Abstract
Background
Prenatal stress is considered a risk factor for several neurodevelopmental disorders including schizophrenia (SZ). An animal model involving restraint stress of pregnant mice suggests that prenatal stress (PRS) induces epigenetic changes in specific GABAergic and glutamatergic genes likely to be implicated in SZ including the gene for brain derived neurotrophic factor (BDNF).
Methods
Studying adult offspring of pregnant mice subjected to PRS, we explored the long-term effect of PRS on behavior and on the expression of key chromatin remodeling factors including DNA methyltransferase 1 (DNMT1), ten-eleven translocation hydroxylases (TETs), methyl CpG binding protein 2 (MeCP2), histone deacetylases (HDACs), histone methyltransferases (MLL1, SETD1, G9A and EZH1) and demethylase (LSD1) in the frontal cortex (FC) and hippocampus (HP). We also measured the expression of BDNF.
Results
Adult PRS offspring demonstrate behavioral abnormalities suggestive of SZ and molecular changes similar to SZ postmortem brain: a significant increase in DNMT1 and TET1 in the FC and HP but not in cerebellum, no changes in HDACs, histone methytransferases/demethylases or MeCP2, and a significant decrease in BDNF variants measured in the FC and HP. The decrease of the corresponding BDNF transcript level was paralleled by an enrichment of 5-methylcytosine and 5-hydroxylmethylcytosine levels at Bdnf gene regulatory regions. In addition, the expression of BDNF transcripts (IV and IX) was positively correlated with social approach in both PRS and non-stressed mice.
Conclusions
Since patients with psychosis and PRS mice show similar epigenetic signature, PRS offspring may be a suitable model for understanding the behavioral and molecular epigenetic changes observed in SZ patients.
Keywords: Prenatal stress, DNA methylation, BDNF, DNMT, TET, Schizophrenia
Introduction
Evidence from clinical and preclinical studies indicate that exposure to stress during pregnancy exerts profound effects on the neurodevelopment and behavior of offspring (1–3). Prenatal stress is considered a risk factor for several neurodevelopmental disorders including schizophrenia (SZ), bipolar (BP) disorder, depression, and anxiety (4–7).
Recently a substantial amount of evidence from our group (3, 8) and other researchers (9–11) suggests that epigenetic modifications of DNA and chromatin induced by environmental factors, including stress, may contribute to the complex phenotypes of neuropsychiatric disorders. Patients with psychosis express an increase in brain DNA methyltransferases (DNMT1 and 3a), and an increase in ten-eleven translocation hydroxylase (TET1) (12–15). DNMT and TET are important components of the DNA-methylation/demethylation dynamic regulating the expression of key molecules involved in brain function (16). Studying postmortem brain of SZ patients, we have shown that overexpression of DNMTs and TETs is associated with an enrichment of 5-methyl cytosine (5MC) and 5-hydroxymethyl cytosine (5HMC) at SZ candidate gene promoters including reelin (Reln), glutamic acid decarboxylase 67 (Gad1) and brain derived neurotrophic factor (Bdnf) exon IX, leading to expression downregulation (14, 16–20). Reduced expression of Bdnf and increased methylation of Bdnf exon IV and IX have been reported in prefrontal cortex (FC) and hippocampus (HP) as well as in cerebrospinal fluid and blood of patients with SZ (21–25), BP disorder (26), depression, and anxiety (27–30).
In recent years we have directed the focus of our research to the study of whether the altered epigenetic signature detected in postmortem brain of SZ patients is also present in offspring of mice stressed during pregnancy (PRS mice). PRS mice, like SZ patients, show a delay of inhibitory neuron progenitor migration in the developing neocortex (31). Adult PRS mice have behavioral deficits reminiscent of behavior observed in psychotic patients (3, 32, 33), and the adult PRS mouse brain, like that of postmortem chronic SZ patients, is characterized by a significant increase in DNMTs, an enrichment of 5MC and 5HMC at GABAergic (Gad1, Reln) and glutamatergic (Bdnf) SZ candidate gene promoters, and a decrease in the expression of these genes (3, 8). Decreased BDNF expression, increased DNMT1 expression, as well an increase in the methylation of Bdnf exon IV, have also been reported recently in hippocampus and amygdala of adult PRS rats (11).
In this study we tested whether: 1) in addition to DNMT1 the expression of TETs is modified in the brain of PRS mice in a manner reminiscent of SZ brain, and whether 2) such an increase in TETs participates in the regulation of the DNA-methylation/hydroxymethylation dynamic of SZ susceptibility genes focusing on BDNF as a target gene. Thus, the goal of our study was to establish, in a mouse model of prenatal exposure to stress, whether the decrease of Bdnf expression related to prenatal stress is associated with changes in methylation/demethylation processes brought about by increased expression of DNMT and TET thereby modeling the epigenetic changes detected in the brain of SZ and BP disorder patients.
Methods and Materials
1. Animals and PRS Procedure
All procedures were performed according to NIH guidelines for animal research (Guide for the Care & Use of Laboratory Animals, NRC, 1996) and were approved by the Animal Care Committee of the University of Illinois at Chicago. Pregnant mice (Swiss albino ND4, Harlan, Indianapolis, IN) were individually housed with a 12-h light-dark cycle, and food and water ad libitum. Control dams were left undisturbed throughout gestation, whereas stressed dams were subjected to repeated episodes of restraint stress, as described previously (3) with slight modification. The stress procedure consisted of restraining the pregnant dam in a transparent tube (12 × 3 cm) under a bright light for 45 min three times per day from the seventh day of pregnancy until delivery. After weaning (postnatal day [PND] 21), male mice were selected for the study and housed four to five per cage separately by condition.
2. Behavioral Tests
Behavioral characteristics, first for locomotor activity and then for social interaction, were examined in PRS male mice and non-stressed controls (NS) on consecutive days at post natal day (PND) 75. We selected PND 75 for behavioral testing because at this post-natal time the performance of the offspring was more reproducible and stable than the performance measured at earlier developmental time points.
2.1) Locomotor activity
A computerized Animal Activity Monitoring System with VersaMax software (AccuScan Instruments, Columbus, OH) was used for the quantification and tracking of locomotor activity in mice as described previously (34). Each activity cage consisted of a Perspex box (20 × 20 × 20 cm divided into quadrants) surrounded by horizontal and vertical infrared sensor beams. The total number of interruptions of the horizontal sensors was taken as a measure of horizontal activity, whereas that of vertical sensors was used as a measure of vertical activity. Activity was recorded for 15 min between 1 and 3 pm.
2.2) Social interaction
Apparatus
Social approach of PRS and NS mice was measured using the “Three-Chambered Apparatus” (35) which is a rectangular, transparent three-chambered box with each chamber measuring 20 cm long × 40.5 cm wide × 22 cm high. Small openings in the clear Plexiglas walls (10 cm wide × 5 cm high) divide the center compartment from the two side compartments. Two identical wire cups were placed in left and right chambers, one for enclosing a stranger (novel) mouse and one as a control object. Between tests the apparatus was thoroughly washed with 70% ethanol and then distilled water. Tests were performed under dim and even lighting between 10 am and 3 pm. Sessions were recorded on video tape for data analysis
Test
The test mouse was first placed in the center chamber and habituated by allowing it to freely explore the entire apparatus for 5 min. Then the mouse was gently coerced into the center chamber and confined by shutting the openings for the side chambers. The stranger mouse was then placed in the wire cup of one of the side chambers and the test was initiated by opening both doorways allowing the test mouse to explore all three chambers freely for 10 min. Social approach or interaction was defined as the ratio of the sniffing time for the empty wire cup vs. the cup enclosing the stranger mouse. Reliability of measurements was assessed by correlating the scores of two raters.
3. Quantitative real-time polymerase chain reaction (qPCR)
QPCR was carried out using the Applied Biosystems Real-Time PCR System with a SYBR green master mix (Fermentas, Glen Burnie, MD). Total RNA from FC and HP, was isolated using TRIZOL reagent (Life Technologies, Grand Island, NY), and was further purified using the Qiagen RNeasy kit (Qiagen, Valencia, CA). The RNA integrity number was measured with an Agilent 2100 bioananlyzer (Agilent Technologies, Santa Clara, CA). The primer sequences for the genes analyzed are summarized in Table S1 in Supplement 1. Each sample was run in duplicate and repeated twice. For normalizing mRNA expression, several housekeeping genes (Nse, NeuN and β-actin) were chosen as the internal control. For each housekeeping gene, we measured the gene stability ranking using the NormFinder algorithm (36). This procedure allows for the identification of the housekeeping gene best suited for normalization. Because each of the genes studied yielded similar results when normalized to either Nse, NeuN, or β-actin, and because β-actin had the highest housekeeping gene stability (NormFinder), we normalized our data to β-actin.
4. Western blot analysis
For protein quantification we conducted measurements as described in detail elsewhere (14) Anti-DNMT1 monoclonal antibodies (0.5ug/ml, Imagenex, San Diego, CA), or anti-TET1 polyclonal antibody (Millipore, Billerica, MA), anti-TET2 and TET3 monoclonal antibodies (Zymo Rearch Irvine, CA) were used to detect DNMT1, TET1, TET2 and TET3 protein. The levels of these proteins in PRS vs NS were normalized by by β-actin protein levels.
5. Methylated DNA immunoprecipitation
Methylated and hydroxymethylated DNA on Bdnf exon I to IX were assessed using MeDIP and hMeDIP kits (Diagenode, Denville, NJ.) followed by qPCR. The procedures for sample treatment and immunoprecipitation are described in kit instruction manuals. We used these procedures because bisulfite and most enzyme-dependent methods are incapable of distinguishing 5MC from the approximately 14% of methylcytosines in the brain that are 5HMC (20). Recent studies demonstrate that 5MC and 5HMC may have very different functions and genomic locations (37, 38). The percentage of methylated (or hydroxymethylated) vs unmethylated (or unhydroxymethylated) promoter was calculated using the following equation: . To test if DNA methylation is gene specific, we also measured the methylation status induced by PRS on Gapdh and Gad2 promoters (primer sequences in Table S1)
6. Chromatin immunoprecipitation assays
We performed chromatin immunoprecipitation (ChIP) assays based on protocols previously described (39). The percentages of immunoprecipitated DNA were calculated as described for MeDIP/hMeDIP. ChIP grade anti-DNMT1 (Imagenex, San Diego, CA), and anti-TET1 polyclonal antibody (Millipore, Billerica, MA) were used to precipitate cross-linked chromatin.
7. Statistical analysis
Results are expressed as mean ± SEM. Experimental differences were assessed by Student’s t test, one-way ANOVA followed by Bonferroni post-hoc comparisons and Pearson correlation analysis using Predictive Analytics SoftWare (PASW) v.18 (SPSS, Chicago, IL). The criterion for significance was p < .05, two tailed.
Results
1. The effect of PRS on the growth of the offspring
We compared the litter size, body weight, and gross behavior at different developmental stages [postnatal day (PND) 1, 14, 21, 42, 75] in a cohort of 12 PRS and 12 NS mice. There were no statistical differences between PRS and NS groups in litter size (8–10 pups per litter). However, PRS mice were born 8–12 hr earlier and had a lower (10–15%) body weight at birth (NS: 4.06 ± 0.4 grams; PRS: 3.47 ± 0.2 grams). We compared the body weights of offspring from PRS and NS groups at PND 75 (late adolescence, early adulthood in mice). No statistically significant differences were found in terms of body weight between the two groups when the offspring reached adulthood (NS: 29.3 ± 0.61 grams; PRS: 29.9 ± 0.35 grams). In addition, no obvious differences in gross behavior (auditory sensitivity, pain sensitivity, or motor coordination) between PRS and NS were apparent [see also (3)].
2. PRS induces behavioral abnormalities in adult offspring
We and others (3, 32, 33) have shown that adult PRS mice express SZ-like behavioral phenotypes such as hyperlocomotion and stereotypic behavior induced by the psychostimulant-MK801 and deficits in attention and information processing including social interaction, fear conditioning and prepulse inhibition of startle (PPI). Consistent with these previous reports, in the present experiment, PRS offspring at PND75 exhibited locomotor hyperactivity (horizontal and vertical) in an open field arena (Figure 1A) and demonstrated a marked social interaction deficit (Figure 1B) measured using the three chamber test method (35). We limited the behavioral study to these two tests because they are considered less stressful than fear conditioning, PPI, or MK-801 response and stress could possibly alter the biochemical parameters per se.
Figure 1.
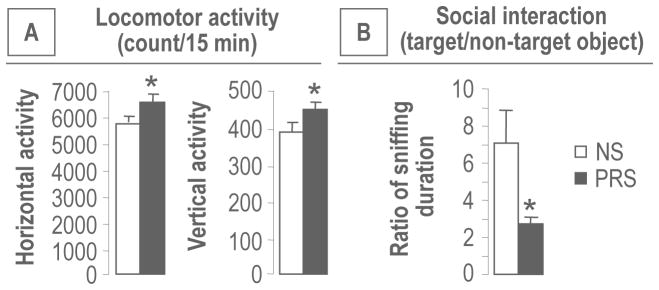
PRS induces behavioral abnormalities in adult [postnatal day (PND) 75] offspring. As compared with NS group, PRS offspring show: A) locomotor (horizontal and vertical) hyperactivity; B) decreased social interaction. Data are presented as mean ± SEM of 12 mice. * p < .05 (Student t-test,) vs. NS group.
3. Increased expression of DNMT1 and TET1 in the FC and HP of PRS mice
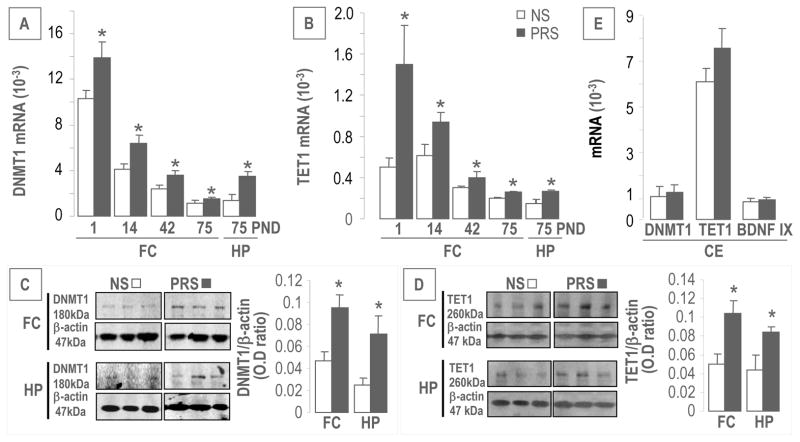
As shown in Figure 2A and 2B, the expression of DNMT1 and TET1 mRNA was significantly higher in the FC and HP of PRS offspring compared to NS mice at PND75. The levels of DNMT1 and TET1 proteins were also increased in both FC and HP of PRS offspring at PND75 compared to controls (Figure 2C and 2D). Interestingly, DNMT1 and TET1 transcripts were high at PND 1 but then decreased dramatically and progressively up to PND 75 in the FC of both PRS and NS mice (Figure 2A and 2B). However, DNMT1 and TET1 expressions were significantly higher in PRS than in NS mice at all of the PNDs studied (Figure 2A and 2B). Similar differences between PRS and NS in the levels of DNMT1 and TET1 were not found in the cerebellum (Figure 2E) even though the cerebellum expresses considerably more TET1 transcript than either the FC or HP. We also measured TET1 isoforms: TET2 and TET3. Both isoforms and their proteins were increased in the FC of PRS offspring at PND75 [TET2 (relative mRNA ×10−3), NS: 0.6 ± 0.07 n = 11, PRS: 0.8 ± 0.06 n = 12, p < .05; TET2 (vs. β-actin), NS: 0.032 ± 0.006, n = 6, PRS: 0.054 ± 0.008 n = 6, p < .05; TET3 (mRNA ×10−3), NS: 0.5 ± 0.06, n = 11, PRS: 0.7 ± 0.06 n = 12, p < .05; TET3 (vs. β-actin), NS: 0.023 ± 0.006 n = 6, PRS: 0.05 ± 0.01 n = 6, p < .05].
Figure 2.
PRS increases expression of DNMT1 (A) and TET1 (B) transcripts in the frontal cortex (FC) at PND 1, 14, 42 and 75. At PND 75, both genes are elevated significantly in the hippocampus (HP) of PRS mice, compared with corresponding controls. Immunoblot data (C and D) normalized by β-actin protein levels shows marked increase in the protein levels of DNMT1 and TET1 in the FC and HP of PRS mice at PND75 compared to NS group (n = 6 mice for each group). 2E shows that the expressions of DNMT1 TET1 and BDNFIX did not change in the cerebellum (CE) between PRS and NS mice. Data are presented as mean ± SEM of 12 mice. * p < .05 (Student’s t-test,) vs. the corresponding NS value.
4. Altered expression of BDNF transcripts in the FC and HP of PRS mice
In previous studies we showed that the promoter hypermethylation of GABAergic genes (Gad1 and Reln) in FC and HP of adult PRS mice was associated with downregulation of the expression of these genes (3). To establish whether a similar association also occurs for non-GABAergic genes, we focused on the expression of Bdnf because this gene is downregulated in SZ (20–23), is mostly expressed in glutamatergic neurons and its promoter is a target of methylation/demethylation processes (25, 40).
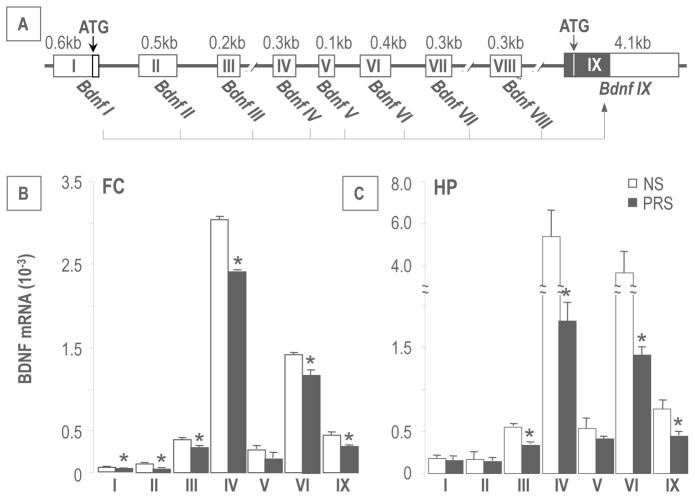
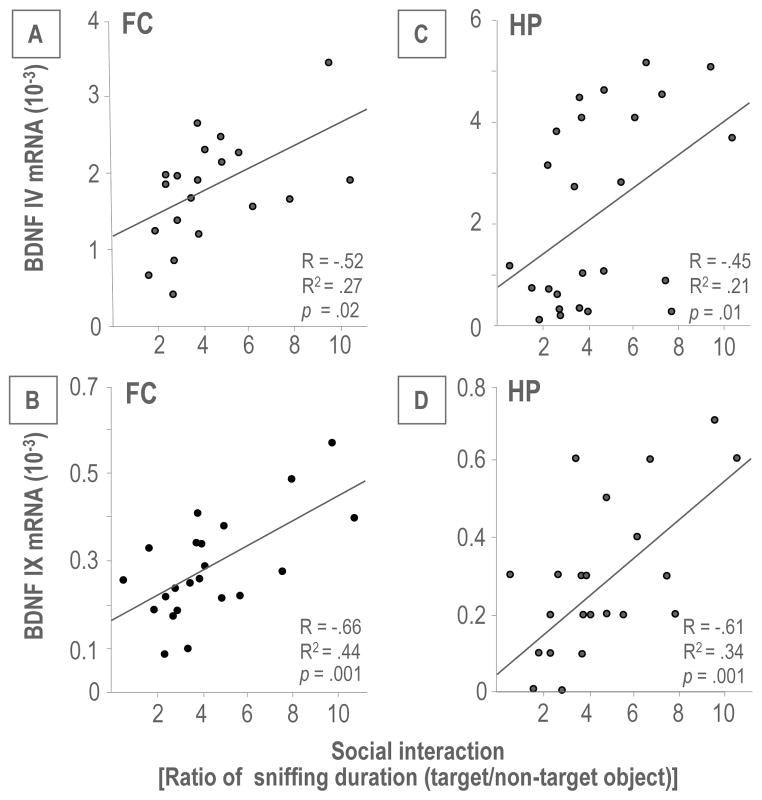
Mouse Bdnf consists of nine-exon driven transcripts (Figure 3A), producing at least nine BDNF splice variants (41). Using qPCR, we measured the expression of seven transcripts (I, II, III, IV, V, IV and IX) in the FC. Transcript VII and VIII were excluded from this study because they lack CpG islands and have low expression levels. Of the seven transcripts measured, BDNF IV and VI were the most abundant, indicating that these two transcripts are the major contributors to steady-state BDNF levels. BDNF III and IX were expressed at an intermediate level whereas BDNF I, II and V transcripts were expressed at low levels (Figure 3B). Importantly, all alternative transcripts measured in FC were significantly decreased in PRS offspring; with the exception of BDNF V. In HP, the expression pattern of BDNF transcripts was similar to that found in FC, and the expression of the transcripts was again lower in PRS mice (Figure 3C). Further, we performed correlation analysis between levels of BDNF IV and IX mRNA expression in FC and HP and measurements of social interaction. As shown in Figure 4, there was a significant positive correlation between BDNF IV and BDNF IX mRNA expression and social interaction.
Figure 3.
PRS downregulates the expression of BDNF transcripts in the FC and HP of adult offspring. A). Mouse Bdnf gene structure and alternative transcripts. Exons are shown as boxes and introns are shown as thick lines. Protein coding regions are shown as solid boxes and untranslated regions are shown as empty boxes. Each of the eight 5′ untranslated exons is spliced to the common 3′ protein coding exon IX (41). In B and C, BDNF transcripts (III, IV, VI and IX) at PND 75 in the FC, and HP of PRS offspring are significantly decreased as compared to the NS group. Data are presented as mean ± SEM of 12 mice. * p < .05 (Student’s t-test) vs. the corresponding NS value.
Figure 4.
Correlations between Bdnf IV or IX mRNA level and social interaction. Pearson correlation analysis shows that there is a positive correlation in the FC (A and B) and HP (C and D) between Bdnf IV or IX mRNA level and social interaction.
5. PRS alters 5MC and 5HMC levels on Bdnf gene regulatory regions
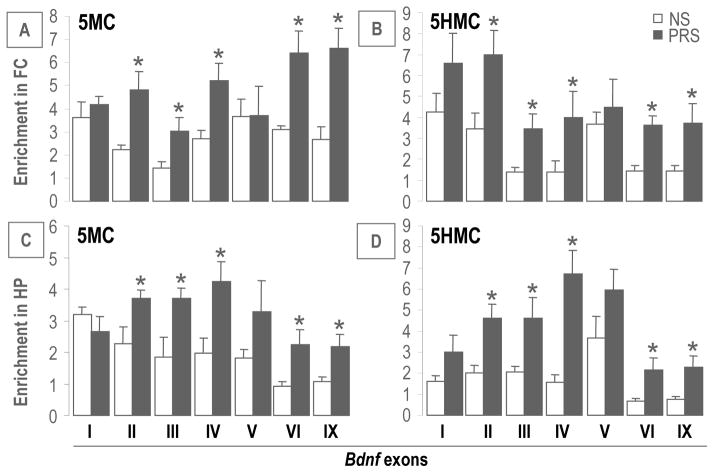
To investigate whether the decreased expression of BDNF transcripts observed in PRS mice are the consequence of epigenetic modification on their corresponding DNA regulatory regions, we measured, using MeDIP and hMeDIP, the enrichment of the two most important epigenetic marks: 5MC and 5HMC on the different BDNF exons corresponding to the transcripts we measured (i.e., I, II, III, IV, V, VI and IX). As shown in Figure 5A and B, 5MC and 5HMC were enriched at Bdnf promoters II, III, IV, IX but not I and V in the FC of PRS compared to NS mice. A similar pattern was also found in the HP area (Figure 5C and 5D). These findings suggest that prenatal stress leads to CpG modification (methylation or demethylation) on specific Bdnf promoters. We also included Gapdh and Gad2 as control genes to check the specificity of methylation induced by PRS at PND75 in the FC. These additional genes have very low 5MC and 5HMC levels in the control mice. Furthermore, they failed to show enrichment of 5MC and 5HMC in PRS mice, indicating that CpG methylation induced by PRS is gene specific. Gad2 [5MC enrichment on promoter (%)], NS: 0.03 ± 0.005 n = 6; PRS: 0.04 ± 0.01 n = 6; Gad2 [5HMC enrichment on promoter (%)], NS: 0.051 ± 0.01 n = 6; PRS: 0.05 ± 0.007 n = 6; Gapdh [5MC enrichment on promoter (%)], NS: 0.036 ± 0.004, n = 6; PRS: 0.041 ± 0.01 n = 6; Gapdh [5HMC enrichment on promoter (%)], NS: 0.16 ± 0.04, n = 6; PRS: 0.21 ± 0.08 n = 6.
Figure 5.
At PND 75, PRS increased the levels of 5MC and 5HMC on Bdnf gene regulatory regions (II, III, IV, VI and IX) in the FC (A and B) and HP (C and D) of the PRS offspring as compared to the NS group. Data are present as mean ± SEM of 10 mice. * p < .05 (Student’s t-test) vs. the corresponding NS values.
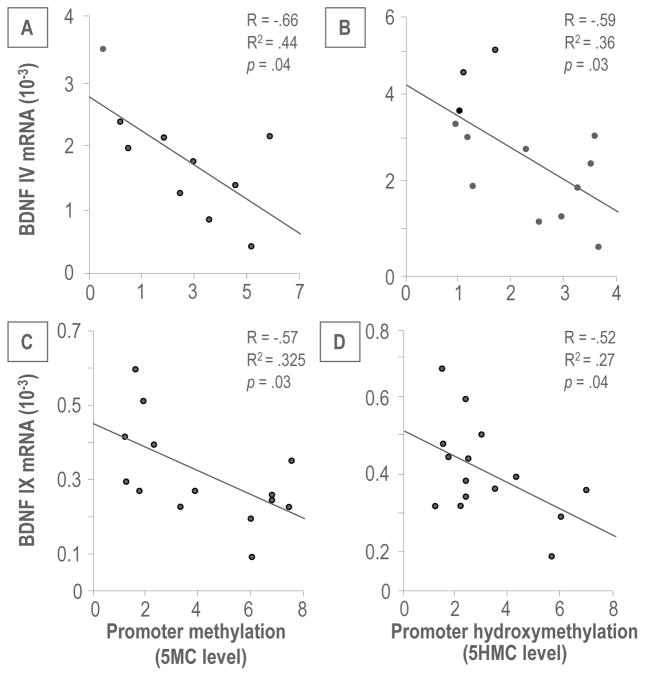
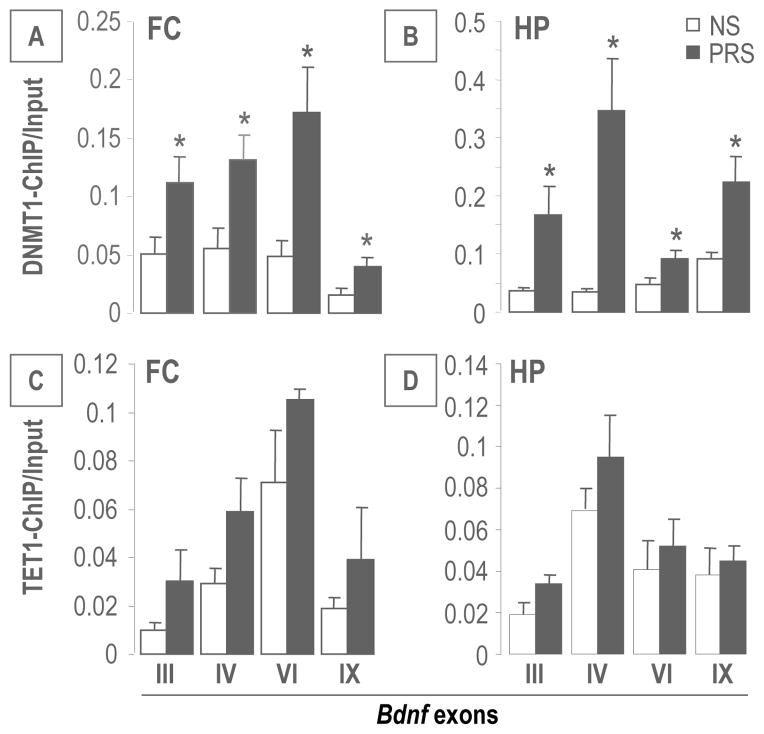
Interestingly, the enrichment of 5MC or 5HMC at Bdnf IV and IX promoters was negatively correlated with the levels of corresponding BDNF transcripts (Figure 6), suggesting an epigenetic mechanism by which promoter methylation/hydroxymethylation may be responsible for the down-regulation of brain BDNF in PRS mice. To elucidate the relationships between global increased DNMT1 and TET1 expressions and elevated Bdnf promoter methylation and hydroxymethylation, we performed ChIP assays with DNMT1 and TET1 specific antibodies in selected Bdnf transcripts: III, IV, VI and IX which were significantly decreased in the FC and HP of PRS offspring. As shown in Figure 7, a significant amount of DNMT1 binds to selected Bdnf III, IV, VI and IX regulatory regions in both FC and HP of PRS mice. In contrast to DNMT1, the binding of TET1 at selected Bdnf promoter regions in the FC and HP of PRS mice was unchanged.
Figure 6.
The enrichment of 5MC and 5HMC on Bdnf exon IV (A and B) and IX (C and D) is negatively correlated with the corresponding transcripts by Pearson correlation analysis.
Figure 7.
PRS causes an increase in DNMT1 and TET1 binding to Bdnf exons (III, IV, VI and IX) in the FC (A and C) and in the HP (B and D) mice compared to controls at PND 75. Data are present as mean ± SEM of 6 mice. * p < .05 (Student’s t-test) vs. the corresponding NS values.
6. PRS fails to alter the expression of other chromatin remodeling factors
In order to probe whether PRS alters the expression of histone tail acetylating or methylating enzymes or other chromatin remodeling factors, we measured the expression of histone deacetylases: HDAC1, HDAC2, HDAC3; histone-lysine N-methyltransferases: MLL1, SETD7, G9A and EZH1; histone lysine-specific demethylase 1A: LSD1 and methyl binding protein MECP2 as these factors are crucial in modulating gene transcription via remodeling chromatin structure. As shown in Figure S1 in Supplement 1, compared to NS, none of these factors showed significant changes in the FC and HP of PRS offspring.
Discussion
The primary objective of this study was to determine whether the SZ-like behavioral phenotype detected in adult PRS mice is accompanied by differences in the expression of epigenetic related biomarkers as previously found in the brain of chronic SZ patients (16). In this study, we report that exposure to stress during gestation leads to a SZ-like behavioral phenotype in adult PRS mice, including locomotor hyperactivity and impairment of sociability, confirming previous reports (3). To establish whether the behavioral abnormalities observed in adult PRS mice may be at least in part related to altered epigenetic mechanisms, we measured the expression of DNMT1 and TETs in FC, HP and cerebellum of adult PRS mice. We focused on DNMT1 and TETs because these enzymes represent epigenetic biomarkers that are increased in the postmortem brain of psychotic patients (13, 14). Furthermore, the altered expression of DNMT1 and TETs is expected to bring about, respectively enrichment of 5MC and 5HMC at regulatory regions of specific SZ candidate genes including Gad1, Reln, and Bdnf, and impact their transcription (3). To establish in detail whether the enrichment of both 5MC and 5HMC at SZ target gene promoters leads to downregulation of their transcripts in PRS mice, we focused on BDNF because this neurotrophin is a significant SZ target gene. There is substantial evidence that Bdnf promoters are hypermethylated and their transcripts are downregulated in the postmortem brain of SZ patients (20, 22, 23, 25).
Our results show that the expressions of DNMT 1 and TETs are elevated in FC and HP but not in cerebellum of adult PRS offspring (which is similar to the brain of SZ patients). The elevation of DNMT1 is associated with increased binding of DNMT1 to regulatory domains of Bdnf promoters along with elevated levels of 5MC. In contrast, the increased levels of TET are associated with an enrichment of 5HMC detected at Bdnf promoters in the absence of increased binding. Although an increase in TETs, which catalyze the oxidation of 5MC to 5HMC, may favor the process of demethylation of hypermethylated CpG rich promoters thereby facilitating transcription, the findings in postmortem SZ brain (14, 20) and in PRS mice suggest that an increase in 5HMC is associated with a downregulation, rather than an upregulation, of BDNF (Figure 3), GAD1 and RELN (3) expression. Therefore, in addition to DNA-methylation (triggered by DNMT) and DNA-demethylation (triggered by TET), chromatin remodeling factors, including repressor proteins such as MECP2 or enzymes that induce covalent modification of histone lysine, may play a role in the epigenetic regulation of BDNF expression. Although the expression of histone factors failed to change in the PRS group, we cannot exclude the possibility that posttranslational modifications of the chromatin remodeling proteins (i.e., MECP2 or HDAC phosphorylation) could contribute to the epigenetic modifications detected in PRS mice.
Taken together, the data suggest that epigenetic modifications of GABAergic and glutamatergic genes detected in SZ and BP disorder patients are also induced by prenatal stress in mice. Hence, the offspring of mice subjected to restraint stress during pregnancy are vulnerable to the development of behavioral abnormalities similar to those observed in psychotic patients. These abnormalities include deficits in social interaction, PPI and fear conditioning (3, 32, 33), and these behavioral abnormalities associate with specific epigenetic changes in the brain (20).
SZ and BP disorder have a natural course, typically starting with a prodromal phase, a first episode during adolescence or early adulthood and repeated episodes eventually leading to a deterioration that ensues over subsequent adult years. Hence, the epigenetic history of such a complex neurodevelopmental disorder changing at each stage of the illness cannot be adequately studied solely in postmortem brain of chronic SZ patients. In other words, each post-mortem sample provides a unique window into a single time point of a chronic illness that is also progressive. To overcome these limitations, we have focused on studying the epigenetic signature and SZ-like behaviors in offspring of PRS mice as a function of postnatal maturation. Since the level of brain DNMT1 and TET are very high at birth and then decrease dramatically with age, it is plausible that DNMT- and TET-induced GABAergic and glutamatergic stress related changes during embryonic life are the basis for a disturbance in the reciprocal interactions between GABAergic, glutamatergic, and monoaminergic neurons that is the likely source of the cognitive and emotional disruptions underlying psychotic symptoms (42). This theory is supported by the fact that psychotic symptoms and SZ-like behaviors in PRS mice can be exacerbated by the administration of NMDA receptor antagonists (3, 42).
To further investigate the hypothesis that prenatal stress, by increasing promoter methylation of GABAergic or glutamatergic genes, may be responsible for the epigenetic alterations of GABA/glutamate neuron interactions in PRS mice, we administered valproic acid (VPA) and clozapine to adult PRS mice in doses that are known to act on chromatin remodeling by inducing Reln and Gad1 promoter demethylation (14, 43). We observed that VPA and clozapine abolish the hyperactivity, stereotypy, and deficits in social interaction and PPI displayed by PRS mice, whereas no effects are observed in vehicle-treated mice. The specificity of drug action in PRS mice is consistent with an epigenetic mechanism underlying PRS behavioral pathology.
These preclinical studies in mice support the concept that the PRS model has construct face validity and pharmacological utility as an experimental epigenetic model of SZ/BP disorder. Furthermore, the PRS model could theoretically be used to predict the course of SZ-like behavioral pathology and more importantly to predict treatment response at different stages of the illness, with particular attention to the early detection of the disease.
Supplementary Material
Acknowledgments
This work was supported by National Institutes Health Grants: R01MN093348 and R01MH101043 to A. Guidotti.
Footnotes
Conflict of Interest
None of the authors have any disclosures to make about actual or potential financial or other conflicts of interests.
References
- 1.Monk C, Spicer J, Champagne FA. Linking prenatal maternal adversity to developmental outcomes in infants: the role of epigenetic pathways. Dev Psychopathol. 2012;24(4):1361–76. doi: 10.1017/S0954579412000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Markham JA, Koenig JI. Prenatal stress: role in psychotic and depressive diseases. Psychopharmacology. 2011;214:89–106. doi: 10.1007/s00213-010-2035-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matrisciano F, Tueting P, Dalal I, Kadriu B, Grayson DR, Davis JM, Nicoletti F, Guidotti A. Epigenetic modifications of GABAergic interneurons are associated with the schizophrenia-like phenotype induced by prenatal stress in mice. Neuropharmacology. 2013;68:184–94. doi: 10.1016/j.neuropharm.2012.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gillott A, Standen PJ. Levels of anxiety and sources of stress in adults with autism. J Intellect Disabil. 2007;11:359–370. doi: 10.1177/1744629507083585. [DOI] [PubMed] [Google Scholar]
- 5.Rice F, Jones I, Thapar A. The impact of gestational stress and prenatal growth on emotional problems in offspring: a review. Acta Psychiatr Scand. 2007;115:171–183. doi: 10.1111/j.1600-0447.2006.00895.x. [DOI] [PubMed] [Google Scholar]
- 6.Walker FR, Knott B, Hodgson DM. Neonatal endotoxin exposure modifies the acoustic startle response and circulating levels of corticosterone in the adult rat but only following acute stress. J Psychiatr Res. 2008;42:1094–1103. doi: 10.1016/j.jpsychires.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 7.Charil A, Laplante DP, Vaillancourt C, King S. Prenatal stress and brain development. Brain Res Rev. 2010;65:56–79. doi: 10.1016/j.brainresrev.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 8.Matrisciano F, Tueting P, Maccari S, Nicoletti F, Guidotti A. Pharmacological activation of group-II metabotropic glutamate receptors corrects a schizophrenia-like phenotype induced by prenatal stress in mice. Neuropsychopharmacology. 2012;37(4):929–38. doi: 10.1038/npp.2011.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McGowan PO, Szyf M. The epigenetics of social adversity in early life: implications for mental health outcomes. Neurobiol Dis. 2010;39(1):66–72. doi: 10.1016/j.nbd.2009.12.026. [DOI] [PubMed] [Google Scholar]
- 10.Zhang TY, Labonté B, Wen XL, Turecki G, Meaney MJ. Epigenetic mechanisms for the early environmental regulation of hippocampal glucocorticoid receptor gene expression in rodents and humans. Neuropsychopharmacology. 2013;38:111–23. doi: 10.1038/npp.2012.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gretha Boersma J, Richard Lee S, Zachary Cordner A, Erin Ewald R, Ryan Purcell H, Alexander Moghadam A, Kellie Tamashiro L. Prenatal stress decreases Bdnf expression and increases methylation of Bdnf exon IV in rats. Epigenetics. 2014;9(3):1–11. doi: 10.4161/epi.27558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Veldic M, Kadriu B, Maloku E, Agis-Balboa RC, Guidotti A, Davis JM, et al. Epigenetic mechanisms expressed in basal ganglia GABAergic neurons differentiate schizophrenia from bipolar disorder. Schizophr Res. 2007;91:51–61. doi: 10.1016/j.schres.2006.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruzicka WB, Zhubi A, Veldic M, Grayson DR, Costa E, Guidotti A. Selective epigenetic alteration of layer I GABAergic neurons isolated from prefrontal cortex of schizophrenia patients using laser-assisted microdissection. Mol Psychiatry. 2007;4:385–397. doi: 10.1038/sj.mp.4001954. [DOI] [PubMed] [Google Scholar]
- 14.Dong E, Gavin D, Chen Y, Davis J. Up-regulation of TET1 and Down-regulation of APOBEC3A and APOBEC3C in the Parietal Cortex of Psychotic Patients. Translational Psychiatry. 2012;2:e159. doi: 10.1038/tp.2012.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhubi A, Chen Y, Dong E, Cook EH, Guidotti A, Grayson DR. Increased binding of MeCP2 to the GAD1 and RELN promoters may be mediated by an enrichment of 5-hmC in autism spectrum disorder (ASD) cerebellum. Transl Psychiatry. 2014;21(4):e349. doi: 10.1038/tp.2013.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grayson DR, Guidotti A. The dynamics of DNA methylation in schizophrenia and related psychiatric disorders. Neuropsychopharmacology. 2013;38:138–66. doi: 10.1038/npp.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abdolmaleky HM, Cheng KH, Russo A, Smith CL, Faraone SV, Wilcox M, et al. Hypermethylation of the reelin (RELN) promoter in the brain of schizophrenic patients: a preliminary report. Am J Med Genet B Neuropsychiatr Genet. 2005;134B:60–66. doi: 10.1002/ajmg.b.30140. [DOI] [PubMed] [Google Scholar]
- 18.Akbarian S. Epigenetics of schizophrenia. Curr Top Behav Neurosci. 2010;4:611–628. doi: 10.1007/7854_2010_38. [DOI] [PubMed] [Google Scholar]
- 19.Auta J, Smith RC, Dong E, Tueting P, Sershen H, Boules S, Lajtha A, Davis J, Guidotti A. DNA-methylation gene network dysregulation in peripheral blood lymphocytes of schizophrenia patients. Schizophr Res. 2013;150(1):312–8. doi: 10.1016/j.schres.2013.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gavin DEP, Sharma RP, Chase KA, Matrisciano F, Dong E, Guidotti A. Growth arrest and DNA-damage-inducible, beta (GADD45b)-mediated DNA demethylation in major psychosis. Neuropsychopharmacology. 2012;2:531–542. doi: 10.1038/npp.2011.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson Ray M, Weickert CS, Wyatt E, Webster MJ. Decreased BDNF, trkB-TK+ and GAD67 mRNA expression in the hippocampus of individuals with schizophrenia and mood disorders. J Psychiatry Neurosci. 2011;36(3):195–203. doi: 10.1503/jpn.100048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong J, Hyde TM, Cassano HL, Deep-Soboslay A, Kleinman JE, Weickert CS. Promoter specific alterations of brain-derived neurotrophic factor mRNA in schizophrenia. Neuroscience. 2010;169(3):1071–84. doi: 10.1016/j.neuroscience.2010.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weickert CS, Hyde TM, Lipska BK, Herman MM, Weinberger DR, Kleinman JE. Reduced brain-derived neurotrophic factor in prefrontal cortex of patients with schizophrenia. Mol Psychiatry. 2003;8(6):592–610. doi: 10.1038/sj.mp.4001308. [DOI] [PubMed] [Google Scholar]
- 24.Gavin DP, Akbarian S. Epigenetic and post-transcriptional dysregulation of gene expression in schizophrenia and related disease. Neurobiol Dis. 2012;46:255–262. doi: 10.1016/j.nbd.2011.12.008. [DOI] [PubMed] [Google Scholar]
- 25.Ikegame T, Bundo M, Murata Y, Kasai K, Kato T, Iwamoto K. DNA methylation of the BDNF gene and its relevance to psychiatric disorders. J Human Genetics. 2013;58:434–438. doi: 10.1038/jhg.2013.65. [DOI] [PubMed] [Google Scholar]
- 26.Autry AE, Monteggia LM. Brain-derived neurotrophic factor and neuropsychiatric disorders. Pharmacol Rev. 2012;64(2):238–58. doi: 10.1124/pr.111.005108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keller S, Sarchiapone M, Zarrilli F, Ferraro A, Carli V, et al. Increased BDNF promoter methylation in the Wernicke area of suicide subjects. Arch Gen Psychiatry. 2010;67:258–267. doi: 10.1001/archgenpsychiatry.2010.9. [DOI] [PubMed] [Google Scholar]
- 28.Karpova NN. Role of BDNF epigenetics in activity-dependent neuronal plasticity. Neuropharmacology. 2014;76(Pt C):709–18. doi: 10.1016/j.neuropharm.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 29.Martinowich K, Manji H, Lu B. New insights into BDNF function in depression and anxiety. Nat Neurosci. 2007;10(9):1089–93. doi: 10.1038/nn1971. [DOI] [PubMed] [Google Scholar]
- 30.Tadić A, Müller-Engling L, Schlicht KF, Kotsiari A, Dreimüller N, Kleimann A, Bleich S, Lieb K, Frieling H. Methylation of the promoter of brain-derived neurotrophic factor exon IV and antidepressant response in major depression. Mol Psychiatry. 2014;19(3):281–3. doi: 10.1038/mp.2013.58. [DOI] [PubMed] [Google Scholar]
- 31.Stevens HE, Su T, Yanagawa Y, Vaccarino FM. Prenatal stress delays inhibitory neuron progenitor migration in the developing neocortex. Psychoneuroendocrinology. 2013;38(4):509–21. doi: 10.1016/j.psyneuen.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fine R, Zhang J, Stevens HE. Prenatal stress and inhibitory neuron systems: implications for neuropsychiatric disorders. Mol Psychiatry. 2014;19(6):641–51. doi: 10.1038/mp.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ignacio-Negron-Oyarzo, Neira D, Espinosa N, Fuentealba, Aboitiz F. Prenatal stress produces persistence of remote memory and disrupts functional connectivity in the hippocampal-prefrontal cortex axis. [Access published May 23];Cerebral Cortex Advance. 2014 doi: 10.1093/cercor/bhu108. [DOI] [PubMed] [Google Scholar]
- 34.Carboni G, Tueting P, Tremolizzo L, Sugaya I, Davis J, Costa E, Guidotti A. Enhanced dizocilpine efficacy in heterozygous reeler mice relates to GABA turnover downregulation. Neuropharmacology. 2004;46(8):1070–81. doi: 10.1016/j.neuropharm.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 35.McFarlane HG, Kusek GK, Yang M, Phoenix JL, Bolivar VJ, Crawley JN. Autism-like behavioral phenotypes in BTBR T+tf/J mice. Genes Brain Behav. 2008;7(2):152–63. doi: 10.1111/j.1601-183X.2007.00330.x. [DOI] [PubMed] [Google Scholar]
- 36.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jin SG, Wu X, Li AX, Pfeifer GP. Genomic mapping of 5-hydroxymethylcytosine in the human brain. Nucleic Acids Res. 2011;39:5015–5024. doi: 10.1093/nar/gkr120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32:4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dong E, Nelson M, Grayson DR, Costa E, Guidotti A. Clozapine and sulpiride but not haloperidol or olanzapine activate brain DNA demethylation. Proc Natl Acad Sci U S A. 2008;105(36):13614–9. doi: 10.1073/pnas.0805493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, Pow-Anpongkul N, et al. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aid T, Kazantseva A, Piirsoo M, Palm K, Timmusk T. Mouse and rat BDNF gene structure and expression revisited. J Neurosci Res. 2007;85(3):525–35. doi: 10.1002/jnr.21139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lisman JE1, Coyle JT, Green RW, Javitt DC, Benes FM, Heckers S, Grace AA. Circuit-based framework for understanding neurotransmitter and risk gene interactions in schizophrenia. Trends Neurosci. 2008;31(5):234–42. doi: 10.1016/j.tins.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guidotti A, Dong E, Kundakovic M, Satta R, Grayson DR, Costa E. Characterization of the action of antipsychotic subtypes on valproate-induced chromatin remodeling. Trends Pharmacol Sci. 2009;30(2):55–60. doi: 10.1016/j.tips.2008.10.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.