Abstract
The Library of Integrated Network-based Cellular Signatures (LINCS) L1000 big data provide gene expression profiles induced by over 10,000 compounds, shRNAs, and kinase inhibitors using the L1000 platform. We developed csNMF, a systematic compound signature discovery pipeline covering from raw L1000 data processing to drug screening and mechanism generation. The csNMF pipeline demonstrated better performance than the original L1000 pipeline. The discovered compound signatures of breast cancer were consistent with the LINCS KINOMEscan data and were clinically relevant. The csNMF pipeline provided a novel and complete tool to expedite signature-based drug discovery leveraging the LINCS L1000 resources.
1 Introduction
Compound profiling, defined as the large-scale screening of candidate compounds for their potential drug-like qualities and toxicity using high-throughput technologies, is the fundamental step of drug discovery [1]. Traditional compound profiling approaches evaluate the pharmacological potential of compounds by measuring their affinities to target enzymes or proteins, inhibitory effects on enzyme activities, or suppressive effects on cancer cell growth [2, 3]. However, compounds that show strong affinity and inhibitory effects on expected targets often also affect the activities or functions of other proteins in a cell-specific way. Lacking the systematic and unbiased profiling of the compound effects at molecular level, candidate drugs suggested by such compound profiling strategies often suffer from a high failure rate in clinical trials [4]. On one hand, such drug targets besides the expected or designed ones are often responsible for the high toxicity to vital organs, a leading cause of clinical trial failures [5]. On the other hand, the unrecognized drug targets sometimes significantly contribute to the success of drugs. For example, compounds that show similar effectiveness against their designed targets in vitro at molecular levels often show dramatically different efficacy at the cellular or patient levels [6]. However, the roles of such “lurking” drug targets of successful drugs under the cellular or in vivo contexts are rarely well known or used for compound profiling. Furthermore, the cell-specific efficacy of different compounds underscores the importance of cell-specific regulatory networks in drug responses, that is, the roles and importance of the unknown drug targets are highly disease-and-cell-type-specific and thus require specific analysis strategies. Thus, there is a critical need in compound profiling and drug discovery to thoroughly examine the impacts of drugs or compounds on cellular functions using a wide panel of essential proteins.
To address the challenges of drug screening coverage, the Library of Integrated Network-Based Cellular Signatures (LINCS) program (http://www.lincsproject.org/) has initiated an effort to generate biomedical big data. LINCS has been systematically exploring the pharmacological roles of more than 3,700 potential drug targets on 15 cancer cell lines at the individual-gene level. Using single-gene knockdown or over-expression of each relevant gene then allows measurement of changes of gene expression patterns. LINCS also contains data on more than 5,000 chemicals at the cellular level, including known drugs and candidate compounds, documented treatment-induced alterations of gene expression on these cell lines. The LINCS program has also performed auxiliary high-throughput assays such as the kinome-wide screening of drug kinase inhibition effects using KINOMEscan® or KiNativTM scan. This is the first time that the targeted proteins by drugs and compounds have been systematically analyzed in the contexts of different cancer cell types in such scope. With LINCS as a reference library, compound profiling can be performed on the panel of more than 3,000 potential drug targets.
Compound profiling using LINCS big data as the reference library is made possible by the first large-scale application of the L1000 platform [7]. As a novel genome-wide gene expression assay platform, the L1000 is highly cost-efficient and robotically automated. It allows the generation of 946,944 profiles of gene expression data testing 5,178 drugs and compounds and perturbations of 3,712 genes across 15 different cancer cell types (http://lincscloud.org/). The LINCS L1000 big data is growing quickly in examined drugs, compounds, genes, dosing, time points, combinations of treatment conditions, and cell lines.
Accompanying such a great opportunity are the new challenges of processing and analyzing data generated from the L1000 platform. The economical usage of the same type of Luminex FLEXMAP 3D® beads [8] by two types of mRNA probes requires a reliable deconvolution approach. Furthermore, biases introduced by batch effects need subtle normalization and quality control methods.
In this work, we present a “compound signature” based approach to profiling the pharmacological potential of compounds by associating these candidates with known drugs in terms of the similarity of their possible targets, using the latest LINCS L1000 data for breast cancer (MCF-7) cell lines. We defined a “compound signature” as a group of small molecule compounds sharing similar target genes. As a member of the LINCS project, we developed a parallel data processing pipeline, the fuzzy c-means guided Gaussian mixture model (GMM), to address the L1000 data processing challenges with superior accuracy and efficiency. We then developed two compound signature discovery approaches using data produced by the GMM pipeline. The first one was the Enrichment of Gene Effects to a Molecule (EGEM) score, which associated a compound with its potential targets. The second approach was the constrained sparse non-negative matrix factorization (csNMF) approach, which used the EGEM scores of drugs, compounds, and genes to reliably detect the compound signatures and associate candidate compounds with known drugs by the shared compound signatures. The LINCS kinomics data for kinome-wide drug inhibitory effects were used to validate discovered signatures. Functional analysis and known mechanisms of the detected signatures further supported the results of compound signature detection. The third approach was quadruple model training, which correlated a drug with its targets, the affected downstream transcription factors, and the transcriptional alterations.
2 Material and Methods
2.1 Datasets
In this paper, we combined the small-molecule compound and shRNA data released from the Broad Institute LINCS Data Generation Center (http://api.lincscloud.org/). Two compound-induced L1000 gene expression datasets were adopted, which included data for treatment effects of 728 and 51 compounds on the MCF-7 breast cancer cell line, respectively. The KINOMEscan® data measured the interactions of compounds and more than 450 kinase assays and disease-relevant mutant variants. Expression patterns after the single-gene knockdown of 3,341 biologically important genes by shRNA treatments were measured on the same cell line. Compounds in the latter dataset were all kinase inhibitors. Thus, we included the auxiliary KINOMEscan® data of these 51 kinase inhibitors released from the Harvard Medical School LINCS Data Generation Center (http://lincs.hms.harvard.edu/db/). This dataset was used to validate the discoveries of compound signatures.
2.2 Work flow
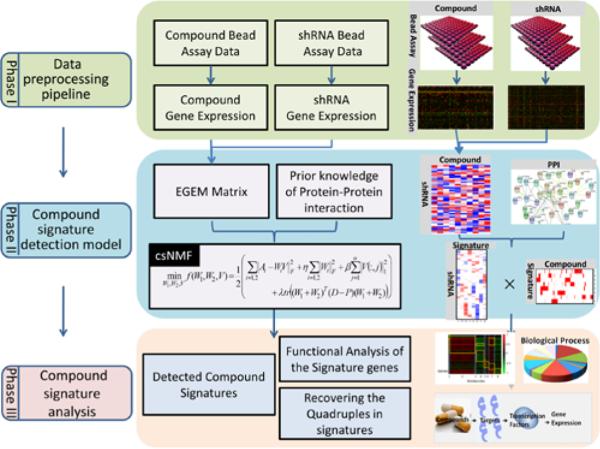
The overall framework of the compound signature discovery pipeline (Figure 1) is composed of three phases:
Fig. 1.
Overview of the compound signature discovery framework. This method requires raw L1000 data after various compounds and gene knockdown treatments. The raw data after the two types of treatments are preprocessed to yield gene expression data in Phase I. In Phase II, the EGEM matrix is constructed based on these gene expression data to measure relationships among compounds and knock-down genes. This matrix is then decomposed to a weight matrix and a coefficient matrix by the csNMF method. Protein-protein interaction data are added in consideration of biological connections. Signatures are identified based on strongly associated genes (i.e., those with larger values in the coefficient matrix).
Phase I
Raw L1000 data processing using the GMM pipeline. At this phase, the L1000 raw data were processed, normalized, cleaned for quality control, and annotated. The GMM pipeline demonstrated better accuracy and efficiency compared to another tool using the k-means method (http://lincscloud.org/exploring-the-data/code-api/, date: 2012/06/27).
Phase II
Compound signature detection using the EGEM-based csNMF model. In this phase, the EGEM method was used to measure the EGEM score for each of the 3,341 perturbed genes, which described the potential of the gene of interest to be the “target” of a small-molecule compound. The targeting potentials of such compound-gene pairs were represented by an EGEM matrix (Figure 1). Then the novel constrained sparse non-negative matrix factorization (csNMF) algorithm was developed and performed on the EGEM matrix to identify compounds of similar targets. Each such compound subgroup is defined as a compound csNMF signature, shares similar targets, and may show similar pharmaceutical potential.
Phase III
csNMF signature analysis and annotation using the Quadruple Model. Since the L1000 gene expression patterns reflect drug effects at the mRNA level, while most drugs directly or indirectly affect protein activities and functions, there is a gap between the actual drug targets at the protein level and the measured drug-induced alterations of gene expressions. We developed the Quadruple Model to reveal how compounds in each csNMF signature, through perturbing the functions of the identified drug targets, altered the downstream transcription factors and caused the differential changes of the apparent gene expression patterns. Quadruple models, composed of the compound – target – transcription factor – gene expression components, provided a novel means to reveal the underlying biological mechanisms shared by similar compounds in each csNMF signature and therefore to systematically annotate csNMF signatures at multiple regulatory levels.
2.3 Phase I. Raw data pre-processing pipeline
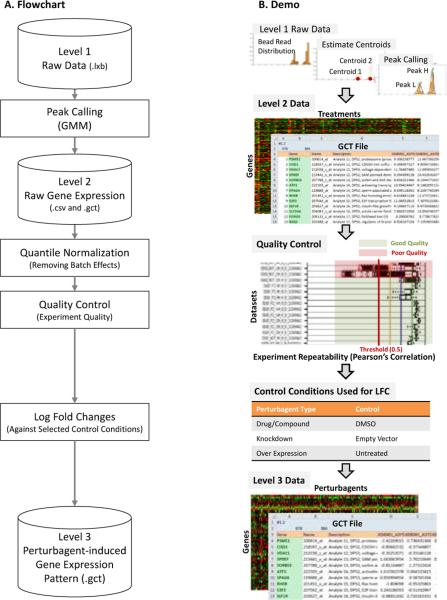
The goal in Phase I was to reliably process, normalize, clean, and annotate the L1000 raw data. The major challenges in this phase were reliable peak calling, normalization and quality control, and the computational burdens for processing big raw data. The GMM pipeline (Figure 2) was developed to address these challenges. The Level 1 raw data in Luminex bead array (LXB) format (untreated controls, the compound, and single-gene knocked down samples) were input into the GMM pipeline following the FCS v3.0 standard [9]. The raw data for each sample were deconvolved and the fluorescent intensity peak corresponding to each mRNA probe was identified using the GMM model, annotated with gene symbol, probe ID, gene description, and the analyte and L1000 probe set information. This information was then outputted in the GCT format, defined as the Level 2 raw gene expression data. After normalization and quality control, each set of perturbation-induced data was compared with its negative control. Differential gene expression (DEG) patterns, in the form of log fold changes (LFCs), were outputted as the Level 3 perturbagen-induced gene expression pattern data in the GCT format.
Fig. 2.
Overview of the data pre-processing framework. The raw Luminex data are transformed to gene expression data by the GMM peak calling
A GMM peak calling approach was developed for reliable peak calling from raw L1000 data [Level 1 to Level 2]. The L1000 approach took advantage of the state-of-art Luminex-bead based flow cytometry multiplex detection technology [10]. Briefly, DNA probes targeting a specific mRNA were immobilized on a distinct type of analyte (Luminex beads filled with a distinct dye). Each type of analyte was composed of a Luminex bead filled with dye of a unique color, and probes for a specific mRNA were immobilized on the surface of the bead. The probes specifically hybridized with the fluorophore-labeled cDNAs derived from the specific mRNAs in cell lysate. The gene expression level was then determined by flow cytometry analysis: the type of an analyte (a bead) was distinguished by the color of the filled dye, and thus the corresponding probe types could be identified according to the designed mapping table of analytes and gene probes. The expression level of the corresponding gene was measured by the sum of intensity from the fluorophore on all beads of the same type. Hundreds of types of analytes were used simultaneously to measure corresponding gene expression in high throughput.
The LINCS project further boosted the throughput of the classical Luminex multiplex technology. About 1,000 “landmark” genes were needed to capture more than 80% of information for expression patterns of about 22,000 genes [7]. However, the current standard LXB platform could only reliably detected about 500 distinct analyte colors. To fill the gap between the number of distinguishable analyte dyes and the number of genes to be measured, the LINCS program utilized a convolution strategy. Totally 1,000 types of analytes were constructed, each immobilized with the mRNA probes of a specific landmark gene. Thus, each distinct dye color i was shared by two types of analytes, namely GeneH(i) and GeneL(i). To distinguish the two types of analytes that shared the same color, analytes GeneH(i) and GeneL(i) were added at a 1.25:0.75 ratio. Thus, the gene expression of the two targeted mRNA types were detected as two peaks on the fluorophore intensity histogram of the same bead color (Figure 2B) with the intensity levels (x-axis) representing the mRNA expression levels and peak sizes corresponding to the amount of analytes. The mRNA types were determined by the sizes of the peaks. Reliable deconvolution of the peaks of the two types of analytes that shared the same color, a process called “peak calling” (Figure 2B), became the critical step for processing raw L1000 data. To deconvolute such overlapped peaks, we assumed that the fluorophore intensities of each analyte type (corresponding to a specific mRNA type) had a Gaussian distribution. The distribution of the mixture of analytes GeneH(i) and GeneL(i) corresponding to the expression levels of GeneH and GeneL, respectively, should be subject to a bimodal Gaussian distribution, with the proportion of 1.25 to 0.75. We initialized the estimations of the two Gaussian distributions using buzzy c-means clustering [11] and estimated the GMM parameters using the Nelder-Mead method [12]. Thus, the overlapped peaks were deconvoluted as the two estimated Gaussian peaks and the expression levels of the two genes sharing the same analyte were extracted. Mathematical details are included in the Supplementary Methods (the GMM model).
As a test of our peak calling method, we introduced another method proposed by Broad Institute based on a k-means algorithm (http://lincscloud.org/exploring-the-data/code-api/, date: 2012/06/27). In this algorithm, the candidate numbers of bead clusters were set as 2, 3, and 4 in the peak calling. This peak calling method chose the numbers of clusters to yielded a ratio of peak areas closest to the expected support proportion, which was 0.65 to 0.35 by default. If more than two clusters were detected, the largest two clusters were defined as corresponding to GeneH(i) and GeneL(i).
Data generation and quality control were realized in the data transforming (normalization and quality control) step [Level 2 to Level 3]. The LINCS L1000 data were generated across several years, and batches of assays were often different in terms of Luminex beads, cells, operators, and environments. Therefore, normalization to remove batch effects and quality control to exclude poor experiments were crucial during data processing. The perturbagen-induced gene expression assays were performed on 384-well plates, each well corresponding to a sample. The controls of perturbagen treatments were on the same plate, and the replicated plates were used for repeated assays. Original gene expression data generated by the GMM-algorithm were quantile-normalized across all assays, and the log fold change (LFC) data were determined by comparing data from treated samples with those from the control samples on the same plate (Figure 2B). Data quality control was performed at multiple levels. At the single-well level, the confidence of the gene expression data were examined by corresponding detectable beads, and those that were supported by less than 20 beads were discarded. At the inter-plate level, data repeatability was examined by Pearson's correlations among replicates, and plates of poor correlations were discarded (Figure 2B).
Data Availability
L1000 data of all three levels, source codes, tutorial, user guide, and the latest updates are available from our website (http://ctsb.is.wfubmc.edu/itNETZ/DPPCSD.html). Processed (Level 2) and transformed (Level 3) data are also available from pLINDAW (the pan-LINCS Data Warehouse. SQL access: metacity.is.wfubmc.edu:3306. Please refer to http://ctsb.is.wfubmc.edu/itNETZ/pLINDAW for more information. Raw L1000 data (Level 1) can also be directly downloaded from the LINCS cloud storage (http://lincscloud.org/) hosted by the Broad Institute.
2.4 Phase II. Compound signature discovery
EGEM score and EGEM matrix
A new metric called the Enrichment of Gene Effect to a Molecule (EGEM) was developed to identify proteins closely related to cellular responses to a small molecule compound, using the LINCS L1000 landmark gene expression data. A small molecule compound affected a cell by directly or indirectly changing the activities and functions of its target proteins, drove downstream biological events, and finally altered cellular gene expression patterns. We hypothesized that the knockdown of a gene that is closely related to the target proteins of a small molecule compounds induces similar gene expression pattern changes. Thus, identification of such genes could reveal the mechanisms of cellular responses to these compounds and predict their pharmaceutical potentials. We defined the “target genes” of a compound in the general meaning: the corresponding proteins of such genes could be either the real drug targets or those at downstream or upstream and were closely related to the real targets. The data for 3,000 single-gene knockdown experiments were used as the target gene reference library, and the data for compound treatments were profiled against this reference library to identify possible target genes of corresponding small molecule compounds.
We defined the EGEM score to describe the similarity between the treatments of a compound and a shRNA targeting a gene using the mutual enrichment of their resultant differential expressed landmark genes. The EGEM metric was derived from the rank-based gene set enrichment analysis (GSEA) [13] and the connectivity analysis [14]. Compound treatments could be taken as “phenotypes” and the differentially expressed genes (DEGs) of a single gene knocking down treatment as a “signature gene set” in the GSEA terminology. The EGEM metric enabled gene set enrichment analysis against the LINCS target gene reference library. The construction of the EGEM score is shown in Figure S1 and details are provided in the Supplementary Data.
We constructed an EGEM matrix A ∈ Rn×m involving n driver genes and m compounds by pairwise calculation of EGEM scores between each compound and each knockdown. Thus, the impacts of these compounds were delineated using the 3000-target-gene reference library.
Compound signature discovery by csNMF
As previously mentioned, a “compound signature” was defined as a group of small molecule compounds sharing similar target genes. We developed a novel method, the constrained sparse non-negative matrix factorization (csNMF), an NMF approach regularized by both the protein-protein-interaction constraint and the sparseness constraint, to effectively detect biomedically meaningful compound signatures from the large EGEM matrix. Non-negative matrix factorization (NMF) [15] is a matrix decomposition method widely used in pattern recognition [16] and has demonstrated its ability in solving various biclustering problems in bioinformatics, including gene pattern recognition, disease module detection, and phenotype classification [17]. Canonically, a non-negative EGEM matrix A ∈ Rn×m would be decomposed into two non-negative matrices W and V, so that A ≈ WV, where W ∈ Rn×k was the weight matrix of target genes, V ∈ Rk×m was the clustering matrix of compounds, and k << min(m, n) was the number of co-clusters. Both weight matrices would be later used to identify the k co-clusters.
We extended the canonical NMF approach to detect
biomedically meaningful co-modules of both compounds and target genes, in which drugs showed similar associations with target genes according to the compound-target EGEM scores. The overall objective function used to solve the csNMF was: and the components were interpreted as described below. The csNMF was optimized using the multiplicative algorithm [15, 17].
Simultaneous clustering of positive and negative EGEM scores
A co-module consisted of both positive and negative EGEM scores as long as they were significant and consistent across compounds in the same module, but canonical NMF approaches could only accept nonnegative values. To simultaneously handle both positive and negative EGEM scores, from the original EGEM matrix A we extracted the positive EGEM scores into the similar EGEM Matrix AS and the absolute values of the negative EGEM scores into the reverse EGEM Matrix AR, both of the same dimensions as A. Both the two EGEM matrices were presented in the overall objective function (Equation 3) and were simultaneously optimized during iterative NMF model training. The corresponding weight matrices of positively and negatively associated target genes, Ws and Wr, and respectively, were achieved at each iteration step, and were merged after optimization.
Sparseness constraint
We introduced a sparseness constraint according to the sparse NMF (sNMF) method proposed by [18]. In sNMF, the L1 norm constraint is added to V, and ||W||F was added to balance the accuracy of the optimization and the sparseness of V. The rationale was that the elements clustered into the co-modules should be a small portion of the matrix. The sparseness constraint was necessary when biclustering a very large EGEM matrix.
PPI constraint
We introduced protein-protein interaction (PPI) constraints according the PPI database [19] to emphasize clusters that were biologically meaningful and thereby control false discovery. The rationale was that in the cellular regulatory network, perturbations of some up- and down-stream proteins (“peers”) of a protein targeted by the compound often also showed similar changes of gene expression patterns. In the PPI constraint component in Equation 3, P was the PPI prior matrix and D was a diagonal matrix, with each row as the sum of the corresponding row of P. The PPI constraint significantly improved both the specificity and the sensitivity of the NMF approach in compound signature discovery. On one hand, false-positive signature genes were often sporadically distributed in the PPI network, and thus their weights downgraded and more likely to be excluded. On the other hand, if in the PPI network a group of “neighbor” genes showed consistent but only moderate EGEM scores with a compound, because of their favorably adjusted weights, they were more likely to be clustered as signature genes of this compound. Introducing prior knowledge of PPI network to the NMF approach thus contributed to more reliable discovery of compound signatures.
Mathematical details (the NMF algorithm) and the pseudo code (Table S1) are provided in the Supplementary Data.
2.5 Phase III. Compound signature analysis
We further examined the biomedical relevance and the pharmaceutical potentials of the detected compound signatures by compound signature analysis using experimental and clinical data.
Biomedical relevance
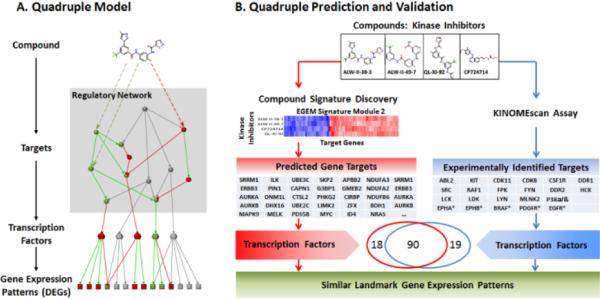
We proposed quadruple models to reveal the molecular events associated with compound signatures and cross-validated the quadruple models using the KINOMEscan experiments. A compound impacts the functions of its target proteins directly or indirectly, triggers regulatory networks, alters the activities of downstream transcription factors, and thus changes the gene expression patterns. To reveal such underlying mechanism of signatures, we proposed a quadruple model (Figure 4A), which included the compound, its direct and indirect targets, downstream transcription factors, and affected genes. Transcription factors for each signature were identified by enrichment analysis according to signature-associated affected genes using ChIP enrichment analysis, setting a p-value of less than 0.05and ratios of the interacting genes to all genes that exceeded 0.1 [20]. The quadruples of compound signatures were thus constructed. The biomedical relevance of a typical signature (Signature 2) was validated by comparing the predicted transcription factors from signature target genes with the enriched transcription factors derived from the direct measurement of kinase targets of four kinase inhibitors (ALW-II-38-3, ALW-II-49-7, QL-XI-92, and CP724714) in this signature.
Fig. 4.
Quadruple models and Signature 2 in a kinome inhibitor study. A quadruple model simultaneously includes a compound, its targets, related transcription factors, and the resulting gene expression pattern. This compound signature discovery method (red) can detect similar quadruples (blue). The similar quadruples include four compounds with similar target sets. 90 in 109 related TFs of the quadruples are covered using the enriched TFs of signature genes.
Pharmaceutical potential
The compound signatures were composed of compounds and their associated target genes. Compounds in a given signature shared similar target genes and thus perturbed the cell functions in similar ways for the corresponding cancer cell line. If some had already demonstrated effectiveness for this type of cancer, other compounds in this signature were more likely to be promising drug candidates. We used FDA-approved chemotherapy drugs for breast cancers to identify breast cancer-specific compound signatures and examined the drug potentials of corresponding drugs. Functions of the signature also could be revealed by enrichment of functions among these target genes. Signatures that demonstrated anti-oncological functions [21] such as reduced cell proliferation, increased cell death, and induced apoptosis, were more likely to be seen in potential drugs. We utilized the DAVID gene functional annotation tool [22] to annotate functions of compound signatures and identify anti-tumor signatures.
3 Results and Discussions
3.1 GMM peak calling pipeline performance
We comprehensively assessed the performance of the GMM peak calling pipeline in accuracy, speed, and scalability for parallel computation using the k-means peak calling approach as the benchmark. The results are summarized in Figure 3.
Fig. 3.
Performance of the GMM peak calling pipeline comparing with the original LINCS pipeline.
Accuracy
We randomly chose 5 raw data sets, 100 analytes each, as the test data set. The accuracy of peak calling was determined by comparing to manually distinguished peaks of GeneH and GeneL based on the bead intensity distributions. During manual peak calling, experts were not able to identify 1.4% of cases; therefore, the maximum accuracy in this assessment would be less than 98.6%. As demonstrated in Figure 3, the GMM approach correctly identified 94.6% of cases, which significantly outperformed the k-means approach (76.4%). Only 4.0% of cases that could be identified by experts were misjudged by the GMM method, compared to the 22.2% false classification rate achieved by the k-means method. Most of the mis-classified cases in the k-means approach were due to the “three-cluster” problem. Figure 3 demonstrates an over-clustering example. When 3 clusters were detected by the k-means peak calling method, only the largest two clusters were retained. If the largest peak (GeneH) was mis-clustered into two small peaks and both were smaller than the small peak (GeneL), the k-means method mistakenly picked the small peak for GeneH. The GMM out-performed the k-means pipeline largely due to avoiding the “three-cluster” problem.
Speed
The R-based GMM approach was about 3.3 times faster than the MATLAB-based k-means approach on Windows-based desktops and Linux clusters, as demonstrated by Figure 3. Vectorization-based code optimization was responsible for the higher calculation efficiency of the GMM approach compared to the KM pipeline.
Scalability
The GMM approach intrinsically encouraged parallel computation on Linux clusters, while the k-means method was basically single-threaded. The GMM method demonstrated good scalability, measured by its efficiency in going from 1 to as many as 32 parallel threads (Figure 3). Parallel processing is critical for L1000 peak calling and raw data processing due to the large sample size. Powered by automatic sample preparation and the high-throughput data acquisition, a typical L1000 profiling experiment involves hundreds of 384-well plates, with 500 analytes in each well. Thus, in regular L1000 raw data processing, millions to tens of millions of peak calling tasks will be accomplished. The ability to conduct parallel data processing on large Linux clusters and good scalability of our approach meets the needs of the high-throughput data processing.
In summary, as a parallel, cross-platform, and high-performance R package, the GMM pipeline is especially suitable for processing large data sets. This pipeline is publically available from our CTSB website (http://www.wakehealth.edu/CTSB/), and will be available from BioConductor (http://www.bioconductor.org/).
3.2 Signatures and quadruples for kinase inhibitors
We used the kinase inhibitor dataset to validate the concept of the compound signatures discovered by the EGEM-based csNMF approach. We chose this dataset because some kinase inhibitors had been experimentally profiled to identify their direct kinase targets, and thus could be used to validate the predictions of the csNMF modeling. The 51 kinase inhibitors were analyzed against the 3,341-target gene reference library. In all, 6 compound signatures were detected (see Supplementary Data File 1).
Validation of predicted target genes using GO similarity
Target genes in the same compound signature should be strongly correlated. We utilized the gene ontology (GO) similarities among the predicted targets within the same signature (95% interaction rate, GOSemSim [23]) to examine whether target genes were biologically associated. Signatures 2, 4, and 5 demonstrated strong GO connectivity (Table S2 in the Supplementary Data). Although Signature 1 did not pass the significance test, three inhibitors in Signature 1 shared the primary target of EGFR and the other two inhibitors shared FGFR3 and p38-alpha. Signatures 3 and 6 demonstrated very high correlations of gene expression patterns among compounds in the signatures (Signature 3: 0.947±0.059, Signature 6: 0.763±0.127).
Validation of predicted target genes using direct kinase targets
We focused on Signature 2 for further analysis because four kinase inhibitors in this signature (ALW-II-38-3, ALW-II-49-7, QL-XI-92, and CP724714), were also experimentally profiled by KINOMEscan® for their direct kinase targets. We first examined whether these kinase inhibitors if they shared kinase targets, as predicted. Three of them (LW-II-38-3, ALW-II-49-7, and QL-XI-92), directly shared the same nominal target, DDR1. We then examined if their kinase targets demonstrated stronger similarity than average by calculating the correlations of interactions of these targets to all 450 kinases in the KINOMEscan® dataset. These three kinase inhibitors were highly related compared to the randomly selected compounds (correlation coefficients around 0.7~0.8 compared to 0.10 for random controls) (Table S3, Supplementary Data). Kinase inhibitor similarity according to EGEM scores also was consistent with the direct kinase target similarity assayed by KINOMEscan® experiments (Figure S2, Supplementary Data). Our results were consistent with previous reports. For example, ALW-II-38-3 and ALW-II-49-7 are known to demonstrat e very similar characteristics [24]. CP724714 did not show similar kinase targets to the other three inhibitors, and was further analyzed using quadruple models.
Validation of predicted target genes using the quadruple model
Compounds that triggered similar molecular cascades might instead share indirect targets, some of which might not be kinases. CP724714, whose major target was HER2, did not show similar kinase targets to the other 3 kinases, but it induced a similar change in gene expression pattern according to the EGEM matrix. Previous literatures suggests a strong co-occurrence between DDR1 and HER2 [25] in breast cancer. We thus examined whether the four kinase inhibitors in Signature 2 instead shared similar downstream signaling pathways and affected activities of transcription factors in the same way. The quadruple models of these four inhibitors were constructed according to predicted target genes (Figure 4B, red) and were compared to those constructed according to direct kinase targets from the KINOMEscan® results (Figure 4B, blue). Among the 108 transcription factors enriched from predicted targets and the 109 from experimental targets, 90 overlapped. Thus, the predicted similarity between CP724714 and the other three compounds could be explained in the quadruple models, reflecting shared patterns of downstream transcription factor activity.
3.3 Functional annotation to determine signature drug potential for breast cancer
Since the csNMF approach was validated for 51 kinase inhibitors, we implemented this approach to screen drug candidates for breast cancer. We studied 728 compounds against the 3,341 target gene reference library screened for the MCF-7 breast cancer cell line and detected eight signatures. As shown in Figure 5A, compounds (columns) belonging to the same signatures were grouped together; red regions denote similar gene expression patterns between the compounds and the target genes (rows), and the green regions denote the reverse effects. In all, 8 compound signatures were identified (Supplementary Data File 2).
Fig. 5.
Breast cancer compound signatures. (A) Eight signatures were detected (yellow rectangles). For each signature, compounds (columns) and genes (rows) corresponding to a red region showed similar gene expression effects, whereas those corresponding to a green region exhibited reverse effects. (B) Degree of yellow represents relative enrichment for related gene ontology (GO) terms. (C) Associations of Signature 4 with drug responses and survival in data from 2,116 breast cancer patients collected from Belgium, England, and Singapore (GEO:GSE45255).
To find the signatures of related compounds that might be beneficial for breast cancer, we focused on functions such as induction of apoptosis and suppression of proliferation. The enrichment of different biological processes of signatures were investigated by DAVID [22] according to the gene ontology (GO) terms of signature target genes. Only terms with a p-value less than 0.05 were considered. To define similar compound-gene effects, we considered the terms with positive regulation of cell death and apoptosis; as to the reverse ones, we considered the negative regulations (cancer treatment-related GO terms). Signatures 7 and 8 were enriched for apoptosis (Figure 5B).
Compounds in Signature 7 demonstrated potential benefit as cancer treatments. Among them, letrozole and megestrol acetate were FDA-approved chemotherapy drugs for breast cancers [26]. Oleoylethanolamide was reported to suppress cell proliferation and was used to treat breast cancer [27]. Calcipotriol exhibited antiproliferative activity in the MCF-7 cell line [28], and leinoleic acid was reported to inhibit cell growth in the same cell line [29]. Dibenzoylmethane and CITCO inhibited cell growth in prostate cancer and brain tumor stem cells [30, 31].
Compounds in Signature 8 were related to antihypertensive and antipsychotic drugs, such as piretanide [32] and benperidol [33], Interestingly, other researchers reported that antipsychotic drugs inhibited the functions of proteins related to breast cancer drug resistance [34]. However, some compounds, such as gabazine [35] and mesulergine [30], demonstrated high toxicity and might not be suitable as drugs.
3.4 Clinical relevance of compound signatures
We examined the associations of the discovered compound signatures with patient survival and other clinical traits. Clinical features and gene expression profiles of 2,116 breast cancer patients collected from Belgium, England, and Singapore (GEO:GSE45255) were examined by the gene set enrichment of the 8 discovered breast cancer related compound signatures. For example, in terms of distant metastasis-free survival, patients the Signature 4Low category responded poorly to chemotherapy compared with those in the Signature 4High category (Figure 5C). Signature 4 was selectively associated with chemotherapy but not hormone therapy (tamoxifen). We performed a univariable and multivariable survival analysis using discovered compound signatures as well as conventional clinical features including patient age, tumor size, PAM50 as well as molecular subtypes, lymph node involvement, the ER status, and the pathological grades (Tables S4, S5 and Data File S3). The results suggested that the compound signatures 4 and 5 are strongly associated with poor prognosis for patients with chemotherapies but not for those with Tamoxifen treatment. The analysis results were consistent with the drug response survival results showed in Figure 5.
Signatures also demonstrated associations with breast cancer subtypes (Signature 2) and receptor status (Signatures 3 and 6 with estrogen receptor status), as shown in Figure S3 in the Supplementary Data section.
Such association results demonstrate the clinical potential of the compound signatures discovered in the MCF-7 breast cancer cell line model. Follow-up investigations could include testing the underlying mechanisms for the poor prognosis of patients in the Signature 4Low category, by further studies of the predicted target genes using the established Signature 4 quadruple model.
CONCLUSIONS
We have developed the csNMF approach, a comprehensive and complete pipeline, for network-based compound signature discovery and drug screening under the target gene reference library. The GMM approach, the L1000 raw data pre-processing module, has demonstrated high accuracy, high efficiency, and high scalability compared with the standard KM pipeline. The EGEM-based csNMF signature discovery module benefited from biological (PPI) and sparseness constraints and simultaneous co-clustering of both positive and negative values. The quadruple model, which incorporates four consequential components along the drug-induced molecular cascade (the drug, drug targets, downstream transcription factors, and affected gene expression), can reveal underlying regulatory mechanisms of similar drugs. The predicted similarity of drug-target genes were validated with experimental profiling. The extracted breast cancer compound signatures also demonstrated strong clinical relevance. Together, as a key module of the itNETZ platform, the csNMF pipeline bridges the gap between the rich resource of the LINCS signature library and biomedical and clinical research needs, and provides biomedical researchers with a systematic drug screening and mechanism discovery framework.
Supplementary Material
Acknowledgements
The authors acknowledge the Texas Advanced Computing Center (TACC) and the DEMON high performance computing (HPC).
Funding: This work was supported by the National Institutes of Health [1U01HL111560]. Open access charges also were supported by the National Institutes of Health.
Footnotes
Electronic Supplementary Information (ESI) available: L1000 data of all three levels, source codes, tutorial, user guide, and the latest updates are available from (http://ctsb.is.wfubmc.edu/itNETZ/DPPCSD.html). Processed (Level 2) and transformed (Level 3) data are also available from pLINDAW (the pan-LINCS Data Warehouse. SQL access: metacity.is.wfubmc.edu:3306. For more information please refer to http://ctsb.is.wfubmc.edu/itNETZ/pLINDAW. Raw L1000 data (Level 1) can also be directly downloaded from the LINCS cloud storage (http://lincscloud.org/) hosted by the Broad Institute.
References
- 1.William Downey C.L.a.J.H. Compound Profiling: size impact on primary screening libraries. 2010. Drug Discovery World Spring. 2010:81. [Google Scholar]
- 2.Hughes JP, et al. Principles of early drug discovery. Br J Pharmacol. 2011;162(6):1239–49. doi: 10.1111/j.1476-5381.2010.01127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hefti F. Requirements for a lead compound to become a clinical candidate. BMC Neuroscience. 2008;9(Suppl 3):1–7. doi: 10.1186/1471-2202-9-S3-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mullard A. Learning lessons from Pfizer's $800 million failure. Nat Rev Drug Discov. 2011;10(3):163–164. doi: 10.1038/nrd3401. [DOI] [PubMed] [Google Scholar]
- 5.Lounkine E, et al. Large-scale prediction and testing of drug activity on side-effect targets. Nature. 2012;486(7403):361–367. doi: 10.1038/nature11159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lehmann BD, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. The Journal of clinical investigation. 2011;121(7):2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duan Q, et al. LINCS Canvas Browser: interactive web app to query, browse and interrogate LINCS L1000 gene expression signatures. Nucleic Acids Research. 2014:gku476. doi: 10.1093/nar/gku476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roth WD, Wayne D. Personal flow cytometers—luminex. The microflow cytometer. Pan Stanford Publishing, Singapore. 2010:37–50. [Google Scholar]
- 9.Seamer L, et al. Proposed new data file standard for flow cytometry, version FCS 3.0. Cytometry. 1997;28(2):118–122. doi: 10.1002/(sici)1097-0320(19970601)28:2<118::aid-cyto3>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 10.Peck D, et al. A method for high-throughput gene expression signature analysis. Genome Biology. 2006;7(7):R61. doi: 10.1186/gb-2006-7-7-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bezdek JC. Pattern Recognition with Fuzzy Objective Function Algorithms. Kluwer Academic Publishers; 1981. p. 256. [Google Scholar]
- 12.Nelder JA, Mead R. A simplex method for function minimization. The computer journal. 1965;7(4):308–313. [Google Scholar]
- 13.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lamb J, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. science. 2006;313(5795):1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 15.Lee DD, Seung HS. Learning the parts of objects by nonnegative matrix factorization. Nature. 1999;401(6755):788–791. doi: 10.1038/44565. [DOI] [PubMed] [Google Scholar]
- 16.Paatero P, Tapper U. Positive matrix factorization: A nonnegative factor model with optimal utilization of error estimates of data values. Environmetrics. 1994;5(2):111–126. [Google Scholar]
- 17.Lee DD, Seung HS. Algorithms for non-negative matrix factorization. Advances in neural information processing systems. 2001 [Google Scholar]
- 18.Kim H, Park H. Sparse non-negative matrix factorizations via alternating non-negativity-constrained least squares for microarray data analysis. Bioinformatics. 2007;23(12):1495–1502. doi: 10.1093/bioinformatics/btm134. [DOI] [PubMed] [Google Scholar]
- 19.von Mering C, et al. STRING: known and predicted protein– protein associations, integrated and transferred across organisms. Nucleic Acids Research. 2005;33(suppl 1):D433–D437. doi: 10.1093/nar/gki005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lachmann A, et al. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. 2010;26(19):2438–2444. doi: 10.1093/bioinformatics/btq466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerl R, Vaux DL. Apoptosis in the development and treatment of cancer. Carcinogenesis. 2005;26(2):263–270. doi: 10.1093/carcin/bgh283. [DOI] [PubMed] [Google Scholar]
- 22.Huang D, et al. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biology. 2007;8(9):R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu G, et al. GOSemSim: an R package for measuring semantic similarity among GO terms and gene products. Bioinformatics. 2010;26(7):976–978. doi: 10.1093/bioinformatics/btq064. [DOI] [PubMed] [Google Scholar]
- 24.Choi Y, et al. Discovery and structural analysis of Eph receptor tyrosine kinase inhibitors. Bioorganic & Medicinal Chemistry Letters. 2009;19(15):4467–4470. doi: 10.1016/j.bmcl.2009.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siddiqa A, et al. Expression of HER-2 in MCF-7 breast cancer cells modulates anti-apoptotic proteins Survivin and Bcl-2 via the extracellular signal-related kinase (ERK) and phosphoinositide-3 kinase (PI3K) signalling pathways. BMC Cancer. 2008;8(1):129. doi: 10.1186/1471-2407-8-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.US Food and Drug Administration Approved drug products with therapeutic equivalence evaluations. 2014 [Google Scholar]
- 27.Yueh-Hsiung Kuo F-YT. Hui-Yi Lin, Inhibition of Cancer Cell Proliferation Using Oleoylethanolamide, in Espacenet. China Medical University; 2013. [Google Scholar]
- 28.Wietrzyk J, et al. Antitumor properties of diastereomeric and geometric analogs of vitamin D3. Anti-cancer drugs. 2007;18(4):447–457. doi: 10.1097/CAD.0b013e3280143166. [DOI] [PubMed] [Google Scholar]
- 29.Cunningham DC, Harrison LY, Shultz TD. Proliferative responses of normal human mammary and MCF-7 breast cancer cells to linoleic acid, conjugated linoleic acid and eicosanoid synthesis inhibitors in culture. Anticancer Res. 1997;17(1A):197–203. [PubMed] [Google Scholar]
- 30.Chakraborty S, Kanakasabai S, Bright JJ. Constitutive androstane receptor agonist CITCO inhibits growth and expansion of brain tumour stem cells. Br J Cancer. 2011;104(3):448–59. doi: 10.1038/sj.bjc.6606064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khor TO, et al. Dietary feeding of dibenzoylmethane inhibits prostate cancer in transgenic adenocarcinoma of the mouse prostate model. Cancer Res. 2009;69(17):7096–102. doi: 10.1158/0008-5472.CAN-09-0597. [DOI] [PubMed] [Google Scholar]
- 32.Clissold SP, Brogden RN. Piretanide. A preliminary review of its pharmacodynamic and pharmacokinetic properties, and therapeutic efficacy. Drugs. 1985;29(6):489–530. doi: 10.2165/00003495-198529060-00002. [DOI] [PubMed] [Google Scholar]
- 33.Bobon J, Collard J, Lecoq R. [Benperidol and Promazine: A “Double Blind” Comparative Study in Mental Geriatrics]. Acta Neurol Belg. 1963;63:839–43. [PubMed] [Google Scholar]
- 34.Wang JS, et al. Antipsychotic drugs inhibit the function of breast cancer resistance protein. Basic Clin Pharmacol Toxicol. 2008;103(4):336–41. doi: 10.1111/j.1742-7843.2008.00298.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Behrens CJ, van den Boom LP, Heinemann U. Effects of the GABA(A) receptor antagonists bicuculline and gabazine on stimulus-induced sharp wave-ripple complexes in adult rat hippocampus in vitro. Eur J Neurosci. 2007;25(7):2170–81. doi: 10.1111/j.1460-9568.2007.05462.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
L1000 data of all three levels, source codes, tutorial, user guide, and the latest updates are available from our website (http://ctsb.is.wfubmc.edu/itNETZ/DPPCSD.html). Processed (Level 2) and transformed (Level 3) data are also available from pLINDAW (the pan-LINCS Data Warehouse. SQL access: metacity.is.wfubmc.edu:3306. Please refer to http://ctsb.is.wfubmc.edu/itNETZ/pLINDAW for more information. Raw L1000 data (Level 1) can also be directly downloaded from the LINCS cloud storage (http://lincscloud.org/) hosted by the Broad Institute.