Abstract
The body is host to a wide variety of microbial communities from which the immune system needs to protect us and which are important for normal immune system development and maintenance of healthy tissues and physiological processes. Investigators have largely focused on the bacterial members of these communities, but an increasing number of studies underscore the presence of fungi as well that may be important for defining the communities and their interactions with immune cells. In this Review we discuss what is currently known about the makeup of fungal communities on the body and features of the immune system that are particularly important for interacting with fungi at these sites.
Largely due to advances in high-throughput sequencing technologies, the past decade or two has seen enormous progress in our understanding of the prevalence and diversity of the microbial communities associated with nearly all of our mucosal surfaces. This microbiota influences processes ranging from digestion to behaviour and is increasingly being appreciated as a fundamental and necessary component of our physiology1-4. The innate and adaptive arms of the immune system are charged with maintaining a healthy relationship with this microbiota and are, in fact, profoundly shaped by the presence of commensal microorganisms. Microbiota has largely been used to refer to the commensal and pathogenic bacteria that inhabit our body surfaces, but recent studies have begun to indicate that other organisms, specifically fungi, are also a significant, although substantially smaller, component of this microbiota. The term mycobiota has been used to refer to this fungal component.
Commensal fungi probably have important roles in health and disease. Fungal infections are increasing in prevalence owing to more people living with suppressed immune systems due to, for example, AIDS, organ transplantation or chemotherapy, and commensal fungal populations can be the source of the opportunistic pathogens that affect these patients. Further, emerging studies suggest that commensal fungal changes may be relevant in diseases that are not primarily fungal such as cystic fibrosis or inflammatory bowel disease. Just as commensal bacteria are important for the development and tuning of the immune system and individual types of bacteria compete with each other for resources in the microenvironment, it is also likely that commensal exposure to fungi influences the immune system and that fungi compete with each other and bacteria for resources. Just as bacterial “dysbiosis” may be an important feature of disease, so too might alterations in the mycobiota.
To understand the impact of interactions between the mycobiota and the host immune system we need to know the makeup of the mycobiota in different locations of the body, and we need to understand the mechanisms by which the immune system interacts with fungi at mucosal surfaces. This Review focuses on efforts to identify the fungal microbiota at different sites, changes in fungal populations that occur together with pathologies, and how the immune system is thought to interact with specific fungi at various sites.
Fungi: an underappreciated part of the microbiota
In the context of the entire microbiota, fungi are generally considered to be a minor component. In the gut, for example, recent shotgun sequencing efforts have suggested that fungi make up approximately 0.1% of the microorganisms5,6. There are several reasons to question whether this underestimates the number and significance of fungi. First, the estimates are based on identifying sequences based on available annotated reference sequences, and fungi are highly underrepresented in these databases compared with bacteria. For example, a recent check (Feb, 2014) of the NCBI Genome database revealed 57 complete fungal genomes compared with >2,700 complete bacterial genomes. This suggests that fungi might be under-detected compared with bacteria in shotgun sequencing efforts. Second, a typical fungal cell (which is ≈5 μm in diameter) is >100 times larger than a typical bacterial cell (which is ≈1 μm in diameter). Fungi are thus a much more substantial mass of biomaterials than simple genome-counting numbers might suggest. Third, fungi, as eukaryotes, are likely to contribute unique metabolic features to the microbiota. Finally, recent studies have clearly demonstrated that even “minor” components of the microbiota that proliferate in response to diet or during dysbiosis can have profound effects on the immune system7-9.
Approaches to studying the mycobiota
To study the mycobiota and its interactions with the immune system, the fungal part of the microbiota needs to be identified and quantified. Early studies relied on culturing fungi from various anatomical sites, but such methods made little progress in really characterizing the net complexity of commensal fungal communities. DNA-based methods have enabled culture-independent detection and identification of fungi. Approaches such as restriction fragment length polymorphism (RFLP) analysis, oligonucleotide fingerprinting of rRNA genes (OFRG), and denaturing gradient gel electrophoresis (DGGE), were good for revealing more complexity than culturing methods, but were relatively poor at identifying specific fungi. The advent of new, affordable high-throughput sequencing technologies has made direct sequencing of fungal DNA the preferred method for characterizing mycobiota. The most sensitive and affordable approach is to amplify a small variable region of fungal genomes in a sample by the polymerase chain reaction (PCR), sequence many thousands or millions of these fragments, and identify each fragment by comparison to a database. Thus which fragment is targeted is an important factor in the efficacy of the approach. Common targets have been the 18S small subunit ribosomal DNA (rDNA), 28S large subunit rDNA or internal transcribed spacer regions (BOX 1).
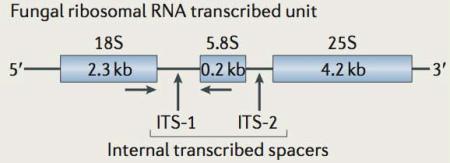
Box 1. Fungal rDNA regions targeted for mycobiome characterization.
The genomic region that encodes the fungal ribosomal DNA (rDNA) is shown in the figure. Certain structural regions of the rDNA are highly conserved and permit the synthesis of primers that will amplify fungal rDNA from nearly all species. There is considerable ongoing debate regarding which primer sets are optimal and which induce the least bias into the amplified sequence pool. As in other eukaryotes and bacteria, the rDNA region typically exists in multiple copies in each genome. In fungi, the locus is typically duplicated 100-200 times. As they are not part of the structural ribosomal RNAs that are transcribed from this region, the ‘internal transcribed spacer’ (ITS) regions are highly divergent between fungi (in sequence and in length), and they are often sufficiently different to classify fungi at the species level. This fungal rDNA region is the most heavily sequenced area in fungal genomes and nearly one-third of publically available fungal sequences are from this region.
The quality of data obtained through sequencing rDNA fragments depends on having a comprehensive, well-annotated database of fungal sequences to compare the fragments to. While several excellent databases have been developed for identifying bacterial 16S rDNA sequences, this is still a formidable challenge for investigators working with fungal sequences. The sequences available in GenBank probably only represent a fraction of the fungi that will be discovered; some estimates suggest that as little as 1% of fungal species are represented10. Furthermore, a large fraction of the available sequences are annotated as “uncultured”, which is not very useful for taxonomical purposes, and sequence annotations are plagued with misclassifications. 10-20% of sequences have been estimated to be misattributed10, and fungal taxonomy is in a state of enormous flux as sequencing data become available and more precise relationships between different fungi are characterized. Finally, it is very common for sexual and asexual forms of a fungus to be classified as different taxa (BOX 2). These are formidable barriers to investigators wanting to describe commensal fungal communities, but many approaches are being developed to address these problems, and the development of better tools will lead to improvement in the detection of fungal species.
Box 2. Sexual dimorphism in fungal taxonomy.
Sexual dimorphism in fungi can be a major problem when classifying fungal sequences. Many fungi have a sexual (teleomorph) and an asexual (anamorph) form. Frequently, the two different forms have been given different names and they are often not even characterized as being in the same genus or family. To complicate matters further, many anamorph-teleomorph pairs have not been identified yet and are only being discovered when sequencing reveals that they are the same organism. When relying on public databases to identify fungi based on nucleotide sequences, this can become a considerable challenge. For example, genetically, Aspergillus glaucus and Eurotium herbariorum are the same organism, although internal transcribed spacer (ITS) sequences that are deposited in the National Center for Biotechnology Information (NCBI) GenBank database are equally split between the two names (see the table for examples). Public databases, including NCBI Taxonomy, can be used to try to resolve some of the known relationships but investigators can often get different taxonomy information depending on which database they use.
Example Anamorph/Teleomorph pairs and the name use in GenBank:
| Anamorph | Teleomorph |
|---|---|
| Fusarium graminearum (326 ITS sequences) | Gibberella zeae (257 ITS sequences) |
| Cladosporium herbarum (236 ITS sequences) | Davidiella tassiana (177 ITS sequences) |
| Aspergillus glaucus (16 ITS sequences) | Eurotium herbariorum (16 ITS sequences) |
| Alternaria infectoria (136 ITS sequences) | Lewia infectoria (93 ITS sequences) |
The intestinal mycobiota
Fungi are normal inhabitants of the mammalian gastrointestinal tract11-16. Candida species have been successfully cultured from the intestines of healthy individuals, and increased Candida colonization of the intestine has been seen in inflammatory bowel disease (IBD) patients17,18. Early culture-independent studies of the mouse mycobiota using OFRG revealed a highly diverse community of fungi in mouse intestines12. In humans, DGGE analysis of 18S fungal rDNA revealed differential fungal profiles between healthy and ulcerative colitis patients19. Whereas initial studies suggested that human and mouse intestines are populated with diverse fungal microbiota, the approaches were labour intensive, marginally quantitative and could not be used to evaluate in-depth fungal variety. More recently, high throughput sequencing approaches have been used to explore fungal communities populating both human and mouse gut. These studies have shown that the gut is home to >50 genera of fungi with Candida, Saccharomyces and Cladosporium species being particularly common (Figure 1)13,16. With respect to Candida species, one study reported that Candida tropicalis is common in mice13, whereas C. albicans, C. glabrata, C. dubliniensis and C. parapsilosis are most prevalent in humans19-21. Only a few of the most common gut fungi were found in mouse food, suggesting that the majority are indigenous to the intestine13,14.
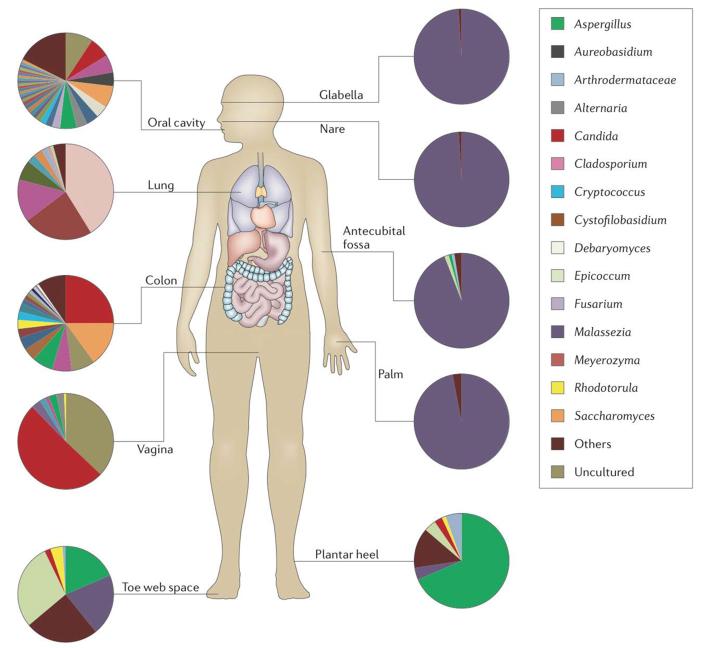
Figure 1. The human mycobiota.
Complex populations of fungi have been found associated with all mucosal surfaces and the skin on the healthy human body. The pie charts indicate the relative proportions of fungal genera reported in representative fungal deep sequencing studies. The legend indicates particularly common fungi associated with the respective sites. Mucosal surfaces tend to be more diverse compared to skin. Healthy lung, as in the reported study, likely reflects largely environmental fungi, and are mostly not included in the legend. “Others” in the legend refers to sequences representing less than 1% of the total recovered sequences at each site. “Uncultured” in the legend refers to sequences identified in GenBank as fungal, but of uncharacterized origin. Pie charts were derived from the following studies: Oral45, Lung83, Colon16, Vagina71, Skin63.
Studies in newborn pups and human infants have suggested that bacterial communities in the gut are initially unstable but become more stable in early childhood and develop more modestly through adulthood22-24. However, this may not be the case with the gut mycobiota. One recent study demonstrated that fungal populations in the mouse gut displayed episodic variation over several months, although the bacterial community remained relatively stable14. This suggests that fungal populations are more variable and may be influenced by fungi in the environment. Another explanation might be that bacteria are more highly abundant than fungi and that a consequence of this is that their communities are more robust than fungal communities. Alterations in diet can also affect the fungal microbiota; in humans, plant-based diet consumption has been linked to an increase of Candida, whereas the consumption of an animal-based diet facilitated the expansion of Penicillium species15.
Influence on bacterial microbiota
Fungal and bacterial communities undoubtedly influence each other. We have observed that commensal fungi in the mouse gut can be found in patches together with gut bacteria13. Germfree mice are highly susceptible to Candida infection25, and antibiotic treatment supports Candida colonization and overgrowth in the mouse gut14,26,27. In one study, long-term antibiotic treatment (76 days) led to robust expansion of fungi, reaching 99% of all intestinal microorganisms detected by deep sequencing analysis14. This was mostly due to an expansion of Candida species which was the single genus found in the faeces by the end of the treatment. Similarly, prolonged antibiotic treatment in humans can predispose to fungal infections, due mostly to expansion of Candida species28,29. In keeping with the idea that fungi can also influence the bacterial composition of the gut microbiota, a recent study has demonstrated that restoration of the bacterial microbiota after treatment with a single antibiotic (cefoperazone) was strongly influenced by colonization with C. albicans30. There were significant decreases in Bacteroidetes and Synergistetes while Firmicutes remained unchanged. However, whether Candida-induced microbiota perturbation is transient or how it affects the host remains to be elucidated.
Interactions with the immune system
We know little about if or how most intestinal commensal fungi interact with the host immune system, although many innate immune receptors have been shown to interact with fungal pathogens (Figure 2). There is ample evidence that host control of intestinal fungi is important, and a variety of specific immune mechanisms have been implicated in the clearance of fungi at mucosal surfaces31,32. Among these, C-type lectin domain family 7 member A (Dectin-1), caspase recruitment domain-containing protein 9 (CARD9), interleukin-17 (IL-17) and IL-22 have emerged as molecules that are responsible for host defence against fungi, and genetic mutations in each of these molecules is associated with susceptibility to fungal infections in humans (Table 1). We have reported that a genetic polymorphism in CLEC7A, the gene encoding the antifungal receptor Dectin-1, is associated with severity of ulcerative colitis severity in humans, and have observed that experimental colitis in mice is more severe in the absence of Dectin-113. The more severe colitis in mice was accompanied by fungal invasion of the colonic mucosa, as well as a general expansion of opportunistic fungi such as Candida and Trichosporon species, and a decrease in non-pathogenic Saccharomyces species. In patients with IBD, the disease has been reported to be associated with an enrichment for Candida species (specifically C. dubliniensis and C. parapsilosis), suggesting that fungal dysbiosis may accompany disease19. However, the DGGE method used was limited in its ability to fully characterize the fungal populations, and further studies applying more sensitive deep sequencing analysis are needed.
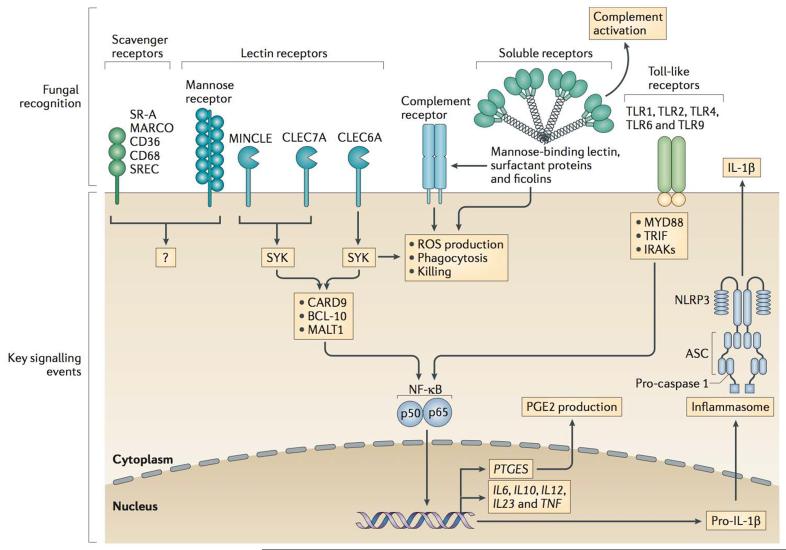
Figure 2. Immune receptors and signalling pathways involved in recognition of fungi.
Innate immune cells utilize a wide variety of membrane-bound and soluble receptors to recognize fungi. Membrane-bound receptors such as lectin receptors (that recognize fungal polysaccharides), Toll-like receptors (TLRs), and scavenger receptor family members can directly recognize a wide variety of fungi or soluble products released from fungi. These receptors trigger phagocytosis, respiratory burst via the NADPH phagocyte oxidase, and killing of fungi, as well as trigger intracellular signaling via pathways leading to activation of transcription factors such as NF-κB that mediate production of many inflammatory cytokines and chemokines that are important for host defence against fungi. Fungi may also be recognized by soluble receptors such as the mannose-binding lectin (MBL) that can direct complement activation for killing and release of inflammatory mediators as well as opsonize fungi for recognition by additional membrane-bound receptors such as complement receptors.
Table 1.
Genetic disorders associated with impaired mucosal immunity to fungi
| Gene | Defects that lead to fungal disease | Disease | Site affected | Mycobiota changes reported |
Refs |
|---|---|---|---|---|---|
| AIRE |
|
|
Skin, nails, oral mucosa and genital mucosa |
|
59 |
| DOCK8 | Defective activation of T cells— including Th17 cells—due to impaired immunological synapse formation |
|
Skin and nails | Increased mycobiota diversity with prevalence of Candida and Aspergillus species |
76, 116, 117 |
| STAT3 |
|
|
Skin, nails and lungs |
|
76, 118-121 |
| STAT1 | Defective IL-17, IL-22 and IFNγ production | CMC | Skin, nails and oral mucosa |
|
53 |
| CLEC7A | Impaired IL-lβ and IL-17 production |
|
Skin, nails and genital mucosa |
|
66, 122 |
| CARD9 |
|
|
Skin, nails and genital mucosa |
|
46,66 |
| IL17RA |
|
CMC | Skin and oral mucosa |
|
57 |
| IL17F |
|
CMC | Skin, nails, oral mucosa and genital mucosa |
|
91 |
| MBL2 | Decreased MBL levels |
|
Genital mucosa and lungs |
|
89,91 |
| IL12RB1 | Abolished responses to IL-12 and IL-23 | CMC | Skin, oral mucosa, oesophageal mucosa and genital mucosa |
|
123 |
A deficiency in CARD9, a signalling molecule downstream of Dectin-1 and other lectins involved in fungal recognition, has been also linked to susceptibility to IBD in humans33-35. Although CARD9 might be involved in signalling by other receptors, its specific importance in antifungal immunity is supported by the observation that individuals with rare null mutations in CARD9 are specifically susceptible to fungal infections and no other type of infection36. CARD9 deficiency in mice enhances susceptibility to experimental colitis, augmented fungal burden in the gut, increased antifungal antibodies in serum and wasting disease. In addition, these mice can be partially rescued by antifungal treatment, further supporting a role for immunity to intestinal fungi being important in colitis37.
IL-22, a cytokine that similar to IL-17 (discussed in more detail below) is closely associated with mucosal immunity, has been directly implicated in controlling gastrointestinal fungi; mice lacking IL-22 are more susceptible to gastrointestinal candidiasis when infected intragastrically with C. albicans38. Further, effective host defence required IL-12 and interferon-γ (IFN-γ, implicating an important contribution of T helper 1 (TH1) cell-mediated immunity. However, whether similar mechanisms are involved during an ongoing commensal relationship with C. albicans, a lifetime colonizer in the human gut, remains to be elucidated. Curiously, mouse models of colitis show that both IL-17A and IL-17F are dispensable for protection against gastrointestinal Candida species38.
Striking evidence suggests that intestinal Candida species can influence immunity at distant body sites through both interactions with immune cells and the production of fungal metabolites. Antibiotic-induced overgrowth of Candida species in the intestines has been shown to promote lung inflammation in a mouse model of allergy26,39. Candida species (as well as many other fungi) directly produce prostaglandin E2 (PGE2), a potent immunomodulator produced by immune cells, from host arachidonic acid, and it has been proposed that Candida-produced PGE2 might be involved in allergic inflammation.40-42. Indeed it has been recently demonstrated that Candida-derived PGE2 can reach the lungs through the bloodstream, act on lung macrophages and promote allergic inflammation43. Similarly, an association between graft-versus-host disease and gut colonization with Candida species has been seen in transplant patients44. Gut colonization was correlated with presence of a defective Dectin-1 gene, suggesting that immune-mediated regulation of colonization may have an effect on peripheral tolerance.
Oral mycobiota
The oral cavity is a well-known environment for microbial growth, although the fungal members of this community have rarely been assessed. The most complete culture-independent evaluation of the mycobiota in the healthy mouth show that the oral cavity is home >75 different fungal genera, with Candida, Cladosporium, Aureobasidium, Aspergillus, and Fusarium being among the most common45. The nature of the host immune response to normal fungal carriage in the mouth has not been evaluated. However, it is well-known that immunosuppressed individuals (such as patients with HIV or undergoing chemotherapy) frequently develop oropharyngeal candidiasis, demonstrating that local immunity is necessary to contain at least Candida species. Whether local immunity influences colonization or growth of other fungal species is not known.
Genetic studies in humans provide some hints as to the types of immunity that are necessary for healthy containment of Candida species in the mouth46. Chronic mucocutaneous candidiasis is a condition characterized by recurrent Candida infections of the mouth, skin, and other mucosal surfaces. Several groups have recently demonstrated that mutations in signal transducer and activator of transcription 1 (encoded by STAT1), a signalling molecule important for responses to IFNγ, IL-17, and IL-22, render patients highly susceptible to chronic mucocutaneous candidiasis with all cases exhibiting oral pathologies47-50. In addition, mutations in the genes encoding IL-17RA, IL-17F, and adapter protein CIKS (ACT1), a protein necessary for IL-17R signalling, have been associated with chronic mucocutaneous candidiasis (Table 1)51,52. This susceptibility to oral Candida infection can also be caused by an autoimmune disorder called autosomal recessive autoimmune polyendocrinopathy candidiasis ectodermal dystrophy (APECED) in which mutations in AIRE lead to the production of autoantibodies against IL-17A, IL-17F, and IL-2253. In mice, deficiencies in the IL-17 pathway have also been shown to be more susceptible to oral candidiasis54,55. Together, these findings highlight the remarkably selective importance of Th17 immunity in restricting pathology caused by Candida colonization.
The innate responses that drive this Th17 immunity can also be inferred from genetic studies. People with genetic deficiencies in CARD9 are highly susceptible to invasive Candida infections, typically accompanied by a history of oral infections36,56,57. Notably patients with CARD9 deficiency showed decreased numbers of Th17 cells36. In a mouse model of oral Candida infection it was recently confirmed that CARD9 is necessary for mounting a Th17 cell response, although IL-17 dependent innate responses do not involve CARD958. Of the receptors that might activate CARD9, the role of Dectin-1 has been explored in a mouse model and was found to be important for effective defence59. A common premature stop mutation in Dectin-1 in humans has been associated with onychomycosis, not oropharyngeal candidiasis60. This might suggest that Dectin-1 is not involved in oral antifungal immunity in humans, although the prevalence of the null mutation is sufficiently high that the penetrance of the effect must be low, even for onychomycosis, and nothing like the severity of the CARD9 mutations. Additional fungal-sensing receptors such as Dectin-2 can signal through CARD9, and this redundancy might explain the more severe oral disease in the absence of CARD9. Further studies will be required to determine the role of additional receptors such as Dectin-2 in oral antifungal immunity.
Skin mycobiota
The skin surface is the first point of contact of the body with bacteria and fungi coming from the environment and is home to billons of microorganisms that have developed a commensal relationship with the host. Deep sequencing approaches have demonstrated that skin harbours its unique bacterial and fungal microbiota61-65. Further, the skin microbiota depends on the nature of the skin site, with certain patterns associated with moist, dry or sebaceous microenvironments61,62. As the skin is a self-renewing organ, dead cells are continuously shed, providing an environment for saprophytic microbial growth.
Culture-dependent approaches have identified many commensal fungi associated with skin, with Malassezia as the most common genus, followed by Penicillium and Aspergillus63,66. Other fungi such as Alternaria, Candida, Rhodotorula, Cladosporium and Mucor have also been cultured, but with a lower frequency63,66. Culture-independent sequencing studies confirmed that the genus Malassezia most commonly represented across different body sites of healthy human skin63,67,68. The higher resolution of these approaches allowed identification of specific species, and certain Malassezia species are associated with specific body sites63. Candida species were also detected across the body sites and were mostly represented by C. tropicalis, C. parapsilosis and C. orthopsilosis, species different from those typically populating the human gut. While sites on the foot revealed the greatest fungal diversity (40 to 80 genera), other sites such as nare, glabella, back, manubrium, and palm had greater bacterial than fungal diversity63. This reverse correlation between bacterial and fungal community diversity might be due to the presence of specific nutrients at different body sites and point to a complex interaction between the two communities.
Patients with primary immunodeficiencies suffer from recurrent fungal infections. Although the immunopathology of each specific immunodeficiency differs, a common hallmark of many is the development of atopic dermatitis-like eczema. One recent study in patients with primary immunodeficiencies revealed a direct link between host immunity and its effect on the skin bacterial and fungal microbiota69. Pathways including STAT3 and dedicator of cytokinesis protein 8 (Dock8) (leading to hyperimmunoglobulin E syndrome) and WAS/WASL-interacting protein family member 1 (WAS, leading to Wiskott-Aldrich syndrome) were affected, and patients exhibited altered bacterial community structure, increased fungal diversity and increased abundance of Candida and Aspergillus species in the skin (Table 1)69. Similar results were obtained in another study showing that non-Malassezia fungal microbiota diversity is increased in patients with atopic dermatitis, with augmented representation of Candida, Cryptococcus and Cladosporium species68. Analysis of co-occurrence of specific bacteria and fungi suggested that the two communities might interact69.
An autosomal recessive CARD9 mutation can lead to deep dermatophytosis, which in contrast to a superficial mucocutaneous disease, affects dermal and subcutaneous tissue, lymph nodes and, occasionally, the central nervous system leading to mortalities56. As noted above, these patients have decreased numbers of Th17 cells36. In vivo studies have shown that mice deficient in IL-17A or IL-23 are more susceptible to C. albicans skin infection70. Although IL-22 deficiency seems to play a role in mucocutaneous dieses in humans, it appears to not be involved in the mouse model of Candida skin infection70. Future studies will be needed to elucidate the effect of the mycobiome composition on the immunity at specific body sites during skin disease.
Vaginal mycobiota
Like other mucosal surfaces, the vagina is home to a pool of microbial occupants that, if not properly contained, can cause pathologies. While yeast infections are common, several recent studies have revealed a more diverse population of resident fungi than previously appreciated71-73. These studies suggest the presence of 11-20 different genera, with commonly detected fungi including Candida, Saccharomyces, Aspergillus, Alternaria, and Cladosporium. Changes in fungal diversity were noted to be associated with diabetes, allergic rhinitis and recurrent vaginal candidiasis72,73. Fungal growth is controlled both by other members of the local microbiota and by host immune defences. Lactobacilli species are the dominant microorganisms in the healthy vaginal microbiota. These bacteria produce lactic acid that contributes to the low pH of the environment, which suppresses fungal growth, and can compete with Candida species for binding to epithelial cells74-76.
The idea that the immune system is responsible for normal surveillance of the vaginal fungal microbiota is suggested by the association of specific immune gene variants with recurrent vaginal candidiasis. Patients with premature stop mutation in Dectin-1 that is associated with onychomycosis (discussed above) also have recurrent vaginal candidiasis, which suggests that Dectin-1 is important for normal containment of vaginal fungi60. Another genetic variant for a length polymorphism in intron 4 of NLRP3 (encoding NACHT, LRR and PYD domains-containing protein 3) has also been associated with recurrent vaginal candidiasis77, suggesting that inflammasomes are important for containment of vaginal fungi. While an animal model of vaginal candidiasis has not been explored in the context of NLRP3, NLRP3-deficient mice are more susceptible to invasive disease in a model of mucosal Candida infection59. The group has also reported a role for another inflammasome containing NLRC4 in defence against oral Candida, but whether this is important in human defence or in vaginal fungal control is not known78.
The mannose-binding lectin (MBL) is a soluble C-type lectin receptor that binds to carbohydrates on microbial cell walls where it activates the complement pathway and opsonizes microorganisms for phagocytosis. A genetic polymorphism in MBL that affects the level of protein expression is associated with recurrent vaginal candidiasis, suggesting that MBL is important for normal containment of vaginal fungi (Table 1)79-81. The low MBL variant has also been reported to be associated with fungal infections at other mucosal sites such as in defence against Aspergillus in the lung82.
Lung mycobiota
There is little evidence of a commensal fungal microbiota in healthy lungs, although the lungs are constantly exposed to oral and environmental fungi83. Thus, the mucosal immune system in the lungs continuously encounters fungi and fungal antigens. When normal lung function is compromised, such as in cystic fibrosis, fungal communities may take hold and persist84,85. Commonly encountered fungi in this setting include Aspergillus sp. and Scedosporium sp., two spore-forming filamentous soil molds that are widespread in the environment. Such spores are inhaled regularly and are normally cleared without pathology in immunocompetent individuals by alveolar macrophages that bind, internalize and kill the spores. Spores that evade killing can grow into hyphae which, if not killed, can invade the tissue, the circulation and disseminate. Disseminated aspergillosis is particularly common in people who lack effective hyphal defence by polymorphonuclear phagocytes. Aspergillus sp. is also a potent allergen, and colonization of the airways has been associated with severe asthma and nasal allergies86.
Recognition of Aspergillus sp. in the lungs is mediated in part by pattern recognition receptors including C-type lectins (i.e. Dectin-1 and Dectin-2) and Toll-like receptors. Resting spores are encased in a waxy shell that shields them from recognition by most receptors, but when the spores break this outer coating to germinate and grow as hyphae, multiple cell wall ligands are revealed87-89. This germination is generally thought to happen in the lumen of the lung, but Dectin-1 can also recognize Aspergillus spores that germinate after ingestion by macrophages in the lung90. In this case, the receptor is localized to phagosomes instead of the cell surface and is activated by the presence of the fungus. The central role of alveolar macrophages in initial clearance of inhaled fungi is illustrated in mice expressing the NADPH oxidase only in monocytes and macrophages. These animals clear Aspergillus normally, whereas NADPH oxidase-deficient mice are highly susceptible91,92.
If fungi survive their first encounter with macrophages in the lungs, inflammatory infiltration of immune cells is initiated, and the adaptive immune system may be engaged. Th1 type T cell responses are generally protective against Aspergillus, whereas Th2-type responses are generally associated with poor outcomes93. However, some reports suggest that Th17 cell responses are even more important than Th1 cell responses. Indeed, it was shown that Dectin-1 signalling specifically is important for decreasing Th1 responses and promoting Th17 cell responses to Aspergillus94. Also, Dectin-1 is important for stimulating the production of IL-22, a Th17-associated cytokine with pro- and anti-inflammatory properties95. Furthermore, Dectin-2 has been shown to have an important role in regulating IL-17 production by a subset of retinoic acid receptor-related orphan receptor-t (RORγt)-expressing neutrophils in response to Aspergillus, indicating that these innate cells may be an important source of IL-17 during infection, although this has yet to be specifically examined in the case of exposure to lung fungi96.
Just as important as mounting effective immunity against fungi in the lungs is the need to prevent allergic inflammation in response to fungal antigens. Allergic responses to Aspergillus in a mouse model are highly dependent on Dectin-1 recognition of the fungus and this receptor’s ability to promote IL-22 production by T cells97. However, the mechanisms promoting allergic inflammation are probably highly dependent on the type of fungus as illustrated by the observation that while Aspergillus responses are promoted by Dectin-1, whereas responses to Cladosporium sp. were independent of Dectin-1 and IL-1798. As noted above, commensal fungi in the gut may have a significant impact on the nature of inflammation in the lungs26,39. Exactly how this connection works and whether it preferentially affects responses to fungal triggers are important questions that have yet to be fully explored. It is possible that intestinal fungal-produced PGE2 influences macrophages in the lungs40-43.
Conclusions
Just as the body is host to diverse populations of bacteria that contribute to health and pathology, so too is the body host to diverse populations of fungi that have been less well studied. Commensal fungal populations vary between body sites and probably vary over time and with disease. It will be interesting in coming years to learn more about how the immune system specifically interacts with the mycobiota in healthy and disease states. The molecular and cellular mechanisms that engage specific fungi during steady state healthy conditions may be quite different from ones that are engaged in diseased tissue or during active fungal invasion. The health of tissues and tissue barriers might influence this, but so too might the fungi themselves. For example, Candida are a dimorphic fungi and can grow in yeast and hyphal forms. This ability to switch between morphologies is important for pathogenesis, and several reports have demonstrated that the yeast and hyphal forms are recognized differently by immune cells99-104. A recent study has demonstrated that commensal C. albicans in the intestines takes on an entirely novel morphology termed “GUT” that is related to the a/α mating type105. GUT Candida cells are metabolically adapted to take advantage of nutrients available in the gut and are attenuated in their ability to induce systemic disease in a mouse bloodstream model of infection, but it is not known if they interact with immune cells in ways that are fundamentally different from the other forms.
Each mucosal surface is a unique environment that shapes the microbiota that inhabits the site. Current studies have documented that, like bacteria, diverse populations of fungi are characteristic to specific sites. The local immune system at each site might interact with commensal fungi in similar or unique ways (Figure 3). Future studies will be required to understand how the mucosal immune system interacts with fungal communities and how we might use these interactions to prevent or treat fungal and inflammatory diseases.
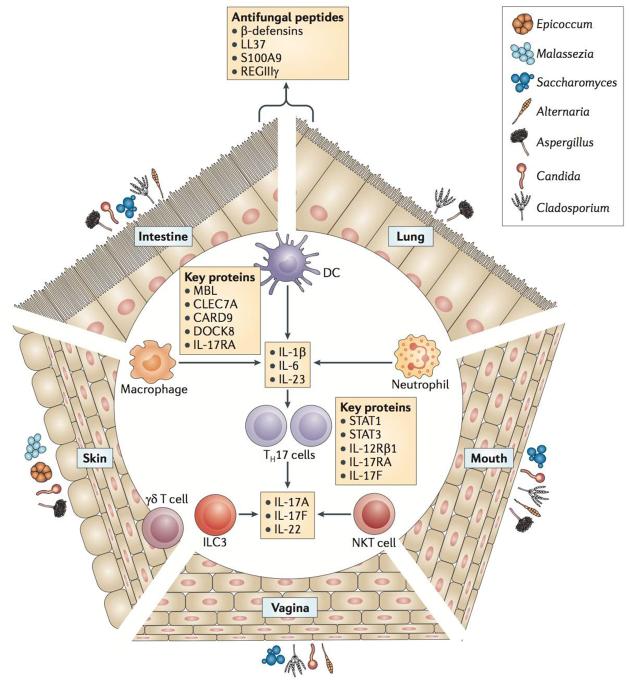
Figure 3. Mucosal immune responses involved in interaction with fungi at different sites.
Different body sites are colonized by diverse groups of fungi, and the communities are shaped by the characteristics of each environment. Epithelial cells at these surfaces produce antimicrobial peptides that directly modulate fungal survival. In response to fungi, cytokines and chemokines that recruit immune cells are also produced. Fungi may also be directly sensed by dendritic cells and γδT cells at epithelial surfaces. When epithelial barriers are breached, macrophages, dendritic cells, and neutrophils kill fungi and produce cytokines that promote adaptive immune responses. Innate lymphoid cells may also respond directly to fungi by producing cytokines. Genetic studies in humans have revealed proteins (indicated in red) that are particularly important for antifungal defence and implicate a crucial role for the IL-17 pathway in this process.
Glossary Terms
- ASCA
Anti-Saccharomyces cerevisiae antibodies (ASCA) are antibodies that are found commonly in serum from IBD patients. They are more common in Crohn’s disease than ulcerative colitis. ASCA cross-react with mannans from cell walls of many fungi (including Candida species), suggesting that the name might be misleading.
- Candida
Candida is a genus of yeasts including common species such as albicans, tropicalis, glabrata, parapsilosis, and krusei. Candida species are normal inhabitants of the skin and mucous membranes, causing disease primarily in immune compromised people. Candidiasis (disease caused by Candida) in the mouth or throat is called “thrush” or “oropharyngeal candidiasis”.
- CARD9
Caspase recruitment domain-containing protein 9 (CARD9) is a signaling adaptor molecule found to be downstream of many immunoreceptor tyrosine-based activation motif (ITAM) receptors including Dectin-1 and Dectin-2. CARD9 associates with BCL10 and the paracaspase MALT1 to facilitate signaling through NF-κB and promote acute inflammatory responses and initiation of adaptive immunity.
- C-type lectin receptor
C-type lectin receptors (CLRs) comprise a large family of receptors that bind to carbohydrates, typically in a calcium-dependent manner. Here the term refers to the set of C-type lectin receptors that act as “pattern recognition receptors” in the detection of microbial threats and activate immune responses including membrane receptors such as Dectin-1 & 2, and the mannose receptor, and soluble receptors such as the mannose-binding lectin (MBL). The binding activity of these receptors is mediated by conserved carbohydrate-recognition domains (CRDs).
- Chronic mucocutaneous candidiasis
Chronic mucocutaneous candidiasis is a condition characterized by recurrent Candida infections of the mouth, skin, and other mucosal surfaces.
- Denaturing gradient gel electrophoresis (DGGE)
A DNA-based technique for generating a genetic profile of a microbial community. A highly variable gene (typically 16SrRNA) is targeted for amplification by PCR and the complex mixture of products is separated by denaturing gradient gel electrophoresis which separates fragments based on sequence.
- Dysbiosis
A term originally coined by 1908 Nobel laureate Eli Metchnikoff to refer to pathogenic alterations of bacterial microflora in the gut. The term is now generally used to refer to any microbial imbalance in or on the body, including the gastrointestinal tract or the skin, or any exposed mucosal surface such as lungs, vagina, or mouth.
- Inflammatory Bowel Disease
Inflammatory Bowel Disease (IBD) is a group of chronic inflammatory conditions of the large and small intestines, the major types being Crohn’s disease and ulcerative colitis.
- Inflammasome
Inflammasomes are multimeric protein complexes that activate caspase-1 to promote inflammatory processes such as processing of pro-interleukin-1β and pro-interleukin-18 into their mature secreted forms.
- Malassezia
Malassezia is a genus of basidiomycetous fungi including species such as dermatis, furfur, and restricta. These yeasts are specifically adapted to growth on mammalian skin. Found on all skin, they are associated with conditions such as dandruff, atopic eczema/dermatitis, pityriasis versicolor, seborrheic dermatitis, and folliculitis.
- Oligonucleotide fingerprinting of rRNA genes (OFRG)
A method for analyzing microbial communities based on hybridization of DNA probes to arrays of cloned DNA fragments representing the ribosomal DNA diversity of the population.
- Onychomycosis
fungal infection of the toenails or fingernails. These infections are most commonly caused by dermatophytes (Microsporum, Epidermophyton and Trichophyton), but can also be caused by Candida and nondermatophytic molds.
- Restriction fragment length polymorphism (RFLP)
A method for characterizing DNA based on sizes of fragments of DNA between restriction enzyme sites. In identification of microbes, “fingerprints” or unique patterns of fragments can be used to distinguish closely-related organisms.
- Shotgun sequencing
A sequencing approach in which a complex pool of DNA is broken up into random small segments that are sequenced en masse. Computational tools are then used to reassemble and characterize the DNA fragments.
- T helper 1 (TH1) cell-mediated immunity
a polarization of the T cell response characterized by production of T cells making IFN-γ. This is generally associated with effective host defence against intracellular bacteria and protozoa.
- Uncultured Fungi
“Uncultured fungi” is a term used in mycobiome sequencing studies to refer to sequences identified in GenBank as fungal, but of yet-uncharacterized origin.
References
- 1.Consortium, H.M.P. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–14. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148:1258–70. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Erturk-Hasdemir D, Kasper DL. Resident commensals shaping immunity. Current opinion in immunology. 2013;25:450–5. doi: 10.1016/j.coi.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tremaroli V, Backhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–9. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- 5.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arumugam M, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–80. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bull-Otterson L, et al. Metagenomic analyses of alcohol induced pathogenic alterations in the intestinal microbiome and the effect of Lactobacillus rhamnosus GG treatment. PLoS One. 2013;8:e53028. doi: 10.1371/journal.pone.0053028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sonnenberg GF, et al. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science. 2012;336:1321–5. doi: 10.1126/science.1222551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Devkota S, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature. 2012;487:104–8. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nilsson RH, et al. Taxonomic reliability of DNA sequences in public sequence databases: a fungal perspective. PLoS One. 2006;1:e59. doi: 10.1371/journal.pone.0000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hube B. From commensal to pathogen: stage- and tissue-specific gene expression of Candida albicans. Curr Opin Microbiol. 2004;7:336–41. doi: 10.1016/j.mib.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 12.Scupham AJ, et al. Abundant and diverse fungal microbiota in the murine intestine. Appl Environ Microbiol. 2006;72:793–801. doi: 10.1128/AEM.72.1.793-801.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iliev ID, et al. Interactions Between Commensal Fungi and the C-Type Lectin Receptor Dectin-1 Influence Colitis. Science. 2012;336:1314–1317. doi: 10.1126/science.1221789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dollive S, et al. Fungi of the murine gut: episodic variation and proliferation during antibiotic treatment. PLoS One. 2013;8:e71806. doi: 10.1371/journal.pone.0071806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.David LA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2013 doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoffmann C, et al. Archaea and fungi of the human gut microbiome: correlations with diet and bacterial residents. PLoS One. 2013;8:e66019. doi: 10.1371/journal.pone.0066019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Odds FC, et al. Candida albicans strain maintenance, replacement, and microvariation demonstrated by multilocus sequence typing. J Clin Microbiol. 2006;44:3647–58. doi: 10.1128/JCM.00934-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Standaert-Vitse A, et al. Candida albicans is an immunogen for anti-Saccharomyces cerevisiae antibody markers of Crohn’s disease. Gastroenterology. 2006;130:1764–75. doi: 10.1053/j.gastro.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 19.Ott SJ, et al. Fungi and inflammatory bowel diseases: Alterations of composition and diversity. Scand J Gastroenterol. 2008;43:831–41. doi: 10.1080/00365520801935434. [DOI] [PubMed] [Google Scholar]
- 20.Scanlan PD, Marchesi JR. Micro-eukaryotic diversity of the human distal gut microbiota: qualitative assessment using culture-dependent and -independent analysis of faeces. ISME J. 2008;2:1183–93. doi: 10.1038/ismej.2008.76. [DOI] [PubMed] [Google Scholar]
- 21.Standaert-Vitse A, et al. Candida albicans colonization and ASCA in familial Crohn’s disease. Am J Gastroenterol. 2009;104:1745–53. doi: 10.1038/ajg.2009.225. [DOI] [PubMed] [Google Scholar]
- 22.Savage DC, Dubos R, Schaedler RW. The gastrointestinal epithelium and its autochthonous bacterial flora. The Journal of experimental medicine. 1968;127:67–76. doi: 10.1084/jem.127.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dominguez-Bello MG, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:11971–5. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Agans R, et al. Distal gut microbiota of adolescent children is different from that of adults. FEMS microbiology ecology. 2011;77:404–12. doi: 10.1111/j.1574-6941.2011.01120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naglik JR, Fidel PL, Jr., Odds FC. Animal models of mucosal Candida infection. FEMS Microbiol Lett. 2008;283:129–39. doi: 10.1111/j.1574-6968.2008.01160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noverr MC, Falkowski NR, McDonald RA, McKenzie AN, Huffnagle GB. Development of allergic airway disease in mice following antibiotic therapy and fungal microbiota increase: role of host genetics, antigen, and interleukin-13. Infect Immun. 2005;73:30–8. doi: 10.1128/IAI.73.1.30-38.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mason KL, et al. Candida albicans and bacterial microbiota interactions in the cecum during recolonization following broad-spectrum antibiotic therapy. Infect Immun. 2012;80:3371–80. doi: 10.1128/IAI.00449-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Samonis G, et al. Prospective evaluation of effects of broad-spectrum antibiotics on gastrointestinal yeast colonization of humans. Antimicrobial agents and chemotherapy. 1993;37:51–3. doi: 10.1128/aac.37.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mulligan ME, Citron DM, McNamara BT, Finegold SM. Impact of cefoperazone therapy on fecal flora. Antimicrobial agents and chemotherapy. 1982;22:226–30. doi: 10.1128/aac.22.2.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Erb Downward JR, Falkowski NR, Mason KL, Muraglia R, Huffnagle GB. Modulation of post-antibiotic bacterial community reassembly and host response by Candida albicans. Sci Rep. 2013;3:2191. doi: 10.1038/srep02191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown GD. Innate antifungal immunity: the key role of phagocytes. Annual review of immunology. 2011;29:1–21. doi: 10.1146/annurev-immunol-030409-101229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Romani L. Immunity to fungal infections. Nature reviews. Immunology. 2011;11:275–88. doi: 10.1038/nri2939. [DOI] [PubMed] [Google Scholar]
- 33.Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–17. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jostins L, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–24. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beaudoin M, et al. Deep resequencing of GWAS loci identifies rare variants in CARD9, IL23R and RNF186 that are associated with ulcerative colitis. PLoS Genet. 2013;9:e1003723. doi: 10.1371/journal.pgen.1003723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Glocker EO, et al. A homozygous CARD9 mutation in a family with susceptibility to fungal infections. N Engl J Med. 2009;361:1727–35. doi: 10.1056/NEJMoa0810719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sokol H, et al. Card9 mediates intestinal epithelial cell restitution, T-helper 17 responses, and control of bacterial infection in mice. Gastroenterology. 2013;145:591–601 e3. doi: 10.1053/j.gastro.2013.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Luca A, et al. IL-22 defines a novel immune pathway of antifungal resistance. Mucosal Immunol. 2010;3:361–73. doi: 10.1038/mi.2010.22. [DOI] [PubMed] [Google Scholar]
- 39.Noverr MC, Noggle RM, Toews GB, Huffnagle GB. Role of antibiotics and fungal microbiota in driving pulmonary allergic responses. Infect Immun. 2004;72:4996–5003. doi: 10.1128/IAI.72.9.4996-5003.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Noverr MC, Phare SM, Toews GB, Coffey MJ, Huffnagle GB. Pathogenic yeasts Cryptococcus neoformans and Candida albicans produce immunomodulatory prostaglandins. Infect Immun. 2001;69:2957–63. doi: 10.1128/IAI.69.5.2957-2963.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Erb-Downward JR, Noverr MC. Characterization of prostaglandin E2 production by Candida albicans. Infect Immun. 2007;75:3498–505. doi: 10.1128/IAI.00232-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Noverr MC, Toews GB, Huffnagle GB. Production of prostaglandins and leukotrienes by pathogenic fungi. Infection and immunity. 2002;70:400–2. doi: 10.1128/IAI.70.1.400-402.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim YG, et al. Gut Dysbiosis Promotes M2 Macrophage Polarization and Allergic Airway Inflammation via Fungi-Induced PGE2. Cell Host Microbe. 2014;15:95–102. doi: 10.1016/j.chom.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van der Velden WJ, et al. Role of the mycobiome in human acute graft-versus-host disease. Biology of blood and marrow transplantation: journal of the American Society for Blood and Marrow Transplantation. 2013;19:329–32. doi: 10.1016/j.bbmt.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 45.Ghannoum MA, et al. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog. 2010;6:e1000713. doi: 10.1371/journal.ppat.1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smeekens SP, van de Veerdonk FL, Kullberg BJ, Netea MG. Genetic susceptibility to Candida infections. EMBO molecular medicine. 2013;5:805–13. doi: 10.1002/emmm.201201678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van de Veerdonk FL, et al. STAT1 mutations in autosomal dominant chronic mucocutaneous candidiasis. The New England journal of medicine. 2011;365:54–61. doi: 10.1056/NEJMoa1100102. [DOI] [PubMed] [Google Scholar]
- 48.Takezaki S, et al. Chronic mucocutaneous candidiasis caused by a gain-of-function mutation in the STAT1 DNA-binding domain. Journal of immunology. 2012;189:1521–6. doi: 10.4049/jimmunol.1200926. [DOI] [PubMed] [Google Scholar]
- 49.Smeekens SP, et al. STAT1 hyperphosphorylation and defective IL12R/IL23R signaling underlie defective immunity in autosomal dominant chronic mucocutaneous candidiasis. PLoS One. 2011;6:e29248. doi: 10.1371/journal.pone.0029248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu L, et al. Gain-of-function human STAT1 mutations impair IL-17 immunity and underlie chronic mucocutaneous candidiasis. The Journal of experimental medicine. 2011;208:1635–48. doi: 10.1084/jem.20110958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Puel A, et al. Chronic mucocutaneous candidiasis in humans with inborn errors of interleukin-17 immunity. Science. 2011;332:65–8. doi: 10.1126/science.1200439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boisson B, et al. An ACT1 mutation selectively abolishes interleukin-17 responses in humans with chronic mucocutaneous candidiasis. Immunity. 2013;39:676–86. doi: 10.1016/j.immuni.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Puel A, et al. Autoantibodies against IL-17A, IL-17F, and IL-22 in patients with chronic mucocutaneous candidiasis and autoimmune polyendocrine syndrome type I. J Exp Med. 2010;207:291–7. doi: 10.1084/jem.20091983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Conti HR, et al. Th17 cells and IL-17 receptor signaling are essential for mucosal host defence against oral candidiasis. The Journal of experimental medicine. 2009;206:299–311. doi: 10.1084/jem.20081463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gladiator A, Wangler N, Trautwein-Weidner K, LeibundGut-Landmann S. Cutting edge: IL-17-secreting innate lymphoid cells are essential for host defence against fungal infection. Journal of immunology. 2013;190:521–5. doi: 10.4049/jimmunol.1202924. [DOI] [PubMed] [Google Scholar]
- 56.Lanternier F, et al. Deep dermatophytosis and inherited CARD9 deficiency. N Engl J Med. 2013;369:1704–14. doi: 10.1056/NEJMoa1208487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Drewniak A, et al. Invasive fungal infection and impaired neutrophil killing in human CARD9 deficiency. Blood. 2013;121:2385–92. doi: 10.1182/blood-2012-08-450551. [DOI] [PubMed] [Google Scholar]
- 58.Bishu S, et al. The adaptor CARD9 is required for adaptive but not innate immunity to oral mucosal Candida albicans infections. Infection and immunity. 2013 doi: 10.1128/IAI.01335-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hise AG, et al. An essential role for the NLRP3 inflammasome in host defence against the human fungal pathogen Candida albicans. Cell host & microbe. 2009;5:487–97. doi: 10.1016/j.chom.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ferwerda B, et al. Human dectin-1 deficiency and mucocutaneous fungal infections. N Engl J Med. 2009;361:1760–7. doi: 10.1056/NEJMoa0901053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grice EA, et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324:1190–2. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Costello EK, et al. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–7. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Findley K, et al. Topographic diversity of fungal and bacterial communities in human skin. Nature. 2013;498:367–70. doi: 10.1038/nature12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tagami H. Location-related differences in structure and function of the stratum corneum with special emphasis on those of the facial skin. Int J Cosmet Sci. 2008;30:413–34. doi: 10.1111/j.1468-2494.2008.00459.x. [DOI] [PubMed] [Google Scholar]
- 65.Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol. 2011;9:244–53. doi: 10.1038/nrmicro2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roth RR, James WD. Microbial ecology of the skin. Annu Rev Microbiol. 1988;42:441–64. doi: 10.1146/annurev.mi.42.100188.002301. [DOI] [PubMed] [Google Scholar]
- 67.Paulino LC, Tseng CH, Strober BE, Blaser MJ. Molecular analysis of fungal microbiota in samples from healthy human skin and psoriatic lesions. J Clin Microbiol. 2006;44:2933–41. doi: 10.1128/JCM.00785-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang E, et al. Characterization of the skin fungal microbiota in patients with atopic dermatitis and in healthy subjects. Microbiol Immunol. 2011;55:625–32. doi: 10.1111/j.1348-0421.2011.00364.x. [DOI] [PubMed] [Google Scholar]
- 69.Oh J, et al. The altered landscape of the human skin microbiome in patients with primary immunodeficiencies. Genome Res. 2013;23:2103–14. doi: 10.1101/gr.159467.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kagami S, Rizzo HL, Kurtz SE, Miller LS, Blauvelt A. IL-23 and IL-17A, but not IL-12 and IL-22, are required for optimal skin host defence against Candida albicans. J Immunol. 2010;185:5453–62. doi: 10.4049/jimmunol.1001153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Drell T, et al. Characterization of the vaginal micro- and mycobiome in asymptomatic reproductive-age Estonian women. PLoS One. 2013;8:e54379. doi: 10.1371/journal.pone.0054379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zheng NN, Guo XC, Lv W, Chen XX, Feng GF. Characterization of the vaginal fungal flora in pregnant diabetic women by 18S rRNA sequencing. European journal of clinical microbiology & infectious diseases: official publication of the European Society of Clinical Microbiology. 2013;32:1031–40. doi: 10.1007/s10096-013-1847-3. [DOI] [PubMed] [Google Scholar]
- 73.Guo R, et al. Increased diversity of fungal flora in the vagina of patients with recurrent vaginal candidiasis and allergic rhinitis. Microbial ecology. 2012;64:918–27. doi: 10.1007/s00248-012-0084-0. [DOI] [PubMed] [Google Scholar]
- 74.Boris S, Barbes C. Role played by lactobacilli in controlling the population of vaginal pathogens. Microbes and infection / Institut Pasteur. 2000;2:543–6. doi: 10.1016/s1286-4579(00)00313-0. [DOI] [PubMed] [Google Scholar]
- 75.Boris S, Suarez JE, Vazquez F, Barbes C. Adherence of human vaginal lactobacilli to vaginal epithelial cells and interaction with uropathogens. Infection and immunity. 1998;66:1985–9. doi: 10.1128/iai.66.5.1985-1989.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kohler GA, Assefa S, Reid G. Probiotic interference of Lactobacillus rhamnosus GR-1 and Lactobacillus reuteri RC-14 with the opportunistic fungal pathogen Candida albicans. Infectious diseases in obstetrics and gynecology. 20122012:636474. doi: 10.1155/2012/636474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lev-Sagie A, et al. Polymorphism in a gene coding for the inflammasome component NALP3 and recurrent vulvovaginal candidiasis in women with vulvar vestibulitis syndrome. American journal of obstetrics and gynecology. 2009;200:303 e1–6. doi: 10.1016/j.ajog.2008.10.039. [DOI] [PubMed] [Google Scholar]
- 78.Tomalka J, et al. A novel role for the NLRC4 inflammasome in mucosal defences against the fungal pathogen Candida albicans. PLoS pathogens. 2011;7:e1002379. doi: 10.1371/journal.ppat.1002379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wojitani MD, de Aguiar LM, Baracat EC, Linhares IM. Association between mannose-binding lectin and interleukin-1 receptor antagonist gene polymorphisms and recurrent vulvovaginal candidiasis. Archives of gynecology and obstetrics. 2012;285:149–53. doi: 10.1007/s00404-011-1920-z. [DOI] [PubMed] [Google Scholar]
- 80.Babula O, Lazdane G, Kroica J, Ledger WJ, Witkin SS. Relation between recurrent vulvovaginal candidiasis, vaginal concentrations of mannose-binding lectin, and a mannose-binding lectin gene polymorphism in Latvian women. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2003;37:733–7. doi: 10.1086/377234. [DOI] [PubMed] [Google Scholar]
- 81.Giraldo PC, et al. Mannose-binding lectin gene polymorphism, vulvovaginal candidiasis, and bacterial vaginosis. Obstetrics and gynecology. 2007;109:1123–8. doi: 10.1097/01.AOG.0000260386.17555.a5. [DOI] [PubMed] [Google Scholar]
- 82.Crosdale DJ, Poulton KV, Ollier WE, Thomson W, Denning DW. Mannose-binding lectin gene polymorphisms as a susceptibility factor for chronic necrotizing pulmonary aspergillosis. The Journal of infectious diseases. 2001;184:653–6. doi: 10.1086/322791. [DOI] [PubMed] [Google Scholar]
- 83.van Woerden HC, et al. Differences in fungi present in induced sputum samples from asthma patients and non-atopic controls: a community based case control study. BMC infectious diseases. 2013;13:69. doi: 10.1186/1471-2334-13-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pihet M, et al. Occurrence and relevance of filamentous fungi in respiratory secretions of patients with cystic fibrosis--a review. Medical mycology. 2009;47:387–97. doi: 10.1080/13693780802609604. [DOI] [PubMed] [Google Scholar]
- 85.Delhaes L, et al. The airway microbiota in cystic fibrosis: a complex fungal and bacterial community--implications for therapeutic management. PLoS One. 2012;7:e36313. doi: 10.1371/journal.pone.0036313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Agarwal R, et al. Allergic bronchopulmonary aspergillosis: review of literature and proposal of new diagnostic and classification criteria. Clinical and experimental allergy: journal of the British Society for Allergy and Clinical Immunology. 2013;43:850–73. doi: 10.1111/cea.12141. [DOI] [PubMed] [Google Scholar]
- 87.Hohl TM, et al. Aspergillus fumigatus triggers inflammatory responses by stage-specific beta-glucan display. PLoS pathogens. 2005;1:e30. doi: 10.1371/journal.ppat.0010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gersuk GM, Underhill DM, Zhu L, Marr KA. Dectin-1 and TLRs permit macrophages to distinguish between different Aspergillus fumigatus cellular states. Journal of immunology. 2006;176:3717–24. doi: 10.4049/jimmunol.176.6.3717. [DOI] [PubMed] [Google Scholar]
- 89.Carrion Sde J, et al. The RodA hydrophobin on Aspergillus fumigatus spores masks dectin-1- and dectin-2-dependent responses and enhances fungal survival in vivo. Journal of immunology. 2013;191:2581–8. doi: 10.4049/jimmunol.1300748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Faro-Trindade I, et al. Characterisation of innate fungal recognition in the lung. PLoS One. 2012;7:e35675. doi: 10.1371/journal.pone.0035675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Grimm MJ, et al. Monocyte- and macrophage-targeted NADPH oxidase mediates antifungal host defence and regulation of acute inflammation in mice. Journal of immunology. 2013;190:4175–84. doi: 10.4049/jimmunol.1202800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Grimm MJ, et al. Role of NADPH oxidase in host defence against aspergillosis. Medical mycology. 2011;49(Suppl 1):S144–9. doi: 10.3109/13693786.2010.487077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lass-Florl C, Roilides E, Loffler J, Wilflingseder D, Romani L. Minireview: host defence in invasive aspergillosis. Mycoses. 2013;56:403–13. doi: 10.1111/myc.12052. [DOI] [PubMed] [Google Scholar]
- 94.Rivera A, et al. Dectin-1 diversifies Aspergillus fumigatus-specific T cell responses by inhibiting T helper type 1 CD4 T cell differentiation. J Exp Med. 2011;208:369–81. doi: 10.1084/jem.20100906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gessner MA, et al. Dectin-1-dependent interleukin-22 contributes to early innate lung defence against Aspergillus fumigatus. Infect Immun. 2012;80:410–7. doi: 10.1128/IAI.05939-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Taylor PR, et al. Activation of neutrophils by autocrine IL-17A-IL-17RC interactions during fungal infection is regulated by IL-6, IL-23, RORgammat and dectin-2. Nature immunology. 2014;15:143–51. doi: 10.1038/ni.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lilly LM, et al. The beta-glucan receptor dectin-1 promotes lung immunopathology during fungal allergy via IL-22. Journal of immunology. 2012;189:3653–60. doi: 10.4049/jimmunol.1201797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mintz-Cole RA, et al. Dectin-1 and IL-17A suppress murine asthma induced by Aspergillus versicolor but not Cladosporium cladosporioides due to differences in beta-glucan surface exposure. Journal of immunology. 2012;189:3609–17. doi: 10.4049/jimmunol.1200589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bi L, et al. CARD9 mediates dectin-2-induced IkappaBalpha kinase ubiquitination leading to activation of NF-kappaB in response to stimulation by the hyphal form of Candida albicans. The Journal of biological chemistry. 2010;285:25969–77. doi: 10.1074/jbc.M110.131300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Saijo S, et al. Dectin-2 recognition of alpha-mannans and induction of Th17 cell differentiation is essential for host defence against Candida albicans. Immunity. 2010;32:681–91. doi: 10.1016/j.immuni.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 101.Moyes DL, et al. Candida albicans yeast and hyphae are discriminated by MAPK signaling in vaginal epithelial cells. PLoS One. 2011;6:e26580. doi: 10.1371/journal.pone.0026580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cheng SC, et al. The dectin-1/inflammasome pathway is responsible for the induction of protective T-helper 17 responses that discriminate between yeasts and hyphae of Candida albicans. Journal of leukocyte biology. 2011;90:357–66. doi: 10.1189/jlb.1210702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gantner BN, Simmons RM, Underhill DM. Dectin-1 mediates macrophage recognition of Candida albicans yeast but not filaments. The EMBO journal. 2005;24:1277–86. doi: 10.1038/sj.emboj.7600594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wheeler RT, Kombe D, Agarwala SD, Fink GR. Dynamic, morphotype-specific Candida albicans beta-glucan exposure during infection and drug treatment. PLoS pathogens. 2008;4:e1000227. doi: 10.1371/journal.ppat.1000227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pande K, Chen C, Noble SM. Passage through the mammalian gut triggers a phenotypic switch that promotes Candida albicans commensalism. Nat Genet. 2013;45:1088–91. doi: 10.1038/ng.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang Q, et al. Combined immunodeficiency associated with DOCK8 mutations. The New England journal of medicine. 2009;361:2046–55. doi: 10.1056/NEJMoa0905506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Engelhardt KR, et al. Large deletions and point mutations involving the dedicator of cytokinesis 8 (DOCK8) in the autosomal-recessive form of hyper-IgE syndrome. The Journal of allergy and clinical immunology. 2009;124:1289–302 e4. doi: 10.1016/j.jaci.2009.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Holland SM, et al. STAT3 mutations in the hyper-IgE syndrome. The New England journal of medicine. 2007;357:1608–19. doi: 10.1056/NEJMoa073687. [DOI] [PubMed] [Google Scholar]
- 109.Milner JD, et al. Impaired T(H)17 cell differentiation in subjects with autosomal dominant hyper-IgE syndrome. Nature. 2008;452:773–6. doi: 10.1038/nature06764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Freeman AF, et al. Causes of death in hyper-IgE syndrome. The Journal of allergy and clinical immunology. 2007;119:1234–40. doi: 10.1016/j.jaci.2006.12.666. [DOI] [PubMed] [Google Scholar]
- 111.Minegishi Y, et al. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature. 2007;448:1058–62. doi: 10.1038/nature06096. [DOI] [PubMed] [Google Scholar]
- 112.Plantinga TS, et al. Early stop polymorphism in human DECTIN-1 is associated with increased candida colonization in hematopoietic stem cell transplant recipients. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2009;49:724–32. doi: 10.1086/604714. [DOI] [PubMed] [Google Scholar]
- 113.Ouederni M, et al. Clinical features of Candidiasis in patients with inherited interleukin 12 receptor beta1 deficiency. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2014;58:204–13. doi: 10.1093/cid/cit722. [DOI] [PMC free article] [PubMed] [Google Scholar]