Abstract
Wilms tumor is genetically heterogeneous, and until recently only one Wilms tumor gene was known, WT1 at 11p13. However, WT1 is altered in only ~20% of Wilms tumors. Recently a novel gene, WTX at Xq11.1, was reported to be mutated in Wilms tumors. No overlap between tumors with mutations in WTX and WT1 was noted, suggesting that WT1 and WTX mutations could account for the genetic basis of roughly half of Wilms tumors. To assess the frequency of WTX mutations and their relationship to WT1 mutations in a larger (n = 125) panel of Wilms tumors which had been thoroughly assessed for mutations in WT1, we conducted a complete mutational analysis of WTX that included sequencing of the entire coding region and quantitative PCR to identify deletions of the WTX gene. Twenty-three (18.4%) tumors carried a total of 24 WTX mutations, a lower WTX mutation frequency than that previously observed. Surprisingly, we observed an equivalent frequency of WTX mutations in tumors with mutations in either or both WT1 and CTNNB1 (20.0%) and tumors with no mutation in either WT1 or CTNNB1 (17.5%). WTX has been reported to play a role in the WNT/β-catenin signaling pathway, and, interestingly, WTX deletion/truncation mutations appeared to be rare in tumors carrying exon 3 mutations of CTNNB1, encoding β-catenin. Our findings indicate that WT1 and WTX mutations occur with similar frequency, that they partially overlap in Wilms tumors, and that mutations in WT1, WTX, and CTNNB1 underlie the genetic basis of about one-third of Wilms tumors.
INTRODUCTION
Wilms tumor is a childhood embryonal tumor of the kidney that arises from embryonal nephric mesenchyme and typically has a triphasic histology with blastemal, epithelial, and stromal elements. Until recently, the only known Wilms tumor gene was the WT1 gene at 11p13, which was identified by virtue of being deleted (germline) in patients with WAGR (Wilms-Aniridia-Genitourinary anomalies-mental Retardation) syndrome and which is mutated in ~20% of sporadic, nonsyndromic Wilms tumors (Huff, 1998). Wilms tumor occurs in both sporadic (98–99%) and familial (1–2%) forms, and the WT1 gene is only rarely altered in familial Wilms tumors (FWT) (Huff, 1998). Genetic linkage analysis studies of several large Wilms tumor families led to the localization of two FWT genes, FWT1 at 17q12-21 (Rahman et al., 1996) and FWT2 at 19q13.33-13.41 (McDonald et al., 1998), but neither gene has yet been identified. The existence of Wilms tumor families for which linkage to both FWT1 and FWT2 has been excluded indicates the existence of at least one additional FWT gene (Ruteshouser and Huff, 2004). Other loci, including 11p15, 1p, 2q, 7p, 9q, 14q, 16q, and 22, have also been implicated in the etiology of Wilms tumor through studies of loss of heterozygosity, loss of imprinting, and constitutional chromosomal defects (Mannens et al., 1990; Kaneko et al., 1991; Maw et al., 1992; Olson et al., 1995; Grundy et al., 1998; Ruteshouser et al., 2005). CTNNB1, the gene encoding β-catenin, is also mutated in about 15% of Wilms tumors (Koesters et al., 1999). Interestingly, most tumors with mutations in WT1 also carry mutations in CTNNB1, and CTNNB1 mutations rarely occur in the absence of WT1 mutation (Maiti et al., 2000). The etiology of Wilms tumor is thus clearly both complex and heterogeneous.
WT1 encodes a zinc finger transcription factor for which many putative target genes have been identified; however, it is still not clear what the biologically relevant target genes are. β-catenin, encoded by the CTNNB1 gene at 3p22.1, is involved in both cell adhesion and the WNT signaling pathway. Protein-stabilizing mutations in exon 3 of β-catenin have been observed in a variety of cancers as well as in Wilms tumors (Fukuzawa et al., 2007; Polakis, 2007). In some Wilms tumors with mutations in WT1, mutations in exons 7 and 8 of CTNNB1 are also seen (Li et al., 2004; Fukuzawa et al., 2007), but the functional significance of these non-exon 3 mutations is not clear.
Recently, a third gene, WTX, was shown to be mutated in Wilms tumors (Rivera et al., 2007). WTX is located at Xq11.1, very close to the centromere, and encodes an 1135 amino acid protein with no conserved functional domains except for a predicted nuclear localization signal. Although WTX orthologs are found in other species, including zebrafish, the protein shows no substantial similarity to other proteins of known or unknown function. WTX appears to participate in the WNT signaling pathway and to promote the ubiquitination and degradation of β-catenin (Major et al., 2007). Previously WTX mutations, both deletions and point mutations, were observed in 15/51 (29.4%) of Wilms tumors, and the mutation frequency was approximately equal in males and females (Rivera et al., 2007). This study also noted that WTX alterations were never seen in tumors carrying mutations in either WT1 or CTNNB1. The implication of these data was that the etiology of approximately half of Wilms tumors involved mutations in either WT1 or WTX.
We have now assessed the frequency and type of WTX mutations in a larger group of Wilms tumors which had been completely analyzed for mutations in all 10 WT1 coding exons, including deletions of all or part of the gene, and for mutations in exons 3, 7, and 8 of CTNNB1. We have assessed 125 tumors (80 with no WT1 or CTNNB1 mutations, 45 with mutations in either or both) for both point mutations and deletions in WTX, and an additional 52 tumors for WTX deletions only. We show that WTX alterations were approximately equally frequent in Wilms tumors with mutations in WT1 and/or CTNNB1 and in tumors with no mutation in either WT1 or CTNNB1, and that WTX mutations occurred with about the same frequency as WT1 mutations. Thus, about one-third of tumors carry mutations at WT1, CTNNB1, and/or WTX; gene mutations have yet to be identified for the remaining two-thirds of tumors.
MATERIALS AND METHODS
Tumor Samples
Wilms tumor samples were obtained from patients following informed consent. All tumor DNAs had been analyzed for mutations in WT1 (all 10 exons) and CTNNB1 (exons 3, 7, and 8), as previously described (Huff et al., 1995; Maiti et al., 2000). Briefly, tumor DNAs were assessed for WT1 point mutations by analysis of PCR products from all 10 exons and for partial or complete deletion of WT1 by Southern blot analysis and, in most cases, quantitative PCR analysis. Of 45 tumors with mutations in WT1 and/or CTNNB1, 24 tumors carried mutations in both genes and 17 carried mutations in WT1 only. Tumor staging ranged from Stages I to V (D’Angio et al., 1989). The age at diagnosis of these children ranged from 4 months to 17 years. Because we were interested in determining whether mutations in WTX and WT1 were in fact mutually exclusive (as reported), the panel of tumors we assessed in this study was enriched for those with WT1 mutations.
WTX Mutational Analysis
The entire coding region of WTX was amplified as a single PCR product from DNA samples from 125 Wilms tumors, including 80 WT1/CTNNB1 wild-type tumors and 45 tumors with mutations in either or both WT1 and CTNNB1. This PCR product was generated using primers F4 5′-AGCAAGCCAAGCATATCGAG-3′ and R1 5′-CTGGGCAGATGCACTTGAGT-3′, then sequenced by the University of Texas M. D. Anderson DNA Sequencing & Analysis Facility using an ABI 3730 DNA Analyzer with the F4 and R1 primers as well as F2 (5′-ACCGGAAGAGCAAGGTC-3′), R2 (5′CCAAGCAGGCCAATCATAG-3′), R3 (5′-GACGGAAGTCCCTCCAGTCT-3′), and R4 (5′-ATGGTCACTAGCCGTTCTTC-3′). Sequence data were generated with ABI Sequencing Analysis software v5.2 and chromatograms were produced using Chromas v1.45. If mutations were observed in the WTX DNA sequence, the analysis was repeated with both tumor and normal tissue DNA from the same patient to confirm the presence of the mutation and to determine if the mutation was present in the patient’s germline.
Quantitative PCR for Genomic DNA
Quantitative PCR (qPCR) was performed using an ABI 7900HT Sequence Detection System thermal cycler (Applied Biosystems, Foster City, California) with SYBR Green. DNA (5 ng) from Wilms tumors was amplified in duplicate with primer sets from the 5′ and 3′ ends of WTX (Rivera et al., 2007) and from two reference amplicons; first, the intron 13 – exon 14 region of NDST1 (N-deacetylase/N-sulfotransferase, located at 5q33.1; intron 13 5′-TCTGAGCTTTCCTTCCCGTTA-3′, exon 14 5′GGAAGTGTCTGGCCCATCTTATA-3′); and second, the intron 4 – exon 5 region of FAH (fumarylacetoacetate hydrolase, located at 15q25.1; intron 4 5′-GGTTGCTGATGGGATCTGTTG-3′, exon 5 5′-TTCTCCTTGTCCCTGAACATGAT-3′). Since both reference amplicons include intronic sequences, these primers would amplify only DNA. Analysis of the dissociation curves for all amplicons demonstrated that only the specific PCR product was generated. These chromosome 5 and 15 genes were used as reference amplicons because chromosomes 5 and 15 are two of the chromosomes most rarely amplified or deleted in Wilms tumors (Wang-Wuu et al., 1990; Kaneko et al., 1991) and both are unique, single-copy genes.
DNA samples were amplified in duplicate using SYBR Green PCR Master Mix (Applied Biosystems). Threshold cycle (Ct) values for duplicate samples were subtracted from the average Ct for each of the reference amplicons to calculate ΔCt. The SD of duplicate measurements of ΔCt was determined and used to generate error bars (Fig. 2). Normal control values for WTX 5′ and 3′ qPCR reactions were determined using the average of five unaffected male or five unaffected female DNA samples, and each experimental sample was normalized using these control values to produce data on relative genomic DNA content. If WTX deletions were observed in tumor samples, the analysis was repeated with both tumor and normal tissue DNA from the same patient to determine if the deletions were present in the patient’s germline. qPCR was also performed on these samples with additional amplicons from the p and q arms of the X chromosome to determine if the deletions involved the entire X chromosome or the entire q arm of the X chromosome. These additional chromosome X amplicons were from the PHEX (phosphate regulating endopeptidase, X-linked) gene at Xp22.11 (intron 19 5′-TCTTCTTCTCTCACCAGGCTTACAG-3′, exon 20 5′-AAGAAGAGCTGGTTGTTGGTGAA-3′) and the OCRL (oculocerebrorenal syndrome of Lowe, phosphatidylinositol polyphosphate 5-phosphatase) gene at Xq25 (intron 19 5′-GACTTCTTTGGTAGGAGGACCTGTTC-3′, exon 20 5′-CGCAAAGGATACGGATTGT-CTC-3′). These amplicons include intronic sequences and hence these primers would amplify only DNA. Analysis of the dissociation curves for these amplicons demonstrated that only the specific PCR product was generated.
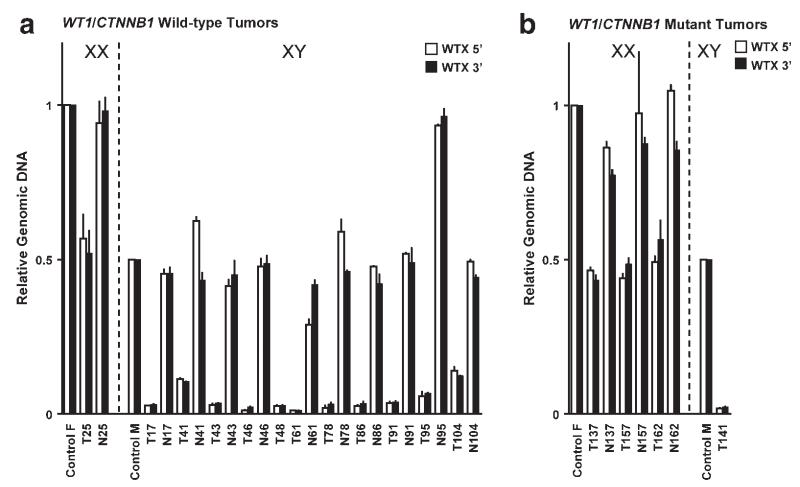
Figure 2.
Quantitative PCR (qPCR) of the 5′ and 3′ ends of the WTX gene. Control samples represent the average of DNA from five normal females or five normal males, set equal to 1.0 or 0.5, respectively. A: comparison of relative WTX genomic DNA in WT1/CTNNB1-wild-type tumors and matched normal tissues from one female and 11 males. For sample T48, no matched normal tissue DNA was available. B: comparison of relative WTX genomic DNA in WT1-mutant tumors and matched normal tissues from three females and one male. For sample T141, no matched normal tissue DNA was available. White bars, amplicon at 5′ end of WTX gene (see Fig. 1B). Black bars, amplicon at 3′ end of WTX gene. Data bars represent the average of duplicate assays normalized for both the chromosome 5 and 15 reference genes for tumor samples from females and the chromosome 5 reference gene alone for males. Error bars indicate SD from the mean. Note: Patient 95 was male but analysis of two normal tissues (N95) yielded results consistent with the presence of two X chromosomes. Both copies of WTX were lost in the tumor (T95); the tumor qPCR results are consistent with the loss of one entire X and the deletion of WTX from the second X.
Quantitative PCR for RNA
cDNAs were prepared from Wilms tumor RNA by reverse transcribing 400 ng of total RNA from tumors using TaqMan Reverse Transcription reagents with random hexamers (Applied Biosystems). cDNAs were then amplified in duplicate in an ABI 7900HT Sequence Detection System thermal cycler using the TaqMan Gene Expression assay for WTX (aka FAM123b; Applied Biosystems), which spans the boundary between exons 1 and 2 of WTX/FAM123b in the region of WTX which is identical to the corresponding region of FAM123b. Expression of glyceraldehyde phosphate dehydrogenase (GAPDH) was determined as an endogenous gene expression control. The SD was determined for duplicate measurements of gene expression and used to generate error bars (Fig. 4). Expression of WTX in Wilms tumors was normalized to the expression in human fetal kidney (Clontech, Mountain View, California; total RNA from pooled fetal kidneys at 12–31 weeks of gestation).
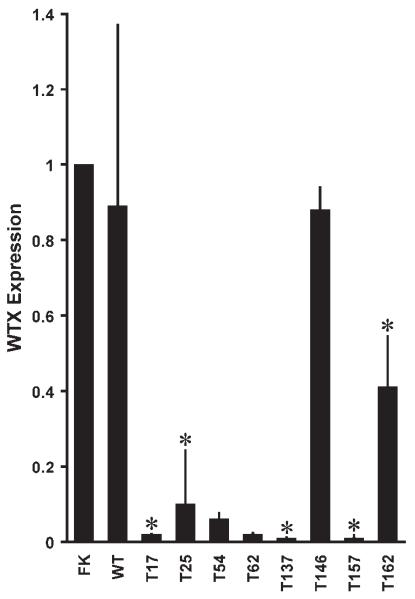
Figure 4.
Quantitative RT-PCR of the WTX gene. Expression of GAPDH, as the endogenous control, was measured in the same cDNA samples. FK, WTX expression in human fetal kidney. WT, the average of WTX expression in 22 Wilms tumors with no identified WTX deletions. Tumors with WTX deletions are indicated by an asterisk (*). Error bars indicate SD from the mean.
RESULTS
Deletions and Mutations of WTX
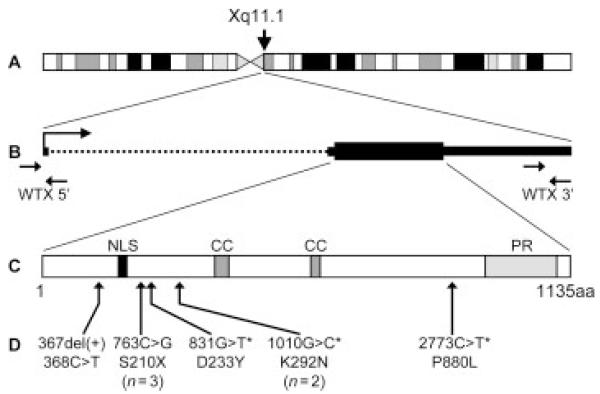
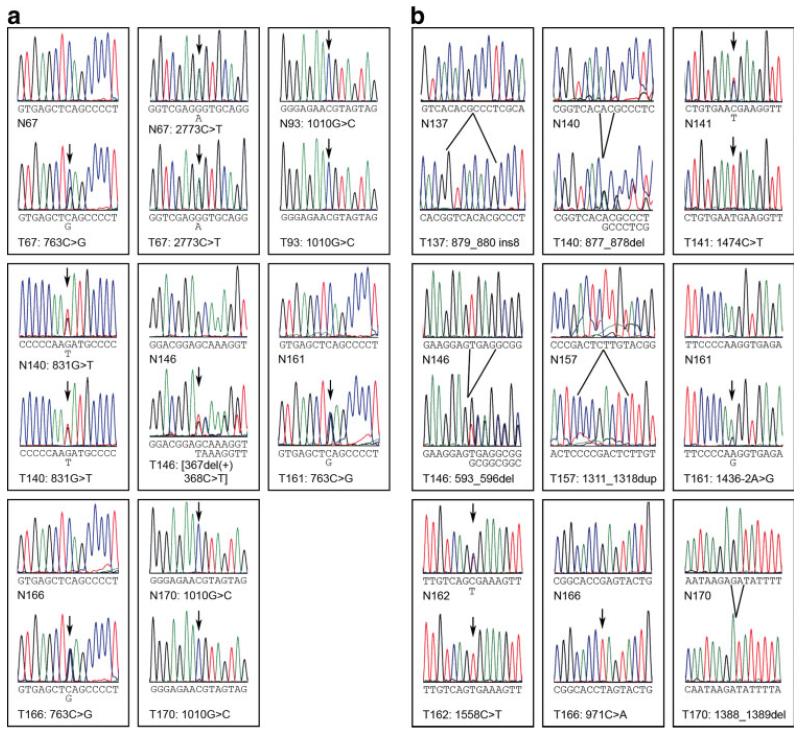
A large panel of 125 Wilms tumors, including 45 tumors with mutations of WT1/CTNNB1 (mutations in WT1 or CTNNB1 or both) and 80 tumors with no detectable mutations in either of these genes, was analyzed for point mutations by sequencing the coding region of the gene and for large deletions of the WTX gene (Fig. 1) using the technique of quantitative real-time PCR with two reference genes, an approach commonly and successfully used to accurately discriminate between zero, one, and two gene copies (Hoebeeck et al., 2005; Howald et al., 2006). WTX was mutated in 23 of these 125 tumors (18.4%); 16 tumors carried deletions of the entire gene and seven tumors carried a total of eight point mutations (Tables 1 and 2, Figs. 2 and 3A). Of the eight point mutations, four were missense alterations (831G>T, D233Y, in one tumor; 1010G>C, K292N, in two tumors; and 2773C>T, P880L, in one tumor), three were nonsense mutations (all 763C>G, encoding S210X), and one was a single nucleotide deletion (367del(+)368C>T) at amino acid 78 that would result in a frameshift and a truncated protein terminating after amino acid 98 (Figs. 1D and 3A). Both deletions and point mutations in WTX were observed in Wilms tumors from both female and male patients (Table 1; Figs. 2A and Fig. 3A). For 20 of the 23 tumors, matched normal tissue from the same patient was available and normal tissue was assessed for the presence of the WTX mutation in the tumor. For all tumors with deletions and nonsense or frameshift mutations, the matched normal tissue showed no alteration in WTX, consistent with a somatic origin for the WTX alteration. For the four WTX missense mutants (one found in a tumor that also carried a nonsense mutation), the alteration was seen also in normal tissue (T67, T93, T140, and T170; see Table 1 and Fig. 3A).
Figure 1.
The X chromosome gene WTX and its protein product. A: the location of WTX near the centromere at Xq11.1. B: the WTX gene. The large arrow indicates the direction of transcription. Small arrows mark the position of 5′ and 3′ primer sets used for quantitative PCR. The WTX coding region is indicated by a thick solid line, the 5′ and 3′ untranslated regions by thin solid lines, and the single intron by a dotted line. The orientation of the gene with respect to the X chromosome has been reversed. C: the 1135 amino acid WTX protein. NLS, nuclear localization signal. CC, coiled-coil domain; PR, proline-rich region. D: the location of point mutations in the WTX protein. Mutations observed more than once in our panel of Wilms tumors are indicated with the number of observations. Mutations present in the germ-line are indicated by an asterisk (*).
TABLE 1.
Wilms Tumors with Alterations of WTX
| Mutations |
|||||||
|---|---|---|---|---|---|---|---|
| Tumor | Sex Age | (m)a | Stageb | WTX c | WT1 c | CTNNB1 c | WTX RNAd |
| T17 | M | 50 | - | Δ e | - | - | 0.02 |
| T25 | F | 36 | - | Δ | - | - | 0.10 |
| T41 | M | 9 | St V | Δ | - | - | - |
| T43 | M | 32 | St I | Δ | - | - | - |
| T46 | M | 16 | - | Δ | - | - | - |
| T48 | M | 35 | St III | Δ | - | - | - |
| T61 | M | 55 | St IV | Δ | - | - | - |
| T67 | F | 83 | St IV | 763C>G, S210X; 2773C>T, P880Lf |
- | - | - |
| T78 | M | 34 | - | Δ | - | - | - |
| T86 | M | 26 | St III | Δ | - | - | - |
| T91 | M | 5 | St III | Δ | - | - | - |
| T93 | M | 54 | St IV | 1010G>C, K292Nf | - | - | - |
| T95 | M | 30 | - | Δ | - | - | - |
| T104 | M | 86 | St II | Δ | - | - | - |
| T137 | F | 26 | - | Δ | 879_880ins8g | - | 0.01 |
| T140 | F | 25 | St II | 831G>T, D233Yg | 877_878dele | - | - |
| T141 | M | 25 | St III | Δ | 1474C>T, R362X | - | - |
| T146 | F | 64 | St III | [367del(1)368C>T] | 593_596del | 343_345del, DS45 | 0.88h |
| T157 | F | 35 | St II | Δ | 1311_1318dupi | - | 0.01 |
| T161 | M | 124 | St III | 763C>G, S210X | 1436-2A>G | 1358G>C, W383S | - |
| T162 | F | 14 | St V | Δ | 1558C>T, R390X | 1357T>G, W383G | 0.41 |
| T166 | F | 36 | St II | 763C>G, S210X | 971C>A, S194X | 1358G>C, W383S | - |
| T170 | M | 9 | - | 1010G>C, K292Nf | 1388_1389del | 1340C>G, P44A | - |
Age in months at diagnosis.
Per the NationalWilms Tumor Study Group staging system (D’Angio et al., 1989).
WTX gene version GI:121556877, WT1 gene version GI:22027472, CTNNB1 gene version GI:40254459.
WTX expression level relative to level (1.00) in fetal kidney.
Deletion.
WTX mutation present in germline.
Insertion.
Mutant WTX allele expressed.
Duplication.
TABLE 2.
WTX Mutation Frequency in Genetically Defined Subsets of Wilms Tumors
| Tumors | Missense | Truncationa | Deletion | All mutations | |
|---|---|---|---|---|---|
| Wilms tumors | 125 | 4 (3.2%) | 4 (3.2%) | 16 (12.8%) | 23b (18.4%) |
| Male | 67 | 1 (1.5%) | 1 (1.5%) | 12 (17.9%) | 14 (20.9%) |
| Female | 58 | 3 (5.2%) | 3 (5.2%) | 4 (6.9%) | 9b (15.5%) |
| Grouped by WT1/CTNNB1 status: | |||||
| Wild-typec | 80 | 2 (2.5%) | 1 (1.3%) | 12 (15.0%) | 14b (17.5%) |
| Male | 44 | 1 (2.3%) | 0 | 11d (25.0%) | 12d (27.3%) |
| Female | 36 | 1 (2.8%) | 1 (2.8%) | 1d (2.8%) | 2b,d (5.6%) |
| Mutante | 45 | 2 (4.4%) | 3 (6.7%) | 4 (8.9%) | 9 (20.0%) |
| Male | 23 | 1 (4.3%) | 1 (4.3%) | 1 (4.3%) | 3 (13.0%) |
| Female | 22 | 1 (4.5%) | 2 (9.1%) | 3 (13.6%) | 6 (27.3%) |
Nonsense or frameshift mutation resulting in a truncated protein.
Includes one tumor with both a missense and a truncation mutation.
No mutations in either WT1 or CTNNB1.
Statistically significant (P ≤ 0.01) difference between frequency of WTX mutations in males and females.
Either or both WT1 and CTNNB1 mutated.
Figure 3.
Sequence chromatograms for WTX and WT1 mutations. The sequence data are shown for the point mutations and small insertions/deletions listed in Table 1. A: WTX mutations. N67/T67 2773C>T sequences are from the non-coding DNA strand. B: WT1 mutations. N166/T166 sequences are from the non-coding DNA strand. N, normal tissue; T, tumor; ins, insertion; del, deletion. Arrows indicate heterozygous or homozygous single-base alterations from the wild-type sequence. Note that patients 93 and 170 were male, with only one X chromosome, and hence were homozygous for germline missense mutations in WTX. Patient 161 was also male, but WTX and WT1 sequencing results both indicate that tumor T161 was likely contaminated with normal tissue. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
On the basis of the previous study, we expected to find no WTX alterations in the group of WT1/CTNNB1 mutant tumors, but when we partitioned the panel of 125 tumors by WT1/CTNNB1 mutation status we were surprised to observe that WTX mutation was equally frequent in the two groups (Table 2). We identified WTX mutations in nine of 45 (20.0%) tumors with mutations in WT1/CTNNB1 (see Fig. 3B) and in 14 of 80 (17.5%) tumors with no WT1/CTNNB1 mutation. Four deletions of the entire WTX gene (Fig. 2B) and five point mutations (Table 1) were found in WT1/CTNNB1 mutant tumors, versus 12 deletions and three point mutations in tumors with no mutation in WT1 or CTNNB1.
The overall frequency of WTX mutations was approximately equivalent in Wilms tumors from males and females (20.9% vs. 15.5%, see Table 2), but the frequencies diverged when the tumors were partitioned into groups with and without WT1 mutations. In the WT1 wild-type tumors, WTX mutations occurred at a significantly higher rate in males than in females (27.3% vs. 5.6%; P = 0.01). In contrast, in WT1-mutant tumors, WTX mutations occurred more frequently in tumors from females (27.3% versus 13.0%), but this difference was not significant (P > 0.05).
DNAs from an additional group of 52 tumors, 49 with no mutation in either WT1 or CTNNB1, two with mutations in both WT1 and CTNNB1, and one with a WT1 mutation only, were analyzed by qPCR only to assess WTX deletion status. We identified WTX deletions (9.6%) in five tumors with no WT1 or CTNNB1 mutation (data not shown), a frequency similar to that seen for the larger group of 125 tumors.
In each case in which we observed a single copy of WTX in a tumor from a female patient, we also carried out qPCR analysis with additional X chromosome amplicons (see Materials and Methods) to determine if the deletion was specific for WTX or encompassed the entire X chromosome or the entire q arm. In four cases of Wilms tumors from female patients we noted the presence of only one copy of X at the WTX, PHEX (Xp22.11), and OCRL (Xq25) genes (data not shown), indicating that one entire X chromosome was lost in these tumors, as has been previously noted in some Wilms tumors (Solis et al., 1988; Wang-Wuu et al., 1990). For all other cases the observed WTX deletion was confined to the WTX amplicons. Tumors from female patients with loss of one entire X chromosome expressed WTX from the remaining, active X chromosome (see below) and so were not included in Tables 1-4 and in Figure 2.
TABLE 4.
Frequency of Mutation of WT1, CTNNB1, and/or WTX in Sporadic, Familial, and Syndromic Wilms Tumors
| Sporadic unilateral | Sporadic bilateral | Sporadic unknown laterality | Familial | Syndromica | All | |
|---|---|---|---|---|---|---|
| Total tumors | 68 | 12 | 13 | 11 | 21 | 125 |
| No mutations | 39 (57.4%) | 6 (50.0%) | 9 (69.2%) | 7 (58.3%) | 5 (23.8%) | 66 (52.8%) |
| WT1 only | 4 (5.9%) | 1 (8.3%) | 1 (7.7%) | 0 | 7 (33.3%) | 13 (10.4%) |
| CTNNB1 only | 1 (1.5%) | 0 | 0 | 2 (16.7%) | 1 (4.8%) | 4 (3.2%) |
| WTX only | 9 (13.2%) | 1 (8.3%) | 2 (15.4%) | 1 (8.3%) | 1 (4.8%) | 14 (11.2%) |
| WT1 + CTNNB1 | 9 (13.2%) | 3 (25.0%) | 1 (7.7%) | 1 (8.3%) | 5 (23.8%) | 19 (15.2%) |
| WT1 + WTX | 3 (4.4%) | 0 | 0 | 0 | 1 (4.8%) | 4 (3.2%) |
| All 3 genes | 3 (4.4%) | 1 (8.3%) | 0 | 0 | 1 (4.8%) | 5 (4.0%) |
Wilms tumors associated with genitourinary anomalies (n = 8), Denys-Drash syndrome (n = 5), WAGR syndrome (n = 4), Beckwith-Wiedemann syndrome (n = 3), or Perlman syndrome (n = 1).
In one patient with bilateral Wilms tumor, a WTX mutation was observed in addition to a germ-line mutation in WT1 in one tumor (T162). Both tumors were reduced to homozygosity for the WT1 mutation (Fig. 3B and data not shown) but were discordant for alterations in WTX and also CTNNB1; tumor T162 carried a deletion of the entire WTX gene but no CTNNB1 exon 3 mutations, and the contralateral tumor from the same patient carried a CTNNB1 exon 3 mutation but no WTX alteration. In another case (T41), a WTX deletion was observed in a tumor from one member of a family with familial predisposition to Wilms tumor linked to FWT2 at 19q13.4 (McDonald et al., 1998), and a tumor from another member of the same family carried no WTX alteration.
WTX Expression Analysis
WTX expression in fetal kidney and in 22 tumors without WTX deletions was approximately equal (Fig. 4, FK and WT). Five Wilms tumors from which RNA was available had been determined to carry deletions of WTX, and, as expected, expression of WTX in four of these tumors (three from females, T25, T137, and T157; one from a male, T17) was correspondingly low (<10% of WTX expression in fetal kidney; see Fig. 4). One tumor (T162) with a WTX deletion from a female patient had moderately low expression (41% of WTX expression in fetal kidney; Fig. 4). This tumor carried a homozygous WT1 missense mutation (Table 1), and sequencing the tumor DNA showed the complete absence of the wild-type nucleotide at the position of the mutation (Fig. 3B), indicating that the tumor preparation was not contaminated with nontumor tissue. The most plausible explanation for the moderately low expression of WTX in T162 is that the inactive (nondeleted) X chromosome had undergone mosaic reactivation in some but not all cells within this tumor, although this was not tested.
Tumor T146, from a female patient, carried a heterozygous WTX frameshift mutation at codon 78 and showed normal expression of WTX. Sequencing of cDNA generated from the T146 WTX mRNA revealed that the expressed allele of WTX carried the frameshift mutation (data not shown). Two additional tumors, T54 and T62, showed very low (<10% of fetal kidney) expression of WTX, possibly indicating that these tumors carried an undetected alteration affecting WTX expression.
Correlation of WTX Status with Clinical Data
Age at diagnosis and assigned staging of tumor for Wilms tumors with alterations of WTX are given in Table 1. The tested panel of Wilms tumors was obtained from a group of patients with an average age at diagnosis of 39.6 months. The average age at diagnosis was 40 months for patients with WTX-mutant tumors and 39.5 months for patients with no detectable WTX mutation, indicating that mutation of WTX was not associated with a change in age at diagnosis. There was a slight preponderance of Stage III tumors in the group of tumors with WTX mutations when compared to the tumors with no mutation in WTX (Table 3), but statistical analysis of this distribution showed no significant difference (P = 0.7 by Fisher’s exact test). Likewise, there was no apparent correlation between WTX expression and age at diagnosis or tumor stage/histology (data not shown).
TABLE 3.
Staging of Genetically Defined Subsets of Wilms Tumors
| Stage I | Stage II | Stage III | Stage IV | Stage V | Total | |
|---|---|---|---|---|---|---|
| All tumors | 24 (24.5%) | 23 (23.5%) | 27 (27.6%) | 11 (11.2%) | 13 (13.3%) | 98 |
| WTX wild-type | 21 (26.6%) | 19 (24.1%) | 19 (24.1%) | 8 (10.1%) | 11 (13.9%) | 79 |
| WTX mutant | 3 (15.8%) | 4 (21.1%) | 7 (36.8%) | 3 (15.8%) | 2 (10.5%) | 19 |
| NWTSa | 40% | 23% | 23% | 10% | 5% |
Mutation Frequency in Sporadic Versus Familial or Syndromic Wilms Tumors
We have previously presented data on the frequency of WT1 mutations in sporadic versus familial and syndromic Wilms tumors (Huff, 1998). In Table 4, we show a similar analysis of the frequency of mutation of WT1, CTNNB1, and WTX, alone or in combination, in our panel of 125 Wilms tumors grouped into categories according to whether the tumors occurred sporadically or within a familial or syndromic context. Sporadic tumors were further grouped by laterality; unilateral, bilateral, or of unknown laterality. The frequency of WTX mutations, alone or in combination with WT1 and/or CTNNB1 mutations, was similar in sporadic unilateral, sporadic bilateral, and sporadic tumors of unknown laterality (15/68, or 22.1%; 2/12, or 16.7%; and 2/13, or 15.4%, respectively, Table 4). WTX mutations were less common in familial tumors (1/11, or 8.3%), but this difference was not significant.
DISCUSSION
We have conducted a thorough mutational analysis of the recently identified Wilms tumor gene, WTX, in a large panel of Wilms tumors which had previously been subjected to a complete mutational analysis for both the WT1 gene and the exon 3 mutational hotspot in the CTNNB1 gene as well as CTNNB1 exons 7 and 8. We identified WTX mutations in 23 of 125 tumors (18.4%) which were subjected to both sequencing and qPCR analyses. We were able to assess matched normal tissue for the presence of WTX germline mutations in 20 of 23 of these cases. Matched normal tissue for all tumors with WTX deletions or nonsense/truncation mutations showed that the WTX alterations were somatic in origin. However, all four patients whose tumors (T67 and T140, from females; T93 and T170, from males) carried WTX missense alterations (831G>T, D233Y; 1010G>C, K292N; 2773C>T, P880L) also showed the WTX alteration in normal tissue, indicating that the variants were present in the germline. The aspartic acid at amino acid 233, the lysine at amino acid 292, and the proline at amino acid 880 are all conserved in most mammals but not in lower vertebrates. The K292N alteration was also previously observed as a somatic alteration (Rivera et al., 2007). None of the missense alterations were present in the National Center for Biotechnology Information (NCBI) SNP database (dbSNP) as known polymorphisms (Sherry et al., 1999). The functional significance of these missense alterations is not clear.
In strong contrast to previous data (Rivera et al., 2007), we identified WTX mutations at the same frequency in tumors with and without mutations in WT1. This result suggests that Wilms tumors with WTX mutations do not comprise a biologically distinct category of tumors. In agreement with this finding is our observation that Wilms tumors with WTX mutations were diagnosed at the same average age and stage as tumors with no mutation in WTX. The discrepancy between our data and the previous findings may be due to the presence of undetected WT1 mutations since the incidence of WT1 mutations in the previous study (<5%) is low relative to that observed when WT1 is assessed for both intragenic mutations in the complete codingregion of the gene and also for exonic deletions. Analysis of the complete coding region of the WT1 gene for both point mutations and also exonic deletions previously demonstrated that WT1 is mutated in ~20% of sporadic, nonsyndromic Wilms tumors (Huff, 1998).
The observed frequency of WTX mutations (18.4%) is lower than the 30% previously reported (Rivera et al., 2007). Although in the present study we enriched our Wilms tumor panel for tumors with mutations in WT1 and/or CTNNB1, this is not expected to introduce a bias since WTX mutations were found to occur with equal frequency in WT1-mutant and WT1-wild-type tumors.
WTX expression analysis revealed, as expected, no or very low WTX expression in the one tested tumor with a WTX deletion from a male patient. WTX expression was also very low in three of four tested tumors with WTX deletions from female patients, indicating that the WTX deletion affected the active X chromosome. One tumor with a WTX deletion from a female patient expressed a moderate level of WTX. In this case, we were able to rule out contamination of the tumor DNA preparation with DNA from nontumor tissue (see Results), which suggests that the WTX expression in this tumor may be due to mosaic reactivation of the inactive (WTX-wild-type) X chromosome. Of 22 tumors with no WTX deletion, two (9%) showed little or no WTX expression, suggesting that in these tumors WTX has been silenced by promoter mutation or epigenetic alteration.
One WT1/CTNNB1 wild-type tumor with a WTX deletion (T41) was from a patient from a Wilms tumor family which showed genetic linkage to 19q13.4. The WTX deletion was not present in the germline, and a tumor obtained from a second affected individual from this same family carried no alteration in WTX. This finding, together with the observations of WTX mutation in one but not both tumors in one bilateral case (T162), no evidence of linkage to the X chromosome in familial Wilms tumor, and the absence of germline WTX alterations that delete the gene or truncate the WTX protein, suggest that WTX mutation is unlikely to be an event predisposing to Wilms tumorigenesis, but may nevertheless play a role in tumorigenesis in predisposed individuals.
The molecular pathogenesis of Wilms tumor is still not well understood, but clearly is genetically heterogeneous. Our previous finding that mutations in CTNNB1 show a highly significant association with mutations in WT1 suggests that the mutation of two different cellular pathways is required for the development of a Wilms tumor (Maiti et al., 2000). The WNT signaling pathway is regulated in large part by modulation of β-catenin protein stability. β-catenin also plays an important role in cell adhesion. Therefore, the observation, in a subset of Wilms tumors, of CTNNB1 mutations that act to stabilize the β-catenin protein implies that dysregulation of the WNT signaling pathway and/or altered cell adhesion is an important step in tumorigenesis in at least some tumors. This notion is further supported by the recent demonstration that the WTX protein forms a complex with members of the WNT pathway and promotes the ubiquitination and degradation of β-catenin (Major et al., 2007). These data also suggest that WTX mutation may have a similar “activating” effect on the WNT pathway as do CTNNB1 mutations. It is therefore of interest to note that five of our Wilms tumors with both WT1 and WTX mutations also carried somatic mutations of CTNNB1. These data would imply that WTX and CTNNB1 mutations are not redundant. However, in three of the tumors the CTNNB1 mutation is an exon 8 missense mutation (1357T>G or 1358G>C) of unknown functional significance. The WTX variant detected in another tumor (T170) was also present in the germline and thus may represent a previously unreported, functionally neutral SNP. Only one tumor (T146) carried a CTNNB1 mutation of known functional significance (343_345del) and also a WTX mutation predicted to be functionally significant (367del(+) 368C>T). With the exception of this single tumor, we found no overlap between functional, expressed WTX mutations and protein-stabilizing mutations in CTNNB1. These data provide further support for our hypothesis that the process of Wilms tumorigenesis requires inactivation of the WNT signaling pathway, via mutation of CTNNB1 or WTX or another WNT pathway protein, as well as a cellular pathway involving WT1, and suggests that those tumors with WT1 mutations but no CTNNB1 or WTX mutation are likely to harbor mutation of another WNT pathway protein. The strong association of CTNNB1 mutations and WT1 mutations suggests that the effect of WT1 ablation on tumor development is either enhanced by β-catenin stabilization and/or is necessary for the viability of CTNNB1-mutant (or, conversely, WT1-mutant) cells.
In contrast to WT1/CTNNB1 mutation, our results indicate that there is no association between WTX and WT1 mutations. The apparently random occurrence of WTX mutation in either a WT1-mutant or WT1-wild-type background sug-gests that WT1 and WTX mutations are not functionally redundant and that there are not distinct mechanisms of Wilms tumorigenesis involving either an initial WT1 mutation or an initial WTX mutation. In fact, our finding of a single case of bilateral Wilms tumors concordant for a germline WT1 mutation but discordant for WTX mutation shows that, in this case, WTX mutation was not an initiating event in tumorigenesis.
Given that the respective mutation frequencies of WT1 and WTX in Wilms tumors are roughly equal at about 20%, that CTNNB1 mutations are found in about 15% of Wilms tumors but are rarely observed in the absence of a WT1 mutation, and that WTX mutations appear to partition randomly between tumors with and without WT1 mutations, alterations in all three of these genes together can underlie the genetic basis of only about one-third of Wilms tumors. Clearly, then, the alteration of additional genes or of their expression must be involved in the process of Wilms tumorigenesis for the remaining two-thirds of tumors. Interestingly, genetic and epigenetic events at chromosome 11p15.5, including loss of heterozygosity at IGF2 and loss of imprinting in the IGF2/H19 region, have been observed in ~70% of Wilms tumors (Ogawa et al., 1993; Rainier et al., 1993; Satoh et al., 2006). However, it is not clear whether these events at 11p15.5 are involved in tumorigenesis or tumor progression, or how alterations at 11p15.5, WT1, WTX, and/or CTNNB1 may interact in the process of forming a Wilms tumor.
ACKNOWLEDGMENT
Authors thank Nargis Alam for technical assistance.
Supported in part by: National Institute of Health; Grant numbers: CA34936, DK69599, CA16672.
REFERENCES
- dbSNP [database on the Internet], build 127. National Center for Biotechnological Information, National Library of Medicine (US); Bethesda (MD): [Google Scholar]
- D’Angio GJ, Breslow N, Beckwith JB, Evans A, Baum E, deLorimier A, Fernbach D, Hrabovsky E, Jones B, Kelalis P, Othersen HB, Tefft M, Thomas PRM. Treatment of Wilms’ tumor. Results of the Third National Wilms’ Tumor Study. Cancer. 1989;64:349–360. doi: 10.1002/1097-0142(19890715)64:2<349::aid-cncr2820640202>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Fukuzawa R, Heathcott RW, More HE, Reeve AE. Sequential WT1 and CTNNB1 mutations and alterations of β-catenin localisation in intralobar nephrogenic rests and associated Wilms tumours: Two case studies. J Clin Pathol. 2007;60:1013–1016. doi: 10.1136/jcp.2006.043083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy RG, Pritchard J, Scambler P, Cowell JK. Loss of heterozygosity for the short arm of chromosome 7 in sporadic Wilms tumour. Oncogene. 1998;17:395–400. doi: 10.1038/sj.onc.1201927. [DOI] [PubMed] [Google Scholar]
- Hoebeeck J, van der Luijt R, Poppe B, De Smet E, Yigit N, Claes K, Zewald R, de Jong GJ, De Paepe A, Speleman F, Vandesompele J. Rapid detection of VHL exon deletions using real-time quantitative PCR. Lab Invest. 2005;85:24–33. doi: 10.1038/labinvest.3700209. [DOI] [PubMed] [Google Scholar]
- Howald C, Merla G, Digilio MC, Amenta S, Lyle R, Deutsch S, Choudhury U, Bottani A, Antonarakis SE, Fryssira H, Dallapiccola B, Reymond A. Two high throughput technologies to detect segmental aneuploidies identify new Williams-Beuren syndrome patients with atypical deletions. J Med Genet. 2006;43:266–273. doi: 10.1136/jmg.2005.034009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huff V. Wilms tumor genetics. Am J Med Genet. 1998;79:260–267. doi: 10.1002/(sici)1096-8628(19981002)79:4<260::aid-ajmg6>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Huff V, Jaffe N, Saunders GF, Strong LC, Villalba F, Ruteshouser EC. WT1 exon 1 deletion/insertion mutations in Wilms tumor patients, associated with di- and trinucleotide repeats and deletion hotspot consensus sequences. Am J Hum Genet. 1995;56:84–90. [PMC free article] [PubMed] [Google Scholar]
- Kaneko Y, Homma C, Maseki N, Sakurai M, Hata J-I. Correlation of chromosome abnormalities with histological and clinical features in Wilms’ and other childhood renal tumors. Cancer Res. 1991;51:5937–5942. [PubMed] [Google Scholar]
- Koesters R, Ridder R, Kopp-Schneider A, Betts D, Adams V, Niggli F, Briner J, von Knebel Doeberitz M. Mutational activation of the beta-catenin proto-oncogene is a common event in the development of Wilms’ tumors. Cancer Res. 1999;59:3880–3882. [PubMed] [Google Scholar]
- Li CM, Kim CE, Margolin AA, Guo M, Zhu J, Mason JM, Hensle TW, Murty VV, Grundy PE, Fearon ER, D’Agati V, Licht JD, Tycko B. CTNNB1 mutations and overexpression of Wnt/β-catenin target genes in WT1-mutant Wilms’ tumors. Am J Pathol. 2004;165:1943–1953. doi: 10.1016/s0002-9440(10)63246-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti S, Alam R, Amos CI, Huff V. Frequent association of beta-catenin and WT1 mutations in Wilms tumors. Cancer Res. 2000;60:6288–6292. [PubMed] [Google Scholar]
- Major MB, Camp ND, Berndt JD, Yi X, Goldenberg SJ, Hubbert C, Biechele TL, Gingras AC, Zheng N, Maccoss MJ, Angers S, Moon RT. Wilms tumor suppressor WTX negatively regulates WNT/β-catenin signaling. Science. 2007;316:1043–1046. doi: 10.1126/science/1141515. [DOI] [PubMed] [Google Scholar]
- Mannens M, Devilee P, Bliek J, Mandjes I, de Kraker J, Heyting C, Slater RM, Westerveld A. Loss of heterozygosity in Wilms’ tumors, studied for six putative tumor suppressor regions, is limited to chromosome 11. Cancer Res. 1990;50:3279–3283. [PubMed] [Google Scholar]
- Maw MA, Grundy PE, Millow LJ, Eccles MR, Dunn RS, Smith PJ, Feinberg AP, Law DJ, Paterson MC, Telzerow PE, Callen DF, Thompson AD, Richards RI, Reeve AE. A third Wilms’ tumor locus on chromosome 16q. Cancer Res. 1992;52:3094–3098. [PubMed] [Google Scholar]
- McDonald JM, Douglass EC, Fisher R, Geiser CF, Krill CE, Strong LC, Virshup D, Huff V. Linkage of familial Wilms’ tumor predisposition to chromosome 19 and a two-locus model for the etiology of familial tumors. Cancer Res. 1998;58:1387–1390. [PubMed] [Google Scholar]
- Ogawa O, Eccles MR, Szeto J, McNoe LA, Yun K, Maw MA, Smith PJ, Reeve AE. Relaxation of insulin-like growth factor II gene imprinting implicated in Wilms’ tumour. Nature. 1993;362:749–751. doi: 10.1038/362749a0. [DOI] [PubMed] [Google Scholar]
- Olson JM, Hamilton A, Breslow NE. Non-11p constitutional chromosome abnormalities in Wilms’ tumor patients. Med Pediatr Oncol. 1995;24:305–309. doi: 10.1002/mpo.2950240507. [DOI] [PubMed] [Google Scholar]
- Polakis P. The many ways of Wnt in cancer. Curr Opin Genet Dev. 2007;17:45–51. doi: 10.1016/j.gde.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Rahman N, Arbour L, Tonin P, Renshaw J, Pelletier J, Baruchel S, Pritchard-Jones K, Stratton MR, Narod SA. Evidence for a familial Wilms’ tumour gene (FWT1) on chromosome 17q12-q21. Nat Genet. 1996;13:461–463. doi: 10.1038/ng0896-461. [DOI] [PubMed] [Google Scholar]
- Rainier S, Johnson LA, Dobry CJ, Ping AJ, Grundy PE, Feinberg AP. Relaxation of imprinted genes in human cancer. Nature. 1993;362:747–749. doi: 10.1038/362747a0. [DOI] [PubMed] [Google Scholar]
- Rivera MN, Kim WJ, Wells J, Driscoll DR, Brannigan BW, Han M, Kim JC, Feinberg AP, Gerald WL, Vargas SO, Chin L, Iafrate AJ, Bell DW, Haber DA. An X chromosome gene, WTX, is commonly inactivated in Wilms tumor. Science. 2007;315:642–645. doi: 10.1126/science.1137509. [DOI] [PubMed] [Google Scholar]
- Ruteshouser EC, Huff V. Familial Wilms tumor. Am J Med Genet. 2004;129C:29–34. doi: 10.1002/ajmg.c.30025. [DOI] [PubMed] [Google Scholar]
- Ruteshouser EC, Hendrickson BW, Colella S, Krahe R, Pinto L, Huff V. Genome-wide loss of heterozygosity analysis of WT1-wild-type and WT1-mutant Wilms tumors. Genes Chromosomes Cancer. 2005;43:172–180. doi: 10.1002/gcc.20169. [DOI] [PubMed] [Google Scholar]
- Satoh Y, Nakadate H, Nakagawachi T, Higashimoto K, Joh K, Masaki Z, Uozumi J, Kaneko Y, Mukai T, Soejima H. Genetic and epigenetic alterations on the short arm of chromosome 11 are involved in a majority of sporadic Wilms’ tumours. Br J Cancer. 2006;95:541–547. doi: 10.1038/sj.bjc.6603302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, Ward M, Sirotkin K. dbSNP-database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res. 1999;9:677–679. [PubMed] [Google Scholar]
- Solis V, Pritchard J, Cowell JK. Cytogenetic changes in Wilms’ tumors. Cancer Genet Cytogenet. 1988;34:223–234. doi: 10.1016/0165-4608(88)90264-6. [DOI] [PubMed] [Google Scholar]
- Wang-Wuu S, Soukup S, Bove K, Gotwals B, Lampkin B. Chromosome analysis of 31 Wilms’ tumors. Cancer Res. 1990;50:2786–2793. [PubMed] [Google Scholar]