Abstract
Background and Aims Salt tolerance has evolved many times independently in different plant groups. One possible explanation for this pattern is that it builds upon a general suite of stress-tolerance traits. If this is the case, then we might expect a correlation between salt tolerance and other tolerances to different environmental stresses. This association has been hypothesized for salt and alkalinity tolerance. However, a major limitation in investigating large-scale patterns of these tolerances is that lists of known tolerant species are incomplete. This study explores whether species’ salt and alkalinity tolerance can be predicted using geochemical modelling for Australian grasses. The correlation between taxa found in conditions of high predicted salinity and alkalinity is then assessed.
Methods Extensive occurrence data for Australian grasses is used together with geochemical modelling to predict values of pH and electrical conductivity to which species are exposed in their natural distributions. Using parametric and phylogeny-corrected tests, the geochemical predictions are evaluated using a list of known halophytes as a control, and it is determined whether taxa that occur in conditions of high predicted salinity are also found in conditions of high predicted alkalinity.
Key Results It is shown that genera containing known halophytes have higher predicted salinity conditions than those not containing known halophytes. Additionally, taxa occurring in high predicted salinity tend to also occur in high predicted alkalinity.
Conclusions Geochemical modelling using species’ occurrence data is a potentially useful approach to predict species’ relative natural tolerance to challenging environmental conditions. The findings also demonstrate a correlation between salinity tolerance and alkalinity tolerance. Further investigations can consider the phylogenetic distribution of specific traits involved in these ecophysiological strategies, ideally by incorporating more complete, finer-scale geochemical information, as well as laboratory experiments.
Keywords: alkalinity tolerance, geochemical modelling, grasses, halophytes, macroevolution, phylogeny, Poaceae, salt tolerance, stress resistance syndrome
INTRODUCTION
Many plant species have developed several ecophysiological strategies to tolerate extreme conditions in challenging environments. For example, species that complete their life cycle in saline environments – known as halophytes – have evolved various mechanisms that have enabled them to survive and reproduce in these environments (Flowers and Colmer, 2008; Munns and Tester, 2008). These mechanisms are related to water uptake and defence against ion toxicity within the plant, such as the accumulation and compartmentalization of saline ions, the ability to limit the entry of these ions into the transpiration stream, the synthesis of compatible solutes for osmoprotection, the ability to accumulate essential nutrients, and the ability to continue to regulate transpiration in the presence of high concentrations of Na+ and Cl− (Flowers and Colmer, 2008; Munns and Tester, 2008; Rozema and Flowers, 2008; Shabala, 2013; Deinlein et al., 2014). Research has unveiled the complex, physiological, molecular and genetic background of these adaptations (e.g. Munns, 2005; Munns and Tester, 2008; Shavrukov, 2012; Ashraf and Foolad, 2013). There are more than 1500 species of halophytes (Aronson, 1989) and salt tolerance is widely distributed across the plant phylogeny, with multiple independent origins (Flowers et al., 1977; Saslis-Lagoudakis et al., 2014). However, some plant groups, such as Caryophyllales and Alismatales, contain more halophytes than others (Flowers et al., 2010; Saslis-Lagoudakis et al., 2014). At a lower hierarchical level, salt tolerance has also evolved multiple times independently. For example, it has evolved over 70 times in the grass family alone, and is phylogenetically non-random, i.e. some clades are more likely than others to contain salt-tolerant species (Bennett et al., 2013).
It has been suggested that tolerance mechanisms and physiological responses to salinity are shared with other types of environmental stresses, such as aridity, flooding and frost (Tuteja, 2007; Munns and Tester, 2008; Rozema and Schat, 2013). For example, a recent study found that salt tolerance in grasses evolves more frequently in C4 than C3 lineages, demonstrating a close association in the evolution of C4 photosynthesis and salt tolerance in these lineages (Bromham and Bennett, 2014). This type of correlation may provide one possible explanation for the repeated evolution of salt tolerance. The stress resistance syndrome hypothesis (Chapin et al., 1993) states that there may be a suite of stress-related traits that allow plants to survive in a variety of stressful environments. Therefore, the presence of ‘enablers’ in some lineages can facilitate the evolution of multiple stress resistance within those lineages (Edwards and Donoghue, 2013). This suggests that traits related to tolerance to one type of stress can facilitate the evolution of another type of stress resistance. For example, salt tolerance, succulence and C4 photosynthesis are associated in chenopods (Kadereit et al., 2012) and occupation of bare environments served as an ‘enabler’ to adaptation to harsh elemental soils in the Brassicaceae (Cacho and Strauss, 2014). Therefore, by studying these ecophysiological traits in a phylogenetic context, we can investigate macroevolutionary patterns of ecophysiological evolution (Ackerly et al., 2000), and explore the correlation between different ecophysiological strategies (Niinemets and Valladares, 2006).
A correlation of this kind has been suggested between salt and alkaline tolerance (Bromham et al., 2013; Bui, 2013b; Bui et al., 2014). Alkalinity (high soil pH) often co-occurs with salinity (high soil NaCl concentrations) in the landscape: many saline soils are also alkaline due to the presence of sodium carbonates (Rengasamy, 2010). Therefore, it is possible that lineages occupying these environments have had to evolve strategies to cope with both alkalinity and salt-stress (Bui, 2013b). Like salinity, alkalinity exacerbates water loss, interfering with stomatal closure due to the accumulation of sodium ions (Bernstein, 1975). Soils of high pH often have poor structure, affecting their hydraulic conductivity and the plants’ water uptake, and causing hypoxia in the root zone (Bernstein, 1975). Both these factors affect water use efficiency, which is also one of the major stresses for plants in saline environments. Plants equipped to deal with salinity and alkalinity employ osmotic adjustments (Farrell et al., 1996; Yang et al., 2007, 2008) that are not found in plants without tolerance to either stresses (Liu et al., 2010; Chen et al., 2011), and which make tolerant plants naturally resistant to water stress (García and Mendoza, 2014). Furthermore, both salinity and alkalinity affect photosynthesis and metabolism through a range of physiological and molecular processes (Yang et al., 2008; Nishiuchi et al., 2010). It is therefore possible that because of the shared challenges, salt and alkaline tolerance have evolved in closely related lineages that possess traits enabling the evolution of mechanisms of tolerance to either stress.
One of the main constraints in exploring large-scale patterns in salt and alkaline tolerance is the lack of exhaustive published lists of halophytes and particularly alkaline-tolerant species. Because field and laboratory observations of plant species’ tolerance to salinity and alkalinity tend to focus on particular species, lists of known halophytes are likely to be incomplete, and there are no comprehensive lists of alkaline-tolerant species. An alternative approach to generating such lists is to predict plant species that are tolerant to these stresses based on their geographical distributions. In the last two decades, inferring species’ environmental niche preferences from their natural distributions and environmental geographical information systems (GIS) data layers has become commonplace in studies of ecology and evolution (Guisan and Thuiller, 2005; Kozak et al., 2008; Warren et al., 2008). By combining distribution data with geochemical observations, we can infer salinity and alkalinity conditions to which species are exposed in their natural distributions. Although microbial studies have combined geochemical data with phylogenetic metrics (Reysenbach and Shock, 2002; Macur et al., 2004; Costa et al., 2009), geochemical modelling has been largely overlooked in studies of macroecology and macroevolution. However, a recent phylogenetic study of Australian Acacia species used geochemical modelling to investigate evolutionary patters of salinity and alkalinity tolerance (Bui et al., 2014).
The aims of this study were two-fold: (1) to evaluate the performance of geochemical modelling using species occurrence data, to identify species’ tolerance to salinity and alkalinity; and (2) to investigate the correlation between salt and alkaline tolerance. We use Australian grasses (Poaceae) as a test case, because they are a group with a continent-wide distribution, occupying a wide range of environmental conditions, including arid, saline and sodic environments. Our dataset included distribution data for 1387 species of mainland Australian grasses, of which 141 are known halophytes.
MATERIALS AND METHODS
We investigated whether we could predict species’ salt and alkaline tolerance based on species distribution modelling. To do that, we used geochemical modelling to generate species’ descriptors for electrical conductivity (EC) and pH at their natural distributions. We evaluated the prediction of salt-tolerant species based on prior knowledge of salt tolerance in Australian grasses. Subsequently, we tested for the correlation between salt and alkalinity tolerance, and we explored if spatial patterns can explain this association.
In the literature, salinity and alkalinity tolerance are often characterized based on EC and pH soil values, respectively. For example, soils with EC over 4000 µS m−1 are characterized as saline (United States Salinity Laboratory Staff, 1969) and plants tolerating 8000 µS m−1 or over are considered halophytes (Aronson, 1989). Similarly, soil pH of 7 or higher is alkaline and most plants prefer pH 5·5–6·5 (Islam et al., 1980). In this study, we do not apply a threshold of EC or pH to characterize soils as saline or alkaline. Instead, we perform a comparative analysis of EC and pH conditions to which Australian grasses are exposed.
Predicting salt and alkalinity tolerance from species distribution modelling
Predicting species salt and alkalinity tolerance from occurrence data.
Because there are no exhaustive databases that describe tolerances of all Australian grasses to salinity and alkalinity, to estimate these tolerances we employed an approach based on species’ distributions. Our approach assumes that conditions of salinity and alkalinity at localities at which species are found naturally reflect their levels of tolerance to these conditions. Although factors other than tolerance affect species’ distributions, such as interspecific competition, we can expect intrinsic tolerance to be correlated with realized tolerances. Therefore, it is possible to describe species’ tolerances if we know: (1) species’ distributions and (2) levels of salinity and alkalinity in these distributions. To generate species’ distributions, we extracted occurrence data from the Atlas of Living Australia (ALA; http://www.ala.org.au), a continent-wide dataset that contains approximately 45 million occurrence records for Australian biodiversity. There are 1387 grass species found in mainland Australia (excluding Tasmania and other islands). Australian grass species are recorded from 354 913 points with unique geographical coordinates in the ALA. We extracted all unique occurrence points for each species and we consider the distribution of each species to be the compilation of all the points at which it is reported.
To infer soil pH and EC at the localities where grass species were reported, we accessed data from the National Geochemical Survey of Australia. This dataset reports the pH and EC on 1 : 5 soil/water extracts from bulk samples at 1315 georeferenced point measurements across the continent, with an average sample density of one site per 5500 km2 (de Caritat and Cooper, 2011). We retrieved indications of EC and pH from the dataset and performed the analyses described below for subsoil (60–80 cm below the surface). Subsoil indications of EC and pH are more likely to reflect tolerance to salinity and alkalinity than shallower samples, as root tips – generally found deeper in the soil – are more highly sensitive to geochemistry than the rest of the root (Shabala, 2013).
From this dataset of subsoil EC and pH indications, we estimated EC and pH at each locality with a reported grass occurrence using Geostatistics in geoR (Diggle and Ribeiro, 2007). Geostatistics are techniques for mapping of surfaces from limited sample data and the estimation of values at unsampled locations in two steps (Clark and Harper, 2000). First, a semi-variogram was constructed to establish the predictability of values from place to place in the study area. The semi-variogram modelled the difference between a value at one location and the value at another according to the distance and between them. Secondly, ‘kriging’ was used to estimate values at unsampled locations. The basic technique of ordinary kriging that we used here used a weighted average of neighbouring samples to estimate the value at an unsampled location. Weights were optimized using the semi-variogram model, given the distance and directional relationships between sampled and unsampled locations. We used the ordinary kriging variance as an estimate of error associated with each prediction (Diggle and Ribeiro, 2007). With this approach, we produced a compilation of EC and pH predictions for each species, given each individual prediction corresponds to an estimate for each location at which the species is recorded. This gives a range of predicted EC and pH values for each species, and from this range we recorded the median and upper quartile (UQ) values. Therefore, for each species, we used four measures to describe soil salinity and alkalinity across its distribution: two describing EC (median and UQ values) and two describing pH (again, median and UQ values). Median values provide species’ central tendency with respect to environmental conditions (EC and pH) in their distributions, while UQ values represent more extreme salinity and alkalinity conditions that species encounter within their geographical ranges.
Evaluating prediction of halophytes.
An ideal way to evaluate how well the geochemical modelling approach performed in predicting species’ salinity and alkalinity tolerance would be to test species’ tolerances experimentally, as well as to take EC and pH measurements at localities where species occur naturally, covering each species range, and then compare those measurements with our predictions. However, to generate these data, even for a single species, would require a considerable amount of time and effort. An alternative way to evaluate the performance of the geochemical modelling is using data that are already available. Although we do not have prior knowledge of alkaline-tolerant species, we have lists of halophytes. These lists might be incomplete, but they are likely to be accurate in the species that are included, as they are based on expert judgment and experimental data. Because halophytes are able to grow in conditions of high salinity, the predicted EC for taxa known to be halophytes should be higher than that for non-halophytes.
Here, we investigated whether known halophytes have been reported to occur at higher predicted EC than non-salt-tolerant species. First, we extracted the species names of known Australian grass halophytes from a recent study (Bennett et al., 2013), which identified 141 Australian grasses as halophytes (Supplementary Data Table S1). Then, we applied a parametric Welch two sample t-test to test if predicted EC values (median and UQ) of known halophytes were significantly higher than the rest of the species in our dataset.
Furthermore, we performed the same analysis (Welch two sample t-test) at the genus level, to see if genera containing halophytes occur in conditions of high predicted EC. There are 234 Australian grass genera in total, 71 of which include at least one known halophyte. We calculated median and UQ soil EC values for each genus, based on the observations for all species within that genus. We also used a phylogeny-corrected two sample t-test. We estimated the phylogenetic correlation matrix among genera using two phylogenies. One is a well-sampled genus-level topology of Poaceae that includes over 800 genera (Bouchenak-Khelladi et al., 2010). This tree included 226 of the 234 Australian genera and 70 of the 71 genera with known haplotypes. We computed the branch lengths of the topology using the method by Grafen (1989), which gives each node on the tree a ‘height’, corresponding to the number of leaves of the subtree minus one. Each height was scaled so that root height is 1, and then raised at power ‘rho’ (Grafen, 1989). Branch lengths were then calculated as the difference between height of lower and upper nodes. The other phylogeny was a smaller, time-calibrated molecular phylogenetic tree with 298 out of approx. 800 genera of Poaceae (Bouchenak-Khelladi et al., 2010). This tree included 146 of 234 Australian genera and 56 of 71 Australian genera with known haplotypes. We performed the analysis using this tree because, although taxon sampling was limited, it was time-calibrated, and we wanted to ensure that the absence of branch lengths in the larger phylogenetic tree did not affect our results. We accounted for phylogenetic relatedness in a two sample t-test using a Generalized Least Squares (GLS) approach. GLS is a generalized approach for estimating parameters in a linear regression model where observations are not homoscedastic or independent from each other (Martins and Hansen, 1997). Phylogenetic relatedness was accounted for by correcting the covariance matrix among observations according to their phylogenetic relatedness (Martins and Hansen, 1997).
The parametric test compared predicted EC values for halophytic taxa with predicted EC values of the rest of the taxa, and evaluated whether halophytic taxa had higher predicted EC than non-halophytic taxa. Because salt tolerance is not randomly distributed in the grass phylogeny (Bennett et al., 2013), by accounting for phylogenetic relatedness, the phylogenetic test ensured that if a relationship was recovered, it was beyond that expected from phylogeny.
Testing the correlation between salt and alkalinity tolerance
Correlation of taxa occurring in high predicted salinity and alkalinity.
We investigated whether the taxa found in conditions of high predicted salinity also tended to be found in conditions of high predicted alkalinity. Similar to the previous section, we first calculated the median and UQ EC and pH values for each taxon. We performed this analysis at the species level, testing the correlation between species’ median or UQ EC values and species’ median or UQ pH values, using the parametric Pearson’s product-moment correlation. The same analysis was performed at the genus level, along with a phylogenetic reduced major axis (RMA) regression (Ives et al., 2007), using the two phylogenies described above to estimate the phylogenetic correlation matrix. RMA regression is a type II regression that does not assume causal directionality between values of salinity and alkalinity. Phylogenetic relatedness is accounted for by a similar approach as in GLS (Martins and Hansen, 1997; Ives et al., 2007). Although the parametric test evaluates the correlation between predicted EC and pH for taxa, the phylogenetic test evaluates whether this correlation is because of covariation due to shared ancestry among taxa.
Geographical correlation of salinity and alkalinity.
We wanted to tease apart whether any association between predicted salinity and alkalinity values was due to geographical correlation between soil EC and pH. First, to assess the degree to which salinity and alkalinity overlapped on the landscape in areas where Australian grasses are found, we fitted a linear model between predicted values of EC and pH for all occurrence points where Australian grasses were reported. If at localities where predicted EC was high, predicted pH was also high (and vice versa), then species exposed to high salinity were also exposed to high alkalinity (and vice versa).
Second, we tested for the correlation between predicted salinity and alkalinity only for known halophytes, using a parametric Pearson’s product-moment correlation. We also tested this relationship at the genus level, only for genera that contain known halophytes, with the parametric Pearson’s product-moment correlation, and a phylogenetic RMA regression (Ives et al., 2007), using the two phylogenies to estimate the phylogenetic correlation matrix. If salt and alkalinity tolerance were functionally associated but conditions of salinity and alkalinity were not geographically associated, then salt-tolerant taxa could be found in conditions of both low and high alkalinity. Under these conditions, we would expect a weaker correlation between predicted EC and pH values in salt-tolerant than non-salt-tolerant taxa. If salt and alkalinity tolerance were functionally associated and conditions of salinity and alkalinity were geographically associated, we would expect a stronger correlation between predicted EC values and pH values in salt-tolerant than non-salt-tolerant taxa.
All statistical analyses used log-transformed EC values for normality and were implemented in R (R Core Team, 2014), with Grafen’s computation of branch lengths (Grafen, 1989) using the ‘compute.brlen’ function in ‘ape’ package (Paradis et al., 2004), the phylogeny-corrected t-test using the ‘gls’ function in ‘nlme’ package (Pinheiro et al., 2014), and the phylogenetic RMA regression using the ‘phyl.RMA’ function in ‘phytools’ package (Revell, 2012).
RESULTS
Predicting salt and alkalinity tolerance from species distribution modelling
Predicting species’ salt and alkalinity tolerance from occurrence data.
Predicted soil EC for all occurrence points where Australian grasses are found ranged between 0·01 and 10·53 dS m−1 and predicted pH ranged from 4·87 to 9·05. The average standard error (as estimated with kriging variance) for predictions across all reported localities was 2·06 dS m−1 for EC and 0·93 for pH.
Evaluating prediction of halophytes
Our results (Table 1) show that halophytic species are not found in significantly higher predicted salinity than non-salt-tolerant species. However, both analyses (parametric and phylogeny-corrected) at the genus level, considering both median and UQ predicted EC, suggest that genera with known halophytes are found in significantly higher predicted soil EC than genera that do not include known halophytes. Although significantly positive, the absolute difference in EC values between genera with and without known halophytes is small. The predicted EC values for genera with known halophytes explains only about 5 % of the variation of EC values in our dataset (R2 in Table 1).
Table 1.
Results of tests for the comparison of predicted EC values for known halophytes versus non salt-tolerant species, and for the correlation between salinity and alkalinity conditions in Australian grass species
| Alternative hypothesis | Variable | Parametric |
|---|---|---|
| Known halophytes are found in conditions of higher predicted salinity than non-salt-tolerant species | Median | t185 = 0·54, R2 = 0·00 |
| UQ | t185 = 1·14, R2 = 0·01 | |
| Species found in conditions of high predicted salinity also tend to be found in conditions of high predicted alkalinity | Median | T1385 = 29·63**, R2 = 0·39 |
| UQ | T1385 = 35·96**, R2 = 0·48 | |
| Known halophytes found in conditions of high predicted salinity tend to be found in conditions of high predicted alkalinity | Median | T139 = 12·33**, R2 = 0·52 |
| UQ | T139 = 17·88**, R2 = 0·70 |
Alternative hypotheses are listed in the first column. The variable tested (median or UQ) for species’ salinity and/or alkalinity is given in the second column. Each hypothesis was tested with a parametric test; t-statistic and R2 values are reported for each test. Asterisks indicate statistical significance at the **0·005 level. Significant statistics support the alternative hypotheses.
Testing the correlation between salt and alkalinity tolerance
Correlation of taxa occurring in high predicted salinity and alkalinity.
Our results indicate that species found in conditions of high predicted salinity also tend to be found in conditions of high predicted alkalinity. This is true when considering species’ median and UQ EC and pH (Table 1). The same result is found at the genus level, including when accounting for phylogenetic relatedness (Table 2).
Table 2.
Results of tests for the comparison of predicted EC values for genera including known halophytes versus those not including halophytes, and for the correlation between salinity and alkalinity conditions in Australian grass genera
| Alternative hypothesis | Variable | Parametric | Phylogeny-corrected |
|
|---|---|---|---|---|
| Complete | Calibrated | |||
| Genera with known halophytes are found in conditions of higher predicted salinity than genera without known halophytes | Median | t186 = 3·25**, R2 = 0·04 | t224 = 3·03**, R2 = 0·04 | t144 = 2·45*, R2 = 0·04 |
| UQ | t209 = 3·89**, R2 = 0·06 | t144 = 4·44**, R2 = 0·08 | t144 = 2·46*, R2 = 0·04 | |
| Genera found in conditions of high predicted salinity tend to be found in conditions of high predicted alkalinity | Median | t232 = 11·18**, R2 = 0·35 | t198 = 12·60**, R2 = 0·28 | t116 = 9·02**, R2 = 0·53 |
| UQ | t232 = 15·60**, R2 = 0·51 | t144 = 16·68**, R2 = 0·56 | t109 = 13·75**, R2 = 0·70 | |
| Genera with known halophytes found in conditions of high predicted salinity tend to be found in conditions of high predicted alkalinity | Median | t69 = 6·71**, R2 = 0·40 | t55 = 3·96**, R2 = 0·35 | t45 = 6·22**, R2 = 0·62 |
| UQ | t69 = 9·05**, R2 = 0·54 | t55 = 5·96**, R2 = 0·56 | t43 = 10·33**, R2 = 0·72 | |
Alternative hypotheses are listed in the first column. The variable tested (median or UQ) for salinity and/or alkalinity of a given taxon is given in the second column. Tests for each hypothesis include a parametric and two phylogeny-corrected analyses. The phylogeny-corrected analyses were performed on a complete genus-level phylogenetic tree of grasses (Complete column) and a smaller, time-calibrated phylogenetic tree (Calibrated column) from a previous study (Bouchenak-Khelladi et al., 2010); t-statistic and R2 values are reported for each test. Asterisks indicate statistical significance at the *0·05 level and **0·005 level. Significant statistics support the alternative hypotheses.
Geographical correlation of salinity and alkalinity.
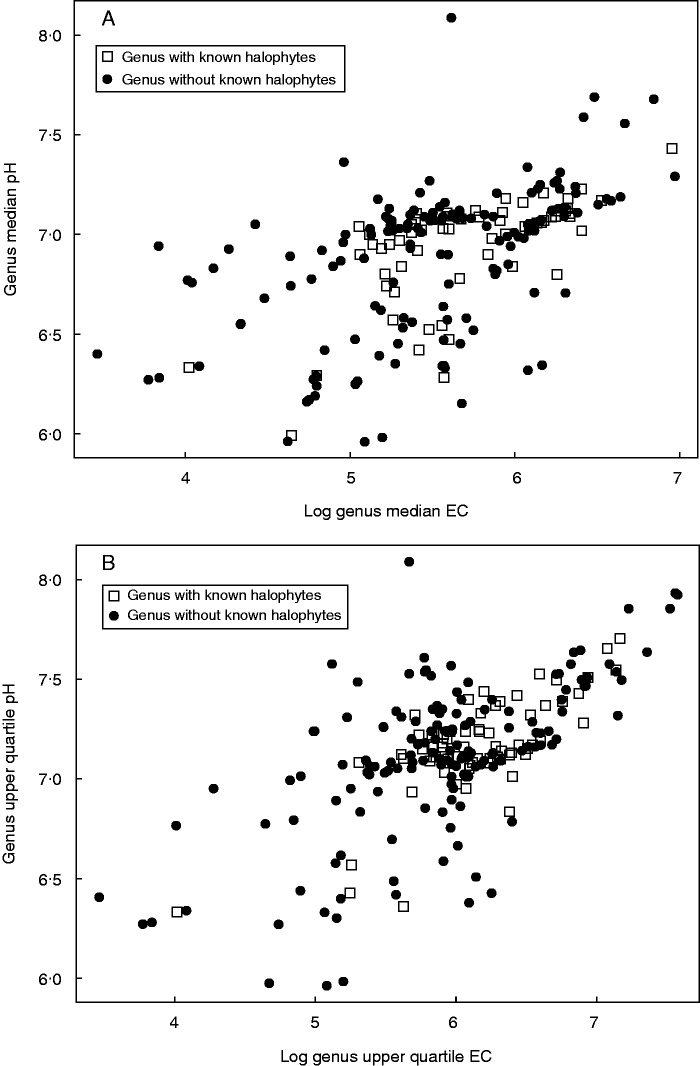
The Pearson correlation coefficient (r) can range between −1 (total negative correlation) and 1 (total positive correlation), with 0 denoting no correlation. The value we recovered for the correlation between predicted EC and pH at localities where species were found is very close to 0 (0·0003), suggesting this correlation is extremely weak. Although we found a significant effect (P < 0·001), this could be due to a weak relationship in a large amount of data points (n = 354 913). We found a stronger correlation between predicted EC values and pH values for salt-tolerant than for non-salt-tolerant taxa, both at the species and at the genus level (Tables 1 and 2, Fig. 1).
Fig. 1.
Correlation between predicted soil salinity and alkalinity for Australian grass genera. The predicted salinity and alkalinity of a given genus is measured as the median (A) and upper quartile (B) value of all predictions of electrical conductivity (EC) or pH, respectively, for all localities where species of that genus occur in mainland Australia. Filled circles are genera that do not include any known halophytes and open squares are genera that include known halophytes.
DISCUSSION
Predicting salt and alkalinity tolerance from species distribution modelling
The motivation for this study was to explore a possible correlation between salt and alkaline tolerance (Bromham et al., 2013; Bui, 2013b; Bui et al., 2014), using Australian grasses as an example. We used a geochemical modelling approach to predict the conditions of salinity and alkalinity in which species occur in their natural distributions (Bui et al., 2014). There are some limitations to this approach. First, our EC predictions were based on measurements in dilute (1 : 5) solutions compared with the salt concentrations that plants would encounter in saline soils. Predicted EC across localities where grass species were found ranged from 0·01 to 10·53 dS m−1, and halophytes are often described as species that complete their life cycles in soils of 8 dS m−1 and above (Aronson, 1989). Very few localities in our dataset were found above that threshold and only four known halophytes are found in these localities. Nevertheless, our geochemical modelling approach was not used to predict species’ absolute tolerances, but rather relative tolerances that can be used in a comparative framework. Second, it is possible that the geochemical modelling does not accurately capture variation in salinity at the scale that is relevant to ecophysiology. Salinity varies on a micro-scale, depending on many factors, such as climate, lithology, topography and vegetation (Bui, 2013a). Plant distributions can be determined by the distribution of salinity at that scale, but that will not necessarily be picked up by these landscape-level estimates.
Because of these possible restrictions, we wanted to evaluate the relevance of our geochemical predictions to plant salt tolerance. To do so, we compared predicted salinity values for known halophytic taxa with the rest of the taxa in our dataset. Using a parametric Welch two-sample t-test, we found that predicted EC for known halophytes is not significantly higher than that for non-halophytes. Nevertheless, when testing this relationship at the genus level, we found that genera with known halophytes have significantly higher predicted soil EC than genera that do not include known halophytes, using a parametric and a phylogeny-corrected approach. This is probably due to the fact that the list of known halophytes in Australian grasses is much more incomplete than the list of genera with known halophytes. Treating unrecognized halophytes that have high predicted EC values as non-halophytes could contribute to the smaller effect size (R2 value) in the species-level analyses compared with the genus-level ones, as we show in Tables 1 and 2. We explored two different values to represent predicted EC for each taxon: median and UQ. Our results show that UQ, representing the more extreme values of EC, is better at predicting clades with halophytes, because the effect size (R) is always larger for UQ values than for median values (Tables 1 and 2). It is problematic that some known halophytes are not found in high predicted EC (Fig. 1), suggesting that our geochemical approach does not identify salt tolerance successfully. However, our predicted EC values have the potential to identify groups of possible halophytes. The main goal of this study was to investigate the correlation between salt and alkaline tolerance. Therefore, as mentioned above, we aimed at generating relative – rather than absolute – tolerances that can be analysed comparatively for all taxa in the dataset.
Patterns of correlation between salt and alkalinity tolerance
Previous studies have found correlations between different types of ecophysiological strategies related to environmental stress tolerance, particularly to water use efficiency. For example, salt-tolerant grasses have evolved more frequently in lineages with C4 photosynthesis, potentially because these lineages can control water loss better than C3 lineages, giving them an advantage to adapt to arid saline environments (Bromham and Bennett, 2014). A correlation was found between salt tolerance, succulence and C4 photosynthesis in chenopods (Kadereit et al., 2012), and a similar evolutionary correlation has been found between CAM photosynthesis and succulence (Ogburn and Edwards, 2010), as well as for occupation of bare environments and to adaptation to harsh elemental soils in the Brassicaceae (Cacho and Strauss, 2014).
Our results suggest that salt and alkaline tolerance are associated: we found that species found in conditions of high predicted salinity tend to be found in conditions of high predicted alkalinity (Table 1). This relationship was also recovered at the genus level, including when correcting for phylogenetic relationships (Table 2). This is in agreement with the recent finding that salt and alkaline tolerance are also linked on the phylogeny of Australian Acacia (Bui et al., 2014). One possible explanation for the association between taxa in high predicted salinity and alkalinity is the presence of ‘enablers’ in some lineages that can facilitate the evolution of multiple stress resistance within those lineages (Edwards and Donoghue, 2013). It could be that some lineages have traits that provide ‘stepping stones’ to developing both salt and alkaline tolerance: that is, lineages may have traits that do not in themselves confer salt tolerance but make it easier for those lineages to evolve tolerance of saline or alkaline conditions.
However, the correlation we find could also be driven by the overlap of salinity and alkalinity in the landscape (Rengasamy, 2010). We assessed the degree to which predicted salinity and alkalinity correlated in localities where Australian grasses are reported. The correlation between EC and pH at species’ localities is significant, but it does not explain much of the variation in our data. Therefore, species exposed to high predicted EC are not necessarily also exposed to high predicted pH at the same localities. For example, as shown in Supplementary Data Fig. S1, the highest predicted EC values are found in both predicted alkaline and acidic soils, and the localities with the highest predicted pH values have low to relatively high predicted EC. Nonetheless, predicted EC values and pH values are more strongly associated for salt-tolerant than for non-salt-tolerant taxa (Tables 1 and 2, Fig. 1). Therefore, we cannot discount the effect of the overlap of salinity and alkalinity in the landscape in shaping the pattern of correlation we found here. Further research is needed to evaluate how much this overlap contributes to the recovered pattern.
As we have demonstrated, geochemical modelling predictions may provide useful starting points for further investigations of macroevolutionary patterns between salt and alkaline tolerance. However, we did not investigate soil ion chemistry across localities where Australian grasses are found, which affects the correlation between salinity and alkalinity in the soil. For example, soil pH between 7 and 10 mainly reflects anions in solution and when neutral salts such as NaCl or Na2SO4 are in solution, sulphate and chloride anions dominate and pH is between 6 and 8 (Rengasamy, 2010). When bicarbonate and carbonate ions dominate, pH rises above 8. At pH above 9, carbonate ions are dominant. Alkaline soils without salt can have pH above 9 but when NaCl is present, pH is lower. However, for pH measured in 1 : 5 soil/water as estimated here, the decrease in pH associated with the presence of NaCl will be diminished by dilution. Also, not all alkaline soils are toxic for plants. For instance, although calcareous soils – abundant in Australia – can be edaphic barriers to plant radiation [e.g. Nullarbor Plain (Crisp and Cook, 2007)], no toxicity has been observed in lime (CaCO3)-dominant soils: although they are alkaline, the solubility of CaCO3 is low and the carbonate concentration is usually around 1 mmol L−1. Given this, we believe more phylogenetic analyses incorporating more complete soil chemistry, as well as testing soil toxicity across sites (Cacho and Strauss, 2014), can lead to more detailed explanations of our reported correlations between salinity and alkalinity for grasses. Furthermore, future investigations could focus on specific traits that might be shared between salinity tolerance and alkalinity tolerance. For example, similar osmotic responses to salinity and alkalinity (Liu et al., 2010; Chen et al., 2011) suggest that some shared mechanisms might be involved in managing water use efficiency under salt and alkaline tolerance. These mechanisms can be investigated experimentally, but a comparative phylogenetic framework may also be useful. For example, species’ geochemical predictions can be analysed in a comparative framework that can reveal the degree to which phylogenetic relatedness or spatial autocorrelation can explain the variation in these datasets (Freckleton and Jetz, 2009).
CONCLUSIONS
In this study, we used distribution data for Australian grasses combined with geochemical modelling to predict the range of values of soil salinity and alkalinity to which species are exposed. The aim of this study was to evaluate the use of geochemical modelling in identifying taxa that can tolerate conditions of high salinity and alkalinity. Therefore, our approach was not used to predict species’ absolute tolerances, but relative tolerances that can be used in a comparative framework. We find that our geochemical predictions, despite their limitations, can identify known halophytic taxa as present in conditions of relatively high salinity. We also found that grass taxa found in areas of high predicted salinity also tend to be found in conditions of high predicted alkalinity. This pattern could suggest a correlation between salt and alkalinity tolerance, for example due to the presence of enabling traits that promote the evolution of salinity and alkalinity tolerance. Our approach provides a valuable test of the use of geochemical modelling to predicting abiotic stress tolerances, beyond those related to temperature and precipitation. Further investigations could consider the phylogenetic distribution of specific traits involved in these ecophysiological strategies, ideally by incorporating more comprehensive and finer scale information on variation of geochemistry in the landscape.
SUPPLEMENTARY DATA
Supplementary data are available online at www.aob.oxfordjournals.org and consist of the following. Figure S1: scatterplot of predicted subsoil EC and pH for each of the 354 913 unique localities where Australian grasses are reported. Table S1: Australian grass halophytes, extracted from Bennett et al. (2013).
ACKNOWLEDGMENTS
We thank Dan Warren for helpful comments during this study. This work was supported by the Australian Research Council.
LITERATURE CITED
- Ackerly DD, Dudley SA, Sultan SE, et al. 2000. The evolution of plant ecophysiological traits: recent advances and future directions. BioScience 50: 979–995. [Google Scholar]
- Aronson J. 1989. HALOPH: a database of salt tolerant plants of the world. Arizona: Office of Arid Land Studies, University of Arizona. [Google Scholar]
- Ashraf M, Foolad MR. 2013. Crop breeding for salt tolerance in the era of molecular markers and marker-assisted selection. Plant Breeding 132: 10–20. [DOI] [PubMed] [Google Scholar]
- Bennett TH, Flowers TJ, Bromham L. 2013. Repeated evolution of salt-tolerance in grasses. Biology Letters 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein L. 1975. Effects of salinity and sodicity on plant growth. Annual Review of Phytopathology 13: 295–312. [Google Scholar]
- Bouchenak-Khelladi Y, Verboom GA, Savolainen V, Hodkinson TR. 2010. Biogeography of the grasses (Poaceae): a phylogenetic approach to reveal evolutionary history in geographical space and geological time. Botanical Journal of the Linnean Society 162: 543–557. [Google Scholar]
- Bromham L, Bennett TH. 2014. Salt tolerance evolves more frequently in C4 grass lineages. Journal of Evolutionary Biology 27: 653–659. [DOI] [PubMed] [Google Scholar]
- Bromham L, Saslis-Lagoudakis CH, Bennett TH, Flowers TJ. 2013. Soil alkalinity and salt tolerance: adapting to multiple stresses. Biology Letters 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui EN. 2013a. Soil salinity: a neglected factor in plant ecology and biogeography. Journal of Arid Environments 92: 14–25. [Google Scholar]
- Bui EN. 2013b. Possible role of soil alkalinity in plant breeding for salt tolerance. Biology Letters 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui EN, Thornhill A, Miller JT. 2014. Salt- and alkaline-tolerance are linked in Acacia. Biology Letters 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacho NI, Strauss SY. 2014. Occupation of bare habitats, an evolutionary precursor to soil specialization in plants. Proceedings of the National Academy of Sciences of the United States of America 111: 15132–15137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapin FS, Autumn K, Pugnaire F. 1993. Evolution of suites of traits in response to environmental stress. American Naturalist 142: S78–S92. [Google Scholar]
- Chen W, Feng C, Guo W, Shi D, Yang C. 2011. Comparative effects of osmotic-, salt- and alkali stress on growth, photosynthesis, and osmotic adjustment of cotton plants. Photosynthetica 49: 417–425. [Google Scholar]
- Clark I, Harper WV. 2000. Practical geostatistics. Columbus, OH: Ecosse North American LLC. [Google Scholar]
- Costa KC, Navarro JB, Shock EL, Zhang CL, Soukup D, Hedlund BP. 2009. Microbiology and geochemistry of great boiling and mud hot springs in the United States Great Basin. Extremophiles 13: 447–459. [DOI] [PubMed] [Google Scholar]
- Crisp MD, Cook LG. 2007. A congruent molecular signature of vicariance across multiple plant lineages. Molecular Phylogenetics and Evolution 43: 1106–1117. [DOI] [PubMed] [Google Scholar]
- de Caritat P, Cooper M. 2011. National Geochemical Survey of Australia: the Geochemical Atlas of Australia. Record 2011/20. Canberra: Geoscience Australia. [Google Scholar]
- Deinlein U, Stephan AB, Horie T, Luo W, Xu G, Schroeder JI. 2014. Plant salt-tolerance mechanisms. Trends in Plant Science 19: 371–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diggle P, Ribeiro P. 2007. Model-based geostatistics. Berlin: Springer. [Google Scholar]
- Edwards EJ, Donoghue MJ. 2013. Is it easy to move and easy to evolve? Evolutionary accessibility and adaptation. Journal of Experimental Botany 64: 4047–4052. [DOI] [PubMed] [Google Scholar]
- Farrell R, Bell D, Akilan K, Marshall J. 1996. Morphological and physiological comparisons of clonal lines of Eucalyptus camaldulensis. II. Responses to waterlogging/salinity and alkalinity. Functional Plant Biology 23: 509–518. [Google Scholar]
- Flowers TJ, Colmer TD. 2008. Salinity tolerance in halophytes. New Phytologist 179: 945–963. [DOI] [PubMed] [Google Scholar]
- Flowers TJ, Troke PF, Yeo AR. 1977. The mechanism of salt tolerance in halophytes. Annual Review of Plant Physiology 28: 89–121. [Google Scholar]
- Flowers TJ, Galal HK, Bromham L. 2010. Evolution of halophytes: multiple origins of salt tolerance in land plants. Functional Plant Biology 37: 604–612. [Google Scholar]
- Freckleton RP, Jetz W. 2009. Space versus phylogeny: disentangling phylogenetic and spatial signals in comparative data. Proceedings of the Royal Society of London. Series B, Biological Sciences 276: 21–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García I, Mendoza R. 2014. Lotus tenuis seedlings subjected to drought or waterlogging in a saline sodic soil. Environmental and Experimental Botany 98: 47–55. [Google Scholar]
- Grafen A. 1989. The phylogenetic regression. Philosophical Transactions of the Royal Society of London B326: 119–157. [DOI] [PubMed] [Google Scholar]
- Guisan A, Thuiller W. 2005. Predicting species distribution: offering more than simple habitat models. Ecology Letters 8: 993–1009. [DOI] [PubMed] [Google Scholar]
- Islam AKMS, Edwards DG, Asher CJ. 1980. pH optima for crop growth. Plant and Soil 54: 339–357. [Google Scholar]
- Ives AR, Midford PE, Garland T., Jr 2007. Within-species variation and measurement error in phylogenetic comparative methods. Systematic Biology 56: 252–270. [DOI] [PubMed] [Google Scholar]
- Kadereit G, Ackerly D, Pirie MD. 2012. A broader model for C4 photosynthesis evolution in plants inferred from the goosefoot family (Chenopodiaceae s.s.). Proceedings of the Royal Society of London. Series B, Biological Sciences 279: 3304–3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak KH, Graham CH, Wiens JJ. 2008. Integrating GIS-based environmental data into evolutionary biology. Trends in Ecology and Evolution 23: 141–148. [DOI] [PubMed] [Google Scholar]
- Liu J, Guo WQ, Shi DC. 2010. Seed germination, seedling survival, and physiological response of sunflowers under saline and alkaline conditions. Photosynthetica 48: 278–286. [Google Scholar]
- Macur RE, Langner HW, Kocar BD, Inskeep WP. 2004. Linking geochemical processes with microbial community analysis: successional dynamics in an arsenic-rich, acid-sulphate-chloride geothermal spring. Geobiology 2: 163–177. [Google Scholar]
- Martins EP, Hansen TF. 1997. Phylogenies and the comparative method: a general approach to incorporating phylogenetic information into the analysis of interspecific data. American Naturalist 149: 646–667. [Google Scholar]
- Munns R. 2005. Genes and salt tolerance: bringing them together. New Phytologist 167: 645–663. [DOI] [PubMed] [Google Scholar]
- Munns R, Tester M. 2008. Mechanisms of salinity tolerance. Annual Review of Plant Biology 59: 651–681. [DOI] [PubMed] [Google Scholar]
- Niinemets Ü, Valladares F. 2006. Tolerance to shade, drought, and waterlogging of temparate Northern Hemisphere trees and shrubs. Ecological Monographs 76: 521–547. [Google Scholar]
- Nishiuchi S, Fujihara K, Liu S, Takano T. 2010. Analysis of expressed sequence tags from a NaHCO3-treated alkali-tolerant plant, Chloris virgata. Plant Physiology and Biochemistry 48: 247–255. [DOI] [PubMed] [Google Scholar]
- Ogburn RM, Edwards EJ. 2010. The ecological water-use strategies of succulent plants. Advances in Botanical Research 55: 179–225. [Google Scholar]
- Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. BioInformatics 20: 289–290. [DOI] [PubMed] [Google Scholar]
- Pinheiro J, Bates D, DebRoy S, Sarkar D, R Core Team. 2014. nlme: linear and nonlinear mixed effects models. R package version 3.1–117. [Google Scholar]
- R Core Team. 2014. R: a language and environment for statistical computing. [Google Scholar]
- Rengasamy P. 2010. Soil processes affecting crop production in salt-affected soils. Functional Plant Biology 37: 613–620. [Google Scholar]
- Revell LJ. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods in Ecology and Evolution 3: 217–223. [Google Scholar]
- Reysenbach A.-L, Shock E. 2002. Merging genomes with geochemistry in hydrothermal ecosystems. Science 296: 1077–1082. [DOI] [PubMed] [Google Scholar]
- Rozema J, Flowers T. 2008. Crops for a salinized world. Science 322: 1478–1480. [DOI] [PubMed] [Google Scholar]
- Rozema J, Schat H. 2013. Salt tolerance of halophytes, research questions reviewed in the perspective of saline agriculture. Environmental and Experimental Botany 92: 83–95. [Google Scholar]
- Saslis-Lagoudakis CH, Moray C, Bromham L. 2014. Evolution of salt tolerance in angiosperms: a phylogenetic approach. In: Rajakaruna N, Boyd B, Harris T, eds. Plant ecology and evolution in harsh environments. New York: Hauppage, 77–95. [Google Scholar]
- Shabala S. 2013. Learning from halophytes: physiological basis and strategies to improve abiotic stress tolerance in crops. Annals of Botany 112: 1209–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shavrukov Y. 2012. Salt stress or salt shock: which genes are we studying? Journal of Experimental Botany 64: 119–127. [DOI] [PubMed] [Google Scholar]
- Tuteja N. 2007. Mechanisms of high salinity tolerance in plants. In: Häussinger D, Sies H, eds. Osmosensing and osmosignaling. San Diego, CA: Elsevier Academic Press, 419–438. [DOI] [PubMed] [Google Scholar]
- United States Salinity Laboratory Staff. 1969. Diagnosis and improvement of saline and alkali soils. Washington, DC: United States Department of Agriculture. [Google Scholar]
- Warren DL, Glor RE, Turelli M. 2008. Environmental niche equivalency versus conservatism: quantitative approaches to niche evolution. Evolution 62: 2868–2883. [DOI] [PubMed] [Google Scholar]
- Yang C, Chong J, Li C, Kim C, Shi D, Wang D. 2007. Osmotic adjustment and ion balance traits of an alkali resistant halophyte Kochia sieversiana during adaptation to salt and alkali conditions. Plant and Soil 294: 263–276. [Google Scholar]
- Yang C, Shi D, Wang D. 2008. Comparative effects of salt and alkali stresses on growth, osmotic adjustment and ionic balance of an alkali-resistant halophyte Suaeda glauca (Bge.). Plant Growth Regulation 56: 179–190. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.