Abstract
Yeast display is a powerful technology for the isolation of monoclonal antibodies (mAbs) against a target antigen. Antibody libraries have been displayed on the surface of yeast as both single-chain variable fragment (scFv) and antigen binding fragment (Fab). Here, we combine these two formats to display well-characterized mAbs as single-chain Fabs (scFabs) on the surface of yeast and construct the first scFab yeast display antibody library. When expressed on the surface of yeast, two out of three anti-human immunodeficiency virus (HIV)-1 mAbs bound with higher affinity as scFabs than scFvs. Also, the soluble scFab preparations exhibited binding and neutralization profiles comparable to that of the corresponding Fab fragments. Display of an immune HIV-1 scFab library on the surface of yeast, followed by rounds of sorting against HIV-1 gp120, allowed for the selection of 13 antigen-specific clones. When the same cDNA was used to construct the library in an scFv format, a similar number but a lower affinity set of clones were selected. Based on these results, yeast-displayed scFab libraries can be constructed and selected with high efficiency, characterized without the need for a reformatting step, and used to isolate higher-affinity antibodies than scFv libraries.
Keywords: immune repertoire, antibody display format, HIV-1, flow cytometry, antibody library
Introduction
Over the past decade, yeast display has emerged as a leading technology for the selection and characterization of monoclonal antibodies (mAbs) from both immune and nonimmune libraries.1–5 The majority of yeast display antibody libraries that have been constructed to date utilize the single-chain variable fragment (scFv) format,1–3 and there are only three examples in the literature of combinatorial antigen binding fragment (Fab) yeast display libraries.6–8 Because both scFv and Fab libraries have unique advantages and disadvantages, there is controversy over which format is better suited for antibody display. ScFvs are generally less stable and more prone to aggregation than Fab fragments,9,10 and due to their decreased stability, scFvs generally bind with lower affinity than Fabs.11 Furthermore, it has been shown that certain scFvs lose antigen binding and/or neutralization activity after conversion into Fabs or immunoglobulin Gs (IgGs).7,12,13 However, because Fab fragments are larger in size and require the assembly of two separate polypeptide chains with a disulfide bond, they generally fold less efficiently than scFvs, which leads to decreased solubility and lower levels of expression in Escherichia coli.14 The most commonly used method to display Fab libraries on the surface of yeast, which employs yeast mating, allows for the display of unpaired heavy chains.7 As a result, nonspecific binding of heavy-chain-only clones could dominate library selection and result in the loss of specific binders.
Recently, a single-chain Fab (scFab) was constructed and shown to be compatible with phage display technology and suitable for soluble expression in E. coli.15,16 By combining the high levels of expression of the scFv with the stability of the Fab fragment, the scFab might represent a useful anti-body display format for yeast display technology. Here, we have displayed three well-characterized anti-human immunodeficiency virus (HIV)-1 antibodies as scFabs on the surface of yeast and constructed the first scFab yeast display library. In order to evaluate the scFab as a selection platform, we used the same cDNA as starting material for the construction of libraries in the scFv and scFab formats, and the libraries were subsequently selected against the same antigens. Our results suggest that scFab libraries displayed on the surface of yeast can be constructed and selected with higher efficiency than Fab libraries and yield higher-affinity antibodies than scFv libraries.
Results
Generation of scFab and scFv yeast display vectors
The scFab yeast display vector was generated from the vector pPNL200.7 Using overlap PCR, a 34-amino-acid linker containing the sequence (SGGG)2(SEGGG)4(SGGG)SG was inserted between the heavy and light chains. Primers were designed to remove the two cysteines involved in the disulfide bond connecting the two chains (Fig. 1a). Removal of this disulfide bond has been shown to increase the solubility of scFabs when expressed in E. coli.15 Next, BssHII and SalI restriction sites were inserted for light-chain gene cloning and NheI and NcoI sites were introduced for heavy-chain gene cloning. These restriction enzyme sites were chosen because they are rarely found in germ line antibody genes. Stuffer sequences containing only the light-chain or heavy-chain constant region were inserted between restriction sites to aid in cloning.
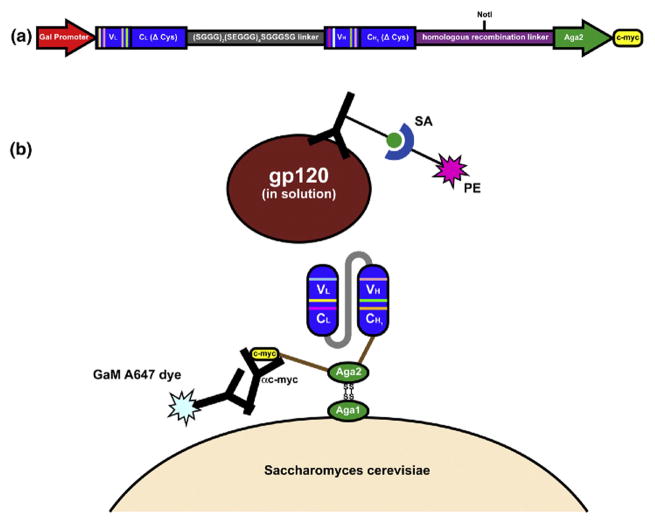
Fig. 1.
scFab yeast display platform. (a) scFab expression is under control of the Gal1 promoter. A 34-amino-acid linker connects the heavy and light chains, and the Aga2 protein is expressed as a C-terminal fusion to the scFab. The cysteines that form the interchain disulfide bond were removed. (b) Monomeric gp120 is labeled with biotinylated HIVIG and detected with SA–PE. Using an anti-c-myc antibody and goat anti-mouse Alexa647 (GaM A647) dye, a C-terminal c-myc tag allows for visualization of yeast expression.
It has been demonstrated that linear plasmid vectors that circularize by homologous recombination transform yeast with higher efficiency than supercoiled plasmids.17 The traditional method for yeast homologous recombination, which involves electroporation of PCR products with 5′ and 3′ homology to a linearized vector,18 was not used because it requires ~100 μg of PCR product to generate large (109) libraries. This quantity of PCR product was not produced from the limited amount of starting material; therefore, we inserted a 51-base-pair linker with homology to the adjacent vector sequence directly after the heavy chain to be used for homologous recombination (Fig. 1a). By digesting the library at a site located between the two homologous sequences, we found that the yeast transformation efficiency was increased by at least 10-fold.
Validation of the new display format using mAbs
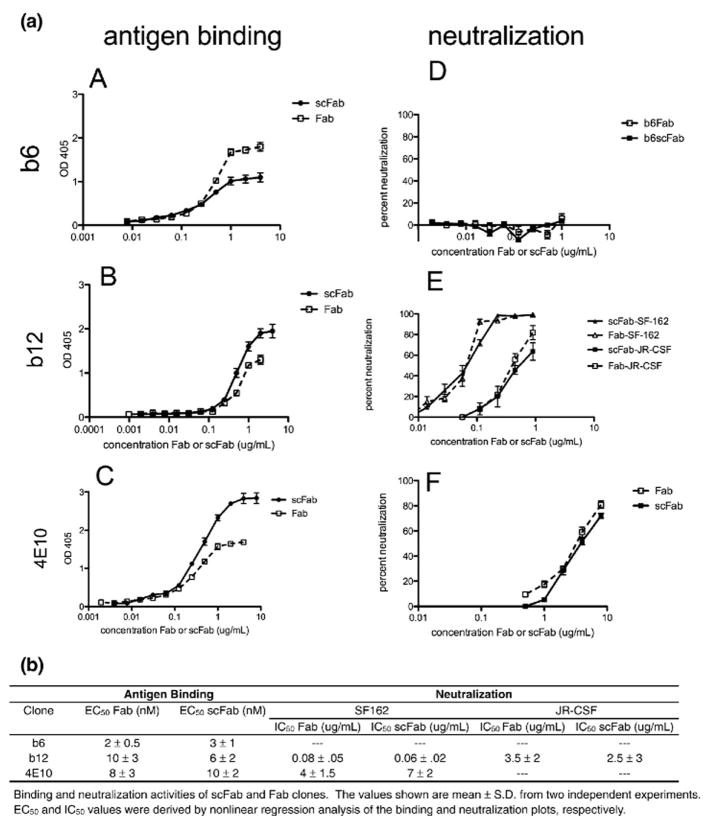
The new display system was validated using three well-characterized anti-HIV-1 antibodies: 4E10, b12, and b6. 4E10 targets the membrane proximal external region on HIV-1 gp41 and both b12 and b6 bind to epitopes overlapping the CD4 binding site on HIV-1 gp120.19 Both 4E10 and b12 exhibit broad neutralization of HIV-1, whereas b6 only neutralizes a few very sensitive viral isolates.19 The mAbs were expressed as both scFabs and scFvs on the surface of yeast, and the binding affinities were calculated by titrating the amount of antigen (gp120 or gp41) and measuring the mean fluorescence intensity (MFI) of antigen binding (Fig. 2a and b). Antibodies b6 and 4E10 bound antigen with 12-fold and 4-fold higher affinity, respectively, in the scFab format. Antibody b12 bound to monomeric gp120 with approximately 15 nM affinity in both formats, although the MFI plateau was higher in the scFv format. This is likely a result of increased levels of b12 surface expression in the scFv format. All three scFab clones were expressed on 50–80% of yeast cells (data not shown), which is comparable to published observations regarding scFv and Fab expression of single clones.
Fig. 2.
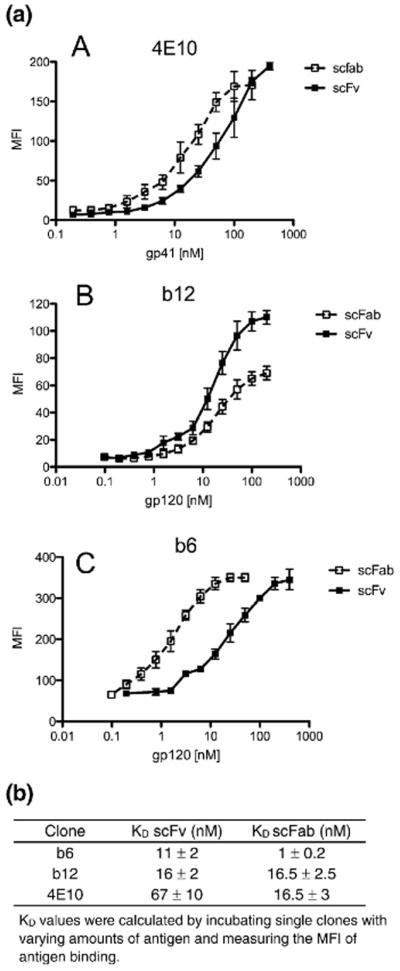
(a) Binding curves for yeast-displayed 4E10, b12, and b6 scFvs and scFabs are shown in (A)–(C). The MFI, determined by flow cytometry, is plotted against increasing concentrations of antigen to determine the Kd values listed in (b). Data are mean±SD from three independent experiments.
Because it has been shown previously that soluble scFabs have a tendency to dimerize and multimerize,15 we sought to determine whether the soluble scFabs would exhibit enhanced levels of binding and neutralization due to avidity effects. b12, b6, and 4E10 were cloned into the vector pComb3X and expressed in E. coli in order to obtain soluble scFabs. Crude scFab supernatants were assayed for binding and neutralization activity and compared to the corresponding crude Fab fragments (Fig. 3a). All three scFabs exhibited similar binding profiles as the corresponding Fab fragments in an enzyme-linked immunosorbent assay (ELISA), although we cannot exclude the possibility that multimerization increases the apparent affinity of soluble scFabs. It should also be noted that b12 and 4E10 scFabs saturate at a higher signal than their Fab counterpart, which also may be due to scFab multimerization. The three scFabs exhibited similar neutralization profiles as the corresponding Fab fragments (Fig. 3b), indicating that soluble scFabs can reliably be used for this functional assay. It is possible that the slight decrease in neutralization for the scFab could be caused by multimerization, but this decrease is not statistically significant and does not interfere with the general trend of neutralizing versus non-neutralizing antibodies.
Fig. 3.
Binding and neutralization profiles of soluble scFabs. b12, b6, and 4E10 were expressed solubly in E. coli, and crude supernatants were assayed for binding and neutralization activity. (a) (A)–(C) represent ELISA binding curves, and (D)–(F) represent neutralization activity of soluble Fabs and scFabs. (b) EC50 and IC50 values derived from the data shown in (a). Data are mean±SD from two experiments.
Construction and selection of scFv and scFab immune HIV-1 libraries
ScFv and scFab immune libraries were cloned from the bone marrow RNA of an HIV-1-infected donor who exhibits broad serum neutralization. Both the scFv and the scFab libraries were cloned from the same pool of cDNA, and the number of independent transformants after electroporation into E. coli was on the order of 5×109 for both libraries. The scFab and scFv library DNA were propagated in E. coli, linearized with NotI, and used to transform EBY100 yeast cells using homologous recombination and the lithium acetate method.20 Dilution plates indicated that the size of both libraries in yeast were ~109. Based on restriction enzyme digestion and sequencing of unselected library clones, 90% of clones from both libraries contained full-length and in-frame inserts.
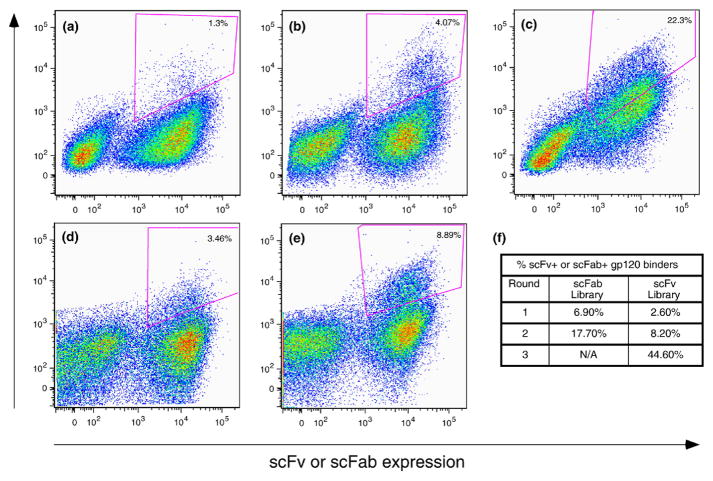
The scFab and scFv libraries were both subject to one round of magnetic bead selection21 followed by two to three rounds of sorting by flow cytometry against monomeric gp120JR-FL and gp120JR-CSF (Fig. 4). No-antigen controls were used to determine sort gates that would only allow for selection of antigen binding clones. Pooled IgG derived from HIV-1-infected donors (HIVIG) was biotinylated and used for gp120 visualization (Fig. 1b). A polyclonal secondary reagent was chosen because it is unlikely to compete with library clones for gp120 binding. Using an anti-c-myc antibody and goat anti-mouse Alexa 647 dye, we monitored antibody expression using a C-terminal c-myc epitope tag (Fig. 1b). After the third round of sorting, 70 clones from each library were selected for sequence analysis and initial antibody characterization.
Fig. 4.
scFv and scFab library sorting by flow cytometry. Yeast cells are double labeled so that antibody expression and antigen binding can be monitored simultaneously. (a)–(c) represent scFv library sorts (rounds 1–3), and (d) and (e) represent scFab library sorts (rounds 1 and 2). The circled populations represent double-positive yeast cells that were sorted, grown, and induced for the next round of selection. The percentages of antigen-positive cells are indicated in the top right corner of each plot. (f) shows the percentage of antigen-positive induced cells for each round of selection.
Comparison of selected scFab and scFv clones
Selected scFab and scFv clones were grouped based on examination of the amino acid sequences of the third complementary determining region of the heavy chain (CDRH3) (Table 1). Clones that appeared to represent somatic variants of the same B-cell were grouped together. Based on this analysis, 13 unique clones were selected from the scFab library and 11 unique clones were selected from the scFv library, with only 1 clone represented in both libraries. Of the selected scFv and scFab clones, 80% and 100% contained full-length inserts, respectively. Because the constant regions of the heavy and light chains were used as stuffer sequences, all of the truncated scFvs contained one full-length chain and only the constant region of the other chain. Selected clones from both libraries exhibited a bias for the VH1–69 family, but it has been previously demonstrated that anti-HIV-1 antibodies commonly use this germ line gene family.23,24
Table 1.
Sequence analysis of selected scFv and scFab clones
| scFab clone | No.a | Germ line IGVLb | Germ line IGVH | CDRL3 | CDRH3c |
|---|---|---|---|---|---|
| 1B3 | 13 | L1–51*02 | 3–23*01 | ETWDSRLSAGVF | GPLGRFLDFW |
| 3C1 | 4 | L2–14*01 | 1–69*01 | SSYTTRGTRIF | EGGRSYGTQVLDPW |
| 1A1 | 2 | L2–23*03 | 3–43*01 | CAYASKSLSFVF | ERHIAGNGGIDYW |
| 1A4 | 4 | K3–20*01 | 1–69*06 | QQYVSSPYTF | SRIVPVAAPQFTYW |
| 1A3 | 6 | L2–11*01 | 1–69*06 | CSYAGSRILWIF | GSDSRSYYHYMDVW |
| J3 | 20 | K1–39*01 | 1–69*12 | QQSYSTPLTF | SRTTIFGVAQDNWFDPW |
| Q10 | 1 | L1–44*01 | 1–2*01 | AAWDGNLDGVVF | DPRPTIAVLPPGMSVWFDPW |
| Q3 | 7 | L3–19*01 | 1–24*01 | NSRDITYNSVVF | AGFDGLKGYYKGFDYW |
| Q7 | 4 | L2–8*01 | 3–33*03 | SSYGGSNNLIF | DPFFKNSPTYYLDFYMDVW |
| J5 | 1 | K3–11*01 | 3–21*01 | QQRSNWPITF | SSGWFFVAKYYFDSW |
| 3J17 | 4 | K3–20*01 | 3–30*01 | QQYSTSITF | DAWGDNYFDYW |
| Q16 | 1 | L3–21*02 | 5–51*01 | QVWDSTFDPWVF | MGPYNDLWSGLPRGGVDPW |
| 2Q10 | 1 | L1–51*01 | 1–3*01 | GTWDSSLGAWVF | YGDYGRAFAIW |
| scFv clone | No.a | Germ line IGVLb | Germ line IGVH | CDRL3 | CDRH3 |
|---|---|---|---|---|---|
| 1R2-17 | 15 | L1–44*01 | 5–51*01 | AAWDDTLHAYVF | PVDYFESSGYYWFDFW |
| 1R2–48 | 6 | L2–23*02 | 1–3*01 | CSDAGIWVF | IGRYFDLW |
| 1R2–20 | 1 | L2–14*01 | 1–2*02 | FSFRSSSIWVF | VSYYDSSGADW |
| 1R2–18 | 6 | L1–47*01 | 3–64*02 | AEWDDLLNGWVF | DDWVGSNHTFSFDVW |
| 2R2–12 | 1 | L1–40*01 | 1–69*01 | QSYDHRVNGYVF | ATWNDDCGTTNCYDWFDPW |
| 2R2–14 | 5 | K3–20*01 | 1–2*02 | QQYDSSGSF | ERRGGFDEYW |
| 2R2–21 | 1 | L1–44*01 | 3–7*01 | AAWDDTLDAYVF | RPCDGAKCDDPPW |
| 2R2–25 | 1 | K3D–15*01 | 1–69*08 | QQYYSSPLTF | GDSGSSSPFDYIYMDVW |
| 2R2–50 | 1 | L3–19*01 | 1–69*01 | YSRDNSGSHNYVF | AVLPFLEWFAAHQYFYMDVW |
| 1R2–46 | 5 | L3–21*03 | 3–30*04 | QVWDGSHDPREVF | DAWGDNYFDYW |
| T13 | 4 | K3–20*01 | 3–43*01 | QQYGSSEGSF | DGASIGKYYDFWSGSSRRGKYYMDVW |
Number of times an individual sequence was observed from selection. Somatic variants (not shown) were grouped together.
Germ line gene sequences were determined using the IMGT database (http://imgt.cines.fr).22 “L” and “K” refer to lambda and kappa, respectively.
Clones were grouped based on analysis of the CDRH3 region.
The affinities of single clones were calculated by incubating each yeast-displayed scFv or scFab with varying amounts of monomeric gp120 (Table 2). The binding constants for the scFab clones ranged from 0.5 to 36 nM, with an average affinity of 10 nM. In contrast, the affinities of selected scFv clones ranged from 0.3 to 100 nM, with an average affinity of 43 nM.
Table 2.
Binding affinities of selected scFv and scFab clones
| Clone | Kd scFab (nM)a | Kd scFv (nM) |
|---|---|---|
| 1B3 | 1 | 34 |
| 3C1 | 6 | — |
| 1A1 | 14 | — |
| 1A4 | 3 | 7 |
| 1A3 | 8.5 | — |
| J3 | 2 | — |
| Q10 | 0.5 | — |
| Q3 | 17 | — |
| Q7 | 36 | 103 |
| J5 | 22 | — |
| 3J17 | 21 | — |
| Q16 | 1 | — |
| 2Q10 | 6 | — |
| 1R2–17 | — | 61 |
| 1R2–48 | — | 9.5 |
| 1R2–20 | — | 46 |
| 1R2–18 | — | 5 |
| 2R2–12 | 10 | 42 |
| 2R2–14 | — | 0.3 |
| 2R2–21 | 30 | 68 |
| 2R2–25 | — | 52 |
| 2R2–50 | 16 | 60 |
| 1R2–46 | — | 29 |
| T13 | — | 100 |
Kd values were calculated by incubating single clones with varying amounts of antigen and measuring the MFI of antigen binding.
Three antibodies from each library were cloned in the alternate display format and assayed for surface expression and antigen binding in order to determine whether the display format had an intrinsic effect on the types of clones selected. We found that the three selected scFab clones exhibited high levels of surface expression as scFvs but bound antigen with lower affinity. Similarly, the three selected scFvs could be displayed on the surface of yeast in the scFab format and bound antigen with 2- to 4-fold higher affinity as scFabs (Table 2). Based on this analysis, the difference in selection of clones is probably not a direct result of the display format, as discussed below.
Discussion
Here, we have constructed the first scFab yeast display library and shown that this format offers distinct advantages over the existing scFv and Fab formats. We found that the majority of antibodies we tested bound with higher affinity as scFabs than as scFvs. This was demonstrated for well-characterized anti-HIV-1 mAbs as well as for selected clones from an immune HIV-1 library. It has been shown that the folding and stability of the Fab fragment are dependent on cooperation between the VH/VL and CH1/CL interfaces25 and stabilization of these domain interactions improves antigen binding.26 The lack of a stabilizing CH1/CL interface in scFv fragments likely increases the flexibility of the binding pocket, which results in an increased entropic cost to binding.27
Surprisingly, when the same cDNA was used to construct an immune library in both the scFab and scFv format and selected against the same antigens, two almost completely different sets of clones were selected. We reasoned that this observation might be a result of the display format and that certain clones only display or bind antigen in one of the formats tested. Six antibodies were cloned in the alternate library format, displayed on the surface of yeast, and assayed for antigen binding in order to evaluate this possibility. We found that all of the antibodies tested could be displayed on the surface of yeast and bind antigen in both formats. Consistent with our previous results, all six antibodies bound antigen with higher affinity in the scFab format. Because sort gates were drawn to select all c-myc-positive antigen binding clones, neither differences in antibody display nor those in antigen binding appear to account for differences in selected antibodies. Remaining explanations for the differences include sampling size and library construction. An explanation based on sampling size would require that the libraries contain an unusually large number of gp120-specific clones for only one clone to be shared among 70 from each library. We therefore favor an explanation based on differences in library construction (PCR priming, chain shuffling, internal restriction sites, etc.).
One observation worth noting concerns the nature of the antibodies selected from each library. We found that the scFab library yielded a much higher proportion of antigen-specific clones over background binders than the scFv library. About 20% of selected scFv clones contained the vector stuffer sequence, and 18% of the full-length clones bound nonspecifically to fluorescence reagents. In contrast, all of the selected scFab clones were full length and only 4% bound to secondary reagents. Because both of the unselected libraries contained 90% full-length clones with similar germ line gene representation, this discrepancy is probably due to the unstable nature of scFvs and not to differences in library construction.
ScFab yeast display libraries also provide advantages over current formats for display of yeast Fab libraries. The most commonly used method requires yeast mating and a two-vector system. This system allows for the display of heavy-chain-only clones, which could potentially interfere with library selection. It has previously been reported that 15% of antigen-specific yeast express only a heavy chain.7 Therefore, although yeast mating allows for the construction of large libraries, heavy-chain-only display results in some loss of selection efficiency. In contrast, we found that our scFab library could be constructed and selected with very high efficiency. Using our homologous recombination system, we were easily able to generate a yeast display library with ~109 members, which is comparable to the size of existing Fab libraries. Also, the scFab linker prevents the display of unpaired heavy or light chains. When expressed in E. coli, we found that the soluble scFabs exhibited similar binding and neutralization profiles as the corresponding Fab fragments, although one caveat in the binding and neutralization assays is the possibility of soluble scFab multimerization. This result suggests that the scFab selection format can also be used for subsequent antibody characterization. Based on these observations, yeast display of scFab libraries is likely a more robust system than Fab display.
In conclusion, we have generated a new yeast display format that offers clear advantages over the existing platforms. Yeast-displayed scFab libraries may allow for the increased recovery of novel antibodies with diagnostic or therapeutic applications.
Materials and Methods
Yeast cell lines and media
Yeast strain EBY100 (GAL1-AGA1::URA3 ura3–52 trp1 leu21 his3200 pep4::HIS2 prb11.6R can1 GAL) was maintained in YPD broth (Difco). Transformation of EBY100 with the vector pYDscFab and pYDscFV was completed using the lithium acetate method20 and maintained in SD-CAA medium, pH 4.5 (6.7 g/L yeast nitrogen base, 5 g/L casamino acids, 20 g/L dextrose, 4.29 g/l citric acid, and 14.7 g/L citric acid monohydrate), and on SD-CAA plates (SD-CAA+17 g/L agar). Yeast surface expression of scFv was induced by transferring to SG/R-CAA medium (6.7 g/L yeast nitrogen base, 5 g/L casamino acids, 20 g/L each galactose and raffinose, 1 g/L dextrose, 9.67 g/L NaH2PO4·2H2O and 10.19 g/L Na2-HPO4·7H2O). After yeast sorting, 0.25 μg/mL ketoconazole (Sigma-Aldrich) was added to the SD-CAA media in order to inhibit growth of Candida parapsilosis.28
Antigens and antibodies
Monomeric gp120JR-FL and soluble CD4 were purchased from Progenics (Tarrytown, NY), and gp120JR-CSF was procured under contract from Advanced Products Enterprises (MD). The human anti-gp120 mAbs used in this study are IgGs b12,29 b6,29 and 4E10.30 Polyclonal human IgG against HIV-1 (HIVIG) was obtained from the AIDS Research Reference Reagent Program, provided by John Mascola. Antibodies were biotinylated using the EZ-Link Sulfo-NHS-Biotinylation Kit from Pierce. Mouse mAb antic-myc (9E10) was obtained from AbD Serotec. Fluorescent reagents goat anti-mouse-Alexa 647 and streptavidin–phycoerythrin (SA–PE) were obtained from Molecular Probes.
Yeast display vector construction
The scFab yeast display vector was generated from the vector pPNL200 (M. Feldhaus, Pacific Northwest National Laboratory). A BssHII cloning site was inserted at the 5′ end of the light chain and a NheI site was inserted at the 3′ end of the heavy chain using the QuikChange Multi Site-Directed Mutagenesis Kit (Stratagene). Overlap PCR was used to insert a 34-amino-acid linker between the heavy and light chains, add an NcoI site at the 5′ end of the heavy chain and a SalI site at the 3′ end of the light chain, and remove the two cysteines involved in the disulfide bond connecting the two chains. Two stuffer sequences, composed of either the constant heavy genes or the constant light genes, were inserted on both sides of the linker. A homologous recombination linker containing the phosphorylated sequence 5′-CTAGTCAGGAACTGACA-ACTATATGCGAGCAAATCCCCTCACCAACTCGC-3′ was ligated in after the heavy chain using the restriction enzyme sites NotI and SpeI. The sequence of the modified vector, pYDscFab, was verified by DNA sequencing.
The yeast display scFv vector, pYDscFv, was generated by modification of pYDscFab. The 34-amino-acid scFab linker was digested out of pYDscFab using the restriction enzymes NcoI and NheI and the digested vector was gel purified using the QIAquick Gel Purification Kit (Qiagen). A 15-amino-acid 5′ phosphorylated oligo containing the scFv linker sequence 5′-TCGA-CAGGGGAGAATTCGGTGGTGGTGGTTCTGGTGGTG-GTGGTTCTGGTGGTGGTGGTTCTCTC-3′ was ligated into the digested vector. The sequence of pYDscFv was verified by DNA sequencing.
scFab and scFv library construction
Total RNA was isolated from the bone marrow mono-nuclear cell RNA from an HIV-infected donor who exhibits broad serum neutralization of HIV-1. Bone marrow was collected, homogenized, and stored in TRI reagent (received from the International AIDS Vaccine Initiative) and placed at −80 °C for long-term storage. Homogenized bone marrow (5 mL) was centrifuged at 5000g for 10 min at 4 °C, supernatant was removed, and the pellet was resuspended in a 10× volume of TRI reagent to isolate total RNA. 1-Bromo-3-chloropropane (3 mL; Sigma) was added to each supernatant, vortexed for 15 s, and incubated for 15 min at room temperature. After centrifugation at 17,000g for 15 min at 4 °C, the upper phase was transferred to a fresh, 50-mL tube. Isopropanol (15 mL) was added to the tube, vortexed for 15 s, and incubated for 10 min at room temperature. Centrifugation was again performed at 17,000g at 4 °C and the supernatant was discarded. The pellet was washed with 30 mL of 75% ethanol, air-dried at room temperature for 5–10 min, and resuspended in 500 μL of RNase-free water. Total RNA was reverse transcribed into cDNA using transcriptor first-strand cDNA synthesis kit (Roche) and an oligo(dT) primer. Heavy- and light-chain antibody genes were PCR amplified from cDNA using a primer set based on that of Sblattero and Bradbury.31 The following oligonucleotides were used as heavy- and light-chain scFab reverse primers to remove the cysteines that form the interchain disulfide bond: scFabHrev: 5′-GACTTGGC-TAGCAGATTTGGGCTCAACTTTC-3′, scFablamdarev: 5′-GCATGT GTCGACTTCYGYAGGGGCMACTGTC-3′, and scFabKaprev: 5′-GCATGTGTCGACCTCTCCCCT-GTTGAAGCTC-3′. Kappa and lambda light-chain genes were gel purified with the QIAquick Gel Extraction Kit (Qiagen), pooled in an equal ratio, digested with the restriction enzymes BssHII and SalI, and ligated into the vector pYDscFab or pYDscFv. After ethanol precipitation, the ligation mixture was used to transform E. coli strain Electro-Ten Blue (Strategene). pYDscFab plasmid DNA containing the light-chain genes was isolated using a Maxiprep kit (Qiagen) and digested with the restriction enzymes NheI and NcoI for ligation of the heavy-chain genes (performed the same as light-chain ligation). The final libraries contained approximately 5×109 independent transformants. The libraries were then linearized with NotI, ethanol precipitated, and used to transform the yeast strain EBY100 using the high-efficiency lithium acetate method,20 and grown in SD-CAA until saturation (about 24 h). The size of both the scFab and the scFv libraries were estimated at 1×109 members.
Library selection, flow cytometric analysis, and single clone characterization
Libraries were grown as described previously.2 Briefly, yeast were grown in SD-CAA media for 12–18 h at 30 °C, pelleted at 3000g for 10 min, and resuspended in SG/R-CAA for 16–18 h at 20 °C in volumes appropriate for the size of the library. For the first round of selection, a magnetic bead sorting technique utilizing the Miltenyi Macs system was used as described previously.32 Yeast cells (1×1010) were incubated with 10 μg/mL gp120 for 1 h at room temperature in FACS wash buffer [phosphate-buffered saline (PBS)/0.5% bovine serum albumin (BSA)/2 mM ethylenediaminetetraacetic acid] followed by 10 min on ice and then washed 3 times with wash buffer. Yeast cells were pelleted and incubated with 15 μg/mL biotinylated HIVIG for 30 min at 4 °C. After washing 3 times with ice-cold wash buffer, the cell pellet was resuspended in 4 mL wash buffer. SA micro-beads (200 μL) were added to the yeast and incubated for 10 min, followed by two 50-mL washes. The yeast were resuspended in 50 mL and loaded onto a Miltenyi LS column at 4 °C in 7-mL aliquots. After each 7 mL, the column was removed from the magnetic field and immediately replaced. The column was then washed with 1 mL of wash buffer before another 7 mL was loaded onto the column. After the entire 50 mL was loaded, the column was washed 3 times with 2 mL of wash buffer with removal and replacement in between. The column was then removed from the magnetic field, and the yeast were eluted with 12 mL of SD-CAA media and grown overnight. The yeast recovered from the magnetic bead sort were induced in 100 mL SG/R-CAA for 20 h at 20 °C. The following two to three rounds of sorting were done using flow cytometry. Approximately 1×108 yeast were pelleted, washed once with wash buffer, and resuspended in 1 mL wash buffer with 10 μg/mL gp120 and a 1:200 dilution of anti-c-myc antibody. After a 1-h incubation at room temperature, yeast were washed 3 times and then resuspended in 1 mL wash buffer. Biotinylated HIVIG (15 μg/mL) was added to the yeast, incubated at 4 °C for 30 min, and washed 3 times with wash buffer. Five micrograms per milliliter of both SA–PE and GaM-647 was added to the cells and incubated for 30 min on ice in the dark, washed 3 times again, and then resuspended in 6 mL wash buffer for flow cytometric sorting. Sorting was performed using a FACS ARIA sorter and sort gates were determined to select only antigen binding clones. Collected cells were grown overnight in SD-CAA media at 30 °C and induced in SG/R-CAA for the next round of sorting. After the final round, yeast were plated on SD-CAA plates and individual colonies were picked for characterization.
Plasmid DNA was isolated from individual yeast clones using the Zymoprep Yeast Plasmid Miniprep Kit (Zymo Research). The recovered plasmids were electroporated in the XL-1 Blue E. coli for amplification of plasmid DNA and miniprepped using the QIAprep Spin Miniprep Kit (Qiagen). After sequencing, clones were grouped based on analysis of the CDRH3 region and a representative clone was chosen from each group for subsequent characterization. To determine approximate binding constants, we incubated ~106 induced yeast cells with a dilution series of gp120 (0–200 nM) and anti-c-myc antibody for 1 h at room temperature. Cells were then washed 3 times and incubated with biotinylated HIVIG at 4 °C for 30 min. After washing, yeast were incubated with fluorescent reagents for 30 min at 4 °C. Cells were pelleted, washed, and resuspended in approximately 100 μL FACS wash buffer. A BD Bioscience FACSArray plate reader was used for flow cytometric analysis, and FlowJo software was used for analysis. Quantitative equilibrium binding constants (Kd values) were calculated by plotting the c-myc-normalized MFI against antigen concentration and fitting the curve using nonlinear regression analysis.
Variable region genes were amplified by PCR, digested with SalI and BssHII (light chains) or NheI and NcoI (heavy chains), gel purified with the Qiagen QIAquick Gel Purification Kit, and ligated into a similarly digested and purified pYDscFv in order to clone scFabs as scFvs. The ligation mixture was heat shocked into Top10 cells and individual colonies were picked for plasmid isolation. Positive clones were verified by DNA sequencing.
An scFab clone that contained ApaI and AvrII restriction sites at the variable–constant region interface of the heavy and light chains, respectively, was used for cloning scFvs as scFabs. Variable region genes were amplified by PCR, digested with AvrII and BssHII (light chains) or ApaI and NheI (heavy chains), gel purified with the QIAquick Gel Extraction Kit (Qiagen), and ligated into a similarly digested vector. The ligation mixture was used to transform Top10 cells, and colonies were picked for plasmid isolation. Positive transformants were verified by DNA sequencing.
Preparation of soluble scFabs and Fabs
Individual scFab clones were digested out of pYDscFab with the restriction enzymes BssHII and NheI, gel extracted and purified with the QIAquick gel extraction kit, and ligated into the similarly digested and gel purified pComb3X vector for soluble expression in E. coli. The ligation reaction was used to transform Top10 cells, and individual colonies were picked for plasmid isolation. Positive inserts were verified by DNA sequencing. ScFab or Fab clones in pComb3X were then used to transform XL-1 Blue cells and grown at 37 °C in 10 mL Superbroth medium (30 g/L bactotryptone, 20 g/L yeast extract, and 10 g/L Mops) supplemented with 25 μg/mL carbenicillin and 10 μg/mL tetracycline until an OD600 (optical density at 600 nm) of 0.6–0.8 was reached. IPTG (1 mM) was then added and cells were grown for an additional 16–18 h at 30 °C. The cells were centrifuged at 3000 RPM for 10 min, and the pellet was resuspended in 0.5 mL 1× PBS. After four rounds of freeze–thawing between 37 and −80 °C, the cells were centrifuged at full speed in a tabletop centrifuge. Supernatants were collected and sterile-filtered for use in ELISAs and neutralization assays.
ELISA binding assays
Ninety-six-well ELISA plates were coated overnight at 4 °C with 50 μL PBS containing 50 ng of goat anti-human IgG F(ab′)2 (Pierce) or 75 ng gp120JR-FL. The wells were washed 4 times with PBS containing 0.05% Tween 20 and blocked with 3% BSA at room temperature for 1 h. The wells were then washed once, and 50 μL of the bacterial supernatants (2× diluted) containing Fab or scFab was added in a 2-fold dilution series. The plates were incubated at room temperature for 1 h and washed 4 times, and goat anti-human IgG F(ab′)2 conjugated to alkaline phosphatase (Pierce), diluted 1:1000 in PBS containing 1% BSA and 0.025% Tween 20, was added to the wells. The plate was incubated at room temperature for 1 h and washed 4 times, and the plate was developed by adding 50 μL of alkaline phosphatase substrate (Sigma) to 5 mL alkaline phosphatase staining buffer (pH 9.8), according to the manufacturer’s instructions. The optical density at 405 nm was read on a microplate reader (Molecular Devices). The concentration of Fab was determined with an anti-Fab ELISA (2-fold dilution series) using simple nonlinear regression. Half maximal effective concentrations (EC50 values) were determined using fixed concentrations of monomeric gp120 or gp41 (20 nM) and titrating the amount of crude Fab or scFab. In general, 12 different scFab or Fab concentrations were used, which covered a range of 0.002–2.0 μg/mL. The concentration of Fab or scFab corresponding to half maximal signal (EC50) was determined by interpolation of the resulting binding curve using nonlinear regression.
HIV-1 neutralization assays
HIV-1 pseudovirus, competent for a single round of replication, was generated by cotransfecting 293T cells with the luciferase reporter plasmid pNL4-3.Luc.R-E,33 an Env-complementation vector, and the polyethyleneimine transfection reagent34 as described previously.35 Neutralization was measured using U87.CD4.CCR5 target cells. Antibodies were diluted in Dulbecco’s modified Eagle’s medium (Gibco) containing 10% fetal bovine serum and mixed 1:1 with pseudovirus (100 μL total volume). The antibody–virus mixture was incubated for 1 h at 37 °C and then transferred (1:1 by volume) to the target cells. After a 72-h incubation at 37 °C, the cells were lysed, luciferase reagent (Promega) was added, and the luminescence in relative light units was measured using an Orion microplate luminometer (Berthold Detection Systems). Virus neutralization was measured as a reduction in viral infectivity compared to an antibody-free control. Neutralization curves were generated by plotting the percentage of virus neutralization against various scFab or Fab concentrations. The concentration of Fab or scFab, which yielded a 50% reduction in viral infectivity (IC50), was determined by interpolation of the resulting neutralization curve using nonlinear regression.
Acknowledgments
We thank Mansun Law, Lars Hangartner, and Michael Huber for helpful discussion; Cody Williams and Christina Corbaci for artwork in Fig. 1; and the members of The Scripps Research Institute Flow Cytometry Facility for assistance. This work was supported by National Institutes of Health Grant RO1 AI33292 and the International AIDS Vaccine Initiative Neutralizing Antibody Consortium.
Abbreviations used
- scFv
single-chain variable fragment
- Fab
antigen binding fragment
- scFab
single-chain Fab
- MFI
mean fluorescence intensity
- CDRH3
heavy-chain third complementary determining region
- mAb
monoclonal antibody
- HIV
human immunodeficiency virus
- IgG
immunoglobulin G
- SA–PE
streptavidin–phycoerythrin
- PBS
phosphate-buffered saline
- BSA
bovine serum albumin
References
- 1.Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nat Biotechnol. 1997;15:553–557. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- 2.Feldhaus MJ, Siegel RW, Opresko LK, Coleman JR, Feldhaus JM, Yeung YA, et al. Flow-cytometric isolation of human antibodies from a nonimmune Saccharomyces cerevisiae surface display library. Nat Biotechnol. 2003;21:163–170. doi: 10.1038/nbt785. [DOI] [PubMed] [Google Scholar]
- 3.Bowley DR, Labrijn AF, Zwick MB, Burton DR. Antigen selection from an HIV-1 immune antibody library displayed on yeast yields many novel antibodies compared to selection from the same library displayed on phage. Protein Eng Des Sel. 2007;20:81–90. doi: 10.1093/protein/gzl057. [DOI] [PubMed] [Google Scholar]
- 4.Lee HW, Lee SH, Park KJ, Kim JS, Kwon MH, Kim YS. Construction and characterization of a pseudo-immune human antibody library using yeast surface display. Biochem Biophys Res Commun. 2006;346:896–903. doi: 10.1016/j.bbrc.2006.05.202. [DOI] [PubMed] [Google Scholar]
- 5.Wang XX, Cho YK, Shusta EV. Mining a yeast library for brain endothelial cell-binding antibodies. Nat Methods. 2007;4:143–145. doi: 10.1038/nmeth993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van den Beucken T, Pieters H, Steukers M, van der Vaart M, Ladner RC, Hoogenboom HR, Hufton SE. Affinity maturation of Fab antibody fragments by fluorescent-activated cell sorting of yeast-displayed libraries. FEBS Lett. 2003;546:288–294. doi: 10.1016/s0014-5793(03)00602-1. [DOI] [PubMed] [Google Scholar]
- 7.Weaver-Feldhaus JM, Lou J, Coleman JR, Siegel RW, Marks JD, Feldhaus MJ. Yeast mating for combinatorial Fab library generation and surface display. FEBS Lett. 2004;564:24–34. doi: 10.1016/S0014-5793(04)00309-6. [DOI] [PubMed] [Google Scholar]
- 8.Blaise L, Wehnert A, Steukers MP, van den Beucken T, Hoogenboom HR, Hufton SE. Construction and diversification of yeast cell surface displayed libraries by yeast mating: application to the affinity maturation of Fab antibody fragments. Gene. 2004;342:211–218. doi: 10.1016/j.gene.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 9.Rippmann JF, Klein M, Hoischen C, Brocks B, Rettig WJ, Gumpert J, et al. Procaryotic expression of single-chain variable-fragment (scFv) antibodies: secretion in L-form cells of Proteus mirabilis leads to active product and overcomes the limitations of periplasmic expression in Escherichia coli. Appl Environ Microbiol. 1998;64:4862–4869. doi: 10.1128/aem.64.12.4862-4869.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marks JD, Hoogenboom HR, Griffiths AD, Winter G. Molecular evolution of proteins on filamentous phage. Mimicking the strategy of the immune system. J Biol Chem. 1992;267:16007–16010. [PubMed] [Google Scholar]
- 11.Bird RE, Walker BW. Single chain antibody variable regions. Trends Biotechnol. 1991;9:132–137. doi: 10.1016/0167-7799(91)90044-i. [DOI] [PubMed] [Google Scholar]
- 12.Labrijn AF, Poignard P, Raja A, Zwick MB, Delgado K, Franti M, et al. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J Virol. 2003;77:10557–10565. doi: 10.1128/JVI.77.19.10557-10565.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quintero-Hernandez V, Juarez-Gonzalez VR, Ortiz-Leon M, Sanchez R, Possani LD, Becerril B. The change of the scFv into the Fab format improves the stability and in vivo toxin neutralization capacity of recombinant antibodies. Mol Immunol. 2007;44:1307–1315. doi: 10.1016/j.molimm.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 14.Skerra A, Pfitzinger I, Plückthun A. The functional expression of antibody Fv fragments in Escherichia coli: improved vectors and a generally applicable purification technique. Biotechnology (NY) 1991;9:273–278. doi: 10.1038/nbt0391-273. [DOI] [PubMed] [Google Scholar]
- 15.Hust M, Jostock T, Menzel C, Voedisch B, Mohr A, Brenneis M, et al. Single chain Fab (scFab) fragment. BMC Biotechnol. 2007;7:14. doi: 10.1186/1472-6750-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jordan E, Al-Halabi L, Schirrmann T, Hust M, Dübel S. Production of single chain Fab (scFab) fragments in Bacillus megaterium. Microb Cell Fact. 2007;6:38. doi: 10.1186/1475-2859-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raymond CK, Mugford VR, Sexson SL. Plasmid topologies that enhance the transformation efficiency of yeast. BioTechniques. 1999;27:892–894. 896. doi: 10.2144/99275bm03. [DOI] [PubMed] [Google Scholar]
- 18.Swers JS, Kellogg BA, Wittrup KD. Shuffled antibody libraries created by in vivo homologous recombination and yeast surface display. Nucleic Acids Res. 2004;32:e36. doi: 10.1093/nar/gnh030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Binley JM, Wrin T, Korber B, Zwick MB, Wang M, Chappey C, et al. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J Virol. 2004;78:13232–13252. doi: 10.1128/JVI.78.23.13232-13252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gietz RD, Schiestl RH. Large-scale high-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc. 2007;2:38–41. doi: 10.1038/nprot.2007.15. [DOI] [PubMed] [Google Scholar]
- 21.Yeung YA, Wittrup KD. Quantitative screening of yeast surface-displayed polypeptide libraries by magnetic bead capture. Biotechnol Prog. 2002;18:212–220. doi: 10.1021/bp010186l. [DOI] [PubMed] [Google Scholar]
- 22.Brochet X, Lefranc MP, Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V–J and V–D–J sequence analysis. Nucleic Acids Res. 2008;36:W503–W508. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorny MK, Wang XH, Williams C, Volsky B, Revesz K, Witover B, et al. Preferential use of the VH5–51 gene segment by the human immune response to code for antibodies against the V3 domain of HIV-1. Mol Immunol. 2009;46:917–926. doi: 10.1016/j.molimm.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choe H, Li W, Wright PL, Vasilieva N, Venturi M, Huang CC, et al. Tyrosine sulfation of human antibodies contributes to recognition of the CCR5 binding region of HIV-1 gp120. Cell. 2003;114:161–170. doi: 10.1016/s0092-8674(03)00508-7. [DOI] [PubMed] [Google Scholar]
- 25.Rothlisberger D, Honegger A, Plückthun A. Domain interactions in the Fab fragment: a comparative evaluation of the single-chain Fv and Fab format engineered with variable domains of different stability. J Mol Biol. 2005;347:773–789. doi: 10.1016/j.jmb.2005.01.053. [DOI] [PubMed] [Google Scholar]
- 26.Acierno JP, Braden BC, Klinke S, Goldbaum FA, Cauerhff A. Affinity maturation increases the stability and plasticity of the Fv domain of anti-protein antibodies. J Mol Biol. 2007;374:130–146. doi: 10.1016/j.jmb.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 27.Thorpe IF, Brooks CL., III Molecular evolution of affinity and flexibility in the immune system. Proc Natl Acad Sci USA. 2007;104:8821–8826. doi: 10.1073/pnas.0610064104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scholler N, Garvik B, Quarles T, Jiang S, Urban N. Method for generation of in vivo biotinylated recombinant antibodies by yeast mating. J Immunol Methods. 2006;317:132–143. doi: 10.1016/j.jim.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burton DR, Barbas CF., III Human antibodies from combinatorial libraries. Adv Immunol. 1994;57:191–280. doi: 10.1016/s0065-2776(08)60674-4. [DOI] [PubMed] [Google Scholar]
- 30.Buchacher A, Predl R, Strutzenberger K, Steinfellner W, Trkola A, Purtscher M, et al. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein–Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res Hum Retroviruses. 1994;10:359–369. doi: 10.1089/aid.1994.10.359. [DOI] [PubMed] [Google Scholar]
- 31.Sblattero D, Bradbury A. A definitive set of oligonucleotide primers for amplifying human V regions. Immunotechnology. 1998;3:271–278. doi: 10.1016/s1380-2933(97)10004-5. [DOI] [PubMed] [Google Scholar]
- 32.Feldhaus MJ, Siegel RW. Yeast display of antibody fragments: a discovery and characterization platform. J Immunol Methods. 2004;290:69–80. doi: 10.1016/j.jim.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 33.Connor RI, Chen BK, Choe S, Landau NR. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mono-nuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 34.Kirschner M, Monrose V, Paluch M, Techodamrongsin N, Rethwilm A, Moore JP. The production of cleaved, trimeric human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein vaccine antigens and infectious pseudoviruses using linear polyethylenimine as a transfection reagent. Protein Expr Purif. 2006;48:61–68. doi: 10.1016/j.pep.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 35.Nelson JD, Kinkead H, Brunel FM, Leaman D, Jensen R, Louis JM, et al. Antibody elicited against the gp41 N-heptad repeat (NHR) coiled-coil can neutralize HIV-1 with modest potency but non-neutralizing antibodies also bind to NHR mimetics. Virology. 2008;377:170–183. doi: 10.1016/j.virol.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]