Abstract
The mammalian placenta exhibits elevated expression of endogenous retroviruses (ERVs), but the evolutionary significance of this feature remains unclear. I propose that ERV-mediated regulatory evolution was, and continues to be, an important mechanism underlying the evolution of placenta development. Many recent studies have focused on the co-option of ERV-derived genes for specific functional adaptations in the placenta. However, the co-option of ERV-derived regulatory elements has the potential to co-opt entire gene regulatory networks, which, I argue, would facilitate relatively rapid developmental evolution of the placenta. I suggest a model in which an ancient retroviral infection led to the establishment of the ancestral placental developmental gene network through the co-option of ERV-derived regulatory elements. Consequently, placenta development would require elevated tolerance to ERV activity, which in turn would expose a continuous stream of novel ERV mutations that may have catalyzed the developmental diversification of the mammalian placenta.
Keywords: conflict, co-option, endogenous retroviruses, evolution, evolvability, placenta, regulatory evolution
Introduction
A fundamental goal in biology is understanding how novel forms evolve [1]. In mammals, the placenta is a recently evolved organ crucial for fetal development, and is responsible for anchoring the embryo to the uterus, invading and remodeling maternal tissue, and mediating maternal-fetal physiological exchange throughout pregnancy [2]. The evolution of this novel organ was accompanied by the evolution of a novel cell lineage. In early mammalian development, before gastrulation, the trophoblast lineage forms and separates from embryonic cells, ultimately giving rise to multiple differentiated trophoblast cell types that compose the fetal placenta. Trophoblast cells are a mammalian innovation, and the placentas of modern eutherians are composed of multiple differentiated trophoblast cell types that serve to mediate all fetal interactions with the maternal uterine environment, including implantation, local immunosuppression, respiration, and nutrient absorption [3].
How did the trophoblast evolve? Identifying conserved features of trophoblast development in modern eutherian mammals can provide insight into ancestral trophoblast evolution. However, unlike most mammalian organs, the development of the placenta is poorly conserved across species, and differentiated trophoblast cell types exhibit little morphological or molecular orthology across major mammalian orders such as rodents and primates [2, 4]. Here I argue that there is one feature that is both conserved and distinctly unique to trophoblast cells: an elevated tolerance to endogenous retroviral (ERV) transcriptional activity. ERVs are “parasitic” genomic elements that may selfishly propagate within host genomes, and their activity is normally repressed in the developing embryo. Why ERV expression is associated with eutherian placentation remains enigmatic, though it is now evident that ERVs have repeatedly been “domesticated” for placental function [5], suggesting that ERV activity has been influential throughout placenta evolution.
Here, I propose that ERV activity was responsible for establishing the ancestral trophoblast cell type through ERV-mediated regulatory recruitment of developmental genes. The primary rationale for this hypothesis is that ERVs serve as a major source of gene regulatory elements, and I suggest that ERV-derived regulatory elements have become incorporated into the gene regulatory network that defines the trophoblast cell state. Under this model, cellular tolerance to ERV activity is an ancestral and essential component of trophoblast development, and continuous invasion of host genomes by novel ERVs promotes lineage-specific evolutionary divergence of the trophoblast gene regulatory network. Overall, this hypothesis provides a plausible mechanism for the evolutionary persistence of placental ERV activity, and advances a model in which ERV co-option facilitated the evolution and diversification of the mammalian placenta.
Overview of trophoblast ERV activity
Upon infection of its host, a retrovirus integrates as a provirus into the nuclear genome of the cell as part of its lifecycle (Fig. 1). Proviruses are replicated along with the host genome during cellular division, and they possess the ability to become transcribed, produce new viruses, and reinfect other cells or retrotranspose within the genome. When retroviruses integrate into the genomes of germline cells, they become inherited by the next generation as endogenous retroviruses (ERVs). Throughout evolution, ERVs have come to occupy around 5–10% of mammalian genomes. Reflecting their retroviral origins, ERVs exhibit inherently selfish behavior, functioning to replicate themselves and consequently promoting insertional mutagenesis, chromosomal and transcriptional instability, and sometimes tumor formation in the host [6].
Figure 1.
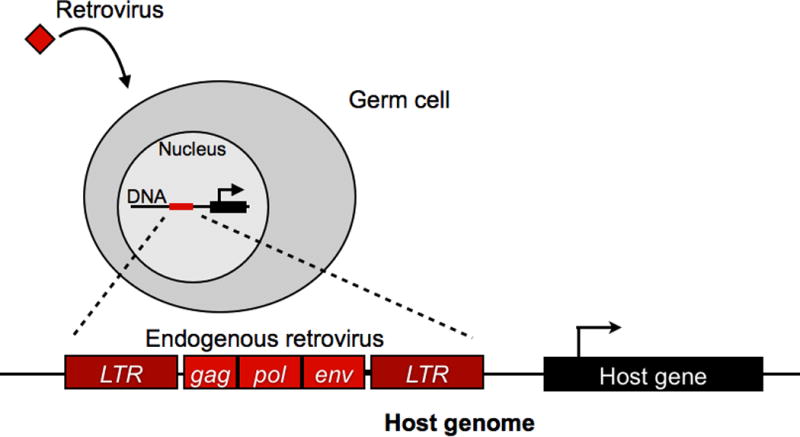
Diagram depicting retroviral endogenization into a host genome.
To repress these deleterious effects, embryonic cells employ a variety of epigenetic and post-transcriptional mechanisms to silence ERV elements in the genome [7]. One of the primary mechanisms of ERV repression is DNA methylation. At the blastocyst stage, embryonic cells undergo a global wave of DNA methylation that silences ERVs and other transposable elements (TEs) throughout the genome [8]. In contrast to the embryo, ERVs tend to be highly transcribed in the fetal placenta. The trophoblast cells in the outer layer of the blastocyst do not undergo a wave of DNA methylation [9], resulting in a transcriptionally permissive epigenetic environment that coincides with hypomethylation of repetitive DNA and elevated genome-wide ERV expression in the placenta [10, 11]. Furthermore, experimentally transfected retroviruses are rapidly silenced in most cell types, but trophoblast cells maintain stable retroviral activity in culture [12]. These data suggest that retroviral-silencing pathways are specifically inactivated in trophoblast cells, and that trophoblast cells feature a distinctively permissive epigenetic environment that tolerates global ERV transcription.
If ERV activity is costly, and it is repressed throughout embryonic development, why would global ERV transcription evolve in the placenta? As a transient organ that serves as a direct physical interface between parent and offspring, the placenta provides a unique context that would theoretically favor the evolution of ERV activity. First, because of its location within the uterus, the placenta presents a novel route for retroviruses to endogenize into the genomes of multiple hosts [13]. Following placental infection by a retrovirus, newly secreted viruses may simultaneously infect the mother, developing siblings, and unfertilized oocytes. Offspring that develop from infected germ cells would harbor these retroviruses in their genomes as novel ERV insertions. This would prove a particularly effective route of transmission, as placental ERVs would be transmitted horizontally to the offspring’s future siblings as well as vertically to the its future progeny. These routes of transmission could be tested by transfecting a blastocyst with a unique replication-competent retrovirus that exhibits placental transcriptional activity, re-implanting the blastocyst into a pseudo-pregnant female, then assaying oocyte genomic DNA for retroviral integration. Subsequent offspring from the same female should contain the newly endogenized retrovirus in their genomes, and exhibit intrinsic retroviral activity in the placenta. Furthermore, mating between infected male offspring with uninfected females should produce similar results, where the newly endogenized retrovirus secreted by the placenta is able to invade uninfected oocytes and thereby transmit horizontally to all subsequent offspring of that female. Though germline endogenization of placental retroviruses has not yet been demonstrated experimentally, infectious retroviruses are clearly capable of intrauterine transmission from the mother to the fetus [14] as well as to the fetal placenta [15]. The combination of both vertical and horizontal transmission would conceivably promote rapid fixation of placental ERVs within the population. Overall, it is possible that placenta ERV activity could be explained purely by the selfish motivations of the retrovirus, without regard to any functional implications for the host.
An emerging trend is that retroviral genes may be readily adaptable for host placental function. During pregnancy, the fetal placenta can be considered as a “parasitic” organ that invades the maternal uterine wall, locally suppresses the maternal immune response, and sequesters maternal nutrients. Viruses, themselves highly successful parasites, may have been recruited by the fetus to assist in some of these basic functions [16, 17]. For example, retroviral expression by the placenta may locally suppress the maternal immune response [16], which would conceivably enable deeper invasion of the placenta and prolonged gestation time. In this way, non-lethal placental ERV activity could plausibly provide a significant fitness advantage to the fetus.
Remarkably, a number of bona fide functional ERV-derived sequences have now been identified in the placentas of a diverse range of mammalian species. Placental retroviral co-option is best exemplified by the “syncytin” genes. The retroviral envelope protein (env) mediates invasion of the virus across the host cell membrane. Most genomic ERV insertions accumulate inactivating mutations in the env genes, presumably because their activity confers no benefit to the host. In humans, a specific env gene contained within a HERV-W insertion has retained an intact open reading frame and is highly expressed in the multinucleate syncytiotrophoblast layer of the placenta. This gene, named “syncytin,” is capable of promoting trophoblast cell fusion in vitro and therefore likely plays a functional role in placenta development [18]. Syncytins have now been identified in at least five mammalian taxa, including rodents [19], ruminants [20], and carnivores [21], and each are independently derived from lineage-specific ERV families [5]. Furthermore, experimental knock-out of syncytin in mouse results in a lethal placental defect [22], demonstrating that ERVs have not only contributed to enhanced placental function, but have become integrated as essential components of placental development.
Additional cases of placental ERV domestication have been identified in sheep [23]. The sheep genome harbors multiple intact endogenous copies of the Jaagsiekte sheep retrovirus (enJSRV), which is related to the exogenous JSRV that infects sheep and causes transmissible lung cancer. The placental expression of replication-incompetent enJSRV transcripts during pregnancy effectively protects the host from exogenous JSRV infection by trans-dominantly interfering with key steps of the JSRV replication cycle [24]. Furthermore, the enJSRV envelope gene is expressed in the trophectoderm and has been co-opted as an essential regulator of early trophoblast growth and differentiation [25]. Overall, the recurrent recruitment, or co-option, of ERV-derived genes suggests that ERVs are indeed well suited for a number of functional roles in the placenta (Fig. 2A).
Figure 2.
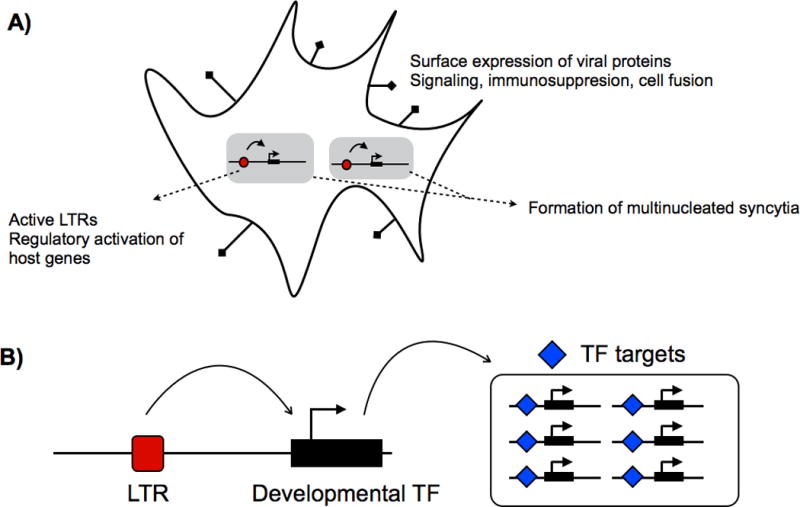
A: Examples of ERV influence on the cell. B: Diagram of LTR cooption resulting in gene network cooption. ERV, endogenous retrovirus; LTR, long terminal repeat.
Taken together, the evolution of placental ERV activity may reflect a highly successful ERV infection strategy, recurrent ERV co-option for placental function, or both. In addition, any costs normally associated with ERV activity, such as elevated mutagenesis or risk of tumor formation, may be mitigated by the transient nature of the placenta. Indeed, the trophoblast lineage appears to be more tolerant of gross chromosomal aberrations such as aneuploidy and polyploidy [26, 27]. Thus, the placenta represents a unique organ that may be primed for the evolution of ERV activity.
LTRs: Architects of the trophoblast cell type?
Given the striking prevalence of ERV co-option across diverse mammalian taxa, it is plausible that trophoblast-specific tolerance to ERV activity originated during the early evolution of the placenta [5]. Such a mutation would fundamentally result in relaxation of ERV-specific repressive epigenetic mechanisms, and result in genome-wide exposure and transcriptional activation of ERVs. An intact ERV insertion contains retroviral genes flanked by long terminal repeats (LTRs), which function to promote transcription of the viral genome. Upon global ERV transcriptional activation, LTRs would be activated throughout the genome, and if these LTRs harbored appropriate binding sites for trophoblast-specific TFs, they may function as active promoters or enhancers with the potential to modulate neighboring gene expression [28]. Thus, any mutation leading to elevated ERV tolerance would significantly alter the regulatory landscape of trophoblast cells.
Though less well studied, the potential for ERVs to facilitate regulatory evolution may be as influential to placenta evolution as their contribution of retroviral protein-coding genes, if not more so. Most co-opted ERV protein-coding genes serve roles in the placenta similar to their original retroviral function. For example, the env-derived “syncytin” genes originally served to facilitate fusion of the virus into the host cell, and have been co-opted to promote fusion of trophoblast cells[5]. Thus, while ERV protein-coding genes are remarkably suitable for use in certain aspects of placentation, there may ultimately be functional limitations to their contribution of novel placental adaptations. By comparison, LTRs are virtually unconstrained in their capacity to change placenta function and development [28]. As regulatory mutations, they may confer placental expression to any existing embryonic gene. For example, a number of placental genes, such as pleiotrophin [29], and leptin [30], have gained trophoblast-specific expression through an LTR promoter. Conceivably, a single LTR regulatory mutation that activates expression of a major transcription factor (TF) could recruit an entire cohort of downstream gene targets. Therefore, whereas co-option of a retroviral gene would confer a specific function such as cell fusion, co-option of an LTR may in turn co-opt an entire gene regulatory network and result in dramatic phenotypic consequences (Fig. 2B).
Modular co-option of gene networks by regulatory mutations is now widely considered a general process underlying the evolution of developmental novelty [31]. Although the trophoblast is a mammal-specific cell type, our current understanding suggests that the ancestral trophoblast gene network was not established by novel mammal-specific genes, but instead evolved primarily by the regulatory recruitment of existing embryonic developmental genes. In mouse, trophoblast stem (TS) cell specification is governed by ancient TFs Tead4, Cdx2, Elf5, and Eomes, all of which hold deeply conserved roles in vertebrate development [32–34]. Furthermore, the transcriptome of the developing placenta is predominated by ancient, rather than mammal-specific genes [35]. Altogether, these findings suggest gene co-option, driven by regulatory mutations, as a major mechanism underlying trophoblast evolution.
Could LTRs have helped to establish the ancestral trophoblast gene network? Upon the evolution of ERV tolerance, trophoblast cells would be exclusively exposed to LTRs as a substantial source of regulatory mutations. Though the individual activity of most of these LTRs would likely be of neutral consequence to the host, it is likely that genome-wide LTR activation would nonetheless have a significant impact on the overall gene regulatory network of the cell. For example, ectopic ERV activation often leads to cancer cell formation in somatic cells, presumably due to stochastic activation of oncogene expression by newly exposed LTR elements [36, 37]. Intriguing parallels have been drawn between placentation and cancer metastasis [38, 39], which raises the possibility that global activation of LTRs may result in certain emergent phenotypes–such as rapid proliferation, invasion, and promotion of vasculogenesis–that are characteristic of cancers as well as placentation. Thus, while ERV-derived genes may have conferred specific functionality to the early placenta, ERV-derived regulatory elements may have orchestrated the evolution of a completely novel cell type–the trophoblast–that is exceptionally adapted for invasive placentation (Fig. 3).
Figure 3.
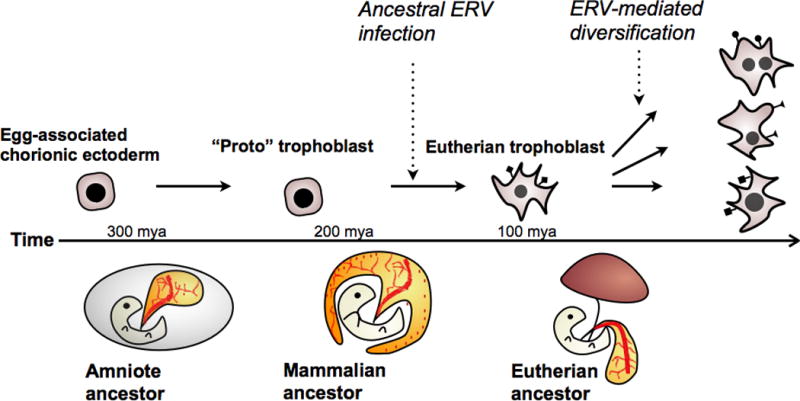
Proposed evolutionary pathway of the mammalian-specific trophoblast cell lineage.
Whether LTRs were actually utilized for the evolution of the ancestral trophoblast gene network remains largely speculative. However, recent studies have provided evidence suggesting that LTRs have an important role in early trophoblast specification {Macfarlan:2012js, [40]. For example, copies of the mouse ERV family MuERV-L function as promoters that activate the transcription of over 100 genes during the pre-blastocyst stage of development, and this activity is required for cells to undergo trophoblast differentiation. MuERV-L activity is silenced in mouse ES cells, which are unable to differentiate into trophoblast [41]. This evidence suggests that LTRs may be required for placental development in mouse, and further investigation in other species should reveal whether co-option of LTR elements as regulators of placental development is a general phenomenon.
ERV co-option promotes rapid trophoblast divergence
One implication of an LTR-dependent gene regulatory network would be strong selection to maintain a permissive epigenetic environment in the cell. Following establishment of global ERV activation in the ancestral placenta, if LTRs became co-opted as essential developmental regulatory elements, then their continued function would strictly depend on the continued tolerance of global ERV activity. Mutations that lead to a more restrictive epigenetic environment that represses ERV activity, as in somatic cells, would pleiotropically silence multiple LTR-derived regulatory elements and prevent proper trophoblast development. In this way, co-option of LTRs as key components of the trophoblast gene regulatory network would eventually establish a trophoblast that is “addicted” to elevated levels of ERV activity. Therefore, I speculate that the ancestral trophoblast cell was formed by an irreversible transition into a cell type that requires tolerance to ERV activity for proper development, and this constraint may explain the apparent conservation of placental ERV transcriptional activity across mammals.
Over time, the trophoblast would be exposed to a continuous stream of novel genes and regulatory mutations as ERVs invade, amplify, and decay throughout the genome. Because these mutations are normally silenced throughout the embryo, I suggest that exposure of ERV mutations would selectively increase the evolvability of the trophoblast cell type. For most organs, an elevated mutation rate would not be intuitively adaptive. However, the placenta is hypothesized to be the site of a coevolutionary arms race between mother and fetus [42]. Theoretically, the optimal strategy for the mother is to fairly allocate resources to maximize the number of her offspring. Conversely, the optimal strategy for the fetus is to “selfishly” maximize its own share of resources at the cost of its mother and siblings. Thus, the fetal placenta would be under constant selection to evolve against maternal counter-adaptations.
The strongest biological evidence for the conflict hypothesis is the prevalence of genomic imprinting, or parent-of-origin- dependent expression, in the mammalian placenta [43]. Most genes are transcribed from both parentally-inherited alleles, but imprinted genes are preferentially transcribed from a specific parent—either the maternal or the paternal allele. Consistent with the conflict hypothesis, many imprinted genes expressed from the paternal allele function to promote fetal and placental growth, such as Insulin-like growth factor 2 (igf2). In contrast, many genes expressed from the maternal allele, such as Insulin-like growth factor 2 receptor (igf2r), act to repress fetal growth [44]. Within vertebrates, genomic imprinting has only been observed in viviparous mammals, which includes marsupials and eutherians. Notably, genomic imprinting has also evolved in the seed endosperm of flowering plants [45]. The endosperm nourishes the plant embryo, contains both maternal and paternal genomes, and is analagous in many ways to the mammalian placenta as a “battleground” for parent-offspring conflict. The convergent evolution of genomic imprinting in flowering plants and viviparous mammals is a strong indication that imprinting is driven in part by parent-offspring conflict. Intriguingly, some evidence suggests that the evolution of imprinted domains was facilitated by differentially methylated retrotransposon insertions [46].
As parent-offspring conflict is contained within each species, the placenta would be expected undergo rapid lineage-specific evolutionary divergence [47]. Consistent with this prediction, the placenta exhibits a high degree of both physiological and morphological diversity. Proteins secreted by the placentas of primates, rodents, and ruminants are encoded by lineage-specific gene families and exhibit elevated amino acid divergence rates [35, 48–50], which is considered a strong signature of genetic conflict [51]. At the developmental level, the spatial patterning of the trophoblast at the blastocyst stage, the mode of attachment and invasion, and the overall architecture of the placenta also exhibit extensive diversity across species [4]. Differentiated trophoblast subtypes are also predominantly species-specific. For example, the mouse placenta features spongiotrophoblast, glycogen trophoblast cells, and multiple subtypes of trophoblast giant cells, none of which have a clear orthologous cell type in non-rodent placentae [52].
Under the context of parent-offspring conflict, increased availability of placental mutations could hypothetically be advantageous for the fetus. The placenta would be under constant selection to outcompete siblings for maternal resources, and also to evolve against repressive maternal counter-adaptations. In such coevolutionary “arms-race” scenarios, the ability to rapidly evolve towards the ever-changing fitness optimum would theoretically be adaptive [53]. For example, retroviruses feature extremely high mutation rates, which allows them to rapidly evolve against host immune systems [54]. In mammals, elevated mutation rates may result in harmful mutation load, but the permissive placental epigenetic environment allows for ERV-derived germline mutations to be specifically exposed in the the transient placenta, without affecting embryonic development. Although most of the exposed placental ERVs would likely be neutral or even slightly deleterious to placental function, they may also serve as a source of mutations that could—over time—facilitate rapid adaptation of the placenta under continuously changing selective pressures.
Whether trophoblast-specific ERVs actually increase the evolvability of the placenta remains speculation and has yet to be functionally tested. Because of the asymmetrical exposure of ERVs in the trophoblast compared to embryonic cells, the placenta would be expected to exhibit consistently greater phenotypic variation than the embryo in species exhibiting sufficient ERV polymorphism. At the genomic level, this would be reflected by individual- or species-specific ERV elements that functionally influence placentation. Functional genomic studies in rodents are beginning to provide evidence for ERV-mediated divergence of the trophoblast gene regulatory network. Maintenance of mouse TS cell self-renewal is controlled by the transcription factors Eomes, Cdx2, and Elf5 [55], and the same factors appear to specify trophoblast stem cells in human as well [56]. Strikingly, when comparing the regulatory networks of mouse and rat TS cells, over a third of all “core” enhancer elements triply bound by Eomes, Cdx2, and Elf5 are derived from a mouse-specific ERV family RLTR13 [57]. RLTR13-derived enhancers are only active in TS cells and not in any somatic cells, suggesting that this source of mutations may only be accessible in a permissive epigenetic environment. While most species-specific LTR elements are not likely to be beneficial to the host upon their initial integration, they collectively serve as an additional source of mutations that have the potential to rapidly rewire the trophoblast gene regulatory network (Figs. 4, 5). Over time, this process could potentially give rise to novel lineage-specific trophoblast developmental traits.
Figure 4.
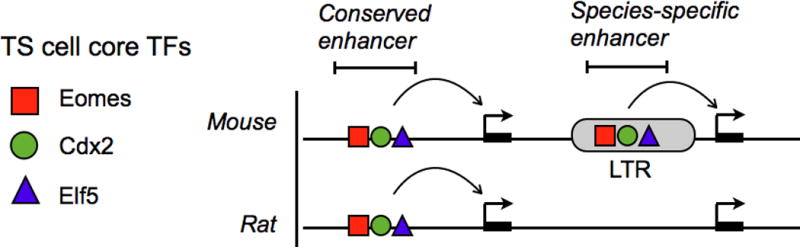
Example of a species-specific gene regulatory enhancer derived from an LTR that contains TS cell-specific TF binding sites. TFs, transcription factors; LTR, long terminal repeat; TS, trophoblast stem.
Figure 5.
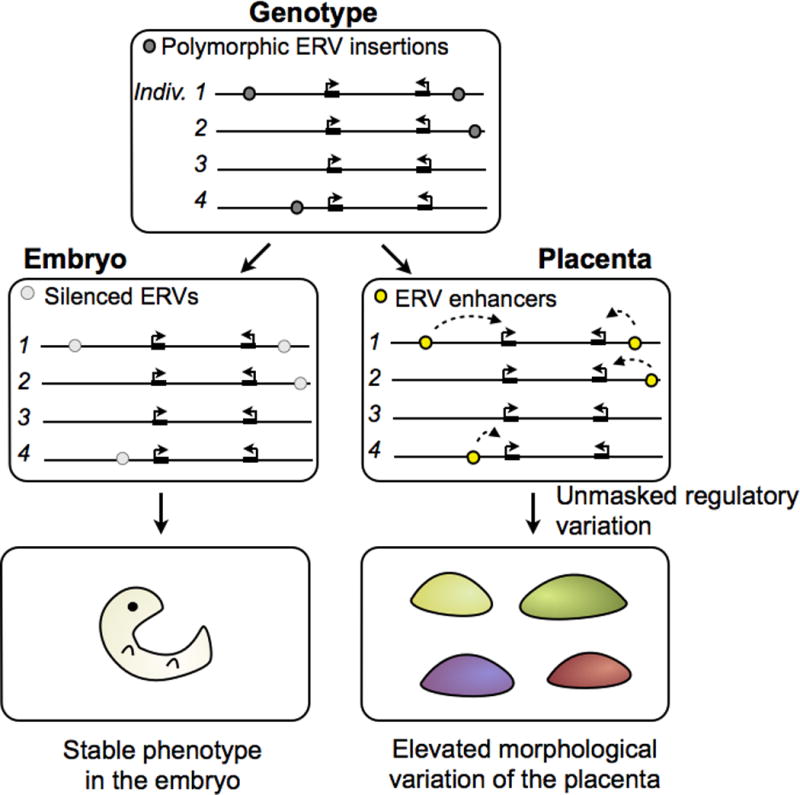
Diagram illustrating how ERVs may be exposed as active regulatory mutations in the placenta, facilitating rapid evolution of placenta development. Adapted from [57].
Open questions
The main hypothesis raised here is that global ERV activity is an ancestral and essential component of trophoblast cell identity. To address this, the molecular bases of trophoblast ERV activity must first be more thoroughly characterized across a wide range of mammalian taxa. If a significant number of species lack trophoblast ERV activity, then it may instead suggest that ERV activity is the result of independent retroviral infections. If this were solely the case, then experimental repression of trophoblast ERV activity should either have no effect or even improve placental efficiency. Though this direct experiment has not yet been performed, animals cloned by somatic cell nuclear transfer (SCNT) often die within in the womb as a result of placental developmental defects. One of the primary conceptual problems of SCNT is that the trophoblast lineage retains the same level of methylation as the original somatic cell nucleus. This results in a placenta where ERVs and LTRs that would normally be exposed in wild-type trophoblast cells are instead silenced. Placental defects in SCNT-cloned embryos have been reported in cattle, sheep, and mice [58], which suggests that a hypomethylated genome is generally important for trophoblast development [11]. However, a more controlled experiment would involve direct experimental ablation of ERV activity by creating mutant trophoblast cells. There are several epigenetic pathways, including LSD1/KDM1A, that are active in ERV-repressing cells but potentially inactive in the trophoblast [12]. Generation of a mouse mutants with ectopic expression of these pathways in the trophoblast lineage should reveal whether defects in placentation are a direct consequence of global ERV repression.
If trophoblast ERV activity is highly conserved across species, and experimental ablation of trophoblast ERV activity consistently results in placental defects, then ERV activity is most likely an ancestral trait and was maintained by selection throughout mammalian evolution. If the ancestral trophoblast cell type was established by early mammalian LTRs, traces of these elements may still be evident in all mammalian genomes. Comparative genomic analyses, combined with functional genomic data from trophoblast cells across diverse species, would reveal whether a conserved set of ancestral, eutherian-specific LTRs drive gene expression in trophoblast development. However, the dynamic nature of ERV evolution theoretically poses several challenges to investigating the role of ERVs in trophoblast evolution. Notably, the absence of conserved ERV elements may be explained by continuous evolutionary turnover as novel ERV insertions functionally replace older ERVs [20]. This appears to be the case with placental ERV gene co-option. For example, the human genome contains multiple conserved ERV-derived genes including syncytin-1, syncytin-2, and erv3, yet erv3 seems to lack any function [59]. If ERV recruitment is driven by parent-offspring conflict, then novel ERVs may be expected to eventually replace old ERVs as maternal defenses evolve to repress placental efficiency [60].
A key question is whether ERV activity evolved before or after eutherians diverged from marsupials. Monotremes, which are egg-laying mammals, would not be expected to exhibit ERV activity in their extraembryonic membranes. Further, the monotreme genome appears to be devoid of LTRs and ERV elements [61]. Marsupials bear live young, but their placenta is relatively short-lived and structurally basic, consisting of a single layer of trophoblast-like cells attached to the yolk sac membrane [62]. Given that marsupial placentation is brief and non-invasive relative to eutherian placentation [63, 64], ERVs might be expected to be silenced in the marsupial placenta. The diversity and complexity of eutherian placental forms would be consistent with a eutherian-specific evolutionary “burst,” possibly coinciding with integration of ERVs into the trophoblast gene regulatory network, followed by LTR-mediated diversification. If ERV activity is absent in the marsupial placenta, this would suggest that the marsupial trophoblast may represent a more primitive non-ERV-dependent trophoblast, and implicate eutherian-specific trophoblast ERV activity as a significant mechanism underlying the evolution of the complex eutherian placenta.
A fundamental prediction of an ancestral ERV-dependent trophoblast model would be evidence for extensive ERV co-option driving placental evolutionary diversification. While ERV co-option does occur in somatic cells [65], elevated tolerance to ERV activity in trophoblast cells would theoretically facilitate a greater frequency of ERV co-option, especially under the context of positive selection driven by parent-offspring conflict. Overall, this model would predict evidence of frequent and continuous co-option of ERVs and LTRs in trophoblast cells in all eutherian species, and that the prevalence of ERV co-option should be elevated compared with somatic cells. Testing this hypothesis would principally involve the identification and functional investigation of LTR-derived regulatory mutations. As a plethora of mammalian genomic data is now available, comparative genomics combined with functional genomic techniques such as chromatin immunopreciptation followed by sequencing (ChIP-Seq) of trophoblast TFs or enhancer-associated chromatin marks should allow for global detection of LTR-derived regulatory elements [57]. However, the most important—and most challenging—step is to demonstrate that specific LTR mutations are functionally important for placentation. Inactivation of species-specific LTRs would be expected to disrupt the development of species-specific features. Exciting developments in genome editing technologies, such as the TALEN [66] and CRISPR [67] systems, should facilitate the experimental perturbation of specific or multiple LTR regulatory mutations. Evidence for increased trophoblast evolvability would suggest that epigenetic suppression of ERVs and other TEs may be modulated throughout evolution to adjust evolvability in a tissue-specific manner. Interestingly, other organs such as the brain or the testis show evidence of unique epigenetic environments that allow for TE activity [68–70], but further investigation will be required to demonstrate whether tissue-specific TE exposure confers any adaptive benefit.
What are the implications of ERV activity in human placentation? Though humans essentially have no currently replicating ERVs [6], ERVs are widely transcribed in the human placenta, including the coopted ERV-derived genes syncytin1 and syncytin2. If successful placentation relied on the activity of ERV-derived genes and regulatory elements, then epigenetic aberrations that result in global ERV repression may be responsible for some placental defects such as pre-eclampsia. Indeed, there is evidence to suggest that mis-regulation of the syncytin genes correlates with increased incidence of pre-eclampsia [71], though further investigation is clearly needed to demonstrate a functional link between ERV activity and placental defects. A straightforward genetic basis for pre-eclampsia has remained elusive [72], but given that environmental or other stochastic factors can influence the placental epigenome during pregnancy [73], it may be possible that incidences of pre-eclampsia with apparently healthy genotypes are a result of inappropriate methylation at co-opted ERV elements. Further, though ERV elements are largely fixed in the population, other TEs such as LINE or SINE elements exhibit polymorphism in the human population [74] and are also hypomethylated and preferentially expressed in the placenta [75]. Thus LINE and SINE elements exposed in the human placenta may introduce an additional source of genetic variation affecting placenta development, and possibly influencing pregnancy outcome.
Finally, though the placenta evolved only once in mammals, the placenta has evolved multiple times in other vertebrates including fish [76], sharks [77], lizards [78], and snakes [79]. The evolutionary pressures that promote ERV activity in the placenta are not necessarily exclusive to mammals, and it would be of particular interest to investigate whether ERVs are also expressed during placentation in non-mammalian species. Such mutualistic relationships with viruses are not unheard of in nature. Parasitic wasps lay their eggs inside live insects, and many species of parasitic wasps exhibit symbiotic relationships with polydnaviruses to influence host behavior and physiology for protection against the host immune system [80, 81].
Conclusions and outlook
Live birth is a definitive feature of modern mammals, made possible by the evolution of the placenta. Though pregnancy may outwardly seem a harmonious symbiosis between parent and offspring, current understanding of placenta evolution suggests that the behavior and evolution of the fetus have certain features in common with parasitism. The prevalence of ERV co-option raises the intriguing possibility that the fetal placenta recruits the activity of its own genomic parasites as part of its arsenal in the parent-offspring coevolutionary arms race. Abundant examples across multiple mammalian taxa now exist where ERV-derived protein-coding genes, such as the syncytins, have been recruited to facilitate diverse placental adaptations. More recent studies have begun to reveal a potentially critical role for ERV-derived LTR regulatory elements, such as RLTR13 or MuERV-L elements, in driving the development and differentiation of the trophoblast lineage. Given that ERVs represent a rapidly evolving source of mutations, and the placenta exhibits extensive divergence across species, future work should seek to elucidate the extent to which ERVs have contributed to the diversification of the eutherian placenta. Overall, it is likely that our understanding of the role of ERVs in placentation is only at its nascent stage, and as more genomic data becomes available and functional genomic experiments become more accessible, ERVs will undoubtedly emerge to be even more intertwined with placenta evolution than is currently appreciated.
Acknowledgments
The author is most grateful for the mentorship and support of Professor Julie Baker. The author also wishes to thank Elizabeth Finn, and Roberta Hannibal whose discussions were invaluable, and Professor Cedric Feschotte for critical feedback. This work was supported by the Stanford Genome Training Grant (E.B.C.; T32 HG000044) and the National Science Foundation Graduate Research Fellowship (E.B.C.; 2008052909).
Abbreviations
- ERV
endogenous retrovirus
- LTR
long terminal repeat
- TF
transcription factor
Footnotes
The author declares no conflicts of interest.
References
- 1.Carroll SB. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- 2.Mossman HW. Vertebrate Fetal Membranes: Comparative Ontogeny and Morphology; Evolution; Phylogenetic Significance; Basic Functions; Research Opportunities. Rutgers University Press; 1987. [Google Scholar]
- 3.Cross J. How to make a placenta: mechanisms of trophoblast cell differentiation in mice–a review. Placenta. 2005;26(Suppl A):S3–9. doi: 10.1016/j.placenta.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 4.Enders A. Reasons for diversity of placental structure. Placenta. 2009;30(Suppl A):S15–8. doi: 10.1016/j.placenta.2008.09.018. [DOI] [PubMed] [Google Scholar]
- 5.Dupressoir A, Lavialle C, Heidmann T. From ancestral infectious retroviruses to bona fide cellular genes: Role of the captured syncytins in placentation. Placenta. 2012;33:663–71. doi: 10.1016/j.placenta.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 6.Feschotte C, Gilbert C. Endogenous viruses: insights into viral evolution and impact on host biology. Nat Rev Genet. 2012;13:283–96. doi: 10.1038/nrg3199. [DOI] [PubMed] [Google Scholar]
- 7.Rowe HM, Trono D. Dynamic control of endogenous retroviruses during development. Virology. 2011;411:273–87. doi: 10.1016/j.virol.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 8.Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013;14:204–20. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 9.Sanford JP, Chapman VM, Rossant J. DNA methylation in extraembryonic lineages of mammals. Trends Genet. 1985;1:89–93. [Google Scholar]
- 10.Chapman V, Forrester L, Sanford J, Hastie N, et al. Cell lineage-specific undermethylation of mouse repetitive DNA. Nature. 1984;307:284–6. doi: 10.1038/307284a0. [DOI] [PubMed] [Google Scholar]
- 11.Hemberger M. Genetic-epigenetic intersection in trophoblast differentiation: Implications for extraembryonic tissue function. Epigenetics. 2010;5:24–9. doi: 10.4161/epi.5.1.10589. [DOI] [PubMed] [Google Scholar]
- 12.Golding MC, Zhang L, Mann MRW. Multiple epigenetic modifiers induce aggressive viral extinction in extraembryonic endoderm stem cells. Cell Stem Cell. 2010;6:457–67. doi: 10.1016/j.stem.2010.03.014. [DOI] [PubMed] [Google Scholar]
- 13.Haig D. Retroviruses and the placenta. Curr Biol. 2012;22:R609–13. doi: 10.1016/j.cub.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 14.Lewis SH, Fox HE, Lewis SH, Lewis SH, et al. HIV-1 in trophoblastic and villous Hofbauer cells, and haematological precursors in eight-week fetuses. Lancet. 1990;335:565–8. doi: 10.1016/0140-6736(90)90349-a. [DOI] [PubMed] [Google Scholar]
- 15.Black SG, Arnaud F, Burghardt RC, Satterfield MC, et al. Viral particles of endogenous betaretroviruses are released in the sheep uterus and infect the conceptus trophectoderm in a transspecies embryo transfer model. J Virol. 2010;84:9078–85. doi: 10.1128/JVI.00950-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris JR. Placental endogenous retrovirus (ERV): structural, functional, and evolutionary significance. BioEssays. 1998;20:307–16. doi: 10.1002/(SICI)1521-1878(199804)20:4<307::AID-BIES7>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 17.Sugimoto J, Schust DJ. Review: human endogenous retroviruses and the placenta. Reprod Sci. 2009;16:1023–33. doi: 10.1177/1933719109336620. [DOI] [PubMed] [Google Scholar]
- 18.Mi S, Lee X, Li X, Veldman GM, et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000;403:785–9. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- 19.Dupressoir A, Marceau G, Vernochet C, Benit L, et al. Syncytin-A and syncytin-B, two fusogenic placenta-specific murine envelope genes of retroviral origin conserved in Muridae. Proc Natl Acad Sci USA. 2005;102:725–30. doi: 10.1073/pnas.0406509102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Esnault C, Cornelis G, Heidmann O, Heidmann T. Differential evolutionary fate of an ancestral primate endogenous retrovirus envelope gene, the EnvV syncytin, captured for a function in placentation. PLoS Genet. 2013;9:e1003400. doi: 10.1371/journal.pgen.1003400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cornelis G, Heidmann O, Bernard-Stoecklin S, Reynaud K, et al. Ancestral capture of syncytin-Car1, a fusogenic endogenous retroviral envelope gene involved in placentation and conserved in Carnivora. Proc Natl Acad Sci USA. 2012;109:E432–41. doi: 10.1073/pnas.1115346109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dupressoir A, Vernochet C, Bawa O, Harper F, et al. Syncytin-A knockout mice demonstrate the critical role in placentation of a fusogenic, endogenous retrovirus-derived, envelope gene. Proc Natl Acad Sci USA. 2009;106:12127–32. doi: 10.1073/pnas.0902925106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varela M, Spencer TE, Palmarini M, Arnaud F. Friendly Viruses. Ann N Y Acad Sci. 2009;1178:157–72. doi: 10.1111/j.1749-6632.2009.05002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murcia PR, Arnaud F, Palmarini M. The transdominant endogenous retrovirus enJS56A1 associates with and blocks intracellular trafficking of Jaagsiekte sheep retrovirus Gag. J Virol. 2007;81:1762–72. doi: 10.1128/JVI.01859-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dunlap KA, Palmarini M, Varela M, Burghardt RC, et al. Endogenous retroviruses regulate periimplantation placental growth and differentiation. Proc Natl Acad Sci USA. 2006;103:14390–5. doi: 10.1073/pnas.0603836103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weier JF, Weier H-UG, Jung CJ, Gormley M, et al. Human cytotrophoblasts acquire aneuploidies as they differentiate to an invasive phenotype. Dev Biol. 2005;279:420–32. doi: 10.1016/j.ydbio.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 27.Simmons DG, Fortier AL, Cross JC. Diverse subtypes and developmental origins of trophoblast giant cells in the mouse placenta. Developmental biology. 2007;304:567–78. doi: 10.1016/j.ydbio.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 28.Cohen CJ, Lock WM, Mager DL. Endogenous retroviral LTRs as promoters for human genes: a critical assessment. Gene. 2009;448:105–14. doi: 10.1016/j.gene.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 29.Schulte AM, Lai S, Kurtz A, Czubayko F, et al. Human trophoblast and choriocarcinoma expression of the growth factor pleiotrophin attributable to germ-line insertion of an endogenous retrovirus. Proc Natl Acad Sci USA. 1996;93:14759–64. doi: 10.1073/pnas.93.25.14759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bi S, Gavrilova O, Gong D-W, Mason M, et al. Identification of a placental enhancer for the human leptin gene. J Biol Chem. 1997;272:30583–8. doi: 10.1074/jbc.272.48.30583. [DOI] [PubMed] [Google Scholar]
- 31.Wittkopp PJ, Kalay G. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat Rev Genet. 2011;13:59–69. doi: 10.1038/nrg3095. [DOI] [PubMed] [Google Scholar]
- 32.Ng RK, Dean W, Dawson C, Lucifero D, et al. Epigenetic restriction of embryonic cell lineage fate by methylation of Elf5. Nat Cell Biol. 2008;10:1280–90. doi: 10.1038/ncb1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niwa H, Toyooka Y, Shimosato D, Strumpf D, et al. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123:917–29. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- 34.Russ AP, Wattler S, Colledge WH, Aparicio SA, et al. Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature. 2000;404:95–9. doi: 10.1038/35003601. [DOI] [PubMed] [Google Scholar]
- 35.Knox K, Baker JC. Genomic evolution of the placenta using co-option and duplication and divergence. Genome Res. 2008;18:695–705. doi: 10.1101/gr.071407.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lamprecht B, Walter K, Kreher S, Kumar R, et al. Derepression of an endogenous long terminal repeat activates the CSF1R proto-oncogene in human lymphoma. Nat Med. 2010;16:571–9. doi: 10.1038/nm.2129. [DOI] [PubMed] [Google Scholar]
- 37.Young GR, Eksmond U, Salcedo R, Alexopoulou L, et al. Resurrection of endogenous retroviruses in antibody-deficient mice. Nature. 2012;491:774–8. doi: 10.1038/nature11599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soundararajan R, Rao AJ. Trophoblast “pseudo-tumorigenesis”: Significance and contributory factors. Reprod Biol Endocrinol. 2004;2:15. doi: 10.1186/1477-7827-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferretti C, Bruni L, Dangles-Marie V, Pecking AP, et al. Molecular circuits shared by placental and cancer cells, and their implications in the proliferative, invasive and migratory capacities of trophoblasts. Hum Reprod Update. 2006;13:121–41. doi: 10.1093/humupd/dml048. [DOI] [PubMed] [Google Scholar]
- 40.Peaston AE, Evsikov AV, Graber JH, de Vries WN, et al. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev Cell. 2004;7:597–606. doi: 10.1016/j.devcel.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 41.Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, et al. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487:57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haig D. Genetic conflicts in human pregnancy. Q Rev Biol. 1993;68:495–532. doi: 10.1086/418300. [DOI] [PubMed] [Google Scholar]
- 43.Renfree MB, Hore TA, Shaw G, Graves JAM, et al. Evolution of genomic imprinting: insights from marsupials and monotremes. Annu Rev Genomics Hum Genet. 2009;10:241–62. doi: 10.1146/annurev-genom-082908-150026. [DOI] [PubMed] [Google Scholar]
- 44.Reik W, Constância M, Fowden A, Anderson N, et al. Regulation of supply and demand for maternal nutrients in mammals by imprinted genes. J Physiol. 2003;547:35–44. doi: 10.1113/jphysiol.2002.033274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feil R, Berger F. Convergent evolution of genomic imprinting in plants and mammals. Trends Genet. 2007;23:192–9. doi: 10.1016/j.tig.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 46.Suzuki S, Ono R, Narita T, Pask AJ, et al. Retrotransposon silencing by DNA methylation can drive mammalian genomic imprinting. PLoS Genet. 2007;3:e55. doi: 10.1371/journal.pgen.0030055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeh DW, Zeh JA. Reproductive mode and speciation: the viviparity-driven conflict hypothesis. BioEssays. 2000;22:938–46. doi: 10.1002/1521-1878(200010)22:10<938::AID-BIES9>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 48.Hughes AL, Green JA, Garbayo JM, Roberts RM. Adaptive diversification within a large family of recently duplicated, placentally expressed genes. Proc Natl Acad Sci USA. 2000;97:3319–23. doi: 10.1073/pnas.050002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hou Z, Romero R, Uddin M, Than NG, et al. Adaptive history of single copy genes highly expressed in the term human placenta. Genomics. 2008;93:33–41. doi: 10.1016/j.ygeno.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chuong EB, Tong W, Hoekstra HE. Maternal-fetal conflict: rapidly evolving proteins in the rodent placenta. Mol Biol Evol. 2010;27:1221–5. doi: 10.1093/molbev/msq034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Swanson W, Wong A, Wolfner M, Aquadro C. Evolutionary expressed sequence tag analysis of Drosophila female reproductive tracts identifies genes subjected to positive selection. Genetics. 2004;168:1457–65. doi: 10.1534/genetics.104.030478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hu D, Cross J. Development and function of trophoblast giant cells in the rodent placenta. Int J Dev Biol. 2009;54:341–54. doi: 10.1387/ijdb.082768dh. [DOI] [PubMed] [Google Scholar]
- 53.van Valen L. The Red Queen. Am Nat. 1977;111:809–10. [Google Scholar]
- 54.Pfeiffer JK, Kirkegaard K. Increased fidelity reduces poliovirus fitness and virulence under selective pressure in mice. PLoS Pathog. 2005;1:e11. doi: 10.1371/journal.ppat.0010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Senner CE, Hemberger M. Regulation of early trophoblast differentiation – Lessons from the mouse. Placenta. 2010;31:944–50. doi: 10.1016/j.placenta.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 56.Hemberger M, Udayashankar R, Tesar P, Moore H, et al. ELF5-enforced transcriptional networks define an epigenetically regulated trophoblast stem cell compartment in the human placenta. Hum Mol Genet. 2010;19:2456–67. doi: 10.1093/hmg/ddq128. [DOI] [PubMed] [Google Scholar]
- 57.Chuong EB, Rumi MAK, Soares MJ, Baker JC. Endogenous retroviruses function as species-specific enhancer elements in the placenta. Nat Genet. 2013;45:325–9. doi: 10.1038/ng.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Palmieri C, Loi P, Ptak G, Salda Della L. Review paper: A review of the pathology of abnormal placentae of somatic cell nuclear transfer clone pregnancies in cattle, sheep, and mice. Vet Pathol. 2008;45:865–80. doi: 10.1354/vp.45-6-865. [DOI] [PubMed] [Google Scholar]
- 59.Nathalie de Parseval TH. Physiological knockout of the envelope gene of the single-copy ERV-3 human endogenous retrovirus in a fraction of the Caucasian population. J Virol. 1998;72:3442. doi: 10.1128/jvi.72.4.3442-3445.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Malik HS. Retroviruses push the envelope for mammalian placentation. Proc Natl Acad Sci USA. 2012;109:2184–5. doi: 10.1073/pnas.1121365109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Warren WC, Hillier LW, Marshall Graves JA, Birney E, et al. Genome analysis of the platypus reveals unique signatures of evolution. Nature. 2008;453:175–83. doi: 10.1038/nature06936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tyndale-Biscoe H, Renfree M. Reproductive physiology of marsupials. Cambridge University Press; 1987. p. 490. [Google Scholar]
- 63.Lillegraven JA, Thompson SD, McNab BK, Patton JL. The origin of eutherian mammals. Biol J Linn Soc. 1987;32:281–336. [Google Scholar]
- 64.Renfree M. Review: Marsupials: Placental mammals with a difference. Placenta. 2010;31:S21–6. doi: 10.1016/j.placenta.2009.12.023. [DOI] [PubMed] [Google Scholar]
- 65.Stoye JP. Studies of endogenous retroviruses reveal a continuing evolutionary saga. Nat Rev Microbiol. 2012;10:395–406. doi: 10.1038/nrmicro2783. [DOI] [PubMed] [Google Scholar]
- 66.Hockemeyer D, Wang H, Kiani S, Lai CS, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29:731–4. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cong L, Ran FA, Cox D, Lin S, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–23. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, et al. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–31. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Molaro A, Hodges E, Fang F, Song Q, et al. Sperm methylation profiles reveal features of epigenetic inheritance and evolution in primates. Cell. 2011;146:1029–41. doi: 10.1016/j.cell.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xie M, Hong C, Zhang B, Lowdon RF, et al. DNA hypomethylation within specific transposable element families associates with tissue-specific enhancer landscape. Nat Genet. 2013 doi: 10.1038/ng.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee X, Keith JC, Jr, Stumm N, Moutsatsos I, et al. Downregulation of placental syncytin expression and abnormal protein localization in pre-eclampsia. Placenta. 2001;22:808–12. doi: 10.1053/plac.2001.0722. [DOI] [PubMed] [Google Scholar]
- 72.Williams PJ, Broughton Pipkin F. The genetics of pre-eclampsia and other hypertensive disorders of pregnancy. Best Pract Res Clin Obstet Gynaecol. 2011;25:405–17. doi: 10.1016/j.bpobgyn.2011.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Novakovic B, Saffery R. The ever growing complexity of placental epigenetics – Role in adverse pregnancy outcomes and fetal programming. Placenta. 2012;33:959–70. doi: 10.1016/j.placenta.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 74.Tuzun E, Sharp AJ, Bailey JA, Kaul R, et al. Fine-scale structural variation of the human genome. Nat Genet. 2005;37:727–32. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 75.Price EM, Cotton AM, Penaherrera MS, McFadden DE, et al. Different measures of “genome-wide” DNA methylation exhibit unique properties in placental and somatic tissues. Epigenetics. 2012;7:652–63. doi: 10.4161/epi.20221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.O’Neill MJ, Lawton BR, Mateos M, Carone DM, et al. Ancient and continuing Darwinian selection on insulin-like growth factor II in placental fishes. Proc Natl Acad Sci USA. 2007;104:12404–9. doi: 10.1073/pnas.0705048104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hamlett WC. Evolution and morphogenesis of the placenta in sharks. J Exp Zool. 1989;252:35–52. [Google Scholar]
- 78.Thompson MB, Speake BK. A review of the evolution of viviparity in lizards: structure, function and physiology of the placenta. J Comp Physiol B. 2005;176:179–89. doi: 10.1007/s00360-005-0048-5. [DOI] [PubMed] [Google Scholar]
- 79.Lynch VJ. Live-birth in vipers (Viperidae) is a key innovation and adaptation to global cooling during the Cenozoic. Evolution. 2009;63:2457–65. doi: 10.1111/j.1558-5646.2009.00733.x. [DOI] [PubMed] [Google Scholar]
- 80.Espagne E, Dupuy C, Huguet E, Cattolico L, et al. Genome sequence of a polydnavirus: insights into symbiotic virus evolution. Science. 2004;306:286–9. doi: 10.1126/science.1103066. [DOI] [PubMed] [Google Scholar]
- 81.Villarreal LP. Origin of Group Identity. Springer; 2009. [Google Scholar]


