Abstract
The basidiomycete Coprinopsis cinerea contains a quinohemoprotein (CcPDH named as CcSDH in our previous paper), which is a new type of pyrroloquinoline-quinone (PQQ)-dependent pyranose dehydrogenase and is the first found among all eukaryotes. This enzyme has a three-domain structure consisting of an N-terminal heme b containing a cytochrome domain that is homologous to the cytochrome domain of cellobiose dehydrogenase (CDH; EC 1.1.99.18) from the wood-rotting basidiomycete Phanerochaete chrysosporium, a C-terminal family 1-type carbohydrate-binding module, and a novel central catalytic domain containing PQQ as a cofactor. Here, we describe the biochemical and electrochemical characterization of recombinant CcPDH. UV-vis and resonance Raman spectroscopic studies clearly reveal characteristics of a 6-coordinated low-spin heme b in both the ferric and ferrous states, as well as intramolecular electron transfer from the PQQ to heme b. Moreover, the formal potential of the heme was evaluated to be 130 mV vs. NHE by cyclic voltammetry. These results indicate that the cytochrome domain of CcPDH possesses similar biophysical properties to that in CDH. A comparison of the conformations of monosaccharides as substrates and the associated catalytic efficiency (k cat/K m) of CcPDH indicates that the enzyme prefers monosaccharides with equatorial C-2, C-3 hydroxyl groups and an axial C-4 hydroxyl group in the 1C4 chair conformation. Furthermore, a binding study shows a high binding affinity of CcPDH for cellulose, suggesting that CcPDH function is related to the enzymatic degradation of plant cell wall.
Introduction
Cellulolytic fungi are known to produce many extracellular oxidoreductases, such as peroxidase and laccases, as well as cellulolytic enzymes [1–4]. Recently, we discovered a pyranose dehydrogenase (CcPDH named as CcSDH in our previous paper) that represents a new type of pyrroloquinoline quinone (PQQ) quinohemoprotein from the basidiomycete Coprinopsis cinerea (formally known as Coprinus cinereus) strain 5338. This CcPDH is the first such protein found among all eukaryotes [5]. The amino acid sequence of CcPDH indicates that the extracellular enzyme contains three domains: an N-terminal cytochrome domain, a C-terminal carbohydrate binding module (CBM), and a central PQQ domain.
The N-terminal cytochrome domain of CcPDH has 32 to 42% identity with the cytochrome b domains of basidiomycete cellobiose dehydrogenases (CDHs). CDH is a major oxidoreductase secreted by many cellulolytic fungi during growth on cellulose, and the genes that code for CDHs have been identified in numerous cellulolytic fungi [6]. CDH is an extracellular flavohemoprotein that consists of a catalytic FAD containing dehydrogenase domain, a heme b containing a cytochrome domain, and a linker region connecting the two domains. CDH catalyzes the dehydrogenation of cellobiose and cello-oligosaccharides to their corresponding δ-lactones. The oxidation of cellobiose takes place in the FAD domain, subsequently followed by inter-domain electron transfer to the cytochrome domain [7]. The cytochrome domain of CDH has unique structural features that fold into an antiparallel ß-sandwich and contains a 6-coordinate low-spin b-type heme with unusual Met-His ligands [8]. Because the cytochrome domain has the ability to communicate electrically with an electrode, CDH exhibits efficient direct electron transfer (DET) on an electrode surface and is an attractive biocatalyst for use in enzymatic biosensors or biofuel cells [9, 10]. According to the carbohydrate-active enzyme database (CAZy; www.cazy.org), the FAD and cytochrome domains of CDH are classified in subfamily 1 of Auxiliary Activity family 3 (AA3_1) and AA8, respectively [11]. CcPDH has an AA8 cytochrome domain but a cellulose-binding domain attached to a PQQ domain instead of an FAD domain.
Several known prokaryotic dehydrogenases contain PQQ as their prosthetic group and mainly catalyze the oxidation of a number of alcohols or aldose sugars in the periplasm of Gram-negative bacteria [12–14]. In these dehydrogenases, PQQ is tightly but not covalently bound to the active site, which contains a divalent metal ion such as calcium [15]. Some of these PQQ quinoproteins contain an additional one or more hemes and are called quinohemoproteins [16, 17]. The known quinohemoproteins containing PQQ or other quinone cofactors almost exclusively have c-type heme cofactors [17, 18]. There are two types of PQQ-dependent glucose dehydrogenases: a membrane-bound form (mGDH) which has been identified in a wide range of bacteria [19, 20] and a soluble form (sGDH) that has been well described in Acinetobacter calcoaceticus [21, 22]. In addition, soluble aldose sugar dehydrogenases (Asd) from Escherichia coli [23] and Pyrobaculum aerophilum [24] are PQQ quinoproteins that have been identified relatively recently as homologues of the A. calcoaceticus sGDH. These Asds have oxidation activity towards a broad range of aldose sugars, with a weak affinity for substrates. The central PQQ domain of CcPDH has low identity with sGDH from A. calcoaceticus (16%). The putative homologues of the PQQ domain of CcPDH are widely distributed in bacteria, archaea, amoebozoa, and fungi. These quinoproteins belong to a different phylogenetic category from the known PQQ-quinoprotein family [5]. Therefore, the PQQ domain of CcPDH is currently identified as a new Auxiliary Activities family 12 (AA12) in the CAZy database.
Many carbohydrate active enzymes including hydrolytic and non-hydrolytic proteins contain one or more non-catalytic CBMs that have functions in recognizing and adhering to carbohydrates [25–27]. Based on amino acid sequence similarities and three-dimensional structures, CBMs are currently grouped into 71 families in the CAZy database [28]. The amino acid sequence of CcPDH indicates the presence of a family 1 CBM (CBM1). The majority of cellulose-binding domains attached to fungal cellulolytic enzymes belong to CBM1. The main role of CBM1 in cellulolytic enzymes is to enhance the hydrolysis of crystalline cellulose by increasing the effective enzyme concentration on the cellulose surface [29, 30]. CBM1s consist of less than 40 residues and usually contain two disulfide bonds and three solvent-exposed aromatic amino acids as critical residues for binding. CBM1s form a flat hydrophobic surface for adsorption to the hydrophobic surface of crystalline cellulose [31–35]. The application of CBMs for development of new carbohydrate-recognition technologies, such as affinity chromatography, have previously been examined [36].
As noted above, CcPDH is an attractive multi-domain oxidoreductase that has three unique domains with different backgrounds. Until now, CcPDH had been overexpressed in the methylotrophic yeast Pichia pastoris, and initial characterization of the recombinant protein has demonstrated the PQQ-dependence of the enzyme reactivity for the first time in a eukaryotic enzyme. Despite its low amino acid sequence homology with known PQQ-binding enzymes, this enzyme binds PQQ strongly and shows oxidation activity towards some sugars. In this study, we thoroughly investigated the enzymatic and electrochemical properties of recombinant CcPDH.
Materials and Methods
Materials
Pyrroloquinoline quinone and equine heart cytochrome c were purchased from Sigma-Aldrich Japan G. K. (Tokyo, Japan). 2-Keto-D-glucose was purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA). l-fucose, l-lyxose, and l-xylose were purchased from Wako Pure Chemical Industries (Osaka, Japan). d-arabinose and l-mannose were purchased from Sigma-Aldrich Japan G. K. (Tokyo, Japan). l-galactose, d-talose, and l-rhamnose were purchased from Funakoshi Co., Ltd. (Tokyo, Japan). l-gulose and l-glucose were purchased from Tokyo Chemical Industry Co., Ltd. (Tokyo, Japan).
Preparation of CcPDH and DHPDH
Recombinant intact CcPDH and PQQ containing dehydrogenase domain (DHPDH) of CcPDH (DHPDH: residues 240–649) were heterologously expressed in the methylotrophic yeast Pichia pastoris and purified as described previously [5]. The enzyme represented as “CcPDH” in this paper means the heterologously expressed wild type. The purity of CcPDH was confirmed by SDS/PAGE and by absorption spectroscopy. The CcPDH concentration in the solution was estimated from the absorbance at 420 nm (ε420 = 130 mM-1 cm-1) [5].
UV-vis spectroscopy
Electronic absorption spectra of CcPDH and DHPDH were recorded at room temperature with a JASCO V-660 spectrophotometer (Tokyo, Japan). CcPDH was diluted in 50 mM HEPES buffer, pH 7.0, containing 1 μM PQQ and 1 mM CaCl2. Ca2+ is ligated with PQQ and several amino acid side chain atoms and is essential for the formation of active holo-enzyme of quinoproteins. The reduced form of CcPDH was prepared by addition of 1 mM ascorbic acid or l-fucose. Apo-DHPDH was stored in 50 mM HEPES buffer, pH 7.0, containing 1 mM CaCl2. Holo-DHPDH was obtained by addition of PQQ to apo-DHPDH according to our previous report [37]. The reduced form of DHPDH was prepared by addition of 1 mM l-fucose.
Resonance Raman spectroscopy
Resonance Raman (RR) spectra were measured on a JASCO NRS-1000 spectrometer using a Kaiser Optical holographic notch-plus filter and a liquid N2-cooled CCD detector (Princeton instrument, Spec-10, Trenton, NJ, U.S.A.). Raman scattering was excited by the 413.1 nm emission from a Kr ion laser (Innova 90C-K, Coherent Inc., Santa Clara, CA, U.S.A.), and the incident powers were 1 mW. The spectra were obtained with data accumulated over 30 sec and a spectral resolution of 1.47 cm-1. The spectra were calibrated against indene and CCl4 as external standards. A two-point baseline was performed with GRAMS/386 software. All measurements were carried out in 50 mM HEPES buffer (pH 7.0) containing 1 μM PQQ and 1 mM CaCl2, at room temperature. The reduced form of CcPDH was prepared by the addition of 1 mM ascorbic acid or l-fucose.
Measurement of midpoint potential
Redox responses of heme in CcPDH were analyzed by cyclic voltammetry (ALS Electrochemical Analyzer 702B, BAS Inc., Tokyo, Japan). The potential was determined by averaging the anodic and cathodic peak potentials. The electrochemical responses of CcPDH were obtained by using a carbon nanoparticle-modified plastic-formed carbon as a working electrode, as described elsewhere [38]. An Ag/AgCl (3 M NaCl) reference electrode (209 mV vs. NHE) and a platinum wire were used as the reference and counter electrodes, respectively. All measurements were performed in 50 mM HEPES buffer (pH 7.0) containing 1 μM PQQ and 1 mM CaCl2 using a three-electrode cell under a nitrogen atmosphere at room temperature.
Enzyme assays and kinetic procedure
The enzyme activities of CcPDH were assayed using cytochrome c as an electron acceptor [5, 39, 40]. The assay was performed by photometrically monitoring the time-dependent reduction of cytochrome c at 550 nm (Δε550 = 17.5 mM-1 cm-1) at 30°C. To obtain the kinetic parameters of oxidation for l-galactose, d-talose, l-xylose, and l-glucose, the assay was performed with various concentrations of monosaccharides (5–200 mM for l-galactose, 2.5–200 mM for d-talose, 5–300 mM for l-xylose, and 10–400 mM for L-glucose) in 50 mM Tris buffer, pH 8.5 containing 1 μM PQQ and 1 mM CaCl2. To determine the Michaelis constant (K m) and turnover number (k cat), experimental data were fitted with the Michaelis-Menten equation with using Origin 8.1 (OriginLab, Inc., Northampton, MA, U.S.A.) The activity of DHPDH was determined using a dye-linked assay with the artificial electron acceptors phenazine methosulfate (PMS) as a primary electron acceptor and 2,6-dichlorophenolindophenol (DCPIP), according to our previous report [37]. The effect of PQQ binding on the activity of DHPDH toward l-fucose was measured in 50 mM MES buffer, pH 6.5, containing 0.1 mM DCPIP, 1 mM PMS, 1 mM CaCl2, 100 mM l-fucose, 50 nM DHPDH, and varying concentrations of PQQ (2.5–100 nM). The reaction rate was measured by following the decrease in the absorbance of DCPIP at 520 nm (Δε520 = 6.9 mM-1 cm-1) [41] at 30°C using UV-vis spectroscopy.
Sequence analysis of CBM1 of CcPDH
The amino acid sequences of CBM1s within carbohydrate-active enzymes were collected from the CAZy database. The CBM1 sequences and the CcPDH sequence were subjected to multiple-alignment analysis using MAFFT (ver. 6.85) [42, 43], and the figure was prepared using ESPript ver 2.3 [44].
Adsorption of CcPDH on celluloses
Adsorption experiments were carried out in 50 mM HEPES buffer, pH 7.0, using crystalline cellulose from Cladophora sp. and phosphoric-acid-swollen cellulose (PASC) prepared from Avicel according to the method described previously [45] as an amorphous substrate. Various concentrations of CcPDH (0.5–32 μM) were incubated with 0.1% w/v celluloses for 1 h at 30°C and then were centrifuged for 20 min at 16,100 g. The absorbance at 420 nm, which corresponded to the Soret band of a b-type heme (ε420 = 130 mM-1 cm-1), was measured with UV-vis spectroscopy to determine the concentration of free enzyme in the supernatants. The equilibrium dissociation constant (K d) and the maximum amount of adsorbed enzyme (A max) were estimated as described previously [46, 47].
Results
UV-visible spectroscopy of CcPDH
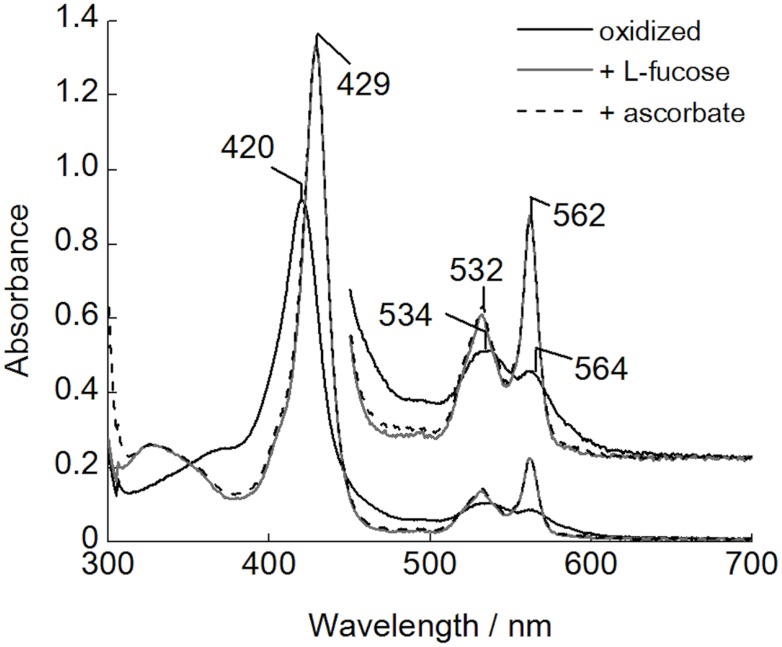
The absorption spectra of the oxidized CcPDH are shown in Fig. 1. The enzyme showed a typical hemoprotein spectrum with an absorption maximum at 420 nm and broad peaks at 534 nm and 564 nm, corresponding to the γ (Soret)-, β-, and α-bands, respectively. The reduction of CcPDH by ascorbic acid, sodium dithionite, or l-fucose, which is a substrate, induced the same behavior, and no difference was observed in the resulting spectra. The absorbances of the γ-, β-, and α-bands were increased and the maxima of the bands shifted slightly to 429 nm, 532 nm, and 562 nm, respectively. The spectra shifts due to the reaction with l-fucose suggests that the electron generated by l-fucose can be transferred from the PQQ domain to the cytochrome domain, like that from the FAD domain to the cytochrome domain in the cellobiose oxidation of CDH.
Fig 1. UV-visible absorption spectra of CcPDH.
Black solid line, oxidized form; gray solid line, reduced form by addition of L-fucose; dotted line, reduced form prepared by addition of ascorbic acid. All spectra were recorded in 50 mM HEPES buffer, pH 7.0, at room temperature.
Resonance Raman spectroscopy
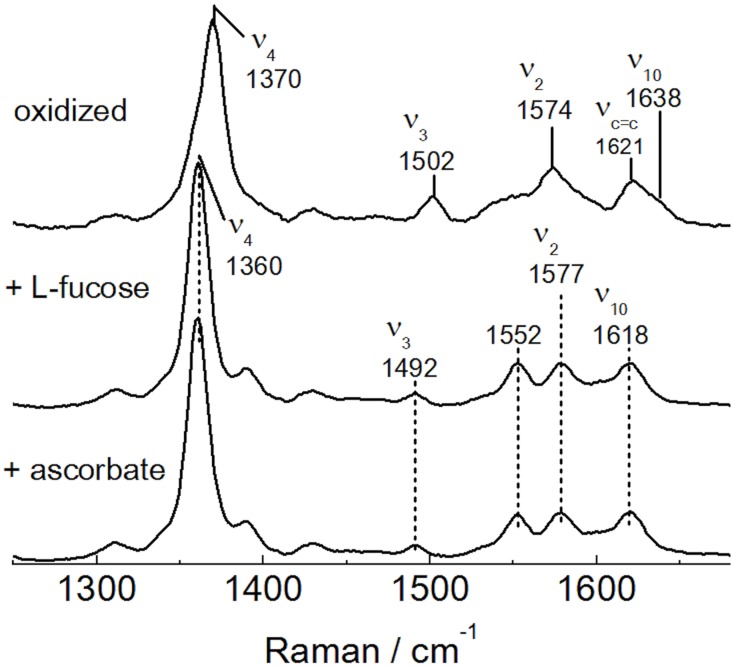
Both the oxidized and reduced heme cofactors are RR active chromophores, and the vibrational modes can be enhanced with laser excitation in the 400–500 nm range. To further investigate the properties of the heme in CcPDH, RR spectra were measured for both the oxidized and reduced forms of CcPDH with excitation at 413 nm as shown in Fig. 2. The oxidation marker ν4 of CcPDH in its oxidized form appears at 1370 cm-1, indicating a ferric heme. The core size marker bonds ν2 and ν3 of the oxidized form, occurring at 1574 and 1502 cm-1, respectively, identifies the ferric heme as having a hexacoordinate low-spin (6cls) heme state. The reduced form of CcPDH prepared with either ascorbic acid or l-fucose showed the same RR spectral bands with the ν2, ν3, and ν4 to 1577, 1492, and 1360 cm-1, respectively. These values indicate that the reduced CcPDHs have a ferrous heme with a 6cls state. The RR spectral data of the oxidized and the reduced forms of CcPDH are summarized in Table 1 and compared with the cytochrome domain of CDH [48] from P. chrysosporium. The RR frequencies of oxidized and reduced CcPDH were near agreement with those of CDH, suggesting that these cytochrome proteins have a similar microenvironment around the heme cofactor.
Fig 2. Resonance Raman spectra of the oxidized and reduced CcPDH.
From the top, oxidized form; reduced form prepared by addition of L-fucose; reduced form prepared with ascorbic acid. The measurements were carried out in 50 mM HEPES buffer, pH 7.0, at room temperature. The excitation wavelength and incident powers are 413.1 nm and 1 mW, respectively. All spectra were obtained with data accumulated over 30 sec with a spectral resolution of 1.47 cm-1.
Table 1. Vibrational frequencies (cm-1) of CcPDH and cytochrome domain of CDH.
| Enzyme | ν4 | ν3 | ν2 | ν10 | |
|---|---|---|---|---|---|
| CcPDH | ferric | 1370 | 1502 | 1574 | 1638 |
| ferrous | 1360 | 1492 | 1577 | 1618 | |
| Cytochrome domain of CDH a | ferric | 1371 | 1505 | 1575 | 1638 |
| ferrous | 1362 | 1494 | 1580 | 1615 |
a Ref. [48]
Redox properties of CcPDH
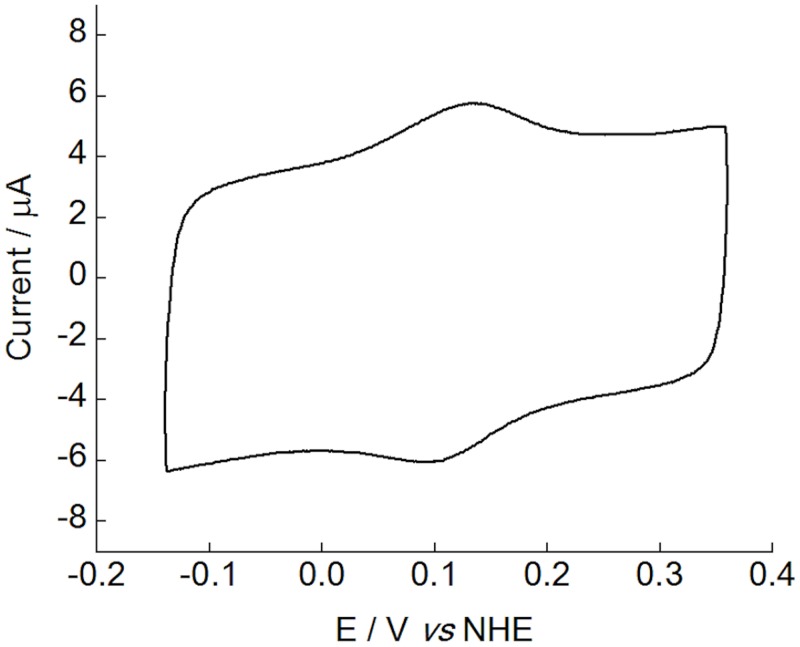
The reduction-oxidation (redox) potential of CcPDH was estimated by cyclic voltammetry. As shown in Fig. 3, well-defined anodic and cathodic peaks can be observed, indicating a direct electron transfer between CcPDH and the electrode. The redox potential determined from the midpoint of the oxidation-reduction peak potentials is 130 mV vs. NHE at pH 7. This is in accordance with previous reports describing the cytochrome domain of CDH from P. chrysosporium in solution (130 mV, pH 7) [39]. Thus, this redox potential was assigned to the Fe3+/Fe2+ redox couple of the heme b center in CcPDH.
Fig 3. Cyclic voltammogram of CcPDH immobilized on a plastic-formed carbon electrode modified with 27 nm carbon nanoparticles.
The voltammogram was obtained in 100 mM HEPES buffer, pH 7.0, at a scan rate of 20 mV/s.
Absorption spectra and PQQ dependent activity of DHPDH
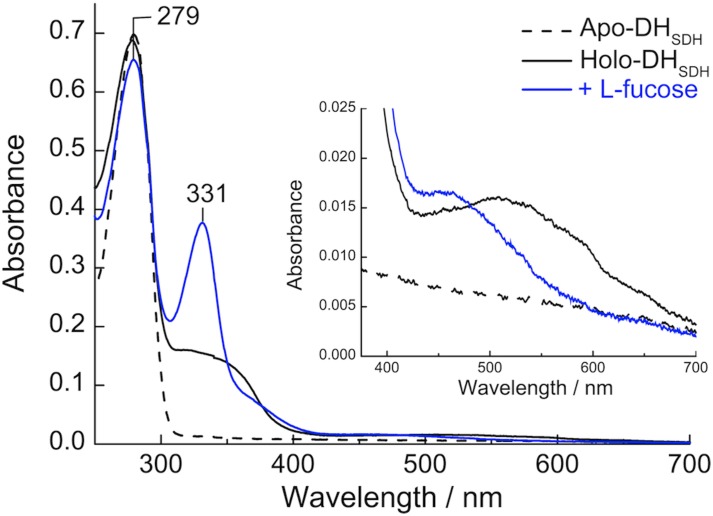
To extend the studies of PQQ binding to CcPDH, we used DHPDH that was separated from a cytochrome domain and a CBM1 by a linker region. The UV-visible spectrum of the purified DHPDH was typical of an enzyme without a cofactor. It had an absorbance maximum at 279 nm with aromatic amino acid residues (Fig. 4). Upon reconstitution with PQQ, the absorption spectrum of holo-DHPDH showed a peak with a maximum at 279 nm, a broad band at around 340 nm with a shoulder at 365 nm, and a less intense and broad band from 420 to 700 nm, centered at around 510 nm. These absorption bands, which overlapped with the α-, β-, and γ-band from cytochrome domain of CcPDH, resulted from using DHPDH. The addition of l-fucose resulted in the reduction of PQQ and the formation of a sharp absorption band at 331 nm; additionally, the maximum of the low broad band shifted from 510 nm to 460 nm. These results were similar to those reported for the quinoprotein sGDH from A. calcoaceticus [15, 21].
Fig 4. UV-visible absorption spectra of the apo- and holo-forms of DHPDH.
Dotted line, apo-form of DHPDH; black solid line, holo-form of DHPDH; blue solid line, reduced form by addition of 1 mM l-fucose. All spectra were recorded in 50 mM HEPES buffer, pH 7.0 at room temperature.
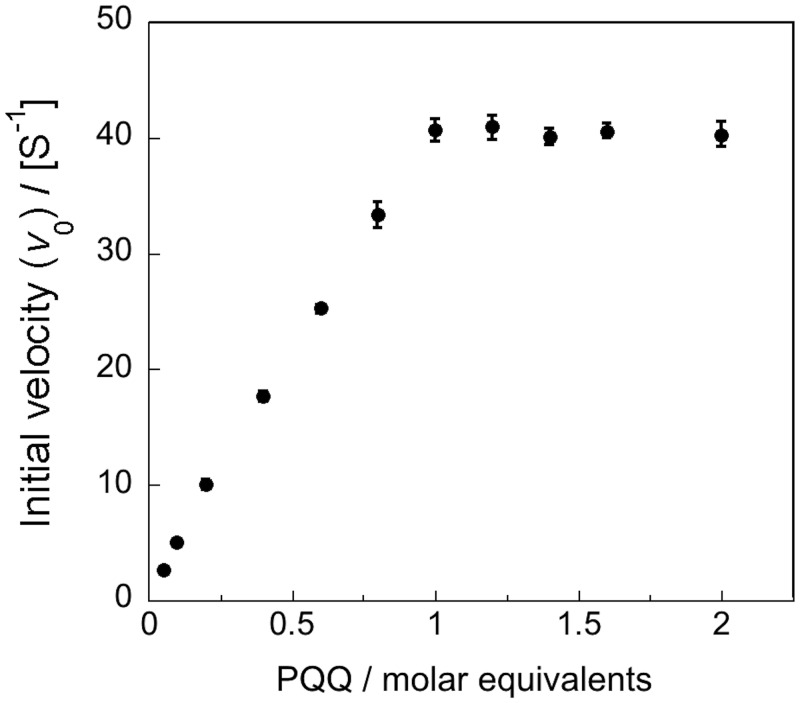
Fig. 5 shows that the catalytic activity of DHPDH increased with stoichiometric amounts of PQQ. It was estimated that full activity was attained at a molar ratio (PQQ: DHPDH) of 0.99. This value increases in linear manner to the equivalence point of the PQQ, indicating that the affinity of apo-DHPDH for PQQ is high, similar to bacterial PQQ quinoproteins [21, 23, 49, 50]. Previous experiments using isothermal titration calorimetry (ITC) have shown that DHPDH binds PQQ with a 1:1 stoichiometry and has strong affinity with a dissociation constant (K d) of 1.11 nM [5]. These results clearly indicate that CcPDH is a PQQ-dependent dehydrogenase.
Fig 5. Titration of the apo-form of DHPDH with PQQ.
The purified apo-form of DHPDH (50 nM) was pre-incubated with various concentrations of PQQ in 50 mM MES buffer, pH 6.5, containing 1 mM CaCl2. After 1 min, the enzyme activity was determined according to the procedure described in the Experimental Procedures.
Catalytic properties of CcPDH
Our previous studies suggest that CcPDH catalyzes the dehydrogenation of monosaccharides having a 1C4 chair conformation, such as l-fucose and d-arabinose [5]. We examined the catalytic activity toward the additional monosaccharides l-galactose, d-talose, l-xylose, l-glucose, l-lyxose, l-mannose, and l-rhamnose (= 6-deoxy- L-mannose), noting the configuration of the substrate as shown in Table 2. The k cat value towards l-galactose was comparable to l-fucose, which is a 6-deoxy sugar of l-galactose, whereas the K m values of l-galactose were twice that of l-fucose. The affinity for d-talose was about the same as for l-fucose, but the k cat value was decreased by 30%. The K m values for l-xylose and l-glucose were approximately 236 mM and 363 mM, respectively, which are markedly greater than the K m obtained for the former monosaccharides. Slight oxidation activities of CcPDH were observed for l-lyxose and l-mannose, but little was observed for l-rhamnose. Thus, the catalytic efficiency was highest for d-glucosone, followed by l-fucose > d-arabinose ≈ l-galactose > d-talose ≈ l-gulose > d-lyxose > l-xylose ≈ l-glucose.
Table 2. Specificity constant values of CcPDH for various monosaccharides.
| Side group orientations in the 1 C 4 conformation a | Kinetic parameters | |||||||
|---|---|---|---|---|---|---|---|---|
| C-2 | C-3 | C-4 | C-5 | k cat (s-1) | K m (mM) | k cat/K m(×103 s-1M-1) | Relative velocity c | |
| d-glucosone b | E (C-3) | E (C-4) | A (C-5) | - (C-6) | 74.1 (±1.4) | 7.9 (±0.3) | 9.37 | 100 |
| l-fucose b | E | E | A | E (-CH3) | 56.4 (±1.8) | 24.8 (±1.2) | 2.27 | 77.7 |
| d-arabinose b | E | E | A | - | 35.5 (±0.6) | 30.3 (±0.8) | 1.17 | 47.5 |
| l-galactose | E | E | A | E (-CH2OH) | 56.1(±1.1) | 49.7 (±1.4) | 1.13 | 65.4 |
| d-talose | E | A | E | A (-CH2OH) | 17.8 (±0.1) | 23.1 (±0.4) | 0.77 | 24.8 |
| l-gulose b | E | A | A | E (-CH2OH) | 53.3 (±1.1) | 84.7 (±2.6) | 0.63 | 52.1 |
| d-lyxose b | E | A | A | - | 12.9 (±0.3) | 66.8 (±2.6) | 0.19 | 13.4 |
| l-xylose | E | E | E | - | 22.4 (±1.2) | 236 (±16) | 0.09 | 11.9 |
| l-glucose | E | E | E | E (-CH2OH) | 24.9 (±1.2) | 363 (±17) | 0.07 | 9.2 |
| l-lyxose | A | E | E | - | n.d. d | n.d. | - | 4.0 |
| l-mannose | A | E | E | E (-CH2OH) | n.d. | n.d. | - | 3.6 |
| l-rhamnose | A | E | E | E (-CH3) | n.d. | n.d. | - | 0.7 |
a Carbon numbers of d-glucosone in brackets; E, equatorial bond; A, axial bond.
b Data for the kinetic parameters are taken from Ref. (5).
c The velocity with each substrate is expressed relative to 100 for the velocity with d-glucosone. The initial substrate concentration was 30 mM d-glucosone and 100 mM the others monosaccharides.
d n.d., not determined.
Carbohydrate-binding properties of CcPDH
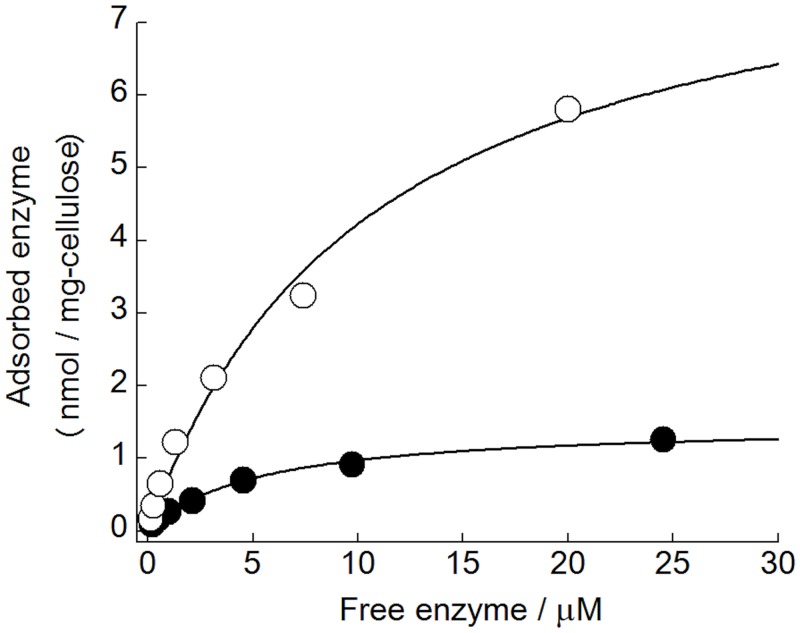
The amino acid sequence of CcPDH indicated the presence of a CBM Family 1 C-terminal domain of CcPDH. The CBM1 homologous sequence of CcPDH was aligned with other CBM1s from fungal cellobiohydrolases (CBHs), ß-glucosidase (BGL), carbohydrate-binding cytochrome b 562 (CBCyt. b 562) and the basidiomycete CDH using MAFFT multiple alignment, as shown in Fig. 6. The multiple sequence alignment shows that the C-terminal CBM1 of CcPDH contains three conserved aromatic residues (Trp693, Trp719, and Tyr720) that are proposed to contribute to CcPDH binding on the surface of carbohydrates. It also shows conserved cysteines (Cys696, 707, 713, and 723) involved in disulfide bond formation. The amount of CcPDH adsorbed on insoluble cellulose was measured at various protein concentrations after incubation for 2 hours. A highly crystalline cellulose Ia from Cladophora and a PASC, which is an amorphous cellulose obtained by phosphoric acid treatment, were used as carbohydrate materials. Fig. 7 shows the plot of the enzyme absorbed on cellulose versus free enzyme concentration. The calculated values for A max and K d from the corresponding plots are shown in Table 3. The A max on PASC showed higher adsorption by CcPDH than on cellulose Ia from Cladophora and the binding efficiency (A max/K d) for PASC was approximately 11 times higher than that for cellulose Ia. This is likely due to the difference in the surface area where the enzyme can adsorb. Therefore, the results suggest CcPDH is a redox enzyme that has the ability to adsorb on cellulose surfaces as well as CDHs.
Fig 6. Multiple alignments of the amino acid sequences of CBM1 of CcPDH and other known CBM1s.

Residues in bold are highly conserved and those in boxes with a black background are perfect matches. Aromatic residues that are candidates for carbohydrate binding are indicated by a filled arrow, and two pairs of cysteines forming disulfide bonds are indicated by filled and open circles, respectively. TrCBHI, cellobiohydrolase I (Cel7A) from Trichoderma reesei (accession no. P62694); PcCBHII, cellobiohydrolase II (Cel6A) from Phanerochaete chrysosporium (Q02321); PcBGL3A, glucan β-1,3-glucosidase (Bgl) from P. chrysosporium (Q8TGC6); PcCBHI, cellobiohydrolase I-2 (Cel7D) from P. chrysosporium (Q09431); TrCBHII, cellobiohydrolase II (Cel6A) from T. reesei (P07987); PcCBCytb562, carbohydrate-binding cytochrome b 562 from P. chrysosporium (Q66NB8); MtCDH, cellobiose dehydrogenase from Myceliophthora thermophila (O74240).
Fig 7. Enzyme concentration dependence of the amount of adsorbed CcPDH.
Closed circle, highly crystalline cellulose from Cladophora; open circle, PASC. The adsorption of CcPDH was measured after incubation for 120 min with 1 mg/mL of cellulose at 30°C as described in the Experimental Procedures.
Table 3. Adsorption parameters of CcPDH for highly crystalline celluloses from Cladophora and PASC.
| K d (μM) | A max (nmol/mg-cellulose) | A max / K d | |
|---|---|---|---|
| Crystalline celluloses | 0.19 | 1.48 | 7.79 |
| PASC | 0.10 | 8.69 | 86.9 |
The parameters were derived from adsorption data plotted as described in Experimental Procedures.
Discussion
In a previous homology search with the cytochrome domain of CDH from P. chrysosporium, CcPDH was discovered in the C. cinerea 5338 strain as a new quinohemoprotein. CcPDH has a three-domain organization, with a catalytic domain containing PQQ as a cofactor (DHPDH), a b-type cytochrome domain as a redox site, and a CBM1. As far as we know, this is the first example of an extracellular quinohemoprotein connected to a cytochrome domain of CDH and CBM1. In this study, the spectroscopic and electrochemical properties, substrate specificity, and carbohydrate-binding properties of this novel PQQ-dependent enzyme from C. cinerea were determined.
The N-terminal cytochrome domain of CcPDH shows significant homology to that of CDH, with well-conserved Met/His ligands for heme binding and a disulfide bond. The structural homology modeling of the cytochrome domain of CcPDH by the Phyre program provided structural folds into an immunoglobulin-like ß-sandwich consisting of a five-stranded and a six-stranded ß-sheet with a short α-helical structure at the C-terminus, which is an unusual module among cytochromes [5]. The electronic absorption and RR spectra of CcPDH in the oxidized and reduced forms are in good agreement with those previously reported for the cytochrome domain of CDH from P. chrysosporium. Those studies confirm a 6cls heme b in both the ferric and ferrous states with Met/His ligands, like CDH. In addition to spectroscopic results, electrochemical studies indicate that the redox potential of the heme is +130 mV, which is comparable to CDH from P. chrysosporium. These observations suggest that the cytochrome domain of CcPDH may have a similar electron transfer function to the cytochrome domain of CDH, i.e., intra- and inter-molecular electron transfer between the cytochrome domain and the catalytic domain and also Fe (III)-reducing ability. Indeed, the electron transfer between CcPDH and cytochrome c can be observed, as it is commonly used as an electron acceptor in the CDH reaction. CDHs are well known as redox enzymes capable of DET between the enzyme and the electrode and the cytochrome domain can act as a built-in electron-transfer mediator [9, 10, 51]. The cytochrome domain of CcPDH was also capable of DET. Thus, CcPDH is an excellent candidate for construction of DET-system bioelectronic devices such as biosensors and biofuel cell anodes, as well as CDHs.
Interestingly, CcPDH exhibited dehydrogenase activity towards monosaccharides in a 1C4 chair conformation, in contrast to the substrate specificity of A. calcoaceticus sGDH, which has an absolute preference for l-arabinose and d-xylose in the 4C1 conformation over the d-arabinose and l-xylose enantiomers [52]. The membrane-bound quinoprotein GDH from E. coli also lacks activity for l-hexoses or pentoses that have a 1C4 chair conformation, such as l-glucose, l-mannose, l-rhamnose, and l-xylose and d-pentoses [53]. Additionally, Asd from E. coli exhibited no preference for different enantiomers with d- and l-arabinose [23].
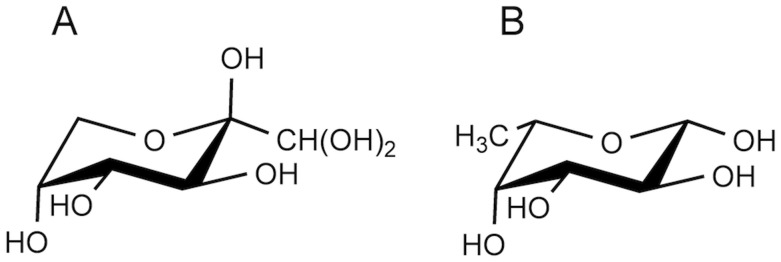
The compared catalytic activity of CcPDH for these substrates and the side group orientation of each monosaccharide when in a 1C4 conformation are summarized in Table 2 and shown for d-glucosone and l-fucose in Fig. 8. d-glucosone (= 2-keto-D-glucose) has four tautomer forms reported by Freimund et al [54]. The 1C4 conformation of 2-keto-D-glucopyranose is the one of the tautomer, at 21% abundance in aqueous solution, as shown by Fig. 8A. The carbons are numbered from the position of the oxygen atom that forms part of the ring. Because these atoms are shifted, the apparent carbon positions of the side group orientations of d-glucosone are shown with the corresponding carbon number of the other monosaccharides in a 1C4 conformation (Table 2). The results shown in Table 2 reveal insights into the substrate recognition dependence on the active site of the catalytic domain in CcPDH. This enzyme showed high activity for aldose substrates with C-2 and C-3 hydroxyl groups in an equatorial configuration and C-4 hydroxyl groups in axial configurations, such as l-fucose, d-arabinose, and l-galactose. In light of these results, we inferred that CcPDH recognizes a 1C4 conformation of d-glucosone (Fig. 8A). The equatorial C-2 hydroxyl group of substrate is essential for binding to the active site of CcPDH, as indicated by a markedly decreased activity for l-lyxose, l-mannose and no activity for l-rhamnose, which have axial C-2 hydroxyl groups. The axial C-4 hydroxyl group is also important for recognition; the K m value increased by an order of magnitude when it the C-4 hydroxyl was replaced with an equatorial bond (in l-xylose and l-glucose compared to d-arabinose and l-galactose). The K m value for l-gulose, which is a C-3 epimer of l-galactose, was approximately 1.7-fold higher than that of l-galactose. This value indicates that the C-3 hydroxyl group in the equatorial conformation is preferred over the axial for CcPDH. The affinity for pentose sugars, which lack a hydroxymethyl group at the C-5 position, or a 6-deoxy-hexose, which has methyl group at C-5 (in L-fucose), increased over hexose sugars that have a C-5 hydroxymethyl group, suggesting that the hydroxymethyl group at this position causes some steric hindrance. However, the result suggests that C-5 methyl group is not critical for binding to CcPDH, as indicated by the oxidation of l-fucose. In light of the stereospecificity of other PQQ-dependent dehydrogenases [20, 52, 55], CcPDH may catalyze the dehydrogenation reaction of regioselective substrates at the C-1 hydroxyl group, although we can not rule out side reactions at other positions. Further studies are necessary to identify a product of CcPDH oxidation and they are currently under consideration.
Fig 8. Structure of d-glucosone (A) and l-fucose (B) in a 1C4 conformation.
Many extracellular fungal carbohydrate hydrolytic enzymes carry a CBM1, which is thought to facilitate binding to the surface of cellulose chains and is related to the catalytic function of these enzymes. Although these CBM1 are not found in basidiomycetous CDHs, the flavocytochrome enzymes are also capable of binding to cellulose by its flavin domain and localizes on cellulose surfaces [56–58]. In the present study, CcPDH shows high affinity toward microcrystalline and amorphous cellulose. This result suggests the possibility of localizing CcPDH on the surface of cellulose, as is the case for CDHs. Further cytochemical analysis will be needed to identify the localization of CcPDH in nature.
Under aerobic conditions, cellulose degradation by cell-free culture filtrates of various fungi has shown much higher rates compared to anaerobic conditions, suggesting that an oxidative reaction may play a crucial role in the fungal degradation of cellulose. CDH is the first example of an oxidoreductase related to the extracellular oxidative process for cellulose degradation. Furthermore, it has been recently reported that the cytochrome domain of CDH can transfer electrons to fungal copper-dependent lytic polysaccharide monooxygenases (LPMOs) that were classified in AA9. The combination of CDHs and LPMOs synergistically enhances cellulase activity [59–62]. The cytochrome domain of CcPDH could allow direct electron transfer for LPMO as well as that of CDH. However the physiological functions remain obscure in vivo. It is not only some redox proteins instead of CDH, but some reductive compounds are the possibly electron donor for LPMO in those fungi. Indeed, l-ascorbic acid is typically used as an electron donor for LPMO in vitro [63]. The oxidation product of d-glucosone by CcPDH could be an intermediate in the l-ascorbic acid biosynthesis [64]. PQQ quinoproteins also play a major role in the bacterial production of l-ascorbic acid [55, 65, 66]. Fugal Penicillium cyaneo-fulvum has an extracellular d-erythorbic acid (isoascorbic acid) synthesis pathway by using glucose oxidase and gluconolactone oxidase [67]. CcPDH, therefore, may play a role in the l-ascorbic acid biosynthesis pathway and then LPMOs could be provided with electrons from these reductive compounds. Anyhow, CcPDH must be related to the enzymatic degradation of the plant cell wall because of its capability of binding to cellulose.
Conclusions
We have demonstrated the full characterization of CcPDH, which was classified as a member of the new AA12 family in CAZy using spectroscopic and electrochemical methods. These enzymatic properties provide new insight into other known PQQ quinoproteins as well as the basidiomycetous oxidoreductase network. The N-terminal cytochrome domain of CcPDH contains a 6-coordinated low-spin heme b in both the ferric and ferrous states with Met/His ligands as well as CDH and it enables DET between CcPDH and the electrode. The catalytic domain shows oxidation activity toward monosaccharides in a 1C4 chair conformation in the presence of PQQ as a cofactor, and the most efficient catalysis was observed for d-glucosone. CcPDH exhibited binding affinity for insoluble cellulose. These results suggest that CcPDH may be involved in the extracellular oxidative degradation as well as CDH. Therefore, these proteins are an attractive component of cellulolytic enzymes for use in biodegradation and biomass conversion or as an anode catalyst for bioelectrochemical applications.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was supported financially by a Grant-in-Aid for Scientific Research (No. 21605004 to N.N.) from the Japan Society for the Promotion of Science (JSPS), by a Grant-in-Aid for Innovative Areas (No. 24114001 and 24114008 to K.I.) from the Japanese Ministry of Education, Culture, Sports, and Technology (MEXT), and by a Grant-in-Aid for JSPS Fellows (Grant No. 268641 to K.T.). H.M. was supported by a Grant-in-Aid for JSPS Fellows (Grant No. 208304) during his postdoc period at the University of Tokyo. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Floudas D, Binder M, Riley R, Barry K, Blanchette RA, et al. (2012) The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 336: 1715–1719. 10.1126/science.1221748 [DOI] [PubMed] [Google Scholar]
- 2. Babot ED, del Rio JC, Kalum L, Martinez AT, Gutiérrez A (2013) Oxyfunctionalization of aliphatic compounds by a recombinant peroxygenase from Coprinopsis cinerea . Biotechnol Bioeng 110: 2323–2332. 10.1002/bit.24904 [DOI] [PubMed] [Google Scholar]
- 3. Kilaru S, Hoegger PJ, Kües U (2006) The laccase multi-gene family in Coprinopsis cinerea has seventeen different members that divide into two distinct subfamilies. Curr Genet 50: 45–60. [DOI] [PubMed] [Google Scholar]
- 4. Yoshida M, Sato K, Kaneko S, Fukuda K (2009) Cloning and transcript analysis of multiple genes encoding the glycoside hydrolase family 6 enzyme from Coprinopsis cinerea . Biosci Biotechnol Biochem 73: 67–73. [DOI] [PubMed] [Google Scholar]
- 5. Matsumura H, Umezawa K, Takeda K, Sugimoto N, Ishida T, et al. (2014) Discovery of a eukaryotic pyrroloquinoline quinone-dependent oxidoreductase belonging to a new auxiliary activity family in the database of carbohydrate-active enzymes. PLoS ONE 9: e104851 10.1371/journal.pone.0104851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Henriksson G, Johansson G, Pettersson G (2000) A critical review of cellobiose dehydrogenases. J Biotechnol 78: 93–113. [DOI] [PubMed] [Google Scholar]
- 7. Igarashi K, Yoshida M, Matsumura H, Nakamura N, Ohno H, et al. (2005) Electron transfer chain reaction of the extracellular flavocytochrome cellobiose dehydrogenase from the basidiomycete Phanerochaete chrysosporium . FEBS J 272: 2869–2877. [DOI] [PubMed] [Google Scholar]
- 8. Hallberg BM, Bergfors T, Bäckbro K, Pettersson G, Henriksson G, et al. (2000) A new scaffold for binding haem in the cytochrome domain of the extracellular flavocytochrome cellobiose dehydrogenase. Structure 8: 79–88. [DOI] [PubMed] [Google Scholar]
- 9. Ludwig R, Harreither W, Tasca F, Gorton L (2010) Cellobiose dehydrogenase: a versatile catalyst for electrochemical applications. Chemphyschem 11: 2674–2697. 10.1002/cphc.201000216 [DOI] [PubMed] [Google Scholar]
- 10. Ludwig R, Ortiz R, Schulz C, Harreither W, Sygmund C, et al. (2013) Cellobiose dehydrogenase modified electrodes: advances by materials science and biochemical engineering. Anal Bioanal Chem 405: 3637–3658. 10.1007/s00216-012-6627-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Levasseur A, Drula E, Lombard V, Coutinho PM, Henrissat B (2013) Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol Biofuels 6: 41 10.1186/1754-6834-6-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Anthony C, Ghosh M (1998) The structure and function of the PQQ-containing quinoprotein dehydrogenases. Prog Biophys Mol Biol 69: 1–21. [DOI] [PubMed] [Google Scholar]
- 13. Anthony C (2001) Pyrroloquinoline quinone (PQQ) and quinoprotein enzymes. Antioxid Redox Signal 3: 757–774. [DOI] [PubMed] [Google Scholar]
- 14. Matsushita K, Toyama H, Yamada M, Adachi O (2002) Quinoproteins: structure, function, and biotechnological applications. Appl Microbiol Biotechnol 58: 13–22. [DOI] [PubMed] [Google Scholar]
- 15. Geiger O, Görisch H (1989) Reversible thermal inactivation of the quinoprotein glucose dehydrogenase from Acinetobacter calcoaceticus. Ca2+ ions are necessary for re-activation. Biochem J 261: 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Oubrie A, Rozeboom HJ, Kalk KH, Huizinga EG, Dijkstra BW (2002) Crystal structure of quinohemoprotein alcohol dehydrogenase from Comamonas testosteroni: structural basis for substrate oxidation and electron transfer. J Biol Chem 277: 3727–3732. [DOI] [PubMed] [Google Scholar]
- 17. Toyama H, Mathews FS, Adachi O, Matsushita K (2004) Quinohemoprotein alcohol dehydrogenases: structure, function, and physiology. Arch Biochem Biophys 428: 10–21. [DOI] [PubMed] [Google Scholar]
- 18. Satoh A, Kim JK, Miyahara I, Devreese B, Vandenberghe I, et al. (2002) Crystal structure of quinohemoprotein amine dehydrogenase from Pseudomonas putida. Identification of a novel quinone cofactor encaged by multiple thioether cross-bridges. J Biol Chem 277: 2830–2834. [DOI] [PubMed] [Google Scholar]
- 19. Ameyama M, Shinagawa E, Matsushita K, Adachi O (1981) D-Glucose dehydrogenase of Gluconobacter suboxydans: solubilization, purification and characterization. Agric Biol Chem 45: 851–861. [Google Scholar]
- 20. Yamada M, Elias MD, Matsushita K, Migita CT, Adachi O (2003) Escherichia coli PQQ-containing quinoprotein glucose dehydrogenase: its structure comparison with other quinoproteins. Biochim Biophys Acta 1647: 185–192. [DOI] [PubMed] [Google Scholar]
- 21. Olsthoorn AJ, Duine JA (1996) Production, characterization, and reconstitution of recombinant quinoprotein glucose dehydrogenase (soluble type; EC 1.1.99.17) apoenzyme of Acinetobacter calcoaceticus . Arch Biochem Biophys 336: 42–48. [DOI] [PubMed] [Google Scholar]
- 22. Oubrie A (2003) Structure and mechanism of soluble glucose dehydrogenase and other PQQ-dependent enzymes. Biochim Biophys Acta 1647: 143–151. [DOI] [PubMed] [Google Scholar]
- 23. Southall SM, Doel JJ, Richardson DJ, Oubrie A (2006) Soluble aldose sugar dehydrogenase from Escherichia coli: a highly exposed active site conferring broad substrate specificity. J Biol Chem 281: 30650–30659. [DOI] [PubMed] [Google Scholar]
- 24. Sakuraba H, Yokono K, Yoneda K, Watanabe A, Asada Y, et al. (2010) Catalytic properties and crystal structure of quinoprotein aldose sugar dehydrogenase from hyperthermophilic archaeon Pyrobaculum aerophilum . Arch Biochem Biophys 502: 81–88. 10.1016/j.abb.2010.08.002 [DOI] [PubMed] [Google Scholar]
- 25. Boraston AB, Bolam DN, Gilbert HJ, Davies GJ (2004) Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem J 382: 769–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shoseyov O, Shani Z, Levy I (2006) Carbohydrate binding modules: biochemical properties and novel applications. Microbiol Mol Biol Rev 70: 283–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guillën D, Sánchez S, Rodríguez-Sanoja R (2010) Carbohydrate-binding domains: multiplicity of biological roles. Appl Microbiol Biotechnol 85: 1241–1249. 10.1007/s00253-009-2331-y [DOI] [PubMed] [Google Scholar]
- 28. Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B (2014) The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42: D490–495. 10.1093/nar/gkt1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tomme P, Van Tilbeurgh H, Pettersson G, Van Damme J, Vandekerckhove J, et al. (1988) Studies of the cellulolytic system of Trichoderma reesei QM 9414. Analysis of domain function in two cellobiohydrolases by limited proteolysis. Eur J Biochem 170: 575–581. [DOI] [PubMed] [Google Scholar]
- 30. Carrard G, Koivula A, Söderlund H, Béguin P (2000) Cellulose-binding domains promote hydrolysis of different sites on crystalline cellulose. Proc Natl Acad Sci U S A 97: 10342–10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kraulis J, Clore GM, Nilges M, Jones TA, Pettersson G, et al. (1989) Determination of the three-dimensional solution structure of the C-terminal domain of cellobiohydrolase I from Trichoderma reesei. A study using nuclear magnetic resonance and hybrid distance geometry-dynamical simulated annealing. Biochemistry 28: 7241–7257. [DOI] [PubMed] [Google Scholar]
- 32. Linder M, Mattinen ML, Kontteli M, Lindeberg G, Ståhlberg J, et al. (1995) Identification of functionally important amino acids in the cellulose-binding domain of Trichoderma reesei cellobiohydrolase I. Protein Sci 4: 1056–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Linder M, Lindeberg G, Reinikainen T, Teeri TT, Pettersson G (1995) The difference in affinity between two fungal cellulose-binding domains is dominated by a single amino acid substitution. FEBS Lett 372: 96–98. [DOI] [PubMed] [Google Scholar]
- 34. Lehtiö J, Sugiyama J, Gustavsson M, Fransson L, Linder M, et al. (2003) The binding specificity and affinity determinants of family 1 and family 3 cellulose binding modules. Proc Natl Acad Sci U S A 100: 484–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nimlos MR, Beckham GT, Matthews JF, Bu L, Himmel ME, et al. (2012) Binding preferences, surface attachment, diffusivity, and orientation of a family 1 carbohydrate-binding module on cellulose. J Biol Chem 287: 20603–20612. 10.1074/jbc.M112.358184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sugimoto N, Igarashi K, Samejima M (2012) Cellulose affinity purification of fusion proteins tagged with fungal family 1 cellulose-binding domain. Protein Expr Purif 82: 290–296. 10.1016/j.pep.2012.01.007 [DOI] [PubMed] [Google Scholar]
- 37. Takeda K, Matsumura H, Ishida T, Samejima M, Igarashi K, et al. (2013) The two-step electrochemical oxidation of alcohols using a novel recombinant PQQ alcohol dehydrogenase as a catalyst for a bioanode. Bioelectrochemistry 94: 75–78. 10.1016/j.bioelechem.2013.08.001 [DOI] [PubMed] [Google Scholar]
- 38. Ortiz R, Matsumura H, Tasca F, Zahma K, Samejima M, et al. (2012) Effect of deglycosylation of cellobiose dehydrogenases on the enhancement of direct electron transfer with electrodes. Anal Chem 84: 10315–10323. 10.1021/ac3022899 [DOI] [PubMed] [Google Scholar]
- 39. Igarashi K, Verhagen MF, Samejima M, Schulein M, Eriksson KE, et al. (1999) Cellobiose dehydrogenase from the fungi Phanerochaete chrysosporium and Humicola insolens. A flavohemoprotein from Humicola insolens contains 6-hydroxy-FAD as the dominant active cofactor. J Biol Chem 274: 3338–3344. [DOI] [PubMed] [Google Scholar]
- 40. Samejima M, Eriksson KE (1992) A comparison of the catalytic properties of cellobiose:quinone oxidoreductase and cellobiose oxidase from Phanerochaete chrysosporium . Eur J Biochem 207: 103–107. [DOI] [PubMed] [Google Scholar]
- 41. Karapetyan KN, Fedorova TV, Vasil’chenko LG, Ludwig R, Haltrich D, et al. (2006) Properties of neutral cellobiose dehydrogenase from the ascomycete Chaetomium sp. INBI 2–26(-) and comparison with basidiomycetous cellobiose dehydrogenases. J Biotechnol 121: 34–48. [DOI] [PubMed] [Google Scholar]
- 42. Katoh K, Misawa K, Kuma K, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30: 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30: 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gouet P, Courcelle E, Stuart DI, Métoz F (1999) ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15: 305–308. [DOI] [PubMed] [Google Scholar]
- 45. Wood TM (1988) Preparation of Crystalline, Amorphous, and Dyed Cellulase Substrates. Method Enzymol 160: 19–25. [Google Scholar]
- 46. Jervis EJ, Haynes CA, Kilburn DG (1997) Surface diffusion of cellulases and their isolated binding domains on cellulose. J Biol Chem 272: 24016–24023. [DOI] [PubMed] [Google Scholar]
- 47. Yoshida M, Igarashi K, Wada M, Kaneko S, Suzuki N, et al. (2005) Characterization of carbohydrate-binding cytochrome b 562 from the white-rot fungus Phanerochaete chrysosporium . Appl Environ Microbiol 71: 4548–4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rotsaert FA, Hallberg BM, de Vries S, Moënne-Loccoz P, Divne C, et al. (2003) Biophysical and structural analysis of a novel heme b iron ligation in the flavocytochrome cellobiose dehydrogenase. J Biol Chem 278: 33224–33231. [DOI] [PubMed] [Google Scholar]
- 49. Groen BW, van Kleef MA, Duine JA (1986) Quinohaemoprotein alcohol dehydrogenase apoenzyme from Pseudomonas testosteroni . Biochem J 234: 611–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mutzel A, Gorisch H (1991) Quinoprotein ethanol dehydrogenase: preparation of the apo-form and reconstitution with pyrroloquinoline quinone and Ca2+ or Sr2+ ions. Agric Biol Chem 55: 1721–1726. [Google Scholar]
- 51. Matsumura H, Ortiz R, Ludwig R, Igarashi K, Samejima M, et al. (2012) Direct electrochemistry of Phanerochaete chrysosporium cellobiose dehydrogenase covalently attached onto gold nanoparticle modified solid gold electrodes. Langmuir 28: 10925–10933. 10.1021/la3018858 [DOI] [PubMed] [Google Scholar]
- 52. Olsthoorn AJ, Duine JA (1998) On the mechanism and specificity of soluble, quinoprotein glucose dehydrogenase in the oxidation of aldose sugars. Biochemistry 37: 13854–13861. [DOI] [PubMed] [Google Scholar]
- 53. Cozier GE, Salleh RA, Anthony C (1999) Characterization of the membrane quinoprotein glucose dehydrogenase from Escherichia coli and characterization of a site-directed mutant in which histidine-262 has been changed to tyrosine. Biochem J 340: 639–647. [PMC free article] [PubMed] [Google Scholar]
- 54. Freimund S, Baldes L, Huwig A, Giffhorn F (2002) Enzymatic synthesis of d-glucosone 6-phosphate (d-arabino-hexos-2-ulose 6-(dihydrogen phosphate)) and NMR analysis of its isomeric forms. Carbohydr Res 337: 1585–1587. [DOI] [PubMed] [Google Scholar]
- 55. Miyazaki T, Sugisawa T, Hoshino T (2006) Pyrroloquinoline quinone-dependent dehydrogenases from Ketogulonicigenium vulgare catalyze the direct conversion of l-sorbosone to l-ascorbic acid. Appl Environ Microbiol 72: 1487–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Renganathan V, Usha SN, Lindenburg F (1990) Cellobiose-oxidizing enzymes from the lignocellulose-degrading basidiomycete Phanerochaete chrysosporium: interaction with microcrystalline cellulose. Appl Microbiol Biotechnol 32: 609–613. [Google Scholar]
- 57. Henriksson G, Pettersson G, Johansson G, Ruiz A, Uzcategui E (1991) Cellobiose oxidase from Phanerochaete chrysosporium can be cleaved by papain into two domains. Eur J Biochem 196: 101–106. [DOI] [PubMed] [Google Scholar]
- 58. Henriksson G, Salumets A, Divne C, Pettersson G (1997) Studies of cellulose binding by cellobiose dehydrogenase and a comparison with cellobiohydrolase 1. Biochem J 324: 833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Langston JA, Shaghasi T, Abbate E, Xu F, Vlasenko E, et al. (2011) Oxidoreductive cellulose depolymerization by the enzymes cellobiose dehydrogenase and glycoside hydrolase 61. Appl Environ Microbiol 77: 7007–7015. 10.1128/AEM.05815-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Phillips CM, Beeson WT, Cate JH, Marletta MA (2011) Cellobiose dehydrogenase and a copper-dependent polysaccharide monooxygenase potentiate cellulose degradation by Neurospora crassa . ACS Chem Biol 6: 1399–1406. 10.1021/cb200351y [DOI] [PubMed] [Google Scholar]
- 61. Beeson WT, Phillips CM, Cate JH, Marletta MA (2012) Oxidative cleavage of cellulose by fungal copper-dependent polysaccharide monooxygenases. J Am Chem Soc 134: 890–892. 10.1021/ja210657t [DOI] [PubMed] [Google Scholar]
- 62. Li X, Beeson WT 4th, Phillips CM, Marletta MA, Cate JH (2012) Structural basis for substrate targeting and catalysis by fungal polysaccharide monooxygenases. Structure 20: 1051–1061. 10.1016/j.str.2012.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Westereng B, Ishida T, Vaaje-Kolstad G, Wu M, Eijsink VG, et al. (2011) The putative endoglucanase PcGH61D from Phanerochaete chrysosporium is a metal-dependent oxidative enzyme that cleaves cellulose. PLoS ONE 6: e27807 10.1371/journal.pone.0027807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hancock RD, Viola R (2002) Biotechnological approaches for l-ascorbic acid production. Trends Biotechnol 20: 299–305. [DOI] [PubMed] [Google Scholar]
- 65. Bremus C, Herrmann U, Bringer-Meyer S, Sahm H (2006) The use of microorganisms in l-ascorbic acid production. J Biotechnol 124: 196–205. [DOI] [PubMed] [Google Scholar]
- 66. Hoshino T, Sugisawa T, Shinjoh M, Tomiyama N, Miyazaki T (2003) Membrane-bound d-sorbitol dehydrogenase of Gluconobacter suboxydans IFO 3255—enzymatic and genetic characterization. Biochim Biophys Acta 1647: 278–288. [DOI] [PubMed] [Google Scholar]
- 67. Salusjärvi T, Kalkkinen N, Miasnikov AN (2004) Cloning and characterization of gluconolactone oxidase of Penicillium cyaneo-fulvum ATCC 10431 and evaluation of its use for production of d-erythorbic acid in recombinant Pichia pastoris . Appl Environ Microbiol 70: 5503–5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.