Abstract
The ISWI family of ATP-dependent chromatin remodelers regulates transcription of coding and noncoding RNA by mobilizing nucleosomes and controlling the length of linker DNA separating nucleosomes (spacing). Nucleosome movement is tightly coupled to the DNA translocation activity of the helicase domain in the catalytic subunit. There may be other domains besides the helicase domain needed to move DNA in and out of nucleosomes. The C terminus of the ISWI catalytic subunit with the conserved HAND, SANT, and SLIDE domains may be involved in nucleosome spacing. There are several models of how the C terminus may facilitate in ISWI remodeling such as regulating the activity of the helicase domain and causing the helicase domain to translocate more efficiently on DNA or to enhance its selectivity for nucleosomes. Another possibility is that domains like SLIDE promote linker DNA entering into nucleosomes in a coordinated manner with the helicase domain.
Introduction
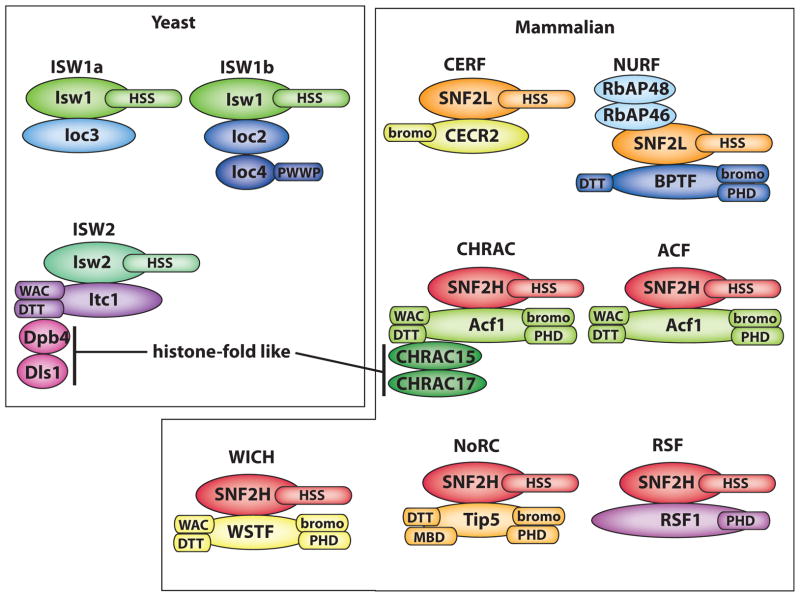
The ISWI (Imitation SWI/SNF) subfamily of ATP-dependent chromatin remodelers is one of five major chromatin remodeler subfamilies (SWI/SNF, CHD, INO80 and ATRX). Remodelers contain a helicase domain conserved among members of the Snf2 family and SF2 superfamily of ATP-dependent DNA and RNA helicases. In Saccharomyces cerevisiae (Isw1 and Isw2) and humans (SNF2H and SNF2L) there are two versions of the ISWI catalytic subunit assembled with one to three accessory subunits to form 3–7 different complexes (Figure 1). Isw1 forms two complexes with distinct auxiliary subunits referred to as ISW1a with subunit Ioc3 and ISW1b with subunits Ioc2 and Ioc4 [1]. Isw2 forms primarily one complex with the accessory subunits Itc1, Dpb4 and Dls1; and is referred to as ISW2 [2]. Dpb4 and Dls1 are two histone-fold like subunits and similar histone-fold like subunits are found in ISWI complexes in Drosophila (CHRAC) and humans (hCHRAC) [3–5]. The ISWI remodelers are involved in transcription regulation of RNA polymerase I, II, and III; and DNA replication and repair[6]. ISW2 binds near promoter regions and moves nucleosomes close to nucleosome free regions (NFR) to repress RNA pol II transcription [7–9]. ISW1a and ISW1b are recruited to the gene body regions and in particular ISW1b is recruited by trimethylation of lysine 36 of histone H3 (H3K36me3) [10–12]. ISW1b promotes the reassembly of chromatin over the coding regions after transcription, reducing cryptic transcription from these sites. ISW1b also has a role in proper termination of RNA Pol II and ISW1a in regulating the early phase of RNA Pol II elongation [13,14]. ISW2 along with the INO80 promotes the progression of DNA replication forks and attenuates S phase checkpoint activity [15,16]. Some of the ISWI complexes assemble nucleosomes onto DNA and can be involved in heterochromatin formation. ACF from Drosophila with its catalytic subunit ISWI and accessory subunit Acf1 was shown to assemble and space nucleosomes in an ATP-dependent manner requiring the histone chaperone Nap1 [17–19]. The complex of SNF2H and RSF-1 assembles nucleosomes without a histone chaperone and facilitates in the formation of centromeric chromatin containing the H3 variant CENP-A [20–22]. A nucleolar form of ISWI called NoRC also appears to be required for formation of heterochromatin over rRNA genes [23,24].
Figure 1. ISWI family of chromatin remodelers in Saccharomyces cerevisiae and mammals.
The subunits are represented as ellipses and conserved protein motifs as rectangles. HSS is the HAND, SANT and SLIDE domains. Some of the conserved proteins domains bind to modified sites in histones such as the bromo, PHD and PWWP domain; and others are involved in subunit-subunit interactions like the WAC and DTT domains.
Model for the interactions of ISW2 with nucleosomes
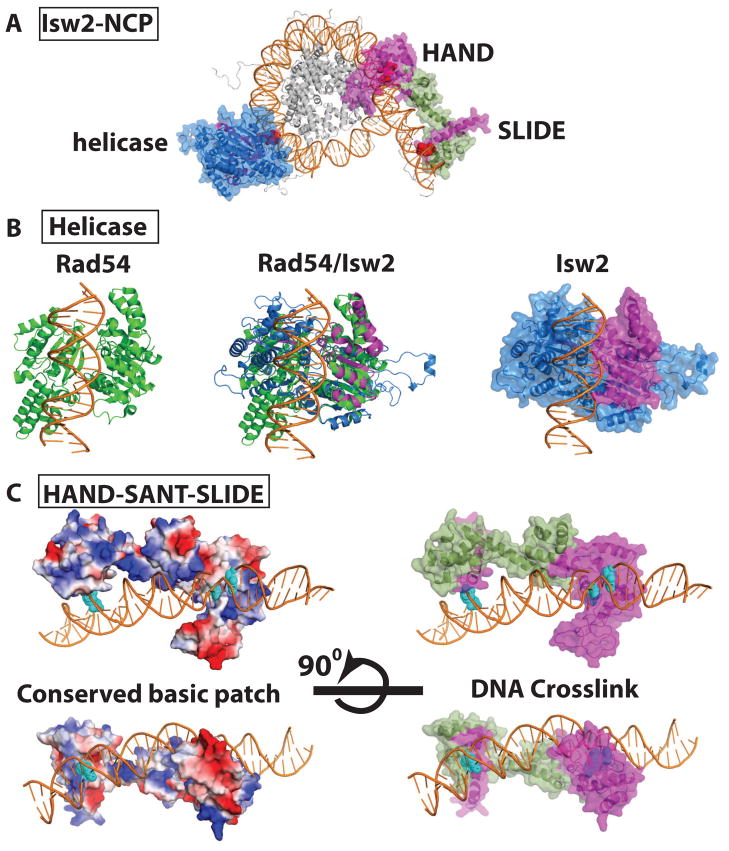
The ISWI subfamily requires a minimal length of linker DNA for mobilizing nucleosomes, a property not shared by the SWI/SNF subfamily. At a mononucleosome level, ISW2 binds to at least 50–60 bp of extranucleosomal DNA shown by DNA footprinting [25]. ISW2 affinity for nucleosomes and its efficiency for mobilizing nucleosomes depend on the length of extranucleosomal DNA. More than 20–23 bp of extranucleosomal DNA is minimally required by ISW2 to mobilize nucleosomes when fully bound [26]. The accessory subunit Itc1 binds 53 bp of linker DNA spanning from the entry site of the nucleosome. In the catalytic subunit Isw2, the SLIDE domain binds DNA 19 bp from the entry site and the HAND domain ~11–13 bp from the entry site inside of nucleosomes [27]. Itc1 significantly contributes to the nucleosome affinity of ISW2 and Isw2 alone binds to nucleosomes with a 5 times lower affinity than native ISW2 [28]. DNA footprinting and site-directed crosslinking showed that the helicase domain of Isw2 was bound at Superhelical Location 2 (SHL2), 17 and 18 bp from the dyad axis. Structures of the helicase domain and C-terminus of Isw2 have not been determined, but models of these have been constructed based on their sequence homology with Rad54 [29,30] and Drosophila ISWI [31] whose structures have been obtained. DNA crosslinking and peptide mapping data were used to dock the structural models of the helicase domain and C-terminus of Isw2 to their nucleosome binding sites. Site-specific DNA crosslinking and structural models together pinpoint the most probable surfaces of the helicase, HAND and SLIDE domains which are in contact with DNA 17–18, 60–62, and 92 bp from the dyad axis, respectively (Figure 2a). Since the direction of translocation relative to the helicase structure is known for some helicases [32], the orientation of the helicase domain on nucleosomes was refined further by correlating it to the direction which ISW2 moves nucleosomal DNA (Figure 2b). One parameter that helps in proper docking of the C-terminus is the requirement of the distance between the HAND and SLIDE domains being consistent with the distance between the DNA sites shown to be crosslinked in nucleosomes. Another parameter that aids in the alignment of the C-terminus is the observation of an extended patch of conserved basic residues on its surface that could bind DNA which aligns well with the binding surface predicted from DNA crosslinking ([31] and Figure 2c). These data together create a model from which structural and functional predictions can be made and tested further.
Figure 2. Interactions of Isw2 with nucleosomes.
(A) Three DNA sites probed by crosslinking are in red and the regions of Isw2 crosslinked to them in magenta. The helicase, HAND and SLIDE domains are labeled. The region in the helicase domain crosslinked to DNA cannot be seen in this orientation. (B) The crystal structure of the Sulfulobus Rad54 helicase domain (green) with DNA (orange) bound is shown on the left. In the middle is the overlay of the model of Isw2 with the known crystal structure of Rad54. On the right is Isw2 (blue) with DNA bound similar to that shown for Rad54 and the region crosslinked to DNA in magenta. (C) On the left is shown the predicted electrostatic surface potential for the HAND-SANT-SLIDE domains and on the right the regions shown to crosslink DNA in magenta. These structures are rotated by 90° to more fully visualize the conserved basic patch in blue.
One aspect examined of this model is the manner in which the helicase domain functions deep inside of nucleosomes. Single nucleotide gaps in DNA at SHL2 interfered with nucleosomal movement through blocking DNA translocation or binding of the helicase domain of ISW2, and similar effects were seen with NURF and SWI/SNF [33,34]. DNA torsional strain in a 10 bp region outside of the helicase bound region appears to be important for ISW2 remodeling with nicks and gaps interfering with remodeling at these positions. Gaps and nicks at these positions are probably releasing DNA twist caused by translocation of the helicase domain since they are outside of the helicase binding site and nicks do not block DNA translocation. Gaps and nicks interfered only on the side of the helicase domain where DNA is pulled into nucleosomes. Torsional strain on the indicated side of the bound helicase domain was required to pull DNA inside nucleosomes as shown by single molecule Fluorescence Resonant Energy Transfer (smFRET) [35]. Initially ~7 bp of DNA exited out of nucleosomes in one base-pair increments before changes occurred at the entry of DNA into nucleosome (Figure 3). After this initial movement, ~3 bp of DNA is required to exit before any additional DNA can enter into nucleosomes and account for the observed sequential kinetic steps of ~7, ~3, and ~3 bp with ISW2. DNA torsional strain between the bound helicase domain and the site where linker DNA enters is required for pulling DNA into nucleosomes.
Figure 3. Nucleosome dynamics and ISW2.
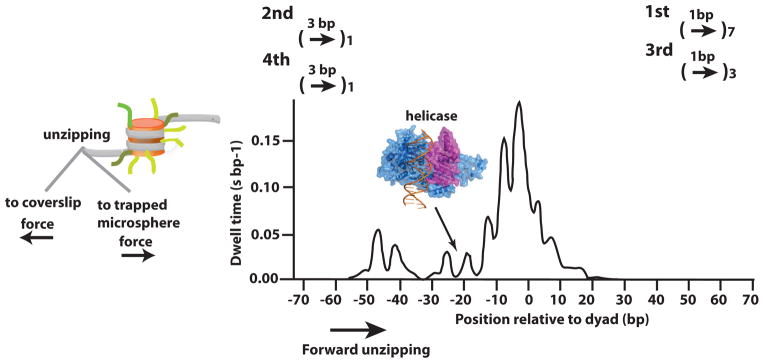
Nucleosome energetics were measured using the single molecule approach shown on the left in which the two ends of DNA are fixed and then progressively unzipped. A typical profile of these experiments is shown on the right and the position where the helicase domain is bound. Above the plot is shown the progressive movement of DNA in (left) and out (right) of nucleosomes by ISW2 using smFRET.
Nucleosome dynamics and ISWI
This uneven movement of DNA through nucleosomes is somewhat unexpected based on nucleosome energetics. There are 121 interactions between histones and DNA in the nucleosome that are mediated through hydrogen bonding with water molecules as well as 116 hydrogen bonds made directly between histones and DNA both of which should equally contribute to the stability of nucleosomes [36,37]. Many of the interactions are with the DNA phosphate backbone and there are 20 critical side-chain interactions that penetrate into the minor groove of DNA, most often mediated by the side-chains of arginine. All histone-DNA contacts are not energetically equivalent as shown by single molecule/optical trap type experiments in which DNA is unzipped from one end. DNA was reversibly peeled off nucleosomes by unzipping DNA and nucleosomes not unwrapped past the dyad axis could be rewrapped once the tension was released [38]. Refinements of this approach showed the energetic profile of DNA peeling off nucleosomes with nearly base pair resolution and found three regions having the strongest energetic barriers for pulling DNA from the histone octamer [39]. The strongest histone octamer binding site to DNA was comprised of contacts from the H3–H4 tetramer spanning 10–15 bp in either direction from the dyad axis (Figure 3). The two next strongest binding sites are ~40–50 bp from the dyad axis with the H2A-H2B dimers. They found a distinct ~5 bp periodicity of contacts, which suggests sequential breaking of a DNA contact first from one strand followed by that of the opposite strand.
The helicase domain binds a region in nucleosomes where energetically it has more pliable histone-DNA contacts due to a low energy valley and is bordered on either side by stronger histone-DNA contacts (Figure 3). Based on energetic constraints, the accumulation of torsional strain on only one side of the helicase domain is not likely to be due to the histone contacts being stronger on that side compared to the other thus requiring extra force to disrupt histone-DNA contacts. It might be that the interactions of the remodeler near the entry site are the “brake” that causes the torsional strain to be required before releasing the DNA into nucleosomes and could be mediated by the C-terminus of ISWI.
Role of the SANT, SLIDE, and HAND domains
The binding of the C-terminus of Isw2 to linker DNA and the entry side of nucleosomes could be important for proper recruitment of the complex to nucleosomes as well as for controlling remodeling. The SLIDE domain from Drosophila ISWI has high affinity for DNA[31] and, based on DNA crosslinking, the HAND and SLIDE domains of ISW2 also bind DNA when ISW2 docks onto nucleosomes [27]. The role of the C-terminus of the catalytic subunit of ISWI has been examined with the catalytic subunit alone and truncation of the entire C-terminus or in the native complex by mutagenesis of the individual domains. In one study loss of the C-terminus of the catalytic subunit of Drosophila ISWI had no effect on the ATPase activity of the helicase domain with free DNA substrate, with or without the addition of H4 peptide, or with nucleosomes [40]. If nucleosomes were subsaturating an order of magnitude difference was observed between full length and truncated ISWI, suggesting the C-terminus increased the selectivity or affinity of the helicase domain. In a different study of the same protein, the C-terminus had the role of stimulating DNA translocation of the helicase domain [41]. Importantly these studies come to two different conclusions of the role of the C-terminus of the ISWI catalytic subunit; namely as a selectivity factor or regulation of helicase activity. In terms of selectivity this might be easiest to explain by the C-terminus of ISWI recruiting the complex to linker DNA and thereby to the core nucleosome.
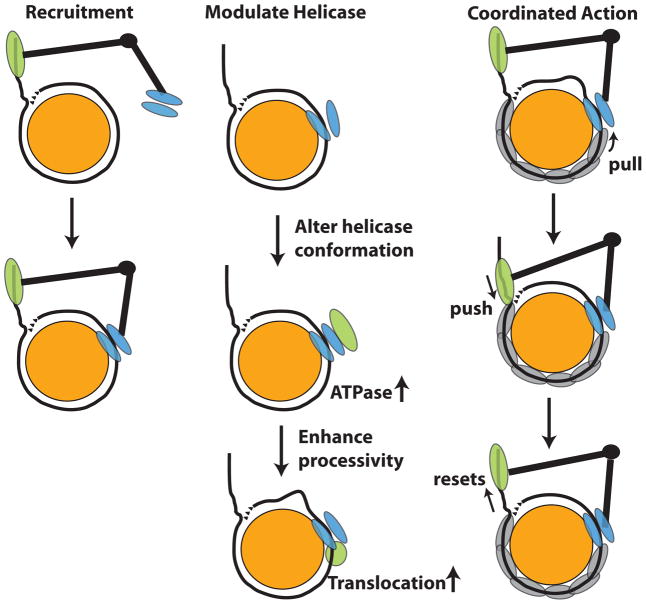
A potential problem with these studies is the activity of the free catalytic subunit being less than the native complex and the accessory subunit(s) being important for the overall activity of the enzyme. Deletion of the entire C-terminus can also have a combination of effects on the ATPase and remodeling activities, and doesn’t address well the separate activities of the HAND, SANT and SLIDE domains. A small subset of the conserved residues was changed to alanine in the SLIDE domain that was predicted to bind DNA based on the model in Figure 2. These changes reduced ISW2 binding ~19 bp from the edge of the nucleosome without disturbing the other interactions of ISW2 with nucleosomal or extranucleosomal DNA [28]. These mutations in SLIDE severely affected the ATPase and nucleosome mobilizing activities of ISW2 under nucleosome saturating conditions and pointed to recruitment not being its sole function. In the absence of SLIDE domain binding DNA, the helicase domain moved only ~10 bp of DNA out of nucleosomes before stalling due to the lack of DNA coming in. The Isw2 subunit alone without the SLIDE domain is unable to remodel nucleosomes even though it still binds nucleosomes [28] and thus has an important role in mobilizing nucleosomes in coordination with the helicase domain (Figure 4).
Figure 4. Roles for the HAND-SANT-SLIDE domains in ISWI remodeling.
The three ways that these domains might be involved in mobilizing nucleosomes are recruitment, modulating the helicase domain activity, and coordinating the movement of DNA in nucleosomes. The helicase domain is represented in blue, histone octamer in orange, and HSS or SLIDE domains in green.
Nucleosome movement: Coordinated action versus solely the helicase domain
ISWI remodeling has been suggested to be dependent exclusively on the helicase domain and other parts of the complex having supporting roles such as facilitating in nucleosome recruitment and regulating the helicase domain activities ([40] and Figure 4). Other suggestions are that domains like the SLIDE domain can have additional roles in remodeling and work in a coordinated manner with the helicase domain to mobilize nucleosomes [28,42]. The interactions of ISW2 with linker DNA promote the stable binding of the helicase domain to DNA at SHL2 as shown in DNA footprinting experiments, supporting a role in recruitment [43], and consequently can regulate the kinetic activity of the helicase domain [44]. There is good evidence that other domains can also be involved in regulating the ATPase and DNA translocation properties of the helicase domain [41]. It is also possible for domains such as those in the C-terminus to facilitate the passage of DNA into nucleosomes and regulate its flow through the nucleosome.
Highlights.
The C terminus of the catalytic subunit of ISWI complexes is evolutionarily conserved and required for efficient nucleosome mobilization
The SANT, SLIDE and HAND domains are in the C-terminus of ISWI
The SLIDE domain binds to linker DNA in the ISW2 complex and facilitates linker DNA movement into nucleosomes
The C-terminus of ISWI may also be required for helicase domain selectivity and efficient DNA translocation.
DNA twist between the helicase domain and the entry site of nucleosomes is required for nucleosome movement
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Vary JC, Jr, Gangaraju VK, Qin J, Landel CC, Kooperberg C, Bartholomew B, Tsukiyama T. Yeast Isw1p forms two separable complexes in vivo. Mol Cell Biol. 2003;23:80–91. doi: 10.1128/MCB.23.1.80-91.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsukiyama T, Palmer J, Landel CC, Shiloach J, Wu C. Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev. 1999;13:686–697. doi: 10.1101/gad.13.6.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McConnell AD, Gelbart ME, Tsukiyama T. Histone fold protein Dls1p is required for Isw2-dependent chromatin remodeling in vivo. Mol Cell Biol. 2004;24:2605–2613. doi: 10.1128/MCB.24.7.2605-2613.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corona DF, Eberharter A, Budde A, Deuring R, Ferrari S, Varga-Weisz P, Wilm M, Tamkun J, Becker PB. Two histone fold proteins, CHRAC-14 and CHRAC-16, are developmentally regulated subunits of chromatin accessibility complex (CHRAC) EMBO J. 2000;19:3049–3059. doi: 10.1093/emboj/19.12.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poot RA, Dellaire G, Hulsmann BB, Grimaldi MA, Corona DF, Becker PB, Bickmore WA, Varga-Weisz PD. HuCHRAC, a human ISWI chromatin remodelling complex contains hACF1 and two novel histone-fold proteins. EMBO J. 2000;19:3377–3387. doi: 10.1093/emboj/19.13.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Erdel F, Rippe K. Chromatin remodelling in mammalian cells by ISWI-type complexes--where, when and why? FEBS J. 2011;278:3608–3618. doi: 10.1111/j.1742-4658.2011.08282.x. [DOI] [PubMed] [Google Scholar]
- 7.Zentner GE, Tsukiyama T, Henikoff S. ISWI and CHD chromatin remodelers bind promoters but act in gene bodies. PLoS Genet. 2013;9:e1003317. doi: 10.1371/journal.pgen.1003317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yen K, Vinayachandran V, Batta K, Koerber RT, Pugh BF. Genome-wide nucleosome specificity and directionality of chromatin remodelers. Cell. 2012;149:1461–1473. doi: 10.1016/j.cell.2012.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whitehouse I, Rando OJ, Delrow J, Tsukiyama T. Chromatin remodelling at promoters suppresses antisense transcription. Nature. 2007;450:1031–1035. doi: 10.1038/nature06391. [DOI] [PubMed] [Google Scholar]
- 10.Maltby VE, Martin BJ, Schulze JM, Johnson I, Hentrich T, Sharma A, Kobor MS, Howe L. Histone H3 lysine 36 methylation targets the Isw1b remodeling complex to chromatin. Mol Cell Biol. 2012;32:3479–3485. doi: 10.1128/MCB.00389-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smolle M, Venkatesh S, Gogol MM, Li H, Zhang Y, Florens L, Washburn MP, Workman JL. Chromatin remodelers Isw1 and Chd1 maintain chromatin structure during transcription by preventing histone exchange. Nat Struct Mol Biol. 2012;19:884–892. doi: 10.1038/nsmb.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tirosh I, Sigal N, Barkai N. Widespread remodeling of mid-coding sequence nucleosomes by Isw1. Genome Biol. 2010;11:R49. doi: 10.1186/gb-2010-11-5-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morillon A, Karabetsou N, O’Sullivan J, Kent N, Proudfoot N, Mellor J. Isw1 chromatin remodeling ATPase coordinates transcription elongation and termination by RNA polymerase II. Cell. 2003;115:425–435. doi: 10.1016/s0092-8674(03)00880-8. [DOI] [PubMed] [Google Scholar]
- 14.Morillon A, Karabetsou N, Nair A, Mellor J. Dynamic lysine methylation on histone H3 defines the regulatory phase of gene transcription. Mol Cell. 2005;18:723–734. doi: 10.1016/j.molcel.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 15.Au TJ, Rodriguez J, Vincent JA, Tsukiyama T. ATP-dependent chromatin remodeling factors tune S phase checkpoint activity. Mol Cell Biol. 2011;31:4454–4463. doi: 10.1128/MCB.05931-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vincent JA, Kwong TJ, Tsukiyama T. ATP-dependent chromatin remodeling shapes the DNA replication landscape. Nat Struct Mol Biol. 2008;15:477–484. doi: 10.1038/nsmb.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fyodorov DV, Kadonaga JT. Dynamics of ATP-dependent chromatin assembly by ACF. Nature. 2002;418:897–900. doi: 10.1038/nature00929. [DOI] [PubMed] [Google Scholar]
- 18.Fyodorov DV, Blower MD, Karpen GH, Kadonaga JT. Acf1 confers unique activities to ACF/CHRAC and promotes the formation rather than disruption of chromatin in vivo. Genes Dev. 2004;18:170–183. doi: 10.1101/gad.1139604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Torigoe SE, Urwin DL, Ishii H, Smith DE, Kadonaga JT. Identification of a rapidly formed nonnucleosomal histone-DNA intermediate that is converted into chromatin by ACF. Mol Cell. 2011;43:638–648. doi: 10.1016/j.molcel.2011.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loyola A, Huang JY, LeRoy G, Hu S, Wang YH, Donnelly RJ, Lane WS, Lee SC, Reinberg D. Functional analysis of the subunits of the chromatin assembly factor RSF. Mol Cell Biol. 2003;23:6759–6768. doi: 10.1128/MCB.23.19.6759-6768.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loyola A, LeRoy G, Wang YH, Reinberg D. Reconstitution of recombinant chromatin establishes a requirement for histone-tail modifications during chromatin assembly and transcription. Genes Dev. 2001;15:2837–2851. doi: 10.1101/gad.937401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perpelescu M, Nozaki N, Obuse C, Yang H, Yoda K. Active establishment of centromeric CENP-A chromatin by RSF complex. J Cell Biol. 2009;185:397–407. doi: 10.1083/jcb.200903088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guetg C, Santoro R. Formation of nuclear heterochromatin: the nucleolar point of view. Epigenetics. 2012;7:811–814. doi: 10.4161/epi.21072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guetg C, Lienemann P, Sirri V, Grummt I, Hernandez-Verdun D, Hottiger MO, Fussenegger M, Santoro R. The NoRC complex mediates the heterochromatin formation and stability of silent rRNA genes and centromeric repeats. EMBO J. 2010;29:2135–2146. doi: 10.1038/emboj.2010.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kagalwala MN, Glaus BJ, Dang W, Zofall M, Bartholomew B. Topography of the ISW2-nucleosome complex: insights into nucleosome spacing and chromatin remodeling. EMBO J. 2004;23:2092–2104. doi: 10.1038/sj.emboj.7600220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zofall M, Persinger J, Bartholomew B. Functional role of extranucleosomal DNA and the entry site of the nucleosome in chromatin remodeling by ISW2. Mol Cell Biol. 2004;24:10047–10057. doi: 10.1128/MCB.24.22.10047-10057.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dang W, Bartholomew B. Domain architecture of the catalytic subunit in the ISW2-nucleosome complex. Mol Cell Biol. 2007;27:8306–8317. doi: 10.1128/MCB.01351-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hota SK, Bhardwaj SK, Deindl S, Lin YC, Zhuang X, Bartholomew B. Nucleosome mobilization by ISW2 requires the concerted action of the ATPase and SLIDE domains. Nat Struct Mol Biol. 2013;20:222–229. doi: 10.1038/nsmb.2486. Mutations in the SLIDE domain of ISW2 show that SLIDE is important for pushing DNA into Nucleosomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thoma NH, Czyzewski BK, Alexeev AA, Mazin AV, Kowalczykowski SC, Pavletich NP. Structure of the SWI2/SNF2 chromatin-remodeling domain of eukaryotic Rad54. Nat Struct Mol Biol. 2005;12:350–356. doi: 10.1038/nsmb919. [DOI] [PubMed] [Google Scholar]
- 30.Durr H, Korner C, Muller M, Hickmann V, Hopfner KP. X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA. Cell. 2005;121:363–373. doi: 10.1016/j.cell.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 31.Grune T, Brzeski J, Eberharter A, Clapier CR, Corona DF, Becker PB, Muller CW. Crystal structure and functional analysis of a nucleosome recognition module of the remodeling factor ISWI. Mol Cell. 2003;12:449–460. doi: 10.1016/s1097-2765(03)00273-9. [DOI] [PubMed] [Google Scholar]
- 32.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 33.Schwanbeck R, Xiao H, Wu C. Spatial contacts and nucleosome step movements induced by the NURF chromatin remodeling complex. J Biol Chem. 2004;279:39933–39941. doi: 10.1074/jbc.M406060200. [DOI] [PubMed] [Google Scholar]
- 34.Zofall M, Persinger J, Kassabov SR, Bartholomew B. Chromatin remodeling by ISW2 and SWI/SNF requires DNA translocation inside the nucleosome. Nat Struct Mol Biol. 2006;13:339–346. doi: 10.1038/nsmb1071. [DOI] [PubMed] [Google Scholar]
- 35.Deindl S, Hwang WL, Hota SK, Blosser TR, Prasad P, Bartholomew B, Zhuang X. ISWI remodelers slide nucleosomes with coordinated multi-base-pair entry steps and single-base-pair exit steps. Cell. 2013;152:442–452. doi: 10.1016/j.cell.2012.12.040. A single molecule study using FRET to examine the dynamics of DNA movement in and out of nucleosomes as mediated by ISW2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1. 9 a resolution. J Mol Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 37.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2. 8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 38.Brower-Toland BD, Smith CL, Yeh RC, Lis JT, Peterson CL, Wang MD. Mechanical disruption of individual nucleosomes reveals a reversible multistage release of DNA. Proc Natl Acad Sci U S A. 2002;99:1960–1965. doi: 10.1073/pnas.022638399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hall MA, Shundrovsky A, Bai L, Fulbright RM, Lis JT, Wang MD. High-resolution dynamic mapping of histone-DNA interactions in a nucleosome. Nat Struct Mol Biol. 2009;16:124–129. doi: 10.1038/nsmb.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mueller-Planitz F, Klinker H, Ludwigsen J, Becker PB. The ATPase domain of ISWI is an autonomous nucleosome remodeling machine. Nat Struct Mol Biol. 2013;20:82–89. doi: 10.1038/nsmb.2457. Focuses on the loss of the C-terminus of ISWI and its effects on the ATPase and remodeling activities of ISWI. [DOI] [PubMed] [Google Scholar]
- 41.Clapier CR, Cairns BR. Regulation of ISWI involves inhibitory modules antagonized by nucleosomal epitopes. Nature. 2012;492:280–284. doi: 10.1038/nature11625. Identifies two regulatory domains in ISWI called AutoN amd NegC that regulate the activity of the helicase domain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gangaraju VK, Prasad P, Srour A, Kagalwala MN, Bartholomew B. Conformational changes associated with template commitment in ATP-dependent chromatin remodeling by ISW2. Mol Cell. 2009;35:58–69. doi: 10.1016/j.molcel.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dang W, Kagalwala MN, Bartholomew B. Regulation of ISW2 by concerted action of histone H4 tail and extranucleosomal DNA. Mol Cell Biol. 2006;26:7388–7396. doi: 10.1128/MCB.01159-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang JG, Madrid TS, Sevastopoulos E, Narlikar GJ. The chromatin-remodeling enzyme ACF is an ATP-dependent DNA length sensor that regulates nucleosome spacing. Nat Struct Mol Biol. 2006;13:1078–1083. doi: 10.1038/nsmb1170. [DOI] [PubMed] [Google Scholar]