Abstract
The purification of recombinant proteins for biochemical assays and structural studies is time-consuming and presents inherent difficulties that depend on the optimization of protein stability. The use of dyes to monitor thermal denaturation of proteins with sensitive fluorescence detection enables the rapid and inexpensive determination of protein stability using real-time PCR instruments. By screening a wide range of solution conditions and additives in 96-well format, the thermal shift assay easily identifies conditions that significantly enhance the stability of recombinant proteins. The same approach can be used as a low cost, initial screen to discover new protein:ligand interactions by capitalizing on increases in protein stability that typically occur upon ligand binding. This unit presents a methodological workflow for the small-scale, high-throughout thermal denaturation of recombinant proteins in the presence of SYPRO Orange dye.
Keywords: differential scanning fluorimetry, ThermoFluor™, thermal denaturation, buffer optimization, ligand screening
INTRODUCTION
The expression and purification of recombinant proteins can be significantly improved with the addition of stabilizing buffers or ligands that reduce the propensity of proteins to unfold or aggregate during purification and storage in vitro. Moreover, increasing stability correlates with the crystallizability of recombinant proteins (Dupeux et al., 2011; Ericsson et al., 2006; Vedadi et al., 2006). The identification of stabilizing buffers and additives is typically established by monitoring increases in the melting temperature of the protein upon thermal denaturation. Differential scanning calorimetry is an excellent, label-free technique used to determine protein melting temperatures, but it requires expensive instrumentation dedicated to this purpose, is time-intensive, and offers low throughput for screening purposes (Bruylants et al., 2005).
Environmentally sensitive dyes have long been used to study protein folding. Commonly used dyes such as 1,8-ANS (1-anilinonaphthalene-8-sulfonate) and 2,6-TNS (naphthalene-6-sulfonic acid) undergo a significant increase in quantum yield upon binding low dielectric/hydrophobic environments that become exposed during protein denaturation, allowing sensitive detection by fluorescence spectroscopy (Hawe et al., 2008). However, 1,8-ANS and related dyes typically have excitation/emission wavelengths outside the range of commonly available real-time PCR instruments that have fluorescence detection capabilities and Peltier-based temperature control. SYPRO Orange dye also undergoes a significant increase in quantum yield upon binding hydrophobic regions in denatured proteins, yet its fluorescence properties (λex 470 nm /λem 570 nm) are compatible with filter sets found on real-time PCR instruments, allowing their adaptation for protein thermal denaturation assays (Niesen et al., 2007).
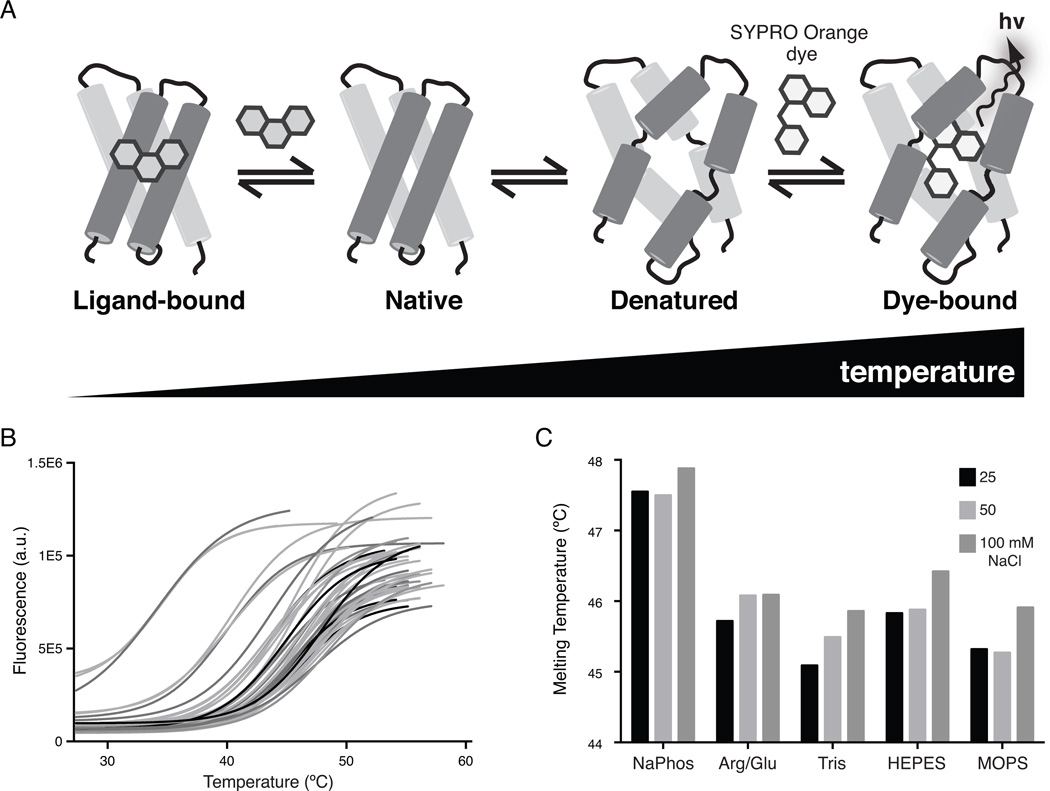
The basic scheme of a thermal shift assay (Figure 1) involves incubation of natively folded proteins with SYPRO Orange dye in a 96-well PCR plate. Through a systematic increase in temperature and concomitant monitoring of SYPRO Orange fluorescence emission, it is possible to monitor thermal denaturation of the protein in many conditions simultaneously. The increase in melting temperatures in different solution conditions gives rise to a ‘thermal shift’ that quantifies the stabilization of the protein under different buffer or additive conditions. The same property is used to identify small molecule ligands, which tend to stabilize proteins upon binding. Therefore, the thermal shift assay (also known as differential scanning fluorimetry or ThermoFluor™) is a rapid and inexpensive assay to identify stabilizing solution conditions, additives and small molecule ligands for purified, recombinant proteins.
Figure 1.
Thermal shift analysis of protein stability and ligand interactions. (A) Starting with a purified recombinant protein in its native (folded) state, the protein is slowly heated to undergo thermal denaturation. The environmentally sensitive dye, SYPRO Orange, interacts with hydrophobic regions in the protein that become exposed upon denaturation. Binding low dielectric, hydrophobic regions increases fluorescence emission of the dye to serve as a read out of thermal denaturation of the protein. Represented on the left: ligand binding tends to rigidify proteins to increase their thermal stability. This general property allows identification of protein-binding ligands through increases in thermal stability. (B) Raw, truncated data from a typical 96-well screen of solution conditions. (C) An analysis of selected denaturation curves from part B shows that for a given pH (pH 7.0), the choice of buffer can impact overall protein stability. Here, the protein is significantly more stable in 50 mM sodium phosphate buffer, pH 7.0 than several others at the same concentration and pH, including: L-Arg/L-Glu, Tris, HEPES, and MOPS.
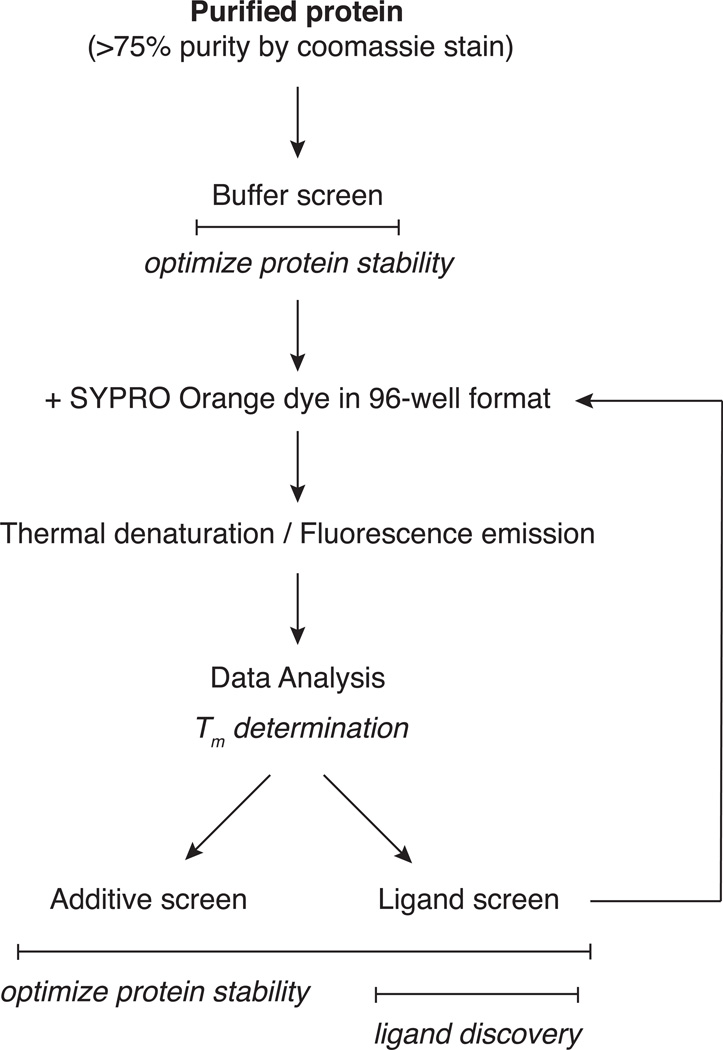
This unit describes the full workflow for a thermal shift assay, starting with a buffer screen to optimize protein stability. Special focus is given to the crucial step of data analysis to derive melting temperatures from the raw thermal denaturation data (Support Protocol 1). The unit also provides alternative protocols for additional screens to discover stabilizing additives or ligand interactions. Figure 2 provides a flow chart of the entire thermal shift assay process.
Figure 2.
Flow chart of a thermal shift assay including an initial buffer optimization and subsequent additive or ligand screens. This figure connects all protocols presented in this unit.
BASIC PROTOCOL 1
BUFFER SCREEN TO OPTIMIZE PROTEIN STABILITY
This protocol is ideal for an initial screen of a wide range of solution conditions (buffer identity, solution pH, ionic strength) to identify an optimal buffer for subsequent thermal shift screens, downstream biochemical assays, or structural biology applications.
Materials
Purified protein (see Critical Parameters)
Dilution buffer (see Recipes)
96-well 2 ml deep well block containing 5× buffer screen of choice (e.g. Hampton Research cat. no. HR2-072 or see Table 1)
Table 1.
Composition of the 96-well buffer optimization screen
| Well | Buffer | Salt | Well | Buffer | Salt |
|---|---|---|---|---|---|
| A1 | 50 mM sodium citrate pH 4.0 | 50 mM NaCl | E1 | 50 mM sodium citrate pH 4.0 | 250 mM NaCl |
| A2 | 50 mM sodium citrate pH 4.5 | 50 mM NaCl | E2 | 50 mM sodium citrate pH 4.5 | 250 mM NaCl |
| A3 | 50 mM sodium citrate pH 5.0 | 50 mM NaCl | E3 | 50 mM sodium citrate pH 5.0 | 250 mM NaCl |
| A4 | 50 mM sodium citrate pH 5.5 | 50 mM NaCl | E4 | 50 mM sodium citrate pH 5.5 | 250 mM NaCl |
| A5 | 50 mM sodium citrate pH 6.0 | 50 mM NaCl | E5 | 50 mM sodium citrate pH 6.0 | 250 mM NaCl |
| A6 | 50 mM sodium citrate pH 6.5 | 50 mM NaCl | E6 | 50 mM sodium citrate pH 6.5 | 250 mM NaCl |
| A7 | 50 mM sodium citrate pH 7.0 | 50 mM NaCl | E7 | 50 mM sodium citrate pH 7.0 | 250 mM NaCl |
| A8 | 50 mM MES pH 5.5 | 50 mM NaCl | E8 | 50 mM MES pH 5.5 | 250 mM NaCl |
| A9 | 50 mM MES pH 6.0 | 50 mM NaCl | E9 | 50 mM MES pH 6.0 | 250 mM NaCl |
| A10 | 50 mM MES pH 6.5 | 50 mM NaCl | E10 | 50 mM MES pH 6.5 | 250 mM NaCl |
| A11 | 50 mM PIPES pH 6.0 | 50 mM NaCl | E11 | 50 mM PIPES pH 6.0 | 250 mM NaCl |
| A12 | 50 mM PIPES pH 6.5 | 50 mM NaCl | E12 | 50 mM PIPES pH 6.5 | 250 mM NaCl |
| B1 | 50 mM PIPES pH 7.0 | 50 mM NaCl | F1 | 50 mM PIPES pH 7.0 | 250 mM NaCl |
| B2 | 50 mM PIPES pH 7.5 | 50 mM NaCl | F2 | 50 mM PIPES pH 7.5 | 250 mM NaCl |
| B3 | 50 mM Bis-Tris pH 6.0 | 50 mM NaCl | F3 | 50 mM Bis-Tris pH 6.0 | 250 mM NaCl |
| B4 | 50 mM Bis-Tris pH 6.5 | 50 mM NaCl | F4 | 50 mM Bis-Tris pH 6.5 | 250 mM NaCl |
| B5 | 50 mM Bis-Tris pH 7.0 | 50 mM NaCl | F5 | 50 mM Bis-Tris pH 7.0 | 250 mM NaCl |
| B6 | 50 mM MOPS pH 6.5 | 50 mM NaCl | F6 | 50 mM MOPS pH 6.5 | 250 mM NaCl |
| B7 | 50 mM MOPS pH 7.0 | 50 mM NaCl | F7 | 50 mM MOPS pH 7.0 | 250 mM NaCl |
| B8 | 50 mM MOPS pH 7.5 | 50 mM NaCl | F8 | 50 mM MOPS pH 7.5 | 250 mM NaCl |
| B9 | 50 mM MOPS pH 8.0 | 50 mM NaCl | F9 | 50 mM MOPS pH 8.0 | 250 mM NaCl |
| B10 | 50 mM L-Arg/50 mM L-Glu pH 6.0 | 50 mM NaCl | F10 | 50 mM L-Arg/50 mM L-Glu pH 6.0 | 250 mM NaCl |
| B11 | 50 mM L-Arg/50 mM L-Glu pH 6.5 | 50 mM NaCl | F11 | 50 mM L-Arg/50 mM L-Glu pH 6.5 | 250 mM NaCl |
| B12 | 50 mM L-Arg/50 mM L-Glu pH 7.0 | 50 mM NaCl | F12 | 50 mM L-Arg/50 mM L-Glu pH 7.0 | 250 mM NaCl |
| C1 | 50 mM L-Arg/50 mM L-Glu pH 7.5 | 50 mM NaCl | G1 | 50 mM L-Arg/50 mM L-Glu pH 7.5 | 250 mM NaCl |
| C2 | 50 mM sodium phosphate pH 6.0 | 50 mM NaCl | G2 | 50 mM sodium phosphate pH 6.0 | 250 mM NaCl |
| C3 | 50 mM sodium phosphate pH 6.5 | 50 mM NaCl | G3 | 50 mM sodium phosphate pH 6.5 | 250 mM NaCl |
| C4 | 50 mM sodium phosphate pH 7.0 | 50 mM NaCl | G4 | 50 mM sodium phosphate pH 7.0 | 250 mM NaCl |
| C5 | 50 mM sodium phosphate pH 7.5 | 50 mM NaCl | G5 | 50 mM sodium phosphate pH 7.5 | 250 mM NaCl |
| C6 | 50 mM sodium phosphate pH 8.0 | 50 mM NaCl | G6 | 50 mM sodium phosphate pH 8.0 | 250 mM NaCl |
| C7 | 50 mM HEPES pH 7.0 | 50 mM NaCl | G7 | 50 mM HEPES pH 7.0 | 250 mM NaCl |
| C8 | 50 mM HEPES pH 7.5 | 50 mM NaCl | G8 | 50 mM HEPES pH 7.5 | 250 mM NaCl |
| C9 | 50 mM HEPES pH 8.0 | 50 mM NaCl | G9 | 50 mM HEPES pH 8.0 | 250 mM NaCl |
| C10 | 50 mM Tris pH 7.0 | 50 mM NaCl | G10 | 50 mM Tris pH 7.0 | 250 mM NaCl |
| C11 | 50 mM Tris pH 7.5 | 50 mM NaCl | G11 | 50 mM Tris pH 7.5 | 250 mM NaCl |
| C12 | 50 mM Tris pH 8.0 | 50 mM NaCl | G12 | 50 mM Tris pH 8.0 | 250 mM NaCl |
| D1 | 50 mM Tris pH 8.5 | 50 mM NaCl | H1 | 50 mM Tris pH 8.5 | 250 mM NaCl |
| D2 | 50 mM Tris pH 9.0 | 50 mM NaCl | H2 | 50 mM Tris pH 9.0 | 250 mM NaCl |
| D3 | 50 mM Bicine pH 7.5 | 50 mM NaCl | H3 | 50 mM Bicine pH 7.5 | 250 mM NaCl |
| D4 | 50 mM Bicine pH 8.0 | 50 mM NaCl | H4 | 50 mM Bicine pH 8.0 | 250 mM NaCl |
| D5 | 50 mM Bicine pH 8.5 | 50 mM NaCl | H5 | 50 mM Bicine pH 8.5 | 250 mM NaCl |
| D6 | 50 mM Bicine pH 9.0 | 50 mM NaCl | H6 | 50 mM Bicine pH 9.0 | 250 mM NaCl |
| D7 | 50 mM CHES pH 8.5 | 50 mM NaCl | H7 | 50 mM CHES pH 8.5 | 250 mM NaCl |
| D8 | 50 mM CHES pH 9.0 | 50 mM NaCl | H8 | 50 mM CHES pH 9.0 | 250 mM NaCl |
| D9 | 50 mM CHES pH 9.5 | 50 mM NaCl | H9 | 50 mM CHES pH 9.5 | 250 mM NaCl |
| D10 | 50 mM CHES pH 10.0 | 50 mM NaCl | H10 | 50 mM CHES pH 10.0 | 250 mM NaCl |
| D11 | open for user-determined buffers | 50 mM NaCl | H11 | open for user-determined buffers | 250 mM NaCl |
| D12 | open for user-determined buffers | 50 mM NaCl | H12 | open for user-determined buffers | 250 mM NaCl |
Concentrations listed represent the final concentration in the thermal shift assay.
Stocks should be made at 5× concentration.
SYPRO Orange dye (e.g. Sigma-Aldrich cat. no. S5692)
15 ml polypropylene conical tubes (e.g. VWR cat. no. 21008-089)
Multichannel pipette capable of handling 1 – 50 µl volumes
Multichannel pipette reservoir trough (e.g. VWR cat. no. 89094-662)
96-well plates specific for real-time PCR instrument (e.g. Life Technologies cat. no. 4346907)
Optically clear sealing film for 96-well plates (e.g. VWR cat. no. 33500-696)
Adhesive aluminum sealing film for 96-well plates (e.g. VWR cat. no. 29445-080)
96-well deep well plates (e.g. VWR cat. no. 37001-518)
Swinging bucket centrifuge with adapters for 96-well plates (e.g. Eppendorf 5810 fitted with adapters A-4-81-MTP)
Real-time PCR instrument (e.g. Applied Biosystems ViiA7)
Protocol steps
Set up 96-well assay plate
Note: Bring 5× buffer screen up to room temperature from 4 °C storage before use (~30 min).
- Add enough dilution buffer (see Recipes) to protein stock in a 15 ml conical tube to obtain a final volume of 5 ml, sufficient to screen one 96-well plate. Add 4 µl of SYPRO Orange dye (stock concentration: 5000×). The final concentrations of protein and dye in the mixture should be 5 µM and 2×, respectively.It may be necessary to optimize SYPRO Orange dye and/or protein concentrations for optimal assay performance, although it is important to note that it is not likely to compensate for poorly shaped denaturation curves that arise from a non-ideal protein substrate (see Critical Parameters). A range of concentrations from 10× to 0.5× SYPRO Orange and 2 to 20 µM protein (final assay concentrations) are usually sufficient for optimization.
Mix protein, dye and dilution buffer thoroughly by inverting the tube several times and place the solution in a multichannel pipette reservoir trough. Use a multichannel pipette to transfer 40 µl of solution into each well of a 96-well assay plate.
Centrifuge room temperature 5× buffer screen plate at 800 × g for 2 min at 25 °C.
Carefully peel off adhesive aluminum sealing film on 5× buffer screen plate. Use the multichannel pipette to add 10 µl of the 5× buffer screen stocks from the 96-well deep well block to the assay plate. Mix the well content using the same pipette and tips by pipetting up and down several times.
Cover the assay plate with a sheet of optically clear adhesive and carefully seal each well. Reseal the buffer screen with new adhesive aluminum foil and store at 4 °C.
Centrifuge the assay plate at 800 × g for 2 min at 25 °C to collect solutions in the bottom of the well and remove bubbles
Perform thermal denaturation in 96-well assay plate
- Place the assay plate into the Applied Biosystems ViiA7 real-time PCR instrument and open the ViiA7 RUO software. Under Experimental Properties, select the following parameters:
- Set up: Fast 96-well block (0.1 ml)
- Experiment type: MELT CURVE
- Reagents used to detect target sequence: OTHER
- Ramp Speed: STANDARD
- Select the Define tab on the left, then select the following parameters:
- Target name: TARGET 1
- Reporter: ROX
- Quencher: NONE
- Passive Reference: NONE
- Select the Assign tab on the left, then perform the following actions:
- Highlight all 96 wells in the assay plate
- Check the box next to ‘Target 1’ on the top left of the plate layout. Note: you do not need to check the ‘Sample’ box on the lower left.
- Select the Run Method tab on the left, then make the following changes to the default Melt Curve profile:
- Delete Step 2 of the default cycle
- Change the run method to “Step and Hold” with a 1:00 time
- Set the following temperatures: an initial 2:00 hold at 25 °C, ramping up in increments of 1 °C to a final temperature of 95 °C (with a 2:00 hold)
- Click on all three cameras to activate fluorescence detection throughout the experiment
- Select total volume per well of 50 µl
Click on the RUN tab to the left to initiate thermal denaturation.
- Once the experiment is done (about 1 hour 45 minutes with the current set-up), export data into a comma-separated value (csv) Excel file.Generalized protocol for any real-time PCR instrument: 1) Open a standard Melt Curve experiment for DNA. 2) Eliminate the initial denaturation step at 95 °C. 3) Create a 2 min initial hold at 25 °C, then ramp temperature in 0.5 – 1.0 °C increments up to 95 °C, reading fluorescence at the end of a 1 min hold at each temperature.
SUPPORT PROTOCOL 1
DETERMINING MELTING TEMPERATURES FROM NON-LINEAR FITTING OF THERMAL DENATURATION DATA
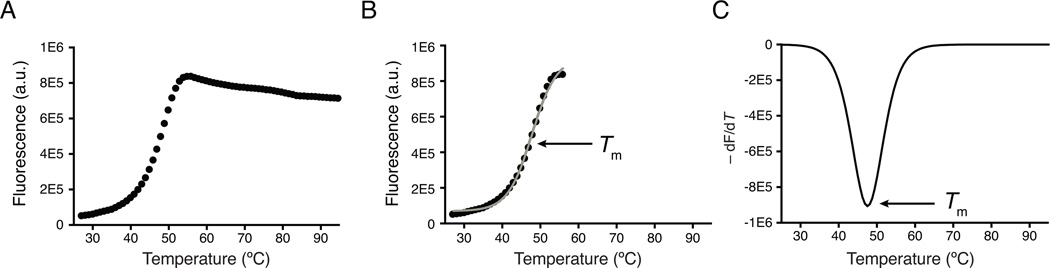
This support protocol describes the steps needed to complete basic non-linear fitting of thermal denaturation curves to derive melting temperatures (Figure 3). An alternative method for estimating melting temperatures involves calculating the first derivative of fluorescence emission with respect to temperature as demonstrated in Figure 3c (not covered in this unit).
Figure 3.
Typical thermal shift assay data and analysis. (A) A typical thermal denaturation profile of a recombinant protein. Low fluorescence at room temperature indicates a well-folded protein. Fluorescence emission increases with increasing temperature, giving rise to a sigmoidal curve that represents cooperative unfolding of the protein. Post-peak aggregation of protein:dye complexes leads to quenching of the fluorescence signal. (B) Automated processing of thermal denaturation curves truncates the dataset to remove post-peak quenching. The resulting sigmoidal curves undergo non-linear fitting to a Boltzmann Equation to identify the melting temperature, or Tm, that occurs at the midpoint of the unfolding transition. The gray line represents the non-linear fit of the fluorescence curve to the Boltzmann Equation. (C) Alternatively, the Tm is easily identified by plotting the first derivative of the fluorescence emission as a function of temperature (−dF/dT). Here, the Tm is represented as the lowest part of the curve.
Materials
Excel software (Microsoft Office, Microsoft, Inc.)
Graphing software (e.g. GraphPad Prism 5.0, GraphPad Software, Inc.)
Truncate post-peak fluorescence
Open csv file containing raw fluorescence data from the real-time PCR instrument in Excel. Data should be in column format where each column represents a different solution condition and each row provides the fluorescence emission at a different temperature from 25 °C to 95 °C. Add labels with solution conditions to data columns, if desired. Label the sheet as ‘Raw Data’.
Create a new sheet within the Excel notebook. Copy raw data and labels to the new sheet. Label the sheet as ‘Truncated Data’.
- Create a new macro ‘deleteaftermax’ in Excel by entering the text in Supplementary File 1 with intact formatting. Execute ‘deleteaftermax’ macro on the truncated dataset following standard Excel protocols.This macro finds the maximum numerical value within a column of data and deletes all data points two rows after this maximum.
Non-linear fitting of truncated fluorescence data
Open a graphing software program that can perform non-linear fitting (e.g. GraphPad Prism). Copy truncated data into a new file for analysis.
- Perform non-linear fitting of the truncated dataset to a Boltzmann Sigmoidal curve with the following equation:
where Y = fluorescence emission in arbitrary units; X = temperature; Bottom = baseline fluorescence at low temperature; Top = maximal fluorescence at top of the truncated dataset; Slope = describes the steepness of the curve, with larger values denoting shallower curves; and Tm = melting temperature of the protein.By deleting post-peak fluorescence quenching with the macro in the previous step, non-linear fitting with the Boltzmann Sigmoidal Equation provides better fits for the Tm based on better estimates of maximal fluorescence. - The graphing software will provide fit values for each of the parameters in the equation, as well as standard errors and the 95% confidence intervals for the data analysis.Thermal denaturation curves that conform to ideal profiles (low baseline fluorescence, sigmoidal transition during denaturation, etc) typically give Tm fits with standard errors ≤ 0.3 °C.
- Plot Tms as a function of solution conditions or perform subsequent analyses to examine the thermal shift as a function of changing solution conditions relative to a standard buffer (i.e. calculate the change in melting temperature, ΔTm).It is helpful to define a baseline solution condition (a previously used buffer condition) as the baseline Tm and examine the Tm of solution conditions relative to this Tm.
ALTERNATE PROTOCOL 1
ADDITIVE SCREEN TO OPTIMIZE PROTEIN STABILITY
This secondary protocol is ideal for screening a wide variety of additives once an optimized buffer has been identified in the Basic Protocol.
Materials
Same materials as for Basic Protocol 1
96-well deep well block containing 5× additive screen of choice (see Table 2)
Table 2.
Composition of the 96-well additive screen
| Well | Additive | Well | Additive |
|---|---|---|---|
| A1 | water | E1 | 5 mM EDTA |
| A2 | water | E2 | 100 mM sodium fluoride |
| A3 | water | E3 | 100 mM potassium fluoride |
| A4 | water | E4 | 100 mM lithium chloride |
| A5 | 100 mM Urea | E5 | 100 mM potassium chloride |
| A6 | 250 mM Urea | E6 | 100 mM ammonium chloride |
| A7 | 500 mM Urea | E7 | 100 mM sodium iodide |
| A8 | 1 M Urea | E8 | 100 mM potassium iodide |
| A9 | 25 mM Guanidine HCl | E9 | 100 mM sodium bromide |
| A10 | 50 mM Guanidine HCl | E10 | 10 mM magnesium chloride |
| A11 | 100 mM Guanidine HCl | E11 | 10 mM calcium chloride |
| A12 | 250 mM Guanidine HCl | E12 | 5 mM manganese chloride |
| B1 | 500 mM Guanidine HCl | F1 | 5 mM nickel chloride |
| B2 | 1% (v/v) DMSO | F2 | 5 mM iron (III) chloride |
| B3 | 2% (v/v) DMSO | F3 | 5 mM zinc chloride |
| B4 | 2.5% (v/v) Glycerol | F4 | 5 mM cobalt chloride |
| B5 | 5% (v/v) Glycerol | F5 | 100 mM sodium formate |
| B6 | 10% (v/v) Glycerol | F6 | 100 mM sodium acetate |
| B7 | 15% (v/v) Glycerol | F7 | 100 mM sodium malonate |
| B8 | 20% (v/v) Glycerol | F8 | 100 mM sodium nitrate |
| B9 | 2.5% (v/v) D-Glucose | F9 | 100 mM sodium thiocyanate |
| B10 | 5% (v/v) D-Glucose | F10 | 100 mM sodium sulfate |
| B11 | 2.5% (v/v) Sucrose | F11 | 100 mM ammonium sulfate |
| B12 | 5% (v/v) Sucrose | F12 | 100 mM ammonium chloride |
| C1 | 2.5% (v/v) PEG400 | G1 | 2 mM AMP + 5 mM MgCl2 |
| C2 | 5% (v/v) PEG400 | G2 | 2 mM ADP + 5 mM MgCl2 |
| C3 | 2.5% (w/v) PEG1000 | G3 | 2 mM ATP + 5 mM MgCl2 |
| C4 | 5% (w/v) PEG1000 | G4 | 2 mM AMPPNP + 5 mM MgCl2 |
| C5 | 2.5% (w/v) PEG4000 | G5 | 2 mM cAMP + 5 mM MgCl2 |
| C6 | 5% (w/v) PEG4000 | G6 | 2 mM GDP + 5 mM MgCl2 |
| C7 | 2.5% (v/v) Ethylene Glycol | G7 | 2 mM GTP + 5 mM MgCl2 |
| C8 | 5% (v/v) Ethylene Glycol | G8 | 2 mM cGMP + 5 mM MgCl2 |
| C9 | 1 mM Octyl Glucoside | G9 | 2 mM NAD + 5 mM MgCl2 |
| C10 | 2 mM CHAPS | G10 | 2 mM NADH + 5 mM MgCl2 |
| C11 | 10 mM L-Proline | G11 | 10 mM Betaine |
| C12 | 50 mM L-Glycine | G12 | 1 mM Spermine |
| D1 | 25 mM L-Histidine | H1 | 1 mM Spermidine (add fresh each time) |
| D2 | 50 mM L-Arginine | H2 | 10 mM β-mercaptoethanol (add fresh each time) |
| D3 | 50 mM L-Glutamate | H3 | 5 mM DTT (add fresh each time) |
| D4 | 50 mM L-Arg/50 mM L-Glu | H4 | 2 mM TCEP (add fresh each time) |
| D5 | 25 mM L-Glutamine | H5 | open for user-determined additives |
| D6 | 50 mM L-Lysine | H6 | open for user-determined additives |
| D7 | 50 mM L-Cysteine | H7 | open for user-determined additives |
| D8 | 50 mM Taurine | H8 | open for user-determined additives |
| D9 | 50 mM Imidazole pH 7.6 | H9 | open for user-determined additives |
| D10 | 100 mM Imidazole pH 7.6 | H10 | open for user-determined additives |
| D11 | 250 mM Imidazole pH 7.6 | H11 | open for user-determined additives |
| D12 | 500 mM Imidazole pH 7.6 | H12 | open for user-determined additives |
Concentrations listed represent the final concentration in the thermal shift assay.
Stocks should be made at 5× concentration.
Note: Bring 5× additive screen up to room temperature from 4 °C storage before use (~30 min).
Add enough of the optimized buffer (at 1× concentration, identified in the initial buffer optimization screen above) to protein stock in a 15 ml conical tube to obtain a final volume of 5 ml, sufficient to screen one 96-well assay plate. Add 4 µl of SYPRO Orange dye (stock concentration: 5000×). The final concentrations of protein and dye in the mixture should be 5 µM and 2×, respectively.
Mix thoroughly by inverting the tube several times and place the solution in a multichannel pipette reservoir trough. Use a multichannel pipette to transfer 40 µl of solution into each well of the assay plate.
Centrifuge room temperature 5× additive screen plate at 800 × g for 2 min at 25 °C.
- Carefully peel off adhesive aluminum sealing film. Use the multichannel pipette to add 10 µl of the 5× additive screen stocks from the 96-well deep well block to the assay plate. Mix the well content using the same pipette and tips by pipetting up and down several times.Design your 96-well additive screen so that there are at least four wells of no additives (i.e. water only in wells A1–A4 in the 96-well additive screen plate) to assess the thermal shift of additives relative to the optimized buffer.
Cover the assay plate with a sheet of optically clear adhesive and carefully seal each well. Reseal the additive screen with adhesive aluminum foil and store at 4 °C.
Centrifuge the assay plate at 800 × g for 2 min at 25 °C to collect solutions in the bottom and remove bubbles from the wells.
Place the assay plate into the real-time PCR instrument and start a temperature gradient program for protein thermal denaturation.
Determine the thermal shift (ΔTm) of all conditions relative to the ‘no additive’ control to identify additives that promote stabilization of the protein.
ALTERNATE PROTOCOL 2
LIGAND SCREEN TO DISCOVER PROTEIN:LIGAND INTERACTIONS
Similar to the additive screen, this secondary protocol is ideal for screening a wide variety of small molecule ligands once an optimized buffer has been identified in the Basic Protocol.
Materials
Same materials as for Basic Protocol 1
96-well plate containing small molecule screen of choice at 50× screening concentration (e.g. 10–100 mM compounds in DMSO or ethanol)
Add enough of the optimized buffer (at 1× concentration, identified from the initial buffer optimization screen) to protein stock in a 15 ml conical tube to obtain a final volume of 5 ml, sufficient to screen one 96-well assay plate. Add 4 µl of SYPRO Orange dye (stock concentration: 5000×). The final concentrations of protein and dye in the mixture should be 5 µM and 2×, respectively.
Mix thoroughly by inverting the tube several times and place the solution in a multichannel pipette reservoir trough. Use a multichannel pipette to transfer 40 µl of solution into each well of the assay plate.
- Use the multichannel pipette to add 1 µl of the 50× small molecule screen stocks and solvent controls to the assay plate.Design your 96-well small molecule screen so that there are four wells of solvent only (i.e. DMSO or ethanol only in wells A1–A4 in the 96-well small molecule screen plate) to assess the thermal shift of solvent relative to the buffer. Selection of small molecules and the screening concentrations are dependent on the protein (See Reagents and Solutions).
Use the multichannel pipette to add 9 µl of water to the assay plate for a final volume of 50 µl. Mix the well content using the same pipette and tips by pipetting up and down several times. This is particularly important for small molecules dissolved in viscous solvents like DMSO.
Cover the assay plate with a sheet of optically clear adhesive and carefully seal each well. Reseal the small molecule screening plate with adhesive aluminum foil and store at −80 °C.
Centrifuge the assay plate at 800 × g for 2 min at 25 °C to collect solutions in the bottom and remove bubbles from the wells.
Place the assay plate into the real-time PCR instrument and start a temperature gradient program for thermal denaturation.
- Determine the thermal shift (ΔTm) of each condition relative to the solvent control to identify small molecules that promote stabilization of the protein.Binding of some small molecules may manifest as decreases in protein stability, particularly for very stable proteins with Tms > 75 °C. For a first pass through the screen, consider any small molecule that gives rise to a ΔTm > 2.0 °C over the solvent control as a potential hit.
REAGENTS AND SOLUTIONS
Dilution buffer
10 mM Sodium Phosphate pH 7.0
50 mM NaCl
Store up to 1 month at room temperature
Other buffers may be used for dilution of the protein, provided that the protein is relatively stable in the buffer when diluted to low concentration. Ideally, the buffer concentration should be maintained as low as possible (10–25 mM) along with low ionic strength to accurately sample solution conditions in buffer screen.
Purified protein
For general information on selection of protein targets that give ideal thermal denaturation curves, see Critical Parameters. For best results, use freshly purified protein with an estimated purity of ≥ 75% as assessed by Coomassie Brilliant Blue staining of SDS-PAGE gels (see protocol in Unit XX).
Buffer and additive screens
Selection of solution conditions (buffer identity, solution pH, ionic strength) and additives to screen are entirely up to the user. Two possible screens for buffers and additives are described in Tables 1 and 2, respectively. General recommendations for screen set-up:
Make up 10–50 ml of each 5× buffer or additive solution.
Aliquot 2 ml of each concentrated stock into a 96-well deep well block for use. Store remaining 5× buffer solutions at 4 °C for up to 6 months. Extra 5× buffer stocks can be diluted to 1× for immediate follow-up in subsequent thermal shift additive screens.
For long-term storage, seal 96-well deep well block with adhesive aluminum foil and store at 4 °C for up to 6 months for optimal stability. Allow 30 min for screens to come to room temperature before use.
Inspect deep well block visually before use for microbial growth or precipitation of buffer components. Replace screen solutions as needed.
Small molecule screens
Selection of small molecule screen composition will vary according to the protein. General recommendations for screen set-up:
Determine an ideal concentration for small molecule stocks based on protein target, small molecule ligand identities and solubilities. Screening at a concentration of 100 – 250 µM will allow identification of target ligands with low micromolar affinity. The use of concentrated DMSO stocks (≥ 10 mM) will keep the total concentration of DMSO ≤ 2% to minimize artifacts.
Use DMSO-resistant 96-well plates (e.g. Thermo Scientific cat. no. 4917) for long-term compound storage at −80 °C and avoid excessive cycles of freeze/thaw by aliquoting libraries out in small volumes.
COMMENTARY
Background Information
Protein stability in vitro is paramount to biochemical activity (Crowther et al., 2010; Sampson et al., 2011) and structural studies, even predicting the ability of well-folded proteins to crystallize with reasonable reliability (Dupeux et al., 2011; Ericsson et al., 2006; Vedadi et al., 2006). Historically, differential scanning calorimetry (DSC) has been the method of choice for characterizing protein stability in vitro. While this technique enables the measurement of thermodynamic properties that give rise to protein stability, the method is time consuming and low throughput, making it difficult to screen for solution conditions that stabilize a given protein (Johnson, 2013). Perhaps most problematic is that DSC requires expensive instrumentation that is dedicated to the sole purpose of denaturing proteins, restricting its use and the applicability of the technique for the common biochemist.
The introduction of the fluorescence-based thermal shift assay has significantly advanced the ability to easily identify conditions that increase protein stability. The use of environmentally sensitive dyes compatible with commonly available real-time PCR machines makes the technique available to essentially any researcher. Moreover, the high-throughput nature of thermal shift assays allows the rapid discovery of solutions that stabilize proteins through sparse matrix screening of solution conditions (buffer identity, solution pH, ionic strength, etc). Recent advances and highlights of this technique are reviewed further in (Crowther et al., 2010; Kranz and Schalk-Hihi, 2011; Mezzasalma et al., 2007).
Ligand identification with thermal shift screening
The small-scale and high-throughput nature of thermal shift assay screens makes them an excellent platform for the discovery of small molecule ligands, provided that the target protein gives an ideal thermal denaturation curve in the assay (Fedorov et al., 2012; Lo et al., 2004; Niesen et al., 2007). In contrast to other ligand screens that require a significant amount of protein, utilize expensive reagents or require time-consuming steps for data acquisition, the thermal shift assay provides many advantages for ligand discovery. Moreover, the assay is easily miniaturized down to 384 or 1536-well format to allow for automation (Mezzasalma et al., 2007).
Critical Parameters
Selection of proteins for thermal shift assay
This technique detects changes in the thermal denaturation of globular proteins to determine how solution conditions and small molecules modulate protein stability. However, not all globular (folded) proteins give ideal profiles upon thermal denaturation in this assay, precluding analysis of protein stability by this technique. It is estimated that 15–25% of recombinant proteins give non-ideal denaturation curves that include high fluorescence at room temperature baseline and/or lack a sigmoidal transition to the unfolded state (Crowther et al., 2010). Typical causes of non-ideal denaturation profiles include: lack of a compact, globular fold (i.e. intrinsic disorder), lack of hydrophobic core and/or hydrophobic patches on the solvent-exposed surface of the folded protein, or poor protein stability at room temperature. In cases such as these, thermal shift assay cannot be used to reliably extract information about protein stability because the environmentally sensitive SYPRO Orange dye gives high fluorescence background at low temperature and/or the protein lacks a cooperative unfolding transition.
Choosing the right solution conditions for your screen
The selection of solution conditions to screen is highly dependent on the protein being screened and specific needs for downstream biochemical assays or structural biological approaches. Suggested buffer and additive screens that have worked well for a variety of proteins are listed in Tables 1 and 2, respectively. In theory, any matrix of solution conditions can be tested by thermal shift assay. Care should be taken to understand how most biological buffers change pH with increasing temperature if a reasonably accurate relationship between pH and protein stability is to be determined (Fukada and Takahashi, 1998; Good et al., 1966).
Collecting thermal shift data on a wide variety of real-time PCR instruments
Many different real-time PCR instruments have been used to collect thermal shift data (Ablinger et al., 2013; Fedorov et al., 2012; Matulis et al., 2005; Sampson et al., 2011). Optimization of protocols for protein thermal denaturation on each real-time instrument will depend on the specific software.
Troubleshooting
Poor melt curves
As indicated above, not every protein will produce an ideal thermal denaturation curve. To determine if a poor melt curve is due to the specific protein target, a few parameters can be screened in an effort to improve assay results. We recommend systematically titrating the concentration of both SYPRO Orange dye (0.5 – 10× final concentration) and the protein being tested (1–20 µM) to determine if optimal conditions for the denaturation assay can be found.
Anticipated Results
Rapid determination of protein melting temperatures can be obtained in a wide range of solution conditions with as little as 2 ml of purified protein at low concentration (5–15 µM). The quality of the thermal denaturation curve is protein-dependent and provides information on protein stability. By screening a variety of solution conditions and additives, it is possible to identify conditions that maximize the stability of recombinant protein fragments. Based on these data, it is possible to significantly improve performance in biochemical assays and structural biology applications. Therefore, the thermal shift technique provides a rapid and inexpensive assay to improve recombinant proteins for in vitro study.
Time Considerations
With purified protein and pre-made buffer or additive screens in hand, less than 3 hours are needed to collect data in a 96-well assay plate, including set-up time. Data analysis for one 96-well assay plate typically takes 1 hour or less using the methods outlined in this unit.
Supplementary Material
ACKNOWLEDGEMENT
Funding sources: NIH GM107069
Contributor Information
Kathy Huynh, Email: kah016@ucsd.edu.
Carrie L. Partch, Email: cpartch@ucsc.edu.
LITERATURE CITED
- Ablinger E, Leitgeb S, Zimmer A. Differential scanning fluorescence approach using a fluorescent molecular rotor to detect thermostability of proteins in surfactant-containing formulations. International journal of pharmaceutics. 2013;441(1–2):255–260. doi: 10.1016/j.ijpharm.2012.11.035. [DOI] [PubMed] [Google Scholar]
- Bruylants G, Wouters J, Michaux C. Differential scanning calorimetry in life science: thermodynamics, stability, molecular recognition and application in drug design. Current medicinal chemistry. 2005;12(17):2011–2020. doi: 10.2174/0929867054546564. [DOI] [PubMed] [Google Scholar]
- Crowther G, He P, Rodenbough P, Thomas A, Kovzun K, Leibly D, Bhandari J, Castaneda L, Hol W, Gelb M, Napuli A, Van Voorhis W. Use of thermal melt curves to assess the quality of enzyme preparations. Analytical biochemistry. 2010;399(2):268–275. doi: 10.1016/j.ab.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupeux F, Röwer M, Seroul G, Blot D, Márquez J. A thermal stability assay can help to estimate the crystallization likelihood of biological samples. Acta crystallographica. Section D, Biological crystallography. 2011;67(Pt 11):915–919. doi: 10.1107/S0907444911036225. [DOI] [PubMed] [Google Scholar]
- Ericsson U, Hallberg B, Detitta G, Dekker N, Nordlund P. Thermofluor-based high-throughput stability optimization of proteins for structural studies. Analytical biochemistry. 2006;357(2):289–298. doi: 10.1016/j.ab.2006.07.027. [DOI] [PubMed] [Google Scholar]
- Fedorov O, Niesen F, Knapp S. Kinase inhibitor selectivity profiling using differential scanning fluorimetry. Methods in molecular biology (Clifton, N.J.) 2012;795:109–118. doi: 10.1007/978-1-61779-337-0_7. [DOI] [PubMed] [Google Scholar]
- Fukada H, Takahashi K. Enthalpy and heat capacity changes for the proton dissociation of various buffer components in 0.1 M potassium chloride. Proteins. 1998;33(2):159–166. [PubMed] [Google Scholar]
- Good N, Winget G, Winter W, Connolly T, Izawa S, Singh R. Hydrogen ion buffers for biological research. Biochemistry. 1966;5(2):467–477. doi: 10.1021/bi00866a011. [DOI] [PubMed] [Google Scholar]
- Hawe A, Sutter M, Jiskoot W. Extrinsic fluorescent dyes as tools for protein characterization. Pharm Res. 2008;25(7):1487–1499. doi: 10.1007/s11095-007-9516-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C. Differential scanning calorimetry as a tool for protein folding and stability. Archives of biochemistry and biophysics. 2013;531(1–2):100–109. doi: 10.1016/j.abb.2012.09.008. [DOI] [PubMed] [Google Scholar]
- Kranz J, Schalk-Hihi C. Protein thermal shifts to identify low molecular weight fragments. Methods in enzymology. 2011;493:277–298. doi: 10.1016/B978-0-12-381274-2.00011-X. This review provides an overview of the thermal shift technique and a nice summary of the thermodynamic principles behind for estimating Kd from thermal shift data in small molecule screens.
- Lo M-C, Aulabaugh A, Jin G, Cowling R, Bard J, Malamas M, Ellestad G. Evaluation of fluorescence-based thermal shift assays for hit identification in drug discovery. Analytical biochemistry. 2004;332(1):153–159. doi: 10.1016/j.ab.2004.04.031. [DOI] [PubMed] [Google Scholar]
- Mashalidis E, Śledź P, Lang S, Abell C. A three-stage biophysical screening cascade for fragment-based drug discovery. Nature protocols. 2013;8(11):2309–2324. doi: 10.1038/nprot.2013.130. This protocol outlines an experimental framework using the thermal shift assay as a first-line screen for small molecule discovery in conjunction with ligand-observed NMR and isothermal titration calorimetry. The protocol also provides information on assembling and maintaining a library of small molecule fragments that are ideal for ligand discovery by thermal shift assay.
- Matulis D, Kranz J, Salemme F, Todd M. Thermodynamic stability of carbonic anhydrase: measurements of binding affinity and stoichiometry using ThermoFluor. Biochemistry. 2005;44(13):5258–5266. doi: 10.1021/bi048135v. [DOI] [PubMed] [Google Scholar]
- Mezzasalma T, Kranz J, Chan W, Struble G, Schalk-Hihi C, Deckman I, Springer B, Todd M. Enhancing recombinant protein quality and yield by protein stability profiling. Journal of biomolecular screening. 2007;12(3):418–428. doi: 10.1177/1087057106297984. [DOI] [PubMed] [Google Scholar]
- Niesen F, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nature protocols. 2007;2(9):2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- Sampson H, Robert R, Liao J, Matthes E, Carlile G, Hanrahan J, Thomas D. Identification of a NBD1-binding pharmacological chaperone that corrects the trafficking defect of F508del-CFTR. Chemistry & biology. 2011;18(2):231–242. doi: 10.1016/j.chembiol.2010.11.016. [DOI] [PubMed] [Google Scholar]
- Vedadi M, Niesen F, Allali-Hassani A, Fedorov O, Finerty P, Wasney G, Yeung R, Arrowsmith C, Ball L, Berglund H, Hui R, Marsden B, Nordlund P, Sundstrom M, Weigelt J, Edwards A. Chemical screening methods to identify ligands that promote protein stability, protein crystallization, and structure determination. Proc Natl Acad Sci U S A. 2006;103(43):15835–15840. doi: 10.1073/pnas.0605224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.