Abstract
Background
The exact mechanisms that underlie the pathological processes of myocardial ischemia in humans are unclear. Cardiopulmonary bypass with cardioplegic arrest allows us to examine the whole transcriptional profile of human left ventricular myocardium at baseline and after exposure to cold cardioplegia induced ischemia as a human ischemia model.
Methods
We obtained biopsies from 45 patients undergoing aortic valve replacement surgery at baseline and after an average of 79 min of cold cardioplegic arrest. Samples were RNA-sequenced and analyzed with the Partek Genomics Suite for differential expression. Ingenuity Pathway Analysis and Biobase Explain systems were used for functional and pathway analysis.
Results
Of the 4,098 genes with a mean expression value greater than 5, 90% were downregulated and 9.1% were upregulated. Of those, 1,241 were significantly differentially expressed. Gene ontology analysis revealed significant downregulation in immune inflammatory response and complement activation categories, and highly consistent was the downregulation of intelectin 1, proteoglycan, and SLPI. Upregulated genes of interest were FBJ murine osteosarcoma viral oncogene homolog (FOS) and the hemoglobin genes hemoglobin 1 (HBA1) and hemoglobin beta (HBB).
In addition, analysis of transcription factor binding sites revealed interesting targets in factors regulating reactive oxygen species production, apoptosis, immunity, cytokine production, and inflammatory response.
Conclusions
We have shown that the human left ventricle exhibits significant changes in gene expression in response to cold cardioplegia induced ischemia during cardiopulmonary bypass which provides great insight into the pathophysiology of ventricular ischemia, and thus, may help guide efforts to reduce myocardial damage during surgery.
Introduction
Myocardial ischemia occurs when there is a mismatch between coronary oxygen delivery and metabolic requirements of the myocardium, which may be clinically manifested during angina, coronary angioplasty or cardiopulmonary bypass (CPB). Myocardial ischemia may lead to a spectrum of myocardial stunning, hibernating myocardium, and ultimately cell death if the ischemic insult is severe. In the human heart, irreversible damage begins after approximately 20 to 40 min of oxygen deprivation.1 Observed molecular and cellular changes of myocardial ischemia are characteristic of an inflammatory response,2 but the exact mechanisms that underlie this pathological process are unclear and may not be full emulated by animal models of ischemia or infarction.
Thus, we felt it valuable to investigate a human ischemia model. During cardiac surgery, CPB with aortic cross-clamping (AoXC) and cardioplegic arrest is associated with excellent clinical outcomes and suitable operative conditions. However, despite the use of cardioprotective strategies, AoXc during CPB is accompanied by a variable, yet obligate ischemic period lasting 1 to 3 h, resulting in hypoxia, metabolic substrate depletion, reperfusion injury, apoptosis, and necrosis.3 Cardiac specific biomarkers of ischemia and infarction, including troponin, are elevated even after routine coronary artery bypass graft surgery and correlate with the duration of ischemia from AoXc.4–6
This process of CPB provides us with the ability to examine the transcriptional profile before and after an expected, consistent, and reproducible human ischemic event, albeit induced by cold cardioplegic arrest and not coronary occlusion. In addition, the absence of reperfusion in this time period allows us to examine the transcriptomic response to intermittent ischemia, without having to account for the perturbations of reperfusion injury. Although various animal models have been used to examine the effects of ischemia on cardiac function, no human data exist which examine the early transcriptomic response to a left ventricular (LV) ischemic insult. We therefore characterized the effect of cold cardioplegia induced acute ischemia on the transcriptional profile of the LV by performing whole transcriptome next-generation RNA-sequencing (RNA-seq) in patients undergoing cardiac surgery by sampling human LV tissue prior to, and after, the obligate ischemia during AoXC. We hypothesized that the cold cardioplegia induced ischemic injury will dramatically alter transcription in the human myocardium, and that we would identify genes and pathways, which will identify interventional targets for pharmacological therapy.
Materials and Methods
Patients presenting for elective aortic valve replacement surgery by a single surgeon provided written informed consent with Institutional Review Board (Boston, Massachusetts) approval.
Human Left Ventricular Samples
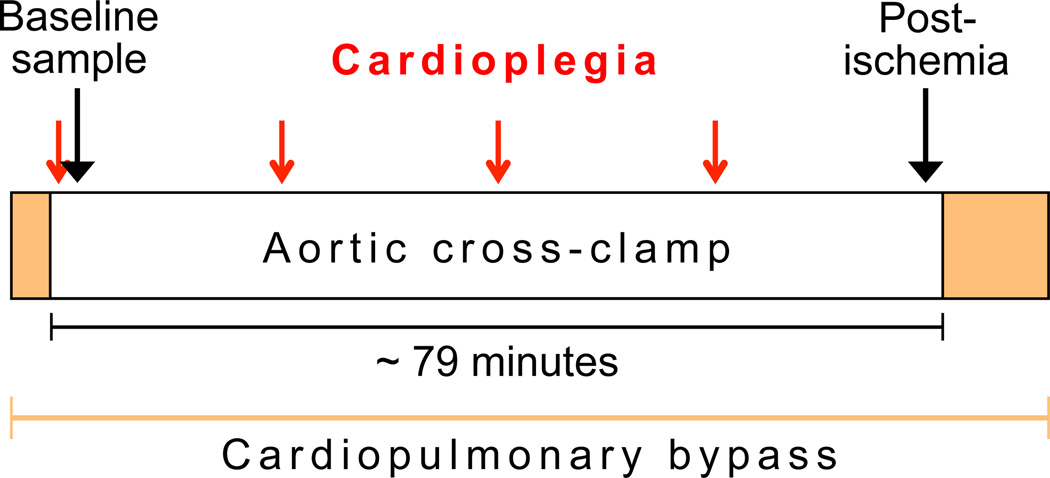
We obtained punch biopsies (~3–5 µg total RNA content) from the site of a routinely placed surgical vent in the anterolateral apical LV wall of patients undergoing elective aortic valve replacement surgery with CPB. After 79 min (interquartile range [IQR] 65–104) of AoXC and intermittent cold blood cardioplegia for myocardial protection every 20 min,7 a second biopsy was obtained in the same manner (fig. 1).
Figure 1. Sample acquisition timeline.
The baseline sample is acquired within minutes of the first dose of cardioplegia and application of the aortic cross-clamp. The postischemia sample is obtained shortly before the end of aortic cross-clamping. The red arrows show the timed application of cardioplegia every 20 min.
RNA sequencing
Tissue samples were immediately placed in RNAlater® (Ambion, ThermoFisher Scientific, Waltham, MA), and after 48 h at +4°C were stored at −80°C until RNA extraction. Total RNA was isolated with Trizol and RNA quality was assessed using the Agilent Bioanalyzer 2100 (Agilent, Santa Clara, CA) with no samples being excluded for poor quality. Library preparation and sequencing has been described previously (see methods, Supplemental Digital Content 1), but briefly, ribosomal RNA was removed by performing 1–2 washings of RNA annealed to poly-T oligos beads (Invitrogen, Life Technologies, Grand Island, NY). RNAs were reverse transcribed using random hexamers (Invitrogen). Double-stranded DNA synthesis was performed using Pol I and RNA-ase H. Short fragments were purified with QiaQuick PCR extraction kit (Qiagen, Hilden, Germany) and resolved with elution buffer for end reparation and poly(A) addition then ligated with sequencing adaptors for cluster generation and sequencing on the Illumina HiSeq 2000 (Illumina, San Diego, CA). As samples were analyzed at different times, different read lengths were employed, initially using single-end reads and then transitioning to paired end reads ranging from 36 – 100 base pairs (See table 1, Supplemental Digital Content 1, which shows the description of samples at the two sequencing centers).
Digital droplet PCR validation
Validation of expression levels was performed in 6 patients, on both the baseline and the post-ischemia samples, using the Bio-Rad QX-100 digital droplet PCR (ddPCR) system (Pleasanton, CA). ddPCR assays were designed to target genes (MGST1,ITLN1, FBJ murine osteosarcoma viral oncogene homolog [FOS], hemoglobin beta [HBB]) and normalized to Gapdh. Absolute concentration of complementary DNA was determined using standard ddPCR protocols. The ratio of target gene to Gapdh concentrations provides normalized gene expression.
Technical replicates
We performed technical replication with two samples (baseline and postischemia) at a single sequencing center using two different library creation methods.
Analysis
Raw reads produced by the Illumina sequencer imaging files were filtered to remove reads containing adaptor sequences, containing >5% unknown nucleotides, or having >50% of reads with base quality scores < 5.
Using Tophat v2.0.58,9 and Bowtie210, reads were aligned to the Homo Sapiens reference genome (UCSC hg19). For paired-end reads, mate inner distance was set at 165bp and mate standard deviation at 37bp. For other parameters, we used the default settings in Tophat and Bowtie2. Final read alignments having more than two mismatches or more than two gaps or more than two edits distances were discarded. The anchor length was 8 without any mismatches being tolerated. The minimum and maximum intron lengths were 70 and 500,000 respectively. The maximum insertion and deletion lengths were 3.
Analysis was performed on aligned BAM files using Partek® Genomics Suite™ 6.6 (Partek Inc., MO). Partek® Genomics Suite™ estimates the most likely relative expression levels of known isoforms and reads that map to the genome using an expectation/maximization algorithm, similar to that described by Xing et al.11 To identify genes with differing relative abundances of isoforms across samples, similar to Marioni et al.12, fold change was calculated as the normalized ratio of mapped reads between baseline and postischemia.
Multivariate testing was performed with a mixed-model analysis of variance (ANOVA) with the computation of p-values based on least-squares means, adjusted for other factors. To build a parsimonious ANOVA model that would be generalizable for other studies, we included only clinical and demographic variables considered to have the greatest importance based on the literature and prior published studies. These variables were age (in years), gender, presence of diabetes mellitus (defined as a HgbA1C > = 6.5 or documented in the chart on oral or parenteral treatment), presence of coronary artery disease, left ventricular ejection fraction, time of sample (baseline vs. postischemia), center where RNA-seq was performed, and minutes of AoXC. Subsequently, these variables were then examined for sources of variation by plotting the mean signal-to-noise ratio (F ratio) of all sequenced genes. Based on their low F ratio, age, diabetes mellitus, left ventricular ejection fraction, coronary artery disease, or gender were not included in the final model. Their exclusion also did not change the significance of the genes included in the final model. With the outcome of absolute gene expression measured in reads mapping to the genome per kilobase of transcript per million reads sequenced (RPKM), we then adjusted for time of sample, center where RNA-seq was performed, and duration of AoXC. Significance of differential expression was defined as expression difference between frequently expressed transcripts (>5 reads per million; RPKM) between postischemia and baseline at a false discovery rate (FDR) of P < 0.05.13
Analysis of transcription factor binding sites
To identify which transcription factors might be important in regulating gene expression, we searched for reported and putative transcription factor binding sites (TFBSs) in the promoter region (−500 to +100 from transcription start site) of differentially expressed genes using Biobases ExPlain MATCH program (Biobase GmbH, Wolfenbuettel, Germany).14,15 We assumed that coexpressed genes are coordinately regulated by a limited number of transcription factors. Overrepresented bindings sites, in our case the top 100 up- and downregulated genes, were compared to an unaltered background set, which in our analysis included 1,000 genes with a nonsignificant fold change of 1.0 ± 0.15. Transcription factor binding sites are predicted from positional weight matrices from the TRANSFAC® (TRANScription FACtor) database.16 Enriched matrices are shown as the ratio of the average number of putative binding sites per 1,000 base pairs for the query and the background sets (yes/no ratio). Values greater than 1 indicate overrepresentation of the motif in the query set. For validation, we performed all analyses three times using a background set of 500 genes picked randomly from all genes with a nonsignificant fold-change of 1.0 and no overlap between sets. Overrepresentation was determined using a one-tailed Fisher exact test. We controlled for multiple testing using a false discovery rate.
Functional analysis of differentially expressed genes
We used Gene Ontology (GO), which provides an extensive ontological description of biological processes, molecular functions and cellular components, to test for enrichment of categories in sets of genes or proteins. GO ANOVA was performed using the same covariate model as for differential expression above. Probe sets were excluded with a maximum signal less then three and a FDR P-value > 0.05, which reflects the differential expression and disruption of GO functional categories instead of individual genes. Analysis was restricted to functional groups between 3–50 genes. To compare individual genes to all other genes in a functional group, we used a Z-test to calculate the disruption score. A low disruption score implies a change in the pattern of gene expression within the functional group, which may or may not be accompanied by differential expression.
A GO-enrichment analysis was performed using Fisher's exact test restricting analyses to functional groups with more than two genes. The enrichment score was calculated using a chi-square test comparing the proportion of the gene list in a group to the proportion of background in the group, with a value of > = 3 corresponding to significant over expression. Furthermore, a gene set enrichment analysis, which compares the frequency of genes detected in the experiment versus random chance, was performed restricting analysis to functional groups between 15–500 genes and using 10,000 permutations.17
The lists of differentially expressed transcripts were uploaded to Ingenuity Pathway Analysis 9.0 (IPA; Ingenuity Systems, Redwood City, CA) and BioBase ExPlain system (BIOBASE GmbH, Germany) to identify regulatory networks operant in human ischemic myocardium. In both applications, the p-value is calculated by determining the probability that a disease or biological function assigned to the data is random. To show the connectivity between proteins, and classify them according to the therapeutic target, biomarker or molecular mechanism, differentially expressed genes and transcripts are categorized by MESH terms and proprietary ontology. The significance of the association is measured by the ratio of the number of proteins from the dataset that map to the category divided by the total number of proteins that map to a canonical pathway.
Results
Forty-five predominantly male subjects with a median age of 72 yr were enrolled (table 1). The patients represent the average of our patient population, who often present with diabetes mellitus (36%), coronary artery disease (46%), and preserved left ventricular systolic function (median ejection fraction 60%). The majority of patients presented with severe aortic stenosis (91%) with minimal aortic regurgitation (64%), mild LV septal thickening, and normal LV dimensions. AoCx time varied depending on whether patients had concomitant coronary artery bypass graft surgery (46%) in addition to their aortic valve replacement. While the median postoperative day 1 creatine kinase MB fraction of 23.0 µg/L (IQR 19.2–27.9 µg/L) was well below the 10-times the 99th percentile of acute myocardial infarction by conventional cardiac biomarker methods (66 µg/L in our laboratory), 2 patients exceeded this value.18
Table 1.
Demographics and Clinical Characteristics of the 45 Paired Subjects
| Age (years) | 72 (67–81) |
| Male gender, N (%) | 30 (67) |
| Body mass index | 30 (27–35) |
| Diabetes, N (%) | 16 (36) |
| CAD, N (%) | 23 (51) |
| Postop Day 1 CKMB (µg/L) | 23.0 (19.2–27.9) |
| Aortic cross-clamp (min) | 79 (65–104) |
| LV ejection fraction (%) | 60 (50–60) |
| Aortic stenosis, N (%) | |
| Severe | 41 (91) |
| None | 4 (9) |
| Aortic insufficiency, N (%) | |
| None | 6 (13) |
| Trace-mild | 23 (51) |
| Moderate | 12 (27) |
| Severe | 4 (9) |
| LV internal diameter (end-diastole) (cm) | 5.1 (4.4–5.7) |
| LV septal thickness (cm) | 1.4 (1.3–1.6) |
Median values with interquartile range (IQR)
CAD = coronary artery disease; CKMB = creatine kinase muscle and brain isoenzyme; LV = left ventricle.
Gene Expression
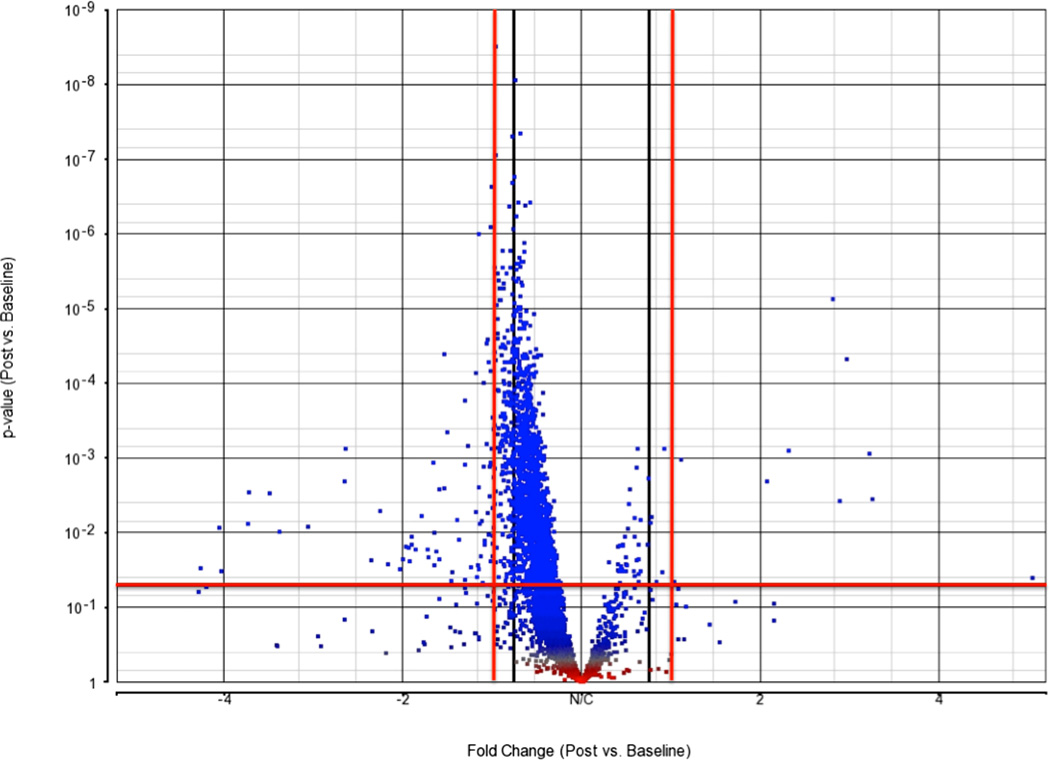
Median RNA integrity score was 7.0 (IQR 6.3–7.5). After quality control measures, we examined 35,470 transcripts from 21,308 genes. When we compared the postischemia sample to the baseline sample, 16,821 (78.9%) genes were downregulated, and 4,460 (20.9%) were upregulated. When we restricted to genes with a mean RPKM of > 5 across all samples (N = 4,098), 3,724 (90.0%) were down-regulated and 374 (9.1%) were upregulated (fig. 2).
Figure 2. Volcano plot.
Fold change on X-axis plotted against P-value on Y-axis. The horizontal line marks the unadjusted P = 0.05 and the red vertical lines mark a fold change of ±1.5. All dots in the upper left and right quadrant are considered significant before adjusting for multiple comparisons.
Of those genes with an RPKM >5, we identified 1,241 genes that were significantly differentially expressed (FDR P-value <0.05) between post-ischemia and baseline after multivariate modeling. The top 16 genes along with their baseline and postischemia expression changes with a mean RPKM >5, fold change >2, and a FDR P-value < 0.05 are shown in table 2.
Table 2.
Shown Are Top Up and Down-regulated Genes between Postischemia and Baseline for the Ventricle at Baseline, Postischemia, and for the Atrium at Baseline
| Gene Symbol | Gene Name | P-value | Fold Change (Post vs. Baseline) |
Baseline (N=45) |
Postischemia (N=45) |
Atrium (N=6) |
|---|---|---|---|---|---|---|
| ITLN1 | intelectin 1 (galactofuranose binding) | 0.009 | −4.08 | 262.3±548.8 | 45.7±121.8 | 242.4 ± 392.1 |
| SLPI | secretory leukocyte peptidase inhibitor | 0.008 | −3.65 | 26.5±52.4 | 5.4±13.7 | 10.5 ± 13.2 |
| PRG4 | proteoglycan 4 | 0.003 | −3.63 | 13.7±23.2 | 3.1±8.2 | 13 ± 20 |
| CCL21 | chemokine (C-C motif) ligand 21 | 0.003 | −3.36 | 21.3±35.6 | 5.2±6.4 | NA |
| HP | haptoglobin | 0.01 | −3.22 | 25.6±51.3 | 5.9±14 | 15 ± 17.2 |
| MGST1 | microsomal glutathione S-transferase 1 | 0.008 | −2.89 | 12.4±22.9 | 3.3±4.3 | 29.8 ± 45.3 |
| C3 | complement component 3 | 0.002 | −2.5 | 53.5±68.8 | 19.3±29.3 | 60 ± 55.3 |
| LYVE1 | lymphatic vessel endothelial hyaluronan receptor 1 | 0.001 | −2.49 | 9.2±14 | 2.8±2.8 | 10.7 ± 7.7 |
| F13A1 | coagulation factor XIII, A1 polypeptide | 0.005 | −2.18 | 12.6±21.4 | 4.5±5.3 | 6.8 ± 5.4 |
| FOS | FBJ murine osteosarcoma viral oncogene homolog | 0.001 | 3.06 | 7.9±12.3 | 27.4±37.5 | 61 ± 80.5 |
| S100A8 | S100 calcium binding protein A8 | 4.76E-05 | 2.8 | 7.3±7.1 | 20.5±20.3 | 41.3 ± 66.1 |
| HBA2 | hemoglobin, alpha 2 | 0.004 | 3.09 | 302.8±242.7 | 917.1±1349.5 | NA |
| HBA1 | hemoglobin, alpha 1 | 7.47E-06 | 2.65 | 262±248.8 | 594.7±506.6 | 321.8 ± 292 |
| HBB | hemoglobin, beta | 0.004 | 2.72 | 237.7±188 | 730.6±1120.1 | 229.7 ± 143.5 |
| S100A9 | S100 calcium binding protein A9 | 0.001 | 2.23 | 16±13 | 36.7±38.1 | NA |
| ZFP36 | ZFP36 ring finger protein | 0.002 | 2.06 | 10.5±11.2 | 22.5±24 | NA |
Shown are all genes with a mean reads per kilobase of transcript per million mapped reads (RPKM) >5, a fold change ≥ 2 and a FDR P<0.05. Genes are in a multivariate model adjusting for time of sample (baseline versus post-ischemia), center where RNA-sequencing was performed, and minutes of aortic cross-clamping (AXC = effective ischemia time). (red, up-regulated; green, down-regulated). Genes are sorted by fold change. FDR = false discovery rate.
" In all analyses, we applied a FDR (Benjamini & Hochberg algorithm) to present the significant P-value after multiple testing. Alternatively, we could have used more stringent multiple testing procedures such as the Bonferroni correction, which can be somewhat conservative if there are a large number of tests and/or the test statistics are positively correlated, as is often the case in genetics where many genes are inherited together and have a strong relationship to each other (strong linkage disequilibrium). Had we used a strict Bonferroni correction, only one gene (hemoglobin α1 [HBA1]) in table 2 would have retained significance."
Functional classification
GO Analysis
To understand their collective biological function, we examined differentially expressed genes for enrichment of GO categories. The majority of gene sets with significant enrichment in a specific GO category were downregulated, and were categorized in cellular components and biological processes, with few identifying with molecular functions (See table 2, Supplemental Digital Content 1, which shows the up- and downregulated GO annotation). The top 5 downregulated GO terms with an FDR significance less than 4.7×10−9 were extracellular component, complement activation, protein activation cascade, immune effector process, and humoral immune response. Highly consistent across almost all downregulated GO groups identified was the presence of complement genes (e.g., C1, C3, C6, C7).
On the other hand, significantly upregulated gene sets were all categorized as biological processes and were associated with GO terms of regulation of catalytic activity and molecular function (See table 2, Supplemental Digital Content 1). In total, of the GO groups that met an FDR threshold of P < 0.01, 55 were downregulated and only 4 were upregulated. Furthermore, the significant P-values were much lower in the downregulated groups as well.
Upon direct comparison between the up- and downregulated gene sets, the highly significant downregulated GO categories were associated with the immune and inflammatory response, and complement activation. Conversely, the only significantly upregulated GO category was vasculature development. (See tables 3 and 4 in Supplemental Digital Content 1 which show the GO annotation of the differential expression and the gene set enrichment analysis respectively, and see figs. 1 and 2 in Supplemental Digital Content 1 which show a graphical representation of the functional analysis of the GO terms).
GO disruption
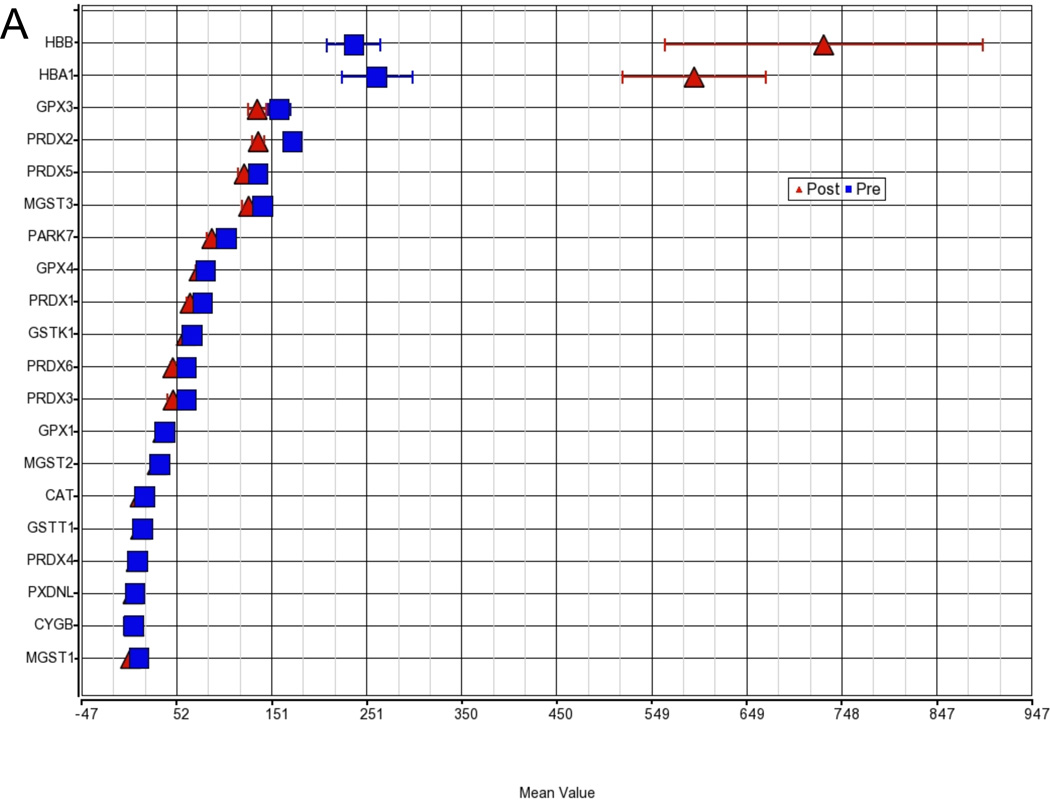
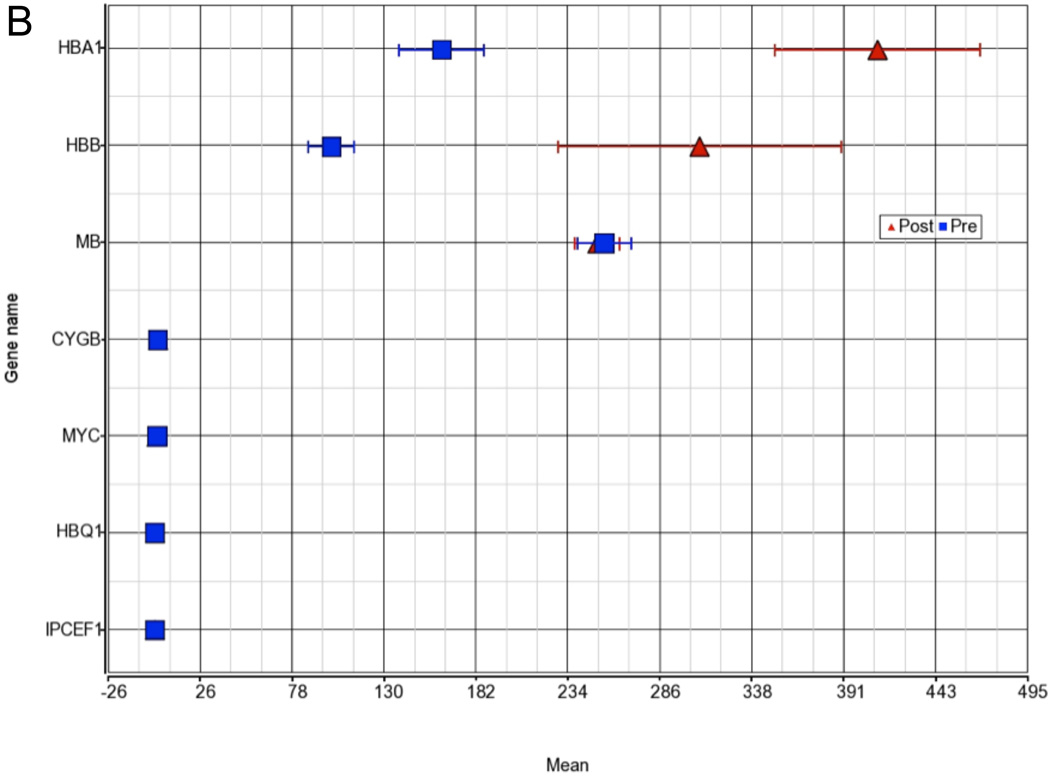
The GO analysis revealed a substantial number of functional categories with low disruption scores, implying changes in the pattern of gene expression within the functional group and potential disruption of normal operation of the group. The most pronounced disruption occurred in the molecular function group encompassing the categories of antioxidant, peroxidase, and oxidoreductase activity containing the hemoglobin genes HBA1 and HBB (fig. 3). While the expression for the majority of genes in this category is very similar between baseline and post-ischemia (23.3 vs. 20.1), HBB and HBA1 are significantly upregulated compared to all other genes in this category (disruption P < 0.001).
Figure 3. Gene Ontology (GO) group disruption a) Oxidoreductase activity and b) Oxygen transport.
Shown are the two groups with the most pronounced disruption of a functional category in the GO ANOVA, implying changes in the pattern of gene expression within the functional group and potential disruption of normal operation of the group. The hemoglobin genes hemoglobin α1 (HBA1) and hemoglobin beta (HBB) disrupt the molecular function group oxidoreductase activity and oxygen transport. While the expression of the majority of genes in these categories is very similar between baseline and postischemia, HBB and HBA1 are significantly upregulated compared to all other genes in this category (disruption P < 0.001).
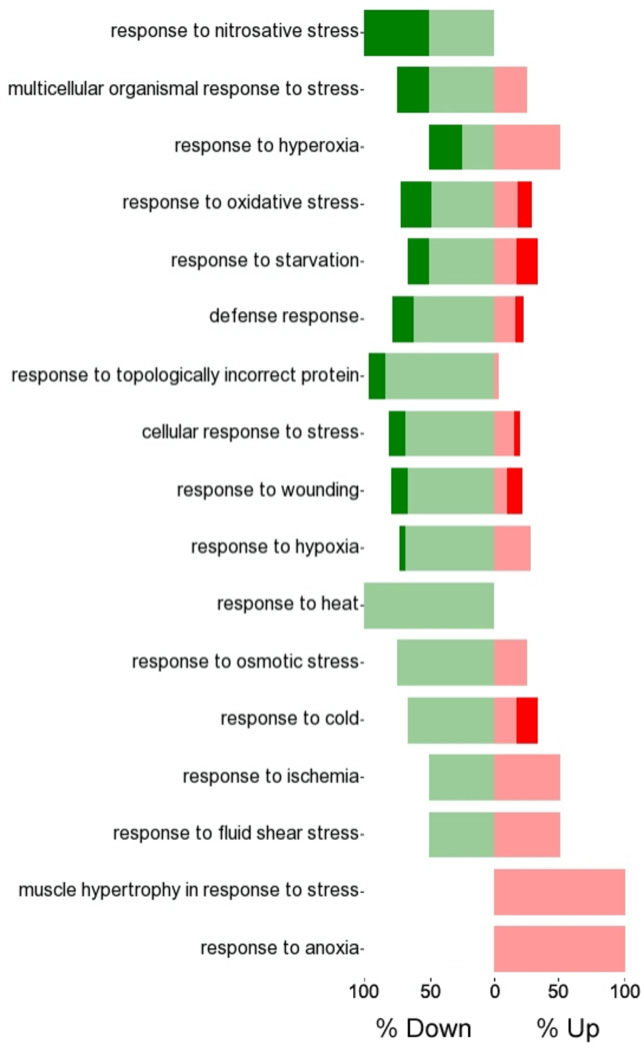
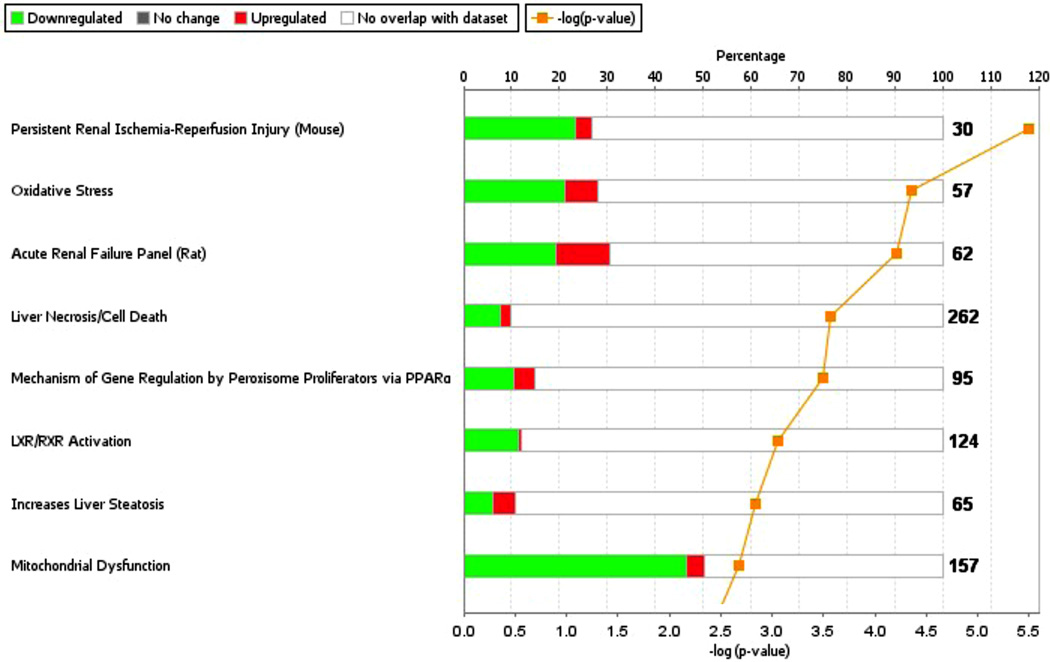
Since the nature of the experiment was to examine a timed insult, a GO category of particular interest was "response to stress", with stressors most commonly associated with oxygen, such as anoxia, hypoxia, or hyperoxia (fig. 4). Oxidative stress was also one of the most significant toxicity lists in Ingenuity Pathway Analysis (P = 4.4×10−5; fig. 5).
Figure 4. Gene Ontology (GO)-Enrichment.
Forest Plot of postischemia compared to baseline in response to stress category. Green bar shows downregulation, red bar show upregulation with dark colors showing percent of significantly down- or upregulated genes (False discovery rate [FDR] < 0.05).
Figure 5. Oxidative stress Tox List in Ingenuity Pathway Analysis.
Shown are groups with percent downregulation (green) and upregulation (red) matching the dataset. The numbers represent the number of genes in each category and the orange line represents the negative logarithmic P-value. PPARα = Peroxisome proliferator-activated receptor alpha.
Transcription factor binding site analysis
To identify which transcription factors are involved in the regulation of gene expression in differentially expressed genes, we scanned the promoter region for overrepresentation of transcription factor binding sites (TFBS). Similar to the GO processes, TFBSs are overrepresented in downregulated genes; we identified 10 TFBS that were significantly overrepresented in the promoter region of upregulated genes, and 40 TFBS that were overrepresented in downregulated genes (table 3).
Table 3.
Transcription Factor Binding Sites Overrepresented in Up- and Down-regulated genes
| Downregulated gene set | Frequency (sites/1000bp) | |||||
| Matrix name | Transcription factor | Down | Background | Ratio | FDR | Matched promoters FDR |
| V$CDP_03 | Cutl1, CUX1, Cux1 | 0.74 | 0.15 | 4.82 | 5.6E-12 | 2.3E-05 |
| V$RORA_Q4 | Rora, RORA | 0.25 | 0.04 | 7.05 | 4.8E-06 | 5.2E-04 |
| V$OCT1_03 | POU2F1, Pou2f1, Pou2f1 | 0.43 | 0.11 | 3.91 | 3.5E-06 | 8.8E-04 |
| V$HMGA2_01 | Hmga2, HMGA2 | 0.38 | 0.10 | 3.97 | 1.1E-05 | 2.0E-03 |
| V$TAL1BETAE47_01 | Tal1, TAL1, Tcf3, TCF3, Tcf4, Tcf4, TCF4, Tcfe2a | 0.29 | 0.09 | 3.25 | 6.2E-04 | 3.4E-03 |
| V$OCT1_06 | POU2F1, Pou2f1, Pou2f1 | 0.42 | 0.11 | 3.74 | 9.1E-06 | 4.7E-03 |
| V$NKX61_03 | Nkx6-1, Nkx6-1, NKX6-1 | 0.27 | 0.07 | 4.17 | 1.4E-04 | 4.7E-03 |
| V$PBX1_04 | Pbx1, Pbx1, PBX1 | 0.27 | 0.08 | 3.61 | 4.2E-04 | 5.1E-03 |
| V$BRN2_01 | Pou3f2, POU3F2, Pou3f2 | 0.22 | 0.05 | 4.53 | 3.7E-04 | 5.3E-03 |
| V$SATB1_Q3 | SATB1, Satb1 | 0.25 | 0.07 | 3.44 | 8.9E-04 | 7.2E-03 |
| V$STAT5A_02 | Stat1, STAT1, Stat2, Stat2, STAT2, Stat3, STAT3, Stat3, Stat4, Stat4, STAT4, Stat4, Stat5a, Stat5a, STAT5A, Stat5b, STAT5B, Stat5b, Stat6, STAT6, Stat6 | 0.40 | 0.13 | 3.10 | 1.1E-04 | 8.6E-03 |
| V$TAL1ALPHAE47_01 | Tal1, TAL1, Tcf3, TCF3, TCF4, Tcf4, Tcf4, Tcfe2a | 0.24 | 0.08 | 3.12 | 2.4E-03 | 8.6E-03 |
| V$CEBPG_Q6_01 | CEBPG, Cebpg, Cebpg | 0.20 | 0.06 | 3.42 | 3.0E-03 | 0.011 |
| V$ATF5_01 | Atf5, Atf5, ATF5 | 0.20 | 0.07 | 3.06 | 5.4E-03 | 0.013 |
| V$HOX13_02 | HOXA5, Hoxa5, Hoxa5, Irf6, IRF6, Irf6 | 0.27 | 0.07 | 3.97 | 2.1E-04 | 0.014 |
| V$NKX61_02 | Nkx6-1, Nkx6-1, NKX6-1 | 0.18 | 0.04 | 5.04 | 6.4E-04 | 0.014 |
| V$POU6F1_03 | POU6F1, Pou6f1, Pou6f1 | 0.24 | 0.05 | 4.44 | 2.5E-04 | 0.014 |
| V$RORA2_01 | Rora, RORA | 0.20 | 0.04 | 4.47 | 6.8E-04 | 0.015 |
| V$CEBPA_01 | CEBPA, Cebpa, Cebpa, Cebpb, Cebpb, CEBPB | 0.18 | 0.05 | 3.78 | 2.8E-03 | 0.016 |
| V$HELIOSA_01 | IKZF1, Ikzf1, Ikzf2, IKZF2 | 0.18 | 0.05 | 3.78 | 2.8E-03 | 0.016 |
| V$BCL6_02 | BCL6, Bcl6, Bcl6 | 0.20 | 0.06 | 3.53 | 2.6E-03 | 0.016 |
| V$POU6F1_02 | Pou6f1, POU6F1, Pou6f1 | 0.25 | 0.06 | 4.35 | 1.7E-04 | 0.018 |
| V$SEF1_C | n/a | 0.18 | 0.06 | 3.02 | 8.2E-03 | 0.018 |
| V$CEBPB_Q6 | Cebpb, Cebpb, CEBPB | 0.20 | 0.05 | 3.88 | 1.5E-03 | 0.018 |
| V$CEBPB_01 | CEBPA, Cebpa, Cebpa, Cebpb, Cebpb, CEBPB | 0.38 | 0.13 | 3.00 | 2.2E-04 | 0.019 |
| V$SATB1_01 | n/a | 0.25 | 0.06 | 4.11 | 2.5E-04 | 0.019 |
| V$LHX3_01 | Lhx3, LHX3, Lhx3 | 0.42 | 0.12 | 3.43 | 2.5E-05 | 0.019 |
| V$STAT1STAT1_Q3 | Stat1, STAT1, Stat4 | 0.25 | 0.05 | 4.63 | 1.1E-04 | 0.021 |
| V$STAT4_Q4 | Stat4, STAT4, Stat4 | 0.18 | 0.05 | 3.53 | 3.9E-03 | 0.021 |
| V$CEBP_Q2 | CEBPA, Cebpa, Cebpa, Cebpb, Cebpb, CEBPB | 0.18 | 0.04 | 4.23 | 1.6E-03 | 0.022 |
| V$PIT1_Q6 | POU1F1, Pou1f1, Pou1f1 | 0.27 | 0.08 | 3.53 | 5.0E-04 | 0.022 |
| V$LHX3_02 | Lhx3, LHX3, Lhx3 | 0.54 | 0.17 | 3.17 | 5.1E-06 | 0.024 |
| V$IPF1_02 | Pdx1, PDX1, Pdx1 | 0.24 | 0.06 | 4.04 | 4.7E-04 | 0.024 |
| V$IPF1_01 | Pdx1, PDX1, Pdx1 | 0.27 | 0.07 | 4.17 | 1.4E-04 | 0.027 |
| V$STAT5A_Q6 | Stat5a, STAT5A, Stat5a | 0.18 | 0.04 | 5.04 | 6.4E-04 | 0.027 |
| V$IPF1_06 | Pdx1, PDX1, Pdx1 | 0.20 | 0.06 | 3.53 | 2.6E-03 | 0.027 |
| V$ZTA_Q2 | n/a | 0.18 | 0.06 | 3.02 | 8.2E-03 | 0.029 |
| V$RP58_01 | Zfp238, Zfp238, ZNF238 | 0.18 | 0.05 | 3.65 | 3.3E-03 | 0.030 |
| V$PBX1_03 | Pbx1, Pbx1, PBX1 | 0.18 | 0.05 | 3.31 | 5.3E-03 | 0.047 |
| V$AFP1_Q6 | ZFHX3, Zfhx3 | 0.18 | 0.06 | 3.11 | 7.1E-03 | 0.047 |
| Upregulated gene set | Frequency (sites/1000bp) | |||||
| Matrix name | Transcription factor | Up | Background | Ratio | FDR | Matched promoters FDR |
| V$SRF_Q4 | Srf, SRF, Srf | 0.19 | 0.01 | 18.64 | 1.83E-06 | 0.021 |
| V$APOLYA_B | n/a | 0.19 | 0.04 | 4.66 | 2.50E-03 | 0.032 |
| V$TATA_01 | BTAF1, TAF1, TAF10, TAF11, TAF12, TAF13, TAF4, TAF5, TAF6, TAF7, TAF9, Tbp, TBP, Tbp | 0.21 | 0.05 | 4.39 | 2.00E-03 | 0.032 |
| V$OCT4_01 | POU5F1, Pou5f1, Pou5f1 | 0.25 | 0.06 | 4.04 | 1.30E-03 | 0.022 |
| V$TBR2_01 | Eomes, EOMES | 0.25 | 0.06 | 4.04 | 1.30E-03 | 0.032 |
| V$MEF2_05 | Mef2a, MEF2A, Mef2a | 0.21 | 0.05 | 3.97 | 3.30E-03 | 0.035 |
| V$MTATA_B | n/a | 0.42 | 0.11 | 3.67 | 4.44E-05 | 0.002 |
| V$ETS2_B | ELF2, Elf2, ETS1, Ets1, Ets1, ETS2, Ets2, Gabpa, GABPA, Gabpb1, GABPB1, Sfpi1, Sfpi1, SPI1 | 0.23 | 0.07 | 3.44 | 4.50E-03 | 0.049 |
| V$MEF2_03 | Mef2a, MEF2A, Mef2a, Mef2c, MEF2C, Mef2d, Mef2d, MEF2D | 0.25 | 0.08 | 3.09 | 5.70E-03 | 0.041 |
| V$NF1_Q6 | Nfia, NFIA, Nfia, Nfib, NFIC, Nfic, Nfic, Nfix | 0.23 | 0.08 | 3.05 | 7.70E-03 | 0.041 |
The top 100 up- and 100 downregulated genes (query set) were compared to the background set of 1,000 genes with a fold-change ~1. Each row contains information on the performance of one matrix of the positional weight matrix. Average number of putative binding sites per 1,000 base pairs is given for the Up/down regulated genes as well as the background set. The ratio of the two values is provided in the "ratio" column where a number greater than 1 indicates overrepresentation of the motif in the query set. Significance of the representation value is measured by the P value derived from a binomial distribution adjusted with false discovery rate (FDR). "Matched promoters FDR" assesses the statistical significance of the number of promoters in the query set that have at least one predicted site compared to that of promoters in the control set.
Matrix name - tissue factor binding site matrix from TRANSFAC database, Transcription factor - TFs that can bind to the matrix element.
Shown is the data for all matrices with a ratio >3, a FDR < 0.05, and a matched promoter FDR<0.05.
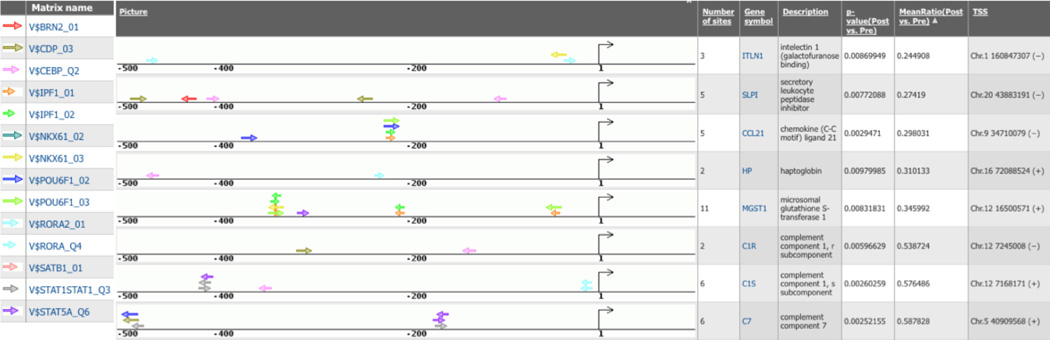
Closer examination of the promoter region of the most significantly downregulated genes (fig. 6) shows multiple matrices matching to similar composition of TFBS, although their location in relevance to the transcription start site can vary widely between different promoters. The transcription factors, such as RORA, STAT1, STAT5a, and CEBP, act in regulation of reactive oxygen species production,19 apoptosis,20 immunity,21 and inflammatory response.22
Figure 6. TRANSFAC® (TRANScription FACtor) database positional weight matrices in promoters.
Shown are maps of predicted binding sites of selected TRANSFAC® (Biobase GmbH, Wolfenbuettel, Germany) positional weight matrices in promoters of genes downregulated in response to ischemia. TSS = transcription start site.
ddPCR Validation and Technical Replication
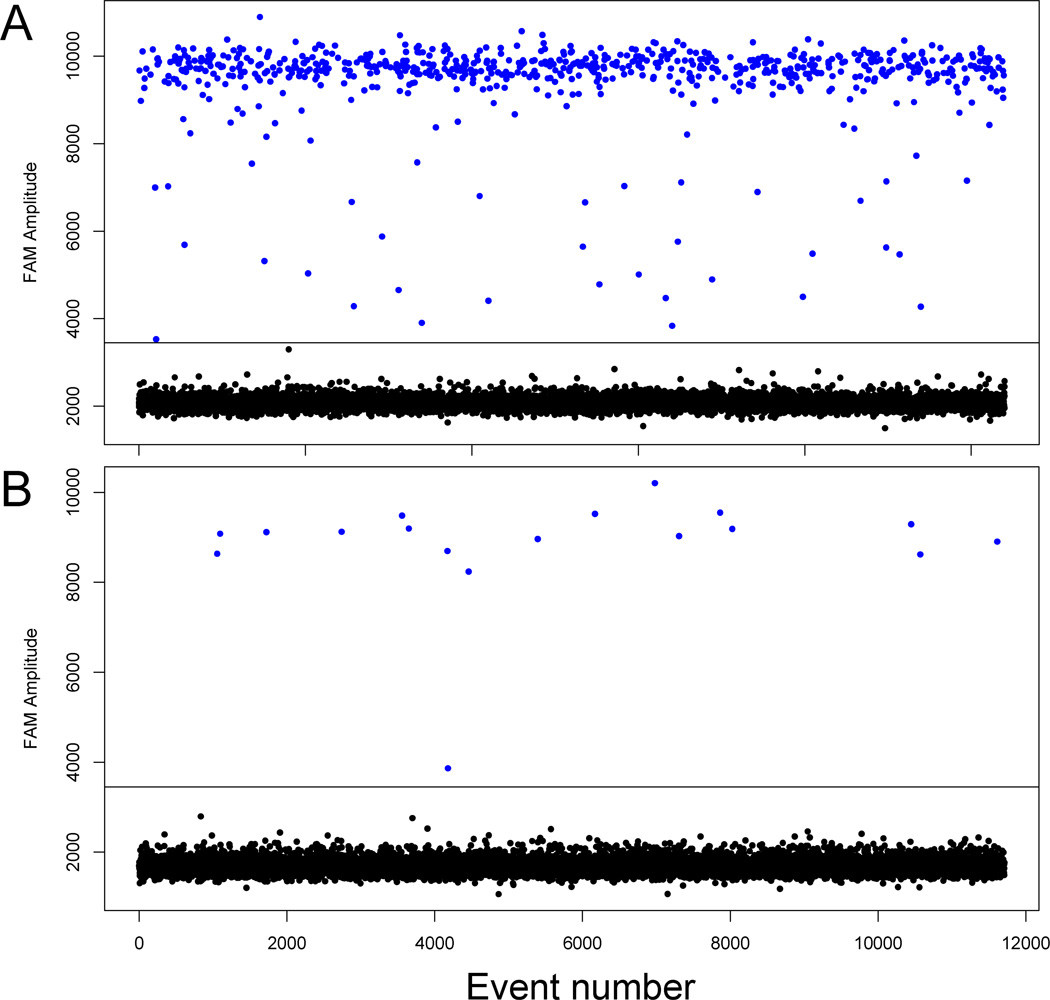
All four genes validated with ddPCR (MGST1, ITLN1, FOS, HBB) showed a similar expression profile to their messenger RNA RNA-seq transcript when normalized to Gapdh. There was a high degree of correlation between the expression values obtained from RNA-seq and the normalized concentration from the ddPCR validation (r2 = 0.83 for baseline values, r2 = 0.76 for postischemia, and r2 = 0.85 for the fold changes). Normalized concentration and log2 fold changes between post-ischemia and baseline were: ITLN1 (mean baseline/post 3,251/14, SD baseline/post 5,068/10, fold change −7.8), MGST1 (mean baseline/post 173/96, SD baseline/post 158/48, fold change −0.9), FOS (mean baseline/post 285/722, SD baseline/post 318/1,062, fold change 1.3), and HBB (mean baseline/post 2,672/3,254, SD baseline/post 2,981/2,579, fold change 0.3). Figure 7 clearly shows the high baseline expression and very low postischemia expression for ITLN1.
Figure 7. Digital droplet (ddPCR) validation.
Fluorescence amplitude plots for Carboxyfluorescein (FAM) labeled droplets in a representative A) baseline and B) postischemia sample for the intelectin 1 (ITLN1) gene. Each dot represents a single droplet, or event. The event number is displayed on the horizontal axis while the fluorescence amplitude is shown on the vertical axis. The blue dots represent high fluorescence intensity (positive droplets) while the black dots represent low intensity (negative droplets).
The results of our technical replication show that for the genes for which we performed the ddPCR, the fold changes show a similar magnitude and the same directionality and the coefficient of variation (CV) is 79%. Likewise, when we compare all genes in both samples, the average CV is 58% (baseline) and 51% (postischemia), and when we restrict to those with an RPKM ≥5, the CV is 49% and 42% respectively.
Discussion
Our comprehensive analysis of whole genome RNA-seq data on human LV myocardium identifies the genes and pathways that are significantly and strongly regulated in cold cardioplegia induced myocardial ischemia. Our results comprehensively delineate gene regulation at the transcriptional level in patients exposed to CPB and suggest that messenger RNA isoform regulation plays a critical role in the cardiac response to cold cardioplegia induced ischemia. However, of the identified pathways that are activated in response to this myocardial injury, it is still not clear what part of the transcriptional response to ischemia is a protective mechanism versus a reflection of genetic activation of maladaptive injury response.
Hallmarks of the transcriptional response to ischemia included changes in genes and pathways associated with inflammation and the oxidative response to stress, which themselves can mediate myocardial injury. Nevertheless, the most profoundly differentially expressed genes have yet to be directly associated with human ischemic myocardial injury.
Downregulation
The 3 most downregulated genes, intelectin 1 (ITLN1), proteoglycan 4 (PRG4), and secretory leukocyte peptidase inhibitor (SLPI) are significantly downregulated by more than 3-fold (P < 0.01).
Omentin/intelectin is a novel adipokine discovered in 2005, codified by two genes, with omentin 1 (ITLN1) as the primary circulating form. This protein stimulates insulin-mediated glucose transport in human adipocytes and is expressed in epicardial and omental adipose tissue. Its role in cardiovascular disease has been well described. Specifically, the actions of omentin are responsible for increasing endothelium-dependent vasodilation (rat ex vivo),23 decreasing vascular inflammation (human in vitro),24 and decreasing extracellular migration and angiogenesis (human in vitro).25 Hence, omentin appears to be a "protective adipokine", with antiinflammatory properties as well as reducing endothelial dysfunction.26
Regulation of omentin in an acute setting has been studied in mice, showing that omentin promotes endothelial cell function and the revascularization process under conditions of ischemic stress, likely through AMP-activated protein kinase (AMPK) and Akt.27
The highly significant and consistent downregulation of ITLN1 in myocardial tissue after exposure to cold cardioplegia induced ischemia suggests ITLN1 also plays an important role in myocardial injury. Additionally, expression in nonischemic right atrial tissue is very similar to baseline values in ventricular myocardium (table 2) further supporting that ITLN1 expression in nonischemic cardiac tissue is high. As with ITLN1, SLPI is also expressed in omental adipose tissue.28 This gene encodes a secreted inhibitor, which protects epithelial tissues from serine proteases. Serine protease inhibitors, such as the withdrawn Aprotinin, have a long clinical history in cardiac surgery and are attractive agents in cardiac surgery to protect the myocardium from ischemia-reperfusion injury.29,30 In summary, this suggests that ITLN1 and SLPI, 2 genes without prior association with ischemic injury in human myocardium, are targets of interest in modulation of ischemia and warrant further investigation.
Proteoglycan 4 (PRG4) produces a protein synthesized by chondrocytes at the surface of articular cartilage and synovial lining cells, although expression in human and mouse cardiac tissue has been described.31 Similar to ITLN1, PRG4 expression at baseline is also comparable to atrial expression levels. The reason for profound downregulation remains to be elucidated, since the only link to cardiac disease is that a homozygous deletion in PRG4 causes camptodactyly-arthropathy-coxa vara-pericarditis syndrome.
Upregulation
In general, upregulation was not as profound in fold-change or in number of genes affected as downregulation. Nevertheless, there are several very intriguing targets, namely FOS and the hemoglobin genes HBA1 and HBB (fold change > 2, P < 0.01).
The gene FOS is part of the FOS gene family, whose proteins can dimerize with proteins of the JUN family, thereby forming the transcription factor complex AP-1.32 These proteins are regulators of cell proliferation, differentiation, transformation and even apoptotic cell death.33 Specifically in cardiac tissue in rats, upregulation of c-Fos by tumor necrosis factor-α has been shown to increase the transcription of TGFβ1, a key cytokine that promotes post-myocardial injury cardiac fibrosis34. This upregulation of FOS goes along with what we observed in our patients and is consistent with an activation of inflammatory mediators after CPB that ultimately can result in postischemia myocardial dysfunction.
We also show for the first time the profound upregulation of HBA1 and HBB in the LV in response to cold cardioplegia induced ischemia. The gene expression atlas shows an increased expression of HBA1 and HBB in heart tissue as compared with mean expression level for all samples35, yet to our knowledge the response to ischemia in human LV myocardium has not been examined. Arab et al showed a rapid increase in hemoglobin expression in right ventricular myocardium in infants and children with congenital heart disease in response to ischemia, with similar findings in isolated cardiomyocytes during simulated ischemia/reperfusion injury.36 Likewise, studies showed increased hemoglobin messenger RNA expression in neurons in embryonic and adult brains in the setting of transient and permanent cerebral artery occlusion in rats.37 Both findings are indicative of nonhematologic origins of hemoglobin gene transcription. A possible mechanism is the myocardium's attempt to minimize nitrosative stress by sequestration of excess nitric oxide generated during ischemia, which is supported by our GO analysis showing the response to nitrosative stress as the top downregulated category (fig. 4). This would imply a protective mechanism.
Functional analysis
Our GO and network pathway analyses identified a number of pathways that are related to oxidative stress, defined as the imbalance between the production of reactive oxygen species and the detoxification of reactive intermediates. While severe oxidative stress can trigger apoptosis and necrosis and is involved in cardiovascular diseases such as atherosclerosis, reactive oxygen species are not necessarily damaging as they are an integral part of the immune systems response to pathogens and part of normal cell signaling.
Complement activation is a key mediator of ischemia induced myocardial tissue damage. Complement inhibitors have been applied successfully in animal models in reducing tissue damage from ischemia-reperfusion injury, yet anticomplement therapy in human myocardial ischemia-reperfusion injury has yielded inconsistent results.38 A direct link exists between the activation of the complement system and the regulation of the immune system and past evidence points to an increase in both in response to ischemia-reperfusion.39 However, the significant downregulation of the humoral immune response concomitantly with both the alternative and classical complement pathway in our cohort would suggest that ischemia, in the absence of reperfusion, may not lead to an immediate increase in these pathways as previously described. This is supported by very low complement deposition (C3d) in brain-dead donor hearts that had ischemic injury without reperfusion,40 by the lag in C3 deposition compared to the loss of plasma membrane integrity during early myocardial ischemia,41 and by the initial decrease in serum C3 levels in pediatric cardiac surgical patients.42
Limitations
The focus of this study was the transcriptional response of the human left ventricular myocardium to cold cardioplegia induced ischemia. We used CPB in patients undergoing cardiac surgery as the model, as exposing normal human myocardium to ischemia in vivo is not feasible. In addition, our model of cold cardioplegia induced myocardial ischemia provides important contributions to the field as many inconsistencies in prior studies using human myocardium stems from the fact that most experiments are inherently uncontrolled using tissue that becomes available only as a result of death or cardiac transplantation. Our prospective study has several features that are novel: 1) every sample is collected from the exact same physical location on the LV apex by the same surgeon at the same time point in the operation, reducing intersample variability; 2) the time between sample collection and aliquoting into RNA preservative is ~ 1 min, resulting in extremely high quality RNA; 3) the tissue sample we collect is from left ventricular tissue (as opposed to atrial or right ventricular tissue used in prior studies) in nonfailing hearts (unlike left-ventricular assist device cores from end-stage heart failure patients). Though the hearts do have concentric hypertrophy from long-standing aortic stenosis, the surgical procedures are all elective and risk factors are optimally medically managed in preparation of the nonurgent operative procedure and adjusted for with multivariable modeling.
At the same time, our experimental design of ischemia induced by cold blood cardioplegia is not exact equivalent of in vivo ischemia and our results might underestimate the true extent of ischemic injury. The downregulation observed in the majority of our samples could also be attributed to hypothermia-induced suppression of transcription, and further work animal studies and in vitro experiments will be necessary to discern biologically relevant differences. In addition, lack of validation at the protein level, secondary to the small amount of tissue obtained, is also a limitation.
Conclusion
We have shown that the human LV exhibits significant, yet variable changes in gene expression in response to cold cardioplegia-induced ischemia during CPB. Investigation of this process on a molecular level by examining whole-genome transcriptional activity provides great insight into the pathophysiology of ventricular ischemia, and thus, may help guide efforts to reduce myocardial damage during surgery as well as to identify interventional targets for pharmacological therapy and biomarkers of injury in the future.
Supplementary Material
Acknowledgments
We acknowledge the outstanding contributory efforts of the Coronary Artery Bypass Graft Genomics research staff: James Gosnell, R.N.; Kujtim Bodinaku, M.D.; Svetlana Gorbatov, M.P.H. (All part of Brigham and Women’s Hospital, Department of Anesthesiology, Perioperative and Pain Medicine, Harvard Medical School, Boston, Massachusetts). We thank all study subjects who participated in the TRANSCRIBE Program.
Funding Sources: The authors acknowledge support for this research by the National Institute of Health grant R01HL118266 to JDM (Bethesda, Maryland), the American Heart Association (Dallas, Texas), The Harvard Catalyst (Boston, Massachusetts), The Watkins Cardiovascular Discovery Award (Brigham and Women’s Hospital, Boston, Massachusetts), the Scholars in Clinical Science Program (Harvard Medical School, Boston, Massachusetts) and a Mercedes Concepcion Faculty Development Fellowship and Scholar in Translational Anesthesia Research grant from the Department of Anesthesiology, Perioperative and Pain Medicine, (Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts).
Footnotes
The authors declare no competing interests.
Contributor Information
Jochen D. Muehlschlegel, Brigham and Women’s Hospital, Department of Anesthesiology, Perioperative and Pain Medicine, Harvard Medical School, Boston, Massachusetts
Danos C. Christodoulou, Department of Genetics, Harvard Medical School, Boston, Massachusetts
David McKean, Department of Genetics, Harvard Medical School, Boston, Massachusetts.
Joshua Gorham, Department of Genetics, Harvard Medical School, Boston, Massachusetts
Erica Mazaika, Department of Genetics, Harvard Medical School, Boston, Massachusetts
Mahyar Heydarpour, Brigham and Women’s Hospital, Department of Anesthesiology, Perioperative and Pain Medicine, Harvard Medical School, Boston, Massachusetts
Grace Lee, Harvard Medical School, Boston, Massachusetts
Steven R. DePalma, Department of Genetics, Harvard Medical School, Boston, Massachusetts
Tjorvi E. Perry, Northwest Anesthesia, Abbott Northwestern Hospital and Minneapolis Heart Institute, Minnesota
Amanda F. Fox, University of Texas Southwestern Medical Center, Department of Anesthesiology, Dallas, Texas
Stanton Shernan, Brigham and Women’s Hospital, Department of Anesthesiology, Perioperative and Pain Medicine, Harvard Medical School, Boston, Massachusetts
Christine E. Seidman, Howard Hughes Medical Institute, Division of Cardiovascular Medicine, Brigham and Women’s Hospital, Boston, Massachusetts
Sary F. Aranki, Brigham and Women’s Hospital, Division of Cardiac Surgery, Harvard Medical School, Boston, Massachusetts
Jon G. Seidman, Department of Genetics, Harvard Medical School, Boston, Massachusetts.
Simon C. Body, Brigham and Women’s Hospital, Department of Anesthesiology, Perioperative and Pain Medicine, Harvard Medical School, Boston, Massachusetts.
References
- 1.Bretschneider HJ, Hubner G, Knoll D, Lohr B, Nordbeck H, Spieckermann PG. Myocardial resistance and tolerance to ischemia: Physiological and biochemical basis. J Cardiovasc Surg (Torino) 1975;16:241–260. [PubMed] [Google Scholar]
- 2.Walsh MC, Bourcier T, Takahashi K, Shi L, Busche MN, Rother RP, Solomon SD, Ezekowitz RA, Stahl GL. Mannose-binding lectin is a regulator of inflammation that accompanies myocardial ischemia and reperfusion injury. J Immunol. 2005;175:541–546. doi: 10.4049/jimmunol.175.1.541. [DOI] [PubMed] [Google Scholar]
- 3.Feng J, Bianchi C, Sandmeyer JL, Li J, Sellke FW. Molecular indices of apoptosis after intermittent blood and crystalloid cardioplegia. Circulation. 2005;112:I184–1189. doi: 10.1161/CIRCULATIONAHA.104.526160. [DOI] [PubMed] [Google Scholar]
- 4.Etievent JP, Chocron S, Toubin G, Taberlet C, Alwan K, Clement F, Cordier A, Schipman N, Kantelip JP. Use of cardiac troponin I as a marker of perioperative myocardial ischemia. Ann Thorac Surg. 1995;59:1192–1194. doi: 10.1016/0003-4975(95)00129-9. [DOI] [PubMed] [Google Scholar]
- 5.Muehlschlegel JD, Perry TE, Liu KY, Nascimben L, Fox AA, Collard CD, Avery EG, Aranki SF, D'Ambra MN, Shernan SK, Body SC, Investigators CG. Troponin is superior to electrocardiogram and creatinine kinase MB for predicting clinically significant myocardial injury after coronary artery bypass grafting. Eur Heart J. 2009;30:1574–1583. doi: 10.1093/eurheartj/ehp134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mohammed AA, Agnihotri AK, van Kimmenade RR, Martinez-Rumayor A, Green SM, Quiroz R, Januzzi JL., Jr Prospective, comprehensive assessment of cardiac troponin T testing after coronary artery bypass graft surgery. Circulation. 2009;120:843–850. doi: 10.1161/CIRCULATIONAHA.108.837278. [DOI] [PubMed] [Google Scholar]
- 7.Matte GS, del Nido PJ. History and use of del Nido cardioplegia solution at Boston Children's Hospital. J Extra Corpor Technol. 2012;44:98–103. [PMC free article] [PubMed] [Google Scholar]
- 8.Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xing Y, Yu T, Wu YN, Roy M, Kim J, Lee C. An expectation-maximization algorithm for probabilistic reconstructions of full-length isoforms from splice graphs. Nucleic Acids Res. 2006;34:3150–3160. doi: 10.1093/nar/gkl396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: An assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 14.Kel A, Voss N, Jauregui R, Kel-Margoulis O, Wingender E. Beyond microarrays: Find key transcription factors controlling signal transduction pathways. BMC Bioinformatics. 2006;7(Suppl 2):S13. doi: 10.1186/1471-2105-7-S2-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kel A, Voss N, Valeev T, Stegmaier P, Kel-Margoulis O, Wingender E. ExPlain: Finding upstream drug targets in disease gene regulatory networks. SAR QSAR Environ Res. 2008;19:481–494. doi: 10.1080/10629360802083806. [DOI] [PubMed] [Google Scholar]
- 16.Wingender E, Chen X, Hehl R, Karas H, Liebich I, Matys V, Meinhardt T, Pruss M, Reuter I, Schacherer F. TRANSFAC: An integrated system for gene expression regulation. Nucleic Acids Res. 2000;28:316–319. doi: 10.1093/nar/28.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thygesen K, Alpert JS, Jaffe AS, Simoons ML, Chaitman BR, White HD. Third universal definition of myocardial infarction. Circulation. 2012;126:2020–2035. doi: 10.1016/j.gheart.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 19.Du W, Wang YC, Hong J, Su WY, Lin YW, Lu R, Xiong H, Fang JY. STAT5 isoforms regulate colorectal cancer cell apoptosis via reduction of mitochondrial membrane potential and generation of reactive oxygen species. J Cell Physiol. 2012;227:2421–2429. doi: 10.1002/jcp.22977. [DOI] [PubMed] [Google Scholar]
- 20.Chen G, Wang H, Xie S, Ma J, Wang G. STAT1 negatively regulates hepatocellular carcinoma cell proliferation. Oncol Rep. 2013;29:2303–2310. doi: 10.3892/or.2013.2398. [DOI] [PubMed] [Google Scholar]
- 21.Lee HG, Minematsu H, Kim KO, Celil Aydemir AB, Shin MJ, Nizami SA, Chung KJ, Hsu AC, Jacobs CR, Lee FY. Actin and ERK1/2-CEBPbeta signaling mediates phagocytosis-induced innate immune response of osteoprogenitor cells. Biomaterials. 2011;32:9197–9206. doi: 10.1016/j.biomaterials.2011.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benderdour M, Fahmi H, Beaudet F, Fernandes JC, Shi Q. Nuclear receptor retinoid-related orphan receptor alpha1 modulates the metabolic activity of human osteoblasts. J Cell Biochem. 2011;112:2160–2169. doi: 10.1002/jcb.23141. [DOI] [PubMed] [Google Scholar]
- 23.Yamawaki H, Tsubaki N, Mukohda M, Okada M, Hara Y. Omentin, a novel adipokine, induces vasodilation in rat isolated blood vessels. Biochem Biophys Res Commun. 2010;393:668–672. doi: 10.1016/j.bbrc.2010.02.053. [DOI] [PubMed] [Google Scholar]
- 24.Yamawaki H, Kuramoto J, Kameshima S, Usui T, Okada M, Hara Y. Omentin, a novel adipocytokine inhibits TNF-induced vascular inflammation in human endothelial cells. Biochem Biophys Res Commun. 2011;408:339–343. doi: 10.1016/j.bbrc.2011.04.039. [DOI] [PubMed] [Google Scholar]
- 25.Tan BK, Adya R, Farhatullah S, Chen J, Lehnert H, Randeva HS. Metformin treatment may increase omentin-1 levels in women with polycystic ovary syndrome. Diabetes. 2010;59:3023–3031. doi: 10.2337/db10-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mattu HS, Randeva HS. Role of adipokines in cardiovascular disease. J Endocrinol. 2013;216:T17–T36. doi: 10.1530/JOE-12-0232. [DOI] [PubMed] [Google Scholar]
- 27.Kataoka Y, Shibata R, Ohashi K, Kambara T, Enomoto T, Uemura Y, Ogura Y, Yuasa D, Matsuo K, Nagata T, Oba T, Yasukawa H, Numaguchi Y, Sone T, Murohara T, Ouchi N. Omentin prevents myocardial ischemic injury through AMP-activated protein kinase- and Akt-dependent mechanisms. J Am Coll Cardiol. 2014;63:2722–2733. doi: 10.1016/j.jacc.2014.03.032. [DOI] [PubMed] [Google Scholar]
- 28.Hoggard N, Cruickshank M, Moar KM, Bashir S, Mayer CD. Using gene expression to predict differences in the secretome of human omental vs subcutaneous adipose tissue. Obesity (Silver Spring) 2012;20:1158–1167. doi: 10.1038/oby.2012.14. [DOI] [PubMed] [Google Scholar]
- 29.Wendel HP, Heller W, Michel J, Mayer G, Ochsenfahrt C, Graeter U, Schulze J, Hoffmeister HM, Hoffmeister HE. Lower cardiac troponin T levels in patients undergoing cardiopulmonary bypass and receiving high-dose aprotinin therapy indicate reduction of perioperative myocardial damage. J Thorac Cardiovasc Surg. 1995;109:1164–1172. doi: 10.1016/S0022-5223(95)70200-8. [DOI] [PubMed] [Google Scholar]
- 30.Buerke M, Pruefer D, Sankat D, Carter JM, Buerke U, Russ M, Schlitt A, Friedrich I, Borgermann J, Vahl CF, Werdan K. Effects of aprotinin on gene expression and protein synthesis after ischemia and reperfusion in rats. Circulation. 2007;116:I121–1126. doi: 10.1161/CIRCULATIONAHA.106.680249. [DOI] [PubMed] [Google Scholar]
- 31.Ikegawa S, Sano M, Koshizuka Y, Nakamura Y. Isolation, characterization and mapping of the mouse and human PRG4 (proteoglycan 4) genes. Cytogenet Cell Genet. 2000;90:291–297. doi: 10.1159/000056791. [DOI] [PubMed] [Google Scholar]
- 32.Matsuo K, Owens JM, Tonko M, Elliott C, Chambers TJ, Wagner EF. Fosl1 is a transcriptional target of c-Fos during osteoclast differentiation. Nat Genet. 2000;24:184–187. doi: 10.1038/72855. [DOI] [PubMed] [Google Scholar]
- 33.Kalra N, Kumar V. c-Fos is a mediator of the c-myc-induced apoptotic signaling in serum-deprived hepatoma cells via the p38 mitogen-activated protein kinase pathway. J Biol Chem. 2004;279:25313–25319. doi: 10.1074/jbc.M400932200. [DOI] [PubMed] [Google Scholar]
- 34.Pan Z, Sun X, Shan H, Wang N, Wang J, Ren J, Feng S, Xie L, Lu C, Yuan Y, Zhang Y, Wang Y, Lu Y, Yang B. MicroRNA-101 inhibited postinfarct cardiac fibrosis and improved left ventricular compliance via the FBJ osteosarcoma oncogene/transforming growth factor-beta1 pathway. Circulation. 2012;126:840–850. doi: 10.1161/CIRCULATIONAHA.112.094524. [DOI] [PubMed] [Google Scholar]
- 35.Lukk M, Kapushesky M, Nikkila J, Parkinson H, Goncalves A, Huber W, Ukkonen E, Brazma A. A global map of human gene expression. Nat Biotechnol. 2010;28:322–324. doi: 10.1038/nbt0410-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arab S, Konstantinov IE, Boscarino C, Cukerman E, Mori A, Li J, Liu PP, Redington AN, Coles JG. Early gene expression profiles during intraoperative myocardial ischemia-reperfusion in cardiac surgery. J Thorac Cardiovasc Surg. 2007;134:74–81. 81, e1–e2. doi: 10.1016/j.jtcvs.2007.01.025. [DOI] [PubMed] [Google Scholar]
- 37.He Y, Hua Y, Liu W, Hu H, Keep RF, Xi G. Effects of cerebral ischemia on neuronal hemoglobin. J Cereb Blood Flow Metab. 2009;29:596–605. doi: 10.1038/jcbfm.2008.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verrier ED, Shernan SK, Taylor KM, Van de Werf F, Newman MF, Chen JC, Carrier M, Haverich A, Malloy KJ, Adams PX, Todaro TG, Mojcik CF, Rollins SA, Levy JH. Terminal complement blockade with pexelizumab during coronary artery bypass graft surgery requiring cardiopulmonary bypass: A randomized trial. JAMA. 2004;291:2319–2327. doi: 10.1001/jama.291.19.2319. [DOI] [PubMed] [Google Scholar]
- 39.Arumugam TV, Magnus T, Woodruff TM, Proctor LM, Shiels IA, Taylor SM. Complement mediators in ischemia-reperfusion injury. Clin Chim Acta. 2006;374:33–45. doi: 10.1016/j.cca.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 40.Kortekaas KA, van der Pol P, Lindeman JH, Baan CC, van Kooten C, Klautz RJ. No prominent role for terminal complement activation in the early myocardial reperfusion phase following cardiac surgery. Eur J Cardiothorac Surg. 2012;41:e117–e125. doi: 10.1093/ejcts/ezs088. [DOI] [PubMed] [Google Scholar]
- 41.Charlagorla P, Liu J, Patel M, Rushbrook JI, Zhang M. Loss of plasma membrane integrity, complement response and formation of reactive oxygen species during early myocardial ischemia/reperfusion. Mol Immunol. 2013;56:507–512. doi: 10.1016/j.molimm.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tarnok A, Hambsch J, Emmrich F, Sack U, van Son J, Bellinghausen W, Borte M, Schneider P. Complement activation, cytokines, and adhesion molecules in children undergoing cardiac surgery with or without cardiopulmonary bypass. Pediatr Cardiol. 1999;20:113–125. doi: 10.1007/s002469900417. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.