Abstract
GAPO syndrome (OMIM#230740) is the acronym for growth retardation, alopecia, pseudoanodontia, and optic atrophy. About 35 cases have been reported, making it among one of the rarest recessive conditions. Distinctive craniofacial features including alopecia, rarefaction of eyebrows and eyelashes, frontal bossing, high forehead, mid-facial hypoplasia, hypertelorism, and thickened eyelids and lips make GAPO syndrome a clinically recognizable phenotype. While this genomic study was in progress mutations in ANTXR1 were reported to cause GAPO syndrome. In our study we performed whole exome sequencing (WES) for five affected individuals from three Turkish kindreds segregating the GAPO trait. Exome sequencing analysis identified three novel homozygous mutations including; one frame-shift (c.1220_1221insT; p.Ala408Cysfs*2), one splice site (c.411A>G; p.Gln137Gln), and one non-synonymous (c.1150G>A; p.Gly384Ser) mutation in the ANTXR1 gene. Our studies expand the allelic spectrum in this rare condition and potentially provide insight into the role of ANTXR1 in the regulation of the extracellular matrix.
Keywords: GAPO syndrome, ANTXR1, whole exome sequencing
INTRODUCTION
GAPO syndrome (OMIM#230740), a complex phenotype consisting of growth retardation, alopecia, pseudoanodontia and progressive optic atrophy, is a well-defined autosomal recessive disorder [Tipton and Gorlin, 1984]. Plagiocephaly, frontal bossing, hypertelorism, depressed nasal bridge, short nose, long philtrum, anteverted nares, thick lips and micrognathia are the typical craniofacial findings of GAPO syndrome and make this disorder a readily recognizable pattern of human malformation. Patients with GAPO syndrome demonstrate abnormalities of the cardiovascular[Kocabay and Mert, 2009], skeletal [Goloni-Bertollo et al., 2008], cerebrovascular [Moriya et al., 1995], pulmonary [Demirgunes et al., 2009], and auditory [Rapsomaniki et al., 2013] systems resulting from involvement of connective tissue. All reported individuals have normal intellectual development and the vast majority demonstrates no remarkable biochemical or endocrinologic abnormalities.
While our exome sequencing work was in progress, homozygous mutations in ANTXR1 located on chromosome 2p13.3 were described to be causative for GAPO syndrome [Stranecky et al., 2013]. Immunofluorescence analysis of cultured skin fibroblasts collected from GAPO cases demonstrated an aberrant pattern of actin cytoskeletal microfilament organization. This observation strongly suggested that loss of ANTXR1 function results in progressive extracellular-matrix accumulation that is observed in patients with GAPO syndrome [Stranecky et al., 2013].
CLINICAL REPORT AND GENETIC STUDIES
Patients
This study was approved by the Institutional Review Board at Baylor College of Medicine and informed consent was obtained from all subjects prior to enrolment in the project. Participants provided venous blood samples, and genomic DNA was extracted from blood based on the manufacturer’s protocol (QIAGEN Sciences, Germantown, MD).
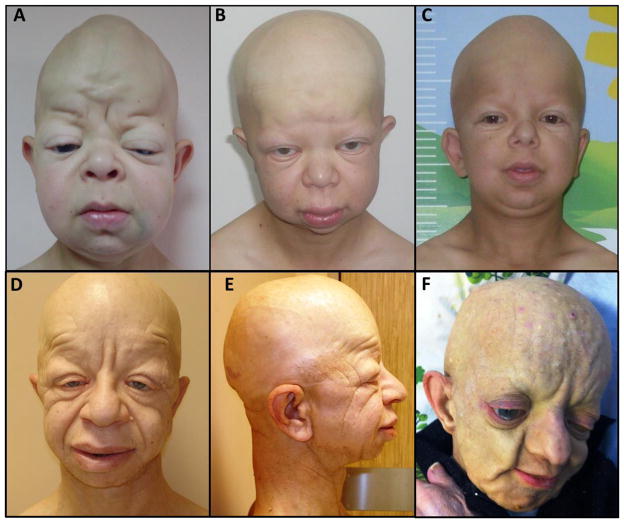
All patients were previously diagnosed with GAPO syndrome and their reported ophthalmologic findings published [Ilker et al., 1999; Bozkurt et al., 2013]. Major clinical findings of these cases are summarized as Table I. The common craniofacial findings in all patients are alopecia, relative macrocephaly, frontal bossing, low set and protruding ears, hypertelorism, thickened eyelids, sparse eyebrows and eyelashes, depressed nasal bridge, long philtrum, thick lips and micrognathia (Fig. 1). Another remarkable finding in our patients is increased subcutaneous accumulations with age that results in an apparent coarsening of facial features (Fig. 1D–F).
TABLE I.
Clinical Findings of the Patients
| Family HOU2001, BAB5033 | Family HOU2034
|
Family HOU2085
|
||||
|---|---|---|---|---|---|---|
| BAB5141 | BAB5142 | BAB5143 | BAB5348 | BAB5349 | ||
| General | ||||||
| Age at evaluation | 12 | 14 | 12 | 7 | 42 | 37 |
| Gender | Male | Male | Male | Female | Male | Male |
| Consanguinity | + | + | + | + | + | + |
| Growth retardation | + | + | + | + | + | + |
| Delayed bone age | + | + | + | + | + | + |
| Craniofacial | ||||||
| Alopecia | + | + | + | + | + | + |
| Relative macrocephaly | + | + | + | + | + | + |
| Delayed closure of anterior fontanel | + | + | + | + | n.d. | n.d. |
| Frontal bossing | + | + | + | + | + | + |
| Low set and protruding ears | + | + | + | + | + | + |
| Hypertelorism | + | + | + | + | + | + |
| Thickened eyelids | + | + | + | + | + | + |
| Sparse eyebrows and eyelashes | + | + | + | + | + | + |
| Optic atrophy | n.d. | − | + | n.d. | − | − |
| Glaucoma | + | − | − | + | + | + |
| Myelinated nerve fiber layer | − | + | + | − | n.d. | n.d. |
| Keratopathy | + | − | − | + | − | + |
| Depressed nasal bridge | + | + | + | + | + | + |
| Short nose, anteverted nares | + | + | + | + | + | − |
| Long philtrum | + | + | + | + | + | + |
| Thick, full lips | + | + | + | + | + | + |
| Pseudoanodontia | + | + | + | + | + | + |
| Micrognathia | + | + | + | + | + | + |
| Other | ||||||
| Breast and nipple hypoplasia | + | + | + | + | n.d. | n.d. |
| Umblical hernia | + | + | + | − | n.d. | n.d. |
| Hepatomegaly | + | − | − | − | n.d. | n.d. |
n.d., not documented.
FIG. 1.
Facial appearance of patients with GAPO syndrome. Note that alopecia, relative macrocephaly, frontal bossing, low set and protruding ears, hypertelorism, thickened eyelids, sparse eyebrows and eyelashes, depressed nasal bridge, long philtrum, thick lips and micrognathia are the common craniofacial findings in all patients. A: Patient BAB5033 at age 12. B,C: Affected brothers in family HOU2034; BAB5141 at age 14 and BAB5142 at age 12. D,E: Frontal and lateral views of patient BAB5348 at age 42. F: Patient BAB5349 at age 37. Note that nodular lesions on scalp probably due to subcutaneous accumulation and extreme senile appearance in comparison with his age. This patient died 2 weeks after this photo was taken.
Exome Sequencing
A total of five affected individuals (BAB5033, BAB5141, BAB5143, BAB5348, and BAB5349) from three different families were sequenced and analyzed through the Baylor-Hopkins Center for Mendelian Genomics research program [Bamshad et al., 2012]. Genomic DNA samples were processed according to protocols previously described [Lupski et al., 2013]. Briefly, DNA was prepared into Illumina paired-end libraries. Capture was performed using the in-house developed BCM Human Genome Sequencing Center (HGSC) Core design and sequenced on the Illumina HiSeq 2000 platform. Data produced was processed through the HGSC developed Mercury pipeline to produce variant call format files (. vcf) using the Atlas2 variant calling method [Shen et al., 2010]. Variants were annotated using the in-house developed “Cassandra” [Bainbridge et al., 2011] annotation pipeline based on ANNOVAR [Wang et al., 2010]. Analysis was performed by comparison of the resulting annotated variants in pairs of affected individuals within the same family and rare variants in shared genes among all affected individuals.
Validation of Mutations With Sanger Sequencing
To confirm the identified exome sequencing candidate variants by an orthologous method and segregate these variants in the families, exons 5, 15, and 16 of the ANTXR1 gene were amplified from genomic DNA by using conventional end-point PCR. Standard PCR was performed in 12 μl of reaction mixture with 0.52 pmol/μl of each primer, 50 ng of genomic DNA, 10×PCR buffer, 0.2 mmol/ L of each deoxynucleotide triphosphate, and 0.6 U of HotStar Taq DNA polymerase (Qiagen, Valencia, CA). The initial denaturation step at 95°C for 15 min was followed by 40 cycles of denaturation at 94°C for 30 sec, annealing at 60°C for 30 sec, and an extension at 72° C for 1 min. A final extension step at 72°C for 7 min was added. Amplification products were electrophoresed on 0.8–1% agarose gels. PCR products were purified using ExoSAP-IT (Affymetrix, Santa Clara, CA) and analyzed by standard Sanger di-deoxy nucleotide sequencing (DNA Sequencing Core Facility at Baylor College of Medicine, Houston, TX).
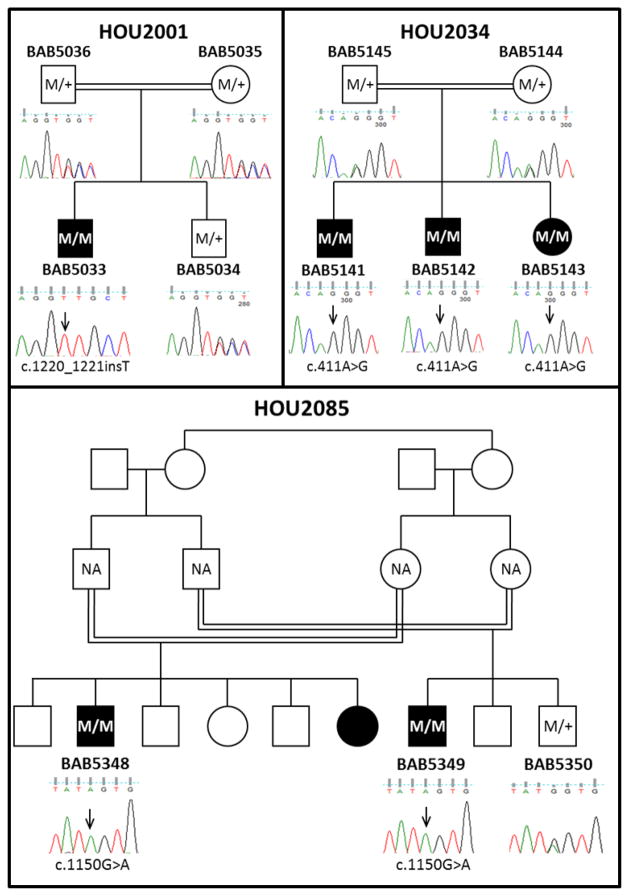
We successfully amplified the target exons in all family members in family HOU2001 (Fig. 2A) and HOU2034 (Fig. 2B). However, in family HOU2085 we were not able to obtain parental samples and only two affected and one unaffected siblings were available to be analyzed by PCR amplification and Sanger sequencing (Fig. 2C).
FIG. 2.
Family pedigrees and segregation of the ANTXR1 mutations. The affected individuals are symbolized with black filled shapes and the locations of mutations are indicated with arrows. All parents are consanguineous. A: Family HOU2001 with one affected and one unaffected child. The affected patient BAB5033 has the homozygous (M/M) c.1220_1221insT variant while the parents and the unaffected sibling are heterozygous carriers (M/+). B: Family HOU2034 with three affected children. All the affected siblings BAB5141, BAB5142 and BAB5143 have the homozygous c.411A>G variant, while the parents are heterozygous carriers. C: Family HOU2085 consists of two related families with cousin marriages. Both affected male cousins BAB5348 and BAB5349 have the homozygous c.1150G>A variant and the unaffected brother of BAB5349 (BAB5350) is a heterozygous carrier. DNA samples from the parents were not available (NA) for segregation analyses.
RESULTS
We studied five individuals from three unrelated Turkish families who met clinical criteria for GAPO syndrome. The comprehensive genomic analyses revealed three homozygous novel deleterious mutations in ANTXR1 that has been recently associated with this rare and distinctive condition.
In family HOU2001, WES analysis revealed a novel homozygous insertion mutation (c.1220_1221insT) in exon 5 of ANTXR1 that is predicted to result in protein truncation two amino acids downstream of the frameshift (p.Ala408Cysfs*2). DNA samples from parents and an unaffected brother were analyzed by Sanger sequencing and found to be heterozygous for the c.1220_1221insT; p. Ala408Cysfs*2 mutation (Fig. 2A).
In family HOU2034, two of the three affected siblings (BAB5141 and BAB5143) were analyzed with WES and found to share a homozygous synonymous mutation (c.411A>G; p.Gln137Gln). This mutation is located in exon5 and is predicted to affect normal splicing and consequently protein structure. For segregation analysis of the candidate variant, Sanger sequencing was performed and the same mutation was also observed in the other affected brother and, consistent with Mendelian expectations, the parents were found to be heterozygous carriers (Fig. 2B).
Combined exome and Sanger sequencing analyses of the two affected cousins in family HOU2085 revealed a homozygous missense mutation (c.1150G>A) in exon15 that results in an amino acid change of Glycine to Serine (p.Gly384Ser) that is predicted to be deleterious. DNA samples from the parents were not available for this family; however one unaffected sibling was analyzed by Sanger sequencing and found to be a heterozygous carrier (Fig. 2C).
DISCUSSION
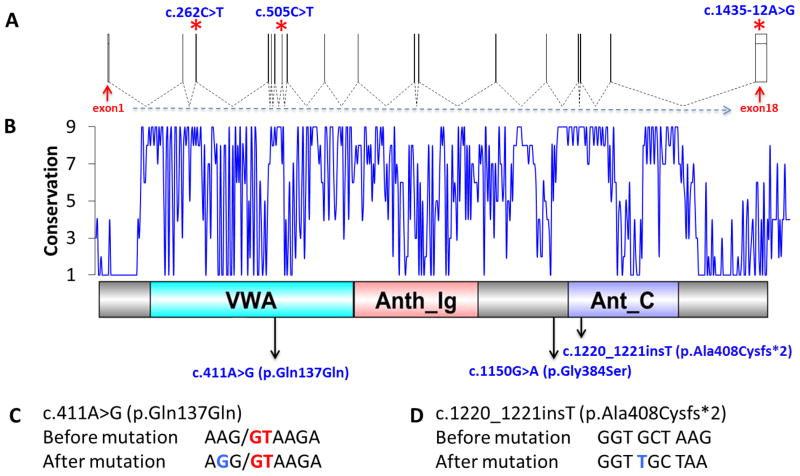
Since the first GAPO syndrome case was reported by Tipton and Gorlin in 1984, nearly 35 clinical cases have been published [Goloni-Bertollo et al., 2008; Demirgunes et al., 2009; Kocabay and Mert, 2009; Castrillon-Oberndorfer et al., 2010; Lei et al., 2010; Nanda et al., 2010; Sinha et al., 2011; Aggarwal et al., 2013; Bozkurt et al., 2013; Karadag et al., 2013; Rapsomaniki et al., 2013; Sharma et al., 2013]. After next-generation sequencing was implemented for identifying the rare variant alleles causing Mendelian disorders, the gene responsible for this well-defined autosomal recessive syndrome was identified using WES and mutations in the ANTXR1 gene were described to be causative for GAPO syndrome [Stranecky et al., 2013]. In our study, we aimed to identify novel mutations in patients with GAPO syndrome by using WES. To date, three ANTRX1 mutations have been identified in 4 unrelated families with GAPO syndrome from three different ethnicities: c.262C>T (p. Arg88*) (2 Egyptian families), c.505C>T (p.Arg169*) (Czech family), and c.1435-12A>G (p.Gly479Phefs*119) (Sri Lankan family) (Fig. 3A) [Stranecky et al., 2013]. Interestingly, all of them produce null alleles as confirmed by functional studies with fibroblasts obtained from GAPO patients. In this study, we have identified one indel, one missense, and one synonymous mutation which is located in an exon–intron boundary predicted to cause a splicing error. Rare variant missense alleles can provide further insights into protein function and a disease process as recently evidenced by the rare p.Arg140His variant in CLP1 [Karaca et al., 2014].
FIG. 3.
Gene structure and diagram of the functional domains of ANTXR1 and localization of the identified mutations. A: Demonstration of ANTXR1 exons. Asterisks indicate localization of previously described mutations in three different ethnicities (c.262C>T = Egyptian, c.505C>T = Czech, c.1435-12A>G = Sri Lankan). B: Conservation pattern and protein domains of ANTXR1 (564 aa). Conservation varies between 1 (variable) and 9 (conserved). Arrows indicate localization of the amino acid changes described in our study in three Turkish families. C: c.411A>G (p.Gln137Gln) mutation. G (blue) and GT (red) are substitution and canonical splicing donor site. D: c.1220_1221insT (p.Ala408Cysfs*2) mutation. Inserted T is highlighted by blue.
The ANTXR1 gene, anthrax toxin receptor 1, contains 18 exons (Fig. 3A) and encodes a type I transmembrane protein that is involved in cell attachment and migration [Hotchkiss et al., 2005]. The protein has a Von Willebrand factor type A domain (VWA), Anthrax receptor extracellular domain (Anth_Ig) and Anthrax receptor C-terminus region (Ant_C) (Fig. 3B) [Punta et al., 2012]. Of the three variants identified in our study, c.411A>G (p.Gln137Gln) and c.1220_1221insT (p.Ala408Cysfs*2) reside within these domains (Fig. 3B). Based on sequence analysis by Consurf [Glaser et al., 2003], c.1150G>A (p.Gly384Ser) and the c.1220_1221insT (p.Ala408Cysfs*2) mutations occurred at highly conserved sites within the protein. The third variant allele we identified, a c.411A>G substitution, is a synonymous change (p. Gln137Gln) that affects a nearby canonical splice donor site (Fig. 3C). Relying on the maximum entropy model, MaxEntScan [Yeo and Burge, 2004] assigns a lower score (9.21 vs 10.57) to the mutated splice site sequence, which suggests that the mutation impairs splicing. As shown in Figure 3D, c.1220_1221insT causes a frame-shift, which introduces a premature stop codon (TAA) downstream of the frameshift insertion at codon position 410. In summary, all three mutations are predicted to give rise to an impaired gene product, which in turn likely causes the phenotype. Additionally, the functional prediction scores of the substitutions were assessed with bioinformatics predictive tools including Poly-Phen-2 [Adzhubei et al., 2010], Mutation Taster [Schwarz et al., 2010], SIFT [Sim et al., 2012] and PROVEAN [Choi et al., 2012], and all variants were predicted as disease causing or damaging.
None of the GAPO associated mutations were reported in the 1000 Genomes Project, NHLBI Exome Sequencing Project (ESP), dbSNP, or our internal exome databases (>2,600 exomes). Additionally, we searched an international multiethnic comparison database containing exome data from 8,602 individuals for the frequency of the rare (Minor allele frequency <1%) missense alleles in ANTXR1. In total, 126 heterozygous missense variants were observed but none of them includes the mutations identified in our patients. In this dataset, heterozygous missense variants in ANTXR1 were detected with the frequency of 42/5748 (0.73%) in Europeans and 84/2854 (2.9%) in African-Americans. A search within a Turkish subpopulation (465 exomes) in our internal exome database revealed four heterozygous missense variants different from the mutations detected in our patients and parents. So, the carrier frequency of ANTXR1 missense alleles in our Turkish exome database was calculated as 0.86%, which is slightly higher than the carrier state in Europeans.
In an Antxr1 knock-out mouse model a mild to moderate increase of extracellular matrix (especially collagen) has been observed in many tissues including: the skin (basal aspects of the hair follicles), endometrium, ovaries, periosteum of femurs and vertebra, cranial sutures of the skull, and the periodontal ligament of the incisors that leads to misalignment and dental dysplasia [Cullen et al., 2009]. Also, in a recently published paper immunohistochemical studies performed on skin fibroblast cell lines obtained from GAPO syndrome patients demonstrated complete loss of the ANTXR1 isoform and remarkable alterations in the actin cytoskeletal network in affected fibroblasts [Stranecky et al., 2013]. These cell biological features match with the tissue involvement observed clinically in GAPO syndrome patients and strongly suggest that loss of function in ANTXR1 is responsible for the progressive extracellular-matrix findings observed in individuals with GAPO syndrome.
From a clinical perspective, keratopathy in three patients (BAB5033, BAB5143, and BAB5349), myelinated retinal nerve fiber in two patients (BAB5141 and BAB5142) and glaucoma in four patients (BAB5033, BAB5143, BAB5348, and BAB5349) were observed as uncommon ophthalmic findings [Ilker et al., 1999; Lei et al., 2010; Bozkurt et al., 2013]. As Ilker et al. noted that optic atrophy is not a consistent feature of this disorder, we also detected optic atrophy in only one of our cases. It is concluded that optic nerve pathologies are observed potentially secondary to physical compression of the optic nerve by the accumulation of extracellular matrix and thickening of the dura matter surrounding the optic nerve [Gagliardi et al., 1984; Wajntal et al., 1990; Ilker et al., 1999]. The other occasional findings of GAPO syndrome published in the literature are bilateral sensorineural deafness [Aggarwal et al., 2013], vestibular dysfunction [Rapsomaniki et al., 2013], pyoderma vegetans [Karadag et al., 2013], dilated cardiomyopathy [Kocabay and Mert, 2009], pulmonary hypertension [Demirgunes et al., 2009], bilateral interstitial keratitis and hypothyroidism [Lei et al., 2010]. Clinical follow up of our families revealed that patient BAB5349 died at age 37 due to central apnea and respiratory insufficiency. Pulmonary involvement is one of the life-span reducing visceral manifestations present in patients with GAPO syndrome and reported in a GAPO patient who had pulmonary hypertension that lead to death at the age of 17 months [Demirgunes et al., 2009].
In this study we identified three novel mutations in the ANTXR1 gene by using exome sequencing. Our data add to the emerging genotype–phenotype correlations in GAPO syndrome patients and potentially provide insight into the role of ANTXR1 in regulation of the extracellular matrix. Rare variants that have recently arisen in a clan and then become rapidly reduced to homozygosity within a population with a high degree of consanguinity again provides further evidence in support of the Clan Genomics hypothesis [Lupski et al., 2011].
Acknowledgments
Grant sponsor: United States National Human Genome Research Institute/National Heart Blood and Lung Institute; Grant number: U54HG006542.
We thank all the families and collaborators that participated in this study. This work was supported by the United States National Human Genome Research Institute/National Heart Blood and Lung Institute grant U54HG006542 to the Baylor-Hopkins Center for Mendelian Genomics.
Footnotes
Conflict of Interest: J.R.L. has stock ownership in 23 and Me and Ion Torrent Systems, and is a co-inventor on multiple United States and European patents related to molecular diagnostics for inherited neuropathies, eye diseases and bacterial genomic fingerprinting. The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from the chromosomal microarray analysis (CMA) and clinical exome sequencing offered in the Medical Genetics Laboratory (MGL; http://www.bcm.edu/geneticlabs/). Other authors have no disclosures relevant to the manuscript.
References
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal S, Uttarilli A, Dalal AB. GAPO syndrome with deafness: New feature or incidental finding? Clin Dysmorphol. 2013;22:161–163. doi: 10.1097/MCD.0000000000000008. [DOI] [PubMed] [Google Scholar]
- Bainbridge MN, Wiszniewski W, Murdock DR, Friedman J, Gonzaga-Jauregui C, Newsham I, Reid JG, Fink JK, Morgan MB, Gingras MC, Muzny DM, Hoang LD, Yousaf S, Lupski JR, Gibbs RA. Whole-genome sequencing for optimized patient management. Sci Transl Med. 2011;3:87re83. doi: 10.1126/scitranslmed.3002243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamshad MJ, Shendure JA, Valle D, Hamosh A, Lupski JR, Gibbs RA, Boerwinkle E, Lifton RP, Gerstein M, Gunel M, Mane S, Nickerson DA Centers for Mendelian G. The Centers for Mendelian Genomics: A new large-scale initiative to identify the genes underlying rare Mendelian conditions. Am J Med Genet Part A. 2012;158A:1523–1525. doi: 10.1002/ajmg.a.35470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozkurt B, Yildirim MS, Okka M, Bitirgen G. GAPO syndrome: Four new patients with congenital glaucoma and myelinated retinal nerve fiber layer. Am J Med Genet Part A. 2013;161A:829–834. doi: 10.1002/ajmg.a.35734. [DOI] [PubMed] [Google Scholar]
- Castrillon-Oberndorfer G, Seeberger R, Bacon C, Engel M, Ebinger F, Thiele OC. GAPO syndrome associated with craniofacial vascular malformation. Am J Med Genet Part A. 2010;152A:225–227. doi: 10.1002/ajmg.a.33192. [DOI] [PubMed] [Google Scholar]
- Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS ONE. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen M, Seaman S, Chaudhary A, Yang MY, Hilton MB, Logsdon D, Haines DC, Tessarollo L, St Croix B. Host-derived tumor endothelial marker 8 promotes the growth of melanoma. Cancer Res. 2009;69:6021–6026. doi: 10.1158/0008-5472.CAN-09-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirgunes EF, Ersoy-Evans S, Karaduman A. GAPO syndrome with the novel features of pulmonary hypertension, ankyloglossia, and prognathism. Am J Med Genet Part A. 2009;149A:802–805. doi: 10.1002/ajmg.a.32686. [DOI] [PubMed] [Google Scholar]
- Gagliardi AR, Gonzalez CH, Pratesi R. GAPO syndrome: Report of three affected brothers. Am J Med Genet. 1984;19:217–223. doi: 10.1002/ajmg.1320190203. [DOI] [PubMed] [Google Scholar]
- Glaser F, Pupko T, Paz I, Bell RE, Bechor-Shental D, Martz E, Ben-Tal N. ConSurf: Identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics. 2003;19:163–164. doi: 10.1093/bioinformatics/19.1.163. [DOI] [PubMed] [Google Scholar]
- Goloni-Bertollo EM, Ruiz MT, Goloni CB, Muniz MP, Valerio NI, Pavarino-Bertelli EC. GAPO syndrome: Three new Brazilian cases, additional osseous manifestations, and review of the literature. Am J Med Genet Part A. 2008;146A:1523–1529. doi: 10.1002/ajmg.a.32157. [DOI] [PubMed] [Google Scholar]
- Hotchkiss KA, Basile CM, Spring SC, Bonuccelli G, Lisanti MP, Terman BI. TEM8 expression stimulates endothelial cell adhesion and migration by regulating cell-matrix interactions on collagen. Exp Cell Res. 2005;305:133–144. doi: 10.1016/j.yexcr.2004.12.025. [DOI] [PubMed] [Google Scholar]
- Ilker SS, Ozturk F, Kurt E, Temel M, Gul D, Sayli BS. Ophthalmic findings in GAPO syndrome. Jpn J Ophthalmol. 1999;43:48–52. doi: 10.1016/s0021-5155(98)00058-6. [DOI] [PubMed] [Google Scholar]
- Karaca E, Weitzer S, Pehlivan D, Shiraishi H, Gogakos T, Hanada T, Jhangiani SN, Wiszniewski W, Withers M, Campbell IM, Erdin S, Isikay S, Franco LM, Gonzaga-Jauregui C, Gambin T, Gelowani V, Hunter JV, Yesil G, Koparir E, Yilmaz S, Brown M, Briskin D, Hafner M, Morozov P, Farazi TA, Bernreuther C, Glatzel M, Trattnig S, Friske J, Kronnerwetter C, Bainbridge MN, Gezdirici A, Seven M, Muzny DM, Boerwinkle E, Ozen M, Clausen T, Tuschl T, Yuksel A, Hess A, Gibbs RA, Martinez J, Penninger JM, Lupski JR. Human CLP1 mutations alter tRNA biogenesis, affecting both peripheral and central nervous system function. Cell. 2014;157:636–650. doi: 10.1016/j.cell.2014.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karadag AS, Calka O, Bilgili SG, Karadag R, Bulut G. GAPO syndrome associated with pyoderma vegetans: An unreported co-existence. Genet Couns. 2013;24:133–139. [PubMed] [Google Scholar]
- Kocabay G, Mert M. GAPO syndrome associated with dilated cardiomyopathy: An unreported association. Am J Med Genet Part A. 2009;149A:415–416. doi: 10.1002/ajmg.a.32638. [DOI] [PubMed] [Google Scholar]
- Lei S, Iyengar S, Shan L, Cherwek DH, Murthy S, Wong AM. GAPO syndrome: A case associated with bilateral interstitial keratitis and hypothyroidism. Clin Dysmorphol. 2010;19:79–81. doi: 10.1097/MCD.0b013e32833593d1. [DOI] [PubMed] [Google Scholar]
- Lupski JR, Belmont JW, Boerwinkle E, Gibbs RA. Clan genomics and the complex architecture of human disease. Cell. 2011;147:32–43. doi: 10.1016/j.cell.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR, Gonzaga-Jauregui C, Yang Y, Bainbridge MN, Jhangiani S, Buhay CJ, Kovar CL, Wang M, Hawes AC, Reid JG, Eng C, Muzny DM, Gibbs RA. Exome sequencing resolves apparent incidental findings and reveals further complexity of SH3TC2 variant alleles causing Char-cot-Marie-Tooth neuropathy. Genome Med. 2013;5:57. doi: 10.1186/gm461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya N, Mitsui T, Shibata T, Yamaguchi K, Kanazawa C, Matsunaga A, Hayasaka K. GAPO syndrome: Report on the first case in Japan. Am J Med Genet. 1995;58:257–261. doi: 10.1002/ajmg.1320580312. [DOI] [PubMed] [Google Scholar]
- Nanda A, Al-Ateeqi WA, Al-Khawari MA, Alsaleh QA, Anim JT. GAPO syndrome: A report of two siblings and a review of literature. Pediatr Dermatol. 2010;27:156–161. doi: 10.1111/j.1525-1470.2010.01100.x. [DOI] [PubMed] [Google Scholar]
- Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer EL, Eddy SR, Bateman A, Finn RD. The Pfam protein families database. Nucleic Acids Res. 2012;40(Database issue):D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapsomaniki M, Chiarella G, Mascaro I, Ceravolo F, Cassandro E, Strisciuglio P, Concolino D. GAPO syndrome associated with vestibular dysfunction and hearing loss. Am J Med Genet Part A. 2013;161A:2102–2104. doi: 10.1002/ajmg.a.35992. [DOI] [PubMed] [Google Scholar]
- Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- Sharma VB, Pandia MP, Raut PP. Anesthetic management of a case of GAPO syndrome for craniosynostosis surgery. J Anaesthesiol Clin Pharmacol. 2013;29:580–581. doi: 10.4103/0970-9185.119174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Wan Z, Coarfa C, Drabek R, Chen L, Ostrowski EA, Liu Y, Weinstock GM, Wheeler DA, Gibbs RA, Yu F. A SNP discovery method to assess variant allele probability from next-generation resequencing data. Genome Res. 2010;20:273–280. doi: 10.1101/gr.096388.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim NL, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. SIFT web server: Predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40(Web Server issue):W452–W457. doi: 10.1093/nar/gks539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha R, Trikha A, Laha A, Raviraj R, Kumar R. Anesthetic management of a patient with GAPO syndrome for glaucoma surgery. Paediatr Anaesth. 2011;21:910–912. doi: 10.1111/j.1460-9592.2011.03550.x. [DOI] [PubMed] [Google Scholar]
- Stranecky V, Hoischen A, Hartmannova H, Zaki MS, Chaudhary A, Zudaire E, Noskova L, Baresova V, Pristoupilova A, Hodanova K, Sovova J, Hulkova H, Piherova L, Hehir-Kwa JY, de Silva D, Senanayake MP, Farrag S, Zeman J, Martasek P, Baxova A, Afifi HH, St Croix B, Brunner HG, Temtamy S, Kmoch S. Mutations in ANTXR1 cause GAPO syndrome. Am J Hum Genet. 2013;92:792–799. doi: 10.1016/j.ajhg.2013.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipton RE, Gorlin RJ. Growth retardation, alopecia, pseudo-anodontia, and optic atrophy—The GAPO syndrome: Report of a patient and review of the literature. Am J Med Genet. 1984;19:209–216. doi: 10.1002/ajmg.1320190202. [DOI] [PubMed] [Google Scholar]
- Wajntal A, Koiffmann CP, Mendonca BB, Epps-Quaglia D, Sotto MN, Rati PB, Opitz JM. GAPO syndrome (McKusick 23074)—A connective tissue disorder: Report on two affected sibs and on the pathologic findings in the older. Am J Med Genet. 1990;37:213–223. doi: 10.1002/ajmg.1320370210. [DOI] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]