Abstract
The ribosome is the target of a large number of antibiotics. Here, we report a 3.4-Å-resolution crystal structure of bactobolin A bound to 70S ribosome–tRNA complex. The antibiotic binds at a previously unseen site in the 50S subunit and displaces tRNA bound at the P-site. It thus likely has a similar mechanism of action as blasticidin S despite binding to a different site. The structure also rationalizes previously identified resistance mutations.
Abbreviations: BlaS, blasticidin S
Keywords: translation, antibiotic, P-site, tRNA
Graphical Abstract
Highlights
-
•
The ribosome is the target of a large number of antibiotics.
-
•
Here, we report a 3.4-Å-resolution crystal structure of bactobolin A bound to 70S ribosome–tRNA complex.
-
•
The antibiotic binds at a previously unseen site in the 50S subunit and displaces tRNA bound at the P-site that inhibits translation.
-
•
The structure also rationalizes previously identified resistance mutations.
Structural studies have been instrumental in revealing the molecular basis of the action of antibiotics [1]. Those studies showed that many antibiotics inhibit various steps of the elongation cycle of translation [2], whereas relatively few clinically useful compounds target initiation [3]. Recently, it was suggested that blasticidin S (BlaS) acts by inhibiting termination [4].
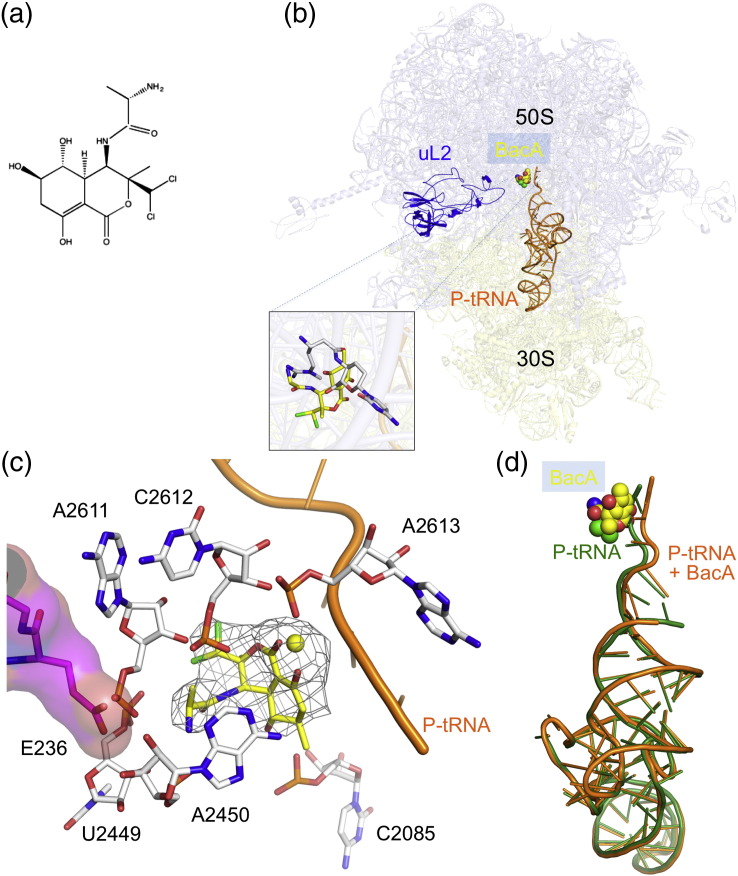
Bactobolin is a member of the polyketide-peptide family of molecules produced by Burkholderia thailandensis [5]. These water-soluble compounds consist of a C6-polyketide fused to a chlorinated hydroxy-valine residue (Fig. 1a). Recently, it was shown to inhibit protein synthesis, and resistance to bactobolin is acquired by mutations in the 50S ribosomal protein uL2 [6]. Bactobolin-resistant mutants remain susceptible to other known ribosome inhibitors, suggesting that it binds to a novel target site in the ribosome.
Fig. 1.
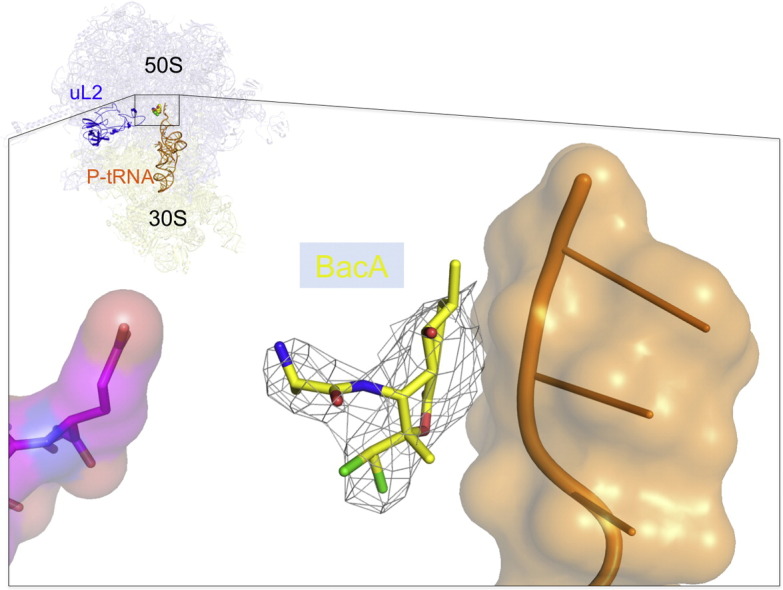
Binding of bactobolin A to the ribosome in the presence of P-tRNA. (a) Chemical structure of bactobolin. (b) Relative positions of bactobolin, tRNA and uL2; overlapping of bactobolin and BlaS binding sites. (c) Unbiased Fo − Fc difference map corresponding to bactobolin A and adjacent Mg2 + ion (yellow sphere) is contoured at 3σ. rRNA forming contacts with bactobolin is shown. E236 of uL2 stabilizes A2450. (d) Superposition of tRNA in the 70S-tRNA-bactobolin complex with that from antibiotic-free 70S-tRNA, demonstrating the shift of the CCA end toward the A-site.
To help understand the molecular basis of its action, we determined the crystal structure of bactobolin A bound to the Thermus thermophilus 70S ribosome in the presence of mRNA and tRNA. The resolution of 3.4 Å allowed us to visualize the antibiotic and deduce its interactions with the ribosome and tRNA. Following refinement of the initial model, we placed bactobolin A in the electron density (Fig. 1b and c). The antibiotic is coordinated primary by helix 73, and its interactions with the ribosome involve a Mg2 + ion that allows it to interact with A2613 through both of its rings via the carbonyl oxygen of the lactone and the hydroxyl of the enol (Fig. 1c). The additional enol hydroxyl is bound by C2085, whereas the chlorides of the lactone ring are coordinated by U2449, A2611 and C2612. The binding of bactobolin is further stabilized by the amine termini of hydroxy-valine interacting with A2450. This holds bactobolin in the orientation that results in direct contacts with the CCA end of P-site tRNA (Fig. 1c and d).
A superposition of our structure with a 70S–tRNA complex without the antibiotic shows that bactobolin would clash sterically with canonical P-site tRNA (Fig. 1d). Thus, in order to accommodate both tRNA and the antibiotic, the CCA backbone of P-site tRNA is displaced in our structure (Fig. 1d). Consequently, A76 of P-tRNA no longer contacts C2063 and is shifted 3.6 Å toward the A-site. Such a conformational change of the P-site tRNA caused by bactobolin A suggests that it would inhibit peptidyl transfer. However, the structural effect of bactobolin A binding is similar to the one recently reported for BlaS [4]. It has been further shown that BlaS-stimulated distortion of CCA P-tRNA would occlude the access of a release factor to the A-site and thus inhibit translation termination, which is also supported by biochemical data [4]. The fact that BlaS and bactobolin A have overlapping binding sites (Fig. 1b) and cause a similar conformational rearrangement of P-site tRNA suggests that both act as translation termination inhibitors.
The crystal structure also rationalizes biochemical data that identified a mutation in E236 (Bacillus subtilis) of uL2 protein as a cause of resistance to bactobolin [6]. The resistance can be explained by disruption of E236:A2450 contacts (Fig. 1c). Since A2450 is directly involved in coordinating bactobolin A, the mutation is likely to disorder the binding site. We note that the previously described bactobolin-resistant mutants remain susceptible to BlaS.
Acknowledgements
We would like to thank the beamline staff on IO3 at Diamond Light Source for help and advice with data collection. This work was supported by grants from the UK Medical Research Council (MC_U105184332), the Wellcome Trust (Senior Investigator Award WT096570), the Agouron Institute and the Jeantet Foundation to V.R.; by United States Public Health Service Grant GM-59026 and National Science Foundation Cooperative Agreement DBI-0939454, to E.P.G.; and by a fellowship from the Human Frontiers Science Program to A.A.
Appendix A. Supplementary data
Supplementary material 1
Supplementary material 2
References
- 1.McCoy L.S., Xie Y., Tor Y. Antibiotics that target protein synthesis. Wiley Interdiscip Rev RNA. 2011;2:209–232. doi: 10.1002/wrna.60. [DOI] [PubMed] [Google Scholar]
- 2.Voorhees R.M., Ramakrishnan V. Structural basis of the translational elongation cycle. Annu Rev Biochem. 2013;82:203–236. doi: 10.1146/annurev-biochem-113009-092313. [DOI] [PubMed] [Google Scholar]
- 3.Wilson D.N. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat Rev Microbiol. 2014;12:35–48. doi: 10.1038/nrmicro3155. [DOI] [PubMed] [Google Scholar]
- 4.Svidritskiy E., Ling C., Ermolenko D.N., Korostelev A.A. Blasticidin S inhibits translation by trapping deformed tRNA on the ribosome. Proc Natl Acad Sci USA. 2013;110:12283–12288. doi: 10.1073/pnas.1304922110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carr G., Seyedsayamdost M.R., Chandler J.R., Greenberg E.P., Clardy J. Sources of diversity in bactobolin biosynthesis by Burkholderia thailandensis E264. Org Lett. 2011;13:3048–3051. doi: 10.1021/ol200922s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandler J.R., Truong T.T., Silva P.M., Seyedsayamdost M.R., Carr G., Radey M. Bactobolin resistance is conferred by mutations in the L2 ribosomal protein. MBio. 2012;3:e00499–e00512. doi: 10.1128/mBio.00499-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1
Supplementary material 2