsis. Plant Cell 19: 1507–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Law JA, Du J, Hale CJ, Feng S, Krajewski K, et al. (2013) Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1. Nature 498: 385–389. 10.1038/nature12178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang H, Ma ZY, Zeng L, Tanaka K, Zhang CJ, et al. (2013) DTF1 is a core component of RNA-directed DNA methylation and may assist in the recruitment of Pol IV. Proc Natl Acad Sci U S A 110: 8290–8295. 10.1073/pnas.1300585110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kasschau KD, Fahlgren N, Chapman EJ, Sullivan CM, Cumbie JS, et al. (2007) Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol 5: e57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fei Q, Xia R, Meyers BC (2013) Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell 25: 2400–2415. 10.1105/tpc.113.114652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Allen E, Xie Z, Gustafson AM, Carrington JC (2005) microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221. [DOI] [PubMed] [Google Scholar]
- 37. Cho SH, Coruh C, Axtell MJ (2012) miR156 and miR390 regulate tasiRNA accumulation and developmental timing in Physcomitrella patens. Plant Cell 24: 4837–4849. 10.1105/tpc.112.103176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhai J, Jeong DH, De Paoli E, Park S, Rosen BD, et al. (2011) MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev 25: 2540–2553. 10.1101/gad.177527.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang X, Lii Y, Wu Z, Polishko A, Zhang H, et al. (2013) Mechanisms of small RNA generation from cis-NATs in response to environmental and developmental cues. Mol Plant 6: 704–715. 10.1093/mp/sst051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ron M, Alandete Saez M, Eshed Williams L, Fletcher JC, McCormick S (2010) Proper regulation of a sperm-specific cis-nat-siRNA is essential for double fertilization in Arabidopsis. Genes Dev 24: 1010–1021. 10.1101/gad.1882810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yu X, Yang J, Li X, Liu X, Sun C, et al. (2013) Global analysis of cis-natural antisense transcripts and their heat-responsive nat-siRNAs in Brassica rapa. BMC Plant Biol 13: 208 10.1186/1471-2229-13-208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK (2005) Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123: 1279–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Paterson AH, Wendel JF, Gundlach H, Guo H, Jenkins J, et al. (2012) Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492: 423–427. 10.1038/nature11798 [DOI] [PubMed] [Google Scholar]
- 44. Li F, Fan G, Wang K, Sun F, Yuan Y, et al. (2014) Genome sequence of the cultivated cotton Gossypium arboreum. Nat Genet 46: 567–572. 10.1038/ng.2987 [DOI] [PubMed] [Google Scholar]
- 45. Jiang C, Wright RJ, El-Zik KM, Paterson AH (1998) Polyploid formation created unique avenues for response to selection in Gossypium (cotton). Proc Natl Acad Sci U S A 95: 4419–4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Guan X, Song Q, Chen ZJ (2014) Polyploidy and small RNA regulation of cotton fiber development. Trends Plant Sci 19: 516–528. 10.1016/j.tplants.2014.04.007 [DOI] [PubMed] [Google Scholar]
- 47. Guan X, Pang M, Nah G, Shi X, Ye W, et al. (2014) miR828 and miR858 regulate homoeologous MYB2 gene functions in Arabidopsis trichome and cotton fibre development. Nat Commun 5: 3050 10.1038/ncomms4050 [DOI] [PubMed] [Google Scholar]
- 48. Liu N, Tu L, Tang W, Gao W, Lindsey K, et al. (2014) Small RNA and degradome profiling reveals a role for miRNAs and their targets in the developing fibers of Gossypium barbadense. Plant J 80: 331–344. 10.1111/tpj.12636 [DOI] [PubMed] [Google Scholar]
- 49. Xue W, Wang Z, Du M, Liu Y, Liu JY (2013) Genome-wide analysis of small RNAs reveals eight fiber elongation-related and 257 novel microRNAs in elongating cotton fiber cells. BMC Genomics 14: 629 10.1186/1471-2164-14-629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang H, Wan Q, Ye W, Lv Y, Wu H, et al. (2013) Genome-wide analysis of small RNA and novel microRNA discovery during fiber and seed initial development in Gossypium hirsutum. L. PLoS One 8: e69743 10.1371/journal.pone.0069743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pang M, Woodward AW, Agarwal V, Guan X, Ha M, et al. (2009) Genome-wide analysis reveals rapid and dynamic changes in miRNA and siRNA sequence and expression during ovule and fiber development in allotetraploid cotton (Gossypium hirsutum L.). Genome Biol 10: R122 10.1186/gb-2009-10-11-r122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang ZM, Xue W, Dong CJ, Jin LG, Bian SM, et al. (2012) A comparative miRNAome analysis reveals seven fiber initiation-related and 36 novel miRNAs in developing cotton ovules. Mol Plant 5: 889–900. 10.1093/mp/ssr094 [DOI] [PubMed] [Google Scholar]
- 53. Kwak PB, Wang QQ, Chen XS, Qiu CX, Yang ZM (2009) Enrichment of a set of microRNAs during the cotton fiber development. BMC Genomics 10: 457 10.1186/1471-2164-10-457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yang X, Wang L, Yuan D, Lindsey K, Zhang X (2013) Small RNA and degradome sequencing reveal complex miRNA regulation during cotton somatic embryogenesis. J Exp Bot 64: 1521–1536. 10.1093/jxb/ert013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yin Z, Li Y, Han X, Shen F (2012) Genome-wide profiling of miRNAs and other small non-coding RNAs in the Verticillium dahliae-inoculated cotton roots. PLoS One 7: e35765 10.1371/journal.pone.0035765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jeong DH, Park S, Zhai J, Gurazada SG, De Paoli E, et al. (2011) Massive analysis of rice small RNAs: mechanistic implications of regulated microRNAs and variants for differential target RNA cleavage. Plant Cell 23: 4185–4207. 10.1105/tpc.111.089045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Meyers BC, Axtell MJ, Bartel B, Bartel DP, Baulcombe D, et al. (2008) Criteria for annotation of plant MicroRNAs. Plant Cell 20: 3186–3190. 10.1105/tpc.108.064311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sato K, Hamada M, Asai K, Mituyama T (2009) CENTROIDFOLD: a web server for RNA secondary structure prediction. Nucleic Acids Res 37: W277–280. 10.1093/nar/gkp367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- 60. Boratyn GM, Camacho C, Cooper PS, Coulouris G, Fong A, et al. (2013) BLAST: a more efficient report with usability improvements. Nucleic Acids Res 41: W29–33. 10.1093/nar/gkt282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tempel S (2012) Using and understanding RepeatMasker. Methods Mol Biol 859: 29–51. 10.1007/978-1-61779-603-6_2 [DOI] [PubMed] [Google Scholar]
- 62. Stocks MB, Moxon S, Mapleson D, Woolfenden HC, Mohorianu I, et al. (2012) The UEA sRNA workbench: a suite of tools for analysing and visualizing next generation sequencing microRNA and small RNA datasets. Bioinformatics 28: 2059–2061. 10.1093/bioinformatics/bts311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chen HM, Li YH, Wu SH (2007) Bioinformatic prediction and experimental validation of a microRNA-directed tandem trans-acting siRNA cascade in Arabidopsis. Proc Natl Acad Sci U S A 104: 3318–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Dai X, Zhao PX (2008) pssRNAMiner: a plant short small RNA regulatory cascade analysis server. Nucleic Acids Res 36: W114–118. 10.1093/nar/gkn297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhou X, Sunkar R, Jin H, Zhu JK, Zhang W (2009) Genome-wide identification and analysis of small RNAs originated from natural antisense transcripts in Oryza sativa. Genome Res 19: 70–78. 10.1101/gr.084806.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lorenz R, Bernhart SH, Honer Zu Siederdissen C, Tafer H, Flamm C, et al. (2011) ViennaRNA Package 2.0. Algorithms Mol Biol 6: 26 10.1186/1748-7188-6-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cuperus JT, Fahlgren N, Carrington JC (2011) Evolution and functional diversification of MIRNA genes. Plant Cell 23: 431–442. 10.1105/tpc.110.082784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Varkonyi-Gasic E, Lough RH, Moss SM, Wu R, Hellens RP (2012) Kiwifruit floral gene APETALA2 is alternatively spliced and accumulates in aberrant indeterminate flowers in the absence of miR172. Plant Mol Biol 78: 417–429. 10.1007/s11103-012-9877-2 [DOI] [PubMed] [Google Scholar]
- 69. Jia X, Yan J, Tang G (2011) MicroRNA-mediated DNA methylation in plants. Front Biol 6: 133–139. [Google Scholar]
- 70. Zhang B, Pan X, Cannon CH, Cobb GP, Anderson TA (2006) Conservation and divergence of plant microRNA genes. Plant J 46: 243–259. [DOI] [PubMed] [Google Scholar]
- 71. Tong YA, Peng H, Zhan C, Fan L, Ai T, et al. (2013) Genome-wide analysis reveals diversity of rice intronic miRNAs in sequence structure, biogenesis and function. PLoS One 8: e63938 10.1371/journal.pone.0063938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yan K, Liu P, Wu CA, Yang GD, Xu R, et al. (2012) Stress-induced alternative splicing provides a mechanism for the regulation of microRNA processing in Arabidopsis thaliana. Mol Cell 48: 521–531. 10.1016/j.molcel.2012.08.032 [DOI] [PubMed] [Google Scholar]
- 73. Visser M, van der Walt AP, Maree HJ, Rees DJ, Burger JT (2014) Extending the sRNAome of Apple by Next-Generation Sequencing. PLoS One 9: e95782 10.1371/journal.pone.0095782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Howell MD, Fahlgren N, Chapman EJ, Cumbie JS, Sullivan CM, et al. (2007) Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell 19: 926–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Fahlgren N, Montgomery TA, Howell MD, Allen E, Dvorak SK, et al. (2006) Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr Biol 16: 939–944. [DOI] [PubMed] [Google Scholar]
- 76. Marin E, Jouannet V, Herz A, Lokerse AS, Weijers D, et al. (2010) miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell 22: 1104–1117. 10.1105/tpc.109.072553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wu MF, Tian Q, Reed JW (2006) Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 133: 4211–4218. [DOI] [PubMed] [Google Scholar]
- 78. Zhang X, Xia J, Lii YE, Barrera-Figueroa BE, Zhou X, et al. (2012) Genome-wide analysis of plant nat-siRNAs reveals insights into their distribution, biogenesis and function. Genome Biol 13: R20 10.1186/gb-2012-13-3-r20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Chen D, Yuan C, Zhang J, Zhang Z, Bai L, et al. (2012) PlantNATsDB: a comprehensive database of plant natural antisense transcripts. Nucleic Acids Res 40: D1187–1193. 10.1093/nar/gkr823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Langmead B (2010) Aligning short sequencing reads with Bowtie. Curr Protoc Bioinformatics Chapter 11: Unit 11 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Axtell MJ, Westholm JO, Lai EC (2011) Vive la différence: biogenesis and evolution of microRNAs in plants and animals. Genome Biol 12: 221 10.1186/gb-2011-12-4-221 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
Data are available from NCBI, accession No: GSM699076 (root) and GSM634227 (fiber).