Abstract
BACKGROUND
Due to their varied outcomes, men with biochemical recurrence (BCR) following radical prostatectomy (RP) present a management dilemma. Here, we evaluate Decipher, a genomic classifier (GC), for its ability to predict metastasis following BCR.
METHODS
The study population included 85 clinically high-risk patients who developed BCR after RP. Time-dependent receiver operating characteristic (ROC) curves, weighted Cox proportional hazard models and decision curves were used to compare GC scores to Gleason score (GS), PSA doubling time (PSAdT), time to BCR (ttBCR), the Stephenson nomogram and CAPRA-S for predicting metastatic disease progression. All tests were two-sided with a type I error probability of 5%.
RESULTS
GC scores stratified men with BCR into those who would or would not develop metastasis (8% of patients with low versus 40% with high scores developed metastasis, P<0.001). The area under the curve for predicting metastasis after BCR was 0.82 (95% CI, 0.76–0.86) for GC, compared to GS 0.64 (0.58–0.70), PSAdT 0.69 (0.61–0.77) and ttBCR 0.52 (0.46–0.59). Decision curve analysis showed that GC scores had a higher overall net benefit compared to models based solely on clinicopathologic features. In multivariable modeling with clinicopathologic variables, GC score was the only significant predictor of metastasis (P = 0.003).
CONCLUSIONS
When compared to clinicopathologic variables, GC better predicted metastatic progression among this cohort of men with BCR following RP. While confirmatory studies are needed, these results suggest that use of GC may allow for better selection of men requiring earlier initiation of treatment at the time of BCR.
Keywords: biochemical recurrence, genomic classifier, prognostic models, metastasis, clinical validation
INTRODUCTION
Prostate cancer remains the most common visceral malignancy in men, with roughly 240 000 patients diagnosed with the disease last year.1 Approximately half these men will undergo local therapy with radical prostatectomy (RP), and among those, at least a third will eventually have a rising serum PSA as a harbinger of disease recurrence.2,3 These men, with rising PSA as their initial indicator for disease recurrence, continue to present a management dilemma to many clinicians as no standardized management protocols exist.4 This is primarily due to the varied outcomes for men presenting with biochemical recurrence (BCR), with some only having local recurrences and others progressing to clinical metastasis. Further, even among men who experience metastasis following BCR, the time to metastasis can vary over an order of magnitude.5
Directing all men with BCR for salvage local (radiation) or systemic (androgen deprivation) therapy would result in over-treatment, exposing them to added urinary and rectal toxicities and adversely affecting their sexual, cardiovascular and bone health respectively.5–7 Despite these concerns, existing studies indicate that early treatment of men with BCR with local therapies may improve progression-free and disease-specific survival,8,9 and that early use of hormonal deprivation may prolong time to metastasis in certain subgroups.10 Indeed, men most likely to benefit from early treatment are those who have aggressive disease but low tumor burden.
There are multiple algorithms that aim to identify men who go on to develop metastatic disease or prostate cancer mortality following local therapy. These have been based on tumor characteristics at the time of prostatectomy as well as the timing of PSA recurrence and kinetics of its rise.11,12 We reasoned that, as is the case with pathologic features of prostate tumors at prostatectomy, molecular characteristics that define these tumors might augment the prediction of disease aggressiveness at the time of BCR and help guide the decision to employ salvage treatments. Previously, we have developed a molecular test that can be run on routinely processed and stored formalin-fixed paraffin-embedded pathologic tissue.13 Here, we evaluate this genomic classifier (GC) for its ability to predict clinical metastasis following BCR.
MATERIALS AND METHODS
Patient cohort
Between 2000 and 2006, 1010 men with high-risk (preoperative PSA >20 ng ml−1, Gleason ≥8, pT3b or GPSM score ≥1014) prostate cancer treated with RP were consented and recruited under an institutional review board approved protocol at the Mayo Clinic. Utilizing this group, we designed a case-cohort study15 that sampled the population of 401 men who experienced BCR, after surgery. A random sample of 18% of the BCR cohort included 71 men, 24 with metastasis (mets) after BCR. Combined with 52 mets cases not in the random sample, the study had 123 men and yielded expression data and GC scores for 110 (with specimens failing to produce GC scores being primarily those older specimens collected between 2000 and 2001). Men who failed to reach an undetectable PSA after surgery or who had missing clinicopathologic variables at the time of diagnosis or surgery were excluded from the analyses (n = 25), leaving a final cohort of 85 men (47 from random sample [13 with metastasis] + 38 metastasis cases not in the random sample.) None of the patient specimens used in this study cohort overlapped with those used in the training cohort from which the GC score was developed.16
Typically, PSA was measured every 3 months for the first year following prostatectomy, every 6 months for the second year and then annually thereafter. BCR was defined as a PSA ≥0.4 ng ml−1. At the time of BCR, men were restaged with a CT or MRI as well as a bone-scan, which were then performed on a yearly basis. Time to BCR (ttBCR) was defined as the time from RP to first detectable PSA ≥0.4 ng ml−1. Metastasis was defined as a positive bone scan or visceral or extra-pelvic nodal metastasis seen on CT scan. Men who experienced metastasis following BCR were designated as ‘Mets’ and men without metastatic progression following BCR were designated as ‘No-Mets’. Adjuvant therapy was defined as any treatment within 90 days after surgery. Salvage therapy was defined as any treatment after 90 days.17
Specimen selection and processing
Specimen selection and processing of extracted RNA with subsequent oligonucleotide microarray data analysis and GC scores were generated as previously described.16 Raw expression data and clinical information for each subject in the study are available from the National Center for Biotechnology Information’s Gene Expression Omnibus database.
Calculation of GC scores, PSAdT and nomogram scores
Previously, we described a validated 22-marker GC that is able to predict the development of metastatic disease following RP.16 The GC outputs a score between 0 and 1 at increments of 0.1. A GC score was calculated on each patient of the cohort described above and was used as a continuous or binary categorical variable. When used as a continuous variable, hazard ratios correlate to 10% increases in score (0.1 increments). For the latter, a GC score cutoff of 0.4 was determined using the discovery dataset and receiver operating characteristic (ROC)-based methods to optimize sensitivity and specificity in this higher risk cohort of patients compared to the original validation dataset18 (Supplementary Figure S1). PSA doubling time (PSAdT) was calculated by log22 divided by slope of least-squares regression line of log2(PSA) measured over time (in months). CAPRA-S scores were calculated as described by Cooperberg et al.19, and Stephenson 5-year survival probabilities were calculated using the nomogram described by Stephenson et al.20
Statistical analyses
All statistical analyses were performed in R v2.14.1.21 All tests were two-sided with a type I error probability of 5%. GC was compared to standard clinicopathologic variables, PSAdT, ttBCR, and clinical nomogram classifiers (CAPRA-S and Stephenson) for predicting metastasis.19,20 The concordance summary index (extension of c-index), an extension of area under the ROC curve (AUC) for censored data (i.e., the survival ROC) was used to compare classifier performance to predict metastasis. For the survival ROC function,22 the nearest-neighbor estimator was used with λ = 0.002 to approximate survival function density. Decision curve analysis for time-dependent data was used to assess the net increase/decrease in the proportion of necessarily/unnecessarily treated patients.23 Survival ROC and decision curves were built for prediction of metastasis within 3 years post-BCR.24
We used weighted Cox proportional hazard regression models for case-cohort design25 to evaluate the prognostic value and significance of GC scores and clinicopathologic risk factors in predicting the development of metastasis after BCR. Proportional hazards assumptions of the Cox model were confirmed by evaluating scaled Schoenfeld residuals.26 GC was used as a continuous variable; pathological Gleason score (GS) was dichotomized into <8 and ≥8 considering the small number of patients who had a score of 6 and below; pre-operative PSA values were log transformed due to their skewed distribution; seminal vesicle invasion (SVI), surgical margins (SM), extra-capsular extension (ECE), and lymph node involvement (LNI) were treated as binary variables. In all Cox models, the estimated risks were adjusted for the administration of adjuvant therapy. Cumulative incidence curves were constructed using Fine-Gray competing risks analysis to estimate the risk of metastasis accounting for death due to other causes.27 Time-dependent analyses were weighted using the scheme suggested by Barlow.28
RESULTS
Characteristics of men in our cohort who experienced BCR following RP are detailed in Table 1. Median ttBCR was 18.8 months (interquartile ranges 11.0–34.8, mean 25.5, s.d. 19.9). Men experiencing metastasis following BCR did so with a median time of 42.6 months (interquartile range 23.7–61.7, mean 45.1, s.d. 26.2). These men had higher pathological grade and stage at prostatectomy, higher pre-operative PSAs, a more rapid time to BCR and shorter PSAdT. They were also more likely to receive adjuvant and salvage androgen deprivation therapy (Table 1).
Table 1.
Patient characteristics
| BCR patients’ characteristics | N |
Mets N (group %) |
No-Mets N (group %) |
Pearson’s Chi-squared, Fisher exact or Wilcoxon-rank significance test (P) |
|---|---|---|---|---|
| Study cohort | 85 | 51 | 34 | |
| Age | 0.822 | |||
| 46–60 | 35 | 21 (41) | 14 (41) | |
| 61–74 | 50 | 30 (59) | 20 (59) | |
| Pre-operative PSA | 0.40 | |||
| <10 ng ml−1 | 38 | 22 (43) | 16 (47) | |
| 10–20 ng ml−1 | 28 | 15 (29) | 13 (38) | |
| >20 ng ml−1 | 19 | 14 (28) | 5 (15) | |
| Pathological Gleason Score | 0.0029 | |||
| ≤6 | 4 | 0 (0.0) | 4 (12) | |
| 7 | 44 | 23 (86) | 21 (62) | |
| ≥8 | 37 | 28 (55) | 9 (26) | |
| Seminal vesicle invasion | 38 | 29 (57) | 9 (27) | 0.011 |
| Positive surgical margin | 50 | 29 (57) | 21 (62) | 0.822 |
| Extra-capsular extension | 50 | 36 (71) | 14 (41) | 0.013 |
| Lymph node involvement | 17 | 13 (25) | 4 (12) | 0.168 |
| Prostate cancer-specific mortality | 22 | 22 (43) | 0 (0) | — |
| Adjuvant radiation therapy | 11 | 8 (16) | 3 (8) | 0.552 |
| Adjuvant androgen deprivation therapy | 37 | 29 (57) | 8 (24) | 0.005 |
| Salvage radiation therapy | 36 | 22 (43) | 14 (41) | 0.182 |
| Salvage androgen deprivation therapy | 43 | 30 (59) | 13 (38) | 0.009 |
| Time to BCR | 0.19 | |||
| ≤2 years | 51 | 34 (67) | 17 (50) | |
| >2 years | 34 | 17 (33) | 17 (50) | |
| PSAdT (NA = 12) | 0.006 | |||
| ≤9 months | 48 | 33 (65) | 15 (44) | |
| >9 months | 25 | 8 (16) | 17 (50) |
Abbreviations: BCR, biochemical recurrence; Mets, metastasis following BCR; PSAdT, PSA doubling time.
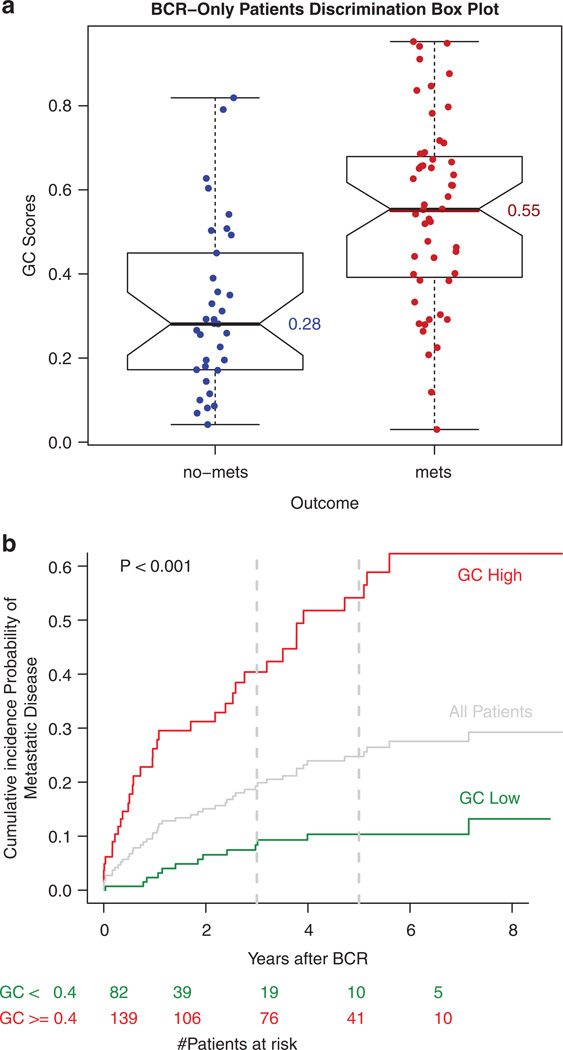
GC scores were significantly higher in men with metastasis following BCR than those without (0.55 versus 0.28, P<0.001, Figure 1a) with a score cut-off of 0.4 best categorizing men into risk groups. For the high-risk group (GC ≥0.4), 73% of men experienced metastasis (sensitivity = 0.73, specificity = 0.74). Furthermore, 40% of men in this group developed metastasis within 3 years of BCR, compared to fewer than 10% of men in the low-risk group (Figure 1b).
Figure 1.
Influence of genomic classifier (GC) score on metastatic progression in patients with biochemical recurrence (BCR) following radical prostatectomy. (a) Box plots demonstrating the distribution of GC among patients who progressed to metastasis following BCR (mets) versus those who did not. (b) Cumulative incidence of clinical metastasis based on GC risk groups.
In univariable analysis, GC predicted metastasis following BCR (HR of 1.62 for every 10% increase in score, 95% CI: 1.33–1.96, P<0.001), as did other clinicopathologic variables commonly associated with poor outcome following prostatectomy, such as GS, SVI, ECE and LNI (Table 2—left). GC was the only significant predictor of metastasis in a multivariable model using clinical information present at the time of BCR (HR of 1.40 for every 10% increase in score, 95% CI: 1.12–1.74, P = 0.003) (Table 2-Right). Furthermore, GC was the dominant predictive risk factor in multivariable analyses involving nomogram scores (Table 3). Though PSAdT values would not be available at the initial time of BCR (like pathological variables and GC score), we also performed regression analysis using PSAdT as a variable. As in previous reports,24 PSAdT was a potent predictor of metastasis in both univariable and multivariable analysis. Even after adjusting for PSAdT, however, GC remained independently predictive of metastatic progression with an HR of 1.49 (95% CI 1.23–1.81, P<0.001) for every 10% increase in score (Supplementary Table 1).
Table 2.
Cox proportional hazards modeling of GC scores
| Univariable | HR (95% CI) | P | Multivariable | HR (95% CI) | P |
|---|---|---|---|---|---|
| GCa | 1.62 (1.33–1.96) | <0.001 | GCa | 1.40 (1.12–1.74) | 0.003 |
| Path Gleason Scoreb ≥8 | 2.55 (1.14–5.69) | 0.023 | Path Gleason Scoreb≥8 | 2.53 (0.94–6.83) | 0.066 |
| Pre-op PSAc (log2) | 1.15 (0.75–1.77) | 0.531 | Pre-op PSAc (log2) | 1.13 (0.78–1.64) | 0.528 |
| SVI | 3.05 (1.36–6.85) | 0.007 | SVI | 1.67 (0.62–4.47) | 0.307 |
| SM | 0.55 (0.25–1.25) | 0.157 | SM | 0.65 (0.27–1.59) | 0.348 |
| ECE | 3.02 (1.31–6.96) | 0.01 | ECE | 1.41 (0.59–3.38) | 0.442 |
| ttBCR | 0.98 (0.749, 1.29) | 0.9 | ttBCR | 1.11 (0.88,1.40) | 0.365 |
| LNI | 5.22 (1.93–14.13) | 0.001 | LNI | 0.59 (0.17–2.09) | 0.417 |
Abbreviations: CI, confidence interval; ECE, extra-capsular extension; GC, genomic classifier; LNI, lymph node involvement; SM, surgical margin; SVI, seminal vesicle invasion; ttBCR, time to biochemical recurrence.
Hazard ratio reported for 10% increase of GC score.
Pathological Gleason Score was dichotomized to compare Gleason Score ≥8 to Gleason Score ≤7.
Hazard ratio reported for 10% increase of log-transformed level.
Table 3.
Cox proportional hazard models of GC scores with clinical-only models
| Univariable | HR (95% CI) | P | Multivariable | HR (95% CI) | P |
|---|---|---|---|---|---|
| GCa | 1.62 (1.33–1.96) | <0.001 | GCa | 1.40 (1.12–1.75) | 0.004 |
| Stephenson 5-Yearsa | 1.51 (1.33–1.72) | <0.001 | Stephenson 5-Yearsa | 1.13 (0.92–1.37) | 0.245 |
| GCa | 1.62 (1.33–1.96) | <0.001 | GCa | 1.44 (1.16–1.78) | <0.001 |
| CAPRA-Sb | 1.58 (1.35–1.85) | <0.001 | CAPRA-Sb | 1.11 (0.89–1.39) | 0.34 |
Abbreviations: CI, confidence interval; GC, genomic classifier.
Hazard ratio reported for 10% increase of GC and Stephenson scores.
Hazard ratio is reported for 1 unit increase in CAPRA-S score.
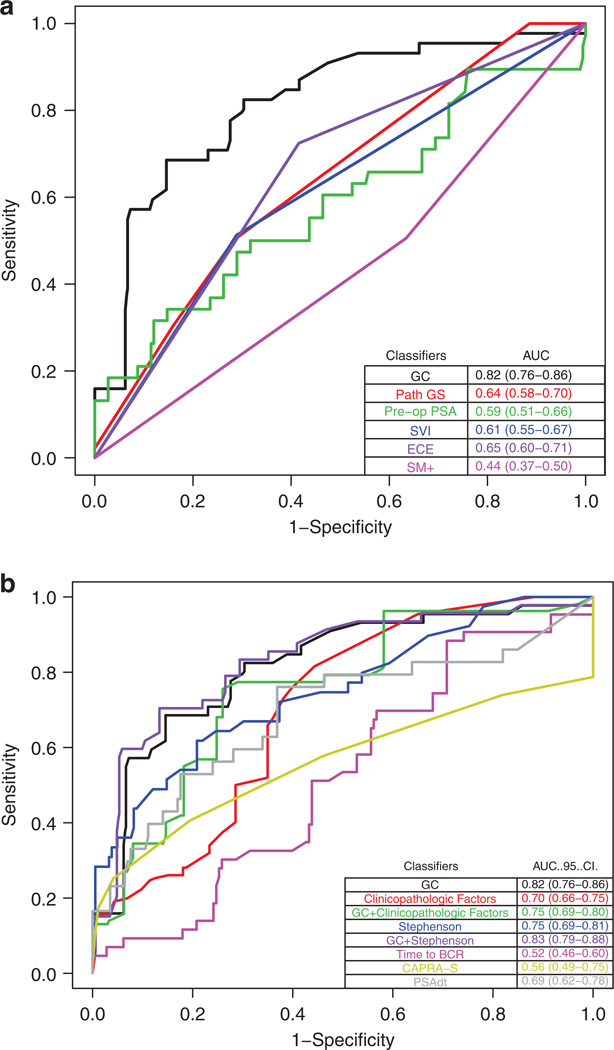
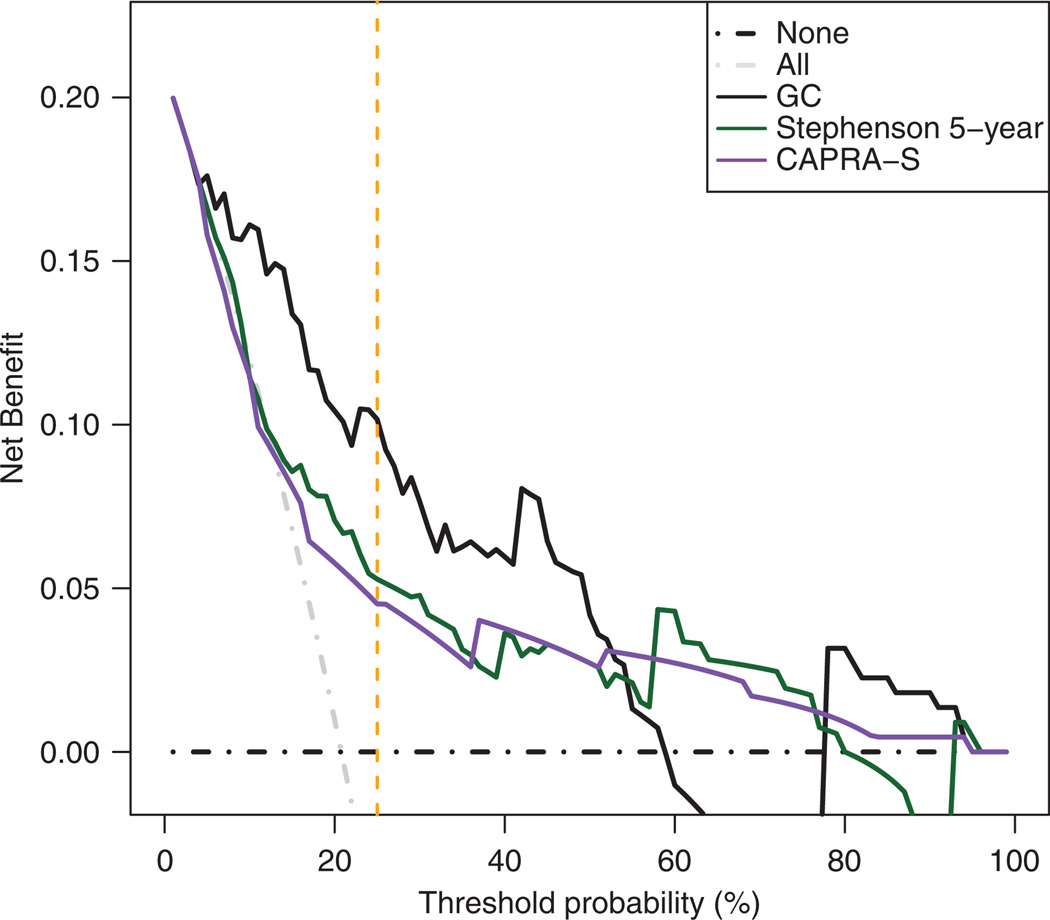
To better determine the performance of GC in identifying men who would go onto metastasis, ROC analysis was performed. GC had a higher AUC than any nomogram or individual clinicopathologic feature (Figure 2a and b). While the AUC of clinicopathologic features taken individually or combined in the Stephenson nomogram could be improved by addition of the GC score, the AUC of the GC score was not improved by adding standard clinicopathologic features (such as pre-operative PSA, tumor stage, grade and surgical margin status) individually or by adding the Stephenson nomogram to GC (AUC 0.82, 95% CI: 0.77–0.86 for GC alone versus 0.75, 95% CI: 0.69–0.80 for GC plus clinicopathologic features or 0.83, 95% CI: 0.79–0.88 for GC plus the Stephenson nomogram; Figure 2b). Finally, decision curve analysis shows that GC was superior to clinical-only models such as CAPRA-S and Stephenson nomograms, with greater net benefit across a wide range of risk (Figure 3). As an example, if the decision to treat was made at a threshold probability of 25% for metastasis, then the reduction in unnecessary treatments using GC and the best clinicopathologic model would be approximately 31% and 16% respectively, when compared to the treat-all model for patients with BCR.
Figure 2.
Receiver operating characteristic (ROC) curve analysis comparing genomic classifier (GC) with (a) individual clinicopathologic factors and (b) clinical models and genomic-clinical models for predicting clinical metastasis at 3 years post biochemical recurrence (BCR). AUC, area under the ROC curve; ECE, extra-capsular extension; GS, Gleason score; SM, surgical margin; SVI, seminal vesicle invasion.
Figure 3.
Decision curve analysis comparing the net benefit of genomic classifier (GC) with clinical-only models.
DISCUSSION
There is clear heterogeneity within the biological spectrum of prostate cancer. While BCR following prostatectomy can be quite concerning to patients and physicians, it has previously been demonstrated that only half of the patients with BCR (followed with observation) will progress to metastatic disease at 10 years.5 Thus, there is a critical need to distinguish those patients who will progress to metastasis versus those who will have an indolent course after PSA recurrence. Currently, standard imaging modalities do not accurately discriminate local from distant metastasis early in the course of PSA recurrence, and promising novel imaging methods await validation.29 In this manuscript, we assess a GC, based on molecular features of the primary tumor, on the prediction of metastatic progression among a cohort of men with BCR following RP. The performance of this GC suggests that it can be used to identify patients who may derive the greatest benefit from initiation of salvage treatments at the time of BCR. Predictors for the development of metastatic disease following BCR have traditionally focused on clinicopathologic variables and PSA-based parameters. These include pathological stage, ttBCR, PSAdT and pathologic Gleason sum, with PSAdT and pathologic Gleason sum being the strongest predictors in multivariable models.5 Based on data generated from our group and that of others, we reasoned that molecular features of the primary tumor might help to better predict metastatic progression.18,30,31 Interestingly, in multivariable models for metastatic outcome, including this molecular information, the GC score was the only independently predictive variable of those available at the time of BCR, though Gleason sum neared statistical significance (P = 0.066). Further, in multivariable models with GC score and CAPRA-S or Stephenson nomograms, only GC retained a significant hazard ratio for predicting metastasis. GC had superior sensitivity and specificity compared to these models with no added benefit to its ROC curve obtained by adding clinicopathologic information. This suggests that the GC score provides independent prognostic information and best captures the metastatic potential of the tumor. Indeed, assessment of the statistical significance of each marker within the group of BCR patients compared against the median (based on 10 000× bootstrapping) of subsets of the same size composed of both BCR and non-BCR patients shows that metastasis within the BCR patients is significantly related to the markers with the greatest significance coming from markers associated with genes involved in cell proliferation and cell cycle progression (such as IQGAP3 and UBE2C), adhesion (ANO7) and control of signaling pathways (TNFRSF19) (a complete list of the markers and their corresponding transcripts can be found in Erho et al.).16.
As with previous reports, PSAdT was highly predictive of metastasis both in univariable and multivariable analysis, but still the GC score remained independently predictive of outcome. Importantly, because the GC score can be determined from analysis of routinely stored, formalin-fixed paraffin-embedded tissue in a time-independent way, it can be utilized regardless of the ttBCR without the need for specialized tissue collection or storage conditions. Additionally, because the GC score can be calculated immediately at the time of BCR, it can guide treatment decisions early, without waiting for the several months that are required to collect PSA measurements necessary to calculate a PSAdT.
While the results of this initial study are promising, there are several important limitations. First, though carefully selected for men with complete clinicopathologic and follow-up information following prostatectomy, the cohort is relatively small and homogeneous, and larger confirmatory studies using specimens from multiple centers reflecting differences in patient management are needed. Second, in order to investigate men who were at risk for metastatic progression, the study population consisted of higher risk men who accordingly had more rapid progression to BCR and metastasis. Thus, the performance of the assay among men with low-grade (Gleason 3 + 3 = 6) disease and BCR cannot be determined. Finally, as this represented a high-risk population, a sizable fraction of men in the study received adjuvant and/or salvage therapy, and this could potentially confound the results despite adjustment for this in multivariable analysis. To further investigate this, studies should be performed using men receiving no adjuvant or salvage therapy until the time of clinical metastasis. It is interesting to note that men with high GC scores were more prone to metastatic progression regardless of aggressive use of additional standard therapies, and this suggests that the GC score may aid in the selection of men for clinical trials in the adjuvant setting.
Currently, an unmet clinical need for patients with post-prostatectomy BCR is the identification of patients who will subsequently develop overt metastatic disease versus those with more indolent disease courses. When compared to clinicopathologic variables and existing nomograms, GC appears to be a better predictor of clinical metastasis among this cohort of men with BCR following RP. While confirmatory studies in additional patient populations are required, these results suggest that GC can be used to better identify men requiring intensification or earlier initiation of treatment at the time of BCR.
Supplementary Material
ACKNOWLEDGEMENTS
AER is supported by the John Hopkins Clinician Scientist Award, EMS is supported by the Howard Hughes Clinician-Scientist Early Careers Award and AUA/Astellas Rising Star Award, and DS is supported by NIH Training Grant T32DK007552.
Footnotes
CONFLICT OF INTEREST
Mercedeh Ghadessi, Nicholas Erho, Anamaria Crisan, Christine Buerki, and Ismael A Vergara are employees of GenomeDx Biosciences. Darby J S Thompson is a consultant for GenomeDx Biosciences. Timothy J Triche and Elai Davicioni are founders of GenomeDx Biosciences. The remaining authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Prostate Cancer and Prostatic Diseases website (http://www.nature.com/pcan)
REFERENCES
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics 2012. Cancer Stat. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Cooperberg MR, Broering JM, Carroll PR. Time trends and local variation in primary treatment of localized prostate cancer. J Clin Oncol. 2010;28:1117–1123. doi: 10.1200/JCO.2009.26.0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moul J. Prostate specific antigen only progression of prostate cancer. J Urol. 2000;163:1632–1642. [PubMed] [Google Scholar]
- 4.Punnen S, Cooperberg MR, D’Amico AV, Karakiewicz PI, Moul JW, Scher HI, et al. Management of biochemical recurrence after primary treatment of prostate cancer: a systematic review of the literature. Eur Urol. 2013 doi: 10.1016/j.eururo.2013.05.025. (e-pub ahead of print). [DOI] [PubMed] [Google Scholar]
- 5.Antonarakis ES, Feng Z, Trock BJ, Humphreys EB, Carducci Ma, Partin AW, et al. The natural history of metastatic progression in men with prostate-specific antigen recurrence after radical prostatectomy: long-term follow-up. BJU Int. 2012;109:32–39. doi: 10.1111/j.1464-410X.2011.10422.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saylor PJ, Smith MR. Metabolic complications of androgen deprivation therapy for prostate cancer. J Urol. 2009;181:1998–2006. doi: 10.1016/j.juro.2009.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patel AR, Stephenson AJ. Radiation therapy for prostate cancer after prostatectomy: adjuvant or salvage? Nature reviews. Urology. 2011;8:385–392. doi: 10.1038/nrurol.2011.80. [DOI] [PubMed] [Google Scholar]
- 8.Stephenson AJ, Scardino PT, Kattan MW, Pisansky TM, Kevin M, Klein EA, et al. Predicting the outcome of salvage radiation therapy for recurrent prostate cancer after radical prostatectomy. J Clin Oncol. 2007;25:2035–2041. doi: 10.1200/JCO.2006.08.9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trock BJ, Han M, Freedland SJ, Humphreys EB, DeWeese TL, Partin AW, et al. Prostate cancer-specific survival following salvage radiotherapy vs observation in men with biochemical recurrence after radical prostatectomy. JAMA. 2008;299:2760–2769. doi: 10.1001/jama.299.23.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moul JW, Wu H, Sun L, McLeod DG, Amling C, Donahue T, et al. Early versus delayed hormonal therapy for prostate-specific antigen only recurrence of prostate cancer after radical prostatectomy. J Urol. 2004;171:1141–1147. doi: 10.1097/01.ju.0000113794.34810.d0. [DOI] [PubMed] [Google Scholar]
- 11.Pound CR, Partin AW, Eisenberger MA, Chan DW, Pearson JD, Walsh PC. Natural history of progression after PSA elevation following radical prostatectomy. JAMA. 1999;281:1591–1597. doi: 10.1001/jama.281.17.1591. [DOI] [PubMed] [Google Scholar]
- 12.Stephenson AJ, Kattan MW, Eastham JA, Bianco FJ, Yossepowitch O, Vickers AJ, et al. Prostate cancer-specific mortality after radical prostatectomy for patients treated in the prostate-specific antigen era. J Clin Oncol. 2009;27:4300–4305. doi: 10.1200/JCO.2008.18.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abdueva D, Wing M, Schaub B, Triche T, Davicioni E. Quantitative expression profiling in formalin-fixed paraffin-embedded samples by affymetrix microarrays. J Mol Diagno. 2010;12:409–417. doi: 10.2353/jmoldx.2010.090155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson RH, Blute ML, Slezak JM, Bergstralh EJ, Leibovich BC. Is the GPSM scoring algorithm for patients with prostate cancer valid in the contemporary era? J Urol. 2007;178:459–463. doi: 10.1016/j.juro.2007.03.124. [DOI] [PubMed] [Google Scholar]
- 15.Barlow WE, Ichikawa L, Rosner D, Izumi S. Analysis of case-cohort designs. J Clin Epidemiol. 1999;52:1165–1172. doi: 10.1016/s0895-4356(99)00102-x. [DOI] [PubMed] [Google Scholar]
- 16.Erho N, Crisan A, Vergara Ia, Mitra AP, Ghadessi M, Buerki C, et al. Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. PLoS ONE. 2013;8:e66855. doi: 10.1371/journal.pone.0066855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blute ML, Bergstralh EJ, Iocca A, Scherer B, Zincke H. Use of Gleason score, prostate specific antigen, seminal vesicle and margin status to predict biochemical failure after radical prostatectomy. J Urol. 2001;165:119–125. doi: 10.1097/00005392-200101000-00030. [DOI] [PubMed] [Google Scholar]
- 18.Karnes RJ, Bergstralh EJ, Davicioni E, Ghadessi M, Buerki C, Mitra AP, et al. Validation of a genomic classifier that predicts metastasis following radical prostatectomy in an at risk patient population. J Urol. 2013 doi: 10.1016/j.juro.2013.06.017. http://linkinghub.elsevier.com/retrieve/pii/S002253471304603X. (e-pub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cooperberg MR, Hilton JF, Carroll PR. The CAPRA-S score: a straightforward tool for improved prediction of outcomes after radical prostatectomy. Cancer. 2011;117:5039–5046. doi: 10.1002/cncr.26169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stephenson AJ, Scardino PT, Eastham JA, Bianco FJ, Jr, Dotan A, Diblasio CJ, et al. Postoperative nomogram predicting the 10-year probability of prostate cancer recurrence after radical prostatectomy. J Clin Oncol. 2008;23:7005–7012. doi: 10.1200/JCO.2005.01.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ihaka R, Gentleman R. R: A language for data analysis and graphics. J Comput Graph Stat. 1996;5:299–314. [Google Scholar]
- 22.Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56:337–344. doi: 10.1111/j.0006-341x.2000.00337.x. [DOI] [PubMed] [Google Scholar]
- 23.Vickers AJ, Cronin AM, Elkin EB, Gonen M. Extensions to decision curve analysis, a novel method for evaluating diagnostic tests, prediction models and molecular markers. BMC Med Inform Decision Making. 2008;8:53. doi: 10.1186/1472-6947-8-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moul JW, Bañez LL, Freedland SJ. Rising PSA in nonmetastatic prostate cancer. Oncology. 2007;21:1436–1445. [PubMed] [Google Scholar]
- 25.Lin DY, Ying Z. Cox regression with incomplete covariate measurements. J Am Statist Assoc. 88:1341–1349. [Google Scholar]
- 26.Grambsch PM, Therneau TM. Proportional hazards tests and diagnostics based on weighted residuals. Biometrika. 1994;81:515–526. [Google Scholar]
- 27.Fine JP, Gray RJ. A proportional hazards model for the subdistribution of a competing risk. J Am Statist Assoc. 1999;94:496–509. [Google Scholar]
- 28.Barlow WE. Robust variance estimation for the case-cohort design. Biometrics. 2013;50:1064–1072. [PubMed] [Google Scholar]
- 29.Murphy RC, Kawashima A, Peller PJ. The utility of 11C-choline PET/CT for imaging prostate cancer: a pictorial guide. Am J Roentgenol. 2011;196:1390–1398. doi: 10.2214/AJR.10.5491. [DOI] [PubMed] [Google Scholar]
- 30.Ding Z, Wu C-J, Chu GC, Xiao Y, Ho D, Zhang J, et al. SMAD4-dependent barrier constrains prostate cancer growth and metastatic progression. Nature. 2011;470:269–273. doi: 10.1038/nature09677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.