Abstract
Objectives
To determine whether rhinovirus (RV) species is associated with more severe clinical illness in adults.
Methods
Seventy-two RV-positive viral respiratory samples from adult patients were sequenced and analyzed phylogenetically after reverse transcriptase polymerase chain reaction of the region spanning the VP4 gene and 5′ terminus of the VP2 gene. The clinical features and severity of illness associated with the different RV species were compared.
Results
Phylogenetic analysis identified three distinct clusters as RV-A (54%), B (11%), or C (35%) species. In an unadjusted model, patients with RV-B infection were significantly more likely to have the composite outcome variable of death or intensive care unit admission (P = .03), but this effect diminished when controlling for patient sex. A logistic model of the relationship between RV species and adverse outcomes produced nonsignificant odds ratios when controlling for patient sex.
Conclusions
Infection with RV-A or RV-B was associated with greater severity of illness in our adult population; however, the association disappeared after controlling for confounders.
Keywords: Rhinovirus, Severity, Species, Genotype
Rhinoviruses (RVs) are nonenveloped, single-stranded RNA viruses belonging to the Enterovirus genus of the family Picornaviridae. Recently, the International Committee on Taxonomy of Viruses changed the nomenclature to remove host species from picornavirus species names, and the species names are currently known as Rhinovirus A, B, and C instead of human rhinovirus A, B and C.1 They are best known for causing the common cold; indeed, they are responsible for the majority of non–influenza-related viral respiratory tract infections.2,3 In spite of the relatively low morbidity associated with most of these infections, they are responsible for $17 billion in direct health care costs and $22 billion in indirect costs each year in the United States.4 Moreover, the clinical impact of RV infections is not limited to their role in causing the common cold; RVs have been implicated in acute otitis media, sinusitis, and lower respiratory tract disease.2,5–7 Furthermore, viral upper respiratory infections, which are most commonly attributable to RV, cause up to 80% of pediatric asthma exacerbations and half of adult asthma and chronic obstructive pulmonary disease (COPD) exacerbations.8,9 RV not only exacerbates reactive airway disease but may also play a role in the pathogenesis of asthma and COPD via upregulation of proinflammatory mediators and airway remodeling.10
In children, the relationship between RV infection and asthma exacerbation is modified by the particular RV species responsible for the respiratory infection.11 Sequencing of RV capsid-coding regions, noncoding regions, and complete genomes has identified three distinct RV species, RV-A (77 types), RV-B (25 types), and RV-C (50 types).12,13 The term type has replaced serotype, and RV-C species has been described by sequencing, because there is no antigenic typing for RV-C. RV-C has been described in some literature as HRV-A2,14,15 HRV-C,16–19 or HRV-X20; however, in this study all of these will be referred to as RV-C.21,22 The more than 150 RV types differ in the amino acid sequences of their viral capsid proteins, resulting in antigenic variation.7,23 Despite this antigenic diversity, all known RV-A and RV-B types bind only two known cell surface receptors, intracellular adhesion molecule 1 and the low-density lipoprotein receptor, which enable entry through the host cell membrane. The receptor for RV-C has not been identified but is thought to be distinct from the receptors for RV-A and RV-B.2,24 Most typing studies that have assessed RV species with clinical outcomes have been in the pediatric population, and have concluded that RV-A and RV-C are associated with severe outcome.3,11,24–28
In adults the association between species and clinical severity has not been as well characterized, because RV infection typically follows a mild course. In elderly patients and adults with chronic lung disease or compromised immune systems, however, severe outcomes have been observed.28–30 Studies have raised the possibility of severe disease in adults related to particular RV species, though the particular strain responsible for the most severe disease has varied across studies.24,31,32 Accordingly, the relationship between RV species and clinical illness in adults has yet to be defined.
The goal of this study is to determine whether, in an adult patient population, there is a particular RV species that is specifically associated with more severe illness, as measured by surrogate markers of disease severity.
Materials and Methods
Study Site
Emory Healthcare (Atlanta, GA) includes four hospitals, two emergency departments, and a large, multispecialty outpatient clinic with almost entirely adult patients. Approval was obtained for retrospective chart review from the Emory University Institutional Review Board.
Study Inclusion Criteria
Patients were included in the study if they were positive for RV/enterovirus between October 2009 and April 2010. Patients younger than 18 years of age were excluded (n = 3). Ten samples could not be amplified and five medical records were inaccessible; these were excluded from the analyses. Patients whose samples sequenced as coxsackievirus (n = 1), echovirus (n = 1), or enterovirus 68 (n = 6) were also excluded because the study was designed to examine the clinical effect from RV.
Laboratory Testing
Patient respiratory samples (nasopharyngeal swabs or bronchoalveolar lavage) in viral transport media underwent routine clinical testing for RV/enterovirus, influenza A, parainfluenza, adenovirus, metapneumovirus, and RSV by xTAG RVP (Luminex Corp, Austin, TX). Positive samples were archived at −80°C and sequenced as described in the next section.
Sequencing of RV
Seventy-two patient specimens were successfully amplified using reverse transcriptase polymerase chain reaction (RT-PCR) and sequenced with the following primers33: RHINO-FOR, 5′-GGGACCAACTACTTTGGGTGTCCGTGT-3′ (forward) and RHINOREV, 5′-GCATCIGGYARYTTCCACCACCANCC-3′ (reverse). The resulting amplicon of 549 nucleotides encompasses the VP4/VP2 region and the hypervariable region in the 5′-noncoding region. The 5′-noncoding hypervariable sequences were discarded and only the remaining VP4/VP2 coding region sequences were analyzed to assign RV species. Reference sequences were obtained from the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/nucleotide) and used for species assignment of the samples. Eight viruses that did not cluster with RV-A, RV-B, or RV-C were identified as enteroviruses with basic local alignment search tool (BLAST) analysis of each sequence, using the NCBI nucleotide BLAST algorithm (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi).
Data Collection
Electronic medical records of patients who were positive for RV/enterovirus were reviewed. The following clinical data were abstracted: age, race, sex, comorbidities, infections at other body sites, antiviral and antibiotic therapy, length of stay, hospitalization status, intensive care unit (ICU) admission, inpatient mortality, signs and symptoms on presentation, radiographic findings, and laboratory values (white blood cell count, hematocrit, platelet count, aspartate aminotransferase, alanine aminotransferase).
Major comorbidities were grouped by organ system and patients with actively treated malignancies, human immunodeficiency virus (HIV) infection, rheumatologic conditions treated with immunosuppressive therapy, or recipients of solid organ or hematopoietic transplants were considered immunocompromised. Patients were considered to have an infection at another site if they had a bacterial, viral, or fungal infection from any source, or a clinical diagnosis of pneumonia or urinary tract infection at the time of respiratory testing. Antiviral therapy included treatment with oseltamivir, zanamivir, or peramivir. Antibiotic therapy included any antibacterial agent given around the time of respiratory testing. Signs and symptoms were abstracted from the medical record on the day of the medical encounter that resulted in ordering the respiratory viral panel testing. Fever was defined as either a subjective fever or a documented temperature more than 37.8°C. The primary end point was a composite variable consisting of death during inpatient stay or admission to the ICU because these individual outcomes were infrequent.
Statistical Analysis
All analyses were performed using SAS version 9.3 software (SAS Institute, Cary, NC). P values less than .05 were considered to be statistically significant. Univariate analyses were performed and variables that followed an approximately normal distribution were treated as numeric variables in subsequent analyses. Variables that did not follow a normal distribution were converted into categorical variables for analysis. Numeric variables were compared using the Student t test for two-sample comparisons and the analysis of variance for comparisons between more than two groups. Categorical variables were compared using the χ2 or Fisher exact test, where appropriate. Logistic regression was performed to model the relationship between RV species and a composite primary endpoint of death or ICU admission. This model was used to generate odds ratios (ORs) with 95% confidence intervals (CIs) for the study population as a whole and in a post hoc analysis among two subsets of patients: immunocompromised patients and those with pulmonary comorbidities.
Results
Excluded Subjects
Subjects from whom no viral sequences were amplified with RT-PCR were excluded from analysis. Of the 98 total samples, 10 (10%) were not typed. Compared with subjects included, those who were excluded because their samples could not be amplified were significantly more likely to have diabetes (P = .05) and less likely to have hypertension (P = .01). Across other demographic and clinical characteristics, patients whose samples could not be amplified were not significantly different from those who were included in the study. The primary outcome measures of death and ICU admission also did not differ between subtyped and nontyped (excluded) patients (P = .22 and P = .34, respectively.)
Results of Phylogenetic Analysis
Of 2,261 samples analyzed between October 2009 and April 2010, 105 were positive for RV. After exclusions, 72 patients were included in the final analysis Figure 1. Eighty-eight discrete patient samples (Figure 1) underwent sequencing and were determined on phylogenetic analysis to be RV-A (n = 39), RV-B (n = 8), or RV-C (n = 25) Figure 2. RV typing beyond species assignment was not attempted, but clusters of infections were caused by very closely related, single RV types in the species clades for RV-A and RV-C. This is consistent with RV sampling over a single season from a single geographic location. Eight viruses that did not cluster with the RVs were identified as enterovirus 68 (n = 6), coxsackievirus A13 (n = 1), and echovirus 6 (n = 1).
Figure 1.
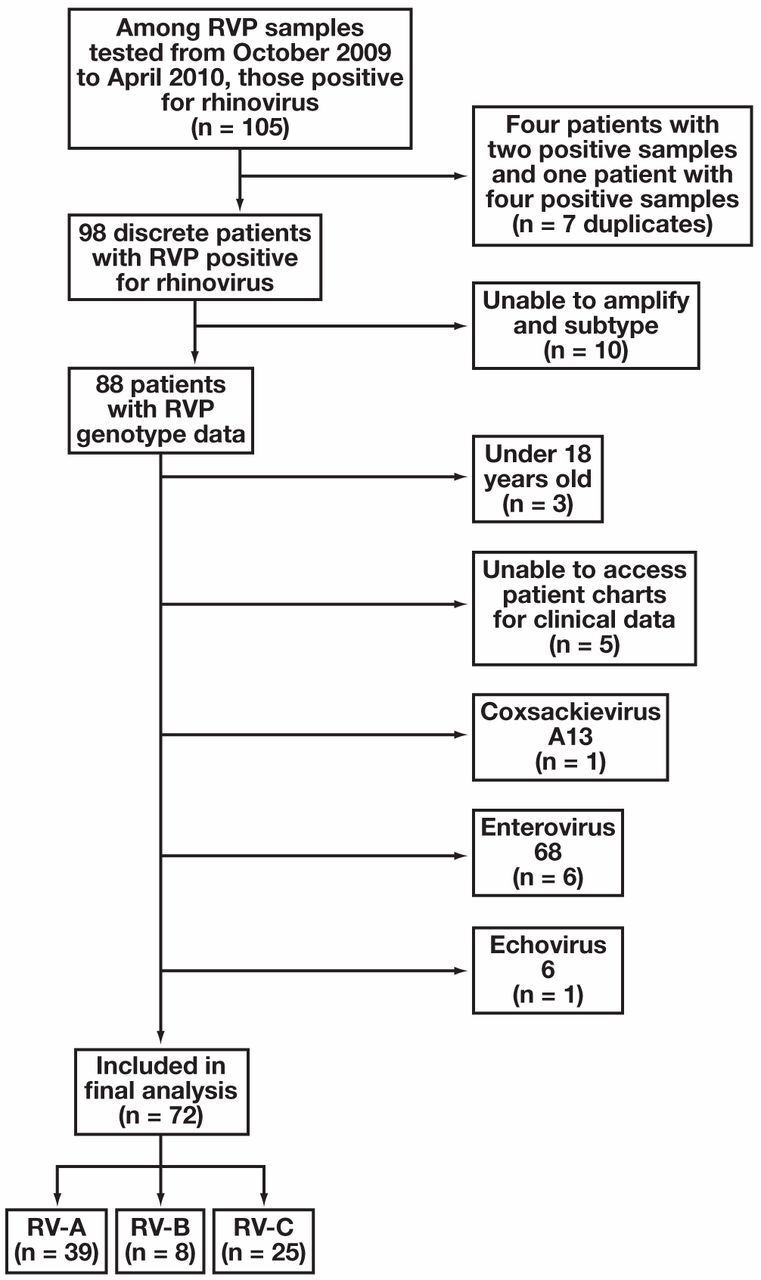
Algorithm of study inclusion beginning with all samples tested within the study time frame. RV, rhinovirus; RVP, respiratory viral panel.
Figure 2.
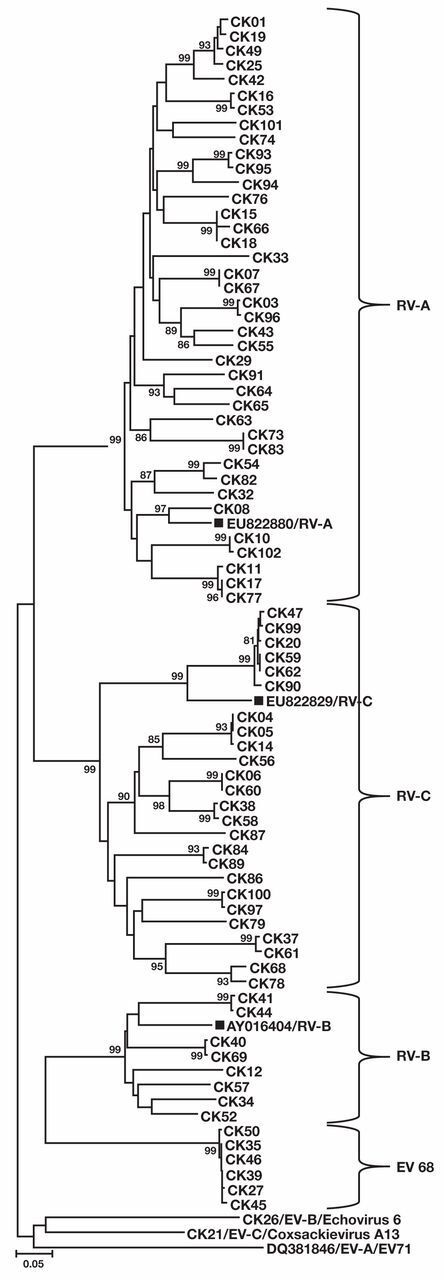
A neighbor-joining tree was constructed, using the VP4/VP2 coding region nucleic-acid sequences, for 72 clinical samples from adults with rhinovirus (RV), grouped in species using reference sequences from the National Center for Biotechnology Information (EU822880 [RV-A], EU822829VP [RV-C], AY016404-B [RV-B] and denoted with solid squares). In addition, three enterovirus (EV) types were identified from patients, including echovirus 6 (species EV-B), coxsackievirus A13 (EV-C), and EV68 (EV-D). EV71 (DQ381846) is included as a representative of the EV-A species. Bootstrap analysis used 1,000 pseudoreplicates. Values greater than 80 are shown on the tree. Scale bar indicates nucleotide changes per site. The tree is unrooted.
Demographic and Clinical Characteristics
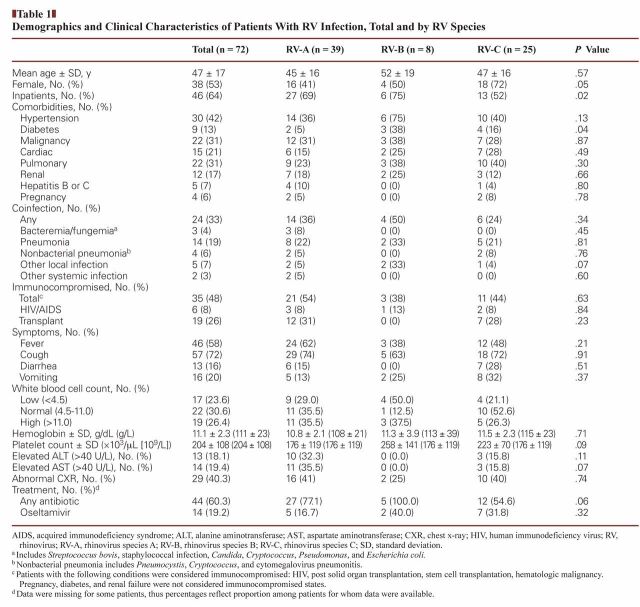
The majority of patients were hospitalized (64%). The mean age was 47 years (range, 18–82 years); 53% were female; 30% had an underlying diagnosis of asthma, bronchitis, COPD, or other pulmonary comorbidity; and 19% had bacterial pneumonia as diagnosed by the clinical team. Forty-nine percent were immunocompromised, of whom 17% were HIV positive and 54% were solid organ transplant recipients. Hypertension (42%), diabetes (13%), and malignancy (30%) were also common. The proportion of female patients was significantly different among the three RV groups (P = .05), as was the proportion of patients with diabetes (P = .04). Otherwise, the groups were demographically similar Table 1.
Table 1.
Demographics and Clinical Characteristics of Patients With RV Infection, Total and by RV Species
RV was the only respiratory virus found in all but two of the 72 subjects. Among RV-positive patients, one (RV-A) also had parainfluenza and one (RV-C) had influenza A. Neither patient died or was admitted to the ICU. None of the patients were infected with more than one type of RV.
Across all RV groups, the majority of patients had a cough and a minority had diarrhea and vomiting. The prevalence of these symptoms did not differ significantly across groups. Laboratory findings, including complete blood counts and liver function tests (transaminases), were similar across groups, as was the prevalence of abnormal chest radiographs.
Patient Outcomes: Univariate Analysis
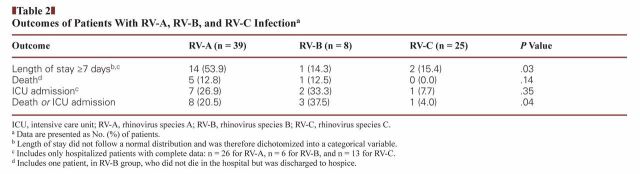
The proportion of patients with a length of stay greater than 7 days was significantly different among the RV species groups (P = .03), with 53.9% of RV-A patients and 14.3% of RV-B patients being hospitalized for longer than 7 days Table 2. The proportion of patients admitted to the ICU was not significantly different among RV species groups, nor was the proportion of patients in each group who died. The proportion of patients whose outcome was either death or ICU admission was significantly different among RV species (P = .04): this outcome occurred in 38% of RV-B patients, 21% of RV-A patients, and 4% of RV-C patients.
Table 2.
Outcomes of Patients With RV-A, RV-B, and RV-C Infectiona
Logistic Model
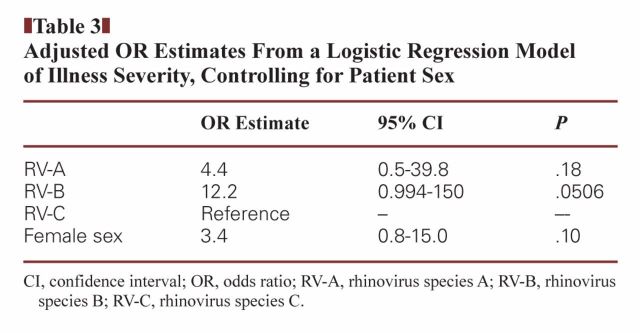
The unadjusted logistic model of the relationship between RV species and clinical disease severity demonstrated that the odds of death or ICU admission among patients with RV-B infection was 15 times that of patients with RV-C infection (OR, 15.0; 95% CI, 1.3–175.3, P = .03). RV-C infection was chosen as the reference group given that greater severity in children had been reported. For RV-A, OR was not significant. An adjusted model Table 3 was constructed, controlling for patient sex, which was identified as the only variable associated with both the RV species and the outcome measure. In the adjusted model, the relationship between RV-A (OR = 4.4, 95% CI = 0.5–39.8) or RV-B (OR = 12.2, 95% CI = 0.994–150) and the composite outcome measure of death or ICU admission was not significant at the 0.05 alpha level.
Table 3.
Adjusted OR Estimates From a Logistic Regression Model of Illness Severity, Controlling for Patient Sex
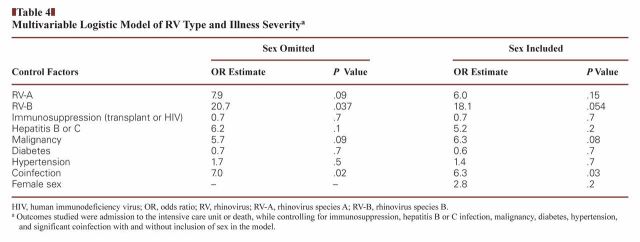
Death and ICU admission in these patients did not result exclusively from respiratory illness; these outcomes tended to be multifactorial. Inclusion of other determinants of health status, however, did not change the results of the model of the relationship between RV subtype and adverse outcome. Additional models were constructed to evaluate the contribution of comorbidities to the outcome; in these models, only the presence of significant coinfection produced a P value less than .05 Table 4.
Table 4.
Multivariable Logistic Model of RV Type and Illness Severitya
Immunocompromised Patients and Patients With Pulmonary Comorbidities
No RV species was associated with greater clinical severity in analyses of subpopulations with pulmonary comorbidities (primarily asthma and COPD). Pulmonary comorbidities were also not independently associated with increased likelihood of severe illness. Additional analyses examining the relationship between RV and severity of clinical illness in immunocompromised patients found no significant relationship between RV species and severity of clinical illness. The relationship between immunocompromised status and severity of illness was also not significant (data not shown).
Discussion
This study has described the relationship between RV species and severity of clinical illness in an adult population. In unadjusted pairwise comparisons, RV-A and RV-B were associated with greater severity of illness compared with RV-C infection, as measured by the composite proxy outcome measure of ICU admission or death. An analysis of potential confounders, considering patient sex, age, immunocompromised status, coinfection, pneumonia, and malignancy, identified only sex as significantly associated with both RV species and the outcomes of interest. Female sex was significantly associated both with severe disease and with RV species and was thus identified as a confounder that should be included in the adjusted model. When patient sex was included in the logistic model, the relationship between RV-A and RV-B and more severe disease was no longer significant.
The key question that emerges from these findings is whether the lack of association that emerged after controlling for other variables reflects a true null result or whether the small sample size in this study limited our ability to detect an existing difference. Previous analysis of RV species and severity of clinical illness in adults has found that RV-A and RV-B species were associated with influenza-like illness among adult patients but RV-C was not.32 The significant findings from our unadjusted model, in which RV-A and RV-B were associated with more severe disease than RV-C, are consistent with these previous studies. In the logistic model controlling for patient sex, however, the estimated ORs for the relationship between RV species and clinical outcome were no longer significant. Of note, these ORs remained in the positive direction, and for RV-B, the P value was .051. It is therefore plausible that, given a larger sample size, the significant association between these species and more severe illness would have persisted even after controlling for potential confounders.
Sex differences in cellular immunity to RV have been described in the literature and are thought to be related to hormonal influences on the immune system.34 Whether this difference in immunity translates into differences in clinical outcomes, however, is unclear. In this study, patient sex was significantly associated with both RV species and the primary composite outcome variable of death or ICU admission. Including sex in the logistic model produced a marginally significant odds ratio of 3.4 for the effect of patient sex on adverse outcomes (P = .10). That is, the odds of adverse outcome (ICU admission or death) are 3.4 times higher for female patients than for male patients. This lends support to the idea that differences in the adaptive immune response to RV infection could in fact translate to dissimilar clinical outcomes in men and women.
Sex differences in the immune response to RV infection also have potential implications for the role of RV infection in the pathogenesis of reactive airway disease. If RV contributes to the development of asthma via the immune/inflammatory response it provokes, and women have a stronger response, perhaps RV infection contributes to sex differences in asthma prevalence. From puberty onward, the incidence, prevalence, and severity of asthma is greater in women than in men; hormonal factors are thought to influence airway hyperresponsiveness.35
This study elucidates some of the unique aspects of the molecular and clinical epidemiology of RV infection in adult populations and highlights some important differences with the pediatric literature, in which RV-C infection has been associated with more severe illness. Our findings suggest that in adults with RV infection, species association differences in clinical outcomes are not the same as in children, and may not exist at all. If these differences do exist in adults they may be related to these patients’ immunocompromised status, sex, and/or coexisting illnesses.
Footnotes
This study was supported by IDSA Medical Scholars Award (D.J.M.) and grants NIH/NCRR KL2 TR000455 and UL1TR000454 from the National Institutes of Health (C.S.K.).
References
- 1. King AMQ, Lefkowitz E, Adams MJ, et al., eds. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. London, England: Academic Press; 2012. [Google Scholar]
- 2. Winther B. Rhinovirus infections in the upper airway. Proc Am Thorac Soc. 2011;8:79–89. [DOI] [PubMed] [Google Scholar]
- 3. Piralla A, Baldanti F, Gerna G. Phylogenetic patterns of human respiratory picornavirus species, including the newly identified group c rhinoviruses, during a 1-year surveillance of a hospitalized patient population in Italy. J Clin Microbiol. 2011;49:373–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fendrick AM, Monto AS, Nightengale B, et al. The economic burden of non-influenza-related viral respiratory tract infection in the United States. Arch Intern Med. 2003;163:487–494. [DOI] [PubMed] [Google Scholar]
- 5. van Piggelen RO, van Loon AM, Krediet TG, et al. Human rhinovirus causes severe infection in preterm infants. Pediatr Infect Dis J. 2010;29:364–365. [DOI] [PubMed] [Google Scholar]
- 6. Kiang D, Yagi S, Kantardjieff KA, et al. Molecular characterization of a variant rhinovirus from an outbreak associated with uncommonly high mortality. J Clin Virol. 2007;38:227–237. [DOI] [PubMed] [Google Scholar]
- 7. Henquell C, Mirand A, Deusebis AL, et al. Prospective genotyping of human rhinoviruses in children and adults during the winter of 2009–2010. J Clin Virol. 2012;53:280–284. [DOI] [PubMed] [Google Scholar]
- 8. Friedlander SL, Busse WW. The role of rhinovirus in asthma exacerbations. J Allergy Clin Immunol. 2005;116:267–273. [DOI] [PubMed] [Google Scholar]
- 9. McManus TE, Marley AM, Baxter N, et al. Respiratory viral infection in exacerbations of COPD. Respir Med. 2008;102:1575–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tacon CE, Wiehler S, Holden NS, et al. Human rhinovirus infection upregulates MMP-9 production in airway epithelial cells via NF-{kappa}B. Am J Respir Cell Mol Biol. 2010;43:201–209. [DOI] [PubMed] [Google Scholar]
- 11. Bizzintino J, Lee WM, Laing IA, et al. Association between human rhinovirus C and severity of acute asthma in children. Eur Respir J. 2011;37:1037–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. International Committee on Taxonomy of Viruses Available from: http://www.picornaviridae.com/enterovirus/enterovirus.htm. Accessed June 12, 2014.
- 13. Palmenberg AC, Spiro D, Kuzmickas R, et al. Sequencing and analyses of all known human rhinovirus genomes reveal structure and evolution. Science. 2009;324:55–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Arden KE, McErlean P, Nissen MD, et al. Frequent detection of human rhinoviruses, paramyxoviruses, coronaviruses, and bocavirus during acute respiratory tract infections. J Med Virol. 2006;78:1232–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McErlean P, Shackelton LA, Lambert SB, et al. Characterisation of a newly identified human rhinovirus, HRV-QPM, discovered in infants with bronchiolitis. J Clin Virol. 2007;39:67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lamson D, Renwick N, Kapoor V, et al. MassTag polymerase-chain-reaction detection of respiratory pathogens, including a new rhinovirus genotype, that caused influenza-like illness in New York State during 2004–2005. J Infect Dis. 2006;194:1398–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lau SK, Yip CC, Tsoi HW, et al. Clinical features and complete genome characterization of a distinct human rhinovirus (HRV) genetic cluster, probably representing a previously undetected HRV species, HRV-C, associated with acute respiratory illness in children. J Clin Microbiol. 2007;45:3655–3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee WM, Kiesner C, Pappas T, et al. A diverse group of previously unrecognized human rhinoviruses are common causes of respiratory illnesses in infants. PLoS One. 2007;2:e966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McErlean P, Shackelton LA, Andrews E, et al. Distinguishing molecular features and clinical characteristics of a putative new rhinovirus species, human rhinovirus C (HRV C). PLoS One. 2008;3:e1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kistler A, Avila PC, Rouskin S, et al. Pan-viral screening of respiratory tract infections in adults with and without asthma reveals unexpected human coronavirus and human rhinovirus diversity. J Infect Dis. 2007;196:817–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Simmonds P, McIntyre C, Savolainen-Kopra C, et al. Proposals for the classification of human rhinovirus species C into genotypically assigned types. J Gen Virol. 2010;91:2409–2419. [DOI] [PubMed] [Google Scholar]
- 22. Rueter K, Bizzintino J, Martin AC, et al. Symptomatic viral infection is associated with impaired response to treatment in children with acute asthma. J Pediatr. 2012;160:82–87. [DOI] [PubMed] [Google Scholar]
- 23. Kennedy JL, Turner RB, Braciale T, et al. Pathogenesis of rhinovirus infection. Curr Opin Virol. 2012;2:287–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lau SK, Yip CC, Lin AW, et al. Clinical and molecular epidemiology of human rhinovirus C in children and adults in Hong Kong reveals a possible distinct human rhinovirus C subgroup. J Infect Dis. 2009;200:1096–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Broberg E, Niemela J, Lahti E, et al. Human rhinovirus C—associated severe pneumonia in a neonate. J Clin Virol. 2011;51:79–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fuji N, Suzuki A, Lupisan S, et al. Detection of human rhinovirus C viral genome in blood among children with severe respiratory infections in the Philippines. PLoS One. 2011;6:e27247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xiang Z, Gonzalez R, Xie Z, et al. Human rhinovirus C infections mirror those of human rhinovirus A in children with community-acquired pneumonia. J Clin Virol. 2010;49:94–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Louie JK, Yagi S, Nelson FA, et al. Rhinovirus outbreak in a long term care facility for elderly persons associated with unusually high mortality. Clin Infect Dis. 2005;41:262–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Falsey AR, Walsh EE, Hayden FG. Rhinovirus and coronavirus infection-associated hospitalizations among older adults. J Infect Dis. 2002;185:1338–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kraft CS, Jacob JT, Sears MH, et al. Severity of human rhinovirus infection in immunocompromised adults is similar to 2009 H1N1 influenza. J Clin Microbiol. 2012;50:1061–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Longtin J, Marchand-Austin A, Winter AL, et al. Rhinovirus outbreaks in long-term care facilities, Ontario, Canada. Emerg Infect Dis. 2010;16:1463–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Watanabe A, Carraro E, Kamikawa J, et al. Rhinovirus species and their clinical presentation among different risk groups of non-hospitalized patients. J Med Virol. 2010;82:2110–2115. [DOI] [PubMed] [Google Scholar]
- 33. Piralla A, Rovida F, Campanini G, et al. Clinical severity and molecular typing of human rhinovirus C strains during a fall outbreak affecting hospitalized patients. J Clin Virol. 2009;45:311–317. [DOI] [PubMed] [Google Scholar]
- 34. Carroll ML, Yerkovich ST, Pritchard AL, et al. Adaptive immunity to rhinoviruses: sex and age matter. Respir Res. 2010;11:184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sears MR. Epidemiology of asthma exacerbations. J Allergy Clin Immunol. 2008;122:662–668; quiz 669–670. [DOI] [PubMed] [Google Scholar]