Abstract
Lung cancer is the leading cause of cancer-related fatalities. Recent success developing genotypically-targeted therapies, with potency only in well-defined subpopulations of tumors, suggests a path to improving patient survival. We utilized a library of oligonucleotide inhibitors to microRNAs, a class of post-transcriptional gene regulators, to identify novel synthetic lethal interactions between miRNA inhibition and molecular mechanisms in NSCLC. Two inhibitors, those for miR-92a and miR-1226*, produced a toxicity distribution across a panel of 27 cell lines that correlated with loss of p53 protein expression. Notably, depletion of p53 was sufficient to confer sensitivity to otherwise resistant telomerase-immortalized bronchial epithelial cells. We found that both miR inhibitors cause sequence-specific down-regulation of the miR-17~92 polycistron, and this down-regulation was toxic only in the context of p53 loss. Mechanistic studies indicated the selective toxicity of miR-17~92 polycistron inactivation was the consequence of derepression of vitamin D signaling via suppression of CYP24A1; a rate limiting enzyme in the 1α,25-dihydroxyvitamin D3 metabolic pathway. Of note, high CYP24A1 expression significantly correlated with poor patient outcome in multiple lung cancer cohorts. Our results indicate that the screening approach utilized in this study can identify clinically relevant synthetic lethal interactions, and that vitamin D receptor agonists may show enhanced efficacy in p53-negative lung cancer patients.
Introduction
The existence of defined genetic abnormalities in NSCLC has enabled the development of targeted therapeutic approaches to NSCLC treatment. In particular, therapies targeting tumors carrying mutations in EGFR or a fusion of the EML4 and ALK genes have been clinically successful as first-line therapies (1–3). Targeted therapies, however, sacrifice breadth of treatable tumors for high efficacy in the presence of a specific biomarker: only 25–35% of NSCLC tumors will respond to the EGFR and EML4/ALK targeted therapies, and the current five-year survival rate remains around 15%.
microRNAs (miRNAs) are a class of post-transcriptional regulators of gene expression. In a sequence-driven process mediated by the RNA-Induced Silencing Complex (RISC), the ~22 nucleotide RNAs associate with 3’ untranslated regions (3’ UTRs), leading to down-regulation of their targets (4, 5). miRNA are found throughout the genome as either individual loci, within introns of host genes, or in polycistrons, single transcripts that produce multiple miRNAs. miRNAs have been implicated in developmental processes, drug response, and cancer initiation and progression (6–10), and can function as both tumor promoters (oncomiRs) or tumor suppressors, with some miRNAs able to play either role, depending on the context (11). In a parallel to oncogene addiction, some cancer cells have been shown to be dependent on the expression of a single oncogenic miRNA. For example, while miR-21 has been shown to lead to a pre-B malignant lymphoid-like phenotype, inactivation of miR-21 leads to rapid and complete regression (12).
miRNAs are readily manipulated both in vitro and in vivo, and both gain and loss of miRNA function have been demonstrated to have substantial effects on tumor initiation and progression in in vivo models (6, 13, 14). Oligonucleotides complementary to a mature miRNA competitively bind the miRNA and prevent it from being loaded into the RISC (15). Such inhibitors have been demonstrated to have therapeutic efficacy in in vivo models due to their high target affinity and bioavailability, even without any packaging or carrier (14, 16, 17).
Our goal is to identify synthetic lethal inhibitor:genotype interactions in NSCLC. Here we used a phased screening approach to identify miRNA inhibitors with selective toxicity across a genetically diverse collection of NSCLC cell lines. We were able to use the diversity of the cell lines in tandem with their mutational and transcriptional profiles to identify a dependency on the miR-17~92 cluster that arises after p53 loss in the lung epithelium.
Materials and Methods
Cell lines
Cell lines were obtained from the Hamon Center for Therapeutic Oncology Research at UT Southwestern Medical Center. All cells were grown in a humidified atmosphere with 5% CO2 at 37°C. HBECs and HCC4017 were grown in ACL-4 medium supplemented with 2% FBS (18, 19). All other cell lines were grown in RPMI-1640 medium (Life Technologies, Rockville, MD) supplemented with 5% FBS (Atlanta Biologicals, Lawrenceville, GA). Cell lines were DNA fingerprinted in October 2013 using the GenePrint PowerPlex 1.2 system (Promega, Madison, WI) and confirmed against libraries maintained by ATCC.
Reagents
The miRCURY LNA™ microRNA Inhibitor Library - Human v14.0, was obtained from Exiqon (Denmark). Inhibitors for miR-92a and miR-1226* were obtained from Exiqon and Dharmacon (Chicago, IL) and mismatch and scrambled derivatives were synthesized by Exiqon. siRNA oligos were obtained from Dharmacon. p53 and β-tubulin antibodies were acquired from Santa Cruz Biotechnology (Dallas, TX) and Sigma Aldrich (St. Louis, MO). 1α,25-dihydroxyvitamin D3 was acquired from Sigma Aldrich.
miRNA inhibitor screen
Cells were plated in 96-well format, transfected with oligos and incubated for 72 h, after which medium was changed, and then incubated for an additional 72 h. Cell viability was determined using the CellTiter-Glo® Luminescent Cell Viability Assay (Promega). Luminescence was quantified on a EnVision plate reader (PerkinElmer, Waltham, MA). Raw values were normalized using R (20) and cellHTS2 (21) to obtain cell viability ratios.
Cell viability assay
Cells were plated in 96-well format, transfected with oligos and incubated for 72 h, after which medium was replaced and, as appropriate, supplemented with 1α,25-dihydroxyvitamin D3. Cells were then incubated for 72 h. Cell viability was determined as above. Luminescence was quantified on a PheraStar FS plate reader (BMG LabTech, Ortenberg, Germany).
1α,25-dihydroxyvitamin D3 toxicity assay
Cells were plated in 96-well plates, allowed to recover for 48 h, and then treated with an addition of growth media, growth media plus carrier (ethanol) or growth media plus 1α,25-dihydroxyvitamin D3 for 6 h. Caspase-Glo 3/7 reagent (Promega) was added at equal volume to the growth media and incubated for 1 h before luminescence was quantified as above.
Quantitative real-time PCR
Total RNA was prepared using the Ambion mirVana™ miRNA Isolation Kit (Life Technologies, Foster City, CA). mRNA and miRNA levels were assessed by qRT-PCR using an ABI PRISM 7900HT using predesigned TaqMan® primer and probe sets (Life Technologies, Foster City, CA). RNU19 was used as a loading control for miRNA assays; ACTB and GAPDH were used as loading controls for pri-miRNA and gene expression assays. Threshold cycle times (Ct) were obtained and relative gene expression was calculated using the comparative Ct method.
Gene expression profiling
HBEC30KT and HBEC30KT-shTP53 cells were reverse transfected with 50 nM miR-92a mismatch oligo or miR-92a inhibitor and plated in triplicate in 6-well plates. 48 h post-transfection, total RNA was prepared as above. RNA quality was assessed by Bio-Rad Experion (Bio-Rad, Hercules, CA). RNA was then transcriptionally profiled on Illumina HumanHT-12 v4 Expression BeadChips (Illumina, San Diego, CA) as described previously. Data have been deposited in the GEO public repository under accession number GSE64007. Additional detail is provided in the Supplementary Materials.
Protein expression profiling
Reverse-phase protein arrays (RPPA) were used to measure protein expression, as described previously (22). Additional detail is provided in the Supplementary Materials.
Survival analysis
Gene expression data from 182 lung adenocarcinoma tumor specimens were obtained from GEO dataset GSE41271. Samples were then ranked by CYP24A1 expression and the highest and lowest quintiles were then compared by Mantel-Cox log-rank test. miRNA expression profiles and corresponding clinical data were on 470 LUAD patients were obtained from The Cancer Genome Atlas (TCGA). Samples were then ranked by expression of miR-17-5p, miR-18a-5p, miR-19a-3p, miR-20a-5p, miR-19b-3p and miR-92a-3p, with the highest and lowest quartiles compared for association with patient survival by Mantel-Cox log-rank test.
Statistical analysis
Correlations were calculated by Pearson product-moment coefficient. p-values were determined by two-tailed, unpaired Student’s t-test with p < 0.05 used as a nominal threshold for significance.
Elastic net for predictive biomarker discovery
The elastic net is a penalized linear regression model that can identify multi-feature biomarker signatures whose additive patterns act to predict a response vector, in this case measurements of viability in response to treatment with the miR-92a inhibitor in a panel of cell lines. The elastic net also allows for weighting of identified features to enable predictions of sensitivities in untested cell lines. Candidate predictive features were selected from measures of gene expression (described above), with replicate measurements for the same gene averaged together. In order to determine the optimal parameter values to use in the model, we did 100 iterations of 10 fold cross-validation where, in each iteration, the cells were randomly re-sampled into different groups. Parameter values were chosen so as to give the minimum mean squared error for each fold. The dataset was then subjected to 100 permutations of bootstrapping. Features were then ranked based on weights and bootstrapped frequency of occurrence. Additional detail is provided in the Supplementary Materials.
Results
A miRNA inhibitor screen on two NSCLC cell lines identifies a potential genetic context-dependent vulnerability to miRNA inhibition
To identify miRNA inhibitors with genotype-driven selective toxicity in NSCLC, we implemented a tiered screen based on a comprehensive library of 919 single-stranded competitive inhibitors targeting 870 of the known human miRNAs (miRbase v.14). Two NSCLC cell lines derived from adenocarcinomas, H358 and H1993, which harbor distinct genetic alterations (Supplementary Table 1), were used for screening.
We observed several patterns of response to individual miRNA inhibitors (Figure 1A), confirming that while histologically similar, the two cell lines can generate divergent responses to a specific perturbation. Specifically, 35 inhibitors (3.79%) decreased viability of H358 cells more than 50%, and 11 (1.19%) decreased viability of H1993 cells to the same extent. Among these, the inhibitors of miR-92a and miR-1226* show the most significant selective cytotoxicity to H358. Notably, the inhibitors are identical in sequence at 10 positions, including a CAGGCC motif at their 5’ ends. Testing across a panel of cell lines with diverse genetic backgrounds (Supplementary Table 2) returned a significantly correlated toxicity profile, indicating that the two inhibitors may converge on a specific NSCLC vulnerability (Figure 1B and 1C). Both inhibitors were benign in normal HBEC30-KT cell lines, indicating selectivity within tumorigenic contexts.
Figure 1. A miRNA inhibitor screen identifies distinct toxicity patterns across NSCLC cell lines.
(A) Results from the 923 miRNA inhibitors used in the first phase of the screen. Each dot represents one inhibitor, with normalized cell viability in H358 on the x-axis and in H1993 on the y-axis. Red dots indicate miR-92a and miR-1226* inhibitors. (B) Cytotoxicity of miR-92a and miR-1226* inhibitors, shown as percent viability relative to negative control. The red bar identifies the representative normal cell line. Error bars represent standard deviations. (C) Viability of 27 NSCLC cell lines after miR-92a inhibitor or miR-1226* inhibitor treatment. Each red dot represents one cell line; the dashed line is the line of best fit. Correlation and p-value were determined by Pearson product moment.
miR-92a is a member of the miR-25 family of miRNAs, which includes miR-25, miR-32, miR-92a, miR-92b, miR-363 and miR-367, all of which share a common seed sequence (Supplementary Figure 1A). miR-92a is also a component of the miR-17~92 polycistron, a cluster of co-transcribed miRNAs, including miR-17, miR-18a, miR-19a, miR-20a miR-19b-1 and miR-92a. However none of the related inhibitors, whether by seed sequence or by co-transcription, exhibited cytotoxic effects similar to the miR-92a or miR-1226* inhibitors (Supplementary Figures 1B, 1C).
p53 levels and activity are inversely correlated with miR-92a inhibitor toxicity, and loss of p53 sensitizes a resistant cell line to the miR-92a inhibitor
Therapeutic approaches based on genotype have shown promising returns in NSCLC (2, 23). We therefore sought a genetic predictor of response to the miR-92a/miR-1226* inhibitors. Interrogating the mutational status and gene expression of known oncogenes and tumor suppressors in NSCLC for correlations with inhibitor toxicity revealed an inverse correlation with TP53 mRNA accumulation (Figure 2A). This was recapitulated by p53 protein abundance as quantified by reverse-phase protein arrays (Figure 2B).
Figure 2. Loss of p53 sensitizes immortalized normal and tumor-derived NSCLC cells to miR-92a inhibitor-induced toxicity.
(A) TP53 mRNA level versus cell viability after transient transfection of 50 nM of miR-92a inhibitor, normalized to viability observed with a negative control oligo. Viability was measured 144 h after transfection of the inhibitor. (B) p53 protein level versus cell viability after transfection with 50 nM miR-92a inhibitor. Each point represents one cell line. The lines of best fit were determined by linear regression. Correlation coefficients and p-values were determined by Pearson product moment. (C) Genes that are decreased in HBEC30KT-shTP53 relative to HBEC30KT, and high in cell lines sensitive to the miR-92a inhibitor, as identified by hypergeometric test. (D) Genes identified as features that alone or in combination are predictive of response to the miR-92a inhibitor in the panel of 22 cell lines. Yellow and blue bars represent positive and negative correlations with viability, respectively. (E) p53 levels in H358 and H1993 cell lines 72h after a 50 nM transfection of either a negative control inhibitor or the miR-92a inhibitor, as determined by western blot. (F) p53 levels in HBEC30-KT and HBEC30KT-shTP53 cells and viability of HBEC30-KT and HBEC30KT-shTP53 72h after transfection with 50 nM of either the miR-92a inhibitor, the miR-1226* inhibitor or a negative control inhibitor. (G) qRT-PCR quantification of miR-92a in H358 and H1993 cells 72 h following transfection with either a LNA- or RNA hairpin-based miR-92a inhibitor or negative control inhibitors. Fold change is relative to the negative control transfection in each panel. (H) Cell viability in H358 and H1993 cells 144 h after transfection with 50 nM of miR-92a inhibitor, miR-1226* inhibitor or negative control inhibitor. (I–J) Cell viability in H358 and HBEC30KT-shTP53 cells 144 h after transfection with either unmodified or sequence-modified versions of the miR-92a and miR-1226* inhibitors, or a negative control inhibitor, as described in Supplementary Table 2. In all panels, results are shown as means and standard deviations of triplicate measurements, relative to the negative control. p-values were determined by two-tailed, unpaired Student’s t-test. ****, p < 0.0001.
To assess the relationship between miR-92a inhibitor response and p53 activity, we employed a genetic model of p53 loss: an immortalized human bronchial epithelial cell line, HBEC30-KT, was modified to constitutively express a short hairpin RNA (shRNA) to TP53 (24), creating a TP53 knockdown cell line HBEC30KT-shTP53. Whole genome transcript arrays from the cell lines on the tails of the viability distribution (the 6 most sensitive and the 6 most resistant lines shown in Figure 1C), obtained from GEO dataset GSE32036, were used to identify the top 1% of differentially expressed genes by their signal-to-noise (S2N) values, calculated as (μsen − μres)/(σsen + σres), where μ and σ represent the mean and standard deviation of expression, respectively, for each class. We next defined p53-dependent genes as those with at least 2-fold differential accumulation in HBEC30KT versus HBEC30KT-shTP53 cells. A hypergeometric distribution test indicated significant overlap between genes that are decreased in HBEC30KT-shTP53 cells and high in the cell lines sensitive to the miR-92a inhibitor (p = 6.2×10−5), as shown in Figure 2C. The 27 genes in the overlap are shown in Supplementary Table 3. We also used an elastic net regularization and variable selection method to identify expression features that alone or in combination are associated with the full viability distribution across all 22 cell lines, and asked if the identified features are part of the p53 response network. 55 genes were identified, as shown in Figure 2D, again with significant over-representation of p53 network genes; p = 0.011 by hypergeometric test. The 55 genes are listed with weights and frequencies in Supplementary Table 4.
These results show that there is a significant 'p53-like' gene signature associated with response to the miR-92a inhibitor and that p53 network genes are sufficient to act as a multi-feature signature whose combinatorial expression can predict cellular response to the miR-92a inhibitor. H358, the sensitive cell line from the first phase of the screen, had the lowest observed TP53 and p53 expression of any of the cell lines in the panel (Figure 2A). However, p53 abundance did not change in response to transfection with miR-92a inhibitor in either the sensitive or resistant cell line as measured by western blot 72 h post-treatment (Figure 2E), indicating that the effect of miR-92a inhibitor on cell survival is not through regulation of TP53 expression.
To address the necessity and sufficiency of p53 loss with respect to cellular response to the miR-92a inhibitor, we returned to the HBEC30KT-shTP53 model. As expected, the unmodified HBEC30-KT did not respond to the miR-92a inhibitor, while the knockdown of p53 was sufficient to render the cell line sensitive to the miR-92a inhibitor (Figure 2F).
Depletion of mature miR-92a is insufficient to induce toxicity in p53-depleted cell lines
One potential pitfall in any experiment involving short oligonucleotides is phenotypes caused by off-target effects (25) of the oligos. We therefore employed a second design, a double-hairpin miRNA inhibitor (miRidian miRNA inhibitors, Dharmacon). We observed successful knockdown of mature miR-92a with inhibitors based on either the LNA or RNA hairpin inhibitors in both cell lines (Figure 2G). However, the hairpin miR-92a inhibitor did not reproduce the toxicity of the LNA inhibitor (Figure 2H). Interestingly, the miR-1226* inhibitor replicated the phenotypes observed with the miR-92a inhibitors, which is consistent with their correlated toxicities.
In order to establish the sequence-dependence of the toxicity of the LNA-based miRNA inhibitors, we designed variants of the miR-92a and miR-1226* inhibitors (Supplementary Table 5). As shown in Figure 2I and 2J, none of the variants displayed significant cytotoxicity in either H358 or HBEC30KT-shTP53 cells.
The miR-17~92 primary transcript is the likely mediator of the cytotoxicity of miR-92a inhibitor
Given the above observation, the sequence similarity between the inhibitors led us to consider sequence-dependent genomic targets other than mature miR-92a that they may both regulate. The transcript with the highest sequence complementarity to the miR-92a and miR-1226* inhibitors is the primary transcript of the miR-17~92 polycistron, a cluster of miRNAs in a single primary transcript (Figure 3A). In order to address whether the levels of the primary transcript of miR-17~92 were altered in response to treatment with the miR-92a inhibitor, we measured its expression in both miR-92a-sensitive and -resistant cells 72 h following transfection of the inhibitor. We observed a decrease in the miR-17~92 primary transcript in all four cell lines (Figure 3B) following transfection of the LNA miR-92a inhibitor, although the extent of the decrease in HBEC30-KT cells was much less than in the other three cell lines. The RNA hairpin inhibitor to miR-92a, however, showed no effect on primary transcript levels in either H358 or H1993 cells (Figure 3C). Treatment with the miR-1226* inhibitor also resulted in depletion of the miR-17~92 primary transcript (Figure 3D), but did not deplete mature miR-92a in H358 cells (Figure 3E), further confirming that inhibition of miR-92a is not required for toxicity. We also observed significantly higher endogenous levels of miR-17~92 primary transcript in tumor cells versus normal cells, and in resistant cell lines relative to sensitive cell lines (Figure 3F), consistent with previous reports demonstrating up-regulation of miR-17~92 in NSCLC (7). The low levels of the primary transcript in HBEC30-KT may explain the lack of toxicity of the inhibitors (Figure 1C). Taken together, these results suggest that the cytotoxic effects of the LNA-based miR-92a and miR-1226* inhibitors are mediated by knock-down of the miR-17~92 primary transcript.
Figure 3. The miR-92a inhibitor down-regulates the miR-17~92 primary transcript and dysregulates the expression of mature miRNAs from the miR-17~92 polycistron.
(A) The miR-17~92a polycistron with predicted interactions between the primary transcript and the miR-92a and miR-1226* inhibitors. (B) qRT-PCR for the primary transcript (pri-miRNA) of the miR-17~92a polycistron in H358, H1993, HBEC30KT-shTP53 and HBEC30-KT cells at two locations: one in the miR-18a locus (miR-18-pri) and one in the miR-92a-1 locus (miR-92a-1-pri). RNA was collected 72 h post-transfection with either the miR-92a inhibitor or a negative control inhibitor. (C) qRT-PCR for miR-18-pri and miR-92a-1-pri in H358 and H1993 cells 72 h post-transfection with either the RNA hairpin miR-92a inhibitor or a negative control inhibitor. (D) qRT-PCR for miR-18-pri and miR-92a-1-pri in H358 cells 72 h post-transfection with either the miR-1226* inhibitor or a negative control inhibitor. (E) Relative abundance of miR-92a following treatment with either miR-92a inhibitor or miR-1226* inhibitor. (F) Relative abundance of pri-miR-18 and pri-miR-92a in H358, H1993, HBEC30KT-shTP53 and HBEC30-KT cells. Ct values were normalized to loading control and then to H358. (G–J) Levels of mature miRNAs from the miR-17~92a polycistron 72 h post-transfection with either the LNA miR-92a inhibitor or the mismatched control inhibitor in H358, H1993, HBEC30KT-shTP53, or HBEC30KT cells, as measured by qRT-PCR. In all panels, results are shown as means and standard deviations of triplicate measurements, relative to the negative control. p-values determined by Student’s t-test. **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
Given the depletion of the miR-17~92 primary transcript, we assayed the four cell lines for the abundance of the mature miRNAs from the miR-17~92 cluster following a 72 h transfection with LNA miR-92a inhibitor. We observed depletion of mature miR-18a, miR-20 and miR-92a in H358 cells (Figure 3G) and of mature miR-19a, miR-19b and miR-92a in H1993 cells (Figure 3H). The miR-92a locus demonstrated the most dramatic knockdown in all cases. All of the mature miRNAs from the locus were depleted in HBEC30KT-shTP53 cells while only miR-92a was depleted in HBEC30KT cells (Figure 3I–J). The down-regulation of mature miRNAs in sensitive cell lines is consistent across both the NSCLC cell lines and HBECs, and is consistent with the synthetic lethal relationship between the miR-92a inhibitor, the miR-17~92 cluster and p53 loss. Notably, in mouse cell models derived from a standard murine model for lung adenocarcinoma, we observed a decrease in levels of mature miR-92a in response to the miR-92a inhibitor, but no effect on the levels of the other mature miRNAs from the cluster or on levels of the primary transcript (Supplementary Figure 2). In addition, treatment with miR-92a inhibitor had no effect on cell viability relative to the mismatch control inhibitor, further indicating the combinatorial participation of the miRNAs in the miR-17~92 locus in support of human p53 hypomorphs.
Expression profiling identifies a group of target genes of the miR-17~92 cluster that are de-repressed by miR-92a inhibitor in p53-depleted cells, but not in p53-wildtype cells
In order to identify the candidate genes responsible for the response of p53-depleted cells to the miR-92a inhibitor, we profiled the two HBEC cell lines for changes in mRNA expression following transfection of the miR-92a inhibitor or control. Consistent with the observation that the miR-92a inhibitor can deplete the miR-17~92 cluster, we observed a corresponding up-regulation of genes in the miR-17~92 target space (Supplementary Figure 3A). Notably, we observe that a substantially larger number of targets were up-regulated in reponse to the miR-92a inhibitor in HBEC30KT-shTP53 cells (141, 64.6%) relative to HBEC30KT cells (29, 35.6%) (Supplementary Figure 3B). These results are consistent with the observation that HBEC30KT is resistant to miR-92a inhibitor-induced cytotoxicity.
The six miRNAs in the miR-17~92 primary transcript (miR-17 and miR-20; miR-19a and miR-19b; miR-92a) include three seed-sequence families and one miRNA with a unique seed sequence which appear to share a combinatorial target space (Supplementary Figure 3C).
miR-92a and p53 converge on CYP24A1, a key element of the vitamin D3 response network, in regulating cell survival
Analysis of the 712 transcripts (representing 675 genes), modulated more than 2-fold by the miR-92a inhibitor in HBEC30KT-shTP53 cells, with the “Upstream Regulators” IPA module identified inflammation-related pathways as potential regulators of the observed differential expression, among which 1α,25-dihydroxyvitamin D3 pathway ranked second (Table 1). 1α,25-dihydroxyvitamin D3 is the active form of vitamin D3, (26–29), and has shown efficacy in reducing proliferation and inducing apoptosis in cancer cells (30–32). We found that 1α,25-dihydroxyvitamin D3 activated Caspase 3/7 by approximately 4-fold compared to control in the miR-92a inhibitor-sensitive H358 cells, but not in the miR-92a inhibitor-resistant H1993 cells (Figure 4B), consistent with the observed activation of Caspase 3/7 by miR-92 inhibitor (Figure 4C). Furthermore, analysis of the miR-17, miR-18, miR-19, miR-20 and miR-92 target genes and the transcriptional network of 1α,25-dihydroxyvitamin D3 shows that there is an overlap between the miR-17~92 targetome and the 1α,25-dihydroxyvitamin D3 response network (Supplementary Figure 3C). Altogether, these results suggest that the 1α,25-dihydroxyvitamin D3 response network is likely a key mediator of the miR-92a inhibitor cytotoxicity.
Table 1.
Twenty most significant upstream regulators in HBEC30KT-shTP53 treated with miR-92a inhibitor relative to mismatch inhibitor.
| Upstream Regulator | Molecule Type | Activation z- score |
p-value of overlap |
|---|---|---|---|
| TGFB1 | growth factor | 5.627 | 7.20×10−18 |
| 1α,25-dihydroxy vitamin D3 | chemical drug | 3.446 | 1.17×10−15 |
| CDKN1A | kinase | 2.173 | 8.85×10−11 |
| IgG | complex | −3.145 | 1.32×10−10 |
| TP53 | transcription regulator | 2.807 | 7.18×10−10 |
| let-7 | microRNA | 2.706 | 1.36×10−09 |
| MAPK1 | kinase | 2.268 | 2.64×10−09 |
| IRF7 | transcription regulator | −4.240 | 2.71×10−09 |
| CCND1 | other | −3.317 | 3.73×10−09 |
| IFNL1 | cytokine | −3.046 | 3.01×10−08 |
| tretinoin | chemical - endogenous mammalian | 2.625 | 4.04×10−08 |
| IFNB1 | cytokine | −2.255 | 1.78×10−07 |
| SMARCB1 | transcription regulator | 3.000 | 3.14×10−07 |
| hydrogen peroxide | chemical - endogenous mammalian | 2.114 | 1.57×10−06 |
| E2F2 | transcription regulator | −2.200 | 1.79×10−06 |
| IL4 | cytokine | 2.647 | 1.94×10−06 |
| IFNA2 | cytokine | −2.915 | 2.42×10−06 |
| IRGM | other | 3.000 | 3.75×10−06 |
| IRF3 | transcription regulator | −2.850 | 6.57×10−06 |
| IL1RN | cytokine | 2.392 | 1.27×10−05 |
Shown are upstream transcriptional regulators consistent with the observed changes in gene expression, activation z-score and overlap p-value. Activation z-score and overlap p-value are derived by comparing the known targets of each regulator with observed differential expression, with respect to number of genes and magnitude and direction of change. Significance is generally attributed to p values < 0.01 and activation z-scores greater than 2 or smaller than −2.
Figure 4. miR-92a inhibitor mimics 1α,25-dihydroxyvitamin D3 signaling and predicts sensitivity to 1α,25-dihydroxyvitamin D3 treatment.
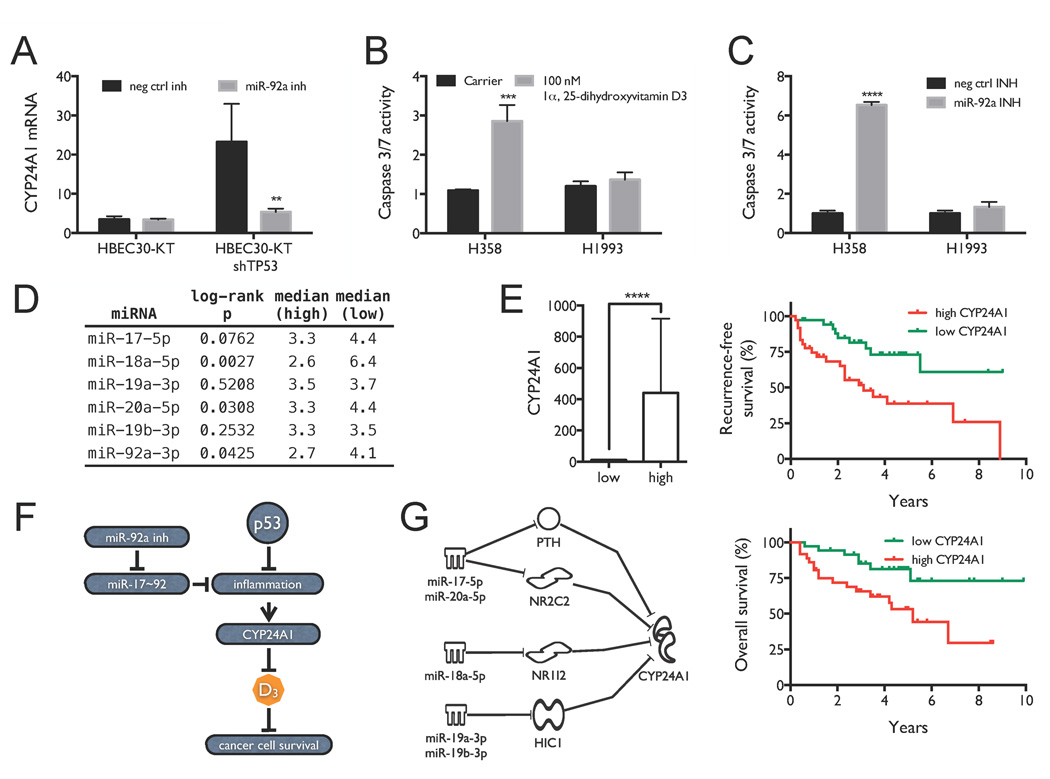
(A) Quantification of CYP24A1 expression. (B) Selective activation of Caspase 3/7 in H358 cells 6 h post treatment with 100 nM 1α,25-dihydroxyvitamin D3. (C) Selective activation of Caspase 3/7 in H358 cells 72 h post-transient transfection with the miR-92a inhibitor or control. Results are shown as means and standard deviations of triplicate measurements. Significance was calculated by Student’s two-tailed t-test. (D) Survival analysis for mature miRNAs from the miR-17~92 cluster. TCGA adenocarcinoma patients were stratified based on tumor levels of miR-17-5p, miR-18a-5p, miR-19a-3p, miR-20a-5p, miR-19b-3p and miR-92a-3p. Shown are log-rank p-values and median survival (y) in the low (n = 116) and high (n = 116) groups. (E) Survival analysis for CYP24A1 in lung adenocarcinoma patients. Patients were stratified based on tumor levels of CYP24A1, taking the top and bottom quintiles. CYP24A1 expression in the two groups is shown on the left. Associations between CYP24A1 levels and recurrence-free and overall survival are shown on the right. (F) Working model for miR-17~92 dependence in p53-depleted NSCLC, showing the interactions between miR-92a inhibitor, miR-17~92, p53 and 1α,25-dihydroxyvitamin D3 in NSCLC cellular growth and survival. (G) Working model for how repression of the miR-17~92 primary transcript by miR-92a and miR-1226* inhibitors can depress target genes that regulate CYP24A1.
Similar analysis of upstream regulators for the 3,842 transcripts (representing 3,586 genes) that are modulated by TP53 knock-down in HBEC30KT cells also returned an inflammation-like response (Table 2). Of the top 20 upstream regulators identified, nine have known roles in inflammation and immune response, including six cytokines (TNF, IFNG, IL1B, OSM, IFNL1 and IFNA2, all of which are consistently activated after p53 loss), lipopolysaccharide, TLR3 and the NFκB complex. This observation suggests that inflammation-related pathways may confer a survival advantage to p53 hypomorphs but also confer collateral addiction to miR-17~92 expression.
Table 2.
Twenty most significant upstream regulators in HBEC30KT-shTP53 versus HBEC30KT.
| Upstream Regulator | Molecule Type | Activation z- score |
p-value of overlap |
|---|---|---|---|
| TNF | cytokine | 7.189 | 6.18×10−55 |
| TGFB1 | growth factor | −2.258 | 1.10×10−50 |
| IFNG | cytokine | 8.041 | 9.68×10−45 |
| lipopolysaccharide | chemical drug | 7.310 | 3.25×10−42 |
| NKX2-3 | transcription regulator | −4.911 | 1.86×10−41 |
| TP53 | transcription regulator | −3.677 | 2.93×10−38 |
| MAPK1 | kinase | −4.847 | 1.46×10−31 |
| OSM | cytokine | 4.194 | 3.00×10−31 |
| IL1B | cytokine | 5.596 | 7.98×10−31 |
| dexamethasone | chemical drug | −6.822 | 1.35×10−29 |
| HRAS | enzyme | 2.353 | 1.05×10−26 |
| NFkB (complex) | complex | 6.191 | 3.39×10−26 |
| IFNL1 | cytokine | 6.570 | 1.40×10−23 |
| IFNA2 | cytokine | 6.380 | 2.42×10−23 |
| poly rI:rC-RNA | chemical reagent | 7.265 | 1.99×10−22 |
| TLR3 | transmembrane receptor | 4.977 | 5.75×10−22 |
| TP63 | transcription regulator | 3.700 | 2.09×10−21 |
| STAT3 | transcription regulator | 2.076 | 3.20×10−21 |
| EZH2 | transcription regulator | 3.200 | 8.20×10−21 |
| CHUK | kinase | 3.898 | 1.46×10−20 |
Within the functional network described above, CYP24A1 was significantly up-regulated in HBEC30KT-shTP53 cells compared to HBEC30KT cells. Notably, CYP24A1 is a rate-limiting regulator of 1α,25-dihydroxyvitamin D3 degradation (32) and directly affects the intracellular half-life of 1α,25-dihydroxyvitamin D3. CYP24A1 is decreased to basal levels by miR-92a inhibitor in HBEC30KT-shTP53 cells (Figure 4A), suggesting that it may participate in miR-92a inhibitor-induced cytotoxicity. Furthermore, the host gene for the miR-17~92 cluster (C13orf25) is over-expressed in human lung cancers, including both NSCLC and SCLC cell lines and patient specimens (7). To further examine the clinical relevance of the miR-17~92-mediated regulation of CYP24A1 expression, we analyzed primary lung adenocarcinoma tumors for associations between expression of miRNAs in the miR-17~92 cluster and patient survival as well as associations between CYP24A1 expression and patient survival and found that high expression of CYP24A1 correlates with poorer overall and recurrence-free survival. Based on stratifying the patients by expression and comparing the highest and lowest quartiles, lower levels of miR-18a, miR-20a and miR-92a are significantly associated with longer overall survival (Figure 4D). In all cases, median survival in patients with low expression is longer than in patients with high expression. In addition, high expression of CYP24A1 correlates with poorer overall and recurrence-free survival. Median recurrence-free survival was 3.1 y in the high CYP24A1 group and > 9 y in the low CYP24A1 group (p = 0.004). Median overall survival was 5.2 y in the high CYP24A1 group and > 9.9 y in the low CYP24A1 group (p = 0.01) (Figure 4E). This was confirmed in both the Director’s Challenge Lung Study, where the top and bottom decile by CYP24A1 expression (n = 45) showed median survival of 3.8 y and 6.6 y, respectively, with p = 0.039 by log-rank test, and the Cancer Genome Atlas (TCGA) study, where the top and bottom deciles (n = 42 for adenocarcinoma; n = 40 for squamous cell carcinoma) showed median survival of 1.7 y and 3.5 y, respectively, for adenocarcinoma, with p = 0.006, and 1.8 and 6.6 y, respectively, for squamous cell carcinoma, with p = 0.05 (33, 34).
Discussion
Here, we find that oncogenotype-selective vulnerability to a miR-92a LNA inhibitor is dependent upon down-regulation of the miR-17~92 primary transcript. The oncogenic function of the miR-17~92 cluster has been well demonstrated in previous studies (7, 35–37), supporting the potential use of a miR-92 inhibitor as an anti-cancer therapeutic agent. Our study identifies p53 as a major mediator of resistance to miR-92a inhibitor-induced cytotoxicity in lung cancers, with low p53 levels a predictor of miR-92a inhibitor toxicity. The quantitative response of NSCLC cell lines to the miR-92a inhibitor in relation to p53 levels in lung cancer cells, coupled with the dramatic sensitization observed in HBEC30-KT cells after p53 knockdown provide an experimental basis for selecting which patients are likely to benefit from the miR-92a inhibition and which are not. Since p53 mutation and loss are among the most common genetic events in tumor biology, identifying a compound with antineoplastic effects in a low p53 context represents a potential avenue to p53-directed therapeutic interventions.
The mechanism for the interaction between the miR-92a and miR-1226* inhibitors and the miR-17~92 primary transcript remains to be determined. LNA-based antisense oligos have demonstrated both in vitro and in vivo potency for silencing long mRNA transcripts, and our observation of miR-17~92 primary transcript depletion could certainly be through a RISC-independent, antisense-like mechanism (38–40). The observed differences in the relative abundance of mature miRNAs from the miR-17~92 cluster by following treatment with miR-92a inhibitor can be attributed to several sources. While mature miRNAs can have half-lives of hours to days in vivo, it has also been demonstrated that mature miRNA species from miR-17~92 do not respond consistently to miR-17~92 depletion (41–43) and that the secondary structure of the miR-17~92 primary transcript affects miRNA processing (44). The lack of response to the miR-92a inhibitor in mouse cells derived from murine KRASmt and KRASmt/p53null lung tumors suggests that the interaction between the miR-92a inhibitor and the miR-17~92 primary transcript may differ between the two organisms. While the mature miRNAs for miR-17-5p, miR-18a-5p, miR-19a-3p, miR-20a-5p, and miR-19b-1-3p are identical, there is only an 85.6% similarity in the region beginning with miR-17 and ending with miR-92a. More importantly, miR-92a differs slightly in sequence between the two organisms. Assessing the energy required to unfold the local region of the primary transcript around miR-92a also shows a difference, with a ΔG of −59.12 kcal/mol for the human transcript and −70.30 kcal/mol for the mouse transcript, reflecting differences in the tertiary structure of the miR-17~92 primary transcript that may affect the processing of the transcript into mature miRNAs (45) or accessibility of the transcript to the miR-92a inhibitor.
Finally, we observed up-regulation of inflammation-related pathways after p53 loss, and activation of a 1α,25-dihydroxy vitamin D3 response in the high-confidence miR-17~92 targetome in HBEC30KT-shTP53 cells. Our results suggest that both an inhibitor of the miR-17~92 polycistron and 1α,25-dihydroxyvitamin D3 are toxic in conjunction with a depletion of p53 in NSCLC, and that these compounds deserve further investigation as avenues for treatment of the many tumors displaying loss of p53 function (Figure 4F). While the benefits of 1α,25-dihydroxyvitamin D3 in lung cancer prevention and treatment are established, the ability of the miR-17~92 cluster to mimic 1α,25-dihydroxyvitamin D3 response in a p53-depleted context is a novel finding with therapeutic implications. The association of p53 loss with a cellular 1α,25-dihydroxyvitamin D3 response suggests that low p53 may be a potential biomarker for 1α,25-dihydroxyvitamin D3 response in NSCLC. Because the presence of hypercalcemia at therapeutic doses of 1α,25-dihydroxyvitamin D3 has limited its potential as an antineoplastic intervention during clinical trials, selecting patients with tumors that are more likely to respond to 1α,25-dihydroxyvitamin D3 or one of the more potent analog compounds under development may lead to a viable therapeutic strategy (31).
A full account of the mechanisms by which miRNAs in the miR-17~92 cluster regulate CYP24A1 expression await clarification in future studies. As shown in Figure 4G, there are signaling pathways that possibly mediate the regulation of CYP24A1 expression by the miRNAs in the miR-17~92 cluster. The miR-92a and miR-1226* inhibitors reduce levels of the miR-17~92 primary transcript and therefore levels of the individual mature miRNAs. The decrease in miR-17-5p/miR-20a-5p can in turn de-repress PTH and NR2C2, which in turn decrease CYP24A1 levels by making the mRNA susceptible to degradation (46) and repressing transcription (47). The decrease in miR-18a-5p can de-repress pregnane X receptor NR1I2, which potentially interferes with VDR activation of CYP24A1 by competing for the same response elements (48). The decrease in miR-19a-3p/miR-19b-3p can de-repress HIC1, a transcriptional repressor known to down-regulate CYP24A1 (49).
In summary, we have described a mechanism for identifying miRNA-based interactions that can mimic the effect of a compound with known positive effects in cancer models. This approach has the potential to allow the leveraging of the biological characterization of other small molecules with known therapeutic potential in model systems, but poor clinical performance, into miRNA and genetic therapeutic approaches that perturb the components of a drug response necessary for a therapeutic effect without modulating the clinically deleterious nodes.
Supplementary Material
Acknowledgments
This work was supported in part by CA129632 (AP), CA071443 (MW), CA176284 (MW), and CA70907 (JM) from the NCI; I-1414 (MW) from the Welch Foundation; RP120732 (JM), RP110708 (JM), and RP120718 (MW) from CPRIT; W81XWH-07-1-0306 (IW) from the Department of Defense; and Cancer Center Support Grants P30 CA054174, P30 CA142543 and P30 CA016672.
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors have no potential conflicts of interest to disclose.
References
- 1.Eberhard DA, Johnson BE, Amler LC, Goddard AD, Heldens SL, Herbst RS, et al. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol. 2005;23:5900–5909. doi: 10.1200/JCO.2005.02.857. [DOI] [PubMed] [Google Scholar]
- 2.Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bunn PA., Jr Worldwide overview of the current status of lung cancer diagnosis and treatment. Arch Pathol Lab Med. 2012;136:1478–1481. doi: 10.5858/arpa.2012-0295-SA. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hatley ME, Patrick DM, Garcia MR, Richardson JA, Bassel-Duby R, van Rooij E, et al. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell. 2010;18:282–293. doi: 10.1016/j.ccr.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 8.Du L, Schageman JJ, Irnov, Girard L, Hammond SM, Minna JD, et al. MicroRNA expression distinguishes SCLC from NSCLC lung tumor cells and suggests a possible pathological relationship between SCLCs and NSCLCs. J Exp Clin Cancer Res. 2010;29:75. doi: 10.1186/1756-9966-29-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Du L, Borkowski R, Zhao Z, Ma X, Yu X, Xie XJ, et al. A high-throughput screen identifies miRNA inhibitors regulating lung cancer cell survival and response to paclitaxel. RNA Biol. 2013;10:1700–1713. doi: 10.4161/rna.26541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao Z, Ma X, Hsiao TH, Lin G, Kosti A, Yu X, et al. A high-content morphological screen identifies novel microRNAs that regulate neuroblastoma cell differentiation. Oncotarget. 2014;5:2499–2512. doi: 10.18632/oncotarget.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Macfarlane LA, Murphy PR. MicroRNA: Biogenesis, Function and Role in Cancer. Curr Genomics. 2010;11:537–561. doi: 10.2174/138920210793175895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 13.Wiggins JF, Ruffino L, Kelnar K, Omotola M, Patrawala L, Brown D, et al. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res. 2010;70:5923–5930. doi: 10.1158/0008-5472.CAN-10-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kota J, Chivukula RR, O'Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Torres AG, Fabani MM, Vigorito E, Gait MJ. MicroRNA fate upon targeting with anti-miRNA oligonucleotides as revealed by an improved Northern-blot-based method for miRNA detection. RNA. 2011;17:933–943. doi: 10.1261/rna.2533811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, et al. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 17.Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 18.Gazdar AF, Oie HK. Re: Growth of cell lines and clinical specimens of human non-small cell lung cancer in a serum-free defined medium. Cancer Res. 1986;46:6011–6012. [PubMed] [Google Scholar]
- 19.Oie HK, Russell EK, Carney DN, Gazdar AF. Cell culture methods for the establishment of the NCI series of lung cancer cell lines. J Cell Biochem Suppl. 1996;24:24–31. doi: 10.1002/jcb.240630504. [DOI] [PubMed] [Google Scholar]
- 20.R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2013. [Google Scholar]
- 21.Boutros M, Bras LP, Huber W. Analysis of cell-based RNAi screens. Genome Biol. 2006;7:R66. doi: 10.1186/gb-2006-7-7-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Du L, Subauste MC, DeSevo C, Zhao Z, Baker M, Borkowski R, et al. miR-337-3p and its targets STAT3 and RAP1A modulate taxane sensitivity in non-small cell lung cancers. PloS one. 2012;7:e39167. doi: 10.1371/journal.pone.0039167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dowell JE, Minna JD. EGFR mutations and molecularly targeted therapy: a new era in the treatment of lung cancer. Nat Clin Pract Oncol. 2006;3:170–171. doi: 10.1038/ncponc0476. [DOI] [PubMed] [Google Scholar]
- 24.Sato M, Larsen JE, Lee W, Sun H, Shames DS, Dalvi MP, et al. Human lung epithelial cells progressed to malignancy through specific oncogenic manipulations. Mol Cancer Res. 2013;11:638–650. doi: 10.1158/1541-7786.MCR-12-0634-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stenvang J, Petri A, Lindow M, Obad S, Kauppinen S. Inhibition of microRNA function by antimiR oligonucleotides. Silence. 2012;3:1. doi: 10.1186/1758-907X-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeLuca HF. Overview of general physiologic features and functions of vitamin D. Am J Clin Nutr. 2004;80:1689S–1696S. doi: 10.1093/ajcn/80.6.1689S. [DOI] [PubMed] [Google Scholar]
- 27.Muller K, Bendtzen K. 1,25-Dihydroxyvitamin D3 as a natural regulator of human immune functions. J Investig Dermatol Symp Proc. 1996;1:68–71. [PubMed] [Google Scholar]
- 28.Peehl DM, Skowronski RJ, Leung GK, Wong ST, Stamey TA, Feldman D. Antiproliferative effects of 1,25-dihydroxyvitamin D3 on primary cultures of human prostatic cells. Cancer Res. 1994;54:805–810. [PubMed] [Google Scholar]
- 29.Bikle DD, Pillai S. Vitamin D, calcium, and epidermal differentiation. Endocr Rev. 1993;14:3–19. doi: 10.1210/edrv-14-1-3. [DOI] [PubMed] [Google Scholar]
- 30.Nakagawa K, Kawaura A, Kato S, Takeda E, Okano T. 1 alpha,25-Dihydroxyvitamin D(3) is a preventive factor in the metastasis of lung cancer. Carcinogenesis. 2005;26:429–440. doi: 10.1093/carcin/bgh332. [DOI] [PubMed] [Google Scholar]
- 31.Chiang KC, Yeh CN, Chen SC, Shen SC, Hsu JT, Yeh TS, et al. MART-10, a New Generation of Vitamin D Analog, Is More Potent than 1alpha,25-Dihydroxyvitamin D(3) in Inhibiting Cell Proliferation and Inducing Apoptosis in ER+ MCF-7 Breast Cancer Cells. Evid Based Complement Alternat Med. 2012;2012:310872. doi: 10.1155/2012/310872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo W, Hershberger PA, Trump DL, Johnson CS. 24-Hydroxylase in cancer: impact on vitamin D-based anticancer therapeutics. J Steroid Biochem Mol Biol. 2013;136:252–257. doi: 10.1016/j.jsbmb.2012.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Director's Challenge Consortium for the Molecular Classification of Lung Adenocarcinoma. Shedden K, Taylor JM, Enkemann SA, Tsao MS, Yeatman TJ, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med. 2008;14:822–827. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olive V, Bennett MJ, Walker JC, Ma C, Jiang I, Cordon-Cardo C, et al. miR-19 is a key oncogenic component of mir-17-92. Genes Dev. 2009;23:2839–2849. doi: 10.1101/gad.1861409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lindholm MW, Elmen J, Fisker N, Hansen HF, Persson R, Moller MR, et al. PCSK9 LNA antisense oligonucleotides induce sustained reduction of LDL cholesterol in nonhuman primates. Mol Ther. 2012;20:376–381. doi: 10.1038/mt.2011.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kauppinen S, Vester B, Wengel J. Locked nucleic acid (LNA): High affinity targeting of RNA for diagnostics and therapeutics. Drug Discovery Today: Technologies. 2005;2:287–290. doi: 10.1016/j.ddtec.2005.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chu Y, Yue X, Younger ST, Janowski BA, Corey DR. Involvement of argonaute proteins in gene silencing and activation by RNAs complementary to a non-coding transcript at the progesterone receptor promoter. Nucleic Acids Res. 2010;38:7736–7748. doi: 10.1093/nar/gkq648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bail S, Swerdel M, Liu H, Jiao X, Goff LA, Hart RP, et al. Differential regulation of microRNA stability. RNA. 2010;16:1032–1039. doi: 10.1261/rna.1851510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Winter J, Diederichs S. Argonaute proteins regulate microRNA stability: Increased microRNA abundance by Argonaute proteins is due to microRNA stabilization. RNA Biol. 2011;8:1149–1157. doi: 10.4161/rna.8.6.17665. [DOI] [PubMed] [Google Scholar]
- 43.Oeztuerk-Winder F, Guinot A, Ochalek A, Ventura JJ. Regulation of human lung alveolar multipotent cells by a novel p38alpha MAPK/miR-17-92 axis. EMBO J. 2012;31:3431–3441. doi: 10.1038/emboj.2012.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chaulk SG, Thede GL, Kent OA, Xu Z, Gesner EM, Veldhoen RA, et al. Role of pri-miRNA tertiary structure in miR-17~92 miRNA biogenesis. RNA Biol. 2011;8:1105–1114. doi: 10.4161/rna.8.6.17410. [DOI] [PubMed] [Google Scholar]
- 45.Chakraborty S, Mehtab S, Patwardhan A, Krishnan Y. Pri-miR-17-92a transcript folds into a tertiary structure and autoregulates its processing. RNA. 2012;18:1014–1028. doi: 10.1261/rna.031039.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zierold C, Mings JA, DeLuca HF. Parathyroid hormone regulates 25-hydroxyvitamin D(3)-24-hydroxylase mRNA by altering its stability. Proc Natl Acad Sci U S A. 2001;98:13572–13576. doi: 10.1073/pnas.241516798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee YF, Young WJ, Lin WJ, Shyr CR, Chang C. Differential regulation of direct repeat 3 vitamin D3 and direct repeat 4 thyroid hormone signaling pathways by the human TR4 orphan receptor. J Biol Chem. 1999;274:16198–16205. doi: 10.1074/jbc.274.23.16198. [DOI] [PubMed] [Google Scholar]
- 48.Zhou C, Assem M, Tay JC, Watkins PB, Blumberg B, Schuetz EG, et al. Steroid and xenobiotic receptor and vitamin D receptor crosstalk mediates CYP24 expression and drug-induced osteomalacia. J Clin Invest. 2006;116:1703–1712. doi: 10.1172/JCI27793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Rechem C, Rood BR, Touka M, Pinte S, Jenal M, Guerardel C, et al. Scavenger chemokine (CXC motif) receptor 7 (CXCR7) is a direct target gene of HIC1 (hypermethylated in cancer 1) J Biol Chem. 2009;284:20927–20935. doi: 10.1074/jbc.M109.022350. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.