Abstract
The ribonome is the total cellular complement of RNAs and their regulatory factors functioning dynamically in time and space within ribonucleoprotein complexes. We theorize that the ribonome is an ancient central co-ordinator that has evolved to communicate on multiple levels to the proteome on the one hand (feed-forward), and the transcriptome and RNA processing machinery on the other (feed-back). Furthermore, the ribonome can potentially communicate to other cells horizontally with implications for biological information transfer and for the evolution of both RNA and DNA operating systems. The post-transcriptional RNA operon theory of co-regulated gene expression accounts for the co-ordinated dynamics of RNA-binding proteins within the cellular ribonome, thus allowing for the recombination and remodelling of the RNPs (ribonucleoproteins) to generate new combinations of functionally related proteins. Thus, post-transcriptional RNA operons form the core of the ribonomic operating system in which both their control and co-ordination govern outcomes. Within the ribonome, RNA-binding proteins control one another’s mRNAs to keep the global mRNA environment in balance. We argue that these post-transcriptional ribonomic systems provide an information management and distribution centre for evolutionary expansion of multicellularity in tissues, organs, organisms, and their communities.
Keywords: exosome particle, post-transcriptional operon, RNA regulon, RNA stability, translational regulation, USER code
The Ribonome
The ribonome operates at multiple levels of gene regulation to control and co-ordinate transcription, splicing, export, stability, and translation (Keene, 2007a; Halbeisen et al., 2008). It is a central regulatory environment that is both self-sustaining and self-limiting. In mammalian cells the ribonome consists of thousands of RBPs (RNA-binding proteins) and their associated mRNAs, non-coding regulatory RNAs and auxiliary proteins that are contained in mRNPs [messenger RNPs (ribonucleoproteins)]. The importance of the ribonome is highlighted by the fact that the steady-state levels of proteins within the proteome of a cell tend to have an imprecise correlation with the steady-state levels of mRNAs within the transcriptome, suggesting that additional regulation must be occurring (reviewed in Keene, 2001; Keene and Tenenbaum, 2002). RBPs are found in all organisms and surprisingly outnumber DNA-binding factors almost two to one in eukaryotes with approx. 2500 or more proteins estimated to function as RNA interactors (Keene, 2001; Gerber et al., 2004). Saccharomyces cerevisiae has hundreds of RBPs but the same ribonomic organizational principles appear to operate (Gerber et al., 2004; Keene, 2007a; Halbeisen et al., 2008; Komili and Silver, 2008; Saint-Georges et al., 2008). Moreover, evidence for coordinately regulated post-transcriptional operons and decay regulons now exists for archaea (Andersson et al., 2006), and decay regulons have been demonstrated in bacteria (Bernstein et al., 2004; Halbeisen et al., 2008), supporting the ribonome’s importance in gene expression across all domains of the Tree of Life.
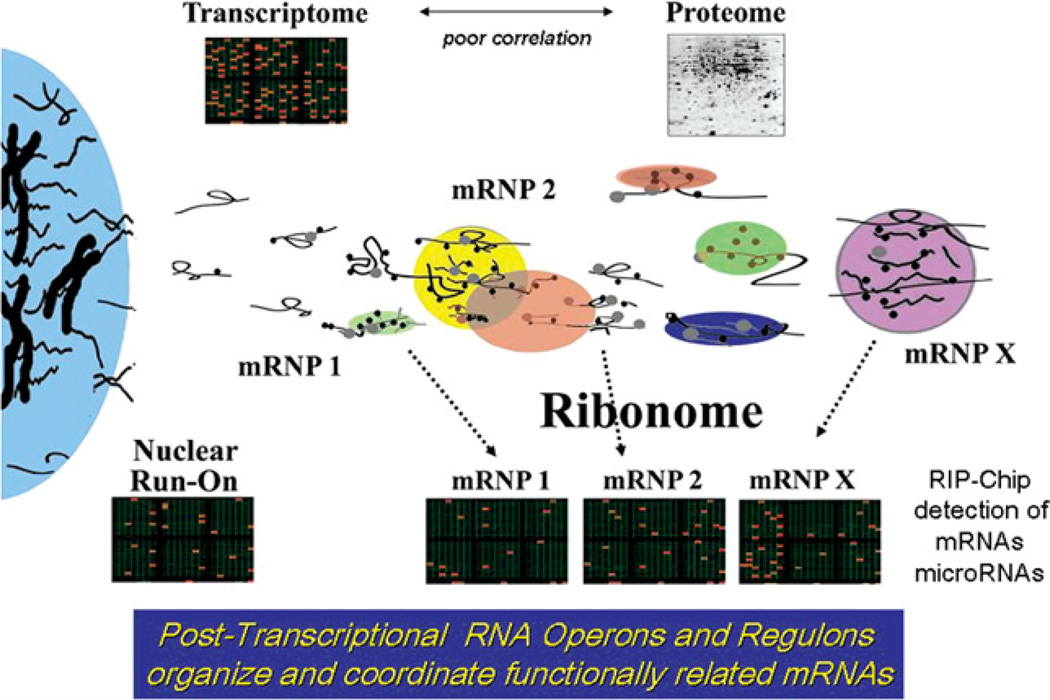
The theory of the ribonome and the PTRO (post-transcriptional RNA operon) is founded on the premise that discrete subsets of mRNAs are assembled and co-regulated in eukaryotic cells via a mRNP-driven process (Figure 1; Keene and Tenenbaum, 2002). RBPs and small non-coding RNAs are multi-targeted to functionally related mRNAs and act as trans-acting factors within the RNP infrastructure to co-ordinate mRNA expression via cis-acting elements contained within the mRNAs (Keene, 2001; Bartel, 2004; Jing et al., 2005; Elemento et al., 2007). Multiple cis-regulatory elements exist on each mRNA and result in a unique, yet modular USER (untranslated sequence element for regulation) code (Keene and Tenenbaum, 2002). These USER codes therefore determine which RNA-binding factors can associate with each message in order to co-operate or compete, and ultimately regulate the fate of that mRNA. Multiple RNA-binding factors can associate with a single message simultaneously or sequentially and a single RBP or non-coding regulator RNA can potentially target a large number of different mRNA species (Gao et al., 1994; Tenenbaum et al., 2000; Bartel, 2004; Keene, 2007a; Komoli and Silver, 2008; Halbeisen et al., 2008; Hendrickson et al., 2008; Hogan et al., 2008; Landthaler et al., 2008). It is the co-operation and competition among these factors which results in the final combinatorial codes of regulation of a given message. However, it is not simply a matter of controlling a single mRNA’s fate, but rather the intricate harmonization of groups of messages that underlie the functioning of PTROs. For example, the stability of multiple mRNAs change together in a co-ordinated fashion following cell perturbations, whether internally or externally derived (Lam et al., 2001; Wang, Y. et al., 2002; Yang et al., 2003; Grigull et al., 2004; Raghavan et al., 2004; Cammas et al., 2008; Lukong et al., 2008; DeGracia et al., 2008). This coordinated orchestration of related mRNAs through turnover, localization and/or translatability provides the cell an agile means to rapidly adapt to its ever-changing environment.
Figure 1. Visualization of the analysis of RNPs within the ribonome.
The organization and co-ordination of RNPs that drive post-transcriptional gene expression within the ribonome as experimentally determined using RIP-Chip to detect mRNA and microRNA components of RNP complexes. The finding that functionally related mRNAs are associated with specific RBPs within mRNPs led to the post-transcriptional RNA operon/regulon model of co-ordinated gene expression. Subsequently, other studies using polysome gradient profiling, RNA stability-microarrays and high throughput deep sequencing demonstrated that subsets of functionally related mRNAs can be co-ordinated at the levels of translation and stability (reviewed in Keene, 2007a). Reproduced from Keene (2001) with permission. © (2001) Proc. Natl. Acad. Sci. U.S.A.
Differences between the ribonome and the transcriptome
The concept of the ribonome was derived as an operational definition. Some may question whether such an environment exists and how it would differ from the transcriptome. The transcriptome is the total mRNA complement of the genome that is present in steady state at a given point of time. The ribonome on the other hand embodies the dynamic organization of mRNAs within the ribonucleoprotein infrastructure (Figure 1). Each mRNA species has multiple lives within the ribonome, and each mRNP exists in a state of activity in which the stability or translatability of mRNA components are changing in a concerted manner within each RNP in response to cellular signals. Indeed, the combinatorial PTRO model provides for each individual mRNA member of a given mRNA species to have a unique life, each of which can be regulated multiple ways as a member of more than one group of co-regulated mRNAs that form an RNA operon or regulon (Keene, 2001; Keene and Tenenbaum, 2002; Hieronymus and Silver, 2004; Moore, 2005; Keene, 2007a). However, transcriptomics does not generally reflect these diverse mRNA activities as determined by the state of the RNP but instead considers the total amount of each accumulated mRNA. For example, if there are 100 mRNAs encoding protein ‘X’, 25 of them may be in a translationally activated state in mRNP #1, another 50 mRNAs may be undergoing temporary translational repression within mRNP #2, and another 25 species may be stored away in a translationally silent compartment. The actual expression state of the total population of mRNAs would then be 25% of the theoretical amount determined by transcriptomic analysis alone since the functional state of each mRNA is not being determined. Many other examples of such a distribution can be imagined for each and every cellular mRNA. Thus, the ribonome is distinctly different from the transcriptome in part because the transcrip-tome does not necessarily provide functional information about the individual or the collective lives of each type of mRNA. Moreover, most transcriptional network studies base their measurements on changes of the steady state levels of mRNAs and do not account for the proportion of RNA-processing events that are acting on these mRNAs within the ribonome (Keene, 2001). Therefore, to gain more functional understanding of gene expression networks, it is essential to account for post-transcriptional co-ordination of RNA components within the ribonome.
There are also many differences between transcription and translation that account for robust regulation at the post-transcriptional level. For example, while the transcriptional machinery is confined to producing immature transcripts in the nucleus, the progression of those transcripts to translation requires many inter-connected steps of RNA processing that occur in various compartments of the cell (Figure 2). Interestingly, many RBPs participate in multiple steps along this maturation pathway. For example, ELAV/HuR, polypyrimidine-tract-binding protein, the U2AF RBPs and many hnRNP (heterogeneous nuclear RNP) proteins have a strong nuclear presence, but are believed to shuttle out of and back into the nucleus with their RNA cargo during normal cellular growth and differentiation. Many of these nucleocytoplasmic shuttling proteins are involved in nuclear RNA processing (splicing, polyadenylation and capping) and nuclear export as well as in subsequent regulation of the stability and translation of the mature mRNA targets in the cytoplasm (Pinñol-Roma and Dreyfuss, 1992; Keene, 1999, 2001; Le Hir et al., 2000; Moore, 2005; Moore et al., 2006; Keene, 2007a, 2007b). Moreover, the recent demonstration that the microRNA miR-124 itself regulates the expression of the polypyrimidine tract binding protein RBP alternative splicing regulator is consistent with the dominance of the ribonome as a regulator of regulators (Makeyev and Maniatis, 2008). Indeed, a tenet of the PTRO model is that decisions regarding the composition of the RNP complex and the co-ordinated fate of mRNA members of a PTRO can be determined very early in the gene expression pathway. In addition, it must be remembered that one species of mRNA is not limited to one particular pathway but instead can mature along different parallel pathways allowing multiple mRNAs to be regulated combinatorially at the levels of RNA stability and/or translation depending on the current needs of the cell (Keene, 2007a).
Figure 2. Diagram of the RNA processing steps between transcription and translation showing the feedback from translation to each step in the pathway.
The flow of both informational mRNAs and regulatory micro-RNAs from transcription and through splicing, export, stability, localization and translation is depicted. The dominance of translation in feeding back and co-ordinating events upstream of the informational flow of mRNA is depicted by green arrows. The combinatorial regulation of translation by RBPs and mi-croRNAs is directed from within RNPs.
Similarities between the ribonome and the transcriptome
Similar to the transcriptome, that is in part determined by the modular state of the chromatin and the action of DNA-binding proteins, the ribonome is defined by the state of the RNPs that exist within the cell at any given time. Just as the RNA synthesis apparatus has modular transcriptional regulatory domains, the protein synthesis apparatus that regulates translation also has modularity. This may be counterintuitive to many biologists because we are taught to think of translation as a passive process that responds to any mRNA on a first come-first served basis in a more or less random Brownian environment. However, translation is a dynamic and selective process that involves modular organization and sorting of multiple transcripts through various mechanisms, including localization to the endoplasmic reticulum and potentially involves special classes of ribosomes for translating specific classes of mRNAs (Mauro and Edelman, 2002; Keene and Tenenbaum, 2002; Lerner et al., 2003; Rajasekhar et al., 2003; Nicchitta et al., 2005; Stephens et al., 2005, Komili et al., 2007; Pyhtila et al., 2008). Furthermore, translation is a highly ordered process that operates in global regulatory states and modular domains not unlike those of transcription. For example, heterochromatin is transcriptionally inactive, and likewise, there are RNPs such as processing bodies and stress granules in which mRNAs are translationally silent or turning over via specific degradative mechanisms. Interestingly, in some cases this can be a dynamically reversible process, as mRNAs may exit these particles to become translationally activated (Brengues et al., 2005; Bhattacharyya et al., 2006). In addition, post-translational modifications of both DNA-binding proteins in chromatin (Orphanides and Reinberg, 2002; Harbison et al., 2004; Aten and Kanaar, 2006; Li et al., 2008), and RNA-binding proteins in RNPs may determine the physical states of modules that co-ordinate the production of RNA and protein, respectively (Intine et al., 2003; Benjamin et al., 2006; Abdelmohsen et al., 2007; Garbarino-Pico et al., 2007; reviewed in Keene, 2007a, 2007b; Cammas et al., 2008).
Since the PTRO theory was proposed, many studies have investigated the global targets of individual RBPs and their results have supported many aspects of the PTRO model, including the main tenet that RBPs function to co-ordinately regulate functionally related messages (Keene, 2007a; Galgano et al., 2008; Hogan et al., 2008; Mazan-Mamczarz et al., 2008; Morris et al., 2008). However, dynamic regulation of the ribonome needs to be examined in order to better understand gene expression networks. As more studies of this type become available it will be important to incorporate dynamic features of the ribonome and the specific functions of PTROs into global gene expression models. Thus, to gain a complete picture of cellular gene expression, it is going to be crucial to fully understand the ribonome’s role in shaping the overall post-transcriptional RNA network.
Intracellular co-ordination of ribonome dynamics
While most studies to date have relied on the measurement of steady-state mRNA levels to determine gene expression, it is becoming clear that these static snapshots fail to account for many regulatory events of gene expression occurring in cells. In fact, many studies looking at the relationship between the transcriptome and the proteome have reported weak to moderate correlation at best (r = 0.2–0.6), suggesting post-transcriptional regulation is playing an important role in co-ordinating outcomes (Gygi et al., 1999; Griffin et al., 2002; Washburn et al., 2003; Tian et al., 2004; Cox et al., 2005; Mijalski et al, 2005; Lu et al., 2006). Many studies have reported RBP target identification through ribonomic methods such as RIP-Chip (RNP immunoprecipitation-microarray) and the related CLIP (cross-linking and immunoprecipitation) procedure (Gao et al., 1994; Ule et al., 2003; reviewed in Keene et al., 2006; Keene, 2007a; Galgano et al., 2008; Halbeisen et al., 2008; Hendrickson et al., 2008; Hogan et al., 2008; Landthaler et al., 2008; Mazan-Mamczarz et al., 2008; Saint-Georges et al., 2008). All of these studies were geared toward defining the RNA contents of cellular RNPs that make the ribonome distinct from the transcriptome.
As more targets of RBPs are identified, the focus is shifting to understanding functional interactions and how RNP interactions are mechanistically manifest, as well as how they change dynamically. Rapid post-transcriptional responses to perturbations require rapid modification of RNP states and reorganization of PTROs during responses to biological stimuli (reviewed in Hieronymus and Silver, 2004). Tenenbaum et al. (2000) induced cellular differentiation of P19 preneuronal cells and noted that the mRNA components associated with the HuB protein changed. This experiment provided the first evidence that interactions between a post-transcriptional coordination factor and a distinct subset of associated mRNAs within the ribonome could be remodelled or crafted by internal responses of the gene expression machinery to external stimulation. It is presumed that the induction of neuronal differentiation in these cells following the addition of retinoic acid caused specific alternations in the pathway from transcription to translation that was captured as a snapshot of the mRNA targets associated with the HuB RNP. In other words, the program of differentiation was changing at the post-transcriptional RNP level. Analysis of these associated mRNAs indicated that their presence in the RNP complex was not the result of increased transcription but instead were probably due to some mRNAs entering and other mRNAs exiting the HuB RNP. The field is now beginning to investigate the mechanisms that drive changes in mRNP composition and their underlying dynamics.
Post-translational modification of RBPs
One process that drives ribonome dynamics is cellular signalling via post-translational modification of RBPs by phosphorylation, acetylation, etc. One recently published example is the regulation of binding of HuR to the SIRT1 [sirtuin (silent mating type information regulation 2 homologue) 1] mRNA and its phosphorylation by the Chk2 kinase (Abdelmohsen et al., 2007). Chk2 is an oxidant-responsive cell cycle checkpoint kinase that is activated during cellular stress. SIRT1 is a deacetylase involved in longevity that promotes cell survival in the face of oxidative and genotoxic stress, and presumably epigenetic changes of the chromatin. SIRT1 has been shown to be a HuR target message and their binding leads to stabilization of the message and enhanced expression of the SIRT1 protein. Previous studies have shown that during cellular stress HuR is activated and stabilizes messages necessary for appropriate response to that stress (Gallouzi et al., 2000; Wang, W. et al., 2002; Gorospe, 2003; Wang et al., 2004; Bhattacharyya et al., 2006; Abdelmohsen et al., 2008). However, in this study, SIRT1 was shown to be down-regulated during cellular stress due to its dissociation from HuR after HuR was phosphorylated by Chk2. Investigation into the global effects of HuR phosphorylation showed that phosphorylation of HuR led to increased binding to certain mRNA targets (prothymosin-α, p21), and reduced binding to other targets (cyclinD, cytochrome C, cyclinA) including the SIRT1 mRNA. Mutation of the phosphorylated residues on HuR led to changes in binding affinity to these targets. In addition to Chk2, HuR has also been shown to be downstream of a number of other kinases, including p38 and protein kinase Cα as well as others (Doller et al., 2007, 2008; Kim and Gorospe, 2008). However, the precise sites and functional consequences of phosphorylation have not been identified. In addition, HuR is methylated by the CARM1 methylase in response to LPS (lipopolysaccharide) stimulation (Li et al., 2002). Thus, although global mRNA analysis of these signaling events is still being investigated, it is clear that post-translational modification of HuR can regulate its binding affinities for a variety of important growth-regulatory protein transcripts and probably affects their intracellular localization (Gallouzi et al., 2000).
Other studies have looked at the effects of phos-phorylation of related RBPs. For example, when the BRF1 (butyrate response factor 1) RBP is in the un-phosphorylated form, it participates in the degradation of associated messages that contain AREs (AU-rich elements). Phosphorylation of BRF1 leads to its association with the 14-3-3 scaffolding proteins, and both the BRF1 protein and its mRNA cargos are stabilized (Schmidlin et al., 2004; Benjamin et al., 2006). Phosphorylation of KRSP, another ARE regulatory RBP, can have differing effects depending on the kinase involved. Phosphorylation of KRSP by p38 kinase leads to loss of mRNA binding and stabilization of myogenic transcripts including myogenin, p21 and myoD, while Akt activation leads to KRSP phosphorylation and stabilization of a subset of messages including those encoding hnRNP F, A/B and A1, β-catenin, and the protein phosphatase PP2A (Briata et al., 2005; Benjamin et al., 2006; Gherzi et al., 2006; Ruggiero et al., 2007). These, and other examples, highlight the fact that not only is the ribonome dynamic, but that different subsets of messages can be differentially regulated within the ribonome depending on the cellular environment and signalling cascades involved (Cammas et al., 2008; Doller et al., 2008; Kim and Gorospe, 2008).
Regulators of regulators
One of the more interesting results to come out of recent ribonomic studies is that predominant among the mRNA targets within a given RNP are those that encode other regulators of gene expression, a phenomena generally described as “regulators of regulators” (Tenenbaum et al., 2000; Lopez de Silanes et al., 2004; Mesarovic et al., 2004; Penalva et al., 2004; Gerber et al., 2006; Keene, 2007a; Pullman et al., 2007; Morris et al., 2008; Galgano et al., 2008). These recognizable regulators of gene expression include factors that affect multiple levels including transcription, splicing, transport, stability, localization, and translation. Moreover, the participation of small non-coding RNAs and RBPs in the combinatorial trans-regulation of mRNAs encoding RBPs offers higher order modular co-ordination (Keene et al., 2006; Galgano et al., 2008; Hendrickson et al., 2008; Hogan et al., 2008; Landthaler et al., 2008). Regulation of these factors includes a series of post-transcriptional feedback loops along the pathway of gene expression (Figure 2). This interconnected network of regulators acting on one another’s mRNAs ensures balanced control of a much broader set of other mRNAs that encode functionally related proteins such as those that form macromolecular complexes and signalling pathways as per the RNA operon theory (Keene and Tenenbaum, 2002).
The concept of the ribonome as a central managerial environment in biology can be visualized as a broad network of regulatory modules. As part of a subnetwork, each RBP interacts with the mRNAs encoding other regulators, thus providing a system in which each regulator can be translated only when the community of bound RBPs allows it (Figure 3). As a result of this mininetwork, these RBPs indirectly regulate the downstream targets of those regulators that it is managing. Thus, in the larger post-transcriptional network, regulators can affect global mRNA composition through their interactions with other RBPs, or by directly affecting the fate of their target mRNAs (Figure 3). Thus each of the RBPs in the system can regulate one another’s mRNAs while also regulating multiple mRNA targets either individually or in concert with other RBPs within the ribonome. When a signal such as phosphorylation is directed at a specific RBP it can affect regulation of its neighbouring RBPs and/or microRNAs within the ribonome (Intine et al., 2003; Benjamin et al., 2006; Abdelmohsen et al., 2007; Garbarino-Pico et al., 2007). Therefore, the direct as well as the indirect properties of the ribonome provide a dynamic quality of multi-level co-ordination that has not been appreciated by biologists in the past.
Figure 3. RBPs functioning as regulators of regulators.
Model of RBPs forming a sub-network in the cytoplasm where they directly regulate the stability or translation of one another’s mRNAs as an interaction balance co-ordination system (Mesarovic et al., 2004). This system is not closed but open to co-ordinating subsets of global mRNA targets (Venn ovals) dynamically in response to incoming signals as determined by the balanced regulation of each RBP in the sub-network.
Highlighting the regulatory potential of an RBP subnetwork, Myriam Gorospe’s group recently addressed whether multiple RBPs can regulate one another’s mRNAs by investigating a group of ARE RBPs they termed “turnover and translation regulatory RBPs” (TTR-RBPs) including AUF1, HuR, KRSP, NF90, TIA-1 (T-cell-restricted intracellu-lar antigen-1) and TIAR (TIA-1-related protein) (Pullman et al., 2007; Abdelmohsen et al., 2008). From this study it was clear that each of these six TTR-RBPs not only bound its own mRNA, but also bound to the messages of many of the other RBPs within this small group. For example, TIA-1 is regulated by mRNA stabilization by HuR, and by repression of its translation by TIAR. This study exemplifies the interconnectivity within the ribonome, as changes in two RBPs, HuR and TIAR, not only affect their own global subsets of mRNA targets, but also regulate TIA-1 protein production as part of a ‘closed’ TTR ribonomic sub-network, thereby affecting TIA-1’s globally targeted mRNA population in the larger ‘open’ system. Thus, the control exerted by each member of this sub-network is proposed to provide a global co-ordination that potentially harmonizes multiple mRNAs during homoeostasis as well as in response to environmental inputs.
In addition to affecting the biosynthesis of one another’s proteins as part of a closed system, translation factors can also mediate regulatory events by affecting other gene expression regulators. For example, post-transcriptional regulation of factors such as the histone deacetylase SIRT1 and other DNA-modifying enzymes could result in epigenetic changes to the genome, resulting in inheritable changes in gene expression. Furthermore, the post-transcriptional regulators of transcription and splicing also have the potential to affect the generation of and contents of a PTRO by activating or inhibiting each specific transcription or splicing factor mRNA (Figure 2). In this way, it would be possible to control the production of new members of PTROs, including microRNAs and alternatively spliced variants in the larger open system of mRNA targets. Indeed, it is well established that splicing events allow RBPs to tag the mRNAs such that subsequent post-transcriptional events, including translatability, are influenced (Le Hir et al., 2000; Moore et al., 2006). This suggests that alternative splicing of specific mRNAs can affect decisions as to which PTRO they would join; one splice variant may mark translational activation, while another might be translationally silenced. Following the appropriate signalling event, the fates of the mRNA components could change, thus feeding back to the transcription and splicing apparatus to further alter the gene expression program in a co-ordinated fashion. While this may be a ribonome-centric view of gene expression, it must be noted that transcriptional regulation of regulators requires that each event pass through the ribonome to either process a regulatory non-coding RNA or to translate the protein regulator (Figure 2).
Intercellular transfer of genetic information between ribonomes
Previous studies have demonstrated that conditioned media and secreted microvesicles derived from several cell types, including T-cells, alter the pheno-types of recipient cells (Hakelien et al., 2002; Fevrier and Raposo, 2004). Moreover, embryonic stem cells were shown to produce RNA-containing microvesicles that could reprogram haematopoietic progenitor cells (Ratajczak et al., 2006). Phenotypic transfer from tumour cells to monocytes by mRNA contained in microvesicles and exosomes was also reported (Baj-Krzyworzeka et al., 2006). In addition, Aliotta et al. (2007) demonstrated that RNAs contained in exosomes released from radiation-treated lung cells could transfer a lung phenotype to bone marrow cells. These findings are relevant to the classical observation of “bystander effects” on surrounding cells following irradiation. Even more interesting is the report by Valadi et al. (2007) demonstrating that both mRNAs and microRNAs are present in exosomal particles purified from mast cells. We have suggested that specific mRNAs and microRNAs organized into modular PTROs may be targeted to exosomes in order to participate in cell-cell exchange during growth, differentiation and environmental assault, including tissue repair and cell death (Keene, 2007a, 2007b). More recently, in studies of mRNAs associated with the HuR RNP in Jurkat T-cells, we found an extensive degree of overlap with mRNAs identified in mast cell exosomes by Valadi et al. (2007) and we detected HuR in exosomes purified from these cells as well (M.A. Thompson and J.D. Keene, unpublished results). These results are consistent with the possibility that exosomes could contain a HuR regulated PTRO. While much more investigation is needed, these findings could have important implications for coherent RNA transfer in many biological systems, including development and pathogenesis in mammals.
The potential of exosomes and other microparticles extruded by one cell to passage RNPs to another cell also has implications for horizontal transfer of genetic information in evolution. The PTRO model proposed that each RNA operon, representing an organized and coherent collection of information, is potentially a meta-representation of a small modular subgenome containing a few hundred mRNAs. To describe the collective functions of PTROs that contain sets of functionally coherent information in the form of RNA, the term “quasi-genome” was suggested in part for comparison with the components of some RNA viruses that contain multiple RNA genes (Keene and Tenenbaum, 2002). While still speculative, if entire PTROs are transferred between cells by modular particles, these quasi-genomes might shape the ribonome of the recipient cell and thereby alter its phenotype by bringing in RNAs and proteins that act on several levels of gene expression (Figures 2 and 4). Further studies of post-transcriptional regulation by RBPs and microRNAs within the ribonome of a single cell type will be needed in order to better understand the effects of their intercellular exchange on recipient cells.
Figure 4. Horizontal transfer of post-transcriptional operons by exosomal particles.
The intercellular transfer of ribonucleoproteins containing specific RNA subsets from a donor cell to a recipient cell is via exosomal particle release from the donor cell. This hypothesis is based upon the transfer of a coherent subset of mRNAs, microRNAs and possibly other components of the ribonome, but it has not been tested. This mechanism would provide the RNA templates and post-transcriptional regulators necessary for translational co-ordination of the transferred mRNAs in the recipient cell.
As suggested previously, the idea of cell-to-cell exchange of PTROs has profound implications for the origins and evolution of viruses as well as cells (Keene, 2007a). Gould et al. (2003) suggested that exosomes augment HIV infection by assisting virus entry and subsequent infectivity. We proposed that PTRO-containing exosomes could be ancestors of RNA viruses by providing groups of mRNAs that encode RBPs that could ultimately serve as viral genes and viral proteins respectively. That RNA operons may be evolutionary precursors of RNA viruses is based in part on the finding in many labs that RNA operons identified to date contain mRNAs encoding many other RBPs (Keene, 2007a, 2007b). Thus, not unlike RNA viruses that contain predominately RNAs encoding RNA-binding and processing factors, RNA operons have the potential to become ‘selfish’ and parasitic in order to gain fitness for an independent existence by evolving toward self-replication (Keene, 2007a, 2007b). This may have helped give rise to retroviruses as ‘non-professional’ mediators of horizontal gene transfer, as opposed to the naturally shed cell entities that we call exosomes from which they may have been derived.
While the idea of exosomes transferring PTROs from cell to cell is intriguing, many questions remain, including the specificity of cell-to-cell transfer, as well as how exosome production and content is regulated. Recently, Arnold Levine and co-workers suggested that a novel function of the p53 tumour suppressor protein is to stimulate exosomal secretion (Yu et al., 2006). Given that p53 responds to stress signals by affecting transcription, genes can be activated that affect secretion, many of which are contained in exosomes. Following H460 cell DNA damage, an analysis of secreted proteins indicated that p53-activated gene products were found in the culture medium. The investigators demonstrated that these p53-activated proteins exited the cells as components of exosomal vesicles. A specific gene product, TSAP6, was found to enhance exosome production from p53-activated cells and these exosomes transferred proteins to other cells. These experiments did not investigate whether RNAs were contained within these exosomes, but as noted above, recent studies from several laboratories suggested that RNAs are cargoes of exosomes as well (Baj-Krzyworzeka et al., 2006; Ratajczak et al., 2006; Aliotta et al., 2007; Valadi et al., 2007). Interestingly, the HuR RBP that forms one of the best documented mammalian PTROs activates the translation of p53 protein in UV-treated cells, making it a good candidate for an intermediate regulator of exosome production (Mazan-Mamczarz et al., 2003; Keene, 2007a).
The classical model for the origins of retroviruses suggested that moveable genetic elements were their ancestors, and many endogenous human retroviruses have since been identified as presumed remnants of exogenous retroviruses (Temin, 1980; Wang et al., 2007). The well-documented exchanges between retroviral sequences and host genomic sequences are believed to be a major driving force of evolution. The recent fascinating discovery that the human p53 transcriptional network may have been shaped during the split between Old-World and New-World monkeys approx. 40 to 60 million years ago by a temporal wave of exchange of endogenous retroviruses raises the question of how these primate exchanges were motivated (Wang et al., 2007). Could it be that particle exchanges of RNAs or DNAs between cells were mediated by exosomal particles and that these events represent evolutionary tracks of retrovirus emergence that accelerated evolution and helped craft the human genome? Elucidating the functional organization of mRNAs and microRNAs within microparticles and exosomes, and the effects of their horizontal transfer to recipient cells has the potential to reveal new mechanisms of cell–cell communication at the level of RNA.
Conclusions
For several years we have maintained that transcriptomic data represents a very incomplete picture of global control and co-ordination of gene expression (Keene, 2001, Keene and Tenenbaum, 2002, Keene, 2007a). Indeed, gene expression networks as derived using transcriptomic data alone do not distinguish transcriptional events from post-transcriptional events, yet both are represented in such data sets, and this can lead to misinterpretation of regulatory networks. The intrinsic modularity of the PTRO model includes both control and co-ordination network qualities that may help determine underlying mechanisms of translation where operational processes cycle back to earlier steps in the pathway of gene expression (Figure 2).
In classical molecular biology, ‘control’ involves dictation of one-on-one events, whereas the term ‘coordination’ describes a distributive process that harmonizes global events by motivating multiple control functions (Mesarovic et al., 2004). However, the term ‘regulation’ has been used to encompass both single and multiple controlling and/or co-ordinating events of unknown configuration that involve any number of combined molecular processes, presumably leading to global gene expression outcomes. Therefore, it is important to understand these separate but inter-related aspects of gene regulation and to account for both kinds of information when interpreting and constructing network models.
It is clear that biogenesis of the proteome determines cell fate, and strongly affects the performance of the surrounding ‘bystander’ cells in multicellular organisms. Moreover, every transcription regulator must be translated and every translation factor must be transcribed. Therefore, at the very least eukaryotic cells have two intercommunicating and interdependent gene expression networks that are likely to be synergistic and they must both be understood before a comprehensive unified model can be formulated. We view the ribonome as a layered system of modules and circuitry that provides control, co-ordination and bounded autonomy across gene expression networks, much of which depends on the cellular localization of RNP particles and granules (Mesarovic et al., 2004; Davidson and Ellington, 2007). Each decision as to when and where to translate discrete subsets of functionally related mRNAs is exceedingly important for co-ordinating coherent outcomes, and is essential for the performance of cellular homoeostasis, responses to environmental signals and differentiation.
Finally, we view the ribonome as the dominant environment of gene regulation. The evidence and reasoning as outlined in this article includes: (i) the high abundance of RNA-binding and -processing factors in eukaryotes (Keene, 2001); (ii) the ability of RBPs to locally determine one another’s biosynthetic role as regulators of regulators (Penalva et al., 2004; Keene, 2007a, 2007b; Pullman et al., 2007; Morris et al., 2008; Galgano et al., 2008; Saint-Georges et al., 2008); (iii) the fact that there are multiple copies of each monocistronic mRNA, each of which can be regulated independently or in combination with others (Keene and Tenenbaum, 2002; Moore, 2005; Halbeisen et al., 2008; Hogan et al., 2008); (iv) the well-established responsiveness of RNA stability and translation to external signals mediated by post-translational modifications (not unlike transcription) (Intine et al., 2003; Abdelmohsen et al., 2007, 2008); and (v) the fact that many regulators of transcription, splicing and RNA export are primary mRNA targets of translation itself (reviewed in Keene, 2007a). This latter point is highlighted by the fact that transcriptional regulators must be translated in the cytoplasm, making a transcriptional ‘regulators of regulators’ model dependent upon the downstream performance of the entire post-transcriptional program that then produces new transcription factors or chromatin-modifying factors that must be transported back to the nucleus. On the contrary, translational RBP regulators can directly determine which regulatory proteins are able to be made for feedback to transcription, splicing, export, stability and translation itself, because their mRNAs are present as mRNPs in the cytoplasm; in many cases these RBPs are bound to one another’s mRNAs, and thus, have an opportunity to directly ‘vote’ on one another’s fate.
While one may argue that this ‘chicken or egg’ scenario of transcription versus translation is not surprising, prevailing dogma places transcriptional control at the centre of gene regulation (control and co-ordination), and relegates translation to a more passive role in which control points operate for individual mRNAs but co-ordination is not considered (Keene, 2001). We have challenged this traditional model of gene expression because data supporting the PTRO model is based on extensive co-ordination of gene expression by RBPs and non-coding regulatory RNAs, suggesting at least an equal, and probably more agile, role for post-transcriptional co-ordination. This less traditional model regarding the importance of post-transcriptional events is consistent with the RNA world view of evolution in that the protein biogenesis machinery is thought to have preceded the acquisition of a nucleus and the appearance of DNA (Margulis, 1970; Martin and Koonin, 2006). Whether post-transcriptional processes overall have retained a dominant operational mode in evolution is yet to be substantiated but at the very least, future models of global gene expression need to account for the extensive amount of post-transcriptional co-ordination that has now been widely demonstrated (Keene, 2001; Keene and Tenenbaum, 2002; Hieronymus and Silver, 2004; Moore, 2005; Keene, 2007a, 2007b; Halbeisen et al., 2008).
Acknowledgements
The authors thank Matthew Friedersdorf, Jordan Komisarow and other members of the Keene Laboratory for their critical reading of the manuscript.
Funding
This work supported in part by a Ruth Kirschstein National Research Service Award from the National Institute of Neurological Disorders and Stroke (KDM) [grant number 5F32NS059100] and by the National Cancer Institute [grant number CA94365].
Abbreviations used
- ARE
AU-rich element
- BRF1
butyrate response factor 1
- PTRO
post-transcriptional RNA operon
- RBP
RNA-binding protein
- RIP-Chip
RNP immunoprecipitation-microarray
- RNP
ribonucleoprotein
- hnRNP
heterogeneous nuclear RNP
- mRNP
messenger RNP
- SIRT
sirtuin (silent mating type information regulation 2 homologue)
- TIA-1
T-cell-restricted intracellular antigen-1
- TIAR
TIA-1-related protein
- TTR-RBP
turnover and translation regulatory RBP
- USER
untranslated sequence element for regulation
- Ribonome
The cellular ribonucleoprotein environment containing mRNAs, non-coding regulatory RNAs and proteins. The ribonome is highly dynamic and responsive to cell signaling
- Messenger ribonucleoproteins (mRNPs)
mRNPs consist of proteins that bind directly and proteins that associate indirectly with RNAs and the RNA components including the mRNAs and non-coding regulatory RNAs. mRNPs are generally very stable in content unless a cellular perturbation or chemical signal are imposed on the cell
- Post-transcriptional RNA operon/regulon (PTRO)
The concept that functionally related groups of mRNAs are co-ordinately regulated by multi-targeted transacting factors in time and space. Many PTROs have been recently demonstrated that mediate co-ordinated mRNA decay, translational activation and RNA localization of multiple mRNAs that encode macromolecular subunits and components of pathways. Eukaryotic mRNAs are used combinatorially as members of distinct PTROs to allow the multifunctional proteins they encode to be co-ordinated in multiple cellular settings
- Untranslated sequence element for regulation (USER)
Modular units of 3′ or 5′untranslated regions of mRNA that embody a complete regulatory unit. USERs are not synonymous with the binding sites of RNA-binding proteins or small regulatory RNAs but consist of such elements in a functional configuration. USERs may occur in coding regions as well but they appear to be rare
- Exosome
Membrane-bound vesicles (including microparticles) containing proteins, mRNAs and small non-coding RNAs that are released by cells and can be taken up selectively by surrounding cells. There is evidence that this horizontal transfer of RNA and protein can affect the phenotype of the recipient cells
- Control and co-ordination
Control is a strong, directed effect of a transacting factor on a single RNA component, whereas coordination is a multiplicity of generally weaker effects that orchestrate or harmonize RNAs. Thus these are different but overlapping aspects of RNA regulation
References
* Articles of special interest
- Abdelmohsen K, Pullmann R, Jr, Lal A, Kim HH, Galban S, Yang X, Blethrow JD, Walker M, Shubert J, Gillespie DA, et al. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol. Cell. 2007;25:543–557. doi: 10.1016/j.molcel.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelmohsen K, Kuwano Y, Kim HH, Gorospe M. Posttranscriptional gene regulation by RNA-binding proteins during oxidative stress: implications for cellular senescence. Biol. Chem. 2008;389:243–255. doi: 10.1515/BC.2008.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aliotta JM, Sanchez-Guijo FM, Dooner GJ, Johnson KW, Dooner MS, Greer KA, Greer D, Pimentel J, Kolankiewicz LM, Puente N, et al. Alteration of marrow cell gene expression, protein production and engraftment into lung by lung-derived microvesicles: a novel mechanism for phenotype modulation. Stem Cells. 2007;25:2245–2256. doi: 10.1634/stemcells.2007-0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson AF, Lundgren M, Ericksson S, Rosenlund M, Bernander R, Nillson P. Global analysis of mRNA stability in the archaeon Sulfolobus . Genome Biol. 2006;7:R99. doi: 10.1186/gb-2006-7-10-r99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aten JA, Kanaar R. Chromosomal organization: mingling with the neighbors. PLoS Biol. 2006;4:e155. doi: 10.1371/journal.pbio.0040155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baj-Krzyworzeka M, Szatanek R, Weglarczyk K, Baran J, Urbanowicz B, Branski P, Ratajczak MZ, Zembala M. Tumour-derived microvesicles carry several surface determinants and mRNA of tumour cells and transfer some of these determinants to monocytes. Cancer Immunol. Immunother. 2006;55:808–818. doi: 10.1007/s00262-005-0075-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Benjamin D, Schmidlin M, Min L, Gross B, Moroni C. BRF1 protein turnover and mRNA decay activity are regulated by protein kinase B at the same phosphorylation sites. Mol. Cell. Biol. 2006;26:9497–9507. doi: 10.1128/MCB.01099-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein JA, Lin PH, Cohen SN, Lin-Chao S. Global analysis of Escherichia coli RNA degradosome function using DNA microarrays. Proc. Natl. Acad. Sci. U.S.A. 2004;101:2758–2763. doi: 10.1073/pnas.0308747101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- Brengues M, Teixeira D, Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briata P, Forcales SV, Ponassi M, Corte G, Chen CY, Karin M, Puri PL, Gherzi R. p38-dependent phosphorylation of the mRNA decay-promoting factor KSRP controls the stability of select myogenic transcripts. Mol. Cell. 2005;20:891–903. doi: 10.1016/j.molcel.2005.10.021. [DOI] [PubMed] [Google Scholar]
- Cammas A, Lewis SM, Vagner S, Holcik M. Post-transcriptional control of gene expression through subcellular relocalization of mRNA binding proteins. Biochem. Pharmacol. 2008;76:1395–1403. doi: 10.1016/j.bcp.2008.05.022. [DOI] [PubMed] [Google Scholar]
- Cox B, Kislinger T, Emili A. Integrating gene and protein expression data: pattern analysis and profile mining. Methods. 2005;35:303–314. doi: 10.1016/j.ymeth.2004.08.021. [DOI] [PubMed] [Google Scholar]
- Davidson EA, Ellington AD. Synthetic RNA circuits. Nat. Chem. Biol. 2007;3:23–28. doi: 10.1038/nchembio846. [DOI] [PubMed] [Google Scholar]
- DeGracia DJ, Jamison JT, Szymanski JJ, Lewis MK. Translation arrest and ribonomics in post-ischemic brain: layers and layers of players. J. Neurochem. 2008;106:2288–2301. doi: 10.1111/j.1471-4159.2008.05561.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doller A, Huwiler A, Muller R, Radeke HH, Pfeilschifter J, Eberhardt W. Protein kinase Cα-dependent phosphorylation of the mRNA-stabilizing factor HuR: implications for posttranscriptional regulation of cyclooxygenase-2. Mol. Biol. Cell. 2007;18:2137–2148. doi: 10.1091/mbc.E06-09-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doller A, Pfeilschifter J, Eberhardt W. Signalling pathways regulating nucleo-cytoplasmic shuttling of the mRNA-binding protein HuR. Cell Signal. 2008;20:2165–2173. doi: 10.1016/j.cellsig.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Elemento O, Slonim N, Tavazoie S. A universal framework for regulatory element discovery across all genomes and data types. Mol. Cell. 2007;28:337–350. doi: 10.1016/j.molcel.2007.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fevrier B, Raposo G. Exosomes: endosomal-derived vesicles shipping extracellular messages. Curr. Opin. Cell Biol. 2004;16:415–421. doi: 10.1016/j.ceb.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Galgano A, Forrer M, Jaskiewicz L, Kanitz A, Zavolan M, Gerber AP. Comparative analysis of mRNA targets for human PUF-family proteins suggests extensive interaction with the miRNA regulatory system. PLoS ONE. 2008;3:e3164. doi: 10.1371/journal.pone.0003164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallouzi IE, Brennan CM, Stenberg MG, Swanson MS, Eversole A, Maizels N, Steitz JA. HuR binding to cytoplasmic mRNA is perturbed by heat shock. Proc. Natl. Acad. Sci. U.S.A. 2000;97:3073–3078. doi: 10.1073/pnas.97.7.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao FB, Carson CC, Levine T, Keene JD. Selection of a subset of mRNAs from combinatorial 3’ untranslated region libraries using neuronal RNA-binding protein Hel-N1. Proc. Natl. Acad. Sci. U.S.A. 1994;91:11207–11211. doi: 10.1073/pnas.91.23.11207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbarino-Pico E, Niu S, Rollag MD, Strayer CA, Besharse JC, Green CB. Immediate early response of the circadian polyA ribonuclease nocturnin to two extracellular stimuli. RNA. 2007;13:745–755. doi: 10.1261/rna.286507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Gerber AP, Herschlag D, Brown PO. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004;2:E79. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber AP, Luschnig S, Krasnow MA, Brown PO, Herschlag D. Genome-wide identification of mRNAs associated with the translational regulator PUMILIO in Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 2006;103:4487–4492. doi: 10.1073/pnas.0509260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Gherzi R, Trabucchi M, Ponassi M, Ruggiero T, Corte G, Moroni C, Chen CY, Khabar KS, Andersen JS, Briata P. The RNA-binding protein KSRP promotes decay of β-catenin mRNA and is inactivated by PI3K–AKT signaling. PLoS Biol. 2006;5:e5. doi: 10.1371/journal.pbio.0050005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gorospe M. HuR in the mammalian genotoxic response: post-transcriptional multitasking. Cell Cycle. 2003;2:412–414. [PubMed] [Google Scholar]
- *.Gould SJ, Booth AM, Hildreth JE. The Trojan exosome hypothesis. Proc. Natl. Acad. Sci. U.S.A. 2003;100:10592–10597. doi: 10.1073/pnas.1831413100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin TJ, Gygi SP, Ideker T, Rist B, Eng J, Hood L, Aebersold R. Complementary profiling of gene expression at the transcriptome and proteome levels in Saccharomyces cerevisiae . Mol. Cell Proteomics. 2002;1:323–333. doi: 10.1074/mcp.m200001-mcp200. [DOI] [PubMed] [Google Scholar]
- Grigull J, Mnaimneh S, Pootoolal J, Robinson MD, Hughes TR. Genome-wide analysis of mRNA stability using transcription inhibitors and microarrays reveals posttranscriptional control of ribosome biogenesis factors. Mol. Cell Biol. 2004;24:5534–5547. doi: 10.1128/MCB.24.12.5534-5547.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol. Cell Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakelien AM, Landsverk HB, Robl JM, Skalhegg BS, Collas P. Reprogramming fibroblasts to express T-cell functions using cell extracts. Nat. Biotechnol. 2002;20:460–466. doi: 10.1038/nbt0502-460. [DOI] [PubMed] [Google Scholar]
- Halbeisen RE, Galgano A, Scherrer T, Gerber AP. Post-transcriptional gene regulation: From genome-wide studies to principles. Cell. Mol. Life Sci. 2008;65:798–813. doi: 10.1007/s00018-007-7447-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson DG, Hogan DJ, Herschlag D, Ferrell JE, Brown PO. Systematic identification of mRNAs recruited to argonaute 2 by specific microRNAs and corresponding changes in transcript abundance. PLoS ONE. 2008;3:e2126. doi: 10.1371/journal.pone.0002126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieronymus H, Silver PA. A systems view of mRNP biology. Genes Dev. 2004;18:2845–2860. doi: 10.1101/gad.1256904. [DOI] [PubMed] [Google Scholar]
- *.Hogan DJ, Riordan DP, Gerber AP, Herschlag D, Brown PO. Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol. 2008;6:e255. doi: 10.1371/journal.pbio.0060255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intine RV, Tenenbaum SA, Sakulich AL, Keene JD, Maraia RJ. Differential phosphorylation and subcellular localization of La RNPs associated with precursor tRNAs and translation-related mRNAs. Mol. Cell. 2003;12:1301–1307. doi: 10.1016/s1097-2765(03)00429-5. [DOI] [PubMed] [Google Scholar]
- Jing Q, Huang S, Guth S, Zarubin T, Motoyama A, Chen J, Di Padova F, Lin SC, Gram H, Han J. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–634. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- Keene JD. Why is Hu where? Shuttling of early-response-gene messenger RNA subsets. Proc. Natl. Acad. Sci. U.S.A. 1999;96:5–7. doi: 10.1073/pnas.96.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene JD. Ribonucleoprotein infrastructure regulating the flow of genetic information between the genome and the proteome. Proc. Natl. Acad. Sci. U.S.A. 2001;98:7018–7024. doi: 10.1073/pnas.111145598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene JD. RNA Regulons: coordination of post-transcriptional events. Nat. Rev. Gen. 2007a;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- Keene JD. Biological clocks and the coordination theory of RNA operons and regulons. Cold Spring Harbor Symp. Quant. Biol. 2007b;72:1–9. doi: 10.1101/sqb.2007.72.013. [DOI] [PubMed] [Google Scholar]
- Keene JD, Tenenbaum SA. Eukaryotic mRNPs may represent posttranscriptional operons. Mol. Cell. 2002;9:1161–1167. doi: 10.1016/s1097-2765(02)00559-2. [DOI] [PubMed] [Google Scholar]
- Keene JD, Komisarow JM, Friedersdorf MB. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat. Protoc. 2006;1:302–307. doi: 10.1038/nprot.2006.47. [DOI] [PubMed] [Google Scholar]
- Kim HH, Gorospe M. Phosphorylated HuR shuttles in cycles. Cell Cycle. 2008;7:3124–3126. doi: 10.4161/cc.7.20.6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komili S, Silver PA. Coupling and coordination in gene expression processes: a systems biology view. Nat. Rev. Gen. 2008;9:38–48. doi: 10.1038/nrg2223. [DOI] [PubMed] [Google Scholar]
- *.Komili S, Farny NG, Roth FP, Silver PA. Functional specificity among ribosomal proteins regulates gene expression. Cell. 2007;131:557–571. doi: 10.1016/j.cell.2007.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam LT, Pickeral OK, Peng AC, Rosenwald A, Hurt EM, Giltnane JM, Averett LM, Zhao H, Davis RE, Sathyamoorthy M, et al. Genomic-scale measurement of mRNA turnover and the mechanisms of action of the anti-cancer drug flavopiridol. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-10-research0041. research0041.1-0041.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landthaler M, Gaidatzis D, Rothballer A, Chen PY, Soll SJ, Dinic L, Ojo T, Hafner M, Zavolan M, Tuschl T. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs, RNA. 2008 doi: 10.1261/rna.1351608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000;19:6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner RS, Seiser RM, Zheng T, Lager PJ, Reedy MC, Keene JD, Nicchitta CV. Partitioning and translation of mRNAs encoding soluble proteins on membrane-bound ribosomes. RNA. 2003;9:1123–1137. doi: 10.1261/rna.5610403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Park S, Kilburn B, Jelinek MA, Henschen-Edman A, Aswad DW, Stallcup MR, Laird-Offringa IA. Lipopolysaccharide-induced methylation of HuR, an mRNA-stabilizing protein, by CARM1. Coactivator-associated arginine methyltransferase. J. Biol. Chem. 2002;277:44623–44630. doi: 10.1074/jbc.M206187200. [DOI] [PubMed] [Google Scholar]
- Li XY, MacArthur S, Bourgon R, Nix D, Pollard DA, Iyer VN, Hechmer A, Simirenko L, Stapleton M, Luengo Hendriks CL, et al. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol. 2008;6:e27. doi: 10.1371/journal.pbio.0060027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez de Silanes I, Fan J, Galban CJ, Spencer RG, Becker KG, Gorospe M. Global analysis of HuR-regulated gene expression in colon cancer systems of reducing complexity. Gene Expr. 2004;12:49–59. doi: 10.3727/000000004783992215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P, Vogel C, Wang R, Yao X, Marcotte EM. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat. Biotechnol. 2006;25:117–124. doi: 10.1038/nbt1270. [DOI] [PubMed] [Google Scholar]
- *.Lukong KE, Chang KW, Khandjian EW, Richard S. RNA-binding proteins in human genetic disease. Trends Genet. 2008;24:416–425. doi: 10.1016/j.tig.2008.05.004. [DOI] [PubMed] [Google Scholar]
- Makeyev EV, Maniatis T. Multilevel regulation of gene expression by microRNAs. Science. 2008;319:1789–1790. doi: 10.1126/science.1152326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulis L. Origin of eukaryotic nucleus. Yale Press; New Haven CT: 1970. [Google Scholar]
- Martin W, Koonin EV. Introns and the origin of nucleus-cytosol compartmentalization. Nature. 2006;440:41–45. doi: 10.1038/nature04531. [DOI] [PubMed] [Google Scholar]
- Mauro VP, Edelman GM. The ribosome filter hypothesis. Proc. Natl. Acad. Sci. U.S.A. 2002;99:12031–12036. doi: 10.1073/pnas.192442499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazan-Mamczarz K, Galban S, Lopez de Silanes I, Martindale JL, Atasoy U, Keene JD, Gorospe M. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc. Natl. Acad. Sci. U.S.A. 2003;100:8354–8359. doi: 10.1073/pnas.1432104100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazan-Mamczarz K, Hagner PR, Dai B, Wood WH, Zhang Y, Becker KG, Liu Z, Gartenhaus RB. Identification of transformation-related pathways in a breast epithelial cell model using a ribonomics approach. Cancer Res. 2008;68:7730–7735. doi: 10.1158/0008-5472.CAN-08-2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesarovic MD, Sreenath SN, Keene JD. Search for organising principles: understanding in systems biology. Syst. Biol. 2004;1:19–27. doi: 10.1049/sb:20045010. [DOI] [PubMed] [Google Scholar]
- Mijalski T, Harder A, Halder T, Kersten M, Horsch M, Strom TM, Liebscher HV, Lottspeich F, de Angelis MH, Beckers J. Identification of coexpressed gene clusters in a comparative analysis of transcriptome and proteome in mouse tissues. Proc. Natl. Acad. Sci. U.S.A. 2005;102:8621–8626. doi: 10.1073/pnas.0407672102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- *.Moore MJ, Schwartzfarb EM, Silver PA, Yu MC. Differential recruitment of the splicing machinery during transcription predicts genome-wide patterns of mRNA splicing. Mol. Cell. 2006;24:903–915. doi: 10.1016/j.molcel.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Morris AR, Mukherjee N, Keene JD. Ribonomic analysis of human Pum1 reveals cis-trans conservation across species despite the evolution of diverse mRNA target sets. Mol. Cell Biol. 2008;28:4093–4103. doi: 10.1128/MCB.00155-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicchitta CV, Lerner RS, Stephens SB, Dodd RD, Pyhtila B. Pathways for compartmentalizing protein synthesis in eukaryotic cells: the template-partitioning model. Biochem. Cell. Biol. 2005;83:687–695. doi: 10.1139/o05-147. [DOI] [PubMed] [Google Scholar]
- Orphanides G, Reinberg D. A unified theory of gene expression. Cell. 2002;108:439–451. doi: 10.1016/s0092-8674(02)00655-4. [DOI] [PubMed] [Google Scholar]
- Penalva LOF, Burdick MD, Lin SM, Sutterluety H, Keene JD. RNA-binding proteins to assess gene expression states of co-cultivated cells in response to tumor cells. Mol. Cancer. 2004;3:24–35. doi: 10.1186/1476-4598-3-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñol-Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- *.Pullmann R, Jr, Kim HH, Abdelmohsen K, Lal A, Martindale JL, Yang X, Gorospe M. Analysis of turnover and translation regulatory RNA-binding protein expression through binding to cognate mRNAs. Mol. Cell. Biol. 2007;27:6265–6278. doi: 10.1128/MCB.00500-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Pyhtila B, Zheng T, Lager P, Keene JD, Reedy M, Nicchitta C. Signal sequence- and translation-independent mRNA localization to the endoplasmic reticulum. RNA. 2008;14:1–9. doi: 10.1261/rna.721108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan A, Dhalla M, Bakheet T, Ogilvie RL, Vlasova IA, Khabar KS, Williams BR, Bohjanen PR. Patterns of coordinate down-regulation of ARE-containing transcripts following immune cell activation. Genomics. 2004;84:1002–1013. doi: 10.1016/j.ygeno.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Ratajczak J, Miekus K, Kucia M, Zhang J, Reca R, Dvorak P, Ratajczak MZ. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia. 2006;20:847–856. doi: 10.1038/sj.leu.2404132. [DOI] [PubMed] [Google Scholar]
- *.Rajasekhar VK, Viale A, Socci ND, Wiedmann M, Hu X, Holland EC. Oncogenic Ras and Akt signaling contribute to glioblastoma formation by differential recruitment of existing mRNAs to polysomes. Mol. Cell. 2003;12:889–901. doi: 10.1016/s1097-2765(03)00395-2. [DOI] [PubMed] [Google Scholar]
- Ruggiero T, Trabucchi M, Ponassi M, Corte G, Chen CY, al-Haj L, Khabar KS, Briata P, Gherzi R. Identification of a set of KSRP target transcripts upregulated by PI3K–AKT signaling. BMC Mol. Biol. 2007;8:28–38. doi: 10.1186/1471-2199-8-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Saint-Georges Y, Garcia M, Delaveau T, Jourdren L, Le Crom S, Lemoine S, Tanty V, Devaux F, Jacq C. Yeast mitochondrial biogenesis: a role for the PUF RNA-binding protein Puf3p in mRNA localization. PLoS ONE. 2008;3:e2293. doi: 10.1371/journal.pone.0002293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidlin M, Lu M, Leuenberger SA, Stoecklin G, Mallaun M, Gross B, Gherzi R, Hess D, Hemmings BA, Moroni C. The ARE-dependent mRNA-destabilizing activity of BRF1 is regulated by protein kinase B. EMBO J. 2004;23:4760–4769. doi: 10.1038/sj.emboj.7600477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens SB, Dodd RD, Brewer JW, Lager PJ, Keene JD, Nicchitta CV. Stable ribosome binding to the endoplasmic reticulum enables compartment-specific regulation of mRNA translation. Mol. Biol. Cell. 2005;16:5819–5831. doi: 10.1091/mbc.E05-07-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temin HM. Origin of retroviruses from cellular moveable genetic elements. Cell. 1980;21:599–600. doi: 10.1016/0092-8674(80)90420-1. [DOI] [PubMed] [Google Scholar]
- Tenenbaum SA, Carson CC, Lager PJ, Keene JD. Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proc. Natl. Acad. Sci. U.S.A. 2000;97:14085–14090. doi: 10.1073/pnas.97.26.14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Q, Stepaniants SB, Mao M, Weng L, Feetham MC, Doyle MJ, Yi EC, Dai H, Thorsson V, Eng J, et al. Integrated genomic and proteomic analyses of gene expression in mammalian cells. Mol. Cell Proteomics. 2004;3:960–969. doi: 10.1074/mcp.M400055-MCP200. [DOI] [PubMed] [Google Scholar]
- Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- *.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- Wang W, Fan J, Yang X, Furer-Galban S, Lopez de Silanes I, von Kobbe C, Guo J, Georas SN, Foufelle F, Hardie DG, et al. AMP-activated kinase regulates cytoplasmic HuR. Mol. Cell. Biol. 2002;22:3425–3436. doi: 10.1128/MCB.22.10.3425-3436.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Yang X, Kawai T, Lopez de Silanes I, Mazan-Mamczarz K, Chen P, Chook YM, Quensel C, Kohler M, Gorospe M. AMP-activated protein kinase-regulated phosphorylation and acetylation of importin α1: involvement in the nuclear import of RNA-binding protein HuR. J. Biol. Chem. 2004;279:48376–48388. doi: 10.1074/jbc.M409014200. [DOI] [PubMed] [Google Scholar]
- Wang Y, Liu CL, Storey JD, Tibshirani RJ, Herschlag D, Brown PO. Precision and functional specificity in mRNA decay. Proc. Natl. Acad. Sci. U.S.A. 2002;99:5860–5865. doi: 10.1073/pnas.092538799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Wang T, Zeng J, Lowe CB, Sellers RG, Salama SR, Yang M, Burgess SM, Brachmann RK, Haussler D. Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53. Proc. Natl. Acad. Sci. U.S.A. 2007;104:18613–18618. doi: 10.1073/pnas.0703637104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang E, van Nimwegen E, Zavolan M, Rajewsky N, Schroeder M, Magnasco M, Darnell JE., Jr Decay rates of human mRNAs: correlation with functional characteristics and sequence attributes. Genome Res. 2003;13:1863–1872. doi: 10.1101/gr.1272403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn MP, Koller A, Oshiro G, Ulaszek RR, Plouffe D, Deciu C, Winzeler E, Yates JR., 3rd Protein pathway and complex clustering of correlated mRNA and protein expression analyses in Saccharomyces cerevisiae . Proc. Natl. Acad. Sci. U.S.A. 2003;100:3107–3112. doi: 10.1073/pnas.0634629100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Yu X, Harris SL, Levine AJ. The regulation of exosome secretion: a novel function of the p53 protein. Cancer Res. 2006;66:4795–4801. doi: 10.1158/0008-5472.CAN-05-4579. [DOI] [PubMed] [Google Scholar]